Abstract
Polyglutamine diseases include at least nine neurodegenerative disorders, each caused by a CAG repeat expansion in a different gene. Accumulation of mutant polyglutamine-containing proteins occurs in patients, and evidence from cell culture and animal experiments suggests the nucleus as a site of pathogenesis. To understand the consequences of nuclear accumulation, we created a cell culture system with nuclear-targeted polyglutamine. In our system, cell death can be mitigated by overexpression of full-length cAMP response element binding protein (CREB)-binding protein (CBP) or its amino-terminal portion alone. CBP is one of several histone acetyltransferases sequestered by polyglutamine inclusions. We found histone acetylation to be reduced in cells expressing mutant polyglutamine. Reversal of this hypoacetylation, which can be achieved either by overexpression of CBP or its amino terminus or by treatment with deacetylase inhibitors, reduced cell loss. These findings suggest that nuclear accumulation of polyglutamine can lead to altered protein acetylation in neurons and indicate a novel therapeutic strategy for polyglutamine disease.
Polyglutamine expansion diseases are caused by mutations in different genes that result in degeneration of different populations of neurons (1–9), but they likely share the same mechanism, in which the expanded polyglutamine tract confers a novel, toxic property on the disease protein. Characterization of that novel property remains a central goal of polyglutamine disease research.
One hypothesis is that expanded polyglutamine causes altered gene transcription. Nuclear accumulation of mutant protein may disrupt the transcriptional machinery by recruiting other polyglutamine-containing proteins, many of which are transcription factors (10–12). Key components of the transcription apparatus are sequestered in polyglutamine-containing inclusions (13–18). Two polyglutamine diseases are caused by expansions in known transcription factors, the androgen receptor (AR) and TATA-binding protein (8, 9). Other nuclear factors with altered distribution in the presence of mutant polyglutamine include the steroid receptor coactivator-1 (SRC-1), cAMP response element binding protein (CREB)-binding protein (CBP), nuclear corepressor, p53, and TAFII130 (13–18). Overexpression of CBP and TAFII130 has been shown to reduce polyglutamine-induced cell loss in cell culture (13, 18, 19). Many of these nuclear factors directly regulate histone acetylation or are in complexes that have acetylase activity. Also, a genetic screen in Drosophila melanogaster identified factors regulating acetylation as modifiers of polyglutamine-induced degeneration (20).
Of the transcription factors implicated in polyglutamine pathogenesis, we have focused on CBP, because it is a coactivator in important signal transduction pathways, for which it is functionally limiting (21). CBP is found in polyglutamine-positive inclusions in patient tissue and in mouse and cell culture models of polyglutamine disease (13, 15, 19, 22). Also, CBP-mediated transcription is impaired in the presence of mutant polyglutamine (13, 19).
In this study, we examined the consequences of CBP disruption by expanded polyglutamine. We found that nuclear-targeted polyglutamine causes cell death that is mitigated by full-length CBP or its amino-terminal domain alone. The cell death is associated with decreased histone acetylation and reduced by histone deacetylase inhibitors. These data implicate transcriptional dysfunction in polyglutamine toxicity and suggest the use of deacetylase inhibitors as therapeutic agents.
Methods
Cells and Plasmids.
A mouse motor neuron-neuroblastoma fusion cell line (MN-1) (23) was maintained in DMEM (Life Technologies, Bethesda, MD) supplemented with penicillin, streptomycin, glutamine, and 10% FBS (Atlanta Biologicals, Norcross, GA). AR constructs encoding normal and expanded polyglutamine tracts (AR16 and AR110, respectively) were derived from pCMV-AR-HA (24), by EcoRI and BamHI digestion. The nuclear localization signal (NLS) and myc tag were derived from the pShooter myc nuc plasmid (Invitrogen). The hemagglutinin (HA) tag from the pCMV-AR-HA constructs was removed by AscI digestion, placing the NLS and myc tags in-frame. Subsequently, the constructs were subcloned into the C2-green fluorescent protein (GFP) plasmid (CLONTECH). The reading frame was confirmed by sequence analysis and detection of the protein by anti-GFP and anti-myc antibodies (CLONTECH and Invitrogen, respectively), which recognize amino- and carboxyl-terminal epitopes on the constructs. All of the CBP constructs, which are fused to the DNA binding domain of Gal4, were a gift from John Chrivia, St. Louis University (25). These constructs include full-length CBP (CBP-FL), its amino and carboxyl termini (respectively, CBP-NT, residues 1–460, and CBP-CT, residues 1678–2441) and the mid-region (CBP-Mid, residues 721-1670). Functionally, these domains include the nuclear hormone receptor binding domain (CBP-NT), histone acetyltransferase activity (CBP-Mid), and a polyglutamine repeat of 15 residues (CBP-CT). Chris Baumann, National Cancer Institute, Bethesda, MD, generously provided the GFP-GRIP1 construct. The Bax-GFP construct was provided by Richard Youle, National Institute of Neurological Disorders and Stroke. Transfections were performed with Lipofectamine plus reagent (Life Technologies) by using 8 μg DNA per 5 ml of medium.
Cell Death.
Cells were harvested 96 h after transfection by trypsin digestion. Care was taken to retain all cells, including those floating in the medium. Fluorescence-activated cell sorting was performed by FAST Systems (Rockville, MD). The samples were spun at 400 g and 4°C, then the supernatant was aspirated to 300 μl, and 300 μl of 10 μg/ml propidium iodide (PI) was added to each sample. After a 10-min incubation at room temperature in the dark, the samples were vortexed and analyzed on an EPICS XL MCL flow cytometer (Beckman Coulter). The cells were identified by forward and orthogonal light scatter signals. Dead cells were identified by PI uptake as measured by fluorescent emission at 620 nm. At least 50,000 cells were counted for each condition. The proportion of dead (PI positive) transfected (GFP positive) cells was determined. For experiments in which deacetylase inhibitors were used, the agents were added to the culture medium every 24 h starting the day after transfection.
Glutathione S-Transferase (GST) Pull-Down.
GST-AR constructs were obtained from Diane Merry (Thomas Jefferson University, Philadelphia) and transformed into DH5α bacteria. GST-AR16 colonies were grown at 37°C until the OD at 600 nm was 0.5. GST was induced by addition of 100 mM isopropyl-β-d-thiogalactopyranoside for 3 h. GST-AR112 colonies were treated identically, except the growth and induction were done at 30°C to limit aggregation. The GST was purified by attachment to glutathione Sepharose 4B beads (Amersham Pharmacia). GST-AR16 and GST-AR112 were quantified by Coomassie stain. Equal amounts were mixed with cell lysates expressing CBP-FL, CBP-NT, CBP-Mid, or CBP-CT at 4°C for 30 min. After washing, lysates were loaded and separated on a 7.5% SDS-polyacrylamide gel. After transfer, the blots were probed with a mouse monoclonal anti-Gal4 antibody (Santa Cruz Biotechnology), which recognizes the Gal4 epitope fused to CBP.
Immunofluorescence and Deconvolution.
MN-1 cells were plated on Permanox chambered slides (Nunc) and transfected as described above. CBP was stained by using either A-22, a rabbit polyclonal antibody that recognizes the amino terminus, or C-1, a mouse monoclonal recognizing the carboxyl terminus (Santa Cruz Biotechnology). Both antibodies were visualized with a Texas red-conjugated secondary antibody (Jackson ImmunoResearch). Images were deconvolved by using deltavision software (Applied Precision, Issaquah, WA).
Deacetylase Inhibitors and Acid Extraction of Histones.
Trichostatin A (TSA, Upstate Biotechnology, Lake Placid, NY) and suberoylanilide hydroxamic acid (SAHA, Calbiochem) were dissolved in DMSO. Sodium butyrate (Sigma) and phenylbutyric acid (PBA) were dissolved in water. The PBA was a generous gift from Kyung-Tai Min, National Institute of Neurological Disorders and Stroke. To detect acetylation, we added 5 ng/ml TSA to inhibit deacetylation. Addition of TSA causes hyperacetylation of histone protein H3 that peaks around 8 h. For experiments measuring H3 acetylation, we added the TSA 30 min before harvesting. Cells were lysed in acid extraction buffer containing 10 mM Hepes (pH 8.0), 10 mM KCl, 1.5 mM MgCl2, 1.5 mM PMSF, 0.5 mM DTT, and 0.2 M hydrochloric acid for 30 min on ice. The lysates were pelleted and the supernatant was dialyzed with two 1-h incubations in cold 0.1 M acetic acid and three incubations in water. Extracted protein was quantified and 10 μg of protein was separated on a 7.5% SDS denaturing gel. The proteins were transferred to an Immobilon-P membrane (Millipore), which was probed with anti-H1, anti-acetylated H3 (Upstate Biotechnology).
Results
Spinal and bulbar muscular atrophy is a polyglutamine disease with motor neuron degeneration caused by a CAG repeat expansion in the AR gene. Previous work has shown toxicity of the truncated mutant AR in cell culture and in vivo (24, 26).
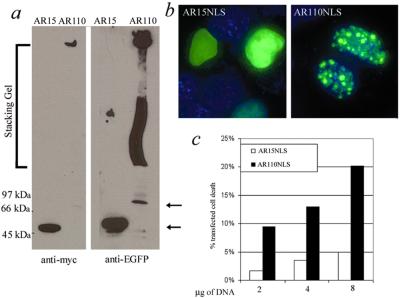
Caspase-dependent formation of a truncated fragment containing the polyglutamine repeat is thought to be an important step in polyglutamine disease pathogenesis (27–29). For this project we restored an NLS to the truncated protein to recreate more accurately the normal localization of mutant AR. In addition, an amino terminal-enhanced GFP tag and a carboxyl-terminal myc tag were added for detection. Expression of these constructs in MN-1 cells caused repeat length-dependent cell death (Fig. 1). Expression peaked around 48 h after transfection, although it was still detectable at 96 h by Western blot and visually by GFP. Both anti-myc and anti-GFP antibodies detected similar bands on Western blot, including an insoluble protein complex that remained in the stacking gel (Fig. 1a). This result is similar to observations in previous cell culture studies of mutant polyglutamine. The inclusions seen in cells expressing expanded polyglutamine with an NLS (AR110NLS) differed in location and number from cells expressing mutant protein without an NLS. In cells expressing the non-NLS constructs, cytoplasmic inclusions predominated and were detectable in roughly 90% of AR-positive cells (data not shown). Those cells that formed nuclear inclusions typically had only one large inclusion, although occasionally two or three inclusions were observed. In contrast, AR110NLS formed many smaller inclusions in the nuclei, with no signal in the cytoplasm (Fig. 1b). Cells with low levels of AR110NLS expression did not form inclusions. No inclusions were observed in cells expressing a construct with normal polyglutamine repeat length (AR15NLS).
Figure 1.
Cell death induced by nuclear accumulation of polyglutamine. Expression of constructs 48 h after transfection. (a) Western blot with antibodies against amino-terminal (GFP) and carboxyl-terminal (myc) tags. AR15NLS is at 50 kDa (lower arrow) and the AR110NLS monomer is at 66 kDa (upper arrow), with an insoluble protein complex in the stacking gel. (b) Construct localization by GFP fluorescence. Expression of both normal and mutant AR is restricted to the nucleus, and only AR110-NLS forms inclusions. (c) Repeat length and dose-dependent cell death in MN-1 cells. Transfected cell death correlates with the quantity of transfected DNA. Cell death was assayed by counting PI- and GFP-positive cells by FACS analysis.
Although expression of the NLS constructs peaked around 48 h after transfection, polyglutamine-dependent cell death was not detected until 72 h. The quantity of cell death in the transfected population correlated with the amount of DNA transfected (Fig. 1c) and the transfection efficiency (data not shown). The corresponding increase in transfected cell death with increased transfection efficiency indicates an increase in cellular levels of the mutant protein, although we have no direct evidence for this increase. Transfection of comparable amounts of NLS and non-NLS constructs resulted in 50% greater toxicity with the NLS.
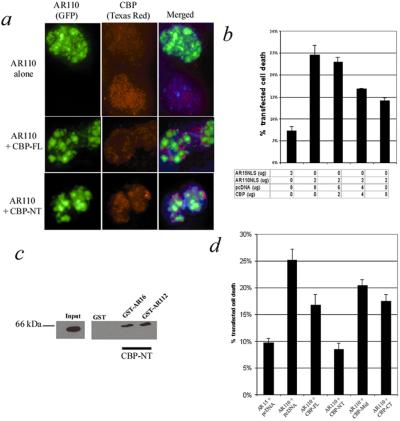
In previous work, we and others have shown that endogenous CBP is recruited to polyglutamine nuclear inclusions (13, 15, 16, 19). In the AR15NLS-expressing cells, as well as noninclusion-bearing AR110NLS cells, endogenous CBP was observed throughout the nucleus (Fig. 2a). In inclusion-bearing cells, only very weak staining for CBP could be seen, perhaps as a consequence of sequestration and epitope masking or CBP depletion by the mutant protein (Fig. 2a).
Figure 2.
Overexpression of CBP's amino terminus is sufficient to block cell death. (a) CBP (Texas red) is depleted in AR110NLS-expressing cells. Cells were transfected with AR110NLS tagged with GFP, either alone or together with CBP-FL or CBP-NT as indicated. Endogenous CBP was labeled with an amino-terminal antibody, except in cells transfected with CBP-NT, in which it was labeled with a carboxyl-terminal antibody. Overexpression of CBP-FL or CBP-NT permits visualization of CBP in the presence of AR inclusions. In the middle row, both endogenous and transfected CBP were labeled. (b) Increasing amounts of transfected CBP progressively reduce cell death caused by AR110-NLS. Transfected CBP was complemented by the vector (pcDNA) to keep the total amount of transfected DNA constant. (c) Western blot of CBP-NT showing retention by GST-AR16 and GST-AR112. The blot was probed with antibody to the Gal4 tag on CBP-NT. CBP-NT is retained by both GST-AR fusion proteins, but not by GST alone. Gal4 alone is not retained by GST-AR (data not shown), indicating that the interaction is mediated directly by AR. (d) Cell death in MN-1 cells transfected with AR15NLS and AR110NLS with or without various CBP constructs. CBP-NT blocks AR110NLS-induced cell death. Both the CBP-Mid and CBP-CT constructs partially reduce cell death. Lower AR expression was routinely observed with cotransfected CBP-NT, so the measurement of cell death is probably an underestimate in this case.
Sequestration of CBP and other coactivators has been proposed as mediating polyglutamine toxicity (13). To test this hypothesis, we transfected increasing amounts of CBP-FL into AR110NLS-expressing cells. We observed a decrease in cell death that corresponded to the amount of transfected CBP (Fig. 2b). Approximately 50% of the AR110NLS-induced toxicity was blocked by CBP. This effect may have been limited because not all AR-positive cells were transfected with CBP, and because other, CBP-independent pathways also may be involved in polyglutamine toxicity.
Cells cotransfected with mutant AR and CBP-FL had strong CBP staining (Fig. 2a), suggesting that overexpression of CBP saturates its binding by AR. This staining does not differentiate between endogenous and exogenous CBP.
Previous investigations have shown that AR interacts primarily with the amino-terminal 460 aa of CBP (30, 31), the same domain through which CBP interacts with other nuclear hormone receptors. In addition, mutant AR may bind to CBP-CT, which contains the polyglutamine repeat. For example, the redistribution of CBP to huntingtin-positive inclusions depends on the polyglutamine repeat of CBP (16, 19).
We sought to determine which regions of CBP bind to our normal and mutant AR constructs. We investigated the interaction by GST pull-down and the mitigation of cell death. When we mixed purified GST-AR protein with cell lysates from cells transfected with CBP-NT, CBP-Mid, or CBP-CT, CBP-NT was consistently retained (Fig. 2c). This interaction was not found to be repeat length-dependent, although the assay was not quantitative. We did not observe any interaction between AR and CBP-Mid. GST alone did not bind to any of the constructs.
If AR binds to a specific domain of CBP, then overexpression of that region should displace the endogenous CBP, permitting it to function normally. To test this theory, we cotransfected the different portions of CBP with AR15NLS or AR110NLS and measured cell death. CBP-NT was most effective at reducing toxicity, decreasing cell death to levels comparable to those induced by AR15NLS (Fig. 2d). CBP-CT and CBP-Mid both had mild effects, neither as potent as the amino terminus. CBP-NT expression had an effect on endogenous CBP distribution similar to CBP-FL (Fig. 2a). The enhanced efficacy of CBP-NT over CBP-FL may have been because of its greater expression level. Furthermore, AR expression levels were consistently lower in cells cotransfected with CBP-NT, contributing to the observed decrease in cell death. After correction for lower AR expression as related to transfection efficiency, cell death in the presence of CBP-NT was still lower than with the other constructs. Other than fewer transfected cells with CBP-NT, we did not observe any consistent change in inclusion number, size, or frequency with transfection of CBP.
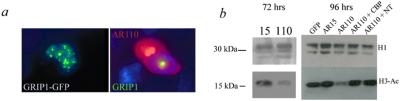
Taken together, these findings support a mechanism in which mutant AR binds to CBP, primarily by means of a domain in the first 460 aa, blocking one or more of its normal functions. This finding raises the question of which function of CBP is critical to polyglutamine toxicity. CBP has two major, interdependent functions: (i) linking DNA-binding proteins to the RNA polymerase II transcription complex, and (ii) acetylating proteins, including histones, as an acetyltransferase. CBP's ability to enhance transcription depends on histone acetylation. We chose to focus on histone acetylation, as other factors with histone acetyltransferase activity also are sequestered in polyglutamine-positive inclusions. SRC-1 can be found in AR-positive inclusions (14). Furthermore, when we cotransfected the glucocorticoid receptor interacting protein-1 (GRIP1) with non-NLS AR110, we found that GRIP1 redistributes from its normal punctuate appearance to the nuclear inclusions (Fig. 3a).
Figure 3.
Decrease in H3 acetylation by AR110NLS is reversed by CBP. (a) GRIP1-GFP redistributes from its normal punctuate distribution in the nucleus to nuclear inclusions formed by AR constructs lacking an NLS. (Left) A cell expressing GRIP1-GFP alone. (Right) A cell expressing both GRIP1-GFP and AR110 (Texas red). Inclusions are present in both the nucleus and cytoplasm of the cell expressing AR110; GRIP1 has been recruited to the nuclear inclusion only. (b) Western blot with antibodies against histone proteins H1 and acetylated H3. AR110NLS reduced H3-Ac at 72 and 96 h after transfection. Cotransfection with either CBP-FL or CBP-NT reversed the hypoacetylation of H3. The amino-terminal portion of CBP that reversed the hypoacetylation also blocked AR110-induced cell loss (Fig. 2d).
Cells were harvested at 48, 72, and 96 h after transfection with AR15NLS or AR110NLS. Although differences were detectable at 72 h, by 96 h there was a major decrease in histone H3 acetylation in the AR110NLS-expressing cells (Fig. 3b). This decrease could be reversed by transfection with either CBP or CBP-NT. Because neither of these constructs induced a measurable increase in acetylation in the absence of AR110NLS (data not shown), and because the acetylation activity of CBP has been mapped to amino acids 1195–1673 (32), which are lacking in our CBP-NT construct, we conclude that the rescue of acetylation in this state is likely caused by increased availability of endogenous CBP.
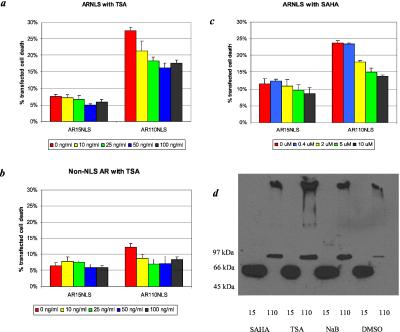
If sequestration of CBP and other histone acetylases leads to cell death by decreasing protein acetylation, then addition of deacetylase inhibitors would be expected to reduce cell loss. We tested the ability of various deacetylase inhibitors to reduce polyglutamine-induced cell death. We transfected cells with AR15NLS or AR110NLS and measured cell death in the presence of TSA. Addition of TSA caused a dose-dependent decrease in AR110NLS-induced toxicity, with a maximal effect at 50 ng/ml (Fig. 4a).
Figure 4.
Histone deacetylase inhibitors reduce AR110NLS-induced cell loss. Cell viability was assayed by FACS analysis of GFP-positive, PI-positive cells at 96 h after transfection. (a and b) Addition of increasing doses of TSA to AR-expressing cells reduced cell loss caused by (a) AR110NLS and (b) AR110 non-NLS. The amount of cell death observed in the non-NLS constructs was consistently less than that observed with the NLS constructs. (c) Increasing doses of SAHA reduced cell death caused by AR110NLS. The reduction of cell loss by SAHA was comparable to TSA. (d) Western blot of AR15NLS and AR110NLS in cells exposed to the highest doses of sodium butyrate (NaB), TSA, SAHA, or DMSO alone. There was no decrease in expression of AR with any of the deacetylase inhibitors.
TSA also partially mitigated cell death induced by AR110 lacking a NLS (Fig. 4b). This finding was unexpected, as the majority of the non-NLS AR110 is found in the cytoplasm. This finding suggests either that the toxicity induced by AR110 expression results from leakage of the protein into the nucleus or that TSA affects acetylation of cytoplasmic proteins.
To determine whether TSA has a general protective effect in MN-1 cells, we measured its ability to protect against cell death induced by the proapoptotic factor Bax. Cell death at 24 h after transfection with Bax was increased, not decreased, in the presence of TSA (data not shown), which suggests that histone deacetylase inhibitors are not general antiapoptotic agents in this system.
We also tested SAHA, a structural relative of TSA (33, 34). Both compounds bind to the same region in histone deacetylase complex-1 (35), but SAHA, unlike TSA, has been found to be active in vivo (34). In our assay, SAHA was comparable to TSA in its ability to reduce cell death induced by AR110NLS (P < 0.01) (Fig. 4c). There was also a significant decrease in death observed with AR15NLS (P < 0.05), but only at the highest concentration of SAHA. SAHA increased histone acetylation in our cells at these concentrations (data not shown). Neither TSA nor SAHA caused morphological changes in the cells.
We tested two other deacetylase inhibitors, sodium butyrate and PBA. These compounds, while inducing histone acetylation, have broader effects on gene expression than TSA. Mariadason et al. (35) showed that sodium butyrate alters the expression of roughly 10 times as many genes as TSA in cultured colon cells. Addition of sodium butyrate to cells expressing AR15NLS or AR110NLS reduced the toxicity (data not shown). The proportional decrease was equivalent with both the normal and mutant AR, suggesting that the protective effect was nonspecific. The reduction in cell death caused by these compounds is not attributable to a decrease in AR expression levels. Indeed, the expression of AR110NLS is slightly increased in the presence of SAHA, TSA, sodium butyrate, and PBA (Fig. 4d).
In contrast to the other deacetylase inhibitors, PBA had no effect on cell survival, although it, too, increases histone acetylation (data not shown). It remains to be determined why PBA failed to block toxicity. One possible explanation is that although all of these agents arrest the cell cycle, they do so at different stages. Cell cycle arrest can increase sensitivity to polyglutamine-induced cell death (36). Although TSA, SAHA, and sodium butyrate all halted the cell cycle at the G2/M boundary in our cells, PBA caused accumulation of cells at the G0/G1 stage (data not shown). Forskolin, which halted MN-1 cells at the same stage as PBA, caused an increase in cell death. It is possible that the protective effects of histone acetylation are countered by the increased sensitivity to cell death caused by cell cycle arrest. Because of the confounding effects of cell cycle arrest, we may be underestimating the true efficacy of the other deacetylase inhibitors.
Discussion
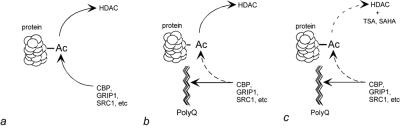
Our findings support nuclear factor sequestration as a mechanism for polyglutamine disease. The cell death caused by expression of mutant polyglutamine in the nucleus can be mitigated in part by increased expression of CBP, which has histone acetylase activity. Our AR constructs interact with the amino terminus of CBP. This portion of CBP is sufficient to reduce cell death, probably by displacing endogenous CBP from the mutant protein. Cell loss correlates with decreased histone acetylation, and both the change in acetylation and the cell loss can be reversed by deacetylase inhibitors or CBP overexpression. These findings are consistent with a mechanism for polyglutamine toxicity in which the mutant protein sequesters CBP and other coactivators, altering protein acetylation. This process, in turn, alters gene expression, leading to cellular dysfunction and death (Fig. 5).
Figure 5.
A deacetylation mechanism for polyglutamine disease. (a) Under physiologic conditions, there is a balance between protein acetylation by coactivators such as CBP, SRC-1, and GRIP1, and deacetylation by histone deacetylase complexes (HDACs). (b) In the presence of mutant polyglutamine, the coactivators responsible for acetylation are sequestered, decreasing the rate of acetylation and causing proteins to be hypoacetylated. (c) The hypoacetylated state can be reversed either by decreasing the rate of deacetylation with HDAC inhibitors such as TSA and SAHA or by increasing acetylation with exogenous coactivator.
Our results were generated with transiently transfected cells and thus require some caveats in their interpretation. The cells had much higher levels of expression, longer repeat lengths, and more truncated protein than are found in patients. Our experience and that of others in the polyglutamine research field has been that such excesses are necessary to model in the laboratory over a period of days a disease process that occurs in patients over a period of decades. We believe our findings to be relevant because key features of the disease process are seen both in our model system and in vivo, including the toxicity of the mutant protein, the binding of nuclear factors, and the accumulation of polyglutamine aggregates. Additionally, two other labs recently identified defects in acetylation with mutant polyglutamine in other systems (37, 38).
Our findings are consistent with a disease mechanism in which CBP and other acetylating nuclear factors are sequestered and inactivated. This sequestration need not depend on an enhanced affinity of the mutant protein for CBP. Instead increased or inappropriate binding could derive from locally increased levels of expanded polyglutamine. The increased levels could arise from impaired clearance of the mutant protein, possibly as a consequence of impaired proteasome function.
We have focused on cell death as an endpoint, partly because it is easily defined. Loss of PI dye exclusion occurs in necrotic and late-stage apoptotic cells. The cell death we observed with PI correlates well with changes in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MIT)-reducing activity (13), and with characteristic features of apoptosis such as nuclear condensation, DNA laddering, caspase activation, and cytochrome c release from mitochondria (unpublished observations). How much cell death directly contributes to the manifestations of polyglutamine diseases is unclear. Although there is evidence of apoptosis in cell culture models of polyglutamine toxicity (39), cell death is not seen in some animal models (40), which implies that neuronal dysfunction may be more important than cell death in the disease process. Altered acetylation may well contribute to neuronal dysfunction as well as death, however.
In a normal cell, protein acetylation reflects a balance between the rate of acetylation by coactivators and other acetyltransferases and the rate of deacetylation by HDACs (Fig. 5a). We propose that nuclear accumulation of polyglutamine perturbs this balance by sequestering coactivators, including CBP, SRC-1, and GRIP1 (Fig. 5b). If our hypothesis is correct, then restoration of the acetylation balance should ameliorate the toxic effects of polyglutamine (Fig. 5c). The decreased acetylation caused by mutant polyglutamine can be rescued either by increasing the rate of acetylation (through addition of an exogenous acetylase or by increasing the availability or activity of an endogenous one) or by reducing the rate of deacetylation.
Our approach focused on nuclear expression and accumulation of a polyglutamine construct derived from AR, the mutant protein of spinal and bulbar muscular atrophy. However, our findings are relevant to the pathogenesis of other polyglutamine diseases, as well. The normal distribution of the proteins causing dentatorubral-pallidoluysian atrophy and spinocerebellar ataxia-1 and -7 is predominantly nuclear (41–44). In spinocerebellar ataxia-1 mouse models, exclusion of ataxin-1 from the nucleus blocks the pathological phenotype (11). Furthermore, flies and mice with transgenic expression of mutant polyglutamine typically feature nuclear accumulation (20, 45, 46), regardless of the protein from which the polyglutamine was derived.
Of the nuclear factors found in AR-positive inclusions, we have focused on CBP for several reasons. CBP functions more broadly than the other coactivators found in polyglutamine inclusions. CBP levels are functionally limiting for a variety of different transcription factors (47), in contrast to SRC-1 and GRIP1, which are thought to function primarily in steroid hormone signaling (48). Evidence from knockout experiments suggests that mice have limited capacity to up-regulate CBP, because serious developmental consequences arise in the hemizygous state (49). In contrast, mice lacking SRC-1 show primarily a loss of steroid hormone responsiveness (50, 51).
We also have examined the ability of GRIP1 and SRC-1 to block polyglutamine-induced cell death, and, although statistically significant, their effect was less than that of CBP (data not shown). These findings are consistent with a model in which mutant polyglutamine may recruit and deplete multiple coactivators, of which CBP is particularly important.
Depletion of coactivator histone acetyltransferases likely accounts for the decrease in histone H3 acetylation we observed. Although we began to observe cell death before the major loss of H3 acetylation, there are indications of changes in acetylation at 72 h. Our sensitivity to changes in acetylation was probably low, as we measured global H3 acetylation in a mixed population of transfected and nontransfected cells. Further confirmation of our hypothesis may come from the identification of candidate genes with expression levels that are both acetylation-dependent and altered in cells expressing mutant polyglutamine.
Our findings that both TSA and SAHA reduce cell loss suggest that the changes in histone acetylation are causative, and not a secondary consequence of cell death. The differences in the efficacy of the other deacetylase inhibitors likely result from differing specificity. These agents could prove useful in differentiating between the protective effects of TSA and those that are not essential.
We have focused on histone acetylation, although acetylation of other factors may be more important. The activity of many components of the transcriptional machinery is modified by acetylation, including other histone acetyltransferases and AR (52). TSA and SAHA likely induce hyperacetylation of proteins other than histones that play important roles in regulating gene expression and cell viability.
Fernandez-Funez et al. (20) reported a P-element screen for modifiers of a mutant polyglutamine phenotype in Drosophila. Several of the reported modifiers were previously described regulators of histone acetylation. The mechanism of their effect on polyglutamine toxicity is not clear. Nevertheless, this investigation also implicates transcriptional disruption and altered protein acetylation in polyglutamine toxicity.
Our finding that deacetylase inhibitors reduce polyglutamine toxicity in cell culture has potential therapeutic implications. Interest in deacetylase inhibitors has grown in recent years for their potential application as tumor-suppressing agents (53). SAHA has been shown to induce histone hyperacetylation and reduce the rate of tumor growth in mice (34) and has been approved for use in clinical trials in humans. These small molecules may be worthy of further testing as candidates for the therapy of polyglutamine disease.
Acknowledgments
We thank B. Howell, A. Lieberman, and J. P. Taylor for helpful discussions. This work was supported by the Intramural Program of the National Institute of Neurological Disorders and Stroke.
Abbreviations
- CBP
cAMP response element binding protein (CREB)-binding protein
- CBP-FL
full-length CBP
- CBP-NT
CBP amino terminus
- CBP-CT
CBP carboxyl terminus
- CBP-Mid
CBP mid-region
- AR
androgen receptor
- SRC-1
steroid receptor coactivator-1
- NLS
nuclear localization signal
- HA
hemagglutinin
- GFP
green fluorescent protein
- PI
propidium iodide
- GST
glutathione S-transferase
- TSA
trichostatin A
- SAHA
suberoylanilide hydroxamic acid
- PBA
phenylbutyric acid
- GRIP1
glucocorticoid receptor interacting protein-1
- HDAC
histone deacetylase complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Orr H T, Chung M Y, Banfi S, Kwiatkowski T J, Jr, Servadio A, Beaudet A L, McCall A E, Duvick L A, Ranum L P, Zoghbi H Y. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 3.Pulst S M, Nechiporuk A, Nechiporuk T, Gispert S, Chen X N, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, et al. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I, et al. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 5.Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton D W, Amos C, Dobyns W B, Subramony S H, Zoghbi H Y, Lee C C. Nat Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 6.Lindblad K, Savontaus M L, Stevanin G, Holmberg M, Digre K, Zander C, Ehrsson H, David G, Benomar A, Nikoskelainen E, et al. Genome Res. 1996;6:965–971. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]
- 7.Nagafuchi S, Yanagisawa H, Sato K, Shirayama T, Ohsaki E, Bundo M, Takeda T, Tadokoro K, Kondo I, Murayama N, et al. Nat Genet. 1994;6:14–18. doi: 10.1038/ng0194-14. [DOI] [PubMed] [Google Scholar]
- 8.La Spada A R, Wilson E M, Lubahn D B, Harding A E, Fischbeck K H. Nature (London) 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura K, Jeong S Y, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, Kanazawa I. Hum Mol Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 10.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klement I A, Skinner P J, Kaytor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 12.Gerber H P, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 13.McCampbell A, Taylor J P, Taye A A, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, et al. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 14.Stenoien D L, Cummings C J, Adams H P, Mancini M G, Patel K, DeMartino G N, Marcelli M, Weigel N L, Mancini M A. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 15.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. . (First Published May 23, 2000; 10.1073/pnas.100110097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutell J M, Thomas P, Neal J W, Weston V J, Duce J, Harper P S, Jones A L. Hum Mol Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 18.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, et al. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 19.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Funez P, Nino-Rosales M L, de Gouyon B, She W C, Luchak J M, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner P J, et al. Nature (London) 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 21.Shikama N, Lyon J, LaThangue N. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 22.Yamada M, Wood J D, Shimohata T, Hayashi S, Tsuji S, Ross C A, Takahashi H. Ann Neurol. 2001;49:14–23. doi: 10.1002/1531-8249(200101)49:1<14::aid-ana5>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 23.Salazar-Grueso E F, Kim S, Kim H. NeuroReport. 1991;2:505–508. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Merry D E, Kobayashi Y, Bailey C K, Taye A A, Fischbeck K H. Hum Mol Genet. 1998;7:693–701. doi: 10.1093/hmg/7.4.693. [DOI] [PubMed] [Google Scholar]
- 25.Hu S C, Chrivia J, Ghosh A. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 26.Abel A, Walcott J, Woods J, Duda J, Merry D E. Hum Mol Genet. 2001;10:107–116. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 27.Wellington C L, Ellerby L M, Hackam A S, Margolis R L, Trifiro M A, Singaraja R, McCutcheon K, Salvesen G S, Propp S S, Bromm M, et al. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi Y, Miwa S, Merry D E, Kume A, Mei L, Doyu M, Sobue G. Biochem Biophys Res Commun. 1998;252:145–150. doi: 10.1006/bbrc.1998.9624. [DOI] [PubMed] [Google Scholar]
- 29.Ellerby L M, Hackam A S, Propp S S, Ellerby H M, Rabizadeh S, Cashman N R, Trifiro M A, Pinsky L, Wellington C L, Salvesen G S, et al. J Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 30.Aarnisalo P, Palvimo J J, Janne O A. Proc Natl Acad Sci USA. 1998;95:2122–2127. doi: 10.1073/pnas.95.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fronsdal K, Engedal N, Slagsvold T, Saatcioglu F. J Biol Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- 32.Chen C-J, Deng Z, Kim A Y, Blobel G A, Lieberman P M. Mol Cell Biol. 2001;21:476–487. doi: 10.1128/MCB.21.2.476-487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen L A, Amin S, Marks P A, Rifkind R A, Desai D, Richon V M. Anticancer Res. 1999;19:4999–5005. [PubMed] [Google Scholar]
- 35.Mariadason J M, Corner G A, Augenlicht L H. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 36.Yoshizawa T, Yamagishi Y, Koseki N, Goto J, Yoshida H, Shibasaki F, Shoji S, Kanazawa I. Hum Mol Genet. 2000;9:69–78. doi: 10.1093/hmg/9.1.69. [DOI] [PubMed] [Google Scholar]
- 37.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y-Z, Greenwald M, et al. Nature (London) 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 38.Hughes R E, Lo R S, Davis C, Strand A D, Neal C L, Olson J M, Fields S. Proc Natl Acad Sci USA. 2001;98:13201–13206. doi: 10.1073/pnas.191498198. . (First Published October 30, 2001; 10.1073/pnas.191498198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez I, Xu C J, Juo P, Kakizaka A, Blenis J, Yuan J. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto A, Lucas J J, Hen R. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 41.Miyashita T, Nagao K, Ohmi K, Yanagisawa H, Okamura-Oho Y, Yamada M. Biochem Biophys Res Commun. 1998;249:96–102. doi: 10.1006/bbrc.1998.9096. [DOI] [PubMed] [Google Scholar]
- 42.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 43.McGinnis M Y, Davis P G, Meaney M J, Singer M, McEwen B S. Brain Res. 1983;275:75–82. doi: 10.1016/0006-8993(83)90418-3. [DOI] [PubMed] [Google Scholar]
- 44.Kaytor M D, Duvick L A, Skinner P J, Koob M D, Ranum L P, Orr H T. Hum Mol Genet. 1999;8:1657–1664. doi: 10.1093/hmg/8.9.1657. [DOI] [PubMed] [Google Scholar]
- 45.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 46.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 47.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 48.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss R E, Xu J, Ning G, Pohlenz J, O'Malley B W, Refetoff S. EMBO J. 1999;18:1900–1904. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi C, Zhu Y, Pan J, Yeldandi A V, Rao M S, Maeda N, Subbarao V, Pulikuri S, Hashimoto T, Reddy J K. Proc Natl Acad Sci USA. 1999;96:1585–1590. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu M, Wang C, Reutens A T, Wang J, Angeletti R H, Siconolfi-Baez L, Ogryzko V, Avantaggiati M-L, Pestell R G. J Biol Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 53.Gore S D, Carducci M A. Expert Opin Invest Drugs. 2000;9:2923–2934. doi: 10.1517/13543784.9.12.2923. [DOI] [PubMed] [Google Scholar]