Abstract
Background: Different studies have provided some evidence for the association between BST1 polymorphisms and Parkinson’s disease (PD). The extent to which these genetic effects are consistent across different populations is unknown.
Methods: A meta-analysis of PD case-control studies using a common set of three variants was conducted. Published reports were obtained from electronic databases including Pubmed, Embase, Chinese National Knowledge Infrastructure (CNKI) and Cochrane Library databases between August 2010 and January 2018.
Results: A total of 11 individual studies with 8,725 cases and 17,079 controls were included. The results showed statistically significant association between the dominant model of rs11931532 and PD risk in Asian populations (P=0.006, OR [95% CI]=1.22 [1.06–1.41]). Significant association was also detected between the allelic, dominant, and recessive models of rs4698412 and PD risk in Asian populations (allelic model: P<0.00001, OR [95% CI]=1.22 [1.16–1.29]; dominant model: P<0.00001, OR [95%CI]=1.35 [1.20–1.52]; recessive model; P=0.0003, OR [95% CI]=1.30 [1.13–1.50]). Nevertheless, the pooled analyses suggested that no significant association was uncovered between rs11724635 and PD risk (P>0.05).
Conclusion: The meta-analysis suggests that the rs11931532 and rs4698412, but not rs11724635 might be risk factors for PD in Asian populations.
Keywords: BST1, Parkinson’s disease, polymorphisms, risk factor
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disease that affects 1–2% of individuals aged ≥65 years.1,2 The progressive loss of the nigro-striatal dopaminergic neurons and presence of Lewy bodies were shown to be the most important histopathological features.3,4 Increasing evidence has supported that multiple genes, environmental factors including smoking, alcohol consumption, well water and pesticide use, as well as the interactions among these aspects, contributed to the etiology of PD.5–7 To date, genome-wide association (GWA) studies and familial PD cases studies have identified Alpha-synuclein (SNCA),8 Leucine rich repeat kinase 2 (LRRK2),9 Parkins (PARK3/9/10/11/15/16/17/18),10–15 HLA-DQB1,16 serine threonine kinase 39 (STK39),17 and bone marrow stromal cell antigen 1 (BST1)18 to be the susceptible loci for PD.
BST1, also referred to as CD157, is located on human chromosome 4 (4p15) and belongs to the NADase/ADP-ribosyl cyclase family, which is also a member of the CD38 gene family.6,19 Recent genetic analysis identified the CD157/BST1 gene as a susceptibility marker for PD. The genetic association between BST1 polymorphisms such as rs11931532, rs12645693, rs4698412, and rs4538475 and PD risk have been reported in several populations by GWA and case-control studies.8,20–24 Although numerous studies have demonstrated the association, inconsistency was presented for different allele frequencies among study populations, particularly in different ethnicity and geographical groups.10,15,25–28
Meta-analysis is an effective tool to compensate the limitations by combining all publications and improve statistical power to obtain potential effects of an individual study with small or moderate sizes of subjects. The purpose of performing this meta-analysis is to reduce heterogeneity and summarize the published evidence on the prevalence of the BST1 polymorphisms among patients diagnosed with PD.
Materials and methods
Identification of eligible studies
This meta-analysis followed the Cochrane collaboration definition and PRISMA 2009 guidelines for meta-analysis and systematic review.29 Literature were searched on electronic databases including Pubmed, Embase, Chinese National Knowledge Infrastructure (CNKI) databases, and the Cochrane Library databases with the following terms: bone marrow stromal cell antigen 1; BST1; CD157; polymorphism; single nucleotide polymorphism; SNP; and Parkinson’s disease; PD. No language was limited. The search deadline for publications was June 1, 2018. Other potentially relevant studies were retrieved by cross-references.
Inclusion criteria: (1) case-control or cohort designed study; (2) refer to the association of BST1 polymorphims and PD risk; (3) the genotype data in the studies were sufficient and available to extract; and (4) the distribution of genotypes in the control group were in Hardy-Weinberg equilibrium (HWE).
Exclusion criteria: (1) repeated studies, letters, dissertations, abstracts or reviews; and (2) publications that violated the inclusion criteria.
Data extraction
Two independent investigators (L.J. and L.L.) manually extracted the data. Any disagreements were resolved through discussion among the authors to achieve a consensus. Information on the first author, published year, ethnicity, number of cases and controls, genotyping methods, mean ages of cases and controls, ratio of male:female, Hardy-Weinberg equilibrium (HWE) for control group, number of alleles and genotype, and SNPs were extracted, and are listed in Table 1. All discrepancies were resolved by a consensus achieved by J.M.L.
Table 1.
Characteristics of included studies
| Reference | Year | Ethnicity/location | Case | Control | Genotyping methods | Age (mean±SD), case/control | Female %, case/control | HWE | SNPs | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Chang et al22 | 2011 | Chinese | 636 | 510 | Massarray | 55.83±11.18/ 52.44±14.01 |
40.6%/43.1% | P>0.05 | rs11931532; rs4698412; rs11724635 | 7 |
| Miyake et al28 | 2012 | Japanese | 229 | 357 | TaqMan | 68.4±8.7/ 66.6±8.5 |
61.6%/61.1% | P>0.05 | rs11931532; rs11724635 | 8 |
| Chang et al23 | 2015 | Chinese | 596 | 597 | Agena Massarray | 68.68±10.82/60.0±12.7 | 45.9%/54.4% | P>0.05 | rs11724635 | 7 |
| Cui et al26 | 2018 | Chinese | 168 | 196 | Massarray | 68.82±10.70/ 67.35±5.51 |
41.7%/45.4% | P>0.05 | rs11931532; rs4698412 | 6 |
| Tian31 | 2012 | Chinese | 1,019 | 1,030 | Sequencing | 54.16±12.50/ 52.16±16.45 |
41.5%/47.6% | P>0.05 | rs11931532; rs4698412 | 7 |
| Satake et al [1]20 | 2011 | Japanese | 1,078 | 2,628 | HumanHap550 array | NA | NA | P>0.05 | rs11931532; rs4698412 | 6 |
| Satake et al [2]20 | 2011 | Japanese | 988 | 2,521 | HumanHap550 array | NA | NA | P>0.05 | rs11931532; rs4698412 | 6 |
| Tan et al10 | 2010 | Chinese | 433 | 916 | Massarray | NA | 44%/40% | P>0.05 | rs11931532; rs4698412 | 6 |
| Simón-Sánchez et al25 | 2011 | the Netherlands | 772 | 2,024 | Human660W-Quad array | 57.5±12.0/ 53.75±13.4 |
39.9%/53.6% | P>0.05 | rs11931532; rs4698412 | 8 |
| Spencer et al8 | 2011 | UK | 2,190 | 5,667 | Human660-Quad array | NA | 27.0%/NA | P>0.05 | rs4698412 | 6 |
| Xie32 | 2011 | Chinese | 744 | 743 | Sequencing | 53.05±10.9/ 52.34±12.1 |
36.7%/41.6% | P>0.05 | rs4698412 | 6 |
| Chen et al27 | 2014 | Chinese | 468 | 487 | TaqMan | 66.02±9.46/ 67.16±9.33 |
49.8%/66.5% | P>0.05 | rs11724635 | 8 |
Abbreviations: HWE, Hardy-Weinberg equilibrium; SNP, single nucleotide polymorphism; NOS, Newcastle-Ottawa scale; NA, not available.
Quality assessment
All included studies were evaluated using the Newcastle-Ottawa Scale (NOS)30 independently by L.L. and F.H. Any discrepancies in the assessment were resolved by the third author (L.T.). Adequacy of case definition, representative of the cases, selection of controls, definition of controls, comparability cases/controls, comparability cases/controls, same method of ascertainment, and non-response rate were taken into account and given a corresponding score. The quality scores ranged from 0–9. Only studies with a score of 6 or higher were included.
Statistical methods
All statistical analyses were performed using the STATA 12.0 software (StataCorp, College Station, TX, USA) and Revman 5 (Cochrane Collaboration, London, UK). The odds ratio (OR) and 95% confidence interval (95% CI) were calculated for evaluating the association between BST1 polymorphisms and PD risk. The pooled ORs were calculated using a genetic model of allelic, recessive, and dominant models. The statistical significance of the OR was determined using the Z-test. Variation and heterogeneity were evaluated using Cochran’s Q-statistic. The random effect model was used when there was significant heterogeneity across studies (I2>50% or P<0.05), otherwise the fixed effect model was used in meta-analysis. Sources of heterogeneity were evaluated by stratification analysis of ethnicity. Sensitivity analysis was performed to assess the effects of individual study on pooled results and the stability of results. The publication bias was detected with Begg’s test and Egger’s test. P<0.05 was considered statistically significant.
Results
Characteristics of the studies
As shown in Figure 1, a total of 1,173 studies were identified through the initial search. After screening the titles and abstracts, 37 studies were excluded for being duplicated studies, and 1,085 studies were excluded for not being related to the genetic association between BST1 polymorphisms and PD risk. After reading the full articles, 35 studies were excluded for being reviews, letters, short communications, or conference abstracts. In addition, five studies were excluded for unavailable data. Overall, a total of 11 studies were finally enrolled in the present meta-analysis.8,10,20,22,23,25–28,31,32 Among these studies, the study conducted by Satake et al20 involved two different groups. We treated each group as an individual study. Therefore, there were 12 studies with 8,725 PD patients and 17,097 healthy controls in the present meta-analysis. Two studies were on Caucasians,8,25 the others were on Asians.10,20,22,23,26–28,31,32 A total of eight, nine, and three studies have reported the association between rs11931532, rs4698412, and rs11724635 and PD risk, respectively. The genetic distributions of the control groups in all studies were all consistent with the Hardy-Weinberg equilibrium (HWE). The Newcastle-Ottawa Scale (NOS) was used for quality assessment, and all of the studies achieved moderately high quality scores above 6 (Tables 1 and S1).
Figure 1.

PRISMA flow chart of studies included and excluded.
Table S1.
Methodological quality of the included studies according to the Newcastle-Ottawa scale
| Reference | Year | Ethnicity/location | Adequacy of case definition |
Representative ness of the cases |
Selection of controls |
Definition of controls |
Comparability cases/controls |
Ascertainment of exposure |
Same method of ascertainment |
Non-response rate |
Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chang | 2011 | Chinese | * | * | * | NA | * | * | * | NA | 7 |
| Miyake | 2012 | Japanese | * | * | * | NA | * | * | * | * | 8 |
| Chang | 2015 | Chinese | * | * | * | NA | * | * | * | NA | 7 |
| Cui | 2018 | Chinese | * | * | * | NA | * | * | * | * | 8 |
| Tian | 2012 | Chinese | * | * | * | NA | * | * | * | NA | 7 |
| Satake | 2011 | Japanese | NA | NA | * | * | * | * | * | NA | 6 |
| Tan | 2010 | Chinese | NA | NA | * | * | * | * | * | NA | 6 |
| Simón-Sánchez | 2011 | the Netherlands | * | * | * | * | * | * | NA | * | 8 |
| Spencer | 2011 | UK | NA | NA | * | NA | * | * | * | * | 6 |
| Xie | 2011 | Chinese | NA | NA | * | * | * | * | NA | * | 6 |
| Chen | 2014 | Chinese | * | * | * | NA | * | * | * | * | 8 |
Notes: This table identifies high quality choices with “an asterisk.” A study can be awarded a maximum of one star for each numbered item within the Selection and Exposure categories. A maximum of two stars can be given for Comparability.
Abbreviations: *, Yes; NA, not applicable.
Results of meta-analysis
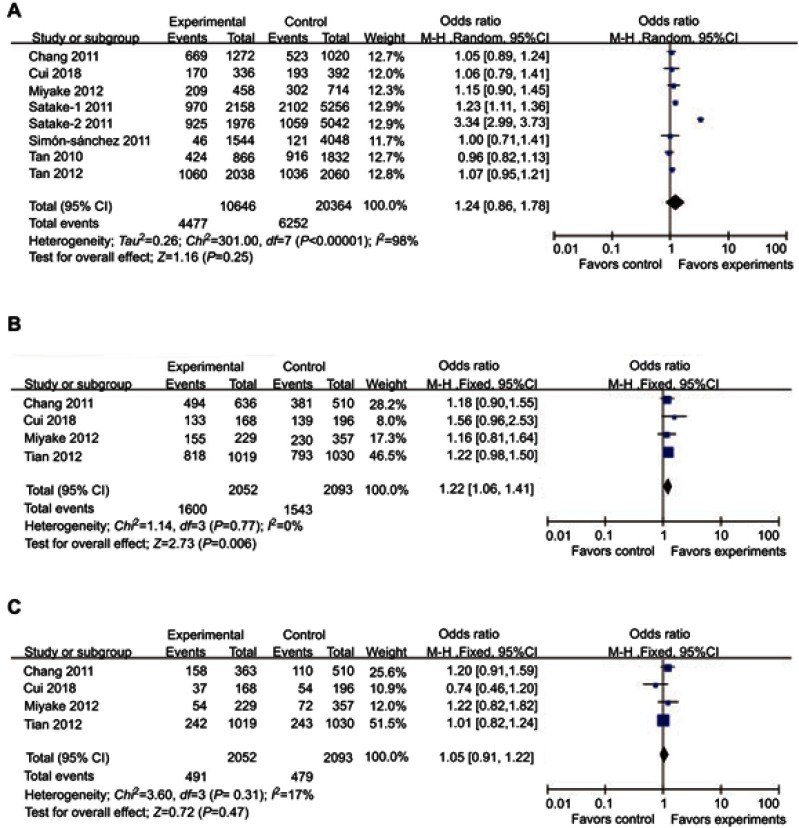
Significant association was observed between the dominant model (TT+CT/CC) of rs11931532 and PD risk (P=0.006, OR [95%CI]=1.22 [1.06–1.41]), but not in allelic (T/C) and recessive models (TT/CT+CC) of rs11931532 (allelic model: P=0.25, OR [95%CI]=1.24 [0.86–1.78]; recessive model: P=0.47, OR [95%CI]=1.05 [0.91–1.22]) (Table 2, Figure 2). Subgroup analysis stratified by ethnicity showed that the significant association between the dominant model (TT+CT/CC) of rs11931532 and PD risk can only be found in the Asian subgroup (P=0.01, OR [95%CI]=1.24 [1.05–1.47]), but not in the Caucasian subgroup (Table 2).
Table 2.
The association between BST1 polymorphisms and Parkinson’s disease
| SNPs (minor allele) |
Genetic model | Subgroup | Number of studies | Numbers | Test of association | Model | Test of heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR [95% CI] | P-value | P-value | I2 (%) | |||||
| rs11931532 (T) | Allelic (T) | Total | 8 | 10,646 | 20,364 | 1.24 [0.86–1.78] | 0.25 | R | <0.00001 | 98 |
| Asian | 7 | 9,102 | 16,316 | 1.28 [0.86–1.88] | 0.22 | R | <0.00001 | 98 | ||
| Caucasian | 1 | 1,544 | 4,048 | 1.00 [0.71–1.41] | 0.25 | – | – | – | ||
| Dominant (TT+CT/CC) | Total | 4 | 2,052 | 2,093 | 1.22 [1.06–1.41] | 0.006 | F | 0.77 | 0 | |
| Asian | 4 | 2,052 | 2,093 | 1.22 [1.06–1.41] | 0.006 | F | 0.77 | 0 | ||
| Caucasian | 0 | – | – | – | – | – | – | – | ||
| Recessive (TT/CT+CC) | Total | 4 | 2,052 | 2,093 | 1.05 [0.91–1.22] | 0.47 | F | 0.31 | 17 | |
| Asian | 4 | 2,052 | 2,093 | 1.05 [0.91–1.22] | 0.47 | F | 0.31 | 17 | ||
| Caucasian | 0 | – | – | – | – | – | – | – | ||
| rs4698412 (A) | Allelic (A) | Total | 9 | 16,056 | 32,470 | 1.16 [1.08–1.25] | <0.0001 | R | 0.003 | 65 |
| Asian | 7 | 10,132 | 17,008 | 1.22 [1.16–1.29] | <0.00001 | F | 0.43 | 0 | ||
| Caucasian | 2 | 5,924 | 15,382 | 1.03 [0.92–1.16] | 0.62 | R | 0.08 | 67 | ||
| Dominant (AA+GA/GG) | Total | 4 | 2,576 | 2,479 | 1.35 [1.20–1.52] | <0.00001 | F | 0.67 | 0 | |
| Asian | 4 | 2,576 | 2,479 | 1.35 [1.20–1.52] | <0.00001 | F | 0.67 | 0 | ||
| Caucasian | 0 | – | – | – | – | – | – | – | ||
| Recessive (AA/GA+GG) | Total | 4 | 2,576 | 2,479 | 1.30 [1.13–1.50] | 0.0003 | F | 0.44 | 0 | |
| Asian | 4 | 2,576 | 2,479 | 1.30 [1.13–1.50] | 0.0003 | F | 0.44 | 0 | ||
| Caucasian | 0 | – | – | – | – | – | – | – | ||
| rs11724635 (A) | Allelic (A) | Total | 3 | 2,586 | 2,882 | 1.07 [0.96–1.20] | 0.19 | F | 0.34 | 7 |
| Asian | 3 | 2,586 | 2,882 | 1.07 [0.96–1.20] | 0.19 | F | 0.34 | 7 | ||
| Dominant (AA+CA/CC) | Total | 3 | 1,293 | 1,441 | 1.08 [0.92–1.27] | 0.33 | F | 0.56 | 0 | |
| Asian | 3 | 1,293 | 1,441 | 1.08 [0.92–1.27] | 0.33 | F | 0.56 | 0 | ||
| Recessive (AA/CA+AA) | Total | 3 | 1,293 | 1,441 | 1.12 [0.92–1.37] | 0.25 | F | 0.42 | 0 | |
| Asian | 3 | 1,293 | 1,441 | 1.12 [0.92–1.37] | 0.25 | F | 0.42 | 0 | ||
Abbreviations: SNP, single nucleotide polymorphism; R, random model; F, fixed model; OR, odds ratios; CIs, confidence intervals.
Figure 2.
Forest plots of odds ratios for the association between BST1 rs11931532 and PD. (A) allelic model; (B) dominant model; (C) recessive model.
Abbreviation: PD, Parkinson’s disease.
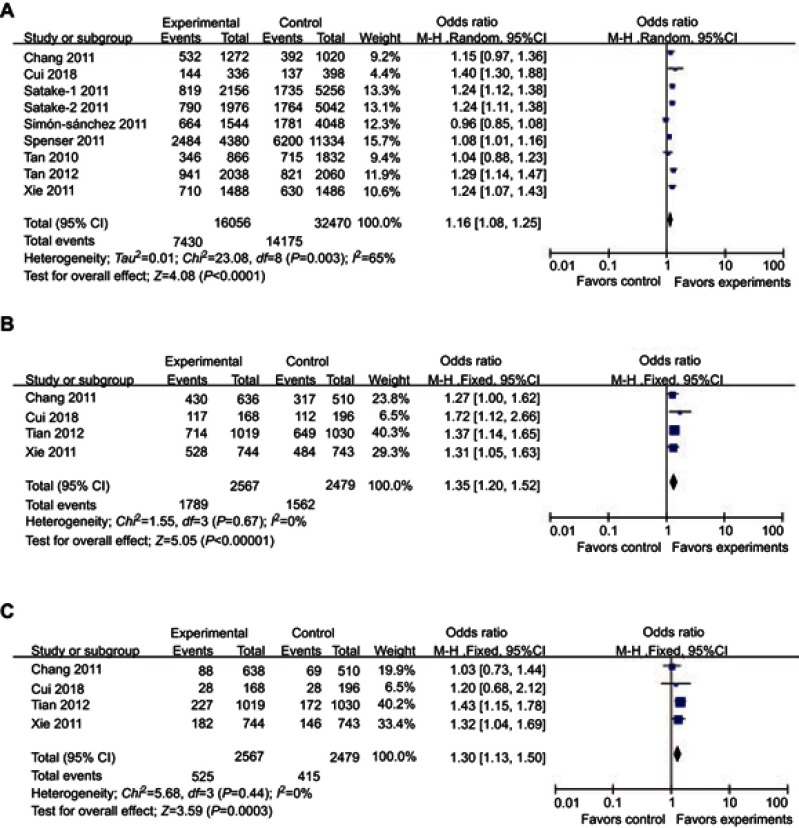
For rs4698412, significantly different distributions in cases and controls were detected between the allelic, dominant, and recessive models and PD risk (allelic model: P<0.0001, OR [95%CI]=1.16 [1.08–1.25]; dominant model: P<0.00001, OR [95%CI]=1.35 [1.20–1.52]; recessive model; P=0.0003, OR [95%CI]=1.30 [1.13–1.50]) (Table 2, Figure 3). Subgroup analysis stratified by ethnicity showed that significant associations between the allelic, dominant, and recessive models of rs4698412 and PD risk can only be found in the Asian subgroup (allelic model: P<0.00001, OR [95%CI]=1.22 [1.16–1.29]; dominant model: P<0.00001, OR [95%CI]=1.35 [1.20–1.52]; recessive model; P=0.0001, OR [95%CI]=1.30 [1.13–1.50]), but not in the Caucasian subgroup (P>0.05) (Table 2).
Figure 3.
Forest plots of odds ratios for the association between BST1 rs4698412 and PD. (A) allelic model; (B) dominant model; (C) recessive model.
Abbreviation: PD, Parkinson’s disease.
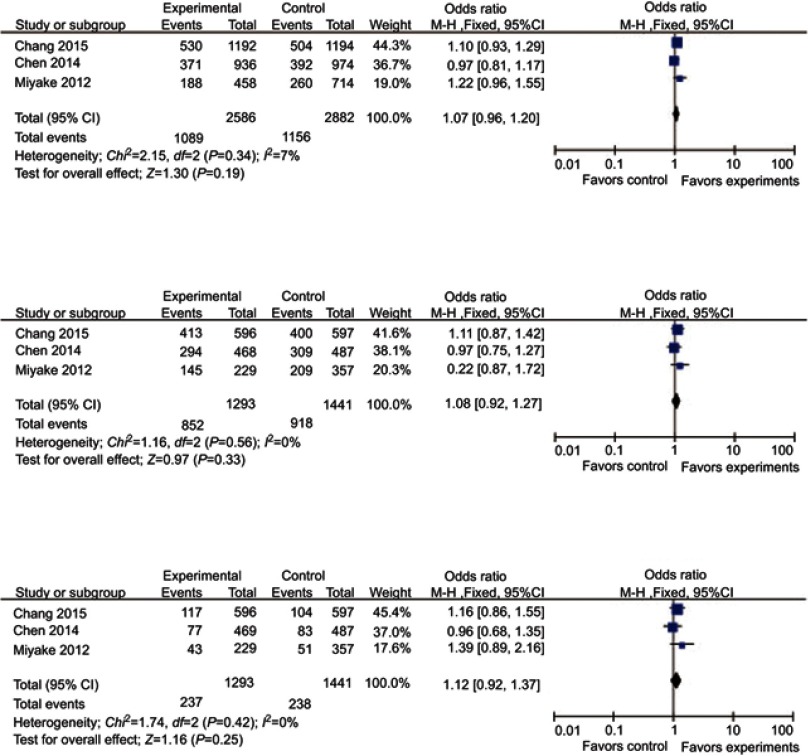
No association was found between genetic models of rs11724635 and PD risk (Table 2, Figure 4). The subgroup analysis stratified by ethnicity was canceled for lack of sufficient data in the Caucasian group.
Figure 4.
Forest plots of odds ratios for the association between BST1 rs11724635 and PD. (A) allelic model; (B) dominant model; (C) recessive model.
Abbreviation: PD, Parkinson’s disease.
Heterogeneity
Significant heterogeneity were detected in the allelic model of rs11931532 (P<0.00001, I2=98%) and the allelic model of rs4698412 (P=0.003, I2=65%). Therefore, subgroup analysis stratified by ethnicity was performed . Notably, the significant heterogeneity of the allelic model of rs11931532 was found in the Asian subgroup (P<0.00001, I2=98%). The significant heterogeneity of the allelic model of rs4698412 disappeared in both Asian (P=0.43, I2=0%) and Caucasian subgroups (P=0.08, I2=67%) (Table 2). For the allelic model of rs11931532 in the Asian subgroup, this heterogeneity was contributed mainly by one positive study conducted by Satake et al6. Removal of this study from meta-analysis gave 0% (P=0.61) heterogeneity.
Sensitive analysis and publication bias
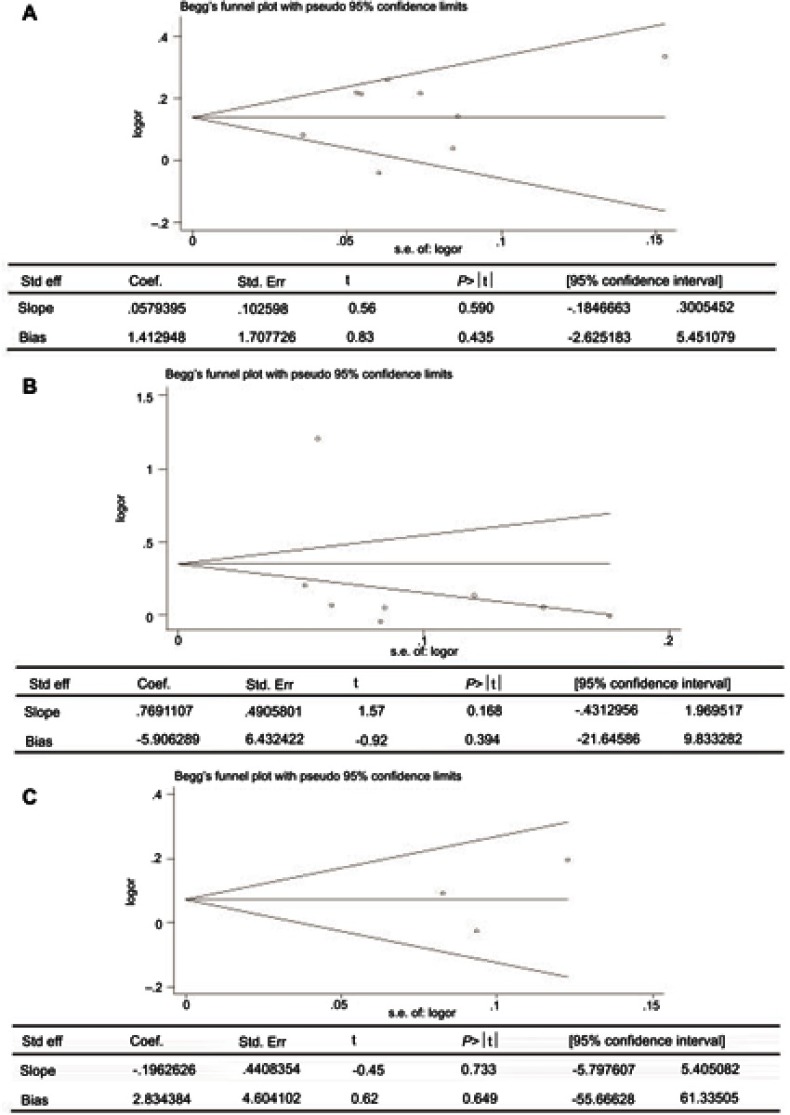
Sensitivity analysis which excluded the influence of a single study on the overall risk estimate by excluding one study at a time was confirmed. The ORs were not significantly altered in genetic models of rs11931532, rs4698412, and rs11724635 in BST1 (Figure 5). Begg’s test and Egger’s test were used to evaluate publication bias. The P-value for Egger’s linear regression test is shown in Figure 6. The results revealed that there was no obvious publication bias in overall analysis for BST1 rs11931532 (pegger=0.435), rs4698412 (pegger=0.394), and rs11724635 (pegger=0.649) (Figure 6).
Figure 5.

Sensitivity analyses between allelic models of BST1 rs11931532, rs4698412, rs11724635, and PD. (A) rs11931532; (B) rs4698412; (C) rs11724635.
Abbreviation: PD, Parkinson’s disease.
Figure 6.
Publication bias of literatures for BST1 rs11931532, rs4698412, and rs11724635 were tested by Begg’ s funnel plot and Egger’ s test. (A) rs11931532; (B) rs4698412; (C) rs11724635.
Abbreviations: BST1, bone marrow stromal cell antigen 1.
Discussion
In the present study, we have detected a significant association between the allelic, dominant, and recessive models of rs4698412, as well as the dominant model of rs11931532 and PD risk. To the best of our knowledge, this is the first study observing the genetic associations between BST1 rs4698412 and rs11931532 and PD risk using a meta-analysis.
CD157/BST-1 plays a variety of roles in humoral immune responses; neutrophil transmigration and hematopoietic stem cell support.33,34 BST-1 produces cyclic ADP-ribose, which is a second messenger that releases Ca2+ from intracellular Ca2+ stores. Ca2+ dyshomeostasis has been proposed a possible cause of selective vulnerability of dopaminergic neurons in PD.35,36 The rs11931532 and rs4698412 were located from intron 8 to 4.1 kb downstream of BST1 separately and may modify ADPribosylcyclase activity, thus leading to Ca2+ dyshomeostasis in dopaminergic neurons.20,37 There are an increasing number of new pathogenic variants located in introns. Recent evidence suggested that many disease-related intronic SNPs have been reported to be responsible for aberrant splice processes.38,39 Satake et al2+.20 have also suggested rs11931532 may modify ADP-ribosylcyclase activity, thus leading to Ca2+ dyshomeostasis in dopaminergic neurons in PD.Strong evidence of an association for the dominant model of rs11931532 and PD risk in Asian populations was found, which might indicate the dominant model of rs11931532 was a risk factor of PD in Asian populations.
The 3ʹ UTR of genes were indicated to play an important role in regulating the expression of genes. Thus, the BST1 rs4698412 may increase the PD risk by changing the expression level of BST1 gene. Significant associations were found in GWAS studies and case-control studies. However, negative results were reported by Tan et al10 and Chuang et al34 in Chinese populations. The inconsistency ofthese researchers may be due to the different ethnicity, study design, sample size, and environmental factors. In our combined meta-analysis, we observed a significant association between BST1 rs4698412 and PD risk in Asian populations, but not in Caucasian populations, which may indicate the genetic background influences the effect of BST1 rs4698412 to the PD risk. However, the sample size in the Caucasian population was relatively small. To identify the association between rs4698412 and PD risk in Caucasian populations, studies with a larger number of subjects from multiple ethnicity are necessary.
When investigating the rs11724635 with genetic models, no significant association with PD was found in the overall and subgroup population-based analysis. A recent logistic regression analysis of data from a multi-center hospital-based case-control study in Japan found no relationship between BST1 rs11724635 and sporadic PD.24 This negative result was replicated in subsequent studies in Chinese populations.23,26 However, a meta-analysis based on the GWAS studies has revealed that the BST1 rs11724635 was a strong susceptible factor for PD in US, European, and Asian populations.25 As there was no available data in the studies conducted by Saad et al21, Spencer et al8, and Simon-Sanchez et al25, we had to exclude these three studies in the present study. In addition, we enrolled three other studies,23,26,28 and found no association between BST1 rs11724635 and PD risk, which brings us to the conclusion that the BST1 rs11724635 may not be a risk factor for PD.
Limitations should be taken into account. First, the polymorphisms included in the present meta-analysis were located in the 5ʹ or 3ʹ noncoding sequences of BST1 gene, and no polymorphism in the coding sequences was included. Although elements such as promoter, enhancer, or silencer were usually at untranslated region, which might be responsible for regulating gene expression, it still cannot be excluded that there are rare forms of mutation in the coding region. Further studies are required to include PD-associated polymorphisms in the coding sequences. Second, the number of subjects in the present study was relatively small, especially in the Caucasian subgroup, which may partly influence the result of the association between the BST1 polymorphisms and PD in the Caucasian population. The lack of a significant association between SNP rs11724635 and sporadic PD is likely to be attributable to insufficient statistical power. Third, ethnic specific effect is an important consideration in meta-analysis. There were only two different ethnicities in the present study. A larger number of studies with more subjects of multiple ethnicities are necessary in the future.
Conclusions
Our meta-analysis suggests that the rs11931532 and rs4698412, but not rs11724635, might increase the risk of PD in Asian populations. However, more large-scale studies with a larger number of subjects are warranted to confirm these findings.
Acknowledgments
The authors thank Professor Yan Zhang for revising the whole manuscript. This study was funded by the National Natural Science Foundation of China (Grant No. 81873780), the Foundation of the Health Department of Hunan (No. B2017041), the Foundation of the Education Department of Hunan (16A027, 13C1115, 13C1116, and 16C0167), The foundation of Hunan national science (The Youngth Item) (No. 2019JJ50697), The Application Characteristic Discipline of Hunan Province, The Hunan Key Laboratory Cultivation Base of the Research and Development of Novel Pharmaceutical Preparations (No. 2016TP1029), and The Hunan Provincial Innovation Platform and Talents Program (No. 2018RS3105).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary material
References
- 1.Nakano N, Matsuda S, Ichimura M, et al. PI3K/AKT signaling mediated by G protein-coupled receptors is involved in neurodegenerative Parkinson‘s disease (Review). Int J Mol Med. 2017;39(2):253–260. [DOI] [PubMed] [Google Scholar]
- 2.Katunina EA, Bezdolny YN. [Epidemiology of Parkinson’s disease]. Zh Nevrol Psikhiatr Im S S Korsakova. 2013;113(12):81–88. [PubMed] [Google Scholar]
- 3.Bonuccelli U, Del DP. New pharmacologic horizons in the treatment of Parkinson disease. Neurology. 2006;67(2):30–38. doi: 10.1212/WNL.67.7_suppl_2.S30 [DOI] [PubMed] [Google Scholar]
- 4.Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H. The Lewy body in Parkinson‘s disease and related neurodegenerative disorders. Mol Neurobiol. 2013;47(2):495–508. doi: 10.1007/s12035-012-8280-y [DOI] [PubMed] [Google Scholar]
- 5.Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013;9:445–454. doi: 10.1038/nrneurol.2013.132 [DOI] [PubMed] [Google Scholar]
- 6.Kasai S, Yoshihara T, Lopatina O, et al. Selegiline ameliorates depression-like behavior in mice lacking the CD157/BST-1 gene, a risk factor for Parkinson’s disease. Front Behav Neurosci. 2017;11:75. doi: 10.3389/fnbeh.2017.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming SM. Mechanisms of gene-environment interactions in Parkinson’s disease. Curr Envir Health Rpt. 2017;4(2):192–199. doi: 10.1007/s40572-017-0143-2 [DOI] [PubMed] [Google Scholar]
- 8.Spencer CC, Plagnol V, Strange A, et al. Dissection of the genetics of Parkinson‘s disease identifies an additional association 5ʹ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20(2):345–353. doi: 10.1093/hmg/ddq469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soto-Ortolaza AI, Heckman MG, Labbé C, et al. GWAS risk factors in Parkinson‘s disease: LRRK2 coding variation and genetic interaction with PARK16. Am J Neurodegener Dis. 2013;2(4):287–299. [PMC free article] [PubMed] [Google Scholar]
- 10.Tan EK, Kwok HK, Tan LC, et al. Analysis of GWAS-linked loci in Parkinson disease reaffirms PARK16 as a susceptibility locus. Neurology. 2010;75(6):508–512. doi: 10.1212/WNL.0b013e3181eccfcd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan JY, Edwards KL, Hutter CM, et al. Association mapping of the PARK10 region for Parkinson‘s disease susceptibility genes. Parkinsonism Relat Disord. 2014;20(1):93–98. doi: 10.1016/j.parkreldis.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma M, Maraganore DM, Ioannidis JP, et al. Role of sepiapterin reductase gene at the PARK3 locus in Parkinson‘s disease-neurobiology of aging. Neurobiol Aging. 2011;32(11):2108.e1-2108.e5. doi: 10.1016/j.neurobiolaging.2011.05.024 [DOI] [PubMed] [Google Scholar]
- 13.Hamza TH, Zabetian CP, Tenesa A, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42(9):781–785. doi: 10.1038/ng.642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maraganore DM, de Andrade AM, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L-H, Luo X-G, Zhou Y-S, et al. Lack of association between three single nucleotide polymorphisms in the PARK9, PARK15, and BST1 genes and Parkinson‘s disease in the northern Han Chinese population. Chin Med J (Engl). 2012;125(4):588–592. [PubMed] [Google Scholar]
- 16.Ahmed I, Tamouza R, Delord M, et al. Association between Parkinson‘s disease and the HLA-DRB1 locus. Mov Disord. 2012;27(9):1104–1110. doi: 10.1002/mds.25035 [DOI] [PubMed] [Google Scholar]
- 17.Li N-N, Tan E-K, Chang X-L, et al. Genetic association study between STK39 and CCDC62/HIP1R and Parkinson‘s disease. PLoS One. 2013;8(11):e79211. doi: 10.1371/journal.pone.0079211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Cheng R, Verbitsky M, et al. Genome-wide association study identifies candidate genes for Parkinson‘s disease in an Ashkenazi Jewish population. BMC Med Genet. 2011;12(1):104. doi: 10.1186/1471-2350-12-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrero E, Buono NL, Morone S, et al. Human canonical CD157/Bst1 is an alternatively spliced isoform masking a previously unidentified primate-specific exon included in a novel transcript. Sci Rep. 2017;7(1):15923. doi: 10.1038/s41598-017-16184-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson‘s disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485 [DOI] [PubMed] [Google Scholar]
- 21.Saad M, Lesage S, Saintpierre A, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson‘s disease in the European population. Hum Mol Genet. 2011;20(3):615–627. doi: 10.1093/hmg/ddq497 [DOI] [PubMed] [Google Scholar]
- 22.Chang X-L, Mao X-Y, Li H-H, et al. Association of GWAS loci with PD in China. Am J Med Genet B Neuropsychiatr Genet. 2011;156(3):334–339. doi: 10.1002/ajmg.b.v156.3 [DOI] [PubMed] [Google Scholar]
- 23.Chang KH, Wu YR, Chen YC, et al. STK39, But Not BST1, HLA-DQB1, and SPPL2B Polymorphism, Is Associated With Han-Chinese Parkinson‘s Disease in Taiwan. Medicine. 2015;94(41):e1690. doi: 10.1097/MD.0000000000000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma M, Ioannidis JPA, Aasly JO, et al. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79(7):659–667. doi: 10.1212/WNL.0b013e318264e353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simón-Sánchez J, van Hilten JJ, van de Warrenburg B, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet. 2011;19(6):655–661. doi: 10.1038/ejhg.2010.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui C, Liu WG, Hua P, et al. Association of BST1 gene polymorphism with Parkinson‘s disease risk in Chinese population. Chin J Clin Neurosci 2018;16(1):1–8. [Google Scholar]
- 27.Chen M-L, Lin C-H, Lee M-J, Wu R-M. BST1 rs11724635 interacts with environmental factors to increase the risk of Parkinson‘s disease in a Taiwanese population. Parkinsonism Relat Disord. 2014;20(3):280–283. doi: 10.1016/j.parkreldis.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 28.Miyake Y, Tanaka K, Fukushima W, et al. Lack of association between BST1 polymorphisms and sporadic Parkinson‘s disease in a Japanese population. J Neurol Sci. 2012;323(1–2):162–166. doi: 10.1016/j.jns.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 29.Harms M. The EQUATOR network and the PRISMA statement for the reporting of systematic reviews and meta-analyses. Physiotherapy. 2009;95(4):237–240. doi: 10.1016/j.physio.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Xia Z, Guo S, Yu X, Li Z, Sung S-Y. Laparoscopically assisted anorectal pull-through versus posterior sagittal anorectoplasty for high and intermediate anorectal malformations: a systematic review and meta-analysis. PLoS One. 2017;12:e0170421. doi: 10.1371/journal.pone.0170421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian JY. Polymorphism Analysis of PARK16, PARK17, PARK18, BST1 and Polygenic Determinants Analysis of Parkinson‘S Disease. Changsha: Central South University; 2012. [Google Scholar]
- 32.Xie D. The Study of Relationship between polymorPhism in BSTI Gene Rs4698412 and Sporadic Parkinson‘S Disease. Hengyang: University of South China; 2011. [Google Scholar]
- 33.Ishihara K, Hirano T. BST-1/CD157 regulates the humoral immune responses in vivo. Chem Immunol. 2004;75(4):235–255. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama S, Mahmuda NA, Munesue T, et al. Association study between the CD157/BST1 gene and autism spectrum disorders in a Japanese population. Brain Sci. 2015;5(2):188–200. doi: 10.3390/brainsci5020188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sassi C. Genetics of Parkinson disease. Neurorx. 2004;1(2):235–242. doi: 10.1602/neurorx.1.2.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung SJ, Jung Y, Hong M, et al. Alzheimer‘s disease and Parkinson‘s disease genome-wide association study top hits and risk of Parkinson‘s disease in Korean population. Neurobiol Aging. 2013;34(11):2695.e. doi: 10.1016/j.neurobiolaging.2012.06.026 [DOI] [PubMed] [Google Scholar]
- 37.Higashida H, Liang M, Yoshihara T, et al. An immunohistochemical, enzymatic, and behavioral study of CD157/BST-1 as a neuroregulator. BMC Neurosci. 2017;18:35. doi: 10.1186/s12868-017-0350-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498 [DOI] [PMC free article] [PubMed] [Google Scholar]