Abstract
Each year, gastric cancer accounts for nearly one million new cases and over 720,000 deaths worldwide. Prognosis is dismal as most patients are diagnosed with advanced gastric cancer. Gastric cancer outcomes will benefit from better ways to identify at risk individuals. An approach to target high-risk populations is to identify those who are genetically predisposed to gastric cancer as up to 15% of all patients report family history of the disease. Based on clinical manifestations, three gastric cancer syndromes have been described but recent findings suggest that the diagnosis of some of these syndromes is sub-optimal and it could benefit from genetic information. Using genome-wide association and next generation sequencing studies, the last decade has seen the identification of low penetrance and high-risk genes that have considerably increased our understanding of gastric cancer risk. PALB2 has emerged as new familial gastric cancer gene from these studies. Furthermore, recent genetic analyses in sporadic patients suggest that >10% of all cases have pathogenic mutations, a finding of great importance for cancer aetiology. In this article, we provide a comprehensive review on the role of genetics in gastric cancer aetiology and the implications of recent genetics findings for the prevention of this malignancy.
Introduction
With nearly one million new cases diagnosed per year, gastric cancer is the fifth most commonly diagnosed cancer worldwide.1–3 After lung and liver cancer, gastric cancer is one of the most deadliest malignancies, each year accounting for >720,000 deaths.3 One third of all gastric cancers are diagnosed in the cardia, the mucosa originating about five centimetres from the gastro-oesophageal junction,4 and two thirds arise in the mucosa distal to the cardia (non-cardia cancers, Figure 1)5,6 Histologically, gastric cancer is classified into intestinal cancers, which resemble the intestinal mucosa and have glandular formations with intercellular junctions; and diffuse cancers, where cell clusters lacking cell-cell cohesion and infiltrated stroma are seen7. Diffuse cancers include signet ring cell adenocarcinomas, where increased intracellular mucosa presses the nucleus to the cell periphery.7 These anatomic and histological classifications have associations with epidemiology. Helicobacter pylori (H. Pylori) infection is closely associated with intestinal non-cardia cancers while obesity or gastro-oesophageal reflux disease (GERD) has stronger effects on cardia cancers.1,2 Due mostly to H. pylori eradication programs, non-cardia cancer incidence is decreasing while cardia cancer incidence is increasing; particularly in countries with high obesity rates.6
Figure 1:
Gastric cancer anatomical classification into cardia and non-cardia cancer and the role of environmental and lifestyle risk factors. Risk factors for both cardia and non-cardia are shown in the middle of figure. They are shown in italics if they have stronger effects for cardia tumours and in bold if they have stronger effects for non-cardia tumours
The limited disparity between incidence and mortality rates highlights the poor prognosis of gastric cancer. Only a small fraction of cases are detected at early stages and, as a consequence, gastric cancer has poor outcomes.1,2,6 Only 5% of metastatic cancer patients are alive five years after diagnosis. 1,2,6 These dismal survival rates underscore the need to identify at-risk individuals who can benefit from early interventions. Major gaps in gastric cancer prevention and treatment remain and it is hoped that recent genetic advances will provide new tools that lead to better outcomes. Here, we provide a review of gastric cancer aetiology, with an emphasis on the role genetic predisposition.
Risk Factors for Gastric Cancer
The major gastric cancer risk factors include age, H. pylori and Epstein-Barr virus (EBV) infection, race, sex, obesity, GERD, tobacco, alcohol and family history (Figure 1). 1,2,6 Data from the U.S. Surveillance, Epidemiology and End Results program (https://seer.cancer.gov/statfacts/html/stomach.html) indicates that >80% of cases are diagnosed in individuals 55 years (y) or older and the median age of diagnosis is 68y.6 Approximately 89% of all non-cardia cancers are attributed to H. pylori8 and 9% to EBV.9 Race/ethnicity is another risk factor. In the United States, the gastric cancer incidence in individuals of Latino and Asian ancestry is about 2-fold higher than in Whites.10 However, when incidence is stratified by anatomic location, cardia cancers are more often diagnosed in Whites.11,12 Indeed, cardia cancer incidence is increasing among White men as this group is enriched for risk factors including obesity and GERD 11,12. Male sex increases cardia cancer risk by ~5-fold while a body mass index >30 and GERD increases risk by 82% 13 and 2–4-fold respectively. 1,2,14,15 Interestingly, a recent study suggested an increase in corpus (non-cardia) cancers among young White women16. Multiple behavioural risk factors have been also identified for gastric cancer. Nearly 17% (95% CI: 10·5–29·5%) of all gastric cancers have been ascribed to smoking; with this risk factor having a stronger effect on cardia cancers (hazard ratio, HR=4·10, 95% CI: 1·76–9·57 for cardia vs. 1·94, 95% CI: 1·05–3·60, for non-cardia).17 Heavy alcohol consumption has an increased relative risk of 1·52 (95% CI: 0·80–2·88)18 and the combined effect of heavy smoking and heavy alcohol consumption increases risk by 4·9-fold (95% CI: 1·90–12·62).19
Besides non-genetic risk factors, ~10–20% of gastric cancer patients show a familial aggregation of the disease and about 2–5% can be classified as hereditary cases. 1,2,20 Twin studies suggested that 28% of the risk is accounted for by inherited factors.21 Despite the important role of genetic factors on risk, until very recently only one gene, E-cadherin (CDH1), 22 had been associated with inherited gastric cancer. The recent advances in high-throughput DNA analysis, however, have led to the identification of several genomic regions associated with an increased risk of gastric cancer.23–32
Hereditary Gastric Cancer Syndromes
The three primary familial gastric cancer syndromes (See Table 1) that have been described in the literature include Hereditary Diffuse Gastric Cancer (HDGC), Familial Intestinal Gastric Cancer (FIGC) and Gastric Adenocarcinoma and Proximal Polyposis of the stomach (GAPPS). Gastric cancer is also reported in cancer syndromes such as Lynch Syndrome (LS), Li-Fraumeni Syndrome (LFS), Juvenile Polyposis Syndrome (JPS), Peutz-Jeghers Syndrome (PJS) and Hereditary Breast and Ovarian Cancer Syndrome (HBOCS). Below, we provide a brief overview of the genetics and clinical characteristics of the most common syndromes where gastric cancer cases are reported.
Table 1.
Genetic heritable syndromes associated with gastric cancer.
| Syndromes | Associated Genes |
Predominant Histology |
Prevalence | Gastric Cancer Risk |
||
|---|---|---|---|---|---|---|
| Primary | Hereditary diffuse gastric cancer (HDGC) |
CDH1 CTNNA1 |
Diffuse | 1:1,000,000 | 56–70% | |
| Familial intestinal gastric cancer (FIGC) | APC | Intestinal | Unknown | Unknown | ||
| Gastric Adenocarcinoma and proximal polyposis of the stomach (GAPPS) | APC | Intestinal | 1:1,000,000 | Unknown | ||
| Secondary | Familial adenomatous polyposis (FAPS) | APC | Intestinal | 1:8,300 | 2.1–4.2% | |
| Hereditary breast and ovarian cancer (HBOC) |
BRCA1
BRCA2 |
Diffuse | 1:400 – 1:500 | 2.6–5.5% | ||
| Juvenile-polyposis syndrome (JPS) |
SMAD4
BMPR1A ENG |
Intestinal | 1:16,000 – 1:100,000 | 21% | ||
| Li-Fraumeni syndrome (LFS) | TP53 | Intestinal | 1:10,000–1:25,000 (U.K.) 1:20,000 (U.S) |
3.1–4.9% | ||
| Lynch syndrome (LS) |
MSH2
MSH6 MLH1 PMS2 EPCAM |
Intestinal | 1:370 to 1:2,000 | 11–30% | ||
| Peutz-Jeghers syndrome (PJS) | STK11 | Intestinal | 1–9 / 1,000,000 | 29% | ||
Hereditary Diffuse Gastric Cancer Syndrome (HDGC)
HDGC is a highly penetrant disease caused by germline mutations in CDH1, which encodes E-Cadherin. 22 E-cadherin is a transmembrane protein that plays an integral role in maintaining cell-cell cohesion, and binds intracellular catenin molecules which mediate cell growth pathways.33 It is also involved in the development and metastases of multiple malignancies through its interactions with multiple signalling pathways.33 Current guidelines classify families as HDGC if they meet one or more of the following criteria: 1) two or more gastric cancer cases, where at least one is the diffuse-type, 2) one case of diffuse gastric cancer before age 40y, and 3) personal or family history of diffuse gastric cancer or lobular breast cancer within a single individual before age 50y. Testing for CDH1 mutations should also be considered for: 1) families or persons with a history of lobular breast cancer under age 50y, 2) individuals with or family history of cleft lip/cleft palate and diffuse gastric cancer, or 3) individuals with signet ring cells in the stomach as diagnosed by an expert pathologist.34
Forty percent of HDGC cases carry CDH1 mutations6,35, which confer a gastric cancer lifetime risk of 70% (95% CI: 59–80%) in men and of 56% (95% CI: 44%−69%) in women.36 Women with CDH1 mutations also have a 42% lifetime risk of developing lobular breast cancer.35,36 The median age of gastric cancer diagnosis in CDH1 mutation carriers is 38y.6,35 Due to poor prognosis, CDH1 mutation carriers are advised to have a prophylactic gastrectomy or to have frequent endoscopic surveillance following a specialised high-risk protocol, as endoscopy is not a fail-safe and may miss cancers.34,35,37 In fact, 90% of prophylactic gastrectomies in CDH1 mutation carriers have signet ring cell gastric adenocarcinoma even after being cleared by endoscopy underscoring the need for a surgical intervention among CDH1 carriers.38 In 2013, evidence for a second HDGC gene was provided when Majewski et al 39 identified a CTNNA1 mutation in a CDH1-negative HDGC pedigree. However, CTNNA1 mutations are rare having not been consistently found in HDGC and not often screened for in the clinical setting.25,28,31,34,36,40–43
Gastric Adenocarcinoma and Proximal Polyposis of the Stomach (GAPPS), now a subset of Familial Adenomatous Polyposis Syndrome
GAPPS was a recently described syndrome characterized by an autosomal dominant inheritance in which patients have over 100 fundic polyps and gastric cancer.6,35 Gastric cancers are associated with such polyps. Owing to its rarity, lifetime gastric cancer risk in GAPPS was unknown and its diagnostic criteria were not derived from international consensus.44–47 The originally proposed GAPPS diagnostic criteria stated that cases with other polyposis syndromes such as Familial Adenomatous Polyposis Syndrome (FAPS), a condition caused by APC mutations, should be excluded.45–47 However, recent studies identified APC mutations in seven families, which suggested that GAPPS is a subset of FAPS44,45 and not a standalone gastric cancer syndrome.
Familial Intestinal Gastric Cancer (FIGC)
FIGC is characterized by intestinal-type gastric cancer and follows an autosomal dominant inheritance pattern.35 Current FIGC diagnostic criteria 48 is stratified on the basis of the population gastric cancer incidence. In high incidence countries, such as Portugal and Japan, FIGC is diagnosed if three criteria are met: 1) at least three relatives with intestinal gastric cancers and one of them is a first-degree relative of the other two, 2) gastric cancer patients occur in at least two generations, and 3) at least one patient is diagnosed with gastric cancer before age 50y. In low-incidence countries, such as the United States, FIGC is diagnosed if at least two first/second-degree relatives have had intestinal gastric cancer, one by age 50y, or if the family includes three or more first/second-degree relatives diagnosed with gastric cancer at any age.49 Unlike HDGC or GAPPS (a subset of FAPS), the genetic basis of FIGC remains unknown. Since there is no clear evidence for the best approach to manage this syndrome, surveillance is recommended in a research environment and should involve regular endoscopy.50
Other Syndromes where Gastric Cancer is Diagnosed
In addition to the primary gastric cancer syndromes discussed above, six cancer syndromes (Table 1), including four gastro-intestinal (GI) cancer syndromes (JPS, PJS, LS and FAPS) and two multi-organ/extra-GI cancer syndromes (LFS and HBOCS), have been associated with an increased gastric cancer risk.
Lynch syndrome (LS), the most common GI cancer syndrome, is an autosomal dominant inheritance condition caused by mutations in DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS1, PMS2) or by EPCAM deletions.35 LS patients have a gastric cancer lifetime risk that ranges from 11–19%.51–53 However, it is thought that incidence rates are decreasing in Europe while remaining higher in Asia suggesting an environmental component.54 Currently, LS patients living in low risk areas are not recommended for gastric cancer surveillance because of low risk and an unproven benefit.54–56 Furthermore, Barrow et al55 found evidence for LS cohort effects and suggested that gastric cancer risk in those born after 1935, a cohort where incidence rates decreased, was not high enough to not justify surveillance. Juvenile Polyposis Syndrome (JPS) is an autosomal dominant disease caused by SMAD4, BMPR1A or ENG mutations 35,57 and is associated with a lifetime gastric cancer risk of 21%.58 Peutz-Jeghers Syndrome (PJS), caused by STK11 mutations35,59, is another syndrome that has been associated with a gastric cancer lifetime risk of 29%.60.Familial Adenomatous Polyposis Syndrome (FAPS) is an autosomal dominant syndrome caused by APC mutations.35,57 About 2% of all APC mutation carriers develop gastric cancer.61
Gastric cancer is also often reported in individuals with Li-Fraumeni Syndrome (LFS) and with Hereditary Breast and Ovarian Cancer Syndrome (HBOCS). LFS follows autosomal dominant inheritance and is caused by TP53 mutations. 35,57 TP53 mutation carriers have a gastric cancer lifetime risk of 2–3%.62 HBOCS is an autosomal dominant condition associated with BRCA1 and BRCA2 mutations 63 and has a gastric cancer lifetime risk of ~5%.64
The role PALB2 and homologous recombination DNA repair genes in gastric cancer
During the last three years, multiple studies have provided evidence for a causal role of PALB2 and other genes involved in homologous recombination DNA repair (HRDR) in gastric cancer risk.25,28,31,36,42,43 PALB2 is a breast and pancreatic cancer gene65,66 and its protein, together with BRCA1, BRCA2, and RAD51C forms a complex that repairs chromosomal breaks during DNA replication.67 Germline HRDR gene mutations generally associate with a somatic tumour phenotype called “BRCAness”, which is characterized by multiple chromosomal alterations. Cancers with BRCAness respond to certain chemotherapies and to poly (adenosine diphosphate [ADP]) ribose polymerase inhibitors (PARPi).68
The first evidence of a role for HRDR genes in gastric cancer was provided by Hansford et al 36, who examined 183 HDGC families for mutations in 55 cancer-associated genes. Of these, 19% (n=34) had CDH1 mutations and five of the 144 (3.5%) CDH1 mutation-negative families had mutations in three HRDR genes. ATM was mutated in three families, BRCA2 in one and PALB2 in another. Lu et al43 used exome sequence data from The Cancer Genome Atlas study (TCGA) in 321 sporadic gastric cancers to identify 8% of such patients as having germline mutations in HRDR genes. Recurrently mutated HRDR genes in TCGA included PALB2 and ATM, mutated in four patients each, and BRCA1 and BRCA2, mutated in three patients each.
In our own recent exome sequencing study in 28 CDH1 mutation-negative HDGC cases we identified two patients with PALB2 mutations and one with a RAD51C mutation.28 After independent replication, we identified a total of 11 patients with mutations in PALB2 (n=7), BRCA1 (n=3), or RAD51C (n=1). Our evaluation of gastric cancers from PALB2 and RAD51C mutation carriers found that they had the BRCAness mutational signature, which provided further causality evidence for these germline HRDR gene mutations.28
Two recent sequencing studies provided further evidence of a role of HRDR genes in gastric cancer.31,42 Slavin et al31 carried out targeted sequencing of 706 cancer-related genes in 43 familial cases and identified several individuals with mutations in the HRDR genes ATM, ATR, BRCA2, BRIP and FANCC. Fewings et al42, carried out exome sequencing in 22 CDH1 mutation-negative HDGC families and reported patients with mutations in PALB2 (n=1), ATR/NBN (n=1) and RECQL5 (n=2). More recently, Huang et al25 evaluated exome sequence data from 443 TCGA cases, a sample which included all patients previously examined by Lu et al 43, and reported several sporadic gastric cancer patients with HRDR gene mutations, including seven cases with ATM mutations, five with PALB2, four with BRCA2 and three with BRCA1.
Age of onset and histology among HRDR gene mutation carriers
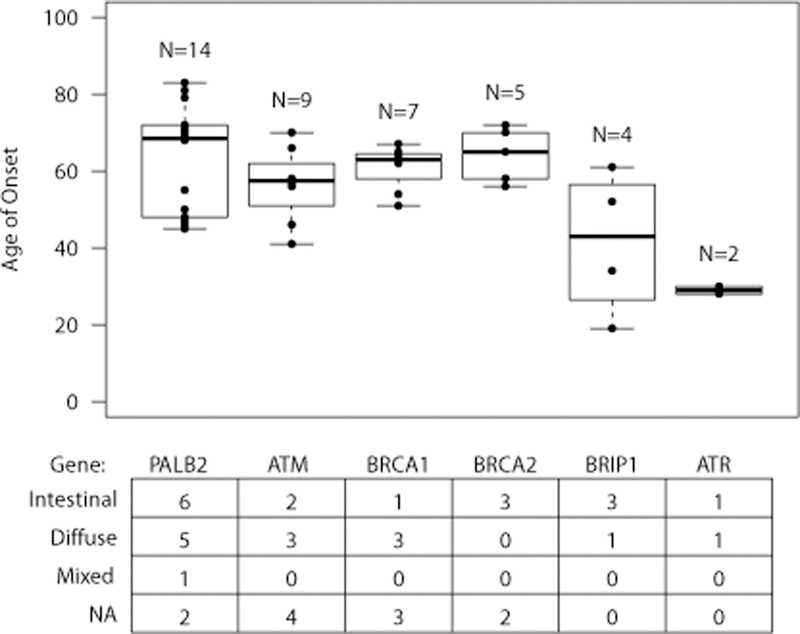
The sequencing studies discussed above have independently identified mutations in six HRDR genes (Table 2). 25,28,31,36,42 PALB2 mutations have been identified in four studies25,28,36,42, patients with ATM25,31,36or BRCA225,31,36 mutations have been reported by three studies each, and BRCA125,28, BRIP1 25,31 and ATR31,42 have been found to be mutated by two studies each. It now feels safe to conclude that PALB2 is a gastric cancer gene69 and that ATM and BRCA1 also represent good candidates. The causal role for BRCA2, BRIP1 and ATR, however, will require further studies. Figure 2 shows box plots with the ages of onset for gastric cancer patients with mutations in these six genes. The average age of onset for the 14 patients that have been reported with PALB2 mutations is 63y (range 46–81y). ATM Mutations, reported in nine patients, are associated with an earlier age of onset (56y, range 41–70y). BRCA1 and BRCA2 mutations have been reported in seven and four patients, respectively, who on average have gastric cancer at ages 61y and 65y, respectively. BRIP1 and ATR mutations have been more rarely reported but seem to be associated with the youngest ages of onset.
Table 2.
Cancer gene panel and whole exome sequencing studies that have independently reported novel candidate causal HRDR genes for gastric cancer.
| Hansford36 | Slavin31 | Huang25 | Fewings42 | Sahasrabudhe28 | ||
|---|---|---|---|---|---|---|
| Study Type | HDGC | Familial | Sporadic | HDGC | HDGC/Sporadic | |
| Design | Gene Panel | Gene Panel | Gene Panel | WES | WES/Gene Panel | |
| # Patients | Total | 183 | 43 | 443 | 22 | 28/331 |
| CDH1 | 34 | 6 | 1 | NA | NA | |
| Other | 16 | 12 | 38 | 6 | 3/9 | |
| PALB2 | 1 | 0 | 6 | 1 | 7 | |
| ATM | 3 | 1 | 7 | 0 | 0 | |
| BRCA2 | 1 | 1 | 4 | 0 | 0 | |
| BRIP1 | 0 | 1 | 3 | 0 | 0 | |
| BRCA1 | 0 | 0 | 3 | 0 | 3 | |
| ATR | 0 | 1 | 0 | 1 | 0 | |
Figure 2 (lower part) also shows the histology of the gastric cancers from HRDR gene mutation carriers. The histology of these cancers is heterogeneous and involves both cancers of intestinal and diffuse histology. For example, six patients with PALB2 mutations developed intestinal cancers, five diffuse cancers and one with a mixed histology. While two of the studies reporting PALB2 mutations had CDH1 mutation-negative HDGC 31,42 as inclusion criteria, and hence all patients included had diffuse cancers, our study and the TCGA-based study25,28 included sporadic gastric cancers, which represent most known mutation carriers with intestinal cancers. Notably, we found instances where patients with the same PALB2 mutation developed cancers with different histology.28 Hence, HRDR-deficient gastric cancers do not fully conform to the histological criteria used for HDGC and current data suggest that they have multiple histological types.
Common genetic variants conferring a high risk of gastric cancer
With the advent of the whole genome genotyping arrays, genome-wide association studies (GWAS) have been used to identify single nucleotide polymorphisms (SNPs) associated with gastric cancer risk. So far, GWAS have identified eight gastric cancer-associated genomic regions (Table 3).23,24,26,27,29,30,32 Below, we briefly describe the main characteristics of these regions.
Table 3.
Gastric cancer susceptibility risk variants discovered by genome-wide association studies (GWAS).
| Region | Gene | SNP | P | OR (95% CI) | Cancer Type | Population | Ref |
|---|---|---|---|---|---|---|---|
| 1q22 | MUC1 | rs4072037-A | 6·28E-17** | 1·35 (1·27–1·45) | 1,4,5 | Asian; European | 23,24,27,29,32 |
| 3q13.31 | ZBTB20 | rs9841504-C | 2·00E-09 | 1·32 (1·20–1·45) | 1 | Asian | 30 |
| 5p13.1 | PRKAA1 | rs13361707-C | 8·00E-29** | 1·41 (1·32–1·49) | 1,2,5 | Asian; European | 30,76 |
| 5q14.3 | lnc-POLR3G-4 | rs7712641-C | 1·21E-11 | 1·19 (1·14–1·25) | 1 | Asian | 32 |
| 6p21.1 | UNC5CL | rs2294693-C | 2·50E-08 | 1·18 (1·12–1·26) | 1 | Asian | 76 |
| LRFN2 | rs2494938-A | 5·00E-09 | 1·18 (1·12–1·25) | 1 | Asian | 26 | |
| 8q24.3 | PSCA | rs2294008-T | 1·00E-20** | 1·33 (1·25–1·41) | 1,3,4 | Asian; European | 29,32 |
| 10q23.33 | PLCE1 | rs2274223-G | 1·00E-20** | 1·57 (1·49–1·65) | 2 | Asian | 23 |
| 11q22.3 | ATM | rs758081262-T rs768362387-A rs777499935-G |
8·00E-10 | 4·74 | 3,4,5 | European | 24 |
Cancer Type: 1) Non-cardia 2) Cardia 3) Diffuse 4) Intestinal 5) Total
Reporting values from largest genome-wide significant study
Chromosome 1q22 (MUC1)
The 1q22 region, tagged by SNP rs4072037-A, was first identified as a diffuse gastric cancer susceptibility variant in Japan29, independently replicated in two East Asian studies. 23,27 rs4072037-A is a variant in the second exon of MUC1, a gene encoding a cell surface mucin that is highly expressed in the gastric epithelium and is commonly mutated in gastric cancers.70 This risk allele causes a shift in MUC1 splicing isoforms from variant 2 to variant 3, which differs by the removal of 9-amino acids from the tandem repeat (TR) domain of the N-terminal signal peptide, due to the reduction of stem-loop stability.71 This change may affect its cellular localization; however, localization of these splice variants have not yet been investigated.72 The MUC1 extracellular domain has been shown to protect the murine stomach epithelial lining from H. pylori colonization.73 Evidence suggests that MUC1 is an oncogene implicated in multiple malignancies through interactions with its intracellular and extracellular domains, as reviewed by Horm et al.74 Recent studies have found that rs4072037-A also increases the risk of non-cardia cancers.24,32
Chromosome 3q13.31 (ZBTB20)
Shi et al 30 identified rs9841504-C as a non-cardia risk variant (OR=1·32), located in an intron of ZBTB20. ZBTB20 encodes a zinc finger protein which regulates alpha-fetoprotein, a tumour marker.30 Little is known about how the rs9841504-C increases cancer risk, but a study showed that ZBTB20 protein expression is associated with poor prognosis in hepatocellular carcinoma.30
Chromosome 5p13.1 (PRKAA1)
rs13361707-C was discovered as a non-cardia gastric cancer susceptibility SNP by Shi et al.30 It increases risk by 41% and maps to a predicted promoter region in the first intron of PRKAA1, a gene that encodes the alpha 1 catalytic subunit of 5’ adenosine monophosphate-activated protein kinase (AMPK), a key regulator of the ATP-consuming biosynthetic pathways and cellular metabolism.75 In another study, Hu et al 76 found that rs10074991-G, a SNP in perfect linkage disequilibrium with rs11361707-C and also located in the first intron of PRKAA1, is associated with the risk of both cardia and non-cardia cancers. Neither rs13361707-C nor rs10074991-G have been functionally characterized and the effect of these variants on PRKAA1 remains unverified. As an AMPK subunit, PRKAA1 plays a key role in cancer development and proliferation as reviewed by Ross et al. 77 TCGA data shows PRKAA1 is highly expressed in gastric cancers.70
Chromosome 5q14.3 (lnc-POLR3G-4)
The 5q14.3 variant (rs7712641-C) is associated with a 19% increased risk of non-cardia gastric cancer32 and located within the intron of lnc-POLR3G-4, a long non-coding RNA (lncRNA). To explore possible effects of rs7712641-C on lnc-POLR3G-4, Wang et al32 carried out expression analyses in 75 matched gastric tumours/normal gastric tissues and found that while lnc-POLR3G-4 was under-expressed in cancers, there was no association between rs7712641 genotypes and lnc-POLR3G-4 expression. These analyses therefore did not support a functional role of rs7712641-C in gastric cancer. As no further functional studies have been carried out to characterize rs7712641-C, the mechanisms by which this allele increases risk remains unknown. Besides being under-expressed in gastric tumours (which suggest a tumour suppressor role), little is known about the lnc-POLR3G-4 role in tumorigenesis.
Chromosome 6p21.1 (UNC5CL, LRFN2)
The 6p12.1 region harbours two independent risk variants. Jin et al26 found that rs2494938-C increases non-cardia gastric cancer risk by 18%. rs2494938 is located in the first intron of the LRFN2 gene, which encodes the leucine rich repeat and fibronectin type III domain containing 2 protein.26 Located 200Kbp upstream, a second independent variant (rs2294693-A) was discovered in Asians.76 rs2294693 is located in the first intron of UNC5CL, a gene that encodes Unc-5 family C-terminal like protein. Neither rs2494938-C nor rs2294693-A have been functionally characterized and hence the mechanisms by which they increase cancer risk remain unknown. However, UNC5CL inhibits NF-kappa-B-dependent (NF-kB) transcription, a nuclear factor that plays a central role in inflammation and cancer progression.78 Little is known about the role of LRFN2 in tumorigenesis.
Chromosome 8q24.3 (PSCA)
Sakamoto et al29 first discovered an association between diffuse gastric cancer and two markers (rs2976392-G and rs2294008-T) in PSCA, a gene encoding the prostate stem cell antigen that is often silenced in gastric cancers. The association of rs2294008-G was discovered in non-cardia cancer, while rs2976392-T was discovered in intestinal cancers.29 rs2294008-G increases risk by 33% and was originally reported as a frameshift mutation in the first methionine leading to an alternate start site. 29 Current PSCA gene models, however, indicate that rs2294008-G occurs 26bp upstream of the start site and creates a binding site for YY1, a transcriptional repressor, causing PSCA promoter methylation, which leads to lower PSCA transcript levels. A recent study found that PSCA expression is reduced in oesophageal squamous cell carcinoma (OSCC). In normal oesophageal cells, PSCA binds, stabilizes, and translocate the retinoblastoma 1-inducible coiled-coil 1 protein, a key regulator of cell cycle arrest and differentiation, to the nucleus thereby promoting cell proliferation.79 Hence the lower PSCA levels associated with rs2294008-G likely lead to increased proliferation. Given the strong effect on PSCA expression, rs2294008-G is therefore the most likely functional 8q24.3 variant.
Chromosome 10q23.33 (PLCE1)
In OSCC and gastric cancer GWAS, Abnet et al discovered a risk association with rs2274223-G.23 Interestingly, when the study stratified cases based upon anatomical location, rs2274223 was only significant in cardia cancers.23 rs2274223-G changes a Histidine to Arginine in the phospholipase C Epsilon 1 (PLCE1) protein.23 The exact mechanism by which rs2274223-G affects PLCE1 still needs to be elucidated. However, phospholipases are activated by tyrosine kinase or G-protein coupled receptors in response to hormones and growth factors. Activated phospholipases hydrolyse phosphoinositides to increase the intracellular level of Ca2+ and produce diacylglycerol, an important mediator for signal transduction. 80
Chromosome 11q22.3 (ATM)
Using an Icelandic cohort, Helgason et al 24 discovered three variants (rs758081262-T, rs768362387-A, rs777499935-G) in the multi-cancer HRDR ATM gene (see above) that were associated with gastric cancer risk. These variants were located in different haplotypes and each had similar effect sizes (OR=4·7).
Future Directions
The role of a genetic predisposition in gastric cancer has been largely under-appreciated. This concept likely stems from the fact that gastric cancer has two strong infection-related risk factors (H. pylori and EBV) and that a limited number of familial gastric cancer syndromes have been genetically characterized. However, the recent analyses in TCGA suggest that an important fraction of patients develop the disease due to inherited factors. In the study of Huang et al25, the prevalence of pathogenic and likely pathogenic mutations in 443 sporadic gastric cancer patients was 13%. This is remarkable given the fact that the average age of onset of these patients was 66y; hence, the study was not enriched by likely genetic cases. Even among mutation carriers, the average age of onset was 61y, an age not typically associated with inherited risk. Should the study of Huang et al be independently replicated, particularly in high-risk regions like China, where most cases are diagnosed81, the implications for gastric cancer epidemiology and prevention will be enormous. At-risk relatives of thousands of patients, of the one million newly diagnosed cases each year, could potentially benefit from genetically informed preventive interventions through cascade genetic counselling and testing that guide early detection and cancer prevention. Therefore, an important priority in future gastric cancer research should be to fully examine the population prevalence of inherited mutations, particularly in high-risk regions such as Asia and Latin America. An examination of mutation prevalence, however, will require well-designed and statistically powered studies with appropriately matched cases and population controls. Even though Huang et al25,found that PALB2 and ATM were enriched (at false discovery rate, FDR, <0.05) for pathogenic variants, FDR significance for BRCA1 and BRCA2 (the other two recurrently mutated genes in TCGA) was not achieved. BRCA1 and BRCA2 were however mutated at 0.6% and 1.9% frequencies, respectively, which is higher than the carrier frequency in EXAC controls (0.2% for BRCA1 and 0.3% for BRCA2). Neither TCGA (a heterogeneous collection of patients from Asia, Europe and North America) nor EXAC (a dataset with individuals from founder populations where BRCA½ carrier frequency is higher than in the general population) are the right populations to compare mutation frequencies and to test for possible associations between BRCA½ mutations and gastric cancer risk. Hence, an effect of BRCA½ on gastric cancer risk in the Huang et al25 study may have been missed by the lack of statistical power (443 cases and 443 well matched controls only provide 12% power to detect an odds ratio=2). More studies are therefore needed to further assess if the prevalence of BRCA½ mutation in gastric cancer patients is significantly higher than that in the general population.
The past decade has seen the identification of several gastric cancer GWAS variants. These studies have been carried out in Asian and European populations, but regions of high incidence such as Latin America have not been examined. Latino populations have a unique genetic demography and gastric cancer epidemiological profiles, which may facilitate the discovery of additional gastric cancer variants.10,82 Hence, additional populations should be included in future studies. This is particularly important as it is clear that many risk variants remain to be identified. Furthermore, only few of the known risk alleles are functional variants (i.e. rs4072037-A/MUC1, rs2294008-T/PSCA and rs2274223-G/PLCE1) and hence, future studies should functionally characterize other risk variants and genes for which little is known about their biology. Functional evaluation of these risk variants will increase our knowledge of gastric tumorigenesis, which will ultimately lead to improved prevention and treatment strategies.
With regards to recent advances in familial genes, another area of great importance in future studies is to determine the risk associated with HRDR gene mutations, particularly among carriers of mutations in PALB2, a gene that represents a bona fide gastric cancer gene. 69 For now, appropriate gastric cancer surveillance among PALB2 mutation carriers remains uncertain. On the basis of the few published data (see Figure 2), patients with family history and other risk factors could be offered, in a research setting, endoscopic surveillance after the third decade of life (i.e. about a decade earlier than the youngest patient reported). Clearly, larger studies will likely need international collaboration that helps provide gastric cancer risk estimates for PALB2 mutation carriers.
The role of non-genetic and genetic (GWAS SNPs) risk factors in gastric cancer is considerable and it is possible that gene-gene and gene-environment interactions mediate risk. For PALB2, BRCA1 and BRCA2 mutation carriers, the average age of gastric cancer diagnosis seems to be around a decade later than reported for breast cancer among carriers of mutations in the same genes.63,65 This suggests that gastric cancer penetrance among HRDR gene mutations could be mediated by additional factors. Although the role of H. Pylori in gastric cancer causality among PALB2 carriers has been previously ruled out, such studies remain small. 28,42 Investigating interactions between inherited mutations and known risk factors should therefore represent another research priority.
Another consensus that is emerging from recent studies of HDGC and other familial syndromes is that these conditions are better defined by genetic rather than by clinical criteria, as rightly suggested by Hansford et al.36 GAPPS should now be classified as FAPS. HDGC should be defined as carrying CDH1 or in CTNNA1 mutations, as the studies by Hansford et al36 and Fewings et al42, which included cases fulfilling HDGC clinical criteria, found that several of them had mutations in genes associated with PJS (STK11), LS (MSH2), HBOCS (BRCA1, BRCA2) or with other hereditary conditions (PALB2, ATM, TP53, BRIP1, SDHB). 36,42 Given the histological heterogeneity reported among the carriers of mutations in these genes (Figure 2), FIGC is likely an amalgamation of cases with mutations in these recently discovered HRDR genes. From what has been reported and in the absence of a clearly associated intestinal gastric cancer-only/predominant syndrome, FIGC now seems unlikely to represent a unique condition.
Given the existence of gastric cancer patients who carry mutations in these multi-organ genes, another important future research area should be to estimate gastric cancer risk among carriers of germline mutations in such genes. Through genetic testing, ascertained in families with breast cancer, thousands of BRCA1, BRCA2 and PALB2 mutation carriers have been identified.63,65 While having gastric cancer family history may aid in refining gastric cancer risk among such mutation carriers, there is clearly a need to identify gastric cancer risk modifiers in such mutation-positive families.
Many of the genes discussed in this review are part of clinical gene panels. Although several gaps must be filled in before most of these candidates can be used in the clinic, a potential gastric cancer panel could include genes with variable levels of causality evidence. In Table 4 we present a list of candidate gastric genes and stratified their causality evidence based on the pre- and post-next generation sequencing (NGS) studies. For pre-NGS evidence, these genes were assessed based on their involvement in cancer syndromes and in gastric cancer-related syndromes in particular. The post-NGS evidence included their mutation status (mutated and/or recurrently mutated) and by belonging to candidate pathways (HRDR or CDH1 pathway). Based on these evidence lines, Tier 1 genes should include CDH1 and PALB2; Tier 2 genes should include CTNNA1, MSH2, TP53, STK11, ATM, BRCA1, BRCA2, BRIP1 and ATR; and Tier 3 genes should include APC, SMAD4, BMPR1A, ENG, MSR1, SDHA, SDHB, RAD51C, NBN, RECQL5 and FANCC. We emphasize that only CDH1 and PALB2 could be used in the clinic now. The remaining genes could be used, within research protocol settings, when gastric cancer patients seeking genetic counselling report personal or family history of gastric cancer associated syndromes (for example, BRCA½ testing could be useful in patients with HBOC history). These genes should also represent the priority list for future gastric cancer genetic studies of mutation prevalence and penetrance. Further, the s clinical genetics community should be encouraged to report the characteristics of gastric cancer cases carrying mutations in these genes.
Table 4.
Genes with evidence for gastric cancer causality
| Gene | Evidence for causality |
Familial Cancer Syndrome Evidence | Evidence from next generation sequencing studies |
|||
|---|---|---|---|---|---|---|
| Cancer syndrome | Gastric cancer related syndrome? |
Mutated? | Recurrently mutated? |
Gastric cancer related pathway? |
||
| Tier 1 | ||||||
| CDH1 | Strong | HDGC | Yes | NA | NA | CDH1 |
| PALB2 | Strong | Breast/ Pancreas cancer | No | Yes25,28,36,42 | Yes | HRDR |
| Tier 2 | ||||||
| CTNNA1 | Moderate | HDGC | Yes | Yes 36 | Yes | CDH1 |
| MSH2 | Moderate | LS | Yes | Yes 31,42 | Yes | No |
| TP53 | Moderate | LFS | Yes | Yes 31 | No | No |
| STK11 | Moderate | PJS | Yes | Yes 36 | Yes | No |
| ATM | Moderate | Breast cancer | No | Yes 25,31,36 | Yes | HRDR |
| BRCA1 | Moderate | HBOC | Yes | Yes 25,28 | Yes | HRDR |
| BRCA2 | Moderate | HBOC | Yes | Yes 25,31,36 | Yes | HRDR |
| BRIP1 | Moderate | Breast cancer | No | Yes 25,31 | Yes | HRDR |
| ATR | Moderate | None | No | Yes 31,42 | HRDR | |
| Tier 3 | ||||||
| APC | Moderate/low | FAPS | Yes | No | No | No |
| SMAD4 | Moderate/low | JPS | Yes | No | No | No |
| BMPR1A | Moderate/low | JPS | Yes | No | No | No |
| ENG | Moderate/low | JPS | Yes | No | No | No |
| MSR1 | Moderate/low | Oeasophageal | No | Yes 42 | Yes | No |
| SDHB | Moderate/low | Paraganglioma- Pheochromocytoma |
No | Yes 36 | No | SDHA/B |
| SDHA | Moderate/low | Paraganglioma- Pheochromocytoma |
No | Yes 25 | No | SDHA/B |
| RAD51C | Moderate/low | Breast cancer | No | Yes 28 | No | HRDR |
| NBN | Moderate/low | Ovarian cancer | No | Yes 42 | No | HRDR |
| RECQL5 | Moderate/low | Breast cancer | No | Yes 42 | No | HRDR |
| FANCC | Moderate/low | Breast cancer | No | Yes 31 | No | HRDR |
The fact that BRCAness has been found in gastric tumors28,83 represents an opportunity to develop new PARPi therapies. Even though a recent trial84 failed to show significant survival improvement with Olaparib (a PARPi) in ATM-negative gastric cancers, it does not necessarily mean a lack of PARPi potential for the treatment of this malignancy. Better biomarkers for the identification of HRDR tumours are however needed, as ATM immunohistochemistry, which was used in the recent trial, is a sub-optimal stratifier.84 As more histological and molecular data in HRDR-deficient gastric cancers is reported, more precise ways to identify such cancers will be developed.
In conclusion, several important advances have been achieved in gastric cancer genetics. These findings will lead to the identification of high-risk groups that can be targeted for early interventions and that will improve our understanding of tumorigenesis. This will ultimately lead to improved outcomes for this common malignancy.
Acknowledgements
We are grateful to Ted Toal and to Ana Estrada for their comments on the content of the manuscript. We are also grateful for research funding from the University of California Davis and the National Cancer Institute (grants R01CA223978, R21CA199631 and P30CA093373) of the National Institutes of Health. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest
The authors declared no conflicts of interest.
Search Strategy and Selection Criteria
We performed a literature search in PubMed, Google Scholar, and GWAS Catalog for Population Risk Factors, Genetic Risk Factors, Hereditary Gastric Cancer and Related Syndromes, and Population based association studies published on or after 2000. For completeness, we also reviewed relevant reviews, meta-analysis, and studies published on gastric cancer. Keywords used in the search included “cardia”, “non-cardia”, “gastric cancer”, “stomach cancer”, “risk factors”, “hereditary gastric cancer”, “BRCAness”, “diffuse gastric cancer”, and “intestinal gastric cancer.” Preference was given to recent primary studies and reviews written or translated in the English language. However, due to reference and text length limitations, reviews were cited and in multiple instances, readers were referred to recent extensive reviews on those subjects that were beyond the scope of this manuscript.
References
- 1.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. The Lancet 2016; 388(10060): 2654–64. [DOI] [PubMed] [Google Scholar]
- 2.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014; 23(5): 700–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136(5): E359–86. [DOI] [PubMed] [Google Scholar]
- 4.Siewert J, Stein H. Carcinoma of the gastroesophageal junction-classification, pathology and extent of resection. Diseases of the Esophagus 1996; 9(3): 173–82. [Google Scholar]
- 5.Colquhoun A, Arnold M, Ferlay J, Goodman KJ, Forman D, Soerjomataram I. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut 2015; 64(12): 1881–8. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Meltzer SJ. Gastric Cancer in the Era of Precision Medicine. Cellular and Molecular Gastroenterology and Hepatology 2017; 3(3): 348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J, H S, Kapesa L, Zeng S Lauren classification and individualized chemotherapy in gastric cancer. Oncology Letters 2016; 11(5): 2959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer 2015; 136(2): 487–90. [DOI] [PubMed] [Google Scholar]
- 9.Khan G, Hashim MJ. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990–2010. Infect Agent Cancer 2014; 9(1): 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Fedewa SA, Miller KD, et al. Cancer statistics for Hispanics/Latinos, 2015. CA Cancer J Clin 2015; 65(6): 457–80. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Rusiecki JA, Zhu K, Potter J, Devesa SS. Stomach carcinoma incidence patterns in the United States by histologic type and anatomic site. Cancer Epidemiol Biomarkers Prev 2009; 18(7): 1945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devessa S, Blot W, Fraumeni JJ. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998; 83(10): 2049–53. [PubMed] [Google Scholar]
- 13.Chen Y, Liu L, Wang X, et al. Body mass index and risk of gastric cancer: a meta-analysis of a population with more than ten million from 24 prospective studies. Cancer Epidemiol Biomarkers Prev 2013; 22(8): 1395–408. [DOI] [PubMed] [Google Scholar]
- 14.Figueroa JD, Terry MB, Gammon MD, et al. Cigarette smoking, body mass index, gastro-esophageal reflux disease, and non-steroidal anti-inflammatory drug use and risk of subtypes of esophageal and gastric cancers by P53 overexpression. Cancer Causes Control 2009; 20(3): 361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel LS, Chow WH, Vaughan TL, et al. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst 2003; 95(18): 1404–13. [DOI] [PubMed] [Google Scholar]
- 16.Anderson WF, Rabkin CS, Turner N, Fraumeni JF, Rosenberg PS, Camargo MC. The Changing Face of Noncardia Gastric Cancer Incidence Among US Non-Hispanic Whites. JNCI Journal of the National Cancer Institute 2018; 42(2): 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez CA, Pera G, Agudo A, et al. Smoking and the risk of gastric cancer in the European Prospective Investigation Into Cancer and Nutrition (EPIC). Int J Cancer 2003; 107(4): 629–34. [DOI] [PubMed] [Google Scholar]
- 18.Bagnardi V, Rota M, Botteri E, et al. Alcohol consumption and site-specific cancer risk: a comprehensive dose-response meta-analysis. Br J Cancer 2015; 112(3): 580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjodahl K, Lu Y, Nilsen TI, et al. Smoking and alcohol drinking in relation to risk of gastric cancer: a population-based, prospective cohort study. Int J Cancer 2007; 120(1): 128–32. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald RC, Hardwick R, Huntsman D, et al. Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet 2010; 47(7): 436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and Heritable Factors in the Causation of Cancer — Analyses of Cohorts of Twins from Sweden, Denmark, and Finland. New England Journal of Medicine 2000; 343(2): 78–85. [DOI] [PubMed] [Google Scholar]
- 22.Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature 1998; 392(6674): 402–5. [DOI] [PubMed] [Google Scholar]
- 23.Abnet CC, Freedman ND, Hu N, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nature genetics 2010; 42(9): 764–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helgason H, Rafnar T, Olafsdottir HS, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nature genetics 2015; 47(8): 906–10. [DOI] [PubMed] [Google Scholar]
- 25.Huang KL, Mashl RJ, Wu Y, et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018; 173(2): 355–70 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin G, Ma H, Wu C, et al. Genetic variants at 6p21.1 and 7p15.3 are associated with risk of multiple cancers in Han Chinese. Am J Hum Genet 2012; 91(5): 928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeki N, Saito A, Choi IJ, et al. A Functional Single Nucleotide Polymorphism in Mucin 1, at Chromosome 1q22, Determines Susceptibility to Diffuse-Type Gastric Cancer. Gastroenterology 2011; 140(3): 892–902. [DOI] [PubMed] [Google Scholar]
- 28.Sahasrabudhe R, Lott P, Bohorquez M, et al. Germline Mutations in PALB2, BRCA1, and RAD51C, Which Regulate DNA Recombination Repair, in Patients With Gastric Cancer. Gastroenterology 2017; 152(5): 983–6 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto H, Yoshimura K, Saeki N, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nature genetics 2008; 40(6): 730–40. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Hu Z, Wu C, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. - PubMed - NCBI. 2011; 43(12): 1215–8. [DOI] [PubMed] [Google Scholar]
- 31.Slavin T, Neuhausen SL, Rybak C, et al. Genetic Gastric Cancer Susceptibility in the International Clinical Cancer Genomics Community Research Network. Cancer Genet 2017; 216–217: 111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Dai J, Hu N, et al. Identification of new susceptibility loci for gastric non-cardia adenocarcinoma: pooled results from two Chinese genome-wide association studies. Gut 2017; 66(4): 581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendonsa AM, Na TY, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Post RS, Vogelaar IP, Carneiro F, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. Journal of Medical Genetics 2015; 52(6): 361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colvin H, Yamamoto K, Wada N, Mori M. Hereditary Gastric Cancer Syndromes. Surgical Oncology Clinics of North America 2015; 24(4): 765–77. [DOI] [PubMed] [Google Scholar]
- 36.Hansford S, Kaurah P, Li-Chang H, et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA oncology 2015; 1(1): 23–32. [DOI] [PubMed] [Google Scholar]
- 37.Lim YC, di Pietro M, O’Donovan M, et al. Prospective cohort study assessing outcomes of patients from families fulfilling criteria for hereditary diffuse gastric cancer undergoing endoscopic surveillance. Gastrointest Endosc 2014; 80(1): 78–87. [DOI] [PubMed] [Google Scholar]
- 38.Hebbard PC, Macmillan A, Huntsman D, et al. Prophylactic total gastrectomy (PTG) for hereditary diffuse gastric cancer (HDGC): the Newfoundland experience with 23 patients. Ann Surg Oncol 2009; 16(7): 1890–5. [DOI] [PubMed] [Google Scholar]
- 39.Majewski IJ, Kluijt I, Cats A, et al. An alpha-E-catenin (CTNNA1) mutation in hereditary diffuse gastric cancer. J Pathol 2013; 229(4): 621–9. [DOI] [PubMed] [Google Scholar]
- 40.van der Post RS, Carneiro F. Emerging Concepts in Gastric Neoplasia: Heritable Gastric Cancers and Polyposis Disorders. Surgical Pathology Clinics 2017; 10(4): 931–45. [DOI] [PubMed] [Google Scholar]
- 41.van der Post RS, Vogelaar IP, Manders P, et al. Accuracy of Hereditary Diffuse Gastric Cancer Testing Criteria and Outcomes in Patients With a Germline Mutation in CDH1. Gastroenterology 2015; 149(4): 897–906.e19. [DOI] [PubMed] [Google Scholar]
- 42.Fewings E AL, Redman J, et al. Whole exome sequencing study to detect germline pathogenic variants in PALB2 and other cancer-predisposing genes in CDH1 mutation negative diffuse gastric cancer families. Lancet Gastroenterology & Hepatology 2018; 3(7): 489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, Xie M, Wendl MC, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 2015; 6: 10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Woods SL, Healey S, et al. Point Mutations in Exon 1B of APC Reveal Gastric Adenocarcinoma and Proximal Polyposis of the Stomach as a Familial Adenomatous Polyposis Variant. The American Journal of Human Genetics 2016; 98(5): 830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Repak R, Kohoutova D, Podhola M, et al. The first European family with gastric adenocarcinoma and proximal polyposis of the stomach: case report and review of the literature. Gastrointestinal Endoscopy 2016; 84(4): 718–25. [DOI] [PubMed] [Google Scholar]
- 46.Worthley DL, Phillips KD, Wayte N, et al. Gastric adenocarcinoma and proximal polyposis of the stomach (GAPPS): a new autosomal dominant syndrome. Gut 2012; 61(5): 774–9. [DOI] [PubMed] [Google Scholar]
- 47.Yanaru-Fujisawa R, Nakamura S, Moriyama T, et al. Familial fundic gland polyposis with gastric cancer. Gut 2012; 61(7): 1103–4. [DOI] [PubMed] [Google Scholar]
- 48.Oliveira C, Seruca R, Carneiro F. Hereditary gastric cancer. Best Practice & Research Clinical Gastroenterology 2009; 23(2): 147–57. [DOI] [PubMed] [Google Scholar]
- 49.Caldas C, Carneiro F, Lynch HT, et al. Familial gastric cancer: overview and guidelines for management. J Med Genet 1999; 36(12): 873–80. [PMC free article] [PubMed] [Google Scholar]
- 50.Kluijt I, Sijmons RH, Hoogerbrugge N, et al. Familial gastric cancer: guidelines for diagnosis, treatment and periodic surveillance. Fam Cancer 2012; 11(3): 363–9. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Wang W, Lee S, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 2006; 296(12): 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005; 352(18): 1851–60. [DOI] [PubMed] [Google Scholar]
- 53.Watson P, Vasen HFA, Mecklin JP, et al. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer 2008; 123(2): 444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasen HF, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 2013; 62(6): 812–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrow E, Robinson L, Alduaij W, et al. Cumulative lifetime incidence of extracolonic cancers in Lynch syndrome: a report of 121 families with proven mutations. Clin Genet 2009; 75(2): 141–9. [DOI] [PubMed] [Google Scholar]
- 56.Therkildsen C, Ladelund S, Smith-Hansen L, Lindberg LJ, Nilbert M. Towards gene- and gender-based risk estimates in Lynch syndrome; age-specific incidences for 13 extra-colorectal cancer types. Br J Cancer 2017; 117(11): 1702–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreira L, Castells A. Surveillance of patients with hereditary gastrointestinal cancer syndromes. Best Pract Res Clin Gastroenterol 2016; 30(6): 923–35. [DOI] [PubMed] [Google Scholar]
- 58.Syngal S, Brand RE, Church JM, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 2015; 110(2): 223–62; quiz 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kopacova M, Tacheci I, Rejchrt S, Bures J. Peutz-Jeghers syndrome: Diagnostic and therapeutic approach. World Journal of Gastroenterology : WJG 2009; 15(43): 5397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Lier MG, Wagner A, Mathus-Vliegen EM, Kuipers EJ, Steyerberg EW, van Leerdam ME. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol 2010; 105(6): 1258–64; author reply 65. [DOI] [PubMed] [Google Scholar]
- 61.Giardiello FM, Hamilton SR, Krush AJ, et al. Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med 1993; 328(18): 1313–6. [DOI] [PubMed] [Google Scholar]
- 62.Masciari S, Dewanwala A, Stoffel EM, et al. Gastric cancer in individuals with Li-Fraumeni syndrome. Genetics in Medicine 2011; 13(7): 651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rebbeck TR, Friebel TM, Friedman E, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat 2018; 39(5): 593–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedenson B BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. MedGenMed 2005; 7(2): 60. [PMC free article] [PubMed] [Google Scholar]
- 65.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371(6): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009; 324(5924): 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harbor Perspectives in Biology 2015; 7(4): a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lord CJ, Ashworth A. BRCAness revisited. Nature Reviews Cancer 2016; 16(2): 110–20. [DOI] [PubMed] [Google Scholar]
- 69.Carvajal Carmona LG. PALB2 as a familial gastric cancer gene: is the wait over? Lancet Gastroenterology & Hepatology 2018; 3(7): 451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Network T Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513(7517): 202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng W, Loh AX, Teixeira AS, Pereira SP, Swallow DM. Genetic regulation of MUC1 alternative splicing in human tissues. Br J Cancer 2008; 99(6): 978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 2014; 20(6): 332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McGuckin MA, Every AL, Skene CD, et al. Muc1 mucin limits both Helicobacter pylori colonization of the murine gastric mucosa and associated gastritis. Gastroenterology 2007; 133(4): 1210–8. [DOI] [PubMed] [Google Scholar]
- 74.Horm TM, Schroeder JA. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh Migr 2013; 7(2): 187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008; 32 Suppl 4: S7–12. [DOI] [PubMed] [Google Scholar]
- 76.Hu N, Wang Z, Song X, et al. Genome-wide association study of gastric adenocarcinoma in Asia: a comparison of associations between cardia and non-cardia tumours. Gut 2016; 65(10): 1611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ross FA, MacKintosh C, Hardie DG. AMP-activated protein kinase: a cellular energy sensor that comes in 12 flavours. FEBS J 2016; 283(16): 2987–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta 2010; 1799(10–12): 775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang LY, Wu JL, Qiu HB, et al. PSCA acts as a tumor suppressor by facilitating the nuclear translocation of RB1CC1 in esophageal squamous cell carcinoma. Carcinogenesis 2016; 37(3): 320–32. [DOI] [PubMed] [Google Scholar]
- 80.Tyutyunnykova A, Telegeev G, Dubrovska A. The controversial role of phospholipase C epsilon (PLCepsilon) in cancer development and progression. J Cancer 2017; 8(5): 716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu X, Li J. Gastric carcinoma in China: Current status and future perspectives (Review). Oncol Lett 2010; 1(3): 407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carvajal-Carmona LG, Soto ID, Pineda N, et al. Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am J Hum Genet 2000; 67(5): 1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nature communications 2015; 6(1): 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bang YJ, Xu RH, Chin K, et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017; 18(12): 1637–51. [DOI] [PubMed] [Google Scholar]


