Abstract
Benchmark small variant calls are required for developing, optimizing and assessing the performance of sequencing and bioinformatics methods. Here, as part of the Genome in a Bottle Consortium (GIAB), we apply a reproducible, cloud-based pipeline to integrate multiple short and linked read sequencing datasets and provide benchmark calls for human genomes. We generate benchmark calls for one previously analyzed GIAB sample, as well as six broadly-consented genomes from the Personal Genome Project. These new genomes have broad, open consent, making this a ‘first of its kind’ resource that is available to the community for multiple downstream applications. We produce 17% more benchmark SNVs, 176% more indels, and 12% larger benchmark regions than previously published GIAB benchmarks. We demonstrate this benchmark reliably identifies errors in existing callsets and highlight challenges in interpreting performance metrics when using benchmarks that are not perfect or comprehensive. Finally, we identify strengths and weaknesses of callsets by stratifying performance according to variant type and genome context.
Introduction
Genome sequencing is increasingly used in clinical applications, making variant calling accuracy of paramount importance. With this in mind, the Genome in a Bottle Consortium (GIAB) developed benchmark small variants for the pilot GIAB genome.1 These benchmark calls have been used for optimization and analytical validation of clinical sequencing,2–4 comparisons of bioinformatics tools,5 and optimization, development, and demonstration of new technologies.6
Here, we build on previous GIAB integration methods to enable the development of highly accurate and reproducible benchmark genotype calls from any genome, using multiple datasets from different sequencing methods (Figure 1). We first generate more comprehensive and accurate integrated SNV, small indel, and homozygous reference calls for HG001—the same sample used in the original GIAB analysis. These calls were made more accurate and comprehensive by using new and higher-coverage short and linked read datasets,7 technology-optimized variant calling methods, and more robust methods to decide when to trust a variant call or homozygous reference region from each technology. Calls supported by two technologies were used to train a model that identified calls from each dataset that were outliers and therefore less trustworthy. Variant calls and regions were included in the benchmark set if at least one input callset was trustworthy and all trustworthy callsets agreed. We compare our callsets to the phased pedigree-based callsets from the Illumina Platinum Genomes Project8 (PGP) and Cleary et al.9, following best practices established by the Global Alliance for Genomics and Health Benchmarking Team,10 and manually examine the differences to evaluate the accuracy of our callsets.
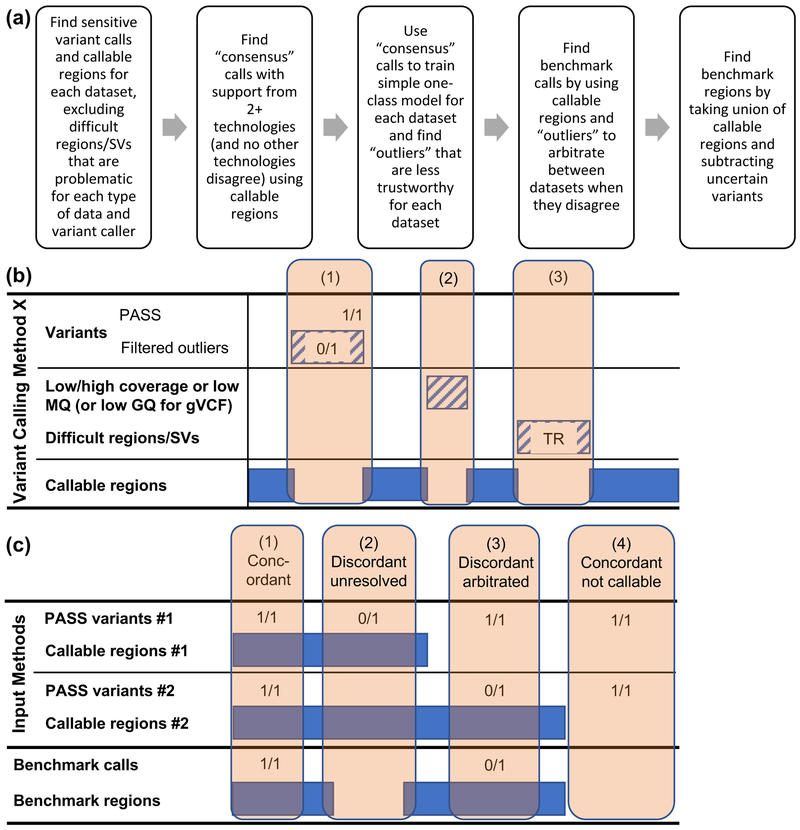
Figure 1:
Arbitration process used to form our benchmark set from multiple technologies and callsets. (a) The arbitration process has two cycles. The first cycle ignores “Filtered Outliers”. Calls that are supported by at least 2 technologies in the first cycle are used to train a model that identifies variants from each callset with any annotation value that is an “outlier” compared to these 2-technology calls. In the second cycle, the outlier variants and surrounding 50bp are excluded from the callable regions for that callset. (b) For each variant calling method, we delineate callable regions by subtracting regions around filtered/outlier variants as at locus (1), regions with low coverage or mapping quality (MQ) as at locus (2), and “difficult regions” prone to systematic miscalling or missing variants for the particular method as at locus (3). For callsets in gVCF format, we exclude homozygous reference regions and variants with genotype quality (GQ) < 60. Difficult regions include different categories of tandem repeats (TR) and segmental duplications. (c) Four arbitration examples with two arbitrary input methods. (1) Both methods have the same genotype and variant and it is in their callable regions, so the variant and region are included in the benchmark set. (2) Method 1 calls a heterozygous variant and Method 2 implies homozygous reference, and it is in both methods’ callable regions, so the discordant variant is not included in the benchmark calls and 50bp on each side is excluded from the benchmark regions. (3) The methods have discordant genotypes, but the site is only inside Method 2’s callable regions, so the heterozygous genotype from Method 2 is trusted and is included in the benchmark regions. (4) The two methods’ calls are identical, but they are outside both methods’ callable regions, so the site is excluded from benchmark variants and regions.
We apply this approach to six broadly consented GIAB genomes from the Personal Genome Project (PGP),11 an Ashkenazi Jewish (AJ) mother-father-son trio and a Han Chinese mother-father-son trio. We also use our methods to form similar benchmark sets with respect to the human GRCh38 reference.
We demonstrate that our benchmark sets reliably identify false positive and false negative variant calls in a variety of high-quality variant callsets, including alternative benchmark sets like Platinum Genomes. The benchmark sets can be used to identify strengths and weaknesses of different methods. Finally, we discuss how strengths and weaknesses of the benchmark sets themselves can bias the resulting performance metrics, particularly for difficult variant types and genome contexts.
In contrast to existing benchmarking efforts, the extensively characterized PGP genomes analyzed here have an open, broad consent and broadly available data and cell lines, as well as consent to re-contact for additional types of samples.11 They are an enduring resource uniquely suitable for diverse research and commercial applications. In addition to homogeneous DNA from NIST Reference Materials and DNA and cell lines from Coriell, a variety of products using these genomes is already available, including induced pluripotent stem cells (iPSCs), cell line mixtures, cell line DNA with synthetic DNA spike-ins with mutations of clinical interest, formalin-fixed paraffin-embedded (FFPE) cells, and circulating tumor DNA mimics.12,13
Previous studies have used more restricted samples to characterize variants and regions complementary to our benchmark sets: for example, the Platinum Genomes pedigree analysis,8 HuRef analysis using Sanger sequencing,14,15 integration of multiple SV calling methods on HS1011,16 and synthetic diploid using long-read sequencing of hydatiform moles17. The recent synthetic diploid method17 has the advantage that long read assemblies of each haploid cell line have orthogonal error profiles to the diploid short and linked read-based methods used in this manuscript, though short reads were used to correct indel errors in the long read assemblies. However the synthetic diploid hydatiform mole cell lines are not available in a public repository.10 The broadly consented and available GIAB samples from PGP have at least three strengths relative to the other samples: (1) we can continue to sequence these renewable cell lines with new technologies and improve our benchmark set over time, (2) any clinical or research laboratory can sequence these samples with their own method and compare their results to our benchmark set, and (3) a wide array of secondary reference samples can be generated from the same cell lines to meet particular community needs.
Results
Design of benchmark calls and regions
Our goal is to design a reproducible, robust, and flexible method to produce a benchmark variant and genotype callset (including homozygous reference regions). When any sequencing-based callset is compared to our benchmark calls requiring stringent matching of alleles and genotypes, the majority of discordant calls in the benchmark regions (i.e., FPs and FNs) should be attributable to errors in the sequencing-based callset. We develop a modular, cloud-based data integration pipeline, enabling diverse data types to be integrated for each genome. We produce benchmark variant calls and regions by integrating methods and technologies that have different strengths and limitations, using evidence of potential bias to arbitrate when methods have differing results (Figure 1). Finally, we evaluate the utility of the benchmark variants and regions by comparing high-quality callsets to the benchmark calls and manually curating discordant calls to ensure most are errors in the other high-quality callsets.
New benchmark sets are more comprehensive and accurate
The new integration method is designed to give more accurate and comprehensive benchmark sets in several ways, which are detailed at the end of the Online Methods: (1) we use newer input data, including 10x Genomics linked reads, and most data were measured from the homogeneous NIST RM batches of DNA; (2) some long homopolymers and difficult-to-map regions are now in the benchmark sets by using PCR-free Illumina GATK gVCF outputs and 10x Genomics LongRanger, respectively; (3) we use a more accurate set of potential structural variants to exclude regions with problematic small variant calls; and (4) we add phasing information. Table 1 shows the evolution of the GIAB/NIST benchmark sets since our previous publication.1 The fraction of non-N bases in the GRCh37 reference covered has increased from 77.4% to 90.8%, and the numbers of benchmark SNVs and indels have increased by 17% and 176%, respectively. The fraction of GRCh37 RefSeq coding regions covered has increased from 73.9% to 89.9%. The gains in benchmark regions and calls are a result of using new datasets and a more accurate and less conservative structural variant (SV) bed file (Online Methods), as well as a new approach where excluded difficult regions are callset-specific depending on read length, error profiles, and analysis methods. GRCh38 is currently characterized less comprehensively characterized than GRCh37 (Supplementary Table 1, Supplementary Figure 1, and Supplementary Note 1). To determine if less input data could be used, we also integrated only two datasets (Illumina and 10x), resulting in a larger number of benchmark indel calls but a five times higher error rate compared to v3.3.2 (Supplementary Note 2). Our callsets are now mostly phased using pedigree or trio analysis, but we did not evaluate the accuracy of this phasing (Supplementary Note 3).
Table 1:
Summary of statistics of GIAB benchmark calls and regions from HG001 from v2.18 to v3.3.2 and their comparison to Illumina Platinum Genomes 2016-v1.0 calls (PG). Note that PG bed files were contracted by 50bp to minimize partial complex variant calls in the PG calls.
| Integration Version | v 2.18 | v 2.19 | v. 3.2 | v. 3.2.2 | v 3.3 | v 3.3.1 | v 3.3.2 | v 3.3.1 | v 3.3.2 |
|---|---|---|---|---|---|---|---|---|---|
| Reference | GRCh37 | GRCh37 | GRCh37 | GRCh37 | GRCh37 | GRCh37 | GRCh37 | GRCh38 | GRCh38 |
| Integration Date | Sep 2014 | Apr 2015 | May 2016 | June 2016 | Aug 2016 | Oct 2016 | Nov 2016 | Oct 2016 | Nov 2016 |
| Number of bases in benchmark regions (chromosomes 1-22 + X) | 2.20 Gb | 2.22 Gb | 2.54 Gb | 2.53 Gb | 2.57 Gb | 2.58 Gb | 2.58 Gb | 2.45 Gb | 2.44 Gb |
| Fraction of non-N bases covered in chromosomes 1-22 + X) | 77.4% | 78.1% | 89.6% | 89.2% | 90.5% | 91.1% | 90.8% | 84.2% | 83.8% |
| Fraction of RefSeq coding sequence covered | 73.9% | 74.0% | 87.8% | 87.9% | 89.9% | 90.0% | 89.9% | 83.4% | 83.3% |
| Total number of calls in benchmark regions | 2915731 | 3153247 | 3433656 | 3512990 | 3566076 | 3746191 | 3691156 | 3617168 | 3542487 |
| SNPs | 2741359 | 2787291 | 3084406 | 3154259 | 3191811 | 3221456 | 3209315 | 3058368 | 3042789 |
| Insertions | 86204 | 172671 | 171866 | 176511 | 171715 | 243856 | 225097 | 269331 | 241176 |
| Deletions | 87161 | 189932 | 169389 | 173976 | 189807 | 266386 | 245552 | 275041 | 247178 |
| Block Substitutions | 1005 | 2532 | 7476 | 7716 | 10364 | 13332 | 11192 | 13976 | 11344 |
| Transition/Transversion ratio for SNVs | 2.12 | 2.12 | 2.14 | 2.14 | 2.11 | 2.10 | 2.10 | 2.10 | 2.11 |
| Fraction phased globally or locally | 0.0% | 0.3% | 3.9% | 3.9% | 8.8% | 99.0% | 99.6% | 98.5% | 99.5% |
| Comparisons of GIAB to Platinum Genomes (PG) calls | |||||||||
| Number of GIAB calls concordant with PG in both PG and GIAB beds | 2825803 | 3030703 | 3312580 | 3391783 | 3441361 | 3550914 | 3529641 | 3459674 | 3431752 |
| Number of PG-only calls in both beds | 194 | 404 | 81 | 52 | 60 | 67 | 61 | 202 | 180 |
| Number of GIAB-only calls in both beds | 49 | 87 | 56 | 57 | 40 | 50 | 47 | 105 | 94 |
| Number of PG-only calls: Total After excluding PG bed |
1223697 605142 | 1018795 493694 |
274671 118810 |
138894 22224 |
550982 172375 |
445563 140857 |
469202 142682 |
659870 386739 |
690887 391523 |
| Number of GIAB-only calls in GIAB benchmark bed | 90722 | 122544 | 121076 | 121207 | 124715 | 195277 | 163467 | 157494 | 111787 |
| Number of concordant calls that are filtered by the GIAB benchmark bed | 12 | 0 | 736918 | 657715 | 608137 | 53460 | 51255 | 48696 | 45779 |
High Concordance with Illumina Platinum Genomes
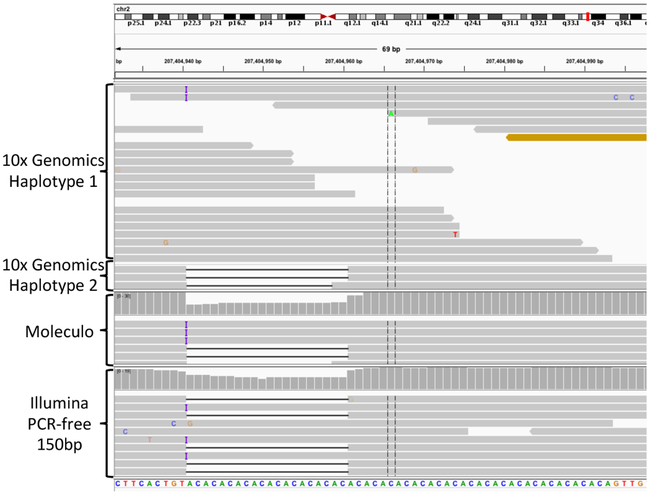
The bottom section of Table 1 shows increasing concordance over multiple versions of our HG001 callset with the Illumina Platinum Genomes 2016-v1.0 callset (PG).8 PG provides orthogonal confirmation of our calls, since the PG phased pedigree analysis may identify different biases from our method. PG contains a larger number of benchmark variant calls, even relative to our newest v3.3.2 calls for HG001. However, when comparing v3.3.2 to PG within both benchmark regions, we found that the majority of differences were places where v3.3.2 was correct and PG had a partial complex or compound heterozygous call (e.g., Figure 2), or where the PG benchmark region partially overlapped with a true deletion that v3.3.2 called correctly. To reduce the number of these inaccurate calls in PG, we contracted each of the PG benchmark regions by 50bp on each side, which is similar to how we remove 50bp on each side of uncertain variants (e.g., locus (1) in Figure 1b). Because PG has many more uncertain variants, contracting the PG benchmark regions by 50bp reduces the number of variants in the PG benchmark region by 32%, but it also eliminates 93% of the differences between v3.3.2 and PG in GRCh37, so that 61 PG-only and 47 v3.3.2-only calls remain in both benchmark regions.
Figure 2:
Complex variant discordant between GIAB and Illumina Platinum Genomes. Compound heterozygous insertion and deletion in HG001 in a tandem repeat at 2:207404940 (GRCh37), for which Illumina Platinum Genomes only calls a heterozygous deletion. When a callset with the true compound heterozygous variant is compared to Platinum Genomes, it is counted as both a FP and a FN. Both the insertion and deletion are supported by PCR-free Illumina (bottom) and Moleculo assembled long reads (middle), and reads assigned haplotype 1 in 10x support the insertion and reads assigned haplotype 2 in 10x support the deletion (top).
After manually curating the remaining 108 differences between v3.3.2 and PG in GRCh37, v3.3.2 has 5 clear FNs, 1 clear FP, 2 unclear potential FNs, and 6 unclear potential FPs in the NIST benchmark regions (Supplementary Data 1, Supplementary Note 4). Based on 3529641 variants concordant between NIST and PG, there would be about 2 FPs and 2 FNs per million true variants. Since these are only in the PG benchmark regions, these error rates are likely lower than the overall error rates, which are difficult to estimate.
Trio Mendelian inheritance analysis
Benchmark callsets were generated for two PGP trios, an Ashkenazi Jewish and Han Chinese trio (Supplementary Table 1, Supplementary Figure 1). As orthogonal benchmark callsets are not available for these genomes a trio Mendelian inheritance analysis was used to evaluate the callsets. The trio analysis allows us to phase some variants in the sons (see below) and determine variants that have a genotype pattern inconsistent with Mendelian inheritance. We separately analyzed Mendelian inconsistent variants that were potential cell-line or germline de novo mutations (i.e., the son was heterozygous and both parents were homozygous reference), and those that had any other Mendelian inconsistent pattern (which are unlikely to have a biological origin). Out of 2038 Mendelian violations in the Ashkenazi son, 1110 SNVs and 213 indels were potential de novo mutations. Out of 425 Mendelian violations in the Chinese son, 103 SNVs and 43 indels were potential de novo mutations. The large difference between the trios may arise from differences in the datasets used to generate the benchmark callsets or mutations arising in the EBV-immortalization process. The number of de novo SNVs is about 10% higher than the 1001 de novo SNVs found by 1000 Genomes for NA12878/HG001, and higher than the 669 de novo SNVs they found for NA19240.18 Upon manual inspection of 10 random de novo SNVs from each trio, 17/20 appeared to be true de novo. Upon manual inspection of 10 random de novo indels from each trio, 10/20 appeared to be true de novo indels (most in homopolymers or tandem repeats), and the remainder were correctly called in the sons but missed in one of the parents. Supplementary Data 1 and Supplementary Note 5 contain additional details about manual curation of each site and Mendelian concordance.
Based on the Mendelian analyses, there may be as many as 16 SNV and 150 indel errors per million variants in one or more of the 3 individuals before excluding these sites. Note that we exclude the errors found by the Mendelian analysis from our final benchmark bed files, but it is likely that the error modality that caused these Mendelian errors also caused other errors that were not found to be Mendelian inconsistent. Many other error modalities can be Mendelian consistent (e.g., systematic errors that cause all genomes in the pedigree to be heterozygous, and systematic errors that cause the wrong allele to be called), so these error estimates should be interpreted with caution.
Performance metrics vary when comparing to different benchmark callsets
We next explore how performance metrics can differ when using different benchmark sets. When comparing a method’s callset against these imperfect and incomplete benchmark callsets, Estimates of a method’s precision and recall/sensitivity computed from imperfect and incomplete benchmark callsets may differ from the true precision and recall of the method for all regions of the genome and all types of variants. Insufficient examples of variants of particular types can result in estimates of precision and recall with high uncertainties. In addition, estimates of precision and recall can be biased by the following: (1) errors in the benchmark callset, (2) use of methods that do not compare differing representations of complex variants, and (3) biases in the benchmark towards easier variants and genome contexts. We demonstrate that these biases are particularly apparent for some challenging variant types and genome contexts by benchmarking a standard bwa-GATK callset against three benchmark sets: GIABv2.18 from our previous publication,1 GIABv3.3.2 from this publication, and Platinum Genomes 2016-1.0 (PG).8
We examine 3 striking examples in coding regions, complex variants, and decoy-associated regions: (1) The bwa-GATK SNV FN rate is 62 times higher when benchmarking against PG vs. against GIAB, and 2/10 manually curated FNs vs. PG appeared to be errors in PG. (2) The bwa-GATK putative FP rate for compound heterozygous indels was only slightly lower for PG vs. GIAB, but upon manual curation 6/10 putative FPs were actually errors in PG whereas only 1/10 putative FPs was an error in GIAB. (3) The 1000 Genomes developed the hs37d5 decoy sequence to remove FPs in some regions with segmental duplications not in GRCh37.19 The precision for bwa-GATK-nodecoy is 67% vs. GIABv3.3.2, whereas it is 91% vs. GIABv2.18 and 93% vs. PG. The much higher precision vs v2.18 or PG is a result of many of the decoy-associated variants being excluded from the benchmark regions of GIABv2.18 and PG. Assessing these variants is important because they make up 7123 of the 16452 FPs (43%) vs. GIABv3.3.2 for this callset. We also found notable differences between benchmarks for large indels (Supplementary Data 1 and Supplementary Note 6). Subtle differences between performance metrics for different GIAB genomes (Supplementary Note 7 and Supplementary Table 2). Since characteristics of the benchmark can affect performance metrics, we have outlined known limitations of our benchmark set (Supplementary Note 8).
Orthogonal “validation” of benchmark set
We have chosen to present detailed results of expert manual curation of multiple short, linked, and long read sequencing alignments in place of the more traditional targeted Sanger sequencing method for “validation”. There are several reasons that confirming some discrepancies by Sanger sequencing is unlikely to add more confidence: (1) We already have multiple technologies supporting many of the calls in our truth set; (2) In our manual curation, we visualized PacBio and moleculo sequencing data, which have longer reads (or pseudo-reads) than Sanger sequencing and were not used to form our truth sets; (3) All 23 of the calls we classified as “Incorrect or partial call by PG at or near a true variant” are in homopolymers or tandem repeats, and 20 of them are heterozygous or compound heterozygous indels that Sanger could not help resolve; (4) We classified 18 SNVs as “Calls that are in PG and appear likely to be FPs despite inheriting properly, either due to a local duplication or a systematic sequencing error that occurs on only one haplotype” (Supplementary Data 1). These regions are unlikely to be easily characterized by Sanger sequencing, and longer reads from PacBio and Moleculo sequencing are more useful orthogonal “validation”. (5) Even when using a single short read sequencing technology, previous work found that discrepancies between Sanger sequencing and their variant calls were mostly errors in Sanger sequencing.20 (6) For 32 of the 47 variants found only in v3.3.2, the RTG phased pedigree calls had an identical call to v3.3.2 even though RTG’s Illumina-only pedigree methods are much more similar to PG’s pedigree methods. For these reasons, we have focused our efforts on expert manual curation of multiple whole genome sequencing technologies rather than performing targeted Sanger sequencing.
Discussion
Trustworthy benchmarks reliably identify errors in the results of any method being comparted to the benchmark. To demonstrate we meet this goal, we show that when using our benchmark calls with best practices for benchmarking established by the Global Alliance for Genomics and Health (Supplementary Note 9),10 the majority of putative FPs and FNs are errors in the callset being benchmarked. This is a moving target, since new methods are continually being developed that may perform better, particularly in challenging regions of the genome.
We form our benchmark callsets to accurately represent genomic variation in regions where we are confident in our technical and analytical performance. A notable limitation in such callsets is that they will tend to exclude more difficult types of variation and regions of the genome. When new methods with different characteristics than those use to form our callsets are applied, they may be more accurate in our regions difficult to access with our methods, and less accurate in regions where we are quite confident. For example, as graph-based variant calling methods are developed, they may be able to make much better calls in regions with many ALT haplotypes like the Major Histocompatibility Complex (MHC) even if they have lower accuracy within our benchmark regions. Therefore, it can be important to consider variant calls outside our benchmark regions when evaluating the accuracy of different variant calling methods.
To assure that our benchmark calls continue to be useful as sequencing and analysis methods improve, we are exploring several approaches for characterizing more difficult regions of the genome. Long reads and linked reads show promise in enabling benchmark calls in difficult-to-map regions of the genome.21,22 The methods presented here are a framework to integrate these and other new data types, so long as variant calls with high-sensitivity in a specified set of genomic regions could be generated, and filters could be applied to give high specificity.
Finally, to ensure these data are an enduring resource that we improve over time, we have made available an online form, where we and others can enter potentially questionable sites in our benchmark regions (https://goo.gl/forms/zvxjRsYTdrkhqdzM2). The results of this form are public and updated in real-time so that anyone can see where others have manually reviewed or interrogated the evidence at any site (https://docs.google.com/spreadsheets/d/1kHgRLinYcnxX3-ulvijf2HrIdrQWz5R5PtxZS-_s6ZM/edit?usp=sharing).
A Life Sciences reporting summary is available.
Online Methods
Sequencing Datasets
In contrast to our previous integration process,1 which used sequencing data that was generated from DNA from various growths of the Coriell cell line GM12878, most of the sequencing data used in this work were generated from NIST RMs, which are DNA extracted from a single large batch of cells that was mixed prior to aliquoting. These datasets, except for the HG001 SOLiD datasets, are described in detail in a Genome in a Bottle data publication.7
For these genomes, we used datasets from several technologies and library preparations:
~300x paired end whole genome sequencing with 2×148bp reads with ~550bp insert size from the HiSeq 2500 in Rapid Mode with v1 sequencing chemistry (HG001 and AJ Trio); the Chinese parents were similarly sequenced to ~100x coverage.
~300x paired end whole genome sequencing with 2×250bp reads with ~550bp insert size from the HiSeq 2500 in Rapid Mode with v2 sequencing chemistry (Chinese son).
~45x paired end whole genome sequencing with 2×250bp reads with ~400bp insert size from the HiSeq 2500 in Rapid Mode with v2 sequencing chemistry (AJ Trio).
~15x mate-pair whole genome sequencing with 2×100bp reads with ~6000bp insert size from the HiSeq 2500 in High Throughput Mode with v2 sequencing chemistry (AJ Trio and Chinese Trio).
~100x paired end 2×29bp whole genome sequencing from Complete Genomics v2 chemistry (all genomes)
~1000x single end exome sequencing from Ion PI™ Sequencing 200 Kit v4 (HG001, AJ Trio, and Chinese son)
Whole genome sequencing from SOLiD 5500W. For HG001, two whole genome sequencing datasets were generated: ~12× 2×50bp paired end and ~12× 75bp single end with error-correction chemistry (ECC). For the AJ son and Chinese son, ~42× 75bp single end sequencing (without ECC) was generated.
10x Genomics Chromium whole genome sequencing (~25x per haplotype from HG001 and AJ son, ~9x per haplotype from AJ parents, and ~13x per haplotype from the Chinese parents). These are the only data not from the NIST RM batch of DNA because longer DNA from the cell lines resulted in better libraries
Implementation of analyses and source code
Most analyses were performed using apps or applets on the DNAnexus data analysis platform (DNAnexus, Inc., Mountain View, CA), except for mapping of all datasets and variant calling for Complete Genomics and Ion exome, since these steps were performed previously. The apps and applets used in this work are included in GitHub (https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2). They run on an Ubuntu 12.04 machine on Amazon Web Services EC2. The apps and applets are structured as:
dxapp.json specifies the input files and options, output files, and any dependencies that can be installed via the Ubuntu command apt.
src/code.sh contains the commands that are run
resources/ contains compiled binary files, scripts, and other files that are used in the applet
The commands were run per chromosome in parallel using the DNAnexus command line interface, and these commands are listed at https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2/DNAnexusCommands/batch_processing_commands.
Illumina analyses
The Illumina fastq files were mapped using novoalign -d <reference.ndx> -f <read1.fastq.gz> <read2.fastq.gz> -F STDFQ --Q2Off -t 400 -o SAM -c 10 (version 3.02.07 from Novocraft Technologies, Selangor, Malaysia). and resulting BAM files were created, sorted, and indexed with samtools version 0.1.18. Bam files are available under:
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NIST_NA12878_HG001_HiSeq_300x/
Variants were called using both GATK HaplotypeCaller v3.5 23,24 and Freebayes 0.9.20 25 with high-sensitivity settings. Specifically, for HaplotypeCaller, special options were “-stand_call_conf 2 -stand_emit_conf 2 -A BaseQualityRankSumTest -A ClippingRankSumTest -A Coverage -A FisherStrand -A LowMQ -A RMSMappingQuality -A ReadPosRankSumTest -A StrandOddsRatio -A HomopolymerRun -A TandemRepeatAnnotator”. The gVCF output was converted to VCF using GATK Genotype gVCFs for each sample independently. For freebayes, special options were “-F 0.05 -m 0 --genotype-qualities”.
For freebayes calls, GATK CallableLoci v3.5 was used to generate callable regions with at least 20 reads with mapping quality of at least 20 (to exclude regions where heterozygous variants may be missed), and with coverage less than two times the median (to exclude regions likely to be duplicated or have mis-mapped reads). Because parallelization of freebayes infrequently causes conflicting variant calls at the same position, 2 variants at the same position are removed from the callable regions.
For GATK calls, the gVCF output from GATK was used to define the callable regions. In general, reference regions and variant calls with GQ<60 were excluded from the callable regions, excluding 50bp on either side of low GQ reference regions and low GQ variant calls. Excluding 50bp minimizes artifacts introduced when integrating partial complex variant calls. The exception to this rule is that reference assertions with GQ<60 are ignored if they are within 10bp of start or end of an indel with GQ>60, because upon manual inspection GATK often calls some reference bases with a low GQ near true indels even when the reference bases should have high GQ, and excluding 50bp regions around these bases excluded many true benchmark indels. The gVCF from GATK is used rather than CallableLoci because it provides a sophisticated interrogation of homopolymers and tandem repeats and excludes regions if insufficient reads completely cross the repeats.
Complete Genomics analyses
Complete Genomics data was mapped and variants were called using v2.5.0.33 of the standard Complete Genomics pipeline.26 Only the vcfBeta file was used in the integration process, because it contains both called variants and “no call” regions similar to gVCF. vcfBeta files are available under:
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/analysis/CompleteGenomics_RMDNA_11272014/
A python script from Complete Genomics (vcf2bed.py) was used to generate callable regions, which exclude regions with no calls or partial no calls in the vcfBeta. To minimize integration artifacts around partial complex variant calls, 50bp padding was subtracted from both sides of callable regions using bedtools slopBed. In addition, the vcfBeta file was modified to remove unnecessary lines and fill in the FILTER field using a python script from Complete Genomics (vcfBeta_to_VCF_simple.py). This process was performed on DNAnexus; an example command for chr20 is
“dx run GIAB:/Workflow/integration-prepare-cg -ivcf_in=/NA12878/Complete_Genomics/vcfBeta-GS000025639-ASM.vcf.bz2 -ichrom=20 --destination=/NA12878/Complete_Genomics/”
Ion exome analyses
The Ion exome data BaseCalling and alignment were performed on a Torrent Suite v4.2 server (ThermoFisher Scientific). Variant calling was performed using Torrent Variant Caller v4.4, and the TSVC_variants_defaultlowsetting.vcf was used as a sensitive variant call file.
GATK CallableLoci v3.5 was used to generate callable regions with at least 20 reads with mapping quality of at least 20.
These callable regions were intersected with the targeted regions bed file for the Ion Ampliseq exome assay available at ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/IonTorrent_TVC_03162015/AmpliseqExome.20141120_effective_regions.bed. In addition, 50bp on either side of compound heterozygous sites were removed from the callable regions, and these sites were removed from the vcf to avoid artifacts around homopolymers. This process was performed on DNAnexus, with the command for chr20:
“dx run GIAB:/Workflow/integration-prepare-ion -ivcf_in=/NA12878/Ion_Torrent/TSVC_variants_defaultlowsetting.vcf -icallablelocibed=/NA12878/Ion_Torrent/callableLoci_output/HG001_20_hs37d5_IonExome_callableloci.bed -itargetsbed=/NA12878/Ion_Torrent/AmpliseqExome.20141120_effective_regions.bed -ichrom=20 --destination=/NA12878/Ion_Torrent/Integration_prepare_ion_output/”
SOLiD analyses
The SOLiD xsq files were mapped with LifeScope v2.5.1 (ThermoFisher Scientific). Bam files are available under:
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NIST_NA12878_HG001_SOLID5500W/
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/HG002_NA24385_son/NIST_SOLiD5500W
ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/ChineseTrio/HG005_NA24631_son/NIST_SOLiD5500W/
Variants were called using GATK HaplotypeCaller v3.5 with high-sensitivity settings. Specifically, for HaplotypeCaller, special options were “-stand_call_conf 2 -stand_emit_conf 2 -A BaseQualityRankSumTest -A ClippingRankSumTest -A Coverage -A FisherStrand -A LowMQ -A RMSMappingQuality -A ReadPosRankSumTest -A StrandOddsRatio -A HomopolymerRun -A TandemRepeatAnnotator”. The gVCF output was converted to VCF using GATK Genotype gVCFs for each sample independently.
For SOLiD, all regions were considered “not callable” because biases are not sufficiently well understood, so SOLiD only provided support from an additional technology when finding training sites and when annotating the benchmark vcf.
10x Genomics analyses
The 10x Genomics Chromium fastq files were mapped and reads were phased using LongRanger v2.0 (10x Genomics, Pleasanton, CA).21 As a new technology, variant calls are integrated in a conservative manner, requiring clear support for a homozygous variant or reference call from reads on each haplotype. Bamtools was used to split the bam file into two separate bam files with reads from each haplotype (HP tag values 1 and 2), ignoring reads that were not phased. GATK HaplotypeCaller v3.5 was used to generate gvcf files from each haplotype separately. A custom perl script was used to parse the gvcf files, excluding regions with DP<6, DP > 2x median coverage, heterozygous calls on a haplotype, or homozygous reference or variant calls where the likelihood was <99 for homozygous variant or reference, respectively (https://github.com/jzook/genome-data-integration/blob/master/NISTv3.3.2/DNAnexusApplets/integration-prepare-10X-v3.3-anyref/resources/usr/bin/process_10X_gvcf.pl). The union of these regions from both haplotypes plus 50bp on either side was excluded from the callable regions.
Excluding challenging regions for each callset
As an enhancement in our new integration methods (v3.3+), we now exclude difficult regions per callset, rather than at the end for all callsets. We used prior knowledge as well as manual inspection of differences between callsets to determine which regions should be excluded from each callset. For example, we exclude tandem repeats and perfect or imperfect homopolymers >10bp for callsets with reads <100bp or from technologies that use PCR (all methods except Illumina PCR-free). We exclude segmental duplications and regions homologous to the decoy hs37d5 or ALT loci from all short read methods (all methods except 10x Genomics linked reads). The regions excluded from each callset are specified in the “CallsetTables” input into the integration process, where a 1 in a column indicates the bed file regions are excluded from that callset’s callable regions, and a 0 indicates the bed file regions are not excluded for that callset. Callset tables are available at https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2/CallsetTables.
Integration process to form benchmark calls and regions
Our integration process is summarized in Figure 1 and detailed in the outline in the Supplementary Note 10 and diagrams in Supplementary Figure 2, Supplementary Figure 3, and Supplementary Figure 4. Similar to our previous work for v2.18/v2.19,1 the first step in our integration process is to use preliminary “concordant” calls to train a machine learning model that finds outliers (Supplementary Figure 5). Callsets with a genotype call that has outlier annotations are not trusted. For training variants, we use genotype calls that are supported by at least 2 technologies, not including sites if another dataset contradicts the call or is missing the call and the call is within that callset’s callable regions. To do this, we normalize variants using vcflib vcfallelicprimitives and then generate a union vcf using vcflib vcfcombine. The union vcf contains separate columns from each callset, and we annotate the union vcf with vcflib vcfannotate to indicate whether each call falls outside the callable bed file from each dataset.
To find outliers, annotations from the vcf INFO and FORMAT fields and which tail of the distribution that we expected to be associated with bias for each callset were selected using expert knowledge (files describing annotations for each caller are at https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2/AnnotationFiles). Then, we used a simple one-class model that treats each annotation independently and finds the cut-off (for single tail) or cut-offs (for both tails) outside which (5/a) % of the training calls lie, where a is the number of annotations for each callset. For each callset, we find sites that fall outside the cut-offs or that are already filtered, and we generate a bed file that contains the call with 50bp added to either side to account for different representations of complex variants. We again annotate the union vcf with this “filter bed file” from each callset, which we next use in addition to the “callable regions” annotations (Figure 1b).
To generate the benchmark calls, we run the same integration script with the new union vcf annotated with both “callable regions” from each dataset and “filtered regions” from each callset (Figure 1c). The integration script outputs benchmark calls that meet all of the following criteria (in the vcf file for each genome ending in _v.3.3.2_all.vcf.gz, an appropriate output vcf FILTER status is given for sites that don’t meet each criterion, summarized in Supplementary Table 3):
Genotypes agree between all callsets for which the call is callable and not filtered. This includes “implied homozygous reference calls”, where a site is missing from a callset and it is within that callset’s callable regions and no filtered variants are within 50bp (otherwise, FILTER status “discordantunfiltered”, e.g., example (2) in Figure 1c).
The call from at least one callset is callable and not filtered (otherwise, FILTER status “allfilteredbutagree” or “allfilteredanddisagree”, e.g., example (4) in Figure 1c).
The sum of genotype qualities for all datasets supporting this genotype call is >=70. This sum includes only the first callset from each dataset and includes all datasets supporting the call even if they are filtered or outside the callable bed (otherwise, FILTER status “GQlessthan70”).
If the call is homozygous reference, then no filtered calls at the location can be indels >9bp. Implied homozygous reference calls (i.e., there is no variant call and it is in the callset’s callable regions are sometimes unreliable for larger indels, because callsets will sometimes miss the evidence for large indels and not call a variant (otherwise, FILTER status “questionableindel”).
The site is not called only by Complete Genomics and completely missing from other callsets, since these sites tended to be systematic errors in repetitive regions upon manual curation (otherwise, FILTER status “cgonly”).
For sites where the benchmark call is heterozygous, none of the filtered calls are homozygous variant, since these are sometimes genotyping errors (otherwise, FILTER status “discordanthet”).
Heterozygous calls where the net allele balance across all unfiltered datasets is <0.2 or >0.8 when summing support for REF and ALT alleles (otherwise, FILTER status “alleleimbalance”).
To calculate the benchmark regions bed file, the following steps are performed:
Find all regions that are covered by at least one dataset’s callable regions bed file.
Subtract 50bp on either side of all sites that could not be determined with high confidence.
We chose to use a simple, one-class model in this work in place of the more sophisticated Gaussian Mixture Model (GMM) used in v2.19 to make the integration process more robust and reproducible. The way we used the GMM in v2.18 and v2.19, GATK Variant Quality Score Recalibration was not always able to fit the data, requiring manual modified parameters, making the integration process less robust and reproducible. At times the GMM also appeared to overfit the data, filtering sites for unclear reasons. The GMM also was designed for GATK, ideally for vcf files containing many individuals, unlike our single sample vcf files. Our one-class model used in v3.3.2 was more easily adapted to filter on annotations in the FORMAT and INFO fields in vcf files produced by the variety of variant callers and technologies used for each sample. Future work could be directed towards developing more sophisticated models that learn which annotations are more useful, how to define the callable regions, and how annotations may have different values for different types of variants and sizes.
The integration process described in this section is implemented in the applet nist-integration-v3.3.2-anyref (https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2/DNAnexusApplets/nist-integration-v3.3.2-anyref). The one-class filtering model to find outliers is a custom perl script (nist-integration-v3.3.2-anyref/resources/usr/bin/VcfOneClassFiltering_v3.3.pl). The integration and arbitration processes to (1) find genotype calls supported by two technologies for the training set and (2) find benchmark genotype calls are both implemented in a perl script (nist-integration-v3.3.2-anyref/resources/usr/bin/VcfClassifyUsingFilters_v3.3.pl). Each line in the benchmark vcf file has FORMAT fields (Supplementary Table 4) and INFO fields (Supplementary Table 5) that give information about the support from each input callset for the final call. Some variants in the benchmark vcf fall outside the benchmark bed file because including these variants when doing the variant comparison helps account for differences in representation of complex variants between the benchmark vcf and the vcf being tested.
Candidate structural variants to exclude from benchmark regions
For HG001, we had previously used all HG001 variants from dbVar, which conservatively excluded about 10% of the genome, because dbVar contained any variants anyone ever submitted for HG001. Since then, several SV callsets from different technologies have been generated for HG001, and we now use these callsets instead of dbVar. Specifically, we include union of all calls <1Mbp in size from:
The union of several PacBio SV calling methods, including filtered sites (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NA12878_PacBio_MtSinai/NA12878.sorted.vcf.gz).
The PASSing calls from MetaSV, which looks for support from multiple types of Illumina calling methods (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/technical/svclassify_Manuscript/Supplementary_Information/metasv_trio_validation/NA12878_svs.vcf.gz).
Calls with support from multiple technologies from svclassify (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/technical/svclassify_Manuscript/Supplementary_Information/Personalis_1000_Genomes_deduplicated_deletions.bed and ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/technical/svclassify_Manuscript/Supplementary_Information/Spiral_Genetics_insertions.bed).
For the AJ trio, we use the union of calls from 11 callers using 5 technologies. Specifically, we include all calls >50bp and <1Mbp in size from:
PacBio callers: sniffles, Parliament, CSHL assembly, SMRT-SV dip, and MultibreakSV
Illumina callers: Cortex, Spiral Genetics, Jitterbug, and Parliament
BioNano haplo-aware calls
10x GemCode
Complete Genomics highConfidenceSvEventsBeta and mobileElementInsertionsBeta
For the Chinese trio, we use the union of calls from 11 callers using 5 technologies. Specifically, we include all calls >50bp and <1Mbp in size from:
Illumina callers: GATK-HC and freebayes
Complete Genomics highConfidenceSvEventsBeta and mobileElementInsertionsBeta
For HG006 and HG007, we also exclude calls from MetaSV in any individual in the trio
For deletions, we add 100bp to either side of the called region, and for all other calls (mostly insertions), we add 1000bp to either side of the called region. This padding helps to account for imprecision in the called breakpoint, complex variation around the breakpoints, and potential errors in large tandem duplications that are reported as insertions. For GRCh38, we remap the GRCh37 SV bed file to GRCh38 using NCBI remap.
GRCh38 integration
To develop benchmark calls for GRCh38, we use similar methods to GRCh37 except for data that could not be realigned to GRCh38. We are able to map reads and call variants directly on GRCh38 for Illumina and 10x, but native GRCh38 pipelines were not available for Complete Genomics, Ion exome, and SOLiD data. For Illumina and 10x data, variant calls were made similarly to GRCh37 but from reads mapped to GRCh38 with decoy but no alts (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/001/405/GCA_000001405.15_GRCh38/seqs_for_alignment_pipelines.ucsc_ids/GCA_000001405.15_GRCh38_no_alt_plus_hs38d1_analysis_set.fna.gz). For Complete Genomics, Ion exome, and SOLiD data, vcf and callable bed files were converted from GRCh37 to GRCh38 using the tool GenomeWarp (https://github.com/verilylifesciences/genomewarp). This tool converts vcf and callable bed files in a conservative and sophisticated manner, accounting for base changes that were made between the two references. Modeled centromere (genomic_regions_definitions_modeledcentromere.bed) and heterochromatin (genomic_regions_definitions_heterochrom.bed) regions are explicitly excluded from the benchmark bed (available under ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/NA12878_HG001/NISTv3.3.2/GRCh38/supplementaryFiles/).
For HG001 Illumina and 10x data, variants were called using GATK HaplotypeCaller v3.5 similarly to all GRCh37 genomes, but for the other genomes’ Illumina and 10x datasets, the Sentieon haplotyper version 201611.rc1 was used instead. (Sentieon, Inc., Mountain View, CA). This tool was designed to give the same results more efficiently than GATK HaplotypeCaller v3.5, except that it does not downsample reads in high coverage regions, so that the resulting variant calls are deterministic.27
Trio Mendelian analysis and phasing
Because variants are called independently in the three individuals, complex variants are sometimes represented differently amongst them; the majority of apparent Mendelian violations found by naive methods are in fact discrepant representations of complex variants in different individuals. We used a new method based on rtg-tools vcfeval to harmonize representation of variants across the 3 individuals, similar to the methods described in a recent publication.28 We performed a Mendelian analysis and phasing for the AJ Trio using rtg-tools v3.7.1. First, we harmonize the representation of variants across the trio using an experimental mode of rtg-tools vcfeval, so that different representations of complex variants do not cause apparent Mendelian violations. After merging the 3 individuals’ harmonized vcfs into a multi-sample vcf, we change missing genotypes for individuals to homozygous reference, since we later subset by benchmark regions (missing calls are implied to be homozygous reference in our benchmark regions). Then, we use rtg-tools mendelian to phase variants in the son and find Mendelian violations. Finally, we subset the Mendelian violations by the intersection of the benchmark bed files from all three individuals. We also generate new benchmark bed files for each individual that exclude 50bp on each side of Mendelian violations that are not apparent de novo mutations. This analysis and the phase transfer below is performed in the DNAnexus applet trio-harmonize-mendelian (https://github.com/jzook/genome-data-integration/tree/master/NISTv3.3.2/DNAnexusApplets/trio-harmonize-mendelian).
Phase transfer
Our integration methods only supply local phasing where GATK HaplotypeCaller (or Sentieon haplotyper) is able to phase the variants. For HG001/NA12878 and the AJ son (HG002), pedigree-based phasing information can supply global phasing of maternal and paternal haplotypes.
For HG001, Real Time Genomics (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/analysis/RTG_Illumina_Segregation_Phasing_05122016/) and Illumina Platinum Genomes (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/technical/platinum_genomes/2016-1.0/) have generated phased variant calls by analyzing her 17-member pedigree. The Platinum Genomes calls are phased in the order paternal∣maternal, and the RTG phasing is not ordered, so the phasing from Platinum Genomes was added first. First, we archive existing local phasing information to the IGT and IPS fields. Then, we use the rtg-tools vcfeval software phase-transfer mode to take the phasing first from Platinum Genomes and add it to the GT field of our HG001 benchmark vcf. We then use the rtg-tools vcfeval’s phase-transfer mode to take the phasing from RTG and add it to variants that were not phased by Platinum Genomes, flipping the phasing to the order paternal∣maternal where necessary. Variants with phasing from PG or RTG are given a PS=PATMAT. Variants that were not phased by either PG or RTG but are homozygous variant are given GT=1∣1 and PS=HOMVAR. Variants that were not phased by either PG or RTG but were phased locally by GATK HaplotypeCaller or Sentieon haplotyper are given the phased GT and PS from the variant caller. This phase transfer is performed in 2 steps with the applet phase-transfer.
For HG002, we apply the phasing from the trio analysis using rtg-tools vcfeval phase-transfer like above. Variants with trio-based phasing are given a PS=PATMAT. Variants that were not phased by the trio but are homozygous variant are given GT=1∣1 and PS=HOMVAR. Variants that were not phased by the trio but were phased locally by GATK HaplotypeCaller or Sentieon haplotyper are given the phased GT and PS from the variant caller. This phase transfer is performed in the applet trio-harmonize-mendelian.
Comparisons to other callsets
The Global Alliance for Genomics and Health formed a Benchmarking Team (https://github.com/ga4gh/benchmarking-tools) to standardize performance metrics and develop tools to compare different representations of complex variants. We have used one of these tools, vcfeval (https://github.com/RealTimeGenomics/rtg-tools) from rtg-tools-3.6.2 with --ref-overlap, to compare our benchmark calls to other vcfs. After performing the comparison, we subset the true positives, false positives, and false negatives by our benchmark bed file and then by the bed file accompanying the other vcf (if it has one). We then manually inspect alignments from a subset of the putative false positives and false negative and record whether we determine our benchmark call is likely correct, if we understand why the other callset is incorrect, if the evidence is unclear, if it is in a homopolymer, and other notes.
For HG001, we compare to these callsets:
NISTv2.18 benchmark calls and bed file we published previously
Platinum Genomes 2016-1.0, with a modified benchmark bed file that excludes an additional 50bp around uncertain variants or regions. This padding eliminates many locations where Platinum Genomes only calls part of a complex or compound heterozygous variant.
In addition, we use the hap.py+vcfeval benchmarking tool (https://github.com/Illumina/hap.py) developed by the GA4GH Benchmarking Team and implemented on precisionFDA website (https://precision.fda.gov/apps/app-F187Zbj0qXjb85Yq2B6P61zb) for use in the precisionFDA “Truth Challenge”. We modified the tool used in the challenge to stratify performance by additional bed files available at https://github.com/ga4gh/benchmarking-tools/tree/master/resources/stratification-bed-files, including bed files of “easier” regions and bed files encompassing complex and compound heterozygous variants. The results of these comparisons, as well as the pipelines used to generate the calls are shared in a Note on precisionFDA (https://precision.fda.gov/notes/300-giab-example-comparisons-v3-3-2). These precisionFDA results can be immediately accessed by requesting a free account on precisionFDA.
The callsets with lowest FP and FN rates for SNVs or indels from the precisionFDA “Truth Challenge” were compared to the v3.3.2 benchmark calls for HG001. From each comparison result, 10 putative FPs or FNs were randomly selected for manual inspection to assess whether they were, in fact, errors in each callset.
Integration with only Illumina and 10x Genomics WGS
To assess the impact of using fewer datasets on the resulting benchmark vcf and bed files, we performed integration for chromosome 1 in GRCh37 using only Illumina 300× 2×150bp WGS and 10x Genomics Chromium data for HG001. We compared these calls to Platinum Genomes as we describe above for v3.3.2 calls, and we manually inspected all differences with Platinum Genomes that were not in v3.3.2.
Differences between old and new integration methods
The new integration methods differ from the previous GIAB calls (v2.18 and v2.19) in several ways, both in the data used and the integration process and heuristics:
1. Only newer datasets were used, which were generated from the NIST RM 8398 batch of DNA (except for 10x Genomics, which used longer DNA from cells).
2. Mapping and variant calling algorithms designed specifically for each technology were used to generate sensitive variant callsets where possible: novoalign + GATK-haplotypecaller and freebayes for Illumina, the vcfBeta file from the standard Complete Genomics pipeline, tmap+TVC for Ion exome, and Lifescope+GATK-HC for SOLID. This is intended to minimize bias towards any particular bioinformatics toolchain.
3. Instead of forcing GATK to call genotypes at candidate variants in the bam files from each technology, we generate sensitive variant call sets and a bed file that describes the regions that were callable from each dataset. For Illumina GATK, we used the GATK-HC gVCF output to find regions with GQ>60. For Illumina freebayes, we used GATK callable loci to find regions with at least 20 reads with MQ >= 20 and with coverage less than 2x the median. For Complete Genomics, we used the callable regions defined by the vcfBeta file and excluded +−50bp around any no-called or half-called variant. For Ion, we intersected the exome targeted regions with the output of GATK CallableLoci for the bam file (requiring at least 20 reads with MQ >= 20). Due to the shorter reads and low coverage for SOLID, it was only used to confirm variants, so no regions were considered callable.
4. A new file with putative structural variants was used to exclude potential errors around SVs. For HG001, these were SVs derived from multiple PacBio callers (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NA12878_PacBio_MtSinai/) and multiple integrated illumina callers using MetaSV (ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/technical/svclassify_Manuscript/Supplementary_Information/metasv_trio_validation/). These make up a notably smaller fraction of the calls and genome (~4.5%) than the previous bed, which was a union of all dbVar calls for HG001 (~10%). For the AJ Trio, the union of >10 submitted SV callsets from Illumina, PacBio, BioNano, and Complete Genomics from all members of the trio combined was used to exclude potential SV regions. For the Chinese Trio, only Complete Genomics and GATK-HC and freebayes calls >49bp and surrounding regions were excluded due to the lack of available SV callsets for this genome at this time, which may result in a greater error rate in this genome. The SV bed files for each genome are under the supplementaryFiles directory.
5. To eliminate some errors from v2.18, homopolymers >10bp in length, including those interrupted by one nucleotide different from the homopolymer, are excluded from all input callsets except PCR-free GATK HaplotypeCaller callsets. For PCR-free GATK HaplotypeCaller callsets, we only include sites with confident genotype calls where HaplotypeCaller has ensured sufficient reads entirely encompass the repeat.
6. A bug that caused nearby variants to be missed in v2.19 is fixed in the new calls.
7. The new vcf contains variants outside the benchmark bed file. This enables more robust comparison of complex variants or nearby variants that are near the boundary of the bed file. It also allows the user to evaluate concordance outside the benchmark regions, but these concordance metrics should be interpreted with great care.
8. We now supply global phasing information from pedigree-based calls for HG001, trio-based phasing for the AJ son, and local phasing information from GATK-HC for the other genomes.
9. We use phased reads to make variant calls from 10x Genomics, conservatively requiring at least 6 reads from both haplotypes, coverage less than 2 times the median on each haplotype, and clear support for either the reference allele or variant allele in each haplotype.
Statistical Methods for Performance Metrics
The Wilson method in the R Hmisc binconf function was used to calculate 95% binomial confidence intervals for the recall and precision statistics in Supplementary Table 2.
Code availability
All code for analyzing genome sequencing data to generate benchmark variants and regions developed for this manuscript are available in a GitHub repository at https://github.com/jzook/genome-data-integration. Publicly available software used to generate input callsets includes novoalign version 3.02.07, samtools version 0.1.18, GATK v3.5, Freebayes 0.9.20, Complete Genomics tools v2.5.0.33, Torrent Variant Caller v4.4, LifeScope v2.5.1, LongRanger v2.0, GenomeWarp, rtg-tools v3.7.1, and sentieon version 201611.rc1.
Data availability
Raw sequence data were previously published in Scientific Data (DOI: 10.1038/sdata.2016.25) and were deposited in the NCBI SRA with the accession codes SRX1049768 to SRX1049855, SRX847862 to SRX848317, SRX1388368 to SRX1388459, SRX1388732 to SRX1388743, SRX852932 to SRX852936, SRX847094, SRX848742 to SRX848744, SRX326642, SRX1497273, and SRX1497276. 10x Genomics Chromium bam files used are at ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/10XGenomics_ChromiumGenome_LongRanger2.0_06202016/. The benchmark vcf and bed files resulting from work in this manuscript are available in the NISTv3.3.2 directory under each genome on the GIAB FTP release folder ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/, and in the future updated calls will be in the “recent” directory under each genome. The data used in this manuscript and other datasets for these genomes are available in ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/, and in the NCBI BioProject PRJNA200694.
Supplementary Material
Acknowledgments
We thank the many contributors to Genome in a Bottle Consortium discussions. We especially thank Rafael Saldana and the Sentieon team for advice on running the Sentieon pipeline, Andrew Carroll and the DNAnexus team for advice on implementing the pipeline in DNAnexus, Fiona Hyland, Srinka Ghosh, Keyan Zhao, and John Bodeau at ThermoFisher for advice integrating Ion exome and SOLiD genome data, Deanna Church and Valerie Schneider for helpful discussions about GRCh38, and many individuals for providing feedback about the current version and previous versions of our calls. Certain commercial equipment, instruments, or materials are identified to specify adequately experimental conditions or reported results. Such identification does not imply recommendation or endorsement by the National Institute of Standards, nor does it imply that the equipment, instruments, or materials identified are necessarily the best available for the purpose. Chunlin Xiao and Stephen Sherry were supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. Justin Zook, Marc Salit, Nathan Olson, and Justin Wagner were supported by the National Institute of Standards and Technology and an interagency agreement with the Food and Drug Administration.
Footnotes
Competing Financial Interests: RT is an employee of, and holds stock in, Invitae. HH was an employee of 10x Genomics. SAI and LT are employees of Real Time Genomics. CYM is an employee of Verily Life Sciences and Google
References
- 1.Zook JM et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol 32, 246–51 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Patwardhan A et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 7, 71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lincoln SE et al. A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. J. Mol. Diagnostics 17, 533–544 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Telenti A et al. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. U. S. A 113, 11901–11906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornish A & Guda C A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. Biomed Res. Int 2015, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poplin R et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol 36, 983 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberle MA et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res 27, 157–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary JG et al. Joint Variant and De Novo Mutation Identification on Pedigrees from High-Throughput Sequencing Data. J. Comput. Biol 21, 405–419 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Krusche P et al. Best Practices for Benchmarking Germline Small Variant Calls in Human Genomes. bioRxiv 270157 (2018). doi: 10.1101/270157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ball MP et al. A public resource facilitating clinical use of genomes. Proc. Natl. Acad. Sci. U. S. A 109, 11920–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudalkar EM et al. Multiplexed Reference Materials as Controls for Diagnostic Next-Generation Sequencing: A Pilot Investigating Applications for Hypertrophic Cardiomyopathy. J. Mol. Diagn 18, 882–889 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lincoln SE et al. An interlaboratory study of complex variant detection. bioRxiv 218529 (2017). doi: 10.1101/218529 [DOI] [Google Scholar]
- 14.Zhou B et al. Extensive and deep sequencing of the Venter/HuRef genome for developing and benchmarking genome analysis tools. bioRxiv 281709 (2018). doi: 10.1101/281709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu JC et al. Leveraging long read sequencing from a single individual to provide a comprehensive resource for benchmarking variant calling methods. Sci. Rep 5, 14493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.English AC et al. Assessing structural variation in a personal genome—towards a human reference diploid genome. BMC Genomics 16, 286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad DF et al. Variation in genome-wide mutation rates within and between human families. Nat. Genet 43, 712–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck TF, Mullikin JC, NISC Comparative Sequencing Program, on behalf of the N. C. S. & Biesecker LG Systematic Evaluation of Sanger Validation of Next-Generation Sequencing Variants. Clin. Chem 62, 647–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks P et al. Resolving the Full Spectrum of Human Genome Variation using Linked-Reads. bioRxiv 230946 (2018). doi: 10.1101/230946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenger AM et al. Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. bioRxiv 519025 (2019). doi: 10.1101/519025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrison E & Marth G Haplotype-based variant detection from short-read sequencing. (2012). [Google Scholar]
- 26.Drmanac R et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Kendig K et al. Computational performance and accuracy of Sentieon DNASeq variant calling workflow. bioRxiv 396325 (2018). doi: 10.1101/396325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toptaş BÇ, Rakocevic G, Kómár P & Kural D Comparing complex variants in family trios. Bioinformatics (2018). doi: 10.1093/bioinformatics/bty443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data were previously published in Scientific Data (DOI: 10.1038/sdata.2016.25) and were deposited in the NCBI SRA with the accession codes SRX1049768 to SRX1049855, SRX847862 to SRX848317, SRX1388368 to SRX1388459, SRX1388732 to SRX1388743, SRX852932 to SRX852936, SRX847094, SRX848742 to SRX848744, SRX326642, SRX1497273, and SRX1497276. 10x Genomics Chromium bam files used are at ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/10XGenomics_ChromiumGenome_LongRanger2.0_06202016/. The benchmark vcf and bed files resulting from work in this manuscript are available in the NISTv3.3.2 directory under each genome on the GIAB FTP release folder ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/release/, and in the future updated calls will be in the “recent” directory under each genome. The data used in this manuscript and other datasets for these genomes are available in ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/, and in the NCBI BioProject PRJNA200694.