Abstract
Macrophages play important and diverse roles during cancer progression. However, cancer therapies based on macrophage modulation are lacking in tools that can recognize and deliver therapeutic payloads to macrophages in a tumor-specific manner. As a result, treatments tend to interfere with normal macrophage functions in healthy organs. We previously identified a macrophage-binding peptide, termed CRV. Here, we show that upon systemic administration into tumor-bearing mice, CRV selectively homes to tumors, extravasates, and preferentially binds to macrophages within. CRV exhibits a higher affinity for tumor macrophages than for other cells in tumors or for other macrophage types elsewhere in the body. We further identified and validated retinoid X receptor beta (RXRB) as the CRV receptor. Intriguingly, although it is known as a nuclear receptor, RXRB shows a prominent cell surface localization that is largely restricted to tumor macrophages. Systemic administration of anti-RXRB antibodies also results in tumor selective binding to macrophages similar to CRV. Lastly, we demonstrate the ability of CRV to improve the delivery of nano carriers into solid tumors and macrophages within. In summary, we describe here a novel cell surface marker and targeting tools for tumor macrophages that may aid in future development of macrophage-modulatory cancer therapies.
Keywords: tumor-associated macrophages, tumor homing, RXR beta targeting, cargo delivery
Introduction
Macrophages exist in virtually all organs and are central players in normal immune functions [1]. They are also key regulators in the pathogenesis of various human diseases including cancer [2, 3]. Tumor-associated macrophages (TAMs) contribute to tumor growth in many ways: stimulating angiogenesis [4, 5], remodeling extracellular matrix, promoting tumor cell migration and metastasis, and mediating drug resistance [6, 7]. Notably, TAMs are instrumental in creating an immunosuppressive microenvironment in many solid tumors [8], which undermines the effectiveness of immunotherapy. Thus, technologies to modulate TAM activities are urgently needed to improve the efficacy of current cancer therapies.
One characteristic feature of macrophages is the plasticity of their functional properties and cell surface markers in response to the local milieu [9]. The binding to some of the macrophage markers by targeting ligands (e.g. peptides and antibodies) can directly modulate TAM functions and generate antitumor effects [10]. Such macrophage markers that are under clinical investigation as targets for cancer therapy include CSF1/CSF1R, CCL2/CCR2, and CD40 [11, 12].
Another way of utilizing cell surface markers and their ligands is ligand-directed delivery of therapeutics, where the ligands are only used to concentrate the drug at the TAM surface. This approach can be used together with generic toxins or therapeutic agents that change the macrophage phenotypes (e.g. from anti- to pro-inflammatory phenotypes), to achieve TAM modulation in tumors Mannose receptor (CD206) and folate receptor β are examples of such TAM markers that have been widely used for ligand-directed delivery [13, 14]. However, the majority of these surface markers are also found on healthy monocytes/macrophages, and more importantly, are critical players in normal immune functions. Targeting these receptors has shown side effects in healthy organs [10]. Therefore, a major task for TAM modulation technologies is to distinguish TAMs from healthy macrophages, which will help confine the modulatory effects in tumors while minimizing the side effects elsewhere.
One possible solution to the above problem is to identify cell surface markers specific for TAMs. Phage display screening has been used to identify peptides that bind selectively to TAMs. It is useful in discovering not only targeting peptides but also potentially novel surface markers of TAMs. Several TAM targeting peptides have been reported in the past decade. A screening performed in vivo using tumor-bearing mice identified a tumor-specific peptide, termed LyP-1 [15], which binds to a protein known as gC1q-R, HABP1 or p32 [16]. LyP-1 primarily accumulates in TAMs in tumors, but cell-surface p32 is also expressed in tumor endothelial cells and tumor cells. Moreover, LyP-1 also recognizes macrophages in atherosclerotic plaques, a chronic disease prevalent in the adult population [17]. A phage screen performed on primary macrophages from tumor mice gave rise to a peptide, termed UNO that also recognizes TAMs [18]. The receptor of this peptide is CD206, which is a known surface protein expressed on TAMs and other macrophages [19]. The apparent TAM specificity of UNO may depend on a modification of the peptide that only happens on the surface of TAMs. Cieslewicz M. et al. used cultured macrophages differentiated into the immune-suppressive M2 phenotype to isolate a peptide that shows a preferential binding to TAMs [20, 21]. However, the receptor for this peptide has not been identified, which prevents the understanding of its TAM-targeting mechanism. Overall, more efforts are needed to identify novel cell surface markers, and targeting tools, that can distinguish TAMs from macrophages in healthy organs and even other pathological tissues.
We previously performed an in vitro phage screen on a murine macrophage cell line, J774A.1 [22]. This screen identified a lead peptide, dubbed CRV (CRVLRSGSC, where the terminal cysteines form a disulfide bond to render the peptide cyclic). Upon intravenous (IV) injection into mice, CRV specifically bound to macrophages in lungs infected by bacteria, but not those in healthy lungs [22]. The ability of CRV to distinguish between macrophages in pathological and healthy tissues led us to evaluate its homing specificity for solid tumors and the TAMs within, and to identify the receptor that accounts for the unique specificity of this peptide toward a macrophage subpopulation.
Materials and methods
Materials.
Carboxyfluorescein-conjugated peptides (FAM-CRV and FAM-GGS, etc.) were purchased from LifeTein, LLC (Somerset, NJ). FAM was attached through an aminohexanoic acid linker to the N-terminal amino group. The C-terminus of peptide was not blocked. Antibodies used in this work are listed in Supplementary Table S2. Aminopropyldimethylethoxysilane and sulforhodamine 101 (SR101) were purchased from Sigma-Aldrich (St. Louis, MO). Maleimide-PEG-succinimidyl valerate (MAL-PEG-SVA, MW: 5,000) was purchased from Laysan Bio, Inc. (Arab, AL). RAW264.7, J774A.1, Py8119, and THP-1 and 4T1 cells were from American Type Culture Collection (Manassas, VA). LL/2 Red-Flue cells were obtained from PerkinElmer, Inc. (Waltham, MA). H1975 cells were kindly provided by Dr. Garth Powis, Cancer Center, Sanford Burnham Prebys Medical Discovery Institute (SBP), KPC cells by Dr. Andre Nel, University of California, Los Angeles, and KRAS-Ink cells by Dr. Douglas Hanahan, Swiss Federal Institute of Technology Lausanne. M1 and M2 macrophages were polarized from bone marrow-derived macrophages (BMDM) of C57BL/6 mice as previously described [23, 24].
Tumor models.
4T1, MCF10CA1a, KRAS-Ink, KPC, LL/2 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. H1975 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. Six to 8-week old BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA) or The Jackson Laboratory (Bar Harbor, ME). C57BL/6 were obtained from the animal facility at SBP. Hsd: Athymic Nude-Foxn1nu were received from Envigo (Indianapolis, IN).
To produce 4T1 tumors, 1 × 106 tumor cells (suspended in 100 μL of PBS) were orthotopically injected into the mammary fat pad of female BALB/c mice. In the MCF10CA1 a model, 2 × 106 tumor cells were injected into the mammary fat pad of female Hsd: Athymic Nude-Foxn1nu mice. The Py8119 model was established by injecting 1 × 106 tumor cells (suspended in 100 μL of PBS) orthotopically into the mammary fat pad of C57BL/6 female mice. Subcutaneous KRAS-Ink tumors were established by injecting 1 × 106 cells in 100 μL of PBS to the lower back of BALB/c mice. For H1975 tumors, 5 × 106 cells were subcutaneously injected to the lower back of female Hsd: Athymic Nude-Foxn1nu mice. LL/2 tumors were generated by injecting 1 × 106 cells in 100 μL of PBS subcutaneously to the lower back of C57BL/6 mice. KPC orthotopic pancreatic tumor model was established by injecting 1 × 106 cells suspended in 20 μL of serum-free medium and Matrigel (1:1 (vol/vol) ratio, Corning) to the pancreas of BALB/c mice. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee at SBP.
Peptide homing in vivo.
Biodistribution of FAM-conjugated peptides was examined after intravenous injection of 100 μL peptide solution (1 mg/mL in PBS) into the tail vein of tumor-bearing mice. The peptide was allowed to circulate for 5 min, 15 min or 1 h. Animals were then sacrificed under deep anesthesia by transcardial perfusion with PBS. Tissues were collected, fixed with a 4% formaldehyde buffer solution overnight, then washed and soaked in 20% sucrose in PBS for two days. The tissues were then frozen in OCT embedding compound (Tissue-Tek) on dry ice and sectioned for immunostaining.
Cell binding studies.
Mice with tumor size about 8 mm in diameter were euthanized by cervical dislocation under deep anesthesia (lack of response to a toe pinch). Target (tumor) and control (spleen, blood) tissues were collected and dissociated into single cells. Tumor cell suspensions were obtained using MACS tumor dissociation kit from Miltenyi Biotec, Inc. (Bergisch Gladbach, Germany). Spleen cells were dissociated by grinding the tissue on a 70 μm cell strainer using a syringe plunger. Cell suspensions from all tissues were treated with red blood cell lysis buffer before they were incubated with peptides or antibodies at 4 °C for 1 h (both primary and secondary antibodies). TruStain fcX™ (rat anti-mouse CD16/32) antibody was included in the primary antibody staining step as the Fc blocking agent. Cells were washed with PBS between steps and analyzed on BD LSRFortessa (BD Biosciences, San Jose, CA). The flow cytometry data were then analyzed using FlowJo.
Homing of anti-RXRB antibody in vivo.
Rabbit polyclonal anti-RXRB antibody from GeneTex or mouse monoclonal anti RXRB antibody from Invitrogen (50 μg in 100 μL of PBS) was intravenously injected to each 4T1 or KPC tumor-bearing mouse. Rabbit IgG or mouse IgG, 50 μg per mouse was injected into control mice. To compare the homing of the antibody and the peptide, FAM-CRV or control FAM-GGS (100 μg in 100 μL PBS) was injected 3 hours after the antibody. After 4-h circulation of the antibody, animals were sacrificed by transcardial perfusion with PBS. Tissues were collected, fixed and sectioned as described above.
Immunofluorescence staining.
Frozen tissue sections were first treated with PBS containing 1% BSA and 0.1% Triton X100 (blocking buffer) at room temperature (RT) for 1 h. The sections were washed three times with PBS and then incubated with primary antibodies diluted (1:200) in blocking buffer at 4 °C overnight, followed by the appropriate secondary antibodies diluted (1:200) in blocking buffer at RT for 1 h. Details about antibodies are summarized in Supplementary Table S2. After washing with PBS, sections were mounted in DAPI-containing mounting medium (Vector Laboratories, Burlingame, CA) with a coverslip and examined under a Zeiss LSM 710 NLO confocal microscope.
RXRB immunohistochemistry staining.
Frozen tissue sections were first treated with peroxidase blocking solution (0.3% Peroxidase in Methanol) at RT for 30 min, and then with goat serum blocking solution at RT for 30 min. Primary rabbit anti-RXRB antibody (GTX79265, dilution 1:500) was then added to the tissue sections for 1-h incubation at RT. After removal of the primary antibody, the sections were treated with secondary anti-rabbit antibody for 30 minutes and then DAB peroxidase substrate for 5 minutes. Counterstaining with Hematoxylin was then performed, followed by dehydration in ethanol and xylene, and mounting with Cytoseal media.
CRV-pSiNP-SR101 in vivo studies.
The synthesis and characterization of CRV-pSiNP-SR101 is described in Supplementary Information. 100 μL of CRV-pSiNP-SR101 or control pSiNP-SR101 solution (400 μg NP in PBS) was injected into the tail vein of 4T1 tumor-bearing mice. After 2 h circulation, the mice were sacrificed by transcardial perfusion with PBS. Tumors and healthy organs were collected and either imaged with Xenogen IVIS or dissociated to single cells and stained for cell markers similar to the cell binding studies described above.
Statistical analysis.
Experiments were performed at least three times. All data represent mean value ± standard error of the mean. The significance analyses were performed using two-tailed Student’s t test as detailed in the figure legends.
Results
CRV selectively homes to tumors and rapidly extravasates upon systemic administration
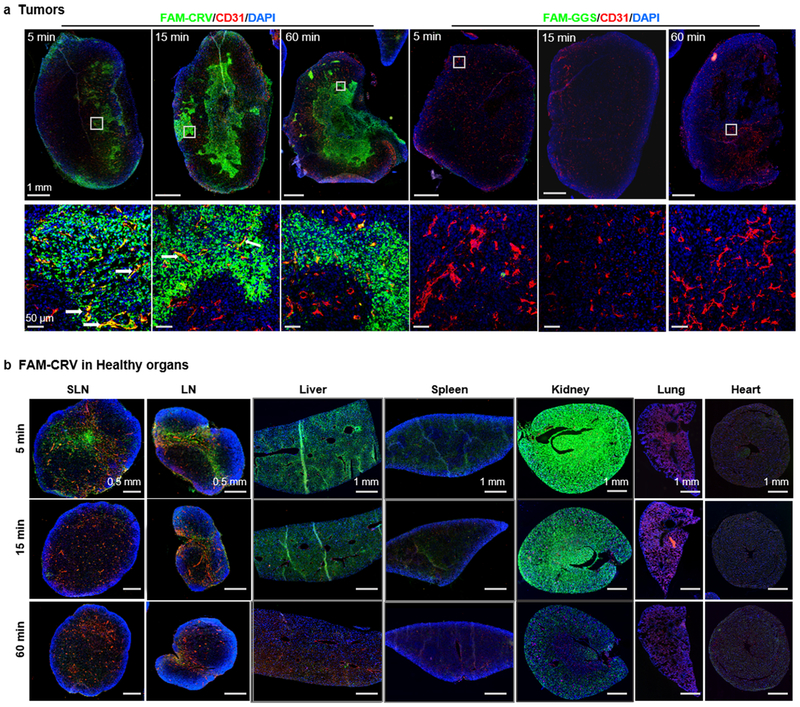
We first evaluated the in vivo distribution of CRV in tumor-bearing mice. To visualize the peptide, a fluorescein (FAM) dye was added to the N-terminus of CRV (FAM-CRV). In our previous report, we showed that biotinylated CRV inhibits the binding of FAM-CRV to macrophages, and FAM-labeled control peptide shows no specific binding to macrophages [22]. Thus, CRV binding to macrophages is independent on the FAM labeling. In this work, FAM-CRV was injected intravenously into tumor-bearing mice and allowed to circulate for 1 h before the major organs were collected for analysis. FAM-GGS (GGSGGSKG) or FAM-ARAL (ARALPSQRSR) [25] was used as a negative control peptide. We found that CRV exhibited tumor-selective accumulation in multiple tumor models including 4T1 mouse mammary carcinoma, MCF10CA1a human mammary carcinoma, KRAS-Ink [26, 27] and KPC mouse pancreatic ductal adenocarcinoma, H1975 human lung adenocarcinoma, and LL/2 mouse Lewis lung carcinoma. Substantially lower fluorescence signals were observed in healthy organs, especially liver and spleen which contain a high number of healthy monocytes/macrophages (Supplementary Fig. S1). The tumor accumulation of CRV was further confirmed by immunofluorescence (IF) staining of FAM in the tumor and healthy tissues (Fig. 1a and Supplementary Fig. S2). These results demonstrate that CRV selectively homes to tumors upon systemic administration.
Fig. 1.
The in vivo distribution of FAM-CRV in tumors and healthy organs over time. The mice were perfused with PBS and tissues were collected at 1 h or at the indicated time points after IV injection of FAM-CRV or FAM-GGS into mice bearing 4T1 orthotopic breast tumors. The tissues were stained with anti-FITC antibody for FAM-labeled peptide (green), anti-CD31 for blood vessels (red), and DAPI for nuclei (blue). (a) Top row: whole tumor scan (scale bar: 1 mm). Bottom row: magnified view of regions marked by white squares in the top row (scale bar: 50 μm). White arrows indicate co-localization of FAM-CRV with CD31. (b) Distribution of FAM-CRV in indicated organs. SLN: sentinel lymph node; LN: lymph node (scale bar: 0.5 mm). Scale bar for liver, spleen, and kidney is 1 mm. These experiments were performed in three animals per group; representative images are shown here.
The dynamics of CRV transport into tumors over time was then investigated. Representative images of IF staining are shown in Fig. 1. FAM-CRV was observed in tumors as early as 5 min after IV injection. Most of CRV had already extravasated (Fig. 1a), but co-localization of CRV and blood vessels was still observed at this time point (white arrows in Fig. 1a). The blood vessel co-localization was less apparent at later time points, and the peptide accumulated and spread extravascularly over time. In comparison, the control peptide showed little accumulation in tumors (Fig. 1a). We observed no CRV in the lungs and heart (Fig. 1b). Some peptide was present in other healthy organs (liver, spleen, kidney and lymph node) at early time points (Fig. 1b, Supplementary Fig. S3). However, the peptide signal was essentially gone at one hour (Fig. 1b, Supplementary Fig. S3). The loss of peptide signal in these organs suggests that the early signal in healthy organs was clearance.
CRV preferentially recognizes macrophages in tumors
In the previous study, we showed that CRV binds to two murine macrophage cell lines, J774A.1 and RAW264.7 (referred to J774 and RAW hereafter) [22]. In the present work, the binding of CRV to macrophages of different origins and activation states was evaluated. Compared to control peptide, FAM-CRV showed specific binding to macrophages differentiated from human monocytic cell line THP-1, as well as bone marrow-derived macrophages polarized to either M1 or M2 phenotype (Supplementary Fig. S4). In contrast, FAM-CRV barely bound to 4T1 and Py8119 breast cancer cell lines (Supplementary Fig. S4). These in vitro results further support the conclusion that CRV binds to macrophages but not to other cell types.
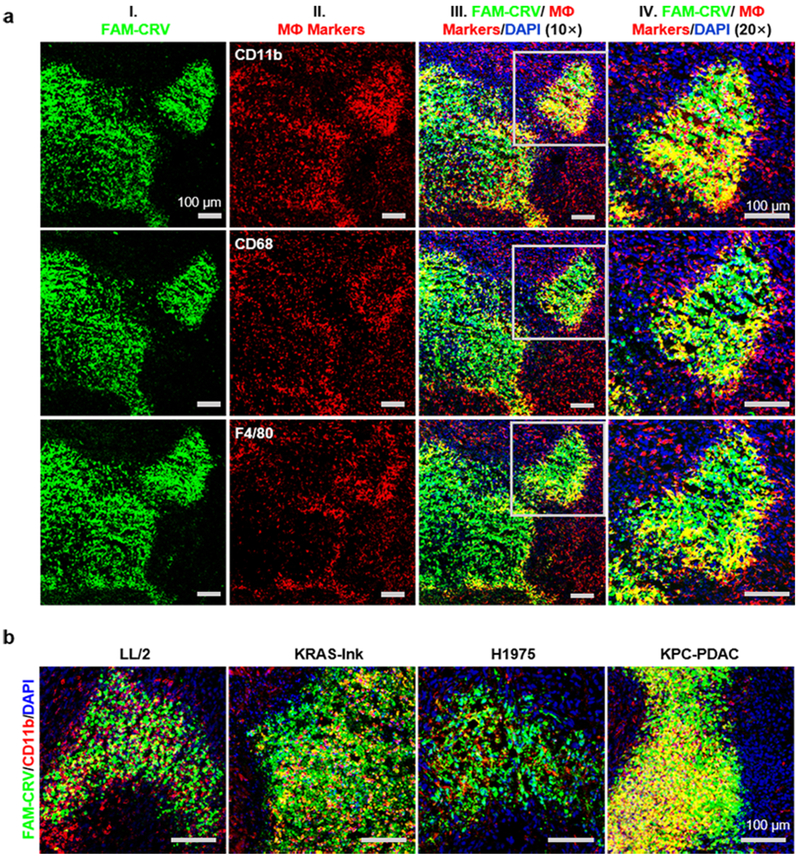
To determine if CRV recognizes TAMs in vivo, mice bearing 4T1 tumors were injected IV with FAM-CRV and tumor tissue was stained for CD11b, F4/80 and CD68, which are all common markers of macrophages [28]. FAM-CRV showed colocalization with all these markers (Fig. 2a; yellow color indicates co-localization); the colocalization was strongest between CRV and CD11b, and CD11b was selected as the primary macrophage marker in subsequent experiments. Similar co-localization between FAM-CRV and macrophage markers was seen in other tumor models positive for CRV homing (Fig. 2b). Tumor staining using antibodies against fibroblast marker fibroblast activation protein (FAP) and epithelial cell marker CD326 (EpCAM) showed no CRV colocalization with these markers, indicating lack of CRV binding to stromal fibroblasts and tumor cells (Supplementary Fig. S5). These results indicate that FAM-CRV mainly associates with TAMs in tumor tissue.
Fig. 2.
Co-localization of FAM-CRV with macrophages in tumors. (a) Co-localization of FAM-CRV and macrophage markers after 1-h in vivo homing of CRV in mice bearing 4T1 breast tumors. IF staining was performed with anti-FITC antibody for FAM-CRV (green) and with antibodies for the indicated macrophage markers (red). Nuclei were stained with DAPI (blue). Scale bars: 100 μm. (b) Co-localization of FAM-CRV and macrophage markers in additional tumor types. After 1-h in vivo homing in the indicated tumor models, tumor sections were stained with anti-FITC antibody for FAM-CRV (green), anti-CD11b for macrophages (red), and DAPI for nuclei (blue). Scale bars: 100 μm. All experiments were performed in three animals per group, and representative images are shown.
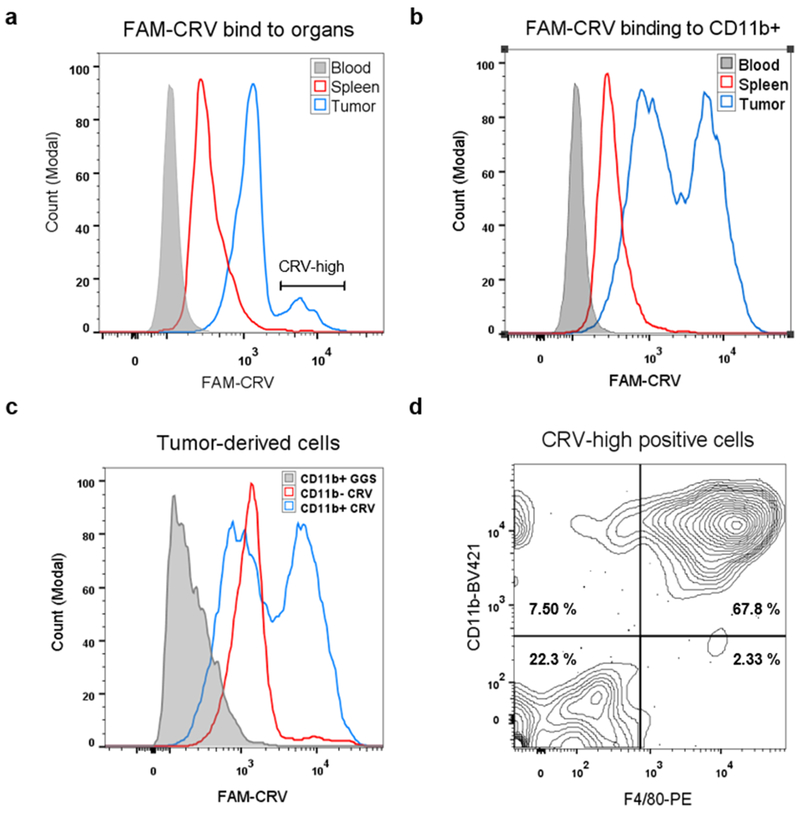
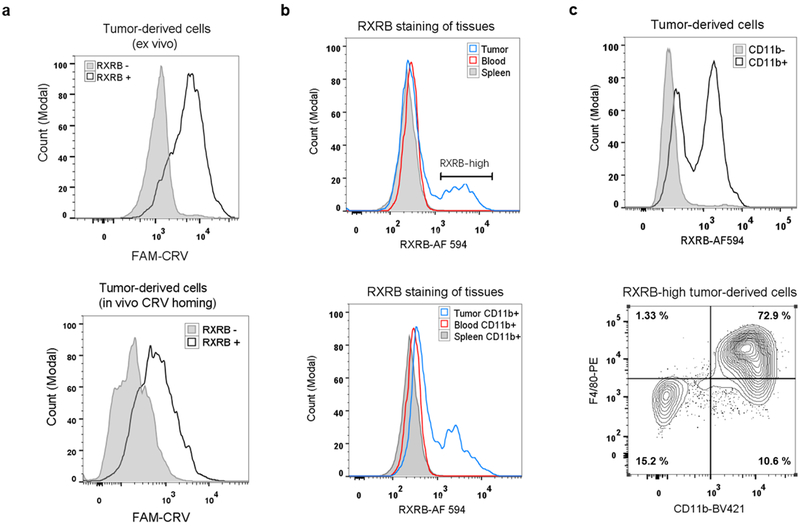
To investigate the intrinsic affinity of CRV for various cell types in tumors, we bypassed the complexity of in vivo transport and performed ex vivo binding studies. Tumor, spleen, and blood were collected from 4T1 tumor-bearing mice and dissociated into live single cells, and the cells were incubated with FAM-CRV. CRV showed a much higher affinity for the whole cell population from tumors than for cells from control organs (Fig. 3a), which is in agreement with the in vivo homing results. We then investigated the affinity of CRV for macrophages in various organs by focusing on CD11b-positive cells. The binding of FAM-CRV to CD11b-positive cells in blood was similar to that of the FAM-GGS control, indicating that there is no substantial binding of CRV to circulating monocytes (Supplementary Fig. S6a). In both spleen and tumor, FAM-CRV showed a higher affinity for CD11b-positive cells than FAM-GGS (Supplementary Fig. S6a, b), while FAM-CRV bound to CD11b-positive cells from the tumor much more strongly than to those from the spleen or blood (Fig. 3b). Among tumor-derived cells, CRV showed a higher affinity for CD11b-positive cells relative to CD11b-negative cells (Fig. 3c), and the majority of CRV-positive cells displayed macrophage markers while those that displayed low affinity for CRV did not (Fig. 3d, Supplementary Fig. S6c), agreeing with the results of IF staining (Fig. 2). Overall, these results demonstrate that the intrinsic affinity of CRV for TAMs is substantially higher than for their macrophage counterparts in healthy organs or for other cell types in tumors.
Fig. 3.
Preferential binding of FAM-CRV to TAMs. Ex vivo binding study of FAM-CRV to cells was performed by isolating single cell suspensions from the indicated tissues of 4T1 tumor-bearing mice. The cells were incubated with peptides and antibodies to the indicated macrophage markers as described in Materials & Methods. (a) Flow cytometry histogram of CRV binding to the cells from the indicated tissues. FAM-CRV showed stronger binding to tumor cells than to blood and spleen cells. (b) Preferential binding of FAM-CRV to tumor-derived CD11b+ cells over CD11b+ cells from the blood and spleen. (c) The binding of FAM-CRV to tumor-derived CD11b+ is stronger than the binding of this peptide to CD11b− cells or the binding of FAM-GGS to CD11b+ cells. (d) Flow cytometry analysis of FAM-CRV-high cells from 4T1 tumors for the indicated macrophage markers. BV421: Brilliant Violet 421. PE: phycoerythrin. These experiments were performed with individual samples from three animals and representative results are shown.
Identification of RXRB as the CRV receptor and a surface marker of TAMs
Next, we proceeded to identify the CRV receptor on macrophages. Membrane proteins of RAW cells were used as the source to isolate putative CRV receptors by affinity chromatography as previously described [29]. Mass spectrometry analysis of proteins that eluted with CRV peptide identified retinoid X receptor beta (RXRB) as the top candidate (Supplementary Table S1).
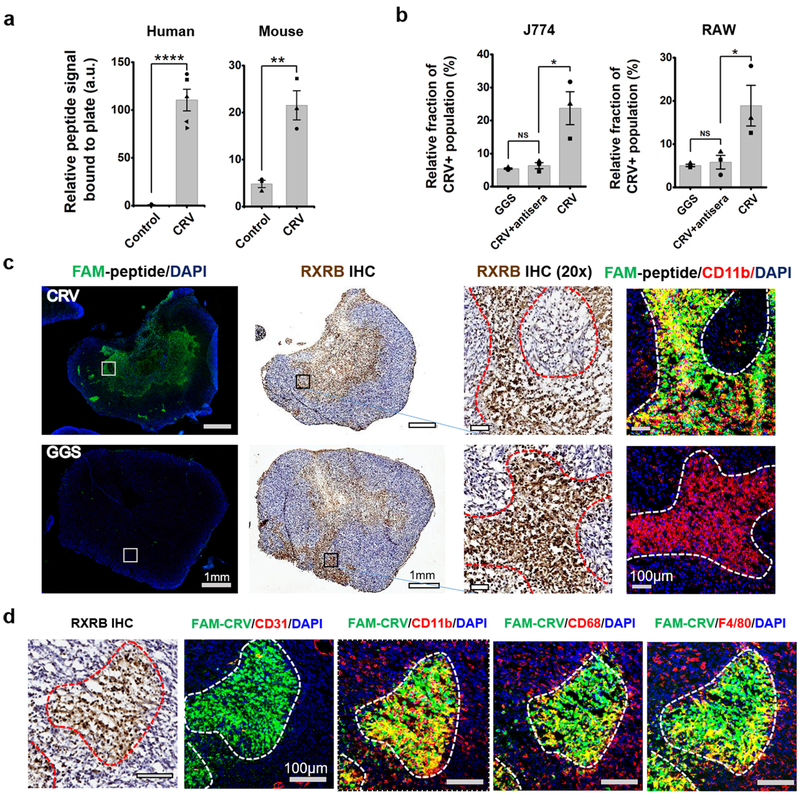
To validate RXRB as the CRV receptor, we first tested the binding of CRV to recombinant RXRB protein. Compared to FAM-GGS control, there was a significantly higher binding of FAM-CRV to both mouse and human RXRB (Fig. 4a), whereas neither peptide bound to the control protein, bovine serum albumin (BSA) (Supplementary Fig. S7a). Both J774 and RAW cells displayed a high surface expression of RXRB, while tumor cell lines had limited RXRB surface expression (Supplementary Fig. S7b), agreeing with the FAM-CRV binding results described above. RXRB belongs to the retinoid X receptor (RXR) family, which are known as nuclear receptors and located within the cell [30]. The RXR family has two other members, RXR alpha and RXR gamma [31]. In contrast to RXRB, there was little presence of these two receptors on the surface of RAW and J774 cells (Supplementary Fig. S7c). We also produced a rabbit polyclonal antiserum against recombinant mouse RXRB protein. Pre-incubation of cells with this antiserum lowered the binding of FAM-CRV to RAW or J774 cells to the level of the FAM-GGS control (Fig. 4b and Supplementary Fig. S7d). Together, these results support the conclusion that RXRB is the CRV receptor at the cell surface of macrophages.
Fig. 4.
Identification of RXRB as the CRV receptor. (a) Strong binding of FAM-CRV compared to FAM-GGS (control) to recombinant human and mouse RXRB or proteins. a.u.: arbitrary units. The experiments were repeated three or more times, and the data shown are mean ± standard error of the mean (SEM) with individual data points overlaid as dots. Two tailed Student’s t-test: t (human) = 9.65, **** P = 0.000011, N = 5. t (mouse) = 5.22, ** P =0.0064, N = 3. (b) Inhibition of CRV binding to J774 and RAW cells by rabbit polyclonal antiserum against recombinant mouse RXRB protein. Cells were first incubated with the antiserum for 15 min, then with FAM-CRV for 30 min at 4 °C. The experiment was repeated three times and the data shown are mean ± SEM with individual data points overlaid as dots. Student’s t-test was performed. NS: not significant. J774: t(NS) = 0.96, P = 0.39. t(*) = 3.42, P = 0.027. RAW: t(NS) = 0.48, P = 0.66. t(*) = 2.64, P = 0.058. (c) Co-localization of RXRB, FAM-CRV and CD11b in 4T1 tumors. After in vivo homing of FAM-CRV, IHC staining was performed on tumor tissue with anti-RXRB antibody (brown). IF staining was performed using anti-FITC antibody for FAM-CRV (green) and anti-CD11b for macrophages (red). Nuclei were stained with DAPI (blue). Top row: 60 min after IV injection of FAM-CRV. Bottom row: 5 min after IV injection of FAM-GGS. The right two columns show magnified views of the regions indicated with the white or black squares in the left two columns, (d) Co-localization of RXRB, FAM-CRV and macrophage markers in 4T1 tumors. Same staining condition as in (c); blood vessels were visualized with anti-CD 31. Scale bar: 100 μm. The staining experiments were performed with samples from three animals per group and representative results are shown.
We next tried to quantify the association constant between CRV and RXRB using Biacore SPR (Surface Plasmon Resonance) system. FAM-CRV and Biotinylated CRV showed similar binding to human recombinant RXRB protein, while FAM-GGS did not (Supplementary Fig. S8). This result further strengthened the notion that the CRV affinity to RXRB is independent of labeling. However, due to the limitation of peptide solubility, we could not provide sufficient peptide concentration to reach a binding equilibrium for accurately measuring the dissociation constant (Kd). Thus, our result can only indicate a Kd of >1mM (Supplementary Fig. S8).
The in vivo distribution of RXRB further supported the above conclusion and revealed an interesting correlation between cell surface-expressed RXRB (hereafter, RXRB refers to the cell surface form unless indicated otherwise) and TAMs. Immunohistochemistry (IHC) staining of RXRB in 4T1 tumor tissue showed that RXRB and FAM-CRV largely co-localized at various time points after the peptide was administered IV (Fig. 4c and Supplementary Fig. S9a). Co-localization was also seen between RXRB and macrophage markers (CD11b, F4/80, and CD68) (Fig. 4d). One caveat for IHC staining is that it cannot fully distinguish the surface and nuclear localization of RXRB due to potential membrane disruption caused by fixation. To exclusively investigate RXRB at the cell surface, weperformed ex vivo binding studies using live cells from tumor, spleen, and blood. FAM-CRV preferentially bound to RXRB-positive cells in tumors and this phenomenon was also observed in dissociated tumor cells after in vivo homing of FAM-CRV (Fig. 5a). Tumor cells showed a subpopulation with a high level of RXRB, and this became more evident when looking into CD11b-positive cells across different organs, or CD11b-positive versus CD11b-negative tumor cells (Fig. 5b, c, and Supplementary Fig. S9b, c). A majority of the cells with high levels of RXRB displayed macrophage markers CD11b and F4/80 (Fig. 5c). Besides TAMs, RXRB was also detectable on the surface of tumor vascular cells (Supplementary Fig. S9d). Collectively, these results support the notion that RXRB (cell surface) is a previously unknown and relatively distinctive marker for TAMs, and that CRV recognizes TAMs in vivo through binding to RXRB.
Fig. 5.
Validation of RXRB as the CRV receptor in TAMs. (a) Flow cytometry histograms show preferential binding of FAM-CRV to RXRB+ over RXRB− 4T1 tumor cells. Top: 4T1 tumor cells incubated with FAM-CRV and anti-RXRB antibody ex vivo. Bottom: After in vivo FAM-CRV 1-h homing, dissociated 4T1 tumor cells were incubated with anti-RXRB antibody. (b) Flow cytometry histograms show RXRB expression in single cells of the indicated tissues. Top: RXRB expression in tumor, spleen, and blood whole cell populations. Bottom: RXRB expression in tumor, spleen, and blood CD11b+ cells. AF 594: Alexa Fluor 594. (c) Flow cytometry analysis showing high RXRB expression on tumor macrophages. Top: Comparison of RXRB expression on CD11b+ and CD11b− cells in tumor-derived cell suspension. Bottom: phenotypic analysis of RXRB-high cells from 4T1 tumors for the indicated macrophage markers. BV421: Brilliant Violet 421. PE: phycoerythrin. All experiments were performed on individual samples from three animals per group, and representative images are shown.
Anti-RXRB antibodies also home to tumors and binds to TAMs within
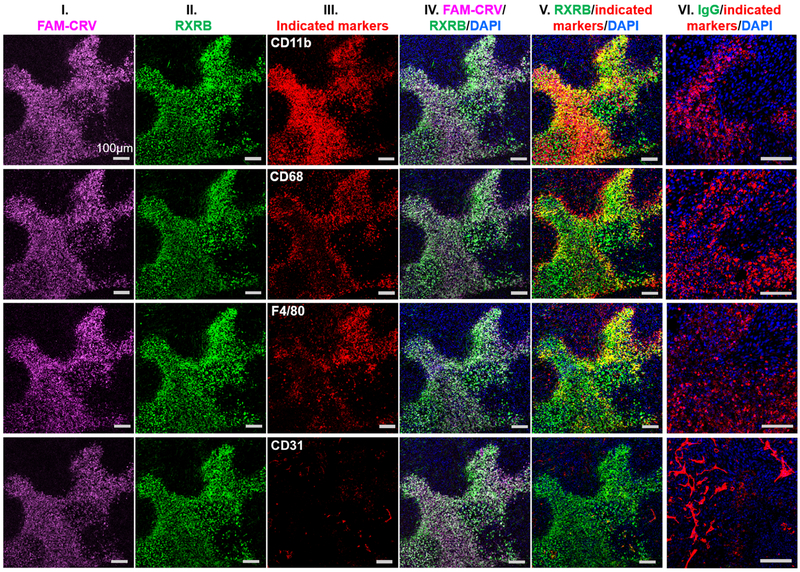
We also investigated the accessibility of RXRB to circulating antibodies in vivo by IV injection of anti-RXRB antibodies into 4T1 tumor-bearing mice. FAM-CRV was co-injected into the same animals to further validate the correlation between CRV and RXRB distribution in vivo. In order not to have the peptide and antibody interfere with one another’s binding in the co-injection experiment, we chose a rabbit polyclonal anti-RXRB antibody that did not block CRV binding in vitro (Supplementary Fig. S10a) The mice were perfused through the heart, similar to the previous CRV homing studies, to eliminate circulating antibody (or rabbit IgG used as a control) and FAM-CRV before analysis. The non-blocking anti-RXRB antibody was seen in the tumors, liver, lymph node, spleen and kidneys, but not in the heart or lungs, whereas IgG control was not detected in any of the tissues (supplementary Fig. S10b). In all healthy organs, anti-RXRB was mainly co-localized with or surrounding the blood vessels except in the spleen, where the antibody was found in the red pulp area with no co-localization with CD31 but somewhat co-localized with CD68 (Supplementary Fig. S10b). In tumors, most of anti-RXRB antibody extravasated across the blood vessels and accumulated in cells positive for macrophage markers (Fig. 6). In line with previous results, FAM-CRV did not accumulate in healthy organs (Supplementary Fig. S10b) and co-localized strongly with anti-RXRB antibody only in tumors (Fig. 6). Similar in vivo distribution was observed when anti-RXRB antibody was injected alone (Supplementary Fig. S11). Experiments with another tumor model (KPC orthotopic pancreatic tumor) also showed similar co-localization of FAM-CRV, anti-RXRB antibody, and macrophage markers in tumors (Supplementary Fig. S12).
Fig. 6.
Co-localization of systemically administered FAM-CRV, rabbit polyclonal anti-RXRB antibody, and macrophages in 4T1 tumors. After in vivo homing of FAM-CRV and anti-RXRB antibody (or control IgG) in mice bearing 4T1 breast tumors, IF staining was performed with anti-FITC (column I, magenta), anti-rabbit IgG secondary antibody for RXRB (column II, green), other indicated antibodies (column III, red) and DAPI (blue). Column IV shows merged images of FAM-CRV, anti-RXRB antibody, and DAPI staining. Column V shows merged images of anti-RXRB antibody with the indicated markers and DAPI. Column VI represents merged images of control IgG, the indicated markers, and DAPI staining of tumors from mice that received the control IgG. Scale bar: 100 μm. The experiments were performed using three animals per group (anti-RXRB or IgG), and representative images are shown.
To further strengthen this observation, we performed the same tumor-homing study with a mouse monoclonal anti-RXRB antibody. To exclude the interference of using mouse-origin antibodies, mouse IgG was used as the control to be co-injected with CRV in parallel. In the tumor tissue (but not healthy organs), we observed fluorescence signals in anti-RXRB group but very little in IgG group (Supplementary Fig S13). This result indicates that the signal in tumors primarily arises from recognizing RXRB but not mouse IgG. Similar tumor-specific extravasation and macrophage co-localization were seen with this anti-RXRB antibody in the tumor tissue (Supplementary Fig S13). Together, the above results indicate that RXRB is mainly present on the cell surface of TAMs but not of most healthy macrophages. RXRB is also present in or around the blood vessels of some tissues, which were positive for anti-RXRB binding but did not retain FAM-CRV. More importantly, we show here that in addition to its ability to distinguish TAMs from healthy counterparts, RXRB can be used for tumor-specific targeting of macrophages in vivo with either CRV or anti-RXRB antibodies.
CRV distinguishes TAMs from macrophages in atherosclerotic plaques
LyP-1 is another peptide that has shown tumor-homing and TAM-binding properties [15, 16, 32]. Upon IV administration into tumor-bearing mice, LyP-1 accumulated in the tumors but not in other organs [32], colocalizing with macrophage/myeloid markers (CD11b, Gr-1, and CD68) similar to CRV [16]. Since CRV and LyP-1 have different receptors, we next investigated whether these two peptides behave differently in recognizing macrophages in pathological tissues other than tumors. LyP-1 has previously been shown to home to atherosclerotic plaques and bind to macrophages within [17, 25]. Here, in vivo homing of FAM-CRV and FAM-LyP1 was performed in ApoE −/− mice bearing atherosclerotic plaques in the aorta. Unlike LyP-1, FAM-CRV showed no accumulation in the plaques and was similar to the negative control peptide (FAM-ARA) in this regard. IHC analysis further demonstrated the lack of RXRB expression in the plaques (Supplementary Fig. S14). These results indicate that CRV targets a subset of macrophages different from LyP-1, and that this subset is differentially expressed in different diseases.
CRV improves the delivery of nano carriers to TAMs
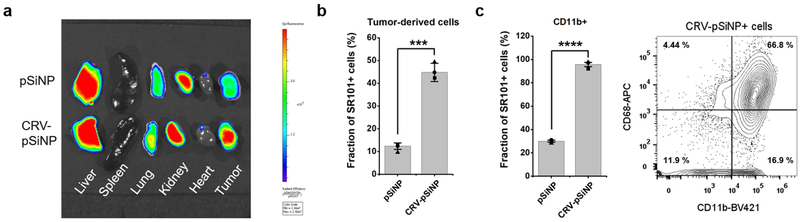
To evaluate the potential of RXRB-targeting for cargo delivery to TAMs, we conjugated the CRV peptide onto porous silicon nanoparticles (CRV-pSiNPs). Several unique advantages of pSiNPs make them desirable cargo carriers: they are nontoxic, compatible with a wide variety of drugs, and have a high loading capacity especially for nucleotide-based drugs [33, 34]. The CRV-pSiNP system was previously shown to improve the delivery of small interfering RNA to macrophages in lungs infected by bacteria, attenuating the acute inflammation and prolonging the survival of the infected animals [22]. Here, we investigated whether CRV can promote the accumulation of pSiNPs in solid tumors and their associated TAMs. A red fluorophore, sulforhodamine 101 (SR101), was loaded into pSiNPs (Supplementary Fig.S15) to track the in vivo fate of the pSiNP cargo and conjugation to pSiNP loaded with different cargo did not affect the specific binding of CRV to macrophages. The SR101-loaded CRV-pSiNPs were intravenously injected into 4T1 tumor-bearing mice, and after 2 h circulation, the major organs were collected for ex vivo fluorescence imaging to quantify the intensity of SR101. The results showed that CRV-pSiNPs delivered more cargo (SR101) into the tumors than pSiNP without CRV (Fig. 7a). There was also substantial accumulation of CRV-pSiNPs in the liver, distinct from the CRV peptide itself. This is due to non-specific clearance of nanoparticles by the liver, rather than specific CRV targeting, supported by the observation that there was no significant difference in intensity from the liver between pSiNP and CRV-pSiNP groups. Flow cytometry analysis showed that CRV-pSiNPs improved the delivery of SR101 to tumor-derived cells, especially to CD11b-positive tumor-derived cells, and the majority of CRV-pSiNP-SR101 positive cells were positive for macrophage markers (Fig. 7b, c). These results show that CRV enhances cargo delivery into solid tumors, and that the target cell type for the delivery is TAMs.
Fig. 7.
CRV facilitates delivery of nanomaterials to TAMs. (a) Biodistribution of CRV-pSiNPs in 4T1 tumor-bearing mice after IV injection. The experiments were performed in three animals per group and a representative fluorescence images of tumors and control tissues are shown. Top row: tissues from a mouse injected with SR101-loaded pSiNPs. Bottom row: tissues from a mouse injected with SR101-loaded CRV-pSiNPs. (b-c) Flow cytometry analysis of tumor cells isolated from 4T1 tumors after in vivo pSiNP or CRV-pSiNP homing. Cells were incubated with the indicated antibodies as described in Materials and Methods and the analysis was performed in three animals per group. (b) Percentages of SR101+ cells in whole tumor-derived cell population. Shown is mean ± SEM with individual data points overlaid as dots. Two tailed Student’s t-test was performed. t= 11.89. *** P = 0.0003. (c) Correlation between pSiNPs and macrophage markers. Left: Percentages of SR101+ cells in CD11b+ tumor-derived cells, t = 32.91, **** P = 0.000005. Right: phenotypic analysis of CRV-pSiNP-SR101+ tumor cells for CD11b and CD68. BV421: Brilliant Violet 421. APC: allophycocyanin.
Discussion
Our study describes a peptide that selectively recognizes TAMs, and establishes RXRB as the target molecule (receptor) for the peptide and as a novel surface marker of TAMs. The tumor homing and TAM association of CRV were observed in a variety of tumor types. One unique feature of CRV homing to tumors is the rapid extravasation (as soon as 5 min after IV injection), which is among the fastest peptides that can penetrate across tumor vasculatures [15, 35]. While the tumor accumulation increased, CRV stayed within blood vessels in healthy organs, and its presence gradually diminished over time. Notably, CRV binding to monocytes in the blood was negligible, suggesting that CRV homing to tumors does not “piggyback” on circulating monocytes as a mode of transport. Instead, CRV may directly bind to its receptor on the tumor vasculature, where extravasation quickly occurs. The extravasation mainly occurred in tumors, suggesting a tumor-specific mechanism for the CRV-RXRB interaction that invokes subsequent transport events once the peptide binds to the vasculature. This notion is further supported by the fact that anti-RXRB antibodies, upon IV injection, also extravasated only in tumors.
Inside tumors, we observed colocalization between CRV and various macrophage markers. The colocalization is to some extent heterogeneous among different markers and tumor types. The possible reason is that these commonly used TAM markers label all or a great majority of macrophages, and sometimes other myeloid cell types [36]. While they are currently the best choice to define TAMs, these markers will not precisely indicate the CRV-positive ones. The presence and expression of these markers also varies from tumor to tumor. The exact molecular nature of CRV-bound, or RXRB-positive, macrophages remains to be further addressed.
The characterization of CRV receptor led us to the intriguing discovery of RXRB as a novel surface marker for TAMs. RXRB is one of RXRs, which belong to a superfamily of nuclear receptors that function as transcription factors [30, 31]. Recent studies have revealed that RXRs regulate macrophage functions and contribute to a number of macrophage-related pathologies such as inflammatory and metabolic disorders [31, 37]. Flowever, to the best of our knowledge, there is no previous study showing the cell surface presence of any RXRs or findings that would link RXRB to a particular cell type. Our identification of RXRB as the CRV receptor in TAMs strongly suggests that RXRB is present on the cell surface of these cells, because it is difficult to see how the peptide would otherwise bind to the cells. Several lines of evidence support our conclusion that cell surface RXRB is the CRV receptor. First, CRV directly bound to purified, recombinant RXRB proteins. Second, blocking RXRB on the cell surface with anti-RXRB antibody abolished the binding of CRV to macrophage cell lines. Third, anti-RXRB antibodies were able to bind to live cells isolated from tumors at 4 °C, which is conditioned to prevent uptake into cells. Fourth, IF staining showed that in vivo distribution of IV injected CRV overlaps with that of RXRB in tumors. Finally, flow cytometry analyses showed that CRV primarily accumulated in RXRB-positive cells of tumor cell suspensions. The presence of intracellular proteins at the cell surface of tumor cells and cells in the tumor microenvironment is not uncommon. Other examples include nucleolin [38], annexin-1 [39], and another TAM marker p32/gC1qR [16]. This aberrant relocation of RXRB sets it apart from other known TAM markers (CCR2, CSF1R, CD206, folate receptors, etc.), which are naturally cell surface receptors. The biological significance of this phenomenon still awaits further investigations.
Our results show that RXRB (but not other RXR isotypes) is present on the surface of macrophages in vitro. IHC staining of tumor tissues revealed a high RXRB signal in tumors, and its distribution pattern overlapped with that of macrophages. Ex vivo staining of live tumor cells indicated that a high expression of cell surface RXRB is largely limited to TAMs and not observed with macrophages from healthy organs. However, IV injection of anti-RXRB antibodies revealed substantial binding of the antibodies to the vessels of several healthy tissues and in tumor vasculature. This pattern is similar to what we observed at the early time points for CRV homing. The disappearance of CRV binding at later time points, which is not the case with anti-RXRB antibodies, may be due to weaker affinity of the peptide for RXRB relative to the antibody. This speculation is supported by the fact that CRV binds with RXRB protein in a much lower affinity than anti-RXRB antibodies (Supplementary Fig. S8). Thus, TAMs may not be the only cell type expressing cell surface RXRB. A critical difference between tumor and normal tissues is that CRV (and anti-RXRB antibodies) penetrate into tumors, whereas in normal tissues they only bind to the vessels. The fact that only RXRB-binding in tumors can invoke a rapid extravasation and allow access to TAMs, makes RXRB a potentially useful target molecule for ligand-directed targeting of tumors.
RXRB targeting by the CRV peptide and antibodies may represent a valuable approach for drug delivery to TAMs. Intensive efforts have been devoted to target macrophages of distinct functional statuses (e.g. M1 and M2) in order to achieve tumor specificity. Instead, we here utilize RXRB-targeting ligands to first home and penetrate into tumors, and then locate the macrophages within. These agents have little or no affinity to circulating monocytes, and can rapidly locate tumors and extravasate. The rapid and tumor-selective extravasation property is particularly useful in improving the access of therapeutic cargo to TAMs, which are generally far from the blood vessels in hypoxic parts of the tumor [40]. Another example of such ligands is LyP-1, while CRV and RXRB antibodies are more selective to TAMs in that unlike LyP-1, they do not recognize macrophages in atherosclerotic plaques. Peptides and antibodies have been widely used with a variety of therapeutic payloads (small molecules, nucleotides, nanoparticles, etc.). Therefore, these RXRB-targeting ligands have the potential to aid in the future development of drug delivery platforms that target TAMs for the purposes of tumor treatment, as was demonstrated in this work with pSiNPs. Moreover, the synergy between RXRB targeting and drugs preferentially modulating TAM activities [41] may greatly improve the tumor specificity and efficacy of macrophage modulation. In summary, our study opens up new possibilities regarding the classification of TAM phenotypes and the role of RXRs in macrophage biology, as well as providing new drug delivery tools for effective and specific TAM modulation.
Conclusions
Tissue-specific recognition of macrophages remains a challenge in terms of both understanding the macrophage biology and developing therapeutics for macrophage modulation. Here we show that CRV, a macrophage-targeting peptide, selectively homes to tumors and accumulates in TAMs. RXRB is identified and validated as the receptor for CRV binding on the surface of TAMs. Intriguingly, while RXRB is known as a nuclear receptor, we find that it also resides at the cell surface of TAMs, and that its surface level in TAMs is much higher than that of their healthy counterpart cells. Upon systemic administration, anti-RXRB antibodies, like the CRV peptide, accumulated in TAMs. CRV is distinct from other macrophage-targeting peptides in that CRV recognizes macrophages in tumors but not in atherosclerotic plaques. Finally, we show that CRV can increase the efficiency of cargo delivery to TAMs. These results establish RXRB as a novel cell surface marker for TAMs that can serve as a target for ligand-directed delivery of compounds to TAMs.
Supplementary Material
Highlights.
A peptide selectively homes to tumors and recognizes macrophages within
Retinoid X receptor beta (RXRB) is the target of the peptide on the macrophage surface
First report of RXRB on the cell surface and as a novel marker for tumor macrophages
Targeting RXRB can distinguish tumor macrophages from those in other tissues
Enables improved cargo delivery to tumor macrophages
Acknowledgements
We thank Drs. Sangeeta Bhatia (Massachusetts Institute of Technology), Klaus Ley (La Jolla Institute for Allergy and Immunology), and Judith Varner (University of California, San Diego) for discussions and advice.
Financial support
This work was supported by grants from the National Cancer Institute (R01CA214550 and R01CA188883) and the National Institutes of Biomedical Imaging and Bioengineering (R21EB022652). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure of conflict of interest
M.J.S. is a scientific founder of Spinnaker Biosciences, Inc., a member of the Board of Directors, and has an equity interest in the company. Although one or more of the grants that supported this research has been identified for conflict of interest management based on the overall scope of the project and its potential benefit to Spinnaker Biosciences, Inc., the research findings included in this particular publication may not necessarily relate to the interests of Spinnaker Biosciences, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murray PJ, Wynn TA, Protective and pathogenic functions of macrophage subsets, Nat. Rev. Immunol, 11 (2011) 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ruffell B, Coussens LM, Macrophages and therapeutic resistance in cancer, Cancer Cell, 27 (2015) 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Moore KJ, Sheedy FJ, Fisher EA, Macrophages in atherosclerosis: a dynamic balance, Nat. Rev. Immunol, 13 (2013) 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, Cheresh DA, Johnson RS, Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis, Nature, 456 (2008) 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rolny C, Mazzone M, Tugues S, Laoui D, Johansson I, Coulon C, Squadrito ML, Segura I, Li X, Knevels E, HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF, Cancer Cell, 19 (2011) 31–44. [DOI] [PubMed] [Google Scholar]
- [6].Kessenbrock K, Plaks V, Werb Z, Matrix metalloproteinases: regulators of the tumor microenvironment, Cell, 141 (2010) 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mason SD, Joyce JA, Proteolytic networks in cancer, Trends Cell Biol, 21 (2011) 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Caux C, Ramos RN, Prendergast GC, Bendriss-Vermare N, Ménétrier-Caux C, A milestone review on how macrophages affect tumor growth, Cancer Res, 76 (2016) 6439–6442. [DOI] [PubMed] [Google Scholar]
- [9].Das A, Sinha M, Datta S, Abas M, Chaffee S, Sen CK, Roy S, Monocyte and macrophage plasticity in tissue repair and regeneration, Am. J. Pathol, 185 (2015) 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Noy R, Pollard JW, Tumor-associated macrophages: from mechanisms to therapy, Immunity, 41 (2014) 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Williams CB, Yeh ES, Soloff AC, Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy, NPJ breast cancer, 2 (2016) 15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yang L, Zhang Y, Tumor-associated macrophages: from basic research to clinical application, J. Hematol. Oncol, 10 (2017) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Paulos CM, Turk MJ, Breur GJ, Low PS, Folate receptor-mediated targeting of therapeutic and imaging agents to activated macrophages in rheumatoid arthritis, Adv. Drug Del. Rev, 56 (2004) 1205–1217. [DOI] [PubMed] [Google Scholar]
- [14].Sun X, Gao D, Gao L, Zhang C, Yu X, Jia B, Wang F, Liu Z, Molecular imaging of tumor-infiltrating macrophages in a preclinical mouse model of breast cancer, Theranostics, 5 (2015) 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E, A tumor-homing peptide with a targeting specificity related to lymphatic vessels, Nat. Med, 8 (2002) 751. [DOI] [PubMed] [Google Scholar]
- [16].Fogal V, Zhang L, Krajewski S, Ruoslahti E, Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma, Cancer Res, 68 (2008) 7210–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hamzah J, Kotamraju VR, Seo JW, Agemy L, Fogal V, Mahakian LM, Peters D, Roth L, Gagnon MKJ, Ferrara KW, Specific penetration and accumulation of a homing peptide within atherosclerotic plaques of apolipoprotein E-deficient mice, Proc. Natl. Acad. Sci. U. S. A, 108 (2011) 7154–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scodeller P, Simón-Gracia L, Kopanchuk S, Tobi A, Kilk K, Säälik P, Kurm K, Squadrito ML, Kotamraju VR, Rinken A, Precision targeting of tumor macrophages with a CD206 binding peptide, Sci. Rep, 7 (2017) 14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nawaz A, Aminuddin A, Kado T, Takikawa A, Yamamoto S, Tsuneyama K, Igarashi Y, Ikutani M, Nishida Y, Nagai Y, CD206+ M2-like macrophages regulate systemic glucose metabolism by inhibiting proliferation of adipocyte progenitors, Nat. Commun, 8 (2017) 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cieslewicz M, Tang J, Jonathan LY, Cao H, Zavaljevski M, Motoyama K, Lieber A, Raines EW, Pun SH, Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival, Proc. Natl. Acad. Sci. U. S. A, 110 (2013) 15919–15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kakoschky B, Pleli T, Schmithals C, Zeuzem S, Brüne B, Vogl TJ, Korf H-W, Weigert A, Piiper A, Selective targeting of tumor associated macrophages in different tumor models, PLoS One, 13 (2018) e0193015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim B, Pang H-B, Kang J, Park J-H, Ruoslahti E, Sailor MJ, Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus, Nat. Commun, 9 (2018) 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marim FM, Silveira TN, Lima DS Jr, Zamboni DS, A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells, PLoS One, 5 (2010) e15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Englen M, Valdez Y, Lehnert N, Lehnert B, Granulocyte/macrophage colony-stimulating factor is expressed and secreted in cultures of murine L929 cells, J. Immunol. Methods, 2 (1995) 281–283. [DOI] [PubMed] [Google Scholar]
- [25].She Z-G, Hamzah J, Kotamraju VR, Pang H-B, Jansen S, Ruoslahti E, Plaque-penetrating peptide inhibits development of hypoxic atherosclerotic plaque, J. Control. Release, 238 (2016) 212–220. [DOI] [PubMed] [Google Scholar]
- [26].Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA, Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma, Genes Dev, 17 (2003) 3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, Belaygorod L, Carpenter D, Collins L, Piwnica-Worms D, Targeting tumor-infiltrating macrophages decreases tumor-Initiating cells, relieves immunosuppression and improves chemotherapeutic response, Cancer Res, (2012) canres. 2731.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu Z, Xiong M, Gong J, Zhang Y, Bai N, Luo Y, Li L, Wei Y, Liu Y, Tan X, Legumain protease-activated TAT-liposome cargo for targeting tumours and their microenvironment, Nat. Commun, 5 (2014) 4280. [DOI] [PubMed] [Google Scholar]
- [29].Teesalu T, Sugahara KN, Ruoslahti E, Mapping of vascular ZIP codes by phage display, in: Methods Enzymol, Elsevier, 2012, pp. 35–56. [DOI] [PubMed] [Google Scholar]
- [30].Dawson MI, Xia Z, The retinoid X receptors and their ligands, BBA-Mol. Cell. Biol. L, 1821 (2012) 21–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rőszer T, Menéndez-Gutiérrez MP, Cedenilla M, Ricote M, Retinoid X receptors in macrophage biology, Trends Endocrinol. Metab, 24 (2013) 460–468. [DOI] [PubMed] [Google Scholar]
- [32].Laakkonen P, Åkerman ME, Biliran H, Yang M, Ferrer F, Karpanen T, Hoffman RM, Ruoslahti E, Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells, Proc. Natl. Acad. Sci. U. S. A, 101 (2004) 9381–9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kang J, Joo J, Kwon EJ, Skalak M, Hussain S, She ZG, Ruoslahti E, Bhatia SN, Sailor MJ, Self - Sealing Porous Silicon - Calcium Silicate Core – Shell Nanoparticles for Targeted siRNA Delivery to the Injured Brain, Adv. Mater, 28 (2016) 7962–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen Y, Ai K, Liu J, Sun G, Yin Q, Lu L, Multifunctional envelope-type mesoporous silica nanoparticles for pH-responsive drug delivery and magnetic resonance imaging, Biomaterials, 60 (2015) 111–120. [DOI] [PubMed] [Google Scholar]
- [35].Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E, Tissue-penetrating delivery of compounds and nanoparticles into tumors, Cancer Cell, 16 (2009) 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards, Nat. Commun, 7 (2016) 12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pérez E, Bourguet W, Gronemeyer H, de Lera AR, Modulation of RXR function through ligand design, BBA-Mol. Cell. Biol. L, 1821 (2012) 57–69. [DOI] [PubMed] [Google Scholar]
- [38].Said EA, Krust B, Nisole S, Svab J, Briand J-P, Hovanessian AG, The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor, J. Biol. Chem, 277 (2002) 37492–37502. [DOI] [PubMed] [Google Scholar]
- [39].Oh P, Testa JE, Borgstrom P, Witkiewicz H, Li Y, Schnitzer JE, In vivo proteomic imaging analysis of caveolae reveals pumping system to penetrate solid tumors, Nat. Med, 20 (2014) 1062. [DOI] [PubMed] [Google Scholar]
- [40].Lewis C, Murdoch C, Macrophage responses to hypoxia: implications for tumor progression and anti-cancer therapies, The American journal of pathology, 167 (2005) 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P, PI3Kγ is a molecular switch that controls immune suppression, Nature, 539 (2016) 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.