Abstract
In response to stress cells must quickly reprogram gene expression to adapt and survive. This is achieved in part by altering levels of mRNAs and their translation into proteins. Recently, the formation of two stress-induced messenger ribonucleoprotein (mRNP) assemblies named stress granules and processing bodies has been postulated to directly impact gene expression during stress. These assemblies sequester and concentrate specific proteins and RNAs away from the larger cytoplasm during stress, thereby providing a layer of post-transcriptional gene regulation with the potential to directly impact mRNA levels, protein translation, and cell survival. The function of these granules has generally been ascribed either by the protein components concentrated into them or, more broadly, by global changes that occur during stress. Recent proteome-wide and transcriptome-wide studies have provided a more complete view of stress-induced mRNP granule composition in varied cell types and stress conditions. However, direct measurements of the phenotypic and functional consequences of stress granule and processing body formation are lacking. This leaves our understanding of their roles during stress incomplete. Continued study into the function of these granules will be an important part in elucidating how cells respond to and survive stressful environmental changes.
Graphical Abstract
Diverse macromolecular interactions lead to the phase separation of protein and RNA during stress. While the identities of many proteins and RNAs contained in these granules (tan spheres) have been elucidated recently, the function of this conserved compartmentalization of the cytoplasm during stress response is still an open question.
Introduction
Cells are frequently exposed to fluctuating, potentially adverse environmental conditions. To survive adverse changes they must rapidly alter gene expression in order to maintain internal homeostasis. The cellular reprogramming that occurs in response to a disruptive or inimical external fluctuation is broadly termed as stress response. Cellular stress response typically includes slowing or ceasing growth that is concomitant with repression of overall translation, though certain genes important for survival and repair are highly induced. Concurrently, while overall translation is repressed, many post-transcriptional regulatory proteins and mRNAs undergo a process called phase separation that results in the formation of concentrated, non-membranous cytoplasmic structures generally described as granules or foci. During stress, this phase separation process might segregate proteins and mRNAs in a way that is functionally important for the cell and that promotes survival. Therefore, these structures are a subject of emergent interest. Although much progress has been made recently to identify the proteins and mRNAs that reside in these granules and the physical characteristics that underlie their formation, there is little known about the phenotypic or functional consequences of their formation during stress and therefore how significantly they contribute to stress response.
There are many different types of cellular granules involved in a wide variety of biological processes such as nucleoli, paraspeckles, PML bodies, and Cajal bodies in the nucleus as well as the stress-induced processing bodies (PBs) and stress granules (SGs) in the cytoplasm. Here we highlight the cytoplasmic PBs and SGs, two well-studied mRNP granules that are present across eukaryotes during a variety of stressful conditions such as exposure to heat shock, oxidative stress, UV irradiation, osmotic stress, and nutrient starvation. The formation of these mRNP granules, which occurs on the scale of minutes after exposure to stress stimuli, is mediated by a physical process called liquid-liquid phase separation (LLPS; see Box 1 for more information). There are common biophysical characteristics and some shared components between SGs and PBs as well as granule-specific features. It should be noted that, while the aptly named SGs are broadly induced during stress, PBs are a bit more organismal specific. S. cerevisiae induces visible PBs primarily during stress response while, in mammalian cells, small, microscopically visible PBs are constitutive but they become much larger and more abundant during stress. It should also be noted that the majority of research into these stress-induced granules is performed with yeast and mammalian cell culture systems. Ultimately, we posit SGs and PBs should be considered as distinct yet closely related mRNP granules; their properties and role in post-transcriptional gene expression during stress response is the focus of this review and we will address them individually, as SGs or PBs, and together more generally, as stress-induced mRNP granules, when appropriate.
Box 1: Liquid-liquid phase separation.
The separation of components in a mixture is termed demixing. The demixing of liquids can take place if the energy of interaction between macromolecules is greater than the entropic energy reduction that arises from their homogeneous mixing. This process drives liquid-liquid phase separation of a molecular mixture into two phases: a smaller volume of a concentrated, condensed phase and a larger volume of a dilute, lower concentration phase. This can allow separation and compartmentalization of biomolecules into organelles without the presence of a membrane. As this is a thermodynamically favorable process, no external cellular energy (ATP) is necessary to maintain this separation within the cell. Biological liquid-liquid phase separation is driven by collective weak interactions between multivalent substrates and intrinsically disordered proteins (IDPs). IDPs or intrinsically disordered regions (IDRs) are proteins or amino acids sequences that fail to form defined structures and instead assume multiple conformations. Many of these IDPs have low complexity domains where a small number of amino acid residues are overrepresented, especially many polar and charged amino acids including glutamine, proline, glutamic acid, lysine, arginine, glycine, and serine. Many proposed weak interactions help to overcome the entropic energy constraints of mixing that include electrostatic, cation-pi, and pi-pi interactions. While IDPs are important for phase separation, nucleic acids, especially RNA molecules, can also serve as multivalent substrates and drive demixing. In fact, RNA has been found sufficient to promote phase separation and alter the internal viscosity of membrane-less compartments. For further detail see Banani, Lee, Hyman, & Rosen, 2017 and X. H. Li, Chavali, Pancsa, Chavali, & Babu, 2018.
Initial characterization of conditions that bring about stress-induced mRNP granules began in the late 1990s when researchers observed that impairment of translation initiation causes SG formation (Bashkirov et al., 1997; Kedersha et al., 1999). A decrease in initiation is one hallmark of stress response; the canonical mechanism of eIF2α-phosphorylation can drive robust granule formation in some stress conditions (Kedersha et al., 1999). Importantly, however, the formation of SGs is not dependent on eIF2α-phosphorylation as the addition of small molecules that block translation initiation through different mechanisms are sufficient to drive granule assembly (Mazroui et al., 2006; Mokas et al., 2009). Moreover, the genetic knockdown of specific translation initiation factor proteins (Mokas et al., 2009) or the overexpression of RNA-binding proteins that function to repress translation (De Leeuw et al., 2007; Gilks, 2004; Kedersha et al., 2005; Mazroui et al., 2006; Wilczynska, 2005) also lead to mRNP granule formation.
The observation that impaired translation drives SG and PB formation spurred further inquiry into their induction and the degree of interrelation between the two granules during stress response. Evidence emerged that SGs and PBs, each lacking membranes, are able to interact, potentially docking and swapping components (Kedersha et al., 2005; Buchan et al., 2008; Wilbertz et al., 2019). Nonetheless, it is known that these granules retain unique protein content and presumably RNA content, though the dynamics and degree of retention of RNAs with an individual granule is more poorly understood than it is for proteins. Interestingly, one study found SG assembly was dependent on and promoted by the presence of pre-existing PBs in yeast cells, demonstrating the akin yet distinct nature of SGs and PBs (Buchan et al., 2008). On the other hand, a different study found that induction of PBs and SGs occurs via independent signaling pathways in yeast and reported no such dependency (Shah et al., 2013). These results highlight the complexity that underlies stress-induced mRNP granule formation. Ultimately, it is not only a loss of translation initiation but also complex networks of signaling pathways, granule-granule interactions, protein-RNA interactions, protein-protein interactions, and RNA-RNA interactions that shape and define both types of distinct but closely related stress-induced mRNP granules.
Understanding when SGs and PBs form is an important first step into recognizing what purpose they have in the broader changes elicited during stress. During the stress response a cell must confront a potential dearth of resources, including of resources necessary to synthesize new proteins, as it responds to the challenge of environmental stress. The direct connection between SG and PB induction coincident with the overall reduction of translation during stress suggests mRNP granule formation may directly control the rapidly changing proteome, yet the extent of this is not clear. The function and molecular relevance of SGs and PBs has been attributed largely based on the protein components concentrated within them. Specifically, the majority of mRNP granule proteins function in translation initiation, translational repression, or mRNA degradation. Consequently, it is generally thought that SGs and PBs function to segregate mRNAs from the larger cytoplasm to regulate their fate, either by storage, decay, or eventual reintroduction to the translating pool. Below we discuss the protein and RNA content of SGs and PBs before we switch to a discussion of what information is known and lacking about their function during stress.
Stress-Induced mRNP Granules: Characteristics and Composition
The function of a stress-induced mRNP granule is presumably related to its molecular composition and so it is important to understand the identity of resident proteins and RNAs in both SGs and PBs. Insight into the ever-expanding catalog of SG and PB residents, discussed below, has enabled informed speculation about granule function; however, it is important to note that simply knowing what is inside these structures has, thus far, been insufficient to clearly elucidate their roles during stress response.
Protein composition of stress-induced mRNP granules
Over the past decade, more and more protein factors residing in PBs and SGs were detected and characterized biochemically and genetically (Buchan & Parker, 2009; Parker & Sheth, 2007). For example, dozens of PB protein components were identified originally in yeast, including Lsm1-7, Xrn1, and Pop2 (Decker et al., 2007). However, due to the challenge of capturing and isolating an intact, membrane-less granule structure in the cytoplasm, a comprehensive profile of the hundreds of resident proteins in mRNP granules under various conditions remained experimentally challenging and therefore elusive. More recently, by taking advantage of mass spectrometry-based high-throughput proteomics and proximity labeling techniques, several studies profiled the larger proteome of PBs and SGs in yeast and different mammalian cell types under various stresses and, for mammalian PBs, native conditions. More than one hundred protein factors were newly identified and extensive interactomes within mRNP granules were characterized (Alberti, 2018; Hubstenberger et al., 2017; Jain et al., 2016; Markmiller et al., 2018; Youn et al., 2018). Overall, mammalian mRNP granule proteomes are larger than yeast proteomes but they have substantial overlap with each other. Therefore, mammalian granules are more complex but stress-induced phase separation is an evolutionarily conserved event. In both mammals and yeast, PBs and SGs only share 10% - 25% of their protein components (5 are shared in yeast and 28 in mammalian cells), leaving the majority of the proteome granule-specific (Table 1A). Intriguingly, even though the majority of the proteome is granule-specific, several protein families and classifications are highly enriched in both types of mRNP granules across species. Most notably, there is very high enrichment in RNA-binding proteins (RBPs), with over 50% of proteins present in human SGs and yeast or human PBs having annotated RNA-binding functionality. There are also many proteins that contain of intrinsically disordered regions (IDRs), which will be discussed in more detail later.
Table 1: Protein composition of stress granules and p-bodies.
A) Properties of yeast and mammalian SG and PB proteomes. Upper: Summary of protein activities of SG and PB proteomes. Lower: Overlap of SG and PB proteome components identified in both granule types for yeast and mammalian systems. Yeast and human SG proteomes and yeast PB proteome are from (Jain et al., 2016). The human PB proteome is from (Hubstenberger et al., 2017). Prion-like domains were predicted by PrionScan (Angarica et al., 2014) and PLAAC (Lancaster et al., 2014). The RNA-binding proteomes are from (Beckmann et al., 2015). ATPase activity annotations are from SGD and NCBI.
B) Representative functional, gene ontology (GO) classification of PB and SG proteomes. Yeast homologs are shown in parentheses. Components that are essential for PB/SG assembly and maintenance are highlighted in bold. GO analysis was performed by GO consortium (Ashburner et al., 2000; Carbon et al., 2017). Citations showing the presence and functions of listed components and those used to generate GO data include Serman et al., 2007; Luo et al., 2018; Andrei et al., 2005; Sheth & Parker, 2003; Hubstenberger et al., 2017; Franks & Lykke-Andersen, 2008; Eulalio et al., 2007; Marnef et al., 2010; Decker et al., 2007; Gilks, 2004; Kedersha et al., 2016; Yang et al., 2014; Buchan & Parker, 2009; Tourriere et al., 2003; Jain et al., 2016; & Markmiller et al., 2018.
| A | Features | Yeast SG (159) | Human SG (411) | Yeast PB (52) | Human PB (109) |
| RNA-binding proteins (RBPs) | 39 | 224 | 36 | 70 | |
| Prion-like domains | 23 | 32 | 21 | 24 | |
| RBP & prion-like domains | 18 | 30 | 11 | 13 | |
| ATPase activity | 10 | 37 | 5 | 19 | |
| SG and PB overlap | Yeast | Human | |||
| Number | 5 | 28 | |||
| Components | Dhhl, Sbpl, Eapl, Dcpl, Scd6 | ACTBL2, AG02, DCP1A, DDX21, DDX50, DDX6, DHX30, EDC4, ELAVL1, ELAVL2, FAM120A, HSPB1, IGF2BP1, IGF2BP2, IGF2BP3, KIF23, LSM14A, LSM14B, MCM7, MEX3A, MOVIO, MSI1, NOP58, PUM1, S100A9, STAU2, UPF1, ZC3HAV1 | |||
| B | Processing Body (P-body/PB) | ||||
| Translation repression | CPEB1, EIF4E-T | ||||
| RNA decay and stabilization | LSM14A/B (Scd6), DDX6 (Dhhl), IGF2BP2 | ||||
| miRNA pathway | Ge-1, GW182, AGOl/2, MOVIO, ZCCHC3, PUM1 | ||||
| Nonsense-mediated mRNA decay (NMD) | UPF1, SMG7 | ||||
| Decapping complex components | DCP1A/1B/2, EDC3/4, PATL1 | ||||
| Deadenylation complex components | LSM1-7, CCR4-NOT | ||||
|
Stress Granule (SG) |
|||||
| Translation repression | TIA-l/TIAR (Publ/Ngrl), Caprin-1, FMRP/FXR1, Ataxin | ||||
| Translation initiation | EIF3, EIF4A/B, EIF4G (Tif4631/Tif4632) | ||||
| RNA decay and stabilization | TDP-43, PAB1, ELAVL1, IGF2BP1, TTP | ||||
| Ribonuclease activity | G3BP, SND1, XRN1, DDX1, CCR-NOT | ||||
| miRNA pathway | TNRC6B, AG02, EIF3A | ||||
| ATPase activity | DDX6 (Dedl), MCM, CCT, RUVBL1/2 (Rvbl/2) | ||||
Amid a general ability to bind RNA, many other sub-categorizations of stress-induced mRNP granule proteins arise from a thorough analysis of their proteomes (Table 1A). For example, in PBs, the majority of protein components are involved in RNA decay and translational repression in yeast and mammals. They include RNA decay factors (EIF4E-T, LSM14A (Scd6 in yeast), LSM14B, and IGF2BP2), decapping complex components (DCP1A/1B, DCP2, EDC4, DDX6 (Dhh1 in yeast), Edc3 and Pat1), factors in the miRNA pathway (Ge-1, GW182, AGO1/2/3, TRNC6A, and ZCCHC3), deadenylation complex components (CCR4-NOT, LSM1-7), ribonucleases (XRN1), nonsense-mediated decay (NMD) factors (UPF1, SMG5/7), and, finally, translation repressors (EIF4E-T, CPEB1) (Andrei et al., 2005; Luo et al., 2018; Serman et al., 2007; Sheth & Parker, 2003). Like PBs, SGs harbor proteins that are related to RNA decay, such as ribonucleases (XRN1, G3BP, SND1) and components in the miRNA pathway (ZFP36, TNRC6B, AGO2), although mRNAs in SGs are not typically considered targets for decay (Lavut & Raveh, 2012). SGs also house translation repressors (CIRP, DDX3 (Ded1 in yeast), FXR1/2, Staufen1). Unlike PBs, SGs contain many components involved in translation including initiation factors (EIF2A/3/4A/4B/4G) and, notably, 40S ribosomal subunits. Transcripts stalled at the initiation step of translation are thought to be enriched in SGs though, to our knowledge, this has not been directly validated. Furthermore, whether these translation factors are directly associated with mRNAs as complete pre-initiation complexes remains to be tested.
The classifications of proteins discussed above are notable in how broad they are. After all, factors involved in general cellular processes like decay and translation have to be able to recognize and regulate the diverse set of mRNAs that comprise a cell’s transcriptome. This raises the question of how specificity arises in targeting the mRNAs that proteins interact with to cytoplasmic granules during stress. To find clues into potential mechanisms one can look to more specific functions in the categories of proteins that arise from classification of PB and SG proteomes. Interestingly, proteins that recognize both RNA secondary structures like G-quadruplexes (FXR1, FMR1) and the epitranscriptional RNA modification N6-methyladenosine (m6A) (YTHDF1/2/3) are enriched in SGs. Relatedly, YTHDF2, a m6A reader, is a recently identified PB component (Luo et al., 2018; Wang et al., 2013). Therefore, RNA structures and modifications recognized by these proteins may provide means to determine specificity in targeting certain mRNAs to mRNP granules, though experimental validation of this potential mechanism remains to be realized.
In addition to enabling classification of the functions of proteins found in SGs and PBs, approaches that combine proximity labeling with mass spectrometry have provided insight into the degree of heterogeneity in the proteomes of stress-induced mRNP granules formed in different cell types and in response to different stresses. Certain proteins are thought to be present in all SGs, particularly those that have been shown to nucleate SG formation, but the protein composition of SGs does vary across different conditions. For example, comparison of SGs formed during arsenite stress with those formed during heat shock showed that 23% of protein components are stress-type specific (Jain et al., 2016; Markmiller et al., 2018). Markmiller and colleagues also reported differences in SG composition in different cell types as well as different stress and disease conditions; SGs in amyotrophic lateral sclerosis (ALS) motor neurons contained particularly distinct proteins. They estimated that up to 20% of the SG proteome is stress and cell type specific. This context-dependence and diversity of SG composition suggests SGs arise and potentially function according to the specific cellular needs and demands brought about by a given stress condition. Similarly, studies of PB composition showed that PB protein content changes during stress compared to native conditions in mammalian systems (Ohn et al., 2008). PBs were found uniquely enriched with ubiquitination-related proteins under arsenite stress (Youn et al., 2018). Understanding how the protein composition of mRNP granules changes in specific conditions may reveal mechanisms of how these granules are fine-tuned to allow the cell to best survive specific stressors and regulate gene expression to meet stress-dependent challenges. This may ultimately help ascertain the function of stress-induced mRNP granules.
Protein-dependent dynamics, assembly, and interactions in stress-induced mRNP granules
Dynamics: liquid and solid states of stress-induced granules
One of the initial observations made about stress-induced mRNP granules was their dynamic nature, reflected in both their rapid formation during stress and dissolution upon recovery. Recently, the biophysical basis of this has been ascribed to LLPS. The biophysical properties inherent to LLPS also lead to dynamic granule states during the duration of stress response as both proteins and mRNAs can rapidly exchange with the non-phase separated cytoplasm while stress-induced granules are present. Much of the insight into stress-induced granule dynamics has come from fluorescence recovery after photobleaching (FRAP) studies. FRAP of mRNP granule proteins showed that these granules can hold liquid-like and therefore dynamic structures, with components moving in and out after photobleaching (Brangwynne et al., 2009; Kroschwald et al., 2015). However, it is important to note that not all stress-induced mRNP granules have been found to be liquid-like.
The type and duration of stress stimuli seems to influence the material state of the granules it induces and can shift them to a more solid-like state in certain contexts. For instance, yeast stress granules induced by heat shock were found to be less dynamic and more solid-like generally (Kroschwald et al., 2015). This is in contrast to mammalian stress granules induced by sodium arsenite, which were very dynamic and liquid-like (Kroschwald et al., 2015). The extent of species-specific or stress-specific influences in determining the material state of SGs is indeed a complicated matter. One cannot simply assume that, in general, yeast SGs are more solid-like and mammalian SGs are more liquid-like. More recently, the same research group has shown that yeast SGs induced by lowered pH are dynamic and behave in a more liquid-like manner akin to mammalian arsenite-induced SGs (Kroschwald et al., 2018). At the same time, ALS-linked mutations that increase protein misfolding drove the material state of heat shock-induced mammalian SGs from fluid to more solid-like (Mateju et al., 2017). This indicates that differences in material state are more strongly influenced by the specific stress stimuli used to induce them rather than species-specific differences (Kroschwald et al., 2018). Moreover, other studies have found that SGs can have differing material states within an individual granule. Specifically, mammalian SGs in lysate were found to be smaller than those in cells, suggesting that individual granules have a distinct, less liquid-like core inside a more soluble, outer shell structure (Jain et al., 2016). Protein components in the shell are more dynamic and fast moving, while components in the core dwell within the structure longer. Taken together, these results indicate that the material states of stress-induced mRNP granules cannot be assumed without direct study and that continued, careful parsing of their dynamics in different contexts will be important to fully understand the nuances of phase separation in biological systems. Assembly: the necessity of certain proteins in stress-induced mRNP granule formation
As previously discussed, proteomic studies revealed hundreds of proteins that reside in PBs and SGs. Though hundreds of proteins reside in these granules, only a fraction of them are considered important for granule assembly or maintenance during stress. Some proteins have been reported to be critical for granule assembly and maintenance as their disruption abolished or decreased the size and number of granules while overexpression had the opposite effect. For instance, in PBs, some translation repressors (CPEB, EIF4E-T), RBPs related to RNA decay and stabilization (LSM14A (Scd6 in yeast), DDX6 (Dhh1 in yeast)), and components in decapping and deadenylation complexes (DCP1/2, EDC3/4, PATL1, LSM1-7, CCR-NOT) were shown to be essential (Ayache et al., 2015; Eulalio et al., 2007; Franks & Lykke-Andersen, 2008; Luo et al., 2018; Sheth & Parker, 2003). In SGs, translation repressors (Caprin-1, TIA-1/TIAR (Pub1/Ngr1 in yeast)), RBPs related to RNA decay and stabilization (G3BP, DDX6 (Ded1 in yeast), TDP-43, PAB1), and enzymes with ATPase activity (RUVBL1/2 (Rvb1/2), MCM, CCT) were all shown to be essential (Buchan & Parker, 2009; Gilks, 2004; Jain et al., 2016; Kedersha et al., 2016; Tourriere et al., 2003). The latter class stands out in particular as it indicates that granule assembly likely depends on ATP. In fact, the ATPase complexes CCT, RVB, and MCM were shown to regulate distinct steps of SG assembly and disassembly, indicating that the properties and functions of SGs are modulated and maintained by active ATPases in an energy-consuming manner (Jain et al., 2016). Similarly, DEAD-box proteins with ATPase activity were also found to be important for maintaining and regulating PB dynamics and turn-over of mRNAs and protein components (Kim & Myong, 2016; Mugler et al., 2016).
Laboratory techniques like genetic screens and knockdown approaches are particularly useful for identifying which proteins mediate granule assembly but are not without caveats. For instance, a screen of yeast SG-defective mutants identified many factors related to translation such as eIF4G2 (Tif4632) and Arc1 (Yang et al., 2014). However, the results of this screen are complicated because the necessity of a given factor for SG nucleation can change in different stress conditions and cell types. For example, G3BP is not required for SG assembly in some osmotic or heat shock stresses but is thought essential for assembly under arsenite-induced oxidative stress (Kedersha et al., 2016). To further add to this complexity, the necessity of these components can be redundant. For example, double-deletion of Edc3 and Lsm4 abolished PB formation in yeast but this was rescued by overexpressing Dhh1 (Rao & Parker, 2017). This complexity calls to mind the complexity that underlies stress-dependent differences in mRNP material state discussed above. Understanding what proteins are necessary for stress-induced mRNP granule formation must be done with proper care; consideration of cell-type and stressor used must be accounted for when one determines what proteins are essential for both SG and PB assembly. Tracking differences in the necessity and redundancy of proteins that mediate stress-induced mRNP granule assembly across stress conditions will likely offer clues into the different functions that these granules have in responding to distinct environmental cues.
Interaction networks between proteins influence stress-induced mRNP granules
Complex networks of interactions mediate mRNP granule assembly, maintenance, and disassembly. Understanding the interactions between macromolecular components may give insight into why some proteins are more important than others in granule assembly and maintenance. Resident protein components can be classified as scaffolds or clients (Ditlev et al., 2018). Scaffolds are proteins required for mRNP granule assembly as described above, while clients are concentrated in the granule via interactions with scaffold components. The distinction between scaffolds and clients can be blurred and condition-dependent in varied biological contexts. Scaffolds are considered to be more concentrated than clients in the granule and are supposed to have higher degrees of interactions. We analyzed the interactions of SG and PB protein components (Figure 1A); the hubs in these interaction networks have higher likelihoods to function as scaffolds that are essential for granule assembly (Hubstenberger et al., 2017; Jain et al., 2016; Youn et al., 2018). It is important to note that many interactions identified during stress were preexistent in non-stressed conditions, although no SGs and only small numbers of PBs were formed. One possible explanation is that interactions during normal growth state are sub-stoichiometric, while interactors become more concentrated in granules during stress. Additionally, the preexisting interactions may drive the pre-assembly of sub-microscopic granules in normal conditions that cannot be observed with conventional microscopy techniques. These possibilities remain to be tested (Youn et al., 2018). Regardless, there is little doubt that understanding protein-protein interaction networks will shed light on the formation of stress-induced mRNP granules and offer potential insight into their function during stress.
Figure 1: Interactions between mammalian PB and SG protein components.
Gene names of proteins with more than 15 interacting protein components in PBs are shown (left network) while those with more than 30 interacting protein components in SGs are shown (right network). Proteins that were identified as essential components for PB or SG assembly are highlighted in red; these tend to have increased numbers of interacting partners. Mammalian interactome datasets of PB and SG components are from Young et al., 2016. The mammalian SG proteome is from Jain et al., 2016 and the PB proteome is from Hubstenberger et al., 2017.
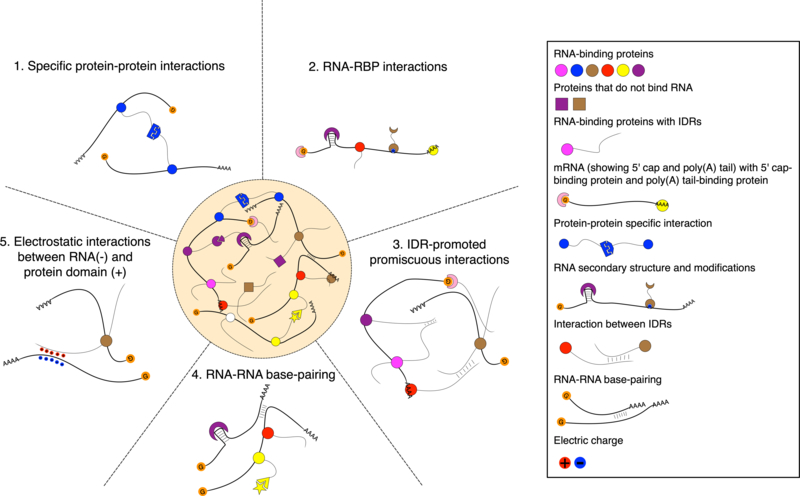
The interactions in mRNP granules can be classified as specific interactions when proteins or mRNAs have limited binding partners or as promiscuous, non-specific interactions when they do not. Specific interactions in granules are usually mediated by well-folded domains or short linear motifs (SLiMs) of IDRs that specifically interact with well-folded domains of other RBPs (Fromm et al., 2014). For example, Edc3 dimerization via a YjeF-N domain is important for PB formation in yeast (Decker et al., 2007). G3BP dimerization and interactions with Caprin-1 are important for mammalian SG formation (Kedersha et al., 2016). Also, post-translational modifications (PTMs) can regulate granule formation by altering specific interactions. For example, methylation of the RGG domains of FUS or EWS recruits Tudor domain-containing proteins (Goulet et al., 2008). NEDDylation of SRSF3 in SGs is required for interaction with TIA-1 (Jayabalan et al., 2016). Banani et al. showed one possibility of how specific interactions drive scaffolds to recruit specific clients and promote LLPS through use of a SUMO/SIM system. Despite these specific interactions, formation of complex, in vivo granules also requires promiscuous interactions mediated by longer IDRs (Figure 2). Since IDRs do not hold well-folded structure they can interact with other proteins non-specifically. It has been shown that in vitro LLPS driven by specific protein-RNA interactions is enhanced by addition of promiscuously interacting IDRs. In yeast, PB formation is promoted by IDRs in cooperation with specific interactions (Protter et al., 2018). It is generally thought that neither specific nor non-specific interactions are individually sufficient to drive stress-induced granule formation, as promiscuous IDRs are not sufficient to form granules in vivo if specific interactions are not also present in certain contexts. For example, high expression levels of fusion proteins hnRNPA1-Cry2 or DDX4-Cry2 cannot phase separate in cells, unless the Cry2 protein is triggered to assemble via light-activated, specific interactions (Shin et al., 2017). Notably, as previously described, RBPs are vastly enriched in both SGs and PBs, suggesting that interactions between RBPs and mRNA transcripts might also play an important role granule assembly that goes beyond protein-protein interactions (Jain et al., 2016). In fact, in addition to multivalent proteins, some longer RNAs can actually serve as scaffolds and thus promote LLPS (Schütz et al., 2017). Overall, synergistic and tuned networks of interactions mediate the formation and maintenance of stress-induced mRNP granules. The identity of these protein components and their interactions potentially drive the specificity of what mRNA transcripts are enriched and excluded from these granules and parsing them might further inform our understanding of how SGs and PBs influence gene expression during cellular stress response.
Figure 2: Diverse sets of interactions drive mRNP granule assembly and LLPS.
Five classes of interactions that contribute to SG and PB formation are modeled. Different protein-protein, protein-RNA, and RNA-RNA interactions contribute to phase separation and drive the formation of stress-induced mRNP granules.
RNA properties and composition in stress-induced mRNP granules
Although a thorough and accurate understanding of the proteins found in SGs and PBs across organisms, cell types, and stressors is necessary to gain deep insight into granule function, one must take an equally deep look at RNA content, particularly at mRNA, to fully realize granule influence on gene expression during stress. The identity and fate of mRNAs in SGs and PBs has remained more elusive than those of proteins for many reasons. First, RNA is more transient and unstable in the cell relative to protein. This makes isolation and subsequent characterization of RNA from a liquid-like, membrane-less granule contained within the larger cytoplasm a challenge. There are also larger varieties of RNAs relative to proteins in SGs and PBs; recent studies identify hundreds of proteins but thousands of RNA species, indicating the necessity of genome-wide approaches for systematic identification (Hubstenberger et al., 2017; Khong et al., 2017). Furthermore, initial studies that provided insight into granule formation and utility during stress paid limited attention to the contribution of individual RNA components in stress-induced granules relative to protein components. Researchers proposed roles based on the functions and identities of proteins, rather than RNAs, identified within them and it was not until recently that studies began to provide genome-wide analyses of enriched RNAs. The protein components of granules enable one to make informed speculations about how these structures influence the fate of mRNAs. However, it is important to directly study the fate of mRNAs that are recruited to RNP granules in order to fully appreciate their function. Fortunately, advances in mRNP granule purification, RNA-sequencing, and single-molecule resolution mRNA imaging have provided valuable insight into the RNAs of SGs and PBs and have advanced and refined our understanding of their regulation. We discuss below a current view of RNA properties relevant to phase separation, RNA components in both SGs and PBs, and how this understanding further informs our perception of the role RNP granules might have in gene expression during stress.
General, biophysical properties of RNAs can influence mRNP granule dynamics
When considering mRNP granule-mediated translation regulation from an RNA-centric perspective, it is prudent to consider how general biophysical properties of RNAs influence phase separation. Unsurprisingly, mRNP granule stability is dependent on RNA concentration and identity, in addition to the presence of previously discussed mRNP-nucleating proteins. For instance, PBs can be dissolved by RNase treatment (Teixeir et al., 2005). Positively charged IDRs on proteins interact with negatively charged mRNAs via electrostatic interactions that influence LLPS propensity (Aumiller & Keating, 2015; Schütz et al., 2017). In addition to charge, RNA secondary structure can influence the properties of phase-separated granules and in fact can control whether LLPS occurs at all. For example, an in vitro reconstitution system shows that recruitment of CLC3 RNA as well as other RNAs into droplets of Whi3, a disordered RBP found in PBs and SGs, is dependent upon CLC3 RNA secondary structure (Langdon et al., 2018). A similar system demonstrated that some RNAs prevent Whi3 droplets from aggregating and help maintain their liquid-like state (Zhang et al., 2015). Conversely, disease-associated RNAs with repeat expansions can serve as templates for multivalent base pairing that drives granule self-assembly and shifts the equilibrium towards phase separation in vitro and in human cells (Jain & Vale, 2017). Ultimately, an appreciation of how RNA’s physical properties affect its capacity to act as a protein scaffold and influence granule formation will be important considerations during analysis of sequence and structural elements both shared and lacking in mRNAs present in SGs and PBs. One must understand and appreciate both RNA-RNA and RNA-protein interactions to determine to what extent a given RNA species may be found in a granule.
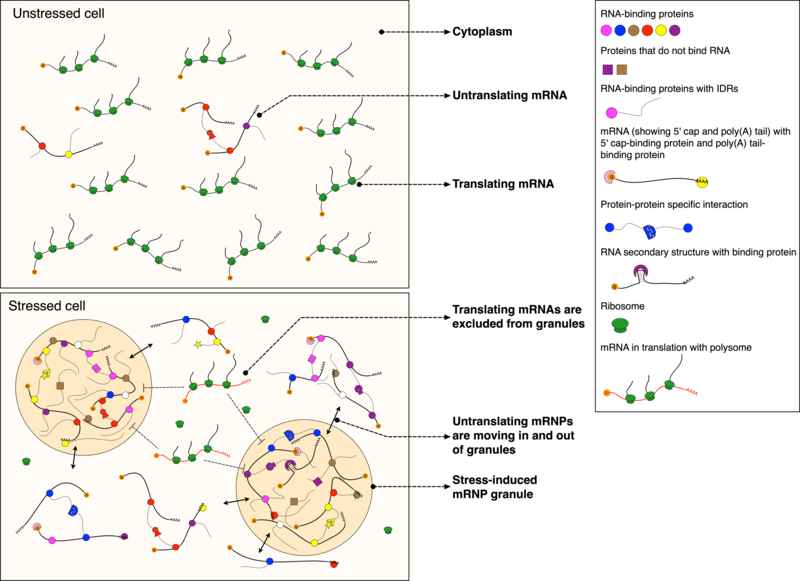
RNA-polysome interactions influence mRNP dynamics
The degree of interaction between ribosomes and mRNAs is of particular importance when considering the relationship between stress-induced mRNP granule formation, protein translation, and changing gene expression during stress. The repression of translation during stress response yields an abundance of nontranslating mRNAs disengaged from polysomes in the cytoplasm within minutes (Kershaw & Ashe, 2017). As previously stated, stress also induces SG and PB formation on the same time scale. Both types of stress-induced mRNP granules accumulate nontranslating mRNAs (Buchan et al., 2008). These observations suggest that there is a direct balance or stoichiometry between levels of polysome engagement, free mRNAs, and stress-induced mRNP granule abundance (Figure 3). This balance, in turn, might help control or limit protein production during a period when overall translation is greatly reduced. Levels of polysome engagement and RNA abundance have been shown to directly influence granule assembly in a RNA-dependent manner. For example, cycloheximide (CHX), an inhibitor of ribosomal translocation that traps mRNAs in polysomes, can repress formation of both PBs and SGs and even dissolve preformed granules (Teixeira et al., 2005). Conversely, addition of puromycin, a drug that dissociates ribosomes from mRNAs actually triggers SG formation (Buchan et al., 2008; Kedersha et al., 2000). This implies that one must consider the translational status of mRNAs as well as the more general biophysical properties of RNAs discussed above to understand RNA’s roles in granule assembly, maintenance, and disassembly as well as broader mRNP function during stress.
Figure 3: Model for composition dynamics and potential function of stress-induced mRNP granules.
Lines with double arrows show that mRNAs associated with RBPs move in and out of stress-induced mRNP granules. Dashed lines with inhibitory arrows show that mRNAs engaged in translation are excluded from stress-induced mRNP granules.
Finally, to fully parse the biological function of SGs and PBs in the context of stress response, one needs to understand not only general changes in polysome engagement but also what transcripts are localized where in the cytoplasm, how transient this localization is, and what distinguishes mRNAs selected for translation into protein during and after stress. It has been suggested the mRNAs can move in and out of mRNP granules due to their liquid-like state. In fact, some mRNAs have been proposed not only to move in and out of a PB or SG but to actually shuttle between granules and polysomes on the timescale of minutes, linking mRNPs to highly controlled regulation of translation during stress even more directly (Brengues et al., 2005; Mollet et al., 2008). These observations, in turn, lead to many questions: what are the proportions and identities of cytoplasmic mRNAs and their bound proteins recruited into granules when translation is downregulated, how dynamically does the mRNA pool actually move into and out of these granules during and after stress in living cells, what proteins accompany such movements, how specific or promiscuous is mRNA recruitment to granules, and how are stress-induced, pro-survival genes excluded from these granules during times of stress to ensure their robust translation. It was not until very recently that we had a genome-wide snapshot of the mRNAs included and excluded from SGs and PBs (Hubstenberger et al., 2017; Namkoong et al., 2018; Wang et al., 2018). The datasets generated by these studies, discussed in more detail below, provide a newfound opportunity to begin to answer to some of the questions outlined above and provide clues or directions for research that can begin to address the others.
Characteristics of mRNAs Targeted to mRNPs During Stress
A surge of recent studies has provided the broadest look into the RNP granule transcriptome to date for both yeast and mammalian models (Hubstenberger et al., 2017; Namkoong et al., 2018; Wang et al., 2018). These studies used varied methods to purify intact RNP granules and report that approximately 10-20% of bulk RNAs in the cytoplasm localize to them; the vast majority (~80%) are mRNAs though ncRNAs are also recruited. A portion of these studies discovered a relationship between certain 3’ UTR features and favorable granule recruitment. For instance, swapping the endogenous 3’UTRs of specific PB-localized transcripts with non-eukaryotic 3’ UTRs was sufficient to halt PB localization of those mRNAs in stressed yeast (Wang et al., 2018). Additionally, a motif search revealed 3’ AU-rich elements (AREs) are most strongly correlated with SG-targeting of mRNAs upon analysis of motifs in the SG-enriched transcriptome isolated during mammalian ER stress (Namkoong et al., 2018). This suggests the possibility that interactions between certain 3’ UTRs and ARE-binding, SG-localized proteins such as TIAR and TIA-1 might contribute to directing and sequestering mRNAs to mRNP granules. More generally, it is likely that 3’ UTR sequences and other untranslated features influence localization of some mRNAs into SGs and PBs.
Although the presence of ARE motifs and the 3’UTR dependence of some granule-localized RNAs is intriguing, it is not sufficient to explain the complex regulatory interactions that target the huge range of mRNAs that localize to granules during stress. Nonetheless, deeper dives into other motifs or sequence features common to stress-induced mRNP granule-localized RNAs have provided very limited insight into what other characteristics intrinsic to mRNAs might drive them to PBs and SGs. Systematic analysis of binding motifs is complicated by the huge variety of RBPs found in these granules, which in turn, have an even larger range of client mRNAs. Furthermore, not all RBPs that localize to PBs or SGs have strong binding motifs. In fact, for many prominent mRNP granule-localized RNA-binding proteins researchers have failed to identify consensus binding motifs (Ishigaki et al., 2012; Mitchell et al., 2013; Rogelj et al., 2012). For instance, for the P-body proteins Pat1, Dhh1, and Lsm1, CLIP-Seq data failed to identify any strong consensus sequence (Mitchell et al., 2013). More generally, the degree of promiscuity and randomness versus tightly regulated specificity of mRNA recruitment into mRNP granules during stress remains unclear. Therefore, there might not be all that many RNA sequence features that cause recruitment to SGs and PBs through recognition by specific RBPs. It is possible that the concentrated environment of a phase-separated granule promotes more nonspecific protein-RNA interactions and so identifying consensus sequences in RNA targets will continue to be elusive. Furthermore, it has been shown that RNA-RNA interactions also contribute to granule formation (Van Treeck & Parker, 2018). Therefore, taken together, not only do some specific RNA-protein interactions contribute to the identity and proportion of mRNAs that are recruited into a PB or SG during a specific stress but so do RNA-RNA interactions and nonspecific RNA-protein interactions. Importantly, the relative extent of each contribution is an open question.
Though the search for sequence commonalities has returned limited results there is another characteristic of mRNAs more strongly correlated with SG and PB enrichment: length. Khong et al. performed RNA-sequencing of purified SG cores from U2OS cells and found that SG-enriched mRNAs are, on average, thousands of base pairs longer than those depleted from SGs. Similarly, bulk purification of all RNP granules (PBs and SGs) from stressed NIH3T3 cells revealed that granule-enriched mRNAs are thousands of base pairs longer than mRNAs not targeted to granules (Namkoong et al., 2018). If the length of a transcript is indeed the most correlative indicator of RNP granule localization during stress this also sheds relatively little light on how sequences within mRNAs target them to granules, thus leaving the specificity in targeting an open question. It might even indicate that longer mRNAs are recruited into SGs and PBs more often simply because a longer mRNA necessarily has a longer primary sequence and this provides more chances for potential nonspecific, promiscuous interactions with RBPs or other RNAs.
The lion’s share of analysis on transcriptome-wide sequencing data from stress-induced mRNP-isolated RNAs has focused on parsing characteristics and identities of genes that reside within SGs and PBs. However, to fully understand how stress impacts translation, one must consider the other side of the data: what mRNAs are not found in SGs and PBs and what mechanisms prevent certain RNAs from recruitment into them? It is known that some transcripts such as those encoding certain heat-shock proteins (HSPs) remain diffusely localized and well translated in the cytoplasm during stress. These are predominantly stress associated mRNAs such as Hsp30, Hsp26, Hsp12, Hsp70, and Hsp90 (Kedersha & Anderson, 2002; Lavut & Raveh, 2012; Stöhr et al., 2006; Zid & O’Shea, 2014). Our previous data implied that during glucose starvation in yeast, it is not the mRNA sequence that determines exclusion of Hsp mRNAs from mRNP granules. Instead, information in the promoter sequence drives cytoplasmic mRNA localization (Zid & O’Shea, 2014). The details of mechanisms that enable this exclusion remain elusive and could likely be informed by a thorough parsing of the SG and PB-excluded transcriptome. In general, we expected that researchers will need to think outside the box and look beyond primary sequences of mRNAs that are recruited to SGs and PBs to solve the puzzle of what directs their recruitment to mRNP granules during stress. Fortunately, some preliminary clues to this puzzle might have already been discovered. For example, it is known that ribosomal proteins, which interact with mRNAs during non-stress conditions, are depleted from PBs while protein-coding mRNAs are enriched. At the same time, noncoding RNAs are depleted from PBs (Hubstenberger et al., 2017). This hints that previous translation and engagement with ribosomes might be a factor in driving mRNA localization to PBs through an unknown mechanism. When it comes to SGs, very recent work has highlighted how the compaction status of a transcript influences its propensity for recruitment. Two separate groups used single-molecule FISH to observe the distance between the 3’ and 5’ ends of mRNAs; both found that compaction increased for mRNAs recruited into granules, indicating that the spatial organization of a transcript influences its localization (Adivarahan et al., 2018; Kong & Parker, 2018). Lastly, a correlation exists between a transcript’s mRNP enrichment and its translational efficiency (TE), as determined by ribosome profiling. Ribosome profiling is a RNA-sequencing based technique that provides a nucleotide-resolution ‘snapshot’ of translation by generating a library of RNA fragments engaged with ribosomes. Traditionally, the occupancy of ribosomes across a gene is quantified and compared to whole transcriptome measurements to generate a measurement of TE (Ingolia et al., 2009). It was found that TE measurements are lower for mRNAs enriched in SGs and PBs relative to depleted mRNAs (Hubstenberger et al., 2017; Khong et al., 2017). This information begets speculation that characteristics that determine the translatability of a mRNA might impact its propensity for ribosome engagement and mRNP localization in an interrelated way, hinting that well-translated mRNAs have characteristics that confer granule exclusion. What exactly these characeristics are remains to be seen.
Stress-Induced mRNP Granules: Function
The stress-inducible formation of SGs and PBs is conserved from yeast to mammals in response to a broad array of stresses. It has also been found that mutants that cannot appropriately form these mRNP granules are more sensitive to stress (Eisinger-Mathason et al., 2008; Kwon et al., 2007; Lavut & Raveh, 2012; Riback et al., 2017; Yang et al., 2014). These data imply that mRNP granules play an important role during stress, yet identifying the actual molecular function of the membrane-less compartments in the cell has proven challenging. One reason for this is that stress in itself has such dramatic effects on gene expression. It can be hard to decouple the formation of mRNP granules from the broad changes induced by stress. While stress-induced mRNP granules were originally posited to have functions related to the function of the proteins concentrated within them, it is unclear if the physical properties of the cytoplasm that govern protein-RNA interactions can be directly compared to those inside phase separated mRNP granules (Helder et al., 2016). Therefore, many established hypotheses on mRNP granule function are currently being reassessed through in vitro models of phase separation that can be tested outside of the context of stress response as well as through modern technological advances that allow higher resolution in imaging and sequencing. Below we discuss current attitudes about SG and PB function, unresolved questions related to function, and potential experimental approaches that might help elucidate them.
Stress Granule Function
As described above, many translation initiation components and translational repressors are concentrated in SGs. While certain mRNAs are enriched in SGs, the impact this sequestration has on gene expression is unclear. Only ~10-20% of bulk mRNA species reside in SGs yet there is a global shutdown of translation during stress. Some speculate that SGs assist with this aspect of stress response but it is unclear how SGs can act as broad and global translational repressors during stress if up to ~80-90% of mRNAs remain excluded and distributed through the cytoplasm. Secondly, inhibiting visible SG formation was shown to have no effect on global translation during stress (Buchan et al., 2008; Kedersha et al., 2016) or on mRNA half-life (Bley et al., 2015; Buchan et al., 2008). While some specific mRNAs have been found to be altered translationally when specific SG proteins are mutated and SG formation is perturbed (Damgaard & Lykke-Andersen, 2011; Gilks, 2004; Mazroui et al., 2007; Moeller et al., 2004; Tsai et al., 2008), it is unclear whether these effects are mediated by aberrant SG localization itself or if they simply reflect changes made by the absence of the wild type protein. An alternative function for SGs may be in helping cells recover upon cessation of the stress response. As much of the translation initiation machinery is present in SGs, it may be that mRNAs enriched in SGs are translationally repressed during stress but some portion of this population is translationally primed for protein synthesis upon stress relief. The knowledge of which mRNAs are enriched in SGs combined with the advent of ribosome profiling provides opportunity for an exciting direction: measurement of the effects that SG dissolution has on mRNA-specific ribosome loading as well as comparison of the timing of ribosome loading onto SG-enriched versus SG-depleted mRNAs after stress ends. Such an experiment would provide insight into the possibility that SGs form to enable rapid engagement of translation machinery with sequestered mRNAs and provide insight into the purpose of SG formation more generally. A very exciting application of this approach would be to isolate SGs and perform profiling specifically on the 40S subunit, as described in Archer et al., 2016. The abundance and identities of transcripts found by this approach would shed light on the presumed but unverified notion that SGs house mRNAs that are engaged with pre-initiation complexes and are primed for reintroduction into the translating pool upon cessation of stress and resumption of growth.
P-Body Function
PBs were historically proposed to be sites of mRNA degradation due to the abundance of decapping factors and exonucleases found within them. Furthermore, yeast strains that had mutations in mRNA degradation machinery showed large increases in the number and size of visible PBs (Sheth & Parker, 2003). Finally, an unstable mRNA that had a polyG sequence inserted to block the exonuclease Xrn1 from fully degrading it accumulated in PBs (Sheth & Parker, 2003). From these results it was concluded that PBs are likely concentrated hubs of mRNA degradation. While these results did point to mRNA decay intermediates, which presumably have very low translatability, accumulating in PBs they did not show whether the actual processing of these mRNAs was taking place inside or outside of the membrane-less compartments. A number of papers have presented contradictory evidence to the notion that PBs serve as hubs of decay and the field is now considering the possibility of a more storage-based role for PBs during stress, leaving active mRNA decay as a process that takes place in the larger cytoplasm. Researchers have found that visible PB formation is not directly necessary for RNA decay; these granules can be disrupted without inhibiting global RNA decay pathways (Ayache et al., 2015; Eulalio et al., 2007). There has also been a lack of degradation intermediates present in recently sequenced, PB-enriched mRNAs (Hubstenberger et al., 2017). This same study reports that depletion of the PB protein DDX6 causes PB dissolution but does not increase levels of PB-enriched mRNAs. Further studies have used fluorescent microscopy to follow the decay of single mRNAs that are labeled at their 5’ and 3’ ends with different fluorescently tagged coat proteins. Over time there was no accumulation of degradation products in PBs (Horvathova et al., 2017). It was also found that there is a general decline in mRNA degradation during stress that is independent of where the reporter mRNA was localized, either in PBs or outside of PBs. This general stabilization of mRNAs during stress has also been previously seen in both yeast and mammalian cells (Gowrishankar et al., 2006; Hilgers et al., 2006). Further microscopy-based studies found that inhibition of reporter mRNA degradation continues for about two hours after stress removal and that the kinetics of degradation appear to be independent of whether an mRNA was localized to a PB or not (Wilbertz et al., 2019). Finally, using purified decapping proteins along with accessory proteins and RNA, studies were able to drive in vitro LLPS, potentially reconstituting PBs (Schütz et al., 2017). It was found that RNA contained within these in vitro reconstituted PBs was protected from endonucleolytic cleavage and that enzymatic activity of the decapping enzyme was greatly decreased (Schütz et al., 2017). Combined, this evidence strongly points towards a storage role for PBs that house a subset of translationally repressed mRNAs during stress and, more generally, demonstrates that caution should be applied when speculating about the function of stress-induced granules.
mRNP Granules: Alternative Functions
While much of the research on mRNP granules has rightly focused on the function of the proteins and mRNAs within the granules, an alternative possibility is that the phase separation of translation initiation factors and mRNA degradation machinery into SGs and PBs, respectively, is to reduce the working concentration of these proteins in the aqueous regions of the cytoplasm. This possibility would help to remedy the contradiction that, though only a small portion of mRNAs in the cell are present in SGs or PBs, the majority of mRNAs present during early stages of stress are from non-stress induced mRNAs. We have observed that after 15 minutes of glucose starvation in yeast, about 90% of the mRNAs present within the cell are from non-stress induced mRNAs (B.M.Z, unpublished data). One factor that must be taken into account when considering this alternative possibility is the intracellular volume that SGs and PBs occupy. Generally, mRNP granules constitute only a minor portion of cellular volume, approximately 1% or less (Banani S. et al 2017). And while proteins are very highly concentrated within granules (for example in mammalian SGs, G3BP1 protein is 13-fold more concentrated in the SG shell than the cytoplasm and about 30-fold more concentrated in the core than it is in the shell; Jain S et al., 2016), it is unclear if this would cause significant enough depletion to have a functional impact. Very recently, quantitative measurements have been performed on yeast PB proteins to compare their concentration inside and outside of PBs (Xing et al., 2018). For Dcp2 protein, the catalytic subunit of the decapping enzyme complex, more than 30% becomes sequestered in PBs. Other accessory decapping proteins such as Edc3 and Pat1 have greater than 20% of their protein content sequestered into P-bodies. The supports the notion that the function of PBs would be to reduce mRNA decapping activity in the bulk cytoplasm, rather than to concentrate it in a granule, which is consistent with the previously mentioned observations that there is a general decline in mRNA degradation during stress regardless of whether a transcript is sequestered into PBs or not.
A different possibility is that visible phase separation during stress is just an indicator or consequence of broader remodeling happening globally to smaller mRNP complexes that exist throughout the cell. Proteins interact with a transcript throughout its life, creating sub-microscopic mRNP complexes that can be translationally active or inactive. At any given point in time, a relatively small number of total cellular protein and RNA is contained in a granule. However, we know that both proteins and mRNAs are dynamically exchanging between mRNP granules and the cytoplasm. Could mRNP complexes that are competent to enter or have previously exited an mRNP granule be in a ‘modified’ state, even outside of the granule? To date, we don’t know if there are changes to the molecular composition of interacting RNAs and proteins as they leave the membrane-less compartment; current techniques only capture granules at a given point in time and don’t reveal if and how many molecules in the general cytoplasm were prior mRNP residents. This alternative proposal is not without some grounding in previous research. For example, during non-stress conditions, the SG component protein Pab1 is predominantly in a soluble, i.e. non-pelletable, state when cell extracts are treated with RNase I (Riback et al., 2017). Yet, during a mild heat stress, about half of the Pab1 molecules transition to insoluble, pelletable quinary assemblies, though no visible SGs form. At higher temperatures when visible SGs do form, over 90% of Pab1 transitions to a pelletable state even though a much smaller portion of total Pab1 resides within phase separated mRNP granules. Therefore, it could be that some mRNAs not directly contained within mRNP granules still interact with their protein partners in “altered”, mRNP-dependent states relevant for survival during stress. For example, post-translational modifications of proteins have been implicated in granule formation. If these proteins are rapidly exchanging with the environment, presumably many proteins not found in the granule at any specific time will still be modified upon leaving the granule. What percent of proteins are modified? How would these modifications affect protein function when they exist as part of mRNP complexes that are sub-microscopic and are no longer contained within the granule? Further investigations into the changes mRNP complexes undergo both within and outside mRNP granules during stress will need to be undertaken to address this possibility. One exciting, relevant direction would be to perform a timecourse experiment that employs a proximity labeling and proteomics approach to see if certain PTMs are upregulated on PB or SG-enriched proteins in both granule and cytoplasmic fractions. If a certain protein is modified in the granule and then released back into the cytoplasm, thereby changing its function, you would expect this to be reflected over the timecourse.
Finally, the role of stress-induced mRNP granules could be entirely passive. As previously described, one proposed function of mRNP granules it to store RNAs throughout the duration of stress to allow optimal growth upon recovery as this provides avoidance of extraneous transcription and nuclear export. A recently considered possible, complementary function of mRNP granules is that instead of only serving as RNA storage depots they may also serve as protein storage depots for growth proteins that are not needed during stressful conditions but that the cell may not want to immediately degrade. After all, stress is transitory and dynamic in nature. As evidence consider the yeast pyruvate kinase protein Cdc19, a key regulator of glycolytic metabolism and cell growth. This protein has been found to form reversible aggregates that co-localize with stress granules during stress. This aggregation was found to be a protective mechanism from stress-induced degradation as, upon stress relief, it proves to be reversible and allows quick re-entry into the cell cycle because these proteins do not have to be re-expressed (Saad et al., 2017). It is possible, therefore, that stress-induced granule formation evolved as a means to coordinate cellular machinery in a way that enables its rapid and efficient deployment in the cell once conditions are more favorable for growth and energy-consumption.
Conclusion
There has been a recent surge in research focusing on phase separation in biological processes. The formation of stress-induced mRNP granules is a broadly conserved example of this phase separation, but for all of this intense study the importance of this phase separation remains unclear. Overall, there has been much progress in understanding the formation and composition of stress-induced mRNP granules. Yet, somewhat surprisingly, this knowledge has only led to marginal increases in our understanding of the function of these membrane-less compartments within the cell. Moving forward, there are a number of distinct directions that may prove fruitful in elucidating the function of stress-induced mRNP granules. In vitro reconstitution has been an important tool for understanding many biochemical and biophysical processes. Several recent advances that helped elucidate mechanisms of biological LLPS have come from reconstitution of these systems in vitro (Banani et al., 2016; Han et al., 2012; Jain & Vale, 2017; Li et al., 2012; Molliex et al., 2015). While progress has been made in making reductionist systems in vitro, some of the properties of mRNP granules may arise because of their complexity. Thus, reduction of granules to limited protein or mRNA components may mask some emergent properties and it would be worth studying if and how an increase in protein and RNA types alters in vitro phase separation to more closely mimic granules in cells. In vitro studies of stress induced mRNP granules will also need to be cognizant of role that ATP plays in the dynamics of mRNP granules (Jain et al., 2016) and ATP’s ability to directly solubilize molecules in aqueous solutions as a biological hydrotrope (Patel et al., 2017). Lastly, a worthwhile avenue would be combined use of techniques like single molecule FISH and SHAPE probing of RNA secondary structure before and after in vitro phase separation to address the relationship between RNA compaction and granule entry to determine the extent to which RNAs undergo compaction before, during, or after entry into granules.
Another exciting direction important to understanding how the assembly of these granules directly affects function is the ability to perturb phase separation in a more controlled, stress-independent manner in vivo. While overexpression of certain proteins is sufficient to drive phase separation without stress, an exciting new direction is using the Arabidopsis thaliana cryptochrome 2 photolyase homology region to drive light-inducible phase separation (Shin et al., 2017; Taylor et al., 2018). This potentially allows experiments to dynamically drive phase separation in cells and thus provide understanding of what effect LLPS has on the physiology of the cell in a way that is decoupled from stress or obscured by consequences of overexpression. Intriguingly, recent experiments have shown that light-induced phase separation of the SG protein G3BP1 is sufficient to recruit many other core SG components and polyadenylated RNA (Taylor et al., 2018). Lastly, more work must be done that employs methods to directly measure the impact of mRNP granules on gene expression. Recent work using microscopy based in vivo reporters of translation and mRNA decay have given interesting insights into what these mRNP granules may and may not be doing (Horvathova et al., 2017; Moon et al., 2019; Pitchiaya et al., 2018; Wilbertz et al., 2019). Further single molecule measurements are needed, particularly in live cells, to increase the diversity of mRNA species analyzed and to directly follow mRNAs that were previously localized to mRNP granule after stress is abated to resolve their fate. Such single molecule approaches could be complemented by a genome-wide approach described in two exciting, recently posted preprints that utilize a novel technique that applies APEX-based proximity labeling to RNA called APEX-seq (Fazal et al., 2018; Padròn et al., 2018). The latter preprint analyzed stress-specific impacts on mRNA localization and offers powerful insight into the relationship between protein localization and RNA localization during stress. Combinations of these in vitro and in vivo approaches will likely help shed light on the elusive function of PBs and SGs and ultimately inform our understanding of cellular function during stress, potentially offering insight into disease states linked to aberrant stress response and granule assembly.
Acknowledgments
The authors would like to thank the Zid lab for helpful discussions on the review. Research on mRNP granules in the Zid lab is supported by grant R35 GM128798 from the National Institutes of Health. A.R.G. is partially supported by National Institutes of Health training grant T32 EB009380 for the multi-scale analysis of biological structure and function.
Key Terms Glossary
- Stress
perturbation that causes reduction in the optimal functionality of a cell
- SG (stress granule)
cytoplasmic membrane-less structure enriched in many translation initiation components and mRNAs
- PB (processing body)
cytoplasmic membrane-less structure enriched in many mRNA processing components and mRNAs
- RBP
RNA-binding protein
- IDR (intrinsically disordered region)
an unstructured sequence of amino acids often found in proteins enriched in SGs and PBs
- LLPS (liquid-liquid phase separation)
the biophysical process that drives formation of SGs and PBs
Contributor Information
Anna R. Guzikowski, Division of Biological Sciences, University of California San Diego, La Jolla, CA, United States of America.
Yang S. Chen, Division of Biological Sciences, University of California San Diego, La Jolla, CA, United States of America.
Brian M. Zid, Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, United States of America.
References
- Alberti S (2018). Guilty by Association: Mapping Out the Molecular Sociology of Droplet Compartments. Molecular Cell, 69(3), 349–351. 10.1016/j.molcel.2018.01.020 [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, & Lührmann R (2005). A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA, 11(5), 717–727. 10.1261/rna.2340405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angarica EV, Ventura S, & Sancho J (2013). Discovering putative prion sequences in complete proteomes using probabilistic representations of Q/N-rich domains. BMC Genomics, 14(1), 1–17. 10.1186/1471-2164-14-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, & Preiss T (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature, 535, 570–574. 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, … Sherlock G (2000). Gene Ontology: tool for the unification of biology. Nature Genetics, 25(1), 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller WM, & Keating CD (2015). Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nature Chemistry, 8(2), 129–137. 10.1038/nchem.2414 [DOI] [PubMed] [Google Scholar]
- Ayache J, Bénard M, Ernoult-Lange M, Minshall N, Standart N, Kress M, & Weil D (2015). P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Molecular Biology of the Cell, 26(14), 2579–2595. 10.1091/mbc.e15-03-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, & Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol, 18(5), 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, & Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell, 166(3), 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, & Heyer WD (1997). A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. Journal of Cell Biology. 10.1083/jcb.136.4.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, … Hentze MW (2015). The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nature Communications, 6(1), 10127 10.1038/ncomms10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaß M, … Hüttelmaier S (2015). Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Research, 43(4), e26–e26. 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, … Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science, 324(5935), 1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, & Parker R (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science, 310(5747), 486–8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, & Parker R (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. Journal of Cell Biology, 183(3), 441–455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, & Parker R (2009). Eukaryotic Stress Granules: The Ins and Outs of Translation. Molecular Cell, 36(6), 932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Dietze H, Lewis SE, Mungall CJ, Munoz-Torres MC, Basu S, … Westerfield M (2017). Expansion of the gene ontology knowledgebase and resources: The gene ontology consortium. Nucleic Acids Research, 45(D1), D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, & Lykke-Andersen J (2011). Translational coregulation of 5’TOP mRNAs by TIA-1 and TIAR. Genes and Development. 10.1101/gad.17355911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, … Liu JO (2006). Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. Journal of Biological Chemistry. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, & Gueydan C (2007). The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Experimental Cell Research. 10.1016/j.yexcr.2007.09.017 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, & Parker R (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. Journal of Cell Biology, 179(3), 437–449. 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Case LB, & Rosen MK (2018). Who’s In and Who’s Out-Compositional Control of Biomolecular Condensates. J Mol Biol. 10.1016/j.jmb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TSK, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, … Lannigan DA (2008). Codependent Functions of RSK2 and the Apoptosis-Promoting Factor TIA-1 in Stress Granule Assembly and Cell Survival. Molecular Cell. 10.1016/j.molcel.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, & Izaurralde E (2007). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol, 27(11), 3970–3981. 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Kaewsapsak P, Parker KR, Xu J, Boettiger AN, Chang HY, & Ting AY (2018) Atlas of Subcellular RNA Localization Revealed by APEX-seq. BioRxiv. https://doi.org/10.110¼54470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, & Lykke-Andersen J (2008). The control of mRNA decapping and P-body formation. Mol Cell, 32(5), 605–615. 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm SA, Kamenz J, Noldeke ER, Neu A, Zocher G, & Sprangers R (2014). In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew Chem Int Ed Engl, 53(28), 7354–7359. 10.1002/anie.201402885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N (2004). Stress Granule Assembly Is Mediated by Prion-like Aggregation of TIA-1. Molecular Biology of the Cell. 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, & Cote J (2008). TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet, 17(19), 3055–3074. 10.1093/hmg/ddn203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar G, Winzen R, Dittrich-Breiholz O, Redich N, Kracht M, & Holtmann H (2006). Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biological Chemistry. 10.1515/BC.2006.043 [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, … McKnight SL (2012). Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Helder S, Blythe AJ, Bond CS, & Mackay JP (2016). Determinants of affinity and specificity in RNA-binding proteins. Current Opinion in Structural Biology, 38, 83–91. 10.1016/j.sbi.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Hilgers V, Teixeira D, & Parker R (2006). Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 10.1261/rna.241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvathova I, Voigt F, Kotrys AV, Zhan Y, Artus-Revel CG, Eglinger J, … Chao JA (2017). The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol Cell. 10.1016/j.molcel.2017.09.030 [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, … Weil D (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell, 68(1), 144–157 e5. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A, … Ohno K (2012). Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Scientific Reports. 10.1038/srep00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, & Vale RD (2017). RNA phase transitions in repeat expansion disorders. Nature. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, & Parker R (2016). ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell, 164(3), 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang GY, Baek JH, … Ohn T (2016). NEDDylation promotes stress granule assembly. Nat Commun, 7, 12125 10.1038/ncomms12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, & Anderson P (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochemical Society Transactions. 10.1042/bst0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, … Anderson P (2000). Dynamic Shuttling of Tia-1 Accompanies the Recruitment of mRNA to Mammalian Stress Granules. J Cell Biol, 151(6), 1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, & Anderson P (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. Journal of Cell Biology. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, … Anderson P (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol, 212(7), 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, … Anderson P (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. Journal of Cell Biology. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw CJ, & Ashe MP (2017). Untangling P-Bodies: Dissecting the Complex Web of Interactions that Enable Tiered Control of Gene Expression. Mol Cell, 68(1), 3–4. 10.1016/j.molcel.2017.09.032 [DOI] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, & Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Molecular Cell. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, & Myong S (2016). RNA Remodeling Activity of DEAD Box Proteins Tuned by Protein Concentration, RNA Length, and ATP. Molecular Cell, 63(5), 865–876. 10.1016/j.molcel.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, … Alberti S (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife, 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, & Matthias P (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes and Development. 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, & King OD (2014). PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics, 30(17), 2501–2502. 10.1093/bioinformatics/btu310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Niaki AG, McLaughlin GA, Weidmann C, Gerbich TM, … Weeks KM (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science, eaar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavut A, & Raveh D (2012). Sequestration of highly expressed mrnas in cytoplasmic granules, p-bodies, and stress granules enhances cell viability. PLoS Genetics. 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, … Rosen MK (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature, 483(7389), 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Chavali PL, Pancsa R, Chavali S, & Babu MM (2018). Function and Regulation of Phase-Separated Biological Condensates. Biochemistry. 10.1021/acs.biochem.7b01228 [DOI] [PubMed] [Google Scholar]
- Luo Y, Na Z, & Slavoff SA (2018). P-Bodies: Composition, Properties, and Functions. Biochemistry, 57(17), 2424–2431. 10.1021/acs.biochem.7b01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, … Yeo GW (2018). Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell, 172(3), 590–604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Maldonado M, Bugaut A, Balasubramanian S, Kress M, Weil D, & Standart N (2010). Distinct functions of maternal and somatic Pat1 protein paralogs. RNA, 16(11), 2094–2107. 10.1261/rna.2295410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, …Alberti S (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J, 361669–1687. 10.15252/embj.201695957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, & Gallouzi I-E (2007). Inhibition of the Ubiquitin-Proteasome System Induces Stress Granule Formation. Molecular Biology of the Cell. 10.1091/mbc.E06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau M-E, Kaufman RJ, Northcote P, Tanaka J, … Pelletier J (2006). Inhibition of Ribosome Recruitment Induces Stress Granule Formation Independently of Eukaryotic Initiation Factor 2α Phosphorylation. Molecular Biology of the Cell. 10.1091/mbc.e06-04-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, & Parker R (2013). Global analysis of yeast mRNPs. Nat Struct Mol Biol, 20(1), 127–133. 10.1038/nsmb.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Li CY, & Dewhirst MW (2004). Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 10.1016/S1535-6108(04)00115-1 [DOI] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, Arya P, … Mazroui R (2009). Uncoupling Stress Granule Assembly and Translation Initiation Inhibition. Molecular Biology of the Cell. 10.1091/mbc.E08-10-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, & Weil D (2008). Translationally Repressed mRNA Transiently Cycles through Stress Granules during Stress. Molecular Biology of the Cell. 10.1091/mbc.E08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, … Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell, 163(1), 123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, & Stasevich TJ (2019). Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nature Cell Biology 10.1038/s41556-018-0263-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler CF, Hondele M, Heinrich S, Sachdev R, Vallotton P, Koek AY, … Weis K (2016). ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. Elife, 5 10.7554/eLife.18746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, & Lee JH (2018). Systematic Characterization of Stress-Induced RNA Granulation. Mol Cell. 10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, & Anderson P (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol, 10(10), 1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padròn A, Iwasaki S, & Ingolia NT (2018). Proximity RNA labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. BioRxiv. https://doi.org/10.110¼54066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, & Sheth U (2007). P bodies and the control of mRNA translation and degradation. Mol Cell, 25(5), 635–646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, & Hyman AA (2017). Biochemistry: ATP as a biological hydrotrope. Science. 10.1126/science.aaf6846 [DOI] [PubMed] [Google Scholar]
- Pitchiaya S, Mourao MDA, Jalihal AP, Xiao L, Jiang X, Chinnaiyan AM, … Walter NG (2018). Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. BioRxiv. 10.1101/375295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, & Parker R (2018). Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep, 22(6), 1401–1412. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BS, & Parker R (2017). Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 10.1073/pnas.1712396114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, & Drummond DA (2017). Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell, 168(6), 1028–1040 e19. 10.1016/j.cell.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, Curk T, … Ule J (2012). Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Scientific Reports. 10.1038/srep00603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, & Dechant R (2017). Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nature Cell Biology. 10.1038/ncb3600 [DOI] [PubMed] [Google Scholar]
- Schütz S, Nöldeke ER, & Sprangers R (2017). A synergistic network of interactions promotes the formation of in vitro processing bodies and protects mRNA against decapping. Nucleic Acids Research, 45(11), 6911–6922. 10.1093/nar/gkx353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, & Weil D (2007). GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Research, 35(14), 4715–4727. 10.1093/nar/gkm491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah KH, Zhang B, Ramachandram V, & Herman PK (2013). Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics, 193(1):109–23. http://doi: 10.1534/genetics.112.146993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, & Parker R (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science, 300(5620), 805–8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, & Brangwynne CP (2017). Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell, 168(1–2), 159–171 e14. 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, & Hüttelmaier S (2006). ZBP1 regulates mRNA stability during cellular stress. Journal of Cell Biology. 10.1083/jcb.200608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Zhang P, Fan B, Yang P, Temirov J, Messing J, & Kim HJ (2018). OptoGranules reveal the evolution of stress granules to ALS-FTD pathology. BioRxiv. 10.1101/348870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, & Parker R (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA, 11(4), 371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, & Tazi J (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol, 160(6), 823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai NP, Ho PC, & Wei LN (2008). Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO Journal. 10.1038/emboj.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, & Parker R (2018). Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell, 174(4), 791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, & Spang A (2018). Context-dependent deposition and regulation of mRNAs in P-bodies. Elife, 7 10.7554/eLife.29815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, … He C (2013). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505(7481), 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbertz JH, Voigt F, Horvathova I, Roth G, Zhan Y, & Chao JA (2019). Single-molecule Imaging of mRNA Localization and Regulation During the Integrated Stress Response. Mol Cell. 10.1016/j.molcel.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Wilczynska A (2005). The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. Journal of Cell Science. 10.1242/jcs.01692 [DOI] [PubMed] [Google Scholar]
- Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijovic M, … Sunnerhagen P (2014). Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet, 10(11), e1004763 10.1371/journal.pgen.1004763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, … Gingras AC (2018). High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol Cell, 69(3), 517–532 e11. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, … Gladfelter AS (2015). RNA Controls PolyQ Protein Phase Transitions. Mol Cell, 60(2), 220–230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, & O’Shea EK (2014). Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 10.1038/nature13578 [DOI] [PMC free article] [PubMed] [Google Scholar]