Abstract
As one of the most important resistance (R) gene families in plants, the NBS–LRR genes, encoding proteins with nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains, play significant roles in resisting pathogens. The published genomic data for cabbage (Brassica oleracea L.) provide valuable data to identify and characterize the genomic organization of cabbage NBS–LRR genes. Ultimately, we identified 105 TIR (N-terminal Toll/interleukin-1 receptor)-NBS–LRR (TNL) genes and 33 CC (coiled-coil)-NBS–LRR (CNL) genes. Further research indicated that 50.7% of the 138 NBS–LRR genes exist in 27 clusters and there are large differences among the gene structures and protein characteristics. Conserved motif and phylogenetic analysis showed that the structures of TNLs and CNLs were similar, with some differences. These NBS–LRRs are evolved under negative selection and mostly arose from whole-genome duplication events during evolution. Tissue-expression profiling of NBS–LRR genes revealed that 37.1% of the TNL genes are highly or specifically expressed in roots, especially the genes on chromosome 7 (76.5%). Digital gene expression and reverse transcription PCR analyses revealed the expression patterns of the NBS–LRR genes upon challenge by Fusarium oxysporum f.sp. conglutinans: nine genes were upregulated, and five were downregulated. The major resistance gene Foc1 probably works together with the other four genes in the same cluster to resist F. oxysporum infection.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1714-8) contains supplementary material, which is available to authorized users.
Keywords: Brassica oleracea, Resistance gene, Bioinformatics, Gene expression, Gene evolution
Introduction
Plants are constantly confronted by pathogens that alter their growth, metabolism, and reproduction. To resist invasion, plants have evolved numerous defense mechanisms (Pennisi 2009; Głowacki et al. 2011; Fujita et al. 2006). Resistant plants have multiple disease-resistance (R) genes, which confer resistance to different pathogens and insects (Van and Kamoun 2008). R proteins can sense the invasion of pathogens by detecting effector molecules generated during infection (Martin et al. 2003; Dangl and Jones 2001). The largest known class of R genes includes those containing a nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains (Deyoung and Innes 2006; Yue et al. 2012). To date, over 150 R genes have been cloned, about 80% of which encode NBS and LRR domains (Shao et al. 2014; Guo et al. 2016). Based on the structure of their N-termini, these NBS–LRR proteins can be further divided into N-terminal Toll/interleukin-1 receptor (TIR)-NBS–LRR (TNL) and non-TNL types. The TNL type possess a domain homologous to the interleukin-1 receptor (TIR) and Toll, while most of the non-TNL proteins having a coiled-coil (CC) are commonly referred to as CC–NBC–LRR (CNL) proteins (Meyers et al. 2010; Blake et al. 2003). Despite both the TIR and CC domains being related to signaling and resistance specificity, their pathways are divergent (Deyoung and Innes 2006; Meyers et al. 2010). The TIR domain plays an important role in pathogen detection (Luck et al. 2000), while the CC domain is associated with protein–protein interactions (Van et al. 2007). The NBS domain consists of a P-loop, Gly–Leu–Pro–Leu (GLPL), kinase-2a, and kinase-3a motifs, and is essential for ATP/GTP binding activity (Saraste 1990; Miller et al. 2008). The LRR domain can interact with pathogens directly or indirectly (Jia et al. 2014). Earlier studies revealed that TNL genes are abundant in dicots, but absent in monocots. Recent whole-genome sequencing data have made it possible to comprehensively analyze NBS–LRR genes in economically important plants.
Cabbage (Brassica oleracea L.) is one of the major members of the Brassicaceae family. However, it is susceptible to infection by numerous fungal and bacterial pathogens, such as Fusarium wilt (FW), Turnip mosaic virus (TuMV), Alternaria brassicicola (Schweinitz) Wilts, and Peronospora parasitica (Pers.) Fr. The FW caused by Fusarium oxysporum f.sp. conglutinans (FOC) is particularly severe. Recently, the TNL-type resistance gene Foc1 (Bo7g104800) was cloned (Lv et al. 2013); however, the defense mechanism involving Foc1 remains unclear (Shimizu et al. 2014). In addition, many cloned R genes (e.g., CRa, CRb, and Crr1a) in Brassica crops are also TNL genes. However, there have been few studies on the cabbage NBS–LRR family (Shazia et al. 2018; Kim et al. 2015), and none of them analyzed the relationship between FOC infection and the NBS–LRR family. Consequently, comprehensive analyses of the relationships between NBS–LRR (especially the TNL genes) and FOC infection are indispensable.
To identify the NBS–LRR genes, their genome-wide encoded protein sequences were analyzed using the Hidden Markov Model (HMM). The distribution, gene cluster, and gene structure of identified genes were compared. Furthermore, their nuclear localization signal (NLS) and cis-elements were predicted. Expression profiles of these genes in six tissues were analyzed. Finally, the TNL family was chosen to elucidate the genes’ response to FOC infection in different stages because the TNL family is much bigger than the CNL family and Foc1 is a TNL-type gene. Fourteen TNL genes were identified as responding significantly to FOC infection in different stages, according to RNA sequencing (RNA-seq) and reverse transcription PCR (RT-PCR) analyses. These results will provide a basis for further identifying, screening, and mapping of cabbage NBS–LRR-encoding genes.
Materials and methods
Identification of NBS genes
To identify the NBS–LRR genes, we downloaded the whole-genome protein sequence of cabbage from the Ensembl Plant database (http://plants.ensembl.org/index.html). Then the sequences were scanned using HMMER v3.1b2 (http://hmmer.org/) with the raw Hidden Markov Model (HMM) corresponding to the Pfam NBS (NB-ARC) family (PF00931) (Finn et al. 2016). Proteins with E value < 1e−10 were selected. High scoring hits were used to construct a cabbage NBS HMM-profile, using HMM-Build to check for any missing hits. The TIR, NBS, and LRR domains of the identified NBS–LRR proteins were confirmed using Pfam (http://pfam.sanger.ac.uk/) and SMART (http://smart.embl-heidelberg.de/) (Zhang et al. 2016a, b; Letunic et al. 2012). CC domains could not be analyzed through Pfam and SMART; therefore, we used Paircoil2 (http://cb.csail.mit.edu/cb/paircoil2/paircoil2.html) with a P-score cutoff of 0.025.
Genomic distribution on chromosomes
According to the gene position, Mapinspect software was used to map the physical location of the NBS–LRR genes. A gene cluster was defined as being present when the distance between two neighboring NBS–LRR genes was < 200 kb and contained ≤ 8 non-NBS genes between the two NBS–LRR genes (Richly et al. 2002; Blake et al. 2003).
Gene characteristics and structure
Information concerning the NBS–LRR genes, including open reading frames (ORFs) and exon numbers were retrieved from Ensembl Plants. The pI (isoelectric point) and MW (molecular weight) of the identified genes were calculated using Pepstats (http://www.ebi.ac.uk/Tools/seqstats/emboss_pepstats). Furthermore, we performed NLS (nuclear localization signal) (http://cello.life.nctu.edu.tw/) analysis, and the promoter sequence (2000 bp upstream of the start codon) of each gene was submitted to the PlantCARE (http://bioinformatics.psb.ugen.be/webtools/plantcare/html/) for cis-element prediction (Lescot et al. 2002). Finally, the gene structure was analyzed using GSDS2.0 (http://gsds.cbi.pku.edu.cn/index.php) by comparing the cDNA and corresponding genomic sequence of each gene (Guo et al. 2007).
Phylogenetic and conserved motif analyses
To determine underlying relationships among family members of various NBS–LRR genes, MEGA 6.0 (http://www.megasoftware.net) was used to construct a phylogenetic tree using the maximum likelihood (ML) method with 1000 bootstrap replicates; excessively short sequences were excluded (Tamura et al. 2011). The conserved motifs were identified using MEME (http://meme-suite.org/tools/meme) and TBtools (http://www.cjchen.name) software (Bailey et al. 2009; Chen et al. 2018).
Gene duplication and Ka/Ks ratio analysis
Tandem duplication and segmental duplication genes were analyzed using CoGe (https://genomevolution.org/coge/) (Lyons and Freeling 2008). The Circos software (http://circos.ca/) was used to draw a circle map of the segmental duplicated genes (Krzywinski et al. 2009). Furthermore, DnaSP (6.12.01) was used to calculate the synonymous (Ks) and non-synonymous (Ka) substitution rates, and duplication events (Yang et al. 2008). A Ka/Ks calculator was used to estimate the Ka/Ks rates of evolution, Ka/Ks values of > 1 and < 1 were deemed to represent positive and purifying selection, respectively. To estimate the evolutionary time, the Ks values were converted to duplication time in millions of years, based on the ratio of one substitution per synonymous site per year. The calculation formula for the duplication events time was T = Ks/2λ × 10−6 Mya (λ = 6.5 × 10−9) (Librado and Rozas 2009; Gaut et al. 1996).
Tissue-expression analysis
The expression data of the 138 NBS–LRR genes in 6 tissues (flower, flower-bud, fruit, leaf, root, and stem) were obtained from the Expression Atlas (https://www.ebi.ac.uk/gxa/home) submitted by Liu et al. (2014). Then, the gene expression data were compiled to construct a heatmap using TBtools to display the expression levels of these NBS–LRR genes.
Plant materials and FOC treatments
The cabbage line ‘R4P1’, which is resistant to FOC, was sown in an artificial growth chamber under a photoperiod of 16-h light/8-h dark at 25 °C/18 °C day/night temperatures. Seedlings at the three-leaf stage were infected with FOC strain GLHW1 (race 1) using the root dip inoculation method (Tian et al. 2009). The suspension inoculation concentration was 1 × 106 spores/ml. Simultaneously, control plants were mock inoculated with distilled water. After inoculation, the seedlings were placed back into the original pots. Both inoculated and mock-inoculated plant roots were collected separately under a various time points (0, 4, 12, 24 and 48 h) using three replications. Briefly, root samples were frozen in liquid nitrogen and stored at − 80 °C until RNA extraction.
RNA isolation, RNA sequencing, and reverse transcription PCR
Total RNA was isolated using a kit (Cat: DP432, TIANGEN, China) following the manufacturer’s instructions. The quality and quantity of the RNA was confirmed using agarose gel electrophoresis and the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), and then sequenced using the Illumina Hiseq 2000 platform (Illumina, San Diego, CA, USA) (Wang et al. 2010). RT-PCR was conducted to measure the expression levels of TNL genes using SYBR Green 1 (Cat: FP205-01, TIANGEN, China) and a Roche LightCycler 480 system (Roche Branchburg, NJ, USA). Contaminating gDNA removal and RNA reverse transcription processes were performed using kits (Cat: AU311-02, Transgen, China). The 20 μL volume contained 10 μL of 2 × SuperReal PreMix Plus, 2 μL of cDNA, 1 μL of each primer, and 6 μL of ddH2O, the constitutively expressed GAPDH gene was used as an internal reference. The amplification procedure comprised: 95 °C for 5 min; followed by 40 cycles of 95 °C for 10 s, then 60 °C for 20 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 7 min. All reactions were performed with three technical and biological replicates. Relative gene expression was calculated using the comparative CT method (Livak and Schmittgen 2001).
Results
Identification and distribution of NBS–LRR genes
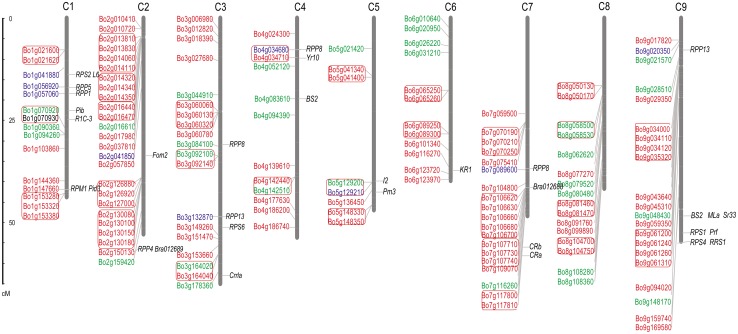
In total, 236 NBS and 172 NBS–LRR genes were identified using HMMER search and verified using Pfam and SMART databases. These NBS–LRR genes could be roughly divided into three classes: TNL, CNL, and others, according to their structure. To facilitate comparative analyses, only the 105 TNL and 33 CNL genes were used in the following study. Among the 138 NBS–LRR genes, 133 were mapped onto the 9 chromosomes of cabbage while other 5 were located on different scaffolds. The ratio of cabbage TNL:CNL genes was almost 3:1 (105:33), which was higher than that in B. rapa (2:1) (90:41) and in A. thaliana (2:1) (83:51) (Yu et al. 2014). According to the criteria defining a gene cluster, we identified 70 (50.7%) genes belonging to 27 clusters (Table S1). Most of them (20) are TNL gene clusters, and it was less common to find TNL and CNL genes in the same cluster. The members in each cluster ranged from 2 to 5, and the cluster sizes ranged from 10,421 to 125,034 bp. Thirty-three cloned functional NBS–LRR genes from other species were mapped on the cabbage chromosomes according to their orthologous genes, which could provide a reference for follow-up gene mapping and the identification of gene function in cabbage (Fig. 1). Among them, three cloned resistance genes (Foc1, CRa, and CRb) in cabbage and Chinese cabbage are all located on chromosome 7. To date, we also mapped a probable Xanthomonas campestris pv. campestris resistance gene on chromosome 7.
Fig. 1.
Physical map of the 133 NBS–LRR genes on chromosomes C1–C9. The sizes of the nine chromosomes are represented by the scale in megabases (Mb) on the left. The TNL, CNL, and certain other types of NBS–LRR genes are distinguished in red, green, and blue, respectively. Gene clusters are marked with red frames. The 33 cloned NBS–LRR genes from other species are shown on the right corresponding to orthologous genes of cabbage
Gene characteristics and structure
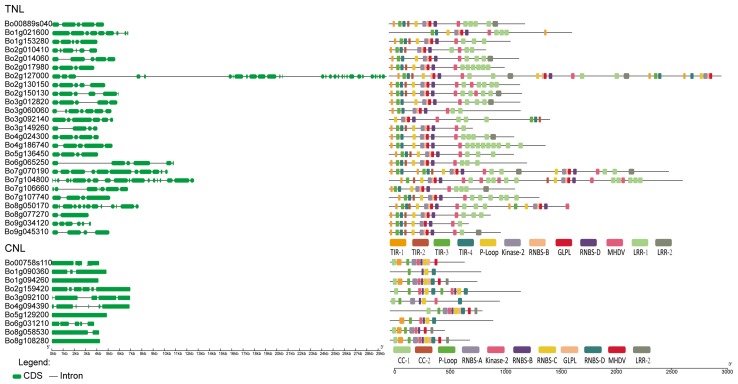
The genomic and coding sequence (CDS) sequences, protein length, MW, and pI of the 138 NBS–LRR genes were comprehensively analyzed (Table S2). The sequence lengths of genomic and CDS ranged from 2036 bp (Bo2g130100) to 28,414 bp (Bo2g127000) and 1644 bp (Bo9g094020) to 8988 bp (Bo3g153660), respectively. The protein lengths ranged from 203 AA (Bo9g094020.1) to 3429 AA (Bo3g153660.1). In addition, there were also significant variations in the MW and pI, which ranged from 30.23 kDa (Bo5g148350.1) to 378.09 kDa (Bo3g153660.1) and from 4.48 to 8.85, respectively. The average MW of TNL (136.39 kDa) and CNL (94.55 kDa) genes were markedly different. Over 88.7% of the NBS–LRR proteins were predicted to be located in the cytoplasm (Table S2), among which the percentage of CNL proteins (93.9%) was higher than that of TNLs (82.9%). The average exon number of the NBS–LRR genes was 5.96, with TNL genes having more exons (6.62) than CNL genes (3.88) (Table S2, Fig. 2). These results suggested that the number of introns may have increased and decreased during the structural evolution of the two types of NBS–LRR resistance genes in cabbage (Sharma et al. 2017).
Fig. 2.
The exon–intron structures and the corresponding conserved motifs of certain TNL and CNL genes. The green bars indicate the exons, and the black lines indicate the introns. Motifs are represented by different colored boxes
Cis-element analysis
Cis-elements are usually involved in gene regulation; therefore, we performed cis-elements prediction for the 138 genes, over 66 CAREs (cis-acting regulatory elements) involved in gene regulation were identified; the conventional elements (TATA-box, CAAT-box) identified in Arabidopsis and common bean were also detected (Meyers et al. 2003; Wu et al. 2017). The remaining CAREs were mainly associated with light responsiveness, hormones, tissue-specific expression, and abiotic stress response (Table S3). The light-responsive elements, such as ACE, Box4, and AE-box were found in many genes. Furthermore, hormone-related CAREs such as abscisic acid (ABRE), auxin (TGA-box and TGA-element), and salicylic acid response (SARE and TCA-element) were also detected. In addition, we found CAT-boxes, RY-elements, AACA_motifs, and As1, which are associated with tissue-specific expression. Among them, As1, which is involved in root-specific expression, was detected in 99 NBS–LRR genes.
Conserved motifs and phylogenetic analysis
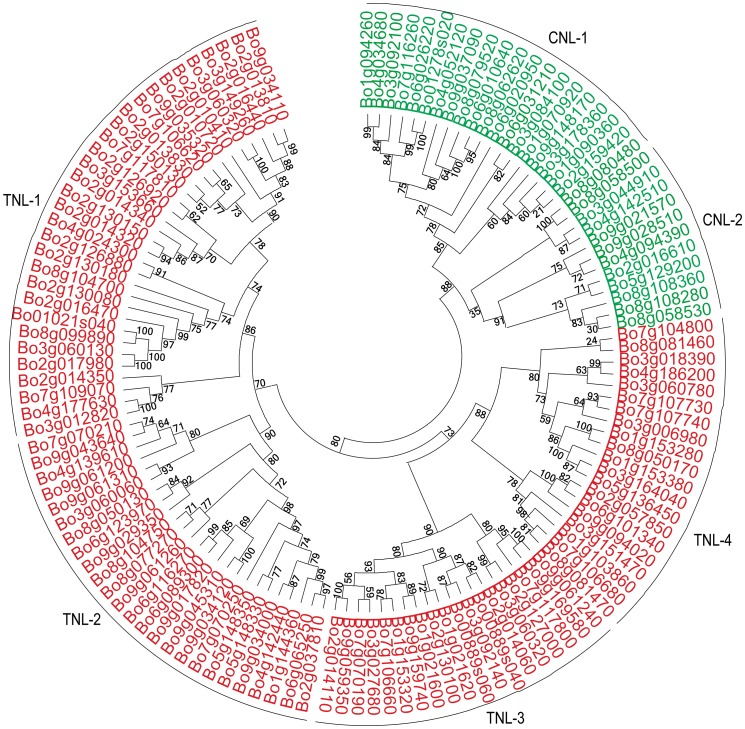
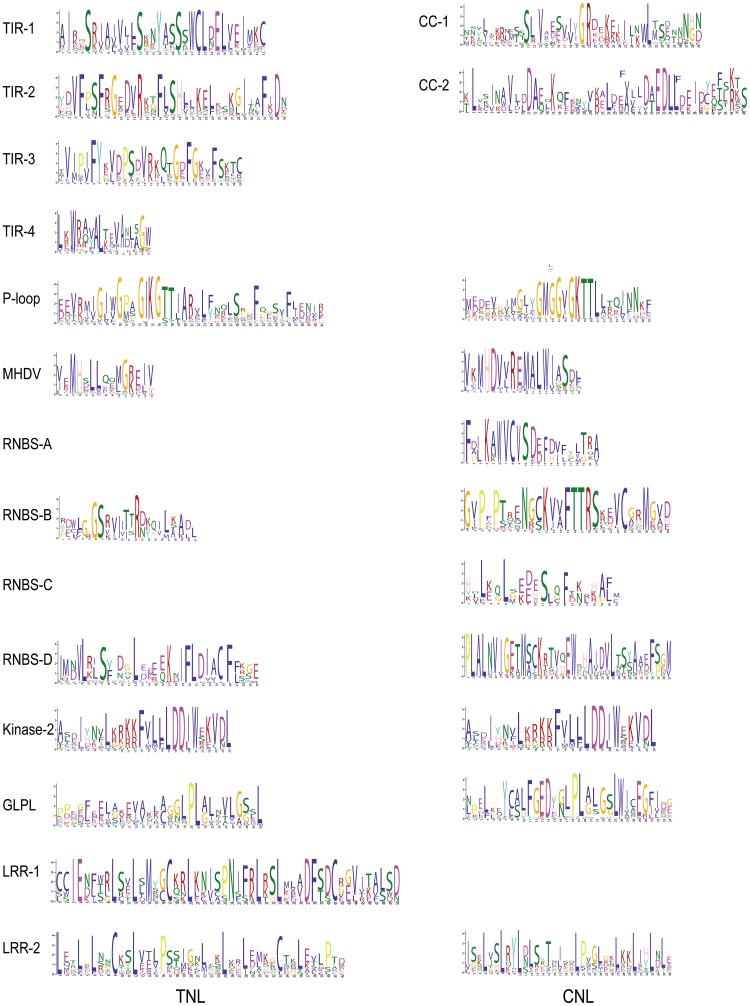
To derive the evolutionary relationships of the NBS–LRR genes, a phylogenetic tree was built using the maximum likelihood (ML) method, as shown in Fig. 3, NBS–LRR genes were clearly divided into two (TNL and CNL) major groups and six subgroups: TNL (1–4), CNL (1–2). TNL-1 was the largest subgroup (31 genes) and accounted for 29.5% of TNL genes, mostly on chromosome 2. In contrast, CNL-2 only harbors 11 members. Using MEME analysis, 12 and 11 different types of common motifs were identified as existing widely in most TNL and CNL proteins. In addition, GLPLA and kinase-3 motifs were detected in some NBS–LRR genes’ NBS-subdomains, LDL was detected in some NBS–LRR genes’ LRR-subdomains, and EDVID was detected in some CNL genes. The eight major motifs (P-loop, Kinase-2, RNBS (A-D), MHDV, and GLPL) found in the Arabidopsis NBS–LRR genes were also detected. P-loop, RNBS, and Kinase-2 were present in all the 138 NBS–LRR genes, and P-loop, kinase-2, and GLPL were highly similar between the CNL and TNL genes (Fig. 4). The NBS motifs (A-D) were identified in all the CNL genes, while there were no NBS-A and NBS-C in the TNL genes. Furthermore, the order of the TIR (1–4) motifs detected for the cabbage TNL proteins was consistent with that in Arabidopsis and common bean (Meyers et al. 2003; Wu et al. 2017). Among the TNL genes, TIR-1 and TIR-4 were missing from the proteins encoded by Bo1g021600, Bo1g021620, and Bo2g127000. TIR-1 was missing from eight proteins (Bo3g092140, Bo3g153660, Bo7g106700, Bo8g081470, Bo9g059350, Bo9g061240, Bo9g061310, and Bo00889s060), while the rest of the TNL proteins contained the four types of TIR motifs (Fig S3). Moreover, we identified two types of CC (CC-1 and CC-2) motifs from 33 CNL proteins, and more than half of these proteins contained both types (Fig S4). These TIR (1–4) and CC (1–2) motifs were also observed in Populus trichocarpa and A. thaliana (Kohler et al. 2008; Meyers et al. 2003). In addition, two (LRR-1 and LRR-2) and one (LRR-2) motifs were detected in the LRR region of the TNL and CNL genes, respectively.
Fig. 3.
Phylogenetic analysis of NBS–LRR proteins using the maximum likelihood method
Fig. 4.
MEME analysis of the TNL and CNL proteins. Different colored letters represent amino acids belonging to the different families
Gene duplication analysis
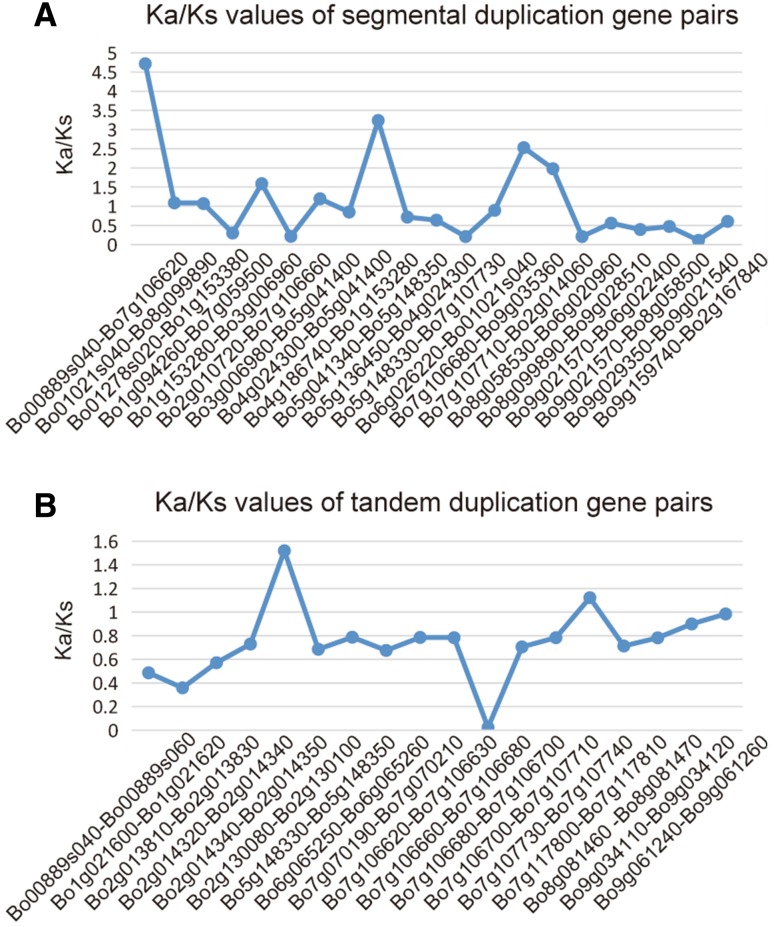
In cabbage, 33 tandem duplicated genes are distributed in 15 tandem arrays (Table S4). Thirty-one genes are distributed on seven chromosomes unevenly, and the remaining two were unanchored on scaffolds. Interestingly, none of the CNL genes were duplicated. Single tandem duplicated genes containing two genes were identified on chromosomes 1, 5, 6 and 8, and no duplicated genes were found on chromosomes 3 and 4 (Table S4, Fig. 5). The highest number of tandem arrays (5), with 12 genes, is located on chromosome 7. The Ka/Ks values of most the 39 and 21 pairs of tandem and segmental duplicated genes were < 1, indicating that these genes have evolved under negative selection (Table S4, Fig. 6). The duplication events of the two types of duplicated genes occurred between 0.62 (Ks = 0.008) and 221.43 Mya (Ks = 2.88), with an average of 104.23 Mya, the tandem duplication events occurred from 2.52 Mya (Ks = 0.033) to 194.93 Mya (Ks = 2.53), with average 97.52 Mya. These results indicated that the expansion of the cabbage NBS–LRR genes mostly arose from whole-genome duplication events during their evolution (Haron et al. 2016; Gaut et al. 1996).
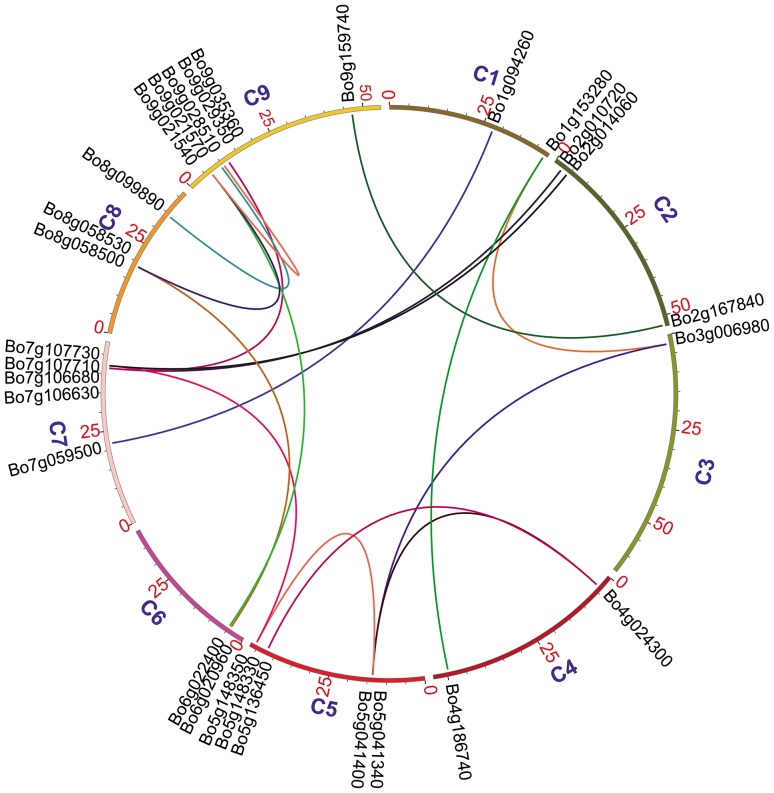
Fig. 5.
Circos diagram of segment duplicated NBS–LRR genes. C1–C9 represent nine chromosomes; the black lines on the chromosomes stand for the location. Colored lines stand for the relationship of the segmental duplicated genes
Fig. 6.
The Ka/Ks values of two types of duplicated genes. a, b Represent Ka/Ks values of segmentally and tandemly duplicated gene pairs, respectively
Tissue-specific expression of NBS–LRR genes
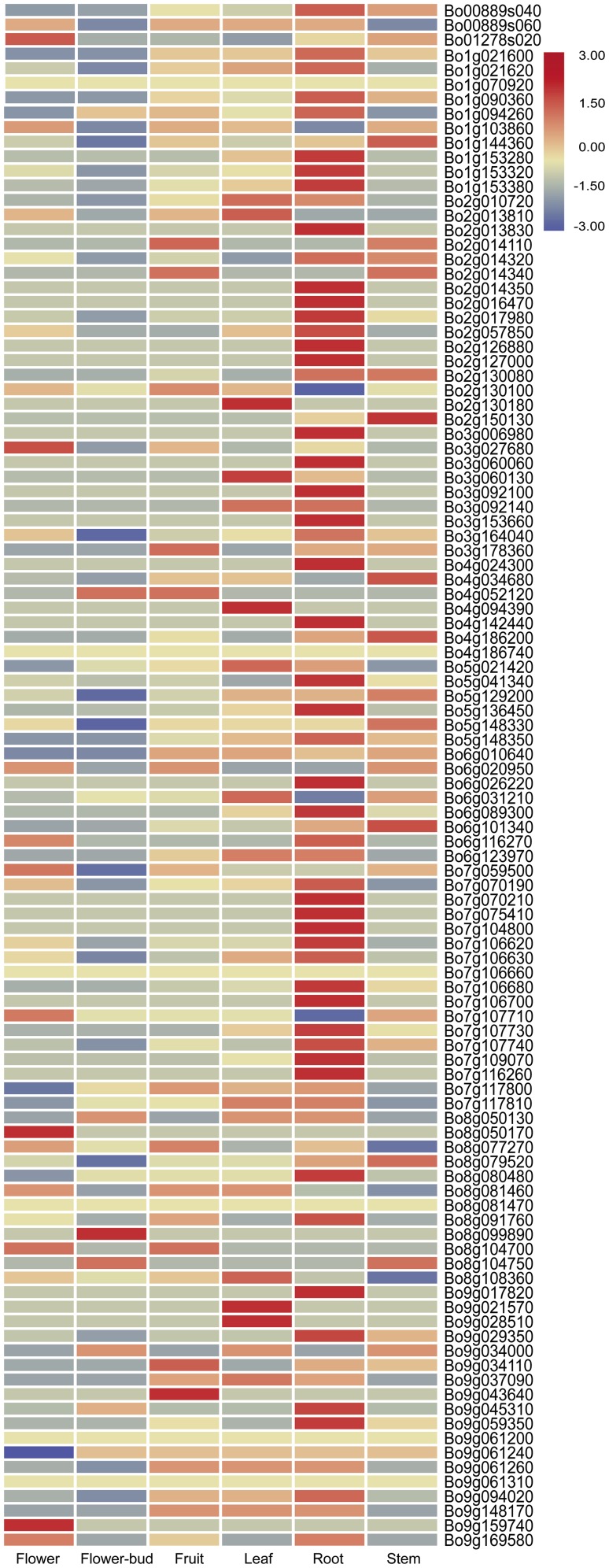
Based on the published transcriptome data, we analyzed the expression levels of 106 NBS–LRR genes (expression data for 25 TNL and 7 CNL genes were not found) in six tissues (Table S5). Among the 80 TNL genes, 39 showed relatively high or specific expression in roots, while only 7 CNL genes were highly or specifically expressed in the roots (Fig. 7). We concluded that over one-third of TNL genes are highly expressed in roots, suggesting functional homology. Combined with the chromosome analysis, we found that, except for four TNL genes (Bo7g059500, Bo7g070250, Bo7g106660, and Bo7g107710), almost all of the 18 NBS–LRR genes on chromosome 7 are highly or specifically expressed in roots. Considering Foc1, Bra012688, CRa, and CRb are the FOC and club-root R genes, suggesting that these genes probably play specific roles in root disease resistance. Meanwhile, there were more TNL genes than CNL genes; therefore, DGE and RT-PCR were conducted to verify the TNL gene expression patterns after FOC infection.
Fig. 7.
Tissue-specific digital expression profiles of 106 genes. Higher expression of each gene is presented in red; otherwise, blue was used. The genes with a TPM equal to 0 were not used in this array
Responses to FOC pathogen inoculations
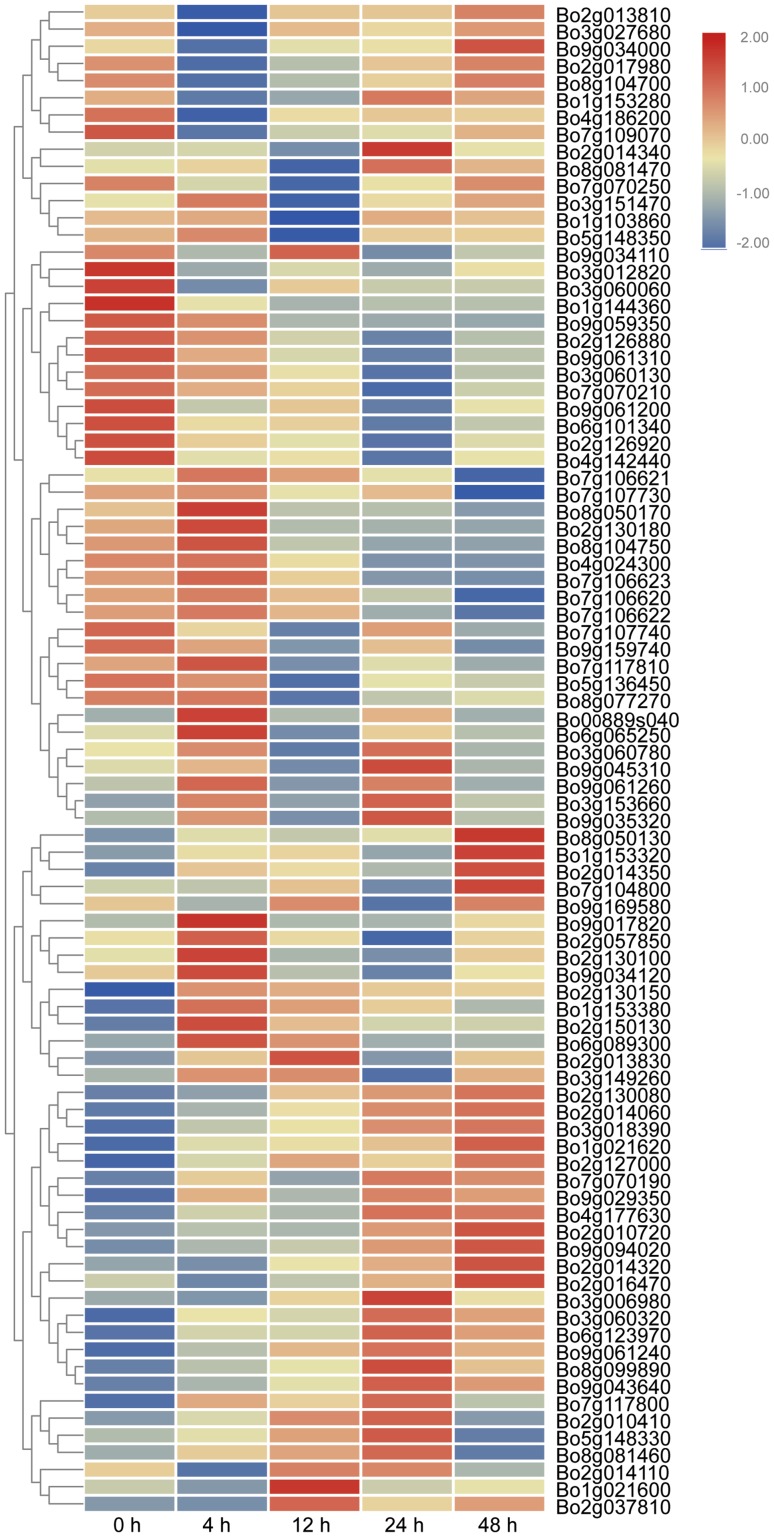
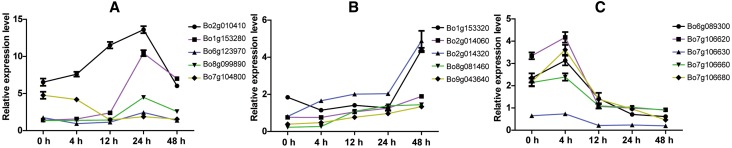
The expression data of 88 out of 105 TNL genes were available, and the expression level of 14 genes significantly decreased or increased after FOC inoculation (Table S6, Fig. 8). These genes could be placed into two categories: upregulated (Fig. 9a, b) and downregulated (Fig. 9c) genes. Overall, three genes (Bo1g153280, Bo6g123970, and Bo8g099890) exhibited higher expression at 24 h after inoculation, whereas four genes (Bo1g153320, Bo2g014060, Bo2g014320, and Bo9g043640) were highly expressed at 48 h. In addition, five genes (Bo6g089300, Bo7g106620, Bo7g106630, Bo7g106660, and Bo7g106680) were initially upregulated after infection, but were then downregulated after 4 h. The expression level of Foc1 after FOC infection was irregular.
Fig. 8.
Expression profiles of 88 TNL-encoding genes under FOC challenge. Cabbage roots were selected in different times (0, 4, 12, 24, and 48 h). The expression color scale is shown at the top right. Higher expression for each gene is presented in red; otherwise, blue was used. The genes with an RPKM equal to 0 were not used in this array
Fig. 9.
The relative expression levels of 18 genes. The vertical axis represents the relative expression level and 0, 4, 12, 24, and 48 h (x-axis) indicate the treatment times. Different lines represent different treatment times. Error bars represent the standard error of the mean based on three replicates
Overall, the DGE data for the 14 TNL genes were basically consistent with RT-PCR results. Besides, through comparative expression profile analysis of these genes, we hypothesized that different genes might have different functions after FOC infection. Taken together, the RNA-seq and RT-PCR expression analyses support the hypothesis that TNL genes are involved in FOC infection at different times.
Discussion
In our research, 172 cabbage NBS–LRR genes were identified, revealing a much higher number compared with the 70 described by Yu et al. (2014). Similar numbers of NBS–LRR genes were also observed in A. thaliana (207) (Yu et al. 2014) and Cucumis sativus L. (233) (Zhang et al. 2016a, b). Compared with B. rapa and A. thaliana, cabbage has more NBS genes, possibly because the cabbage genome (630 Mb) (Liu et al. 2014) is larger than that of B. rapa (485 Mb) (Wang et al. 2014) and A. thaliana (123 Mb) (Dennis and Surridge 2000). The NBS proteins of cabbage account for 0.39% of the total proteome, which is similar to chickpea (0.36%) (Sharma et al. 2017).
NBS genes evolve in concert with pathogens, and rapid evolution of NBS genes among different cultivars of a species exposed to different biotic stresses is frequently observed. Many studies indicate that NBS proteins carrying a TIR domain originated earlier than the non-TIR type (Yue et al. 2012) and the TNL genes are less diversified in most monocots (Bai et al. 2003). The ratio of cabbage TNL to CNL genes is 3:1, which is higher than that in B. rapa (2:1), A. thaliana (2:1) (Yu et al. 2014), and in other monocot crops, indicating that cabbage NBS–LRR genes might have evolved more slowly (Tarr and Alexander 2009; Zhang et al. 2016a, b). The greater contribution of TNL genes than CNL genes to resistance might be one of the main explanations for the large difference in the number of TNL and CNL genes in cabbage.
Similar to other species, the distribution of cabbage NBS–LRR genes is uneven and they mainly exist in clusters, as a result of rapid gene evolution (Leister 2004; Friedman and Baker 2007; Wan et al. 2013; Lv et al. 2015; Lozano et al. 2015; Die et al. 2018). However, only 51% of the cabbage NBS–LRR genes are present in 27 clusters (Yang et al. 2008), whereas over 70% of the NBS–LRR genes in rice are present in 104 clusters. This small number of gene clusters may be another explanation for the slower evolution of cabbage compared with other monocots.
Gene duplication is considered to be the source of plant diversity and complexity, allowing them to adapt to changing circumstances. Segmental duplication and tandem duplication are considered to represent the two principal evolutionary patterns that cause gene family expansion in plants. In many plants, segmental duplication appears to be the dominant process that generates gene families, including the NBS–LRR and other R gene families. Moreover, the proportion of gene clusters in cabbage is small, and the probability of generating new NBS–LRR genes through tandem duplications is low. Therefore, we concluded that most NBS–LRR genes in cabbage arose from whole-genome duplication events during evolution (Haron et al. 2016). Overall, the 33 tandemly duplicated genes are distributed in 15 arrays located in the identified 27 clusters. Similar to B. rapa, the Ka/Ks values of most tandemly and segmentally duplicated genes are < 1, indicating that these NBS–LRR genes have evolved under negative selection (Yu et al. 2014).
Although most tandem duplicated genes in the same clusters showed high similarity, some of them were located in different phylogenetic clades (Lozano et al. 2015). These NBS–LRR genes were clearly divided into two major groups and six main subgroups (four TNL and two CNL genes) with different numbers of members. The motif analysis indicated that four and two types of TIR and LRR motifs could be detected among the TNL genes, while only two and one types of CC and LRR motifs were detected among the CNL genes. Furthermore, the order of the four types of TIR motifs in cabbage TNL genes was consistent with that in the common bean (Wu et al. 2017). The P-loop, RNBS, and Kinase-2 motifs were detected in all 138 NBS–LRR proteins. In addition, the P-loop, kinase-2, and GLPL motifs showed high similarity between the CNL and TNL proteins. Combined with the inference that NBS–LRRs with TIR domains may have originated earlier than non-TIR proteins, we hypothesized that some TIR motifs in TNL genes had been replaced by CC or other motifs via gene recombination and duplication.
Similar to the specific expression of NBS–LRR genes in Arabidopsis, the obtained public expression data of cabbage NBS–LRR genes also showed that numerous TNL genes were highly or specifically expressed in the roots, including Foc1 and the homologous genes of CRa and CRb, which correlates with their designation as root disease R genes (Tan et al. 2007). Analysis of the cis-elements showed that As1, which is involved in root-specific expression, was detected in over 70% of NBS–LRR genes, including Foc1 and the homologous genes CRa and CRb. Therefore, we inferred that the presence of As1 may be one reason explanation for the higher expression of these genes in the root than in other tissues.
To further study the resistance mechanism involving Foc1 and other TNL genes, RNA was extracted at different times after infection to conduct DGE and qRT-PCR analyses. The results suggested that nine upregulated and five downregulated genes probably participate in FOC resistance (Conesa et al. 2016; Meyers et al. 2010) the expression of the Foc1 was irregular during infection (Xing et al. 2016). In addition, four genes (Bo7g106620, Bo7g106630, Bo7g106660, and Bo7g106680) were all downregulated at 4 h after infection. Previous studies have shown that a series of different NBS–LRR genes work together to resist the invasion of the same pathogenic bacteria (Van and Kamoun 2008). The orthologous genes of Foc1 and the four genes in A. thaliana were analyzed because genes in same cluster are often similar in structure and function (Landolfo et al. 2018; Kozák et al. 2018). We found that Foc1, Bo7g106630, and Bo7g106680 are orthologous or have relatively high similarity with AT4G19500, while the orthologous gene of Bo7g106620 and Bo7g106660 is AT4G19510, and both AT4G19500 and AT4G19510 encode TNL type proteins (Peele et al. 2014; Iyer and Aravind 2012). In addition, the protein interaction relationship analysis in the STRING database (https://string-db.org/cgi/input.pl) showed that the products of AT4G19500 and AT4G19510 have known interactions and are co-expressed. The NLS prediction indicated that the proteins encoded by Foc1, Bo7g106620, and Bo7g106680 are located in the cytoplasm, while the proteins of Bo7g106630 and Bo7g106660 are in the outer-membrane. Based on these results, we hypothesized that the four genes participate in FOC resistance with Foc1. Further experiments will be performed to verify this inference.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Exon-intron structures of CNLs. The green bars indicate the exons, and the black lines indicate the introns (TIFF 6142 kb)
Exon-intron structures of TNLs. The green bars indicate the exons, and the black lines indicate the introns (TIFF 7235 kb)
MEME analysis of the TNL proteins. The detected motifs are shown in the top right corner (TIFF 3657 kb)
MEME analysis of the CNL proteins. The detected motifs are shown in the top right corner (TIFF 1986 kb)
Identified clusters of NBS–LRR genes (XLS 35 kb)
Characteristics of NBS–LRR genes and proteins (XLS 47 kb)
Predicted cis-elements of identified NBS–LRR genes (XLS 96 kb)
Gene duplication result, and Ka, Ks and Ka/Ks values (XLS 51 kb)
The expression of identified NBS–LRR genes in six tissues (XLS 34 kb)
DGE data, primer sequences, and qRT-PCR data (XLS 42 kb)
Acknowledgements
This work was supported financially by grants from the National key research and development program (2016YFD0101804, 2016YFD0101702), the National Natural Science Foundation of China (31171958), and the Science & Technology Innovation Program of BAAFS (KJCX20180427, KJCX20170102, KJCX20170710).
Abbreviations
- DGE
Digital gene expression
- R
Resistance
- NBS
Nucleotide-binding site
- LRR
Leucine-rich repeat
- TIR
N-terminal Toll/interleukin-1 receptor
- CC
Coiled-coil
Author contributions
ZCL, JHY, JGK, JMX, and HPW designed the research. ZCL and HPW completed the experiments. JMX and HLL performed the data analysis and prepared the manuscript. ZCL, XHZ, JGK, and JX revised the manuscript. All authors had read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to disclose.
Contributor Information
Jihua Yu, Email: yujihua@gsau.edu.cn.
Jungen Kang, Email: kangjungen@nercv.org.
References
- Bai J, Pennill L, Ning J, Weon L, Jegadeesan R, Webb C, Zhao B, Sun Q, Nelson J, Leach J, Hulbert S. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2003;12:1871–1884. doi: 10.1101/gr.454902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T, Bodén M, Buske F, Frith M, Grant C, Clementi L, Ren J, Li WS, Noble W. Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Xia R, Chen H, He YH. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. 2018 doi: 10.1101/289660. [DOI] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MJ, Gaffney D, Elo L, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:181. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dennis C, Surridge C. Arabidopsis thaliana genome. Introduction. Nature. 2000;408(6814):791. doi: 10.1038/35048677. [DOI] [PubMed] [Google Scholar]
- Deyoung BJ, Innes RW. Plant NBS–LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Die JV, Román B, Qi X, Rowland LJ. Genome-scale examination of NBS-encoding genes in Blueberry. Sci Rep. 2018;8:3429. doi: 10.1038/s41598-018-21738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Penelope C, Eberhardt RY. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17:493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, Mccaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. PNAS. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głowacki S, Macioszek VK, Kononowicz AK. R proteins as fundamentals of plant innate immunity. Cell Mol Biol Lett. 2011;16:1–24. doi: 10.2478/s11658-010-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Guo CJ, Sun XG, Chen X, Yang SH, Li J, Wang L, Zhang XH. Cloning of novel rice blast resistance genes from two rapidly evolving NBS–LRR gene families in rice. Plant Mol Biol. 2016;90:95–105. doi: 10.1007/s11103-015-0398-7. [DOI] [PubMed] [Google Scholar]
- Haron S, Gong W, He S, Sun G, Sun J, Du X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016;17:129. doi: 10.1186/s12863-016-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L. ALOG domains: provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol Direct. 2012;7:39. doi: 10.1186/1745-6150-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Mcadams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2014;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Jung HJ, Park JI, Hur Y, Nou IS. Response of NBS encoding resistance genes linked to both heat and Fungal stress in Brassica oleracea. Plant Physiol Biochem. 2015;86:130–136. doi: 10.1016/j.plaphy.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Kohler A, Guinet C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers B, Boerjan W, Martin F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- Kozák L, Szilágyi Z, Vágó B, Kakuk A, Tóth L, Molnár I, Pócsi I. Inactivation of the indole-diterpene biosynthetic gene cluster of Claviceps paspali, by Agrobacterium-mediated gene replacement. Appl Microbiol Bio. 2018;102:3255–3266. doi: 10.1007/s00253-018-8807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman DJ, Jones S, Marra M. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolfo S, Ianiri G, Camiolo S, Porceddu A, Mulas G, Chessa R, Zara G, Mannazzu I. CAR gene cluster and transcript levels of carotenogenic genes in Rhodotorula mucilaginosa. Microbiology. 2018;164:78–87. doi: 10.1099/mic.0.000588. [DOI] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet Tig. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin I, Zhao M, Ma J, Yu J, Shunmou H, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang TJ, Hu Q, Wang X, Yue ZH, Paterson A. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lozano R, Hamblin MT, Prochnik S, Jannink JL. Identification and distribution of the NBS–LRR gene family in the Cassava genome. BMC Genom. 2015;16:360. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck JE, Lawrence GJ, Dodds PN, Shepherd KW, Ellis JG. Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell. 2000;12:1367–1377. doi: 10.1105/tpc.12.8.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv HH, Yang LM, Kang JG, Wang QB, Wang XW, Fang ZY, Liu YM, Zhuang M, Zhang YY, Lin Y, Yang YH, Xie BY, Liu B, Liu JS. Development of Indel markers linked to Fusarium wilt resistance in cabbage. Mol Breed. 2013;32:961–967. doi: 10.1007/s11032-013-9925-x. [DOI] [Google Scholar]
- Lv S, Zhang CW, Tang J, Li Y, Wang Z, Jiang D, Hou XL. Genome-wide analysis and identification of TIR-NBS–LRR genes in Chinese cabbage (Brassica rapa, ssp. pekinensis) reveal expression patterns to TuMV infection. Physiol Mol Plant P. 2015;90:89–97. doi: 10.1016/j.pmpp.2015.04.001. [DOI] [Google Scholar]
- Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313x.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS–LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW. TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS–LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2010;32:77–92. doi: 10.1046/j.1365-313x.2002.01404.x. [DOI] [PubMed] [Google Scholar]
- Miller R, Bertioli D, Baurens FC, Santos C, Alves PC, Martins N, Togawa R, Souza JM, Pappas G. Analysis of non-TIR-NBS–LRR resistance gene analogs in Musa acuminata Colla: isolation, RFLP marker development, and physical mapping. BMC Plant Biol. 2008;8:15. doi: 10.1186/1471-2229-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peele HM, Guan N, Fogelqvist J, Dixelius C. Loss and retention of resistance genes in five species of the Brassicaceae family. BMC Plant Bio. 2014;14:1–11. doi: 10.1186/s12870-014-0298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Stressed Out Over a Stress Hormone. Science. 2009;324:1012–1013. doi: 10.1126/science.324-1012. [DOI] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol. 2002;19:76–84. doi: 10.1093/oxfordjournals.molbev.a003984. [DOI] [PubMed] [Google Scholar]
- Saraste M. The P-loop-a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-F. [DOI] [PubMed] [Google Scholar]
- Shao ZQ, Zhang YM, Hang YY, Xue JY, Zhou GC, Wu P, Wu X, Wu XZ, Wang Q, Wang B, Chen J. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol. 2014;166:217–234. doi: 10.1104/pp.114.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Rawat V, Suresh CG. Genome-wide identification and tissue-specific expression analysis of nucleotide binding site-leucine rich repeat gene family in Cicer arietinum (kabuli chickpea) Genom Data. 2017;14:24–31. doi: 10.1016/j.gdata.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shazia AK, Abdur RM, Jong-In P, et al. Identification of NBS-encoding genes linked to black rot resistance in cabbage (Brassica oleracea var. capitata) Mol Biol Rep. 2018;45:773–778. doi: 10.1007/s11033-018-4217-5. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Pu ZJ, Kawanabe T, Kitashiba H, Matsumoto S, Ebe Y, Sano M, Funaki T, Fukai E, Fujimoto R, Okazaki K. Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor Appl Genet. 2014;128:119–130. doi: 10.1007/s00122-014-2416-6. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. Mega 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Meyers B, Kozik AAL, West M, Morgante MA, St CD, Bent A, Michelmore R. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM. TIR-NBS–LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes. 2009;2:197. doi: 10.1186/1756-0500-2-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian RP, Kang JG, Geng LH, Xie JM, Jian YC, Ding YH. Study on the method of Fusarium wilts resistance in cabbage. CASB. 2009;5:39–42. [Google Scholar]
- Van RDH, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van OG, Ha VDB, Cornelissen BJ, Takken F. Structure and function of resistance proteins in Solanaceous plants. Annu Rev Phytopathol. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- Wan H, Yuan W, Bo K, Shen J, Pang X, Chen J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013;14:109. doi: 10.1186/1471-2164-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2014;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhu J, Wang L, Wang S. Genome-wide association study identifies NBS–LRR-encoding genes related with anthracnose and common bacterial blight in the common bean. Front Plant Sci. 2017;8:1398. doi: 10.3389/fpls.2017.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing MM, Lv HH, Ma J, Xu DH, Li HL, Yang LM, Kang JG, Wang X, Fang ZY. Transcriptome Profiling of Resistance to Fusarium oxysporum f.sp. conglutinans in Cabbage (Brassica oleracea) Roots. Plos One. 2016;11:e0148048. doi: 10.1371/journal.pone.0148048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Xi Zhang, Yue JX, Tian D, Chen JQ. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genom. 2008;280:187–198. doi: 10.1007/s00438-008-0355-0. [DOI] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Zhang F, Tong C, Huang J, Cheng X, Dong C, Zhou Y, Qin R, Hua W, Liu S. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014;15:3. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Meyers B, Chen JQ, Tian DC, Yang SH. Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS–LRR) genes. New Phytol. 2012;193:1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liang P, Ming R. Genome-wide identification and characterization of nucleotide-binding site (NBS) resistance genes in pineapple. Trop Plant Biol. 2016;9:187–199. doi: 10.1007/s12042-016-9178-z. [DOI] [Google Scholar]
- Zhang YM, Sha ZQ, Wang Q, Hang YY, Xue JY, Wang B, Chen JQ. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS–LRR) genes in Brassicaceae. J Integr Plant Biol. 2016;58:165–177. doi: 10.1111/jipb.12365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exon-intron structures of CNLs. The green bars indicate the exons, and the black lines indicate the introns (TIFF 6142 kb)
Exon-intron structures of TNLs. The green bars indicate the exons, and the black lines indicate the introns (TIFF 7235 kb)
MEME analysis of the TNL proteins. The detected motifs are shown in the top right corner (TIFF 3657 kb)
MEME analysis of the CNL proteins. The detected motifs are shown in the top right corner (TIFF 1986 kb)
Identified clusters of NBS–LRR genes (XLS 35 kb)
Characteristics of NBS–LRR genes and proteins (XLS 47 kb)
Predicted cis-elements of identified NBS–LRR genes (XLS 96 kb)
Gene duplication result, and Ka, Ks and Ka/Ks values (XLS 51 kb)
The expression of identified NBS–LRR genes in six tissues (XLS 34 kb)
DGE data, primer sequences, and qRT-PCR data (XLS 42 kb)