Abstract
Xenopus laevis is an amphibian (frog) species widely used in developmental biology and genetics. To unravel the molecular machinery regulating sex differentiation of Xenopus gonads, we analyzed for the first time the transcriptome of developing amphibian gonads covering sex determination period. We applied microarray at four developmental stages: (i) NF50 (undifferentiated gonad during sex determination), (ii) NF53 (the onset of sexual differentiation of the gonads), (iii) NF56 (sexual differentiation of the gonads), and (iv) NF62 (developmental progression of differentiated gonads). Our analysis showed that during the NF50, the genetic female (ZW) gonads expressed more sex-specific genes than genetic male (ZZ) gonads, which suggests that a robust genetic program is realized during female sex determination in Xenopus. However, a contrasting expression pattern was observed at later stages (NF56 and NF62), when the ZW gonads expressed less sex-specific genes than ZZ gonads, i.e., more genes may be involved in further development of the male gonads (ZZ). We identified sexual dimorphism in the expression of several functional groups of genes, including signaling factors, proteases, protease inhibitors, transcription factors, extracellular matrix components, extracellular matrix enzymes, cell adhesion molecules, and epithelium-specific intermediate filaments. In addition, our analysis detected a sexually dimorphic expression of many uncharacterized genes of unknown function, which should be studied further to reveal their identity and if/how they regulate gonad development, sex determination, and sexual differentiation. Comparison between genes sex-specifically expressed in developing gonads of Xenopus and available transcriptome data from zebrafish, two reptile species, chicken, and mouse revealed significant differences in the genetic control of sex determination and gonad development. This shows that the genetic control of gonad development is evolutionarily malleable.
Electronic supplementary material
The online version of this article (10.1007/s00427-019-00630-y) contains supplementary material, which is available to authorized users.
Keywords: Testis, Ovary, Sex determination, Gonad development, Xenopus, Transcriptome
Introduction
Xenopus laevis is a good model to study molecular mechanisms of gonad development because the structural changes in developing gonads and the master gene determining sex, the W-linked DM-domain gene (dm-w), are well known. The dm-w is located on W chromosome and thus is present only in the genetic females (ZW) (Yoshimoto et al. 2008). At the earliest stage of gonad development, the gonads are undifferentiated and bipotential. The expression of dm-w triggers ovary development, while its absence promotes testis development. It is believed that the DM-W protein blocks the DMRT1 (doublesex and mab-3-related transcription factor 1) involved in male sex determination (Yoshimoto et al. 2010). In addition to the dm-w, many other genes, which act independently or downstream of dm-w, are involved in the development of bipotential gonads into the ovaries or the testes (Piprek et al. 2016). However, the expression and role of many genes involved in gonadal development is still vague. At the initial stage of gonadogenesis (NF50, Nieuwkoop-Faber stage 50), the gonads consist of the gonadal cortex and the medulla. The gonadal cortex contains coelomic epithelium and the germ cells, which adhere to the interior face of the epithelium. The medulla is sterile and contains medullar cells only (Piprek et al. 2016, 2017). At this stage, the sex-determining genes (dm-w and dmrt1) are expressed in the somatic cells of the gonads. In the absence of dm-w, i.e., in the differentiating testis (ZZ), around stage NF53, the cortex and medulla fuse. Subsequently, around stage NF56, the germ cells become enclosed by the somatic cells, which results in the formation of testis cords (Piprek et al. 2017). The typical structure of the testis, i.e., fully differentiated testis cords separated by the interstitium, is established at stage NF62. In contrast, in differentiating ovaries, which express dm-w, the germ cells remain in the cortical position, and at stage NF56, the ovarian cavity forms inside the gonad. Around NF62, the ovaries are fully differentiated, with the oocytes located in the cortex (Piprek et al. 2017; Yoshimoto et al. 2008). This divergent development of the female and male gonads has to be controlled by differential gene expression. A global analysis of Xenopus gonad transcriptome, which we performed in this study, is the step in obtaining a broad database of gene expression pattern in developing male and female Xenopus gonads.
Among vertebrates, the transcriptome of developing gonads has been studied in the mouse (Beverdam and Koopman 2006; Chen et al. 2012; Gong et al. 2013; Jameson et al. 2012; Nef et al. 2005; Small et al. 2005), chicken (Ayers et al. 2015; Scheider et al. 2014), slider Trachemys scripta (Czerwinski et al. 2016), Alligator mississippiensis (Yatsu et al. 2016), and in several teleost fish species (Bar et al. 2016; Lin et al. 2017; Sreenivasan et al. 2008; Sun et al. 2018; Xu et al. 2016). These studies provided valuable insights into the genes involved in gonad development and identified new sex-determining gene candidates.
Among anurans, a transcriptome analysis was performed only in Silurana (Xenopus) tropicalis and only on already sexually differentiated gonads (from stage NF58) (Haselman et al. 2015). Thus, the genes expressed before and during the sexual differentiation of amphibian gonads are still unknown. The aim of our study was to examine the transcriptome of developing Xenopus gonads from the earliest stage of gonad development. We studied the gene expression pattern in four different stages of gonad development: the undifferentiated gonad during the period of sex determination (NF50), gonads at the onset of sexual differentiation (NF53), the differentiating gonads (NF56), and during the developmental progression of differentiated gonads (NF62) (Fig. 1).
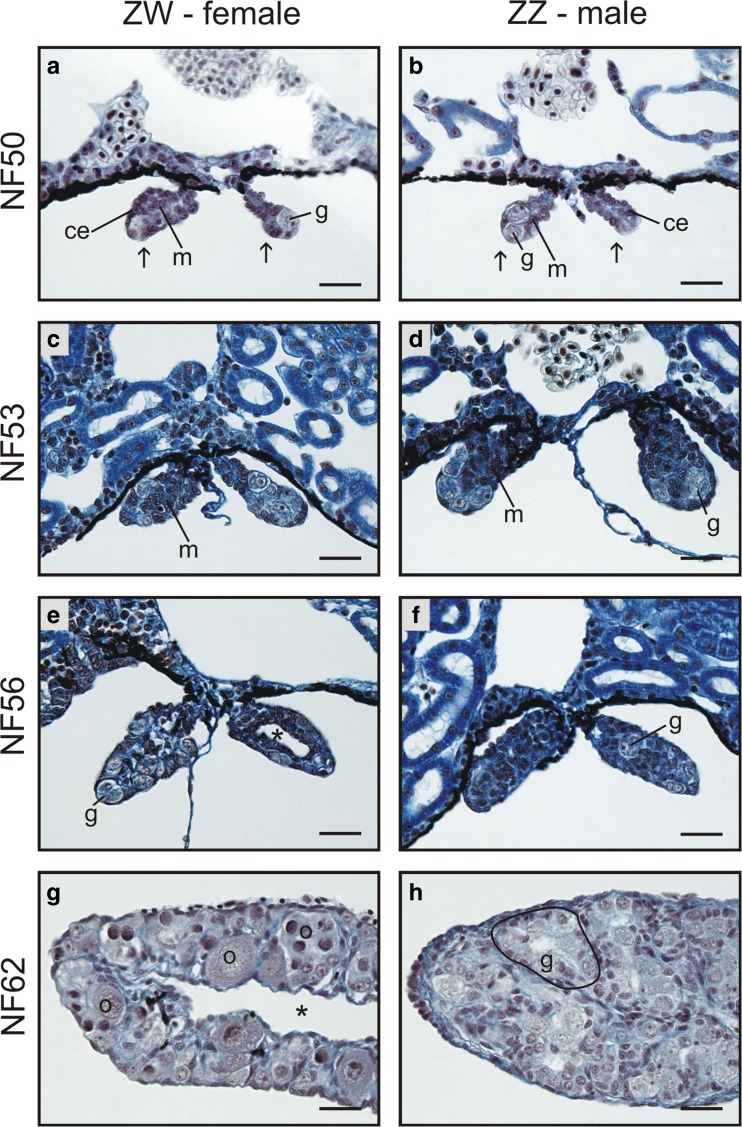
Fig. 1.
Structural changes in developing gonads. a, b At stage NF50, there is no difference in the gonad structure between genetic sexes (ZW and ZZ). Such undifferentiated gonads (arrows) are composed of the somatic cells of coelomic epithelium (ce) covering the gonad, and germ cells (g) located inside; the germ cells are attached to the coelomic epithelium. The somatic cells gather in the gonad center forming gonadal medulla (m). At stage NF53, the first sexual differences appear in the gonad structure; in the differentiating ovaries (c, ZW), the germ cells remain in the peripheral position forming the ovarian cortex, whereas the centrally located medulla remains sterile. In the ZZ (male) gonads at the onset of sexual differentiation (d, the onset of the testis differentiation), the germ cells (g) detach from the coelomic epithelium and move towards the gonad center (medulla, m). At stage NF56, the differentiating ovaries (e) becomes compartmentalized into cortex and medulla; all germ cells (g) are located in the cortex and are attached to the coelomic epithelium; an ovarian cavity forms in the medulla (asterisk). In the differentiating testes (f), the germ cells (g) are dispersed and the cortex and medulla are absent. At stage NF62, the ovaries (g) contain large ovarian cavity (asterisk); the ovarian cortex contains meiotic cells (o). In the testes (h), the germ cells (g) are located within the testis cords (encircled). Scale bar, 25 μm
Results and discussion
Sex-specific changes in the level of gene expression
In developing Xenopus laevis gonads (stages NF50, NF53, NF56, and NF62 combined), we detected the expression of 63,084 transcripts in total. We found that while the expression level of the majority of genes was similar between stages and between male and female gonads, a subpopulation of genes showed distinct changes in the expression level between stages and sexes, which suggested that they may play a role in sex determination and/or sexual differentiation (Figs. 2A, B and 3, Tables 1 and 2).
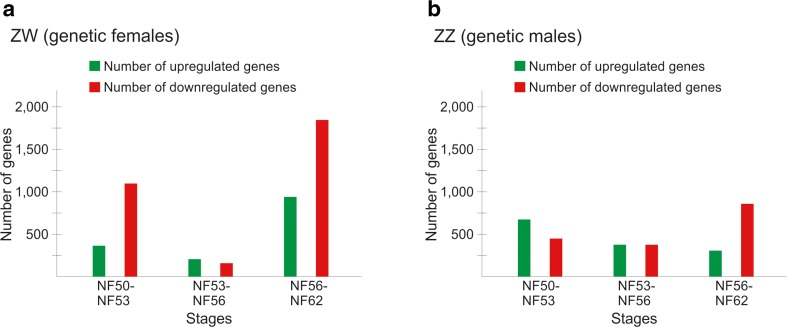
Fig. 2.
Diagram of changes in the number of genes upregulated and downregulated (≥ 2-fold change) between different stages in ZW gonads (a) and ZZ gonads (b)
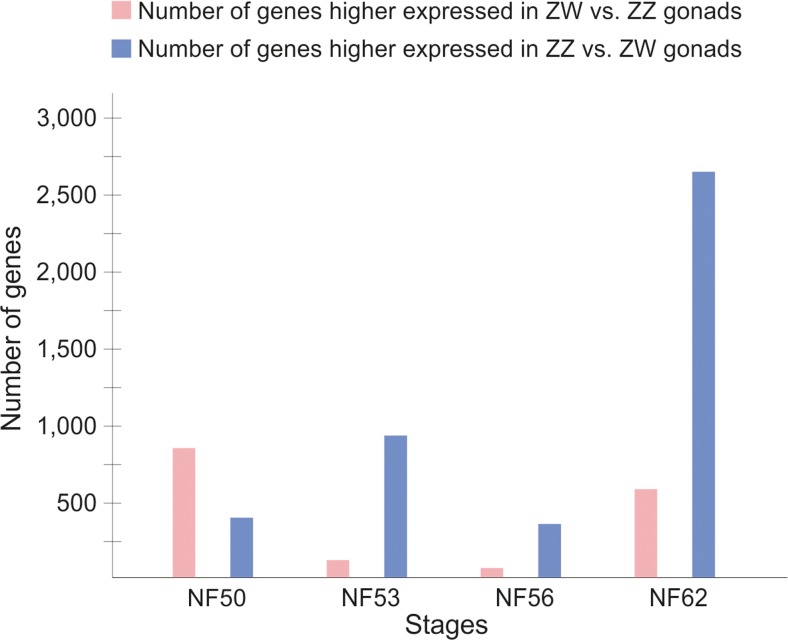
Fig. 3.
Diagram of changes in the number of genes with higher expression in ZW or ZZ gonads (≥ 2-fold change)
Table 1.
Number of genes with up- and downregulated (≥ 2-fold change) expression in ZW and ZZ gonads
| Compared stages | ZW (females) | ZZ (males) | ||
|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | |
| NF53 vs. NF50 | 376 | 1078 | 659 | 436 |
| NF56 vs. NF53 | 143 | 128 | 340 | 340 |
| NF62 vs. NF56 | 918 | 1834 | 334 | 831 |
Table 2.
Number of genes with up- and downregulated (≥ 2-fold change) expression in ZW versus ZZ gonads
| ZW vs. ZZ compared at stages | Upregulated in ZW | Downregulated in ZW |
|---|---|---|
| NF50 | 820 | 372 |
| NF53 | 193 | 890 |
| NF56 | 75 | 346 |
| NF62 | 594 | 2630 |
Analysis of gene expression level in the gonads showed that in the genetic females (ZW), the gonads at the onset of sexual differentiation (NF53) had 376 genes with upregulated expression and 1078 genes with downregulated expression in comparison to the undifferentiated gonad during sex determination period (NF50) (Fig. 2, Table 1). In the differentiating ovaries (NF56), only 143 genes were upregulated and 128 genes were downregulated in comparison to NF53 (Table 1). In differentiated ovaries (NF62), there were 918 genes with upregulated expression and 1834 genes with downregulated expression in comparison to NF56 (Table 1).
The genetic male (ZZ) gonads at the onset of sexual differentiation (NF53) had 659 genes with upregulated expression and 436 genes with downregulated expression in comparison to NF50 stage (Fig. 2, Table 1). In differentiating testes (NF56), 340 genes were up-, and 340 downregulated in comparison to NF53 stage. The differentiated testes at stage NF62 had 334 genes with upregulated expression and 831 genes with downregulated expression in comparison to NF56 stage.
Altogether, these data indicate that in both sexes, the transcriptional regulation is more robust during early gonadal development, i.e., at the onset of sexual differentiation of the gonad (NF50-NF53) and in the already differentiated gonads NF56-NF62 than in the differentiating gonads (NF53-NF56).
The comparison of gene expression level in between ZW and ZZ gonads showed significant differences between the sexes and revealed sexually dimorphic pattern of gene expression. At the initial phase of gonad development, i.e., in the undifferentiated gonads during sex determination (NF50), there were 1192 genes (i.e., 3.4%) with sexually dimorphic expression (≥ 2-fold change). Eight hundred twenty genes showed higher expression in ZW (genetic females), and only 372 showed higher expression in ZZ (genetic males) gonads (Fig. 3, Table 2). This indicates that female sex determination in Xenopus involves a robust transcriptional regulation. In contrast, in mice, during the sex determination period (between embryonic day E10.5 and E12.5), a higher number of genes were upregulated in the XY (genetic males) than in the XX (genetic females) gonads (Nef et al. 2005), which suggested that programs of sex determination may be diverse among vertebrates.
Our analysis showed that at NF53, i.e., at the beginning of sexual differentiation of Xenopus gonads, 1083 genes (i.e., 3%) showed sexually dimorphic expression (≥ 2-fold change), which was slightly lower number than at NF50 (during sex determination). One hundred ninety-three genes showed higher expression in ZW gonads, and 890 in ZZ gonads (Fig. 3, Table 2). Thus, at the onset of sexual differentiation, more genes were specifically expressed in ZZ (male) gonads than in ZW (female) gonads in Xenopus, which was opposite to the mouse, where more genes were specifically expressed in XX (female) than XY (male) gonads at the beginning of sexual differentiation (E13.5) (Nef et al. 2005). This again indicates differences in the molecular programs of gonad development among vertebrates.
At NF56, i.e., in the differentiating gonads, only 421 genes (i.e., 1.2%) showed sexually dimorphic expression (≥ 2-fold change). This stage showed the lowest percentage of genes with sexually dimorphic expression among all stages. Seventy-five genes had higher expression in ZW, and 346 in ZZ gonads (Fig. 3, Table 2). Thus, more genes were highly expressed in ZZ gonads (differentiating testes) than in ZW (differentiating ovaries). We previously showed that the testis differentiation in Xenopus is a complex process during which the basement membranes between gonadal cortex and medulla disintegrate, the cortex and medulla fuse, and the germ cells and somatic cells gather to form the testis cords (Piprek et al. 2017). This sequence of profound structural changes certainly requires an involvement of a number of different genes, which is reflected in the high number of genes expressed in ZZ gonads at this stage.
At stage NF62, the sexual dimorphism of gene expression is the most pronounced. At this stage, 3224 genes (i.e., 5%) showed sexually dimorphic expression (≥ 2-fold change). However, only 594 genes showed higher expression in ZW (ovaries), and as many as 2630 in ZZ (testes) gonads. This is the stage when the gonads of both sexes are already differentiated and fully prepared to perform their sex-specific functions, and therefore the sexual dimorphism is evident not only at structural but also at molecular level.
The expression of genes during different stages of ovary development
We found that in ZW gonads at stage NF53, in comparison to stage NF50, 376 genes had upregulated expression. The list of genes is presented in Suppl. Table 1, and chosen genes are presented in Table 3. Functional analysis grouped these genes in several distinct categories shown in Table 4. Among the upregulated genes, monoacylglycerol O-acyltransferase 2 gene 1 (mogat2.1) is involved in synthesis of diacylglycerol (DAG) that acts as a messenger lipid in cell signaling (Toker 2005); retinol-binding protein 2 (rbp2) is involved in retinoic acid regulation; extracellular proteins: collagen 2 and collagen 9, cysteine protease cathepsin K, epithelium-specific intermediate filaments: keratin 14 and keratin 19, estrogen receptor 1 (esr1), and synuclein gamma. At this early stage, the germ and somatic cells proliferate, and somatic cells start gathering in the gonad center forming medulla (Fig. 1A, C). Collagens accumulate between the gonad cortex and medulla (Piprek et al. 2017). Importantly, around stage NF50, a sex determination period takes place and gene expression analysis suggest that DAG, retinol, and estradiol may be involved in Xenopus sex determination.
Table 3.
Chosen genes up- and downregulated in ZW (genetic females) gonads at NF53 in relation to NF50 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF53 than at NF50) | |||
| A_10_P009259 | mogat2.1 | Monoacylglycerol O-acyltransferase 2.1 | 6.53907 |
| A_10_P079665 | rbp2 | Retinol-binding protein 2 | 5.67257 |
| A_10_P002950 | col9a1 | Collagen, type IX, alpha 1 | 4.86313 |
| A_10_P005551 | srpx2 | Sushi repeat–containing protein, X2 | 4.74263 |
| A_10_P000515 | bcan | Brevican | 4.008 |
| A_10_P136703 | krt14 | Keratin 14 | 3.56258 |
| A_10_P007276 | aldh3b1 | Aldehyde dehydrogenase 3 B1 | 3.5464 |
| A_10_P143593 | ctsh | Cathepsin H | 3.39144 |
| A_10_P004976 | matn4 | Matrilin 4 | 3.3843 |
| A_10_P027124 | col2a1b | Collagen, type II, alpha 1 | 3.12339 |
| A_10_P002931 | matn2 | Matrilin 2 | 3.10895 |
| A_10_P041821 | sncg-b | Synuclein, gamma b | 2.79753 |
| A_10_P032181 | sncg-a | Synuclein, gamma a | 2.7756 |
| A_10_P046256 | ctsk | Cathepsin K | 2.75751 |
| A_10_P165493 | krt19 | Keratin 19 | 2.48345 |
| A_10_P006607 | col9a3 | Collagen, type IX, alpha 3 | 2.41836 |
| A_10_P033056 | esr1-a | Estrogen receptor 1 | 2.36005 |
| A_10_P224323 | racgap1 | Rac GTPase activating protein 1 | 2.29739 |
| A_10_P036156 | dcn | Decorin | 2.25563 |
| A_10_P065984 | itga11 | Integrin, alpha 11 | 2.17377 |
| Genes downregulated (higher expression at NF50 than at NF53) | |||
| A_10_P174228 | chrd | Chordin | 11.53231 |
| A_10_P030946 | rbp4 | Retinol-binding protein 4 | 6.862097 |
| A_10_P056207 | vtn | Vitronectin | 6.558013 |
| A_10_P075910 | serpini2 | Serpin peptidase inhibitor, clade I .2 | 5.739304 |
| A_10_P008816 | serpina3 | Serpin peptidase inhibitor, clade A .3 | 4.968027 |
| A_10_P065884 | wnt10b | Wingless-type MMTV integration site 10B | 4.090623 |
| A_10_P002182 | serpinc1 | Serpin peptidase inhibitor, clade C .1 | 3.044408 |
| A_10_P009298 | igf3 | Insulin-like growth factor 3 | 3.030882 |
| A_10_P043816 | dmrt2 | Doublesex and mab-3 related transcription factor 2 | 2.872563 |
| A_10_P178123 | mafb | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B | 2.66641 |
Table 4.
Number of genes assigned to functional groups up- and downregulated in ZW (genetic female) gonads
| Functional gene groups | ZW (genetic females) | |||||
|---|---|---|---|---|---|---|
| NF53 vs. NF50 | NF56 vs. NF53 | NF62 vs. NF56 | ||||
| Up | Down | Up | Down | Up | Down | |
| Signaling factors | 20 | 61 | 7 | 8 | – | 103 |
| Calcium-binding proteins | 6 | – | 3 | – | – | – |
| Iron-binding proteins | 4 | – | – | – | – | – |
| Monooxygenases | 4 | – | – | – | – | 11 |
| Oxidoreductases | 5 | 11 | – | – | – | 22 |
| Sushi domain–containing proteins | 2 | – | – | – | – | – |
| Metalloproteinases | 3 | – | – | – | – | 8 |
| Intermediate filaments | 3 | – | – | – | – | – |
| EGF-like domain–containing proteins | 3 | – | – | – | – | – |
| ECM-receptor interaction pathway | 3 | – | – | – | – | – |
| Progesterone-mediated oocyte maturation pathway | 4 | – | – | – | 11 | – |
| Proteases | – | 12 | – | – | – | 18 |
| Hydrolases | – | 27 | – | – | – | 33 |
| Disulfide bond–containing proteins | – | – | 5 | – | – | 45 |
| Extracellular matrix components | – | – | – | 5 | – | – |
| Markers of epithelial differentiation | – | – | – | 2 | – | – |
| Meiosis regulation factors | – | – | – | – | 8 | – |
| RNA-binding proteins | – | – | – | – | 15 | – |
| Phosphoproteins | – | – | – | – | 16 | – |
| Proteins involved in development | – | – | – | – | 22 | – |
| Proteins involved in oogenesis | – | – | – | – | 3 | – |
| Cytoplasmic proteins | – | – | – | – | 35 | – |
| Cytoskeletal proteins | – | – | – | – | 12 | – |
| Proteins involved in differentiation | – | – | – | – | 9 | – |
| Nuclear proteins | – | – | – | – | 45 | – |
| Transcriptional repressors | – | – | – | – | 8 | – |
| DNA-binding proteins | – | – | – | – | 3 | – |
| Oocyte meiosis | – | – | – | – | 10 | – |
| p53 signaling | – | – | – | – | 6 | – |
| Basal transcription factors | – | – | – | – | 4 | – |
| Proteins involved in DNA Replication | – | – | – | – | 4 | – |
| Proteins involved in the formation of dorso-ventral axis | – | – | – | – | 3 | – |
| Secreted proteins | – | – | – | – | – | 23 |
| Transport proteins | – | – | – | – | – | 36 |
We also found that in ZW gonads at stage NF53, there were 1078 genes with a downregulated expression in comparison to stage NF50. All these genes are listed in Suppl. Table 2, and chosen genes are presented in Table 3. Functional analysis grouped these genes in four categories shown in Table 4. Among these downregulated genes, there were signaling protein chordin (chrd), retinol-binding protein (rbp4), several protease inhibitors serpins, signaling proteins wnt10b and igf3 (insulin-like growth factor 3), transcription factors dmrt2, and mafb (Table 3).
In developing ZW gonad at stage NF56, in comparison to stage NF53, there were 143 genes with upregulated expression (Suppl. Table 3, and chosen genes are presented in Table 5). Functional analysis grouped these genes in three categories shown in Table 4. One of important genes upregulated in this period is a neurotrophin receptor a-1 (p75NTRa) (Table 2); its role in gonad development has never been studied; however, its upregulation suggests that neurotrophins (ligands of this receptor) can play a role in ovarian differentiation. We also found that in ZW gonad at stage NF56, in comparison to stage NF53, there were 128 genes with downregulated expression (Suppl. Table 4, and chosen genes are presented in Table 5). Functional analysis grouped these genes in several categories shown in Table 4. At NF56 stage, more genes responsible for reorganization of extracellular matrix and epithelial differentiation in ZW gonads are expressed than at stage NF53. Between stages NF53 and NF56, the medulla cells disperse, which results in the formation of the cavity in the ovary center (Fig. 1E). The mechanism of this event is not known and would be interesting to study how the neurotrophins, extracellular matrix, and epithelial differentiation are involved in this process.
Table 5.
Chosen genes up- and downregulated in ZW (genetic females) gonads at NF56 in relation to NF53 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF56 than at NF53) | |||
| A_10_P259017 | sag | Arrestin | 3.6903 |
| A_10_P000364 | p75NTRa | p75 neurotrophin receptor a-1 | 3.24871 |
| Genes downregulated (higher expression at NF53 than at NF56) | |||
| A_10_P000515 | bcan | Brevican | 3.073221 |
| A_10_P136703 | krt14 | Keratin 14 | 2.897822 |
| A_10_P004976 | matn4 | Matrilin 4 | 2.768856 |
| A_10_P002950 | col9a1 | Collagen, type IX, alpha 1 | 2.713584 |
| A_10_P140568 | krt5.6 | Keratin 5, gene 6 | 2.618006 |
| A_10_P006607 | col9a3 | Collagen, type IX, alpha 3 | 2.601338 |
| A_10_P084685 | krt14 | Keratin 14 | 2.530431 |
| A_10_P038721 | col2a1b | Collagen, type II, alpha 1 | 2.509659 |
| A_10_P032181 | sncg-a | Synuclein, gamma a | 2.497221 |
In developing ZW gonad at stage NF62, in comparison to stage NF56, there were 918 genes with upregulated expression (Suppl. Table 5, and chosen genes are presented in Table 6). Functional analysis grouped these genes in the many categories (Table 4). Among known genes upregulated in the ovaries at stage NF62 are genes involved in meiosis and oocyte development, such as poly(A)-binding protein, oocyte-specific pou5f3.3, zygote arrest 1, zona pellucid proteins (zp2, zpd, zpy1), sycp3 (synaptonemal complex protein 3),and lhx8 (LIM homeobox 8). This reflects the onset of meiosis at stage NF62 and appearance of first oocytes (Fig. 1G). Also, more genes involved in the regulation of development, such as genes encoding the following: vegt protein, growth differentiation factor (gdf1), foxh1, foxr1, wnt11b, ddx25, and the survivin which prevents apoptosis, were upregulated at stage NF62 than at stage NF56.
Table 6.
Chosen genes up- and downregulated in ZW (genetic females) gonads at NF62 in relation to NF56 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF62 than at NF56) | |||
| A_10_P000661 | spdyc-b | Speedy/RINGO cell cycle regulator C | 5.91483 |
| A_10_P041271 | pabpn1l-a | Poly(A) binding protein, nuclear 1-like | 5.78779 |
| A_10_P078660 | rnf138 | Ring finger protein 138 | 5.43076 |
| A_10_P004355 | pou5f3.3 | POU class 5 homeobox 3, gene 3 | 4.82381 |
| A_10_P002029 | zar1 | Zygote arrest 1 | 4.7962 |
| A_10_P038461 | LOC398389 | Survivin | 4.75826 |
| A_10_P027361 | vegt-a | vegt protein | 4.68137 |
| A_10_P007276 | aldh3b1 | Aldehyde dehydrogenase 3 family, B1 | 4.65557 |
| A_10_P032511 | cldn6.1 | Claudin 6, gene 1 | 4.50308 |
| A_10_P162298 | zp2 | Zona pellucida glycoprotein 2 | 4.43055 |
| A_10_P009533 | gdf1 | Growth differentiation factor 1 | 4.40831 |
| A_10_P002027 | velo1 | velo1 protein | 4.36483 |
| A_10_P027280 | zpd | Zona pellucida protein D | 4.2713 |
| A_10_P205908 | foxh1 | Forkhead box H1 | 4.2256 |
| A_10_P031016 | foxr1 | Forkhead box R1 | 4.10517 |
| A_10_P008731 | wnt11b | Wingless-type MMTV integration site family, member 11B | 4.0833 |
| A_10_P033516 | zpy1 | Zona pellucida protein Y1 | 4.00754 |
| A_10_P117061 | ddx25 | DEAD box helicase 25 | 3.89223 |
| A_10_P040816 | sycp3 | Synaptonemal complex protein 3 | 3.70889 |
| A_10_P071715 | lhx8 | LIM homeobox 8 | 3.56271 |
| A_10_P056732 | dppa2 | Developmental pluripotency-assoc 2 | 3.51303 |
| A_10_P027350 | adam21 | ADAM metallopeptidase domain 21 | 2.89064 |
| Genes downregulated (higher expression at NF56 than at NF62) | |||
| A_10_P047196 | LOC100037217 | Uncharacterized LOC100037217 | 6.582348 |
| A_10_P180718 | hrg | Histidine-rich glycoprotein | 6.249551 |
| A_10_P004053 | rbp4 | Retinol-binding protein 4 | 5.794043 |
| A_10_P034336 | serpina1 | Serpin peptidase inhibitor, A1 | 5.541168 |
| A_10_P006319 | sag | Arrestin | 5.153979 |
| A_10_P075910 | serpini2 | Serpin peptidase inhibitor, I2 | 4.285183 |
| A_10_P030976 | LOC398504 | Villin-1-like | 3.897723 |
| A_10_P068493 | fetub | Fetuin B | 3.871496 |
| A_10_P110124 | krt12 | Keratin 12 | 3.5294 |
| A_10_P006916 | emx1.2 | Empty spiracles homeobox 1, gene 2 | 3.507484 |
| A_10_P002103 | mmp7 | Matrix metallopeptidase 7 | 3.50358 |
| A_10_P153143 | igf3 | Insulin-like growth factor 3 | 3.452683 |
| A_10_P003788 | igfbp1-a | Insulin-like growth factor–binding 1 | 3.06992 |
| A_10_P005507 | ctsl | Cathepsin L | 2.882569 |
| A_10_P137683 | gata2 | GATA binding protein 2 | 2.5687 |
| A_10_P053899 | cdh26 | Cadherin 26 | 2.529154 |
| A_10_P126889 | rdh16 | Retinol dehydrogenase 16 (all-trans) | 2.355785 |
| A_10_P174228 | chrd | Chordin | 2.347432 |
| A_10_P007857 | timp2 | TIMP metallopeptidase inhibitor 2 | 2.066979 |
In developing ZW gonad at stage NF62, in comparison to stage NF56, there were 1834 genes with downregulated expression (Suppl. Table 6, and chosen genes are presented in Table 6). Functional analysis grouped these genes into several categories (Table 4). Also, many (24) pathways were downregulated, including metabolic pathways, steroid hormone biosynthesis, retinol metabolism, PPAR signaling pathway, and adipocytokine signaling pathway (Table 4). Among known genes downregulated in the ovaries at stage NF62 are the following genes: retinol-binding protein 4 (rbp4), rdh16 (retinol dehydrogenase 16), several serpins, emx1.2 (empty spiracles homeobox 1), igf3 (insulin-like growth factor 3), igfbp1-a (insulin-like growth factor–binding protein 1), gata2 (GATA binding protein 2), and chordin. This indicates that retinol pathway and insulin-like growth factor pathway are downregulated at a later stage of ovarian development (NF62), and that these two pathways may be important for earlier stages of ovarian development. The PPAR signaling pathway and adipocytokine signaling pathway are involved in fat tissue differentiation (Ogunyemi et al. 2013) and are probably important for the development of corpora adiposa (fat tissue) at the anterior edges of the developing gonads at stages before NF62. Thus, after the fat tissue had been formed, these pathways are downregulated at stage NF62.
Another interesting gene expressed at the onset of gonadogenesis (NF50), showing upregulation at NF53 and downregulated at NF62 is chordin (chrd). Several studies showed that this gene is crucial for early organogenesis (dorsalization, gastrulation, and head development (Pappano et al. 1998; Bachiller et al. 2000), but its role in gonad development is unknown. Overall, our gene expression analysis showed that the later development of the ovary (NF62) is a very transcriptionally active period (many genes become upregulated and downregulated between NF56 and NF62), which may be related to the initialization of meiosis and oocyte formation during this developmental period.
The expression of genes during different stages of testis development
Our analysis showed that in the genetic male (ZZ) gonads at stage NF53, i.e., at the beginning of sexual differentiation, there were 659 genes with upregulated expression in comparison to the stage NF50 gonad (Suppl. Table 7, and chosen genes are presented in Table 7). Functional analysis grouped these genes into several categories (Table 8). There were the following genes with known function: igf3 (insulin-like growth factor 3), rbp4 (retinol-binding protein 4), vtn (vitronectin), several serpins, esr2 (estrogen receptor 2), several components of extracellular matrix (collagen 9, matrilin 2), and extracellular matrix (timp3) enzymes. A role of these genes in the early phase of ZZ gonad development is not known, and it would be interesting to study if retinol and/or igf3 are involved in male sex determination in Xenopus. Upregulation of PPAR and adipocytokine signaling pathways, characteristic for fat tissue, possibly reflects the onset of the development of the fat bodies at the anterior edge of the gonad.
Table 7.
Chosen genes up- and downregulated in ZZ (genetic males) gonads at NF53 in relation to NF50 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF53 than at NF50) | |||
| A_10_P030946 | rbp4 | Retinol-binding protein 4 | 4.97523 |
| A_10_P056207 | vtn | Vitronectin | 4.51992 |
| A_10_P075910 | serpini2 | Serpin peptidase inhibitor, clade I. 2 | 4.4381 |
| A_10_P041856 | igf3 | Insulin-like growth factor 3 | 4.34284 |
| A_10_P003882 | timp3 | TIMP metallopeptidase inhibitor 3 | 2.37097 |
| A_10_P007964 | serpinf2 | Serpin peptidase inhibitor, F2 | 2.32024 |
| A_10_P030126 | esr2 | Estrogen receptor 2 (ER beta) | 2.28938 |
| A_10_P058537 | col9a1-b | Collagen, type IX, alpha 1 | 2.24147 |
| A_10_P048579 | ocln-b | Occludin | 2.22878 |
| A_10_P002931 | matn2 | Matrilin 2 | 2.06945 |
| Genes downregulated (higher expression at NF50 than at NF53) | |||
| A_10_P017957 | ocm | Oncomodulin | 6.204741 |
| A_10_P140568 | krt5.6 | Keratin 5, gene 6 | 4.866387 |
| A_10_P138508 | krt15 | Keratin 15 | 4.154655 |
| A_10_P126949 | mmp1 | Matrix metallopeptidase 1 | 2.745253 |
| A_10_P008082 | fgfbp1 | Fibroblast growth factor–binding 1 | 2.66819 |
| A_10_P203798 | lum | Lumican | 2.460273 |
| A_10_P222743 | isyna1-b | Inositol-3-phosphate synthase 1 | 2.399986 |
| A_10_P002391 | capn8-a | Calpain 8 | 2.388139 |
| A_10_P040276 | wnt7b | Wingless-type MMTV integration site family, member 7B | 2.038541 |
Table 8.
Number of genes assigned to functional groups up- and downregulated in ZZ (genetic male) gonads
| Functional gene groups | ZZ (genetic males) | |||||
|---|---|---|---|---|---|---|
| NF53 vs NF50 | NF56 vs NF53 | NF62 vs NF56 | ||||
| Up | Down | Up | Down | Up | Down | |
| Signaling factors | 48 | – | 13 | 43 | 17 | – |
| Calcium-binding proteins | – | – | 5 | – | – | – |
| Metal-binding proteins | 30 | – | – | 21 | – | – |
| Monooxygenases | – | – | – | – | 3 | 8 |
| Oxidoreductases | 9 | – | – | 8 | 5 | 14 |
| Metalloproteinases | 4 | – | – | 3 | – | – |
| EGF-like domain–containing proteins | 4 | – | – | – | – | – |
| Proteases | 13 | – | – | 14 | 9 | – |
| Hydrolases | 20 | – | – | 25 | 12 | – |
| Disulfide bond–containing proteins | 35 | – | – | 31 | 13 | – |
| Secreted proteins | – | – | – | 12 | – | – |
| Transport proteins | – | 3 | – | – | – | – |
| Steroid hormone synthesis pathway | 4 | – | – | – | 2 | – |
| Insulin signaling pathway | 4 | – | – | 7 | – | – |
| PPAR signaling pathway | 4 | – | – | 3 | – | – |
| Adipocytokine signaling pathway | 5 | – | – | 6 | – | – |
| Mitochondrial proteins | – | 7 | – | – | – | – |
| Ion transport | – | 5 | – | – | – | – |
| Terpenoid backbone biosynthesis pathway | – | 3 | – | – | – | 3 |
| ER protein processing pathway | – | 5 | – | – | – | – |
| Receptors | – | – | 6 | – | – | – |
| Metabolic pathway | – | – | – | 23 | 8 | – |
| FoxO signaling pathway | – | – | – | 7 | – | 45 |
| Cell membrane proteins | – | – | – | – | – | 48 |
| Intercellular transport | – | – | – | – | – | 21 |
| Retinol metabolism | – | – | – | – | – | 5 |
Our analysis also showed that in the genetic male (ZZ) gonads at stage NF53, there were 436 genes with downregulated expression (Suppl. Table 8, and chosen genes are presented in Table 7). Functional analysis grouped these genes in the categories shown in Table 8.
Comparison of gene expression level in the ZZ gonads between stage NF56 and NF53 showed that at stage NF56, there were 340 genes with upregulated expression (Suppl. Table 9, and chosen genes are presented in Table 9). Functional analysis grouped these genes in categories shown in Table 8. Some of these upregulated genes are rbp2 (retinol-binding protein 2), receptor of prostaglandin E (ptger3), stromal cell-derived factor 2-like 1 (sdf2l1), and neurotrophin receptor (p75NTRa). Further, studies are necessary to establish what is the exact role of the prostaglandin E, retinol, and neurotrophins in testis differentiation. Importantly, around NF53-NF56, the cortex and medulla fuse in differentiating testes, and the germ cells lose their connection with the superficial coelomic epithelium and disperse in the whole testis (Fig. 1F). There were also 340 genes downregulated at stage NF56 ZZ gonad in comparison to stage NF53 gonad (Suppl. Table 10, and chosen genes are presented in Table 9). Functional analysis grouped these genes into several categories (Table 8).
Table 9.
Chosen genes up- and downregulated in ZZ (genetic males) gonads at NF56 in relation to NF53 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF56 than at NF53) | |||
| A_10_P079665 | rbp2 | Retinol-binding protein 2, cellular | 6.44222 |
| A_10_P043951 | ptger3 | Prostaglandin E receptor 3 | 2.75218 |
| A_10_P036706 | sdf2l1 | Stromal cell-derived factor 2-like 1 | 2.32111 |
| A_10_P000364 | p75NTRa | p75 neurotrophin receptor a-1 | 2.02373 |
| Genes downregulated (higher expression at NF53 than at NF56) | |||
| A_10_P036346 | LOC100189571 | Uncharacterized LOC100189571 | 8.899836 |
| A_10_P102465 | rbp4 | Retinol-binding protein 4 | 7.963364 |
| A_10_P056207 | vtn | Vitronectin | 7.657184 |
| A_10_P027027 | ptx | Pentraxin | 7.367071 |
| A_10_P041856 | igf3 | Insulin-like growth factor 3 | 3.440657 |
| A_10_P002182 | serpinc1 | Serpin peptidase inhibitor C1 | 2.826371 |
| A_10_P094993 | krt12 | Keratin 12 | 2.032246 |
Comparison of gene expression level in the ZZ gonads between stages NF62 and NF56 showed that at stage NF62 gonad, there were 334 genes with the upregulated expression (Suppl. Table 11, and chosen genes are presented in Table 10). Functional analysis grouped these genes into several categories (Table 8). Around stage NF56-NF62, cells group into the testis cords (Fig. 1H). Genes involved in this process are not known, and presumably, the genes upregulated at this stage may be responsible for the formation of testis cords. There were also 831 genes downregulated in ZZ gonad at stage NF62 in comparison to stage NF56 (Suppl. Table 12, and chosen genes are presented in Table 10, and the gene categories, which were analyzed functionally are shown in Table 8).
Table 10.
Chosen genes up- and downregulated in ZZ (genetic males) gonads at NF62 in relation to NF56 stage
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| Genes upregulated (higher expression at NF62 than at NF56) | |||
| A_10_P049320 | prss1 | Protease, serine, 1 | 6.7897 |
| A_10_P045961 | prss3 | Protease, serine, 3 | 6.64558 |
| A_10_P259137 | tfip11 | Tuftelin-interacting protein 11 | 6.14112 |
| A_10_P027545 | mmp11 | Matrix metallopeptidase 11 | 3.31378 |
| A_10_P027246 | klf9-a | Kruppel-like factor 9 | 2.7011 |
| A_10_P203798 | lum | Lumican | 2.10476 |
| Genes downregulated (higher expression at NF56 than at NF62) | |||
| A_10_P032408 | ocm.2 | Oncomodulin | 7.542298 |
| A_10_P004053 | rbp4 | Retinol-binding protein 4 | 5.656075 |
| A_10_P000084 | krt5.5 | Keratin 5, gene 5 | 4.410092 |
| A_10_P003972 | mmp28-b | Matrix metallopeptidase 28 | 3.191715 |
| A_10_P044151 | fgfr4-b | Fibroblast growth factor receptor 4 | 3.091348 |
| A_10_P002657 | isyna1-a | Inositol-3-phosphate synthase 1 | 3.030967 |
| A_10_P094993 | krt12 | Keratin 12 | 2.838533 |
Genes with sexual dimorphism of expression in ZW and ZZ gonads in different developmental stages
The master sex-determining gene in Xenopus the dm-w was discovered in 2008 (Yoshimoto et al. 2008), but the molecular machinery of sex determination is certainly very complex and contains many other genes. We previously published the expression profile of known genes involved in sex determination and sexual differentiation in the Xenopus gonads (Piprek et al. 2018). We showed that the gata4, sox9, dmrt1, amh, fgf9, ptgds, pdgf, fshr, and cyp17a1 had upregulated expression in testes, while dm-w, fst, foxl2, and cyp19a1 had upregulated expression in the ovary (Piprek et al. 2018).
Here, we compared gene expression level between ZW and ZZ gonads at different stages of gonad development. These analyses showed that at stage NF50 (undifferentiated gonads during sex determination period), there were 820 genes with upregulated expression in ZW gonad (Suppl. Table 13, and chosen genes are shown in Table 12). Functional analysis grouped these genes into several categories (Table 11). Many genes upregulated in this period are uncharacterized. Among known genes upregulated in ZW gonad at stage NF50 is chordin (chrd). Chordin is a secreted protein responsible for several developmental processes such as dorsalization, head development, and gastrulation (Sasai et al. 1994; Pappano et al. 1998; Bachiller et al. 2000); our study indicates that it may play a crucial role in female sex determination (Table 12, Suppl. Table 13). Other genes upregulated in ZW gonad at NF50 are two protease inhibitors, serpin A3 and serpin I2, extracellular glycoprotein vitronectin, metalloproteinases mmp7 and adam27, retinol-binding protein rbp4, signaling molecules wnt10b, wnt11b, and igf3, helicase ddx25, and transcription factors foxa2 and lhx8. A role of these factors in sex determination in Xenopus is unknown and requires further study.
Table 12.
Chosen genes upregulated in ZW (genetic females) in relation to ZZ (genetic males) gonads at NF50 stage [higher gene expression level in ZW than in ZZ gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P174228 | chrd | Chordin | 11.30213 |
| A_10_P007346 | MGC85508 | MGC85508 protein | 8.151194 |
| A_10_P008816 | serpina3 | Serpin peptidase inhibitor, clade A3 | 6.774417 |
| A_10_P075910 | serpini2 | Serpin peptidase inhibitor, clade I2 | 6.378762 |
| A_10_P233398 | vtn | Vitronectin | 5.368433 |
| A_10_P187778 | wnt11b | Wingless-type MMTV integration site family, member 11B | 5.00604 |
| A_10_P004053 | rbp4 | Retinol-binding protein 4, plasma | 4.876474 |
| A_10_P065884 | wnt10b | Wingless-type MMTV integration site family, member 10B | 4.20504 |
| A_10_P027350 | adam21 | ADAM metallopeptidase domain 21 | 3.851196 |
| A_10_P009298 | igf3 | Insulin-like growth factor 3 | 3.848738 |
| A_10_P202038 | MGC69070 | Matrix metalloproteinase 7 | 3.690095 |
| A_10_P006376 | anxa13 | Annexin A13 | 3.483353 |
| A_10_P003549 | MGC69070 | Matrix metalloproteinase 7 | 3.459862 |
| A_10_P000388 | ddx25 | DEAD box helicase 25 | 3.239557 |
| A_10_P082395 | foxa2 | Forkhead box A2 | 3.049031 |
| A_10_P003648 | lhx8 | LIM homeobox 8 | 2.965778 |
Table 11.
Number of genes assigned to functional groups expressed at higher level in ZW and ZZ gonads
| Functional gene groups | NF50 | NF53 | NF56 | NF62 | ||||
|---|---|---|---|---|---|---|---|---|
| ZW | ZZ | ZW | ZZ | ZW | ZZ | ZW | ZZ | |
| Signaling factors | 64 | 18 | – | 50 | 18 | – | – | 73 |
| Calcium-binding proteins | – | – | – | – | 3 | – | – | – |
| Metal-binding proteins | 28 | – | – | 26 | – | – | – | – |
| Metalloproteinases | 7 | – | – | – | – | – | – | – |
| Progesterone-mediated oocyte maturation pathway | – | – | – | – | – | – | 8 | – |
| Proteases | 20 | – | – | 9 | – | – | – | – |
| Hydrolases | 28 | – | – | 21 | – | – | – | 25 |
| Disulfide bond–containing proteins | 42 | – | – | 34 | 10 | 6 | – | 52 |
| Extracellular matrix components | – | 3 | – | – | – | – | – | – |
| Markers of epithelial differentiation | – | 2 | – | – | – | – | – | – |
| Meiosis regulation factors | – | – | – | – | – | – | 4 | – |
| Oocyte meiosis | – | – | – | – | – | – | 7 | – |
| RNA-binding proteins | – | – | – | – | – | – | 11 | – |
| Phosphoproteins | – | – | – | – | – | – | 11 | – |
| Proteins involved in development | – | – | – | – | – | – | 18 | 19 |
| Cytoplasmic proteins | – | – | – | – | – | – | 30 | – |
| Cytoskeletal proteins | – | – | – | – | – | – | 10 | – |
| Nuclear proteins | – | – | – | – | – | – | 35 | – |
| p53 signaling | – | – | – | – | – | – | 6 | – |
| Secreted proteins | 15 | 7 | – | 14 | 6 | – | – | 19 |
| Transport proteins | – | – | – | – | – | 5 | – | – |
| Metabolic pathway | 14 | – | – | 33 | – | – | – | – |
| Intermediate filaments | – | 3 | – | – | – | – | – | – |
| Mitochondrial proteins | – | 5 | – | – | – | – | – | – |
| Insulin signaling pathway | – | – | – | 7 | – | – | – | – |
| Steroid hormone synthesis | – | – | – | 3 | – | – | – | 3 |
| Adipocytokine signaling pathway | – | – | – | 4 | – | – | – | – |
| FoxO signaling pathway | – | – | – | 8 | – | – | – | – |
| Cell membrane proteins | – | – | – | – | – | 5 | – | 63 |
| Cell junction proteins | – | – | – | – | – | 4 | – | – |
| Ion channel proteins | – | – | – | – | – | 4 | – | – |
| Cell division proteins | – | – | – | – | – | – | 10 | – |
| Mitotic proteins | – | – | – | – | – | – | 6 | – |
| Wnt signaling pathway | – | – | – | – | – | – | – | 5 |
There were 372 genes with higher expression level in the ZZ (genetic males) gonads at stage NF50 (Suppl. Table 14, and chosen genes are shown in Table 13, and the functional groups are shown in Table 11). Among these upregulated genes are known genes such as epithelium markers keratin 5, 12, and 14, coiled-coil domain containing 50 (ccdc50) that acts as an effector in EGF signaling and negative regulator of NF-kB factor (Tsukiyama et al. 2012), signaling molecules: wnt3a, wnt7b, growth differentiation factor 3 (gdf3), fibroblast growth factor–binding protein 1 (fgfbp1), proteases cathepsin K and H, extracellular matrix molecules lumican, collagen IX and I, and decorin. A role of these genes in male sex determination and early testis development remains unknown.
Table 13.
Chosen genes downregulated in ZW in relation to ZZ gonads at NF50 stage [higher gene expression level in ZZ than in ZW gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P136703 | krt14 | Keratin 14 | 7.50258 |
| A_10_P183185 | ccdc50 | Coiled-coil domain containing 50 | 6.57626 |
| A_10_P140568 | krt5.6 | Keratin 5, gene 6 | 5.84154 |
| A_10_P003366 | lum | Lumican | 5.75697 |
| A_10_P193923 | krt14 | Keratin 14 | 5.1494 |
| A_10_P008082 | fgfbp1 | Fibroblast growth factor–binding protein 1 | 3.66899 |
| A_10_P002950 | col9a1 | Collagen, type IX, alpha 1 | 3.07046 |
| A_10_P046256 | ctsk | Cathepsin K | 2.60816 |
| A_10_P036156 | dcn | Decorin | 2.60212 |
| A_10_P244713 | col1a1 | Collagen, type I, alpha 1 | 2.56712 |
| A_10_P040276 | wnt7b | Wingless-type MMTV integration site family, member 7B | 2.3506 |
| A_10_P026995 | wnt3a | Wingless-type MMTV integration site family, member 3A | 2.25544 |
| A_10_P094993 | krt12 | Keratin 12 | 2.23416 |
| A_10_P000272 | gdf3 | Growth differentiation factor 3 | 2.07545 |
| A_10_P046876 | ctsh | Cathepsin H | 2.01352 |
There are 193 genes with a higher expression in ZW (genetic females) gonad at stage NF53 (the onset of sexual differentiation of gonads) (Suppl. Table 15, and chosen genes are shown in Table 14). Functional analysis did not link these genes to any specific pathway. Among these upregulated genes, there are the following known genes: retinol-binding protein 2 (rbp2), protease calpain 8, synuclein gamma with unknown function, cell adhesion gene claudin 6, metalloproteinases mmp1 and adam21, and galectin-la involved in cell adhesion and signaling.
Table 14.
Chosen genes upregulated in ZW in relation to ZZ gonads at NF53 stage [higher gene expression level in ZW than in ZZ gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P079665 | rbp2 | Retinol-binding protein 2 | 5.229889 |
| A_10_P032636 | LOC100101274 | Uncharacterized LOC100101274 | 3.804135 |
| A_10_P062524 | lgalsia-a | Galectin-Ia | 2.939513 |
| A_10_P008579 | krt5.2 | Keratin 5, gene 2 | 2.846329 |
| A_10_P057292 | sncg-a | Synuclein, gamma | 2.52171 |
| A_10_P002391 | capn8-a | Calpain 8 | 2.404349 |
| A_10_P032511 | cldn6.1 | Claudin 6, gene 1 | 2.207111 |
| A_10_P027350 | adam21 | ADAM metallopeptidase domain 21 | 2.177794 |
| A_10_P126949 | mmp1 | Matrix metallopeptidase 1 | 2.07391 |
There were 890 genes with higher expression in ZZ (genetic males) gonad at stage NF53 (the onset of sexual differentiation of gonads) (Suppl. Table 16, and chosen genes are shown in Table 15). Functional analysis grouped these genes into several categories (Table 11). The upregulated known genes are coiled-coil domain containing 50 (ccdc50), retinol-binding protein 4 (rbp4), signaling molecules igf1 and igf3, estrogen receptor 2 (esr2), transcription factors, Kruppel-like factor 9 (klf9), Kruppel-like factor 15 (klf15), and foxo1 (forkhead box O1), enzyme glycerophosphodiester phosphodiesterase 1 (gde1) responsible for synthesis of signaling molecule lysophosphatidic acid (LPA), cell adhesion proteins gap junction protein alpha 3 (gja3), occluding (ocln), and extracellular matrix component vitronectin (vtn).
Table 15.
Chosen genes downregulated in ZW in relation to ZZ gonads at NF53 stage [higher gene expression level in ZZ than in ZW gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P183185 | ccdc50 | Coiled-coil domain containing 50 | 7.79896 |
| A_10_P009082 | gde1 | Glycerophosphodiester phosphodiesterase 1 | 6.52222 |
| A_10_P030946 | rbp4 | Retinol-binding protein 4, plasma | 5.8968 |
| A_10_P233398 | vtn | Vitronectin | 5.85368 |
| A_10_P009298 | igf3 | Insulin-like growth factor 3 | 3.50579 |
| A_10_P002488 | gja3 | Gap junction protein, alpha 3, 46 kDa | 3.2409 |
| A_10_P001965 | klf15 | Kruppel-like factor 15 | 2.9746 |
| A_10_P027246 | klf9-a | Kruppel-like factor 9 | 2.74415 |
| A_10_P030126 | esr2 | Estrogen receptor 2 (ER beta) | 2.42444 |
| A_10_P027093 | igf1 | Insulin-like growth factor 1 | 2.40431 |
| A_10_P000763 | foxo1 | Forkhead box O1 | 2.14949 |
| A_10_P048579 | ocln-b | Occludin | 2.12035 |
There were 75 genes with higher expression in ZW (genetic females) gonad at stage NF56 (Suppl. Table 17, and chosen genes are shown in Table 16, and the functional groups are shown in Table 11). Among known genes are retinoic binding protein 4 and vitronectin.
Table 16.
Chosen genes upregulated in ZW versus ZZ gonads at NF56 stage [higher gene expression level in ZW than in ZZ gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P036346 | LOC100189571 | Uncharacterized LOC100189571 | 5.456322 |
| A_10_P056207 | vtn | Vitronectin | 4.026518 |
| A_10_P030946 | rbp4 | Retinol-binding protein 4, plasma | 3.555976 |
There were 346 genes with higher expression in ZZ (genetic males) gonad at stage NF56 (Suppl. Table 18, and chosen genes are shown in Table 17, and the functional groups are shown in Table 11). Among known genes are keratin 14 and 15, cell molecule gap junction protein, alpha (gja3), endophilin B2 (sh3glb2) and coiled-coil domain containing 50 (ccdc50).
Table 17.
Chosen genes downregulated in ZW in relation to ZZ gonads at NF56 stage [higher gene expression level in ZZ than in ZW gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P084685 | krt14 | Keratin 14 | 3.53568 |
| A_10_P171263 | sh3glb2 | SH3-domain GRB2-like endophilin B2 | 3.50581 |
| A_10_P183185 | ccdc50 | Coiled-coil domain containing 50 | 3.3565 |
| A_10_P138508 | krt15 | Keratin 15 | 3.13432 |
| A_10_P002488 | gja3 | Gap junction protein, alpha 3, 46 kDa | 2.0898 |
There were 594 genes with higher expression in ZW (genetic females) gonad at stage NF62 (Suppl. Table 19, and chosen genes are shown in Table 18, and the functional groups are shown in Table 11). Many genes expressed at this stage such as zona pellucida glycoprotein 4 (zp4) and zona pellucida C glycoprotein (xlzpc) are involved in ovarian follicles and oocytes formation and development. Other genes with upregulated expression at this stage were enzyme arachidonate 12-lipoxygenase 12R type (alox12b) responsible for metabolism of a signal compound—arachidonic acid (ARA), signaling factors such as growth differentiation factor 1 (gdf1), Wnt11b, cell adhesion molecules claudin 6 and connexin 38, transcription factors foxr1 and foxh1, and survivin—an inhibitor of apoptosis.
Table 18.
Chosen genes upregulated in ZW versus ZZ gonads at NF62 stage [higher gene expression level in ZW than in ZZ gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P009488 | alox12b | Arachidonate 12-lipoxygenase, 12R | 5.326002 |
| A_10_P031553 | zp4-a | Zona pellucida glycoprotein 4 | 4.350343 |
| A_10_P032511 | cldn6.1 | Claudin 6, gene 1 | 4.081705 |
| A_10_P038461 | LOC398389 | Survivin | 3.781997 |
| A_10_P034497 | kpna2 | Importin alpha 1b | 3.634424 |
| A_10_P048511 | foxh1 | Forkhead box H1 | 3.486778 |
| A_10_P009533 | gdf1 | Growth differentiation factor 1 | 3.42356 |
| A_10_P031016 | foxr1 | Forkhead box R1 | 3.378015 |
| A_10_P005051 | xlzpc | Zona pellucida C glycoprotein | 2.893829 |
| A_10_P205908 | foxh1 | Forkhead box H1 | 2.859015 |
| A_10_P004066 | LOC397866 | Connexin 38 | 2.845325 |
| A_10_P008731 | wnt11b | Wingless-type MMTV integration site family, member 11B | 2.404891 |
There were 2630 genes with upregulated expression in ZZ (genetic males) gonad at stage NF62 (Suppl. Table 20, and chosen genes are shown in Table 19). Functional analysis grouped these into many categories (Table 11). Among known genes with upregulated expression were factors involved in signaling and signaling pathways: igf1, desert hedgehog (dhh), sonic hedgehog (shh), indian hedgehog (ihh), wnt3a, wnt8b, wnt7b, Janus kinase 2 (jak2), frizzled receptor 4 and 10 (fzd4, fzd10), cellular retinoic acid–binding protein 2 (crabp2), SMAD family member 4 (smad4); proteases: serine protease 3 (prss3), cathepsin H (ctsh), peptidase inhibitor—serpini2; transcription factors: LIM homeobox 1 (lhx1), homeobox a9, d10, and d13 (hoxa9, hoxd10, hoxd13), foxf1, foxa2, gata2; extracellular matrix components: collagen III (col3a1), collagen I (col1a1), fibrillin 3 (fbn3); extracellular matrix enzymes: mmp2, mmp16, cell adhesion molecule 3 (cadm3); and intermediate filaments: keratin 15 and nestin (nst).
Table 19.
Chosen genes downregulated in ZW in relation to ZZ gonads at NF62 stage [higher gene expression level in ZZ than in ZW gonads]
| Probe name | Gene symbol | Gene name | Log FC |
|---|---|---|---|
| A_10_P077615 | MGC116439 | Uncharacterized protein MGC116439 | 8.36828 |
| A_10_P045961 | prss3 | Protease, serine, 3 | 7.894 |
| A_10_P075910 | serpini2 | Serpin peptidase inhibitor, clade I2 | 4.04603 |
| A_10_P143593 | ctsh | Cathepsin H | 3.91699 |
| A_10_P041916 | smad4.1 | SMAD family member 4, gene 1 | 3.54869 |
| A_10_P186858 | lhx1 | LIM homeobox 1 | 3.43296 |
| A_10_P067362 | igf1 | Insulin-like growth factor 1 | 3.21852 |
| A_10_P037301 | dhh-b | Desert hedgehog | 3.07896 |
| A_10_P004008 | hoxd10 | Homeobox D10 | 2.92652 |
| A_10_P036201 | krt15 | Keratin 15 | 2.86458 |
| A_10_P027055 | shh | Sonic hedgehog | 2.84344 |
| A_10_P047936 | hoxd13 | Homeobox D13 | 2.7812 |
| A_10_P026995 | wnt3a | Wingless-type MMTV integration site family, member 3A | 2.78056 |
| A_10_P002038 | mmp16 | Matrix metallopeptidase 16 | 2.77842 |
| A_10_P137013 | col3a1 | Collagen, type III, alpha 1 | 2.75957 |
| A_10_P143748 | crabp2 | Cellular retinoic acid–binding protein 2 | 2.74791 |
| A_10_P116556 | wnt8b | Wingless-type MMTV integration site family, member 8B | 2.72865 |
| A_10_P139638 | nes | Nestin | 2.71329 |
| A_10_P000674 | foxf1-a | Forkhead box F1 | 2.69515 |
| A_10_P232633 | fbn3 | Fibrillin 3 | 2.64504 |
| A_10_P002666 | cadm3 | Cell adhesion molecule 3 | 2.53779 |
| A_10_P040276 | wnt7b | Wingless-type MMTV integration site family, member 7B | 2.52685 |
| A_10_P016774 | foxa2 | Forkhead box A2 | 2.48915 |
| A_10_P050489 | jak2 | Janus kinase 2 | 2.48864 |
| A_10_P000087 | fzd10-a | Frizzled class receptor 10 | 2.46006 |
| A_10_P267657 | col1a1 | Collagen, type I, alpha 1 | 2.43227 |
| A_10_P162773 | gata2 | GATA binding protein 2 | 2.41128 |
| A_10_P141938 | hoxa9 | Homeobox A9 | 2.3827 |
| A_10_P000694 | fzd4 | Frizzled class receptor 4 | 2.3253 |
| A_10_P027230 | ihh | Indian hedgehog | 2.11478 |
| A_10_P164973 | mmp2 | Matrix metallopeptidase 2 | 2.05927 |
Genes identified here that showed sexual dimorphism of expression can be categorized into several functional groups: (1) signaling molecules: chordin (upregulated in ♀), wnt3a (upregulated in ♂), wnt7b (♂), wnt8b (♂), wnt10b (♀), wnt11b (♀), igf1 (♂), igf3 (♀ and ♂), gdf1 (♀), gdf3 (♂), ccdc50 (effector in EGF pathway) (♂), including hedgehog factors (♂): dhh, shh, ihh; (2) retinoic binding proteins: rbp2 (♀), rbp4 (♀ and ♂); (3) enzymes involved in signaling: enzyme glycerophosphodiester phosphodiesterase 1 (gde1) responsible for synthesis of signaling molecule lysophosphatidic acid (LPA) (♂), enzyme arachidonate 12-lipoxygenase 12R type (alox12b) responsible for metabolism of a signal compound—arachidonic acid (♀); (4) receptors of wnt signaling: fzd4 (♂), fzd10 (♂); (5) proteases: cathepsin H (♂), cathepsin K (♂), calpain 8 (♀); (6) protease inhibitors: serpin A3 (♀), serpin I2 (♀ and ♂); (7) transcription factors: foxa2 (♀), foxf1 (♂), foxh1 (♀), foxo1 (♂), foxr1 (♀), lhx1 (♂), lhx8 (♀), gata2 (♂), Kruppel-like factor 9 (klf9) (♂), Kruppel-like factor 15 (klf15) (♂); (8) helicase: ddx25 (♀); (9) cell adhesion molecules: occludin (♂), claudin 6 (♀),galectin-a (♀); (10) extracellular matrix components (mainly in ♂): collagens 1,3,9 (♂), vitronectin (♂), decorin (♂), lumican (♂), fibrillin 3 (♂); (11) extracellular matrix enzymes: mmp1 (♀), mmp2 (♂), mmp7 (♀), mmp16 (♂), adam21 (♀), adam27 (♀); (12) oocyte-specific proteins (♀): zp4, xlzpc; (13) epithelium-specific intermediate filaments (♂): keratins 5, 12, 14, 15.
The changes in the level of the expression of several genes listed above indicate that EGF signaling and lysophosphatidic acid (LPA) signaling may be involved in testis differentiation, arachidonic acid signaling may be involved in ovarian differentiation, while the wnt signaling, insulin-like growth factor signaling, and retinol signaling may be involved in gonad development in both sexes.
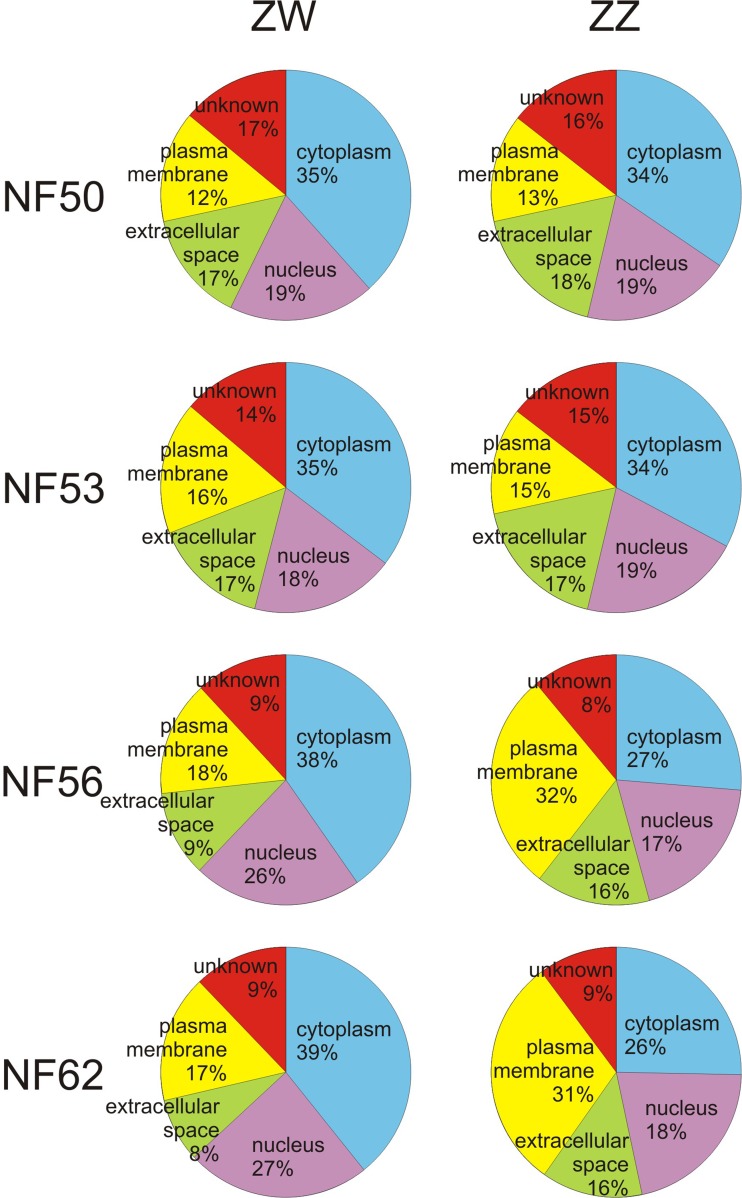
Interestingly, from the moment of sexual differentiation (after stage NF53), the genes encoding cytoplasmic and nuclear proteins are upregulated in ZW gonads (developing ovaries), while the genes encoding cell membrane proteins are upregulated in ZZ gonads (developing testes) (Fig. 4). The same trend was noted during gonad development in Silurana tropicalis (Haselman et al. 2015). This indicates that there are important molecular differences between developing ovaries and testes.
Fig. 4.
Subcellular distribution of gene products (obtained from the Ingenuity Pathway Analysis)
Comparison of sex-specifically expressed genes in developing gonads of Xenopus and other vertebrates
We compared Xenopus microarray data to the published microarray data of developing gonads in other vertebrates: mouse (Jameson et al. 2012), chicken (Ayers et al. 2015), a red-eared slider Trachemys scripta (Czerwinski et al. 2016), American alligator (Yatsu et al. 2016)—both species with temperature-dependent sex determination, and zebrafish (Sreenivasan et al. 2008). The comparison is shown in Tables 20, 21, and 22.
Table 20.
Comparison of sex-specifically expressed genes in developing gonads of Xenopus and mouse
| Gene | Xenopus laevis (this paper) | Mouse (Jameson et al. 2012) |
|---|---|---|
| Wnt3, Wnt7, Wnt8, Wnt10, Wnt11, chordin | Sexual dimorphism | No sexual dimorphism |
| Igf1 | Higher in ZZ | Higher in XX |
| Gdf1 | Higher in ZW | Not expressed |
| Igf3, Gdf3 | Higher in ZZ | Not expressed |
| Ccdc50 | Higher in ZZ | No sexual dimorphism |
| Dhh, Shh, Ihh | Higher in ZZ | Only Dhh expressed |
| Rbp | rbp2 higher in ZW and rbp4 in ZZ and ZW | Rbp1 (in XX) and Rbp4 (in XY) |
| Gde1 | Higher in ZZ | No sexual dimorphism |
| Alox12b | Higher in ZW | Not expressed |
| serpins | Several expressed | Not expressed |
| Cathepsin H (ctsh) | ctsh higher in ZZ | Only Ctsh higher in XY |
| Foxo1 | Higher in ZZ | Higher in XY |
| Lhx1 | Higher in ZZ | Higher in XY |
| Col9 | Higher in ZZ | Higher in XY |
| MMP2 | Higher in ZZ | Higher in XY |
| calpain 8 (Capn8) | Higher in ZW | Not expressed |
Table 21.
Comparison of sex-specifically expressed genes in developing gonads of Xenopus and chicken
| Gene | Xenopus laevis (this paper) | Chicken (Ayers et al. 2015) |
|---|---|---|
| calpain 5 (Capn5) | No sexual dimorphism | Higher in ZW |
| gpr56 | Not expressed | Higher in ZW |
| fgfr3 | Not expressed | Higher in ZW |
Table 22.
Comparison of sex-specifically expressed genes in developing gonads of Xenopus and red-eared slider (Trachemys scripta), American alligator, and zebrafish
| Gene | Xenopus laevis (this paper) | Red-eared slider (Czerwinski et al. 2016) | American alligator (Yatsu et al. 2016) | Zebrafish (Sreenivasan et al. 2008) |
|---|---|---|---|---|
| fdxr2 | Slightly higher in ZZ | Higher at male-producing temperature | – | – |
| hspb6 | Slightly higher in ZZ | Higher at female-producing temperature | – | – |
| twist1 | Slightly higher in ZZ | Higher at female-producing temperature | – | – |
| nov, pcsk6 | No sexual dimorphism | Higher at male-producing temperature | – | – |
| vwa2, rbm20 | Not expressed | Higher at male-producing temperature | – | – |
| frank1, avil | Not expressed | Higher at female-producing temperature | – | – |
| kdm6b | Not expressed | Higher at male-producing temperature | – | – |
| wnt11 | Higher in ZW | – | Higher at male-producing temperature | – |
| Estrogen receptor 2 esr2 |
Higher in ZZ | – | – | Higher in testes |
The transcriptome of developing mouse gonad did not show the expression of Wnt3, Wnt7, Wnt8, Wnt10, Wnt11, and chordin (Jameson et al. 2012), which were expressed in Xenopus developing gonads. The Igf1 was expressed in XX (genetic females) mouse gonads at a higher level than in XY gonads (Jameson et al. 2012); however, in Xenopus, this gene was expressed in ZZ developing gonads (genetic males). In mouse, in contrast to Xenopus (data presented in this study), the developing gonads did not express the Igf3, Gdf1, and Gdf3 (Jameson et al. 2012). The Ccdc50 was expressed in the developing mouse gonads but did not show sexual dimorphism of expression (Jameson et al. 2012). In Xenopus, this gene had an upregulated expression in ZZ gonads. Among hedgehog growth factors, in developing mouse gonads, only the dhh was expressed (Jameson et al. 2012). In Xenopus, gonads dhh and also shh and ihh were expressed. In mice, the Rbp1 (in XX) and Rbp4 (in XY gonads) were expressed (Jameson et al. 2012). In Xenopus, the rbp2 was expressed in ZW and rbp4 in ZZ and ZW gonads. Gde1 gene was expressed in developing mouse gonads; however, it did not show sexual dimorphism of expression (Jameson et al. 2012) In Xenopus, this gene had an upregulated expression in ZZ gonads. Alox12b gene was not expressed in the developing mouse gonads (Jameson et al. 2012) but was upregulated in Xenopus ZW gonads. A subpopulation of fzd receptors was expressed in the developing mouse gonads. In Xenopus, fzd4 and fzd10 had an upregulated expression in developing ZZ gonads. The calpain 8 (Capn8) was not expressed in developing mouse gonads (Jameson et al. 2012) but was upregulated in Xenopus ZW gonads. The serpins were not expressed in developing mouse gonad (Jameson et al. 2012), but they were expressed in Xenopus developing gonads. In developing mouse gonads, several cathepsins (Cts) were expressed; however, only cathepsin H (ctsh) was upregulated in XY gonads (Jameson et al. 2012), and this gene was also upregulated in ZZ Xenopus gonads. Among forkhead box factors, only Foxo1 was expressed in XY developing mouse gonads (Jameson et al. 2012) and in ZZ Xenopus gonads. Similarly, Lhx1 was expressed in XY developing mouse gonads (Jameson et al. 2012) and ZZ Xenopus gonads. Considering proteins of extracellular matrix, only collagen 9 and metalloproteinase Mmp2 were expressed in a similar manner in XY developing mouse gonads (Jameson et al. 2012) and ZZ Xenopus gonads.
Analysis of transcriptome of developing chicken gonads showed that calpain 5 (Capn5), Gpr56, and Fgfr3 were upregulated in ZW (female) gonads, which suggested that they may be involved in sexual differentiation (Ayers et al. 2015). Calpain 5 was expressed in developing Xenopus gonads, but not in a sex dimorphic manner. We showed the upregulation of calpain 8 in ZW (females) Xenopus gonads, which suggests a role of this group of proteases in sexual differentiation of vertebrate gonads. However, calpain 5 or 8 was not expressed in developing mouse gonads (Jameson et al. 2012). Gpr56 was upregulated in XY mouse and ZW chicken gonads (Ayers et al. 2015; Jameson et al. 2012), but it was not expressed in Xenopus developing gonads. Fgfr3 showed sexual dimorphism of expression in developing chicken gonads (upregulated in ZW) (Ayers et al. 2015) and was also expressed, equally in both sexes, in mouse (Jameson et al. 2012) and Xenopus gonads.
Analysis of transcriptome of a red-eared slider (T. scripta) developing gonads showed that Vwa2, Fdxr, Nov, Kdm6b, Rbm20, and Pcsk6 were upregulated in the male-producing temperature, while Fank1, Avil, Twist1, and Hspb6 were upregulated in the female-producing temperature (Czerwinski et al. 2016). Fdxr2 and Hspb6 were also upregulated in ZW (male) developing gonads of Xenopus, but the sexual dimorphism in the level of expression was not statistically significant. Twist1 gene was slightly upregulated in ZZ gonads of Xenopus, but the sexual dimorphism in the level of expression was also not significant. We detected the expression of Nov and Pcsk6 in Xenopus gonads but these genes did not show a sexual dimorphism of expression. Among Kdms genes, we detected only the expression of kdm6a but it did not show sexual dimorphism. We did not detect the expression of Vwa2, Rbm20, Frank1, or Avil in developing Xenopus gonads.
In American alligator, the expression of Wnt11 was shown at male-producing temperature, which induces the development of the testes (Yatsu et al. 2016). We detected the expression of this gene in ZW (female) developing gonads in Xenopus. Analysis of transcriptome of zebrafish developing gonads showed that the estrogen receptor 2 (esr2) was upregulated in developing testes (Sreenivasan et al. 2008). The ZZ developing Xenopus gonads also upregulated the expression of this gene.
This comparison indicates that there is a profound difference in the pattern of gene expression and sexual dimorphism of gene expression between Xenopus and other vertebrates. Only few genes indicated above show a similar pattern of expression between Xenopus and other vertebrates. This shows how complex and fast-evolving is a molecular regulation of gonad development.
Conclusion
In this study, we revealed genes representing many functional groups, which showed sexual dimorphism of expression in developing Xenopus gonads. Some of these genes are probably involved in sex determination and sexual differentiation of the gonads. We also detected a sexual dimorphism of expression of many uncharacterized and unnamed genes. These genes should be characterized and studied further to discover if they are involved in sex determination and sexual differentiation. Comparative analysis of genes expressed in developing gonads of different classes of vertebrates showed striking inter-specific differences. Only few genes showed similarities of expression pattern between the species. This indicates how little we know and how complex, diversified, and evolutionary malleable are molecular mechanisms driving gonad development in vertebrates.
Material and methods
Animals
Tadpoles of the African clawed frog (Xenopus laevis Daudin, 1802) were raised in 10-L aquaria (30 tadpoles per 10 L) at 22 °C, fed daily with powder food Sera Micron (Sera), and staged according to Nieuwkoop and Faber (1956). The tadpoles at four stages (NF50, NF53, NF56, and NF62) were anesthetized with 0.1% MS222 solution, and the gonads were manually dissected under the dissecting microscope. All individuals used in the experiments were handled according to Polish legal regulations concerning the scientific procedures on animals (Dz. U. nr 33, poz. 289, 2005) and with the permission from the First Local Commission for Ethics in Experiments on Animals.
Sex determination by PCR
The genetic sex of each tadpole was determined using PCR detection of female-specific dm-w gene. DNA was isolated from tadpole tails using NucleoSpin Tissue Kit (Macherey-Nagel, 740952.240C). The dm-w gene (W-linked female-specific marker) and dmrt1 gene (positive control) were used to determine ZZ or ZW status of tested animals. PCR was performed as previously described (Yoshimoto et al. 2008). Following pairs of primers were used: for dm-w, 5′-CCACACCCAGCTCATGTAAAG-3′ and 5′-GGGCAGAGTCACATATACTG-3′, and for dmrt1, 5′-AACAGGAGCCCAATTCTGAG-3′ and 5′-AACTGCTTGACCTCTAATGC-3′.
Histological analysis
Bouin’s solution-fixed and paraffin-embedded samples were sectioned at 4 μm. Sections were deparafinated, rehydrated, and stained with hematoxylin and picroaniline according to Debreuill’s procedure (Piprek et al. 2012). Sections were viewed under the Nikon Eclipse E600 microscope.
RNA isolation
Total RNA was isolated using Trizol and purified with Direct-zol RNA kit according to the manufacturer’s protocol (Zymo Research, R2061). The total RNA was quantified using NanoDrop 2000, and RIN (RNA Integrity Number) was assessed with Bioanalyzer 2100. All samples used in the study had RIN above 8. In order to obtain a sufficient amount of RNA, the samples from 10 individuals were pooled in each experiment as previously described (Piprek et al. 2018). Total RNA in RNase-free water was frozen at − 80 °C until further use.
Microarray analysis
Microarray analysis was performed as previously described (Piprek et al. 2018). Total RNA was labeled with fluorescent dyes using Agilent One-Color Quick Amp Labeling Protocol. RNA isolated from ZW gonads were labeled with Cy3, and RNA from ZZ gonads with Cy5. Fluorescently labeled RNA samples were mixed with Agilent Hi-RPM Hybridization Buffer, and hybridized at 65 °C for 17 h in HybArray12 hybridization station (Perkin Elmer). RNA from ZW and ZZ were mixed together and hybridized to the same chip. The RNA isolated from the gonads in different stages of development was labeled with the same fluorochrome (either Cy3 or Cy5) and hybridized individually to the separate chips. Samples were washed in Gene Expression Wash Buffer 1 (6X SSPE, 0.005% N-lauroylsarcosine; at RT) and Gene Expression Wash Buffer (0.06X SSPE, 0.005% N-lauroylsarcosine; at RT) for 1 min each and immersed in a solution of acetonitrile. Air-dried slides (custom-commercial Agilent-070330 X. laevis Microarray slides) were scanned in the Agilent Technologies G2505C Microarray Scanner at a 5-μm resolution. The microarray experiment was repeated three times.
Data processing
Data processing was performed as previously described (Piprek et al. 2018). TIF files obtained in microarray scanner were processed using Agilent Feature Extraction software version 10.5.1.1. Control and non-uniform features were removed; remaining values for each unique probe sequence were averaged. Log base 2 intensities were median centered between arrays. Differential gene expression was filtered using a statistical significance threshold (FDR < 0.05) and a fold change threshold (2-fold). The data were published in Gene Expression Omnibus (accession number GSE105103). Functional analysis and gene ontology were carried out using DAVID 6.8 (https://david.ncifcrf.gov/tools.jsp) and IPA (Ingenuity Pathway Analysis, Qiagen). First, we compared the level of gene expression between gonads in different stages of development within each sex. The gene expression level at each stage of gonad development was compared to the gene expression level at the previous developmental stage, i.e., the stage NF53 was compared to the stage NF50, the stage NF56 was compared to the stage NF53, and the stage NF62 was compared to the stage NF56. In each comparison, the level of gene expression in the younger stage of gonad development was arbitrarily designated as the reference level of expression. The results of these analyses gave us an overview of the pattern of gene expression in consecutive stages of gonad development. Subsequently, we compared the level of gene expression between genetic female (ZW) versus male (ZZ) gonads at each studied developmental stage.
Electronic supplementary material
(XLSX 38 kb)
(XLSX 88 kb)
(XLSX 19 kb)
(XLSX 19 kb)
(XLSX 83 kb)
(XLSX 151 kb)
(XLSX 60 kb)
(XLSX 45 kb)
(XLSX 36 kb)
(XLSX 35 kb)
(XLSX 33 kb)
(XLSX 74 kb)
(XLSX 69 kb)
(XLSX 38 kb)
(XLSX 25 kb)
(XLSX 76 kb)
(XLSX 15 kb)
(XLSX 34 kb)
(XLSX 58 kb)
(XLSX 182 kb)
Funding information
The study was conducted within the project financed by the National Science Centre assigned on the basis of the decision number DEC-2013/11/D/NZ3/00184.
Compliance with ethical standards
All individuals used in the experiments were handled according to Polish legal regulations concerning the scientific procedures on animals (Dz. U. nr 33, poz. 289, 2005) and with the permission from the First Local Commission for Ethics in Experiments on Animals.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ayers KL, Lambeth LS, Davidson NM, Sinclair AH, Oshlack A, Smith CA. Identification of candidate gonadal sex differentiation genes in the chicken embryo using RNA-seq. BMC Genomics. 2015;16:704. doi: 10.1186/s12864-015-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- Bar I, Cummins S, Elizur A. Transcriptome analysis reveals differentially expressed genes associated with germ cell and gonad development in the Southern bluefin tuna (Thunnus maccoyii) BMC Genomics. 2016;17:217. doi: 10.1186/s12864-016-2397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A, Koopman P. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet. 2006;15:417–431. doi: 10.1093/hmg/ddi463. [DOI] [PubMed] [Google Scholar]
- Chen H, Palmer JS, Thiagarajan RD, Dinger ME, Lesieur E, Chiu H, Schulz A, Spiller C, Grimmond SM, Little MH, Koopman P, Wilhelm D. Identification of novel markers of mouse fetal ovary development. PLoS One. 2012;7:e41683. doi: 10.1371/journal.pone.0041683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwinski M, Natarajan A, Barske L, Looger LL, Capel B. A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev Biol. 2016;420:166–177. doi: 10.1016/j.ydbio.2016.09.018. [DOI] [PubMed] [Google Scholar]
- Gong W, Pan L, Lin Q, Zhou Y, Xin C, Yu X, Cui P, Hu S, Yu J. Transcriptome profiling of the developing postnatal mouse testis using next-generation sequencing. Sci China Life Sci. 2013;56:1–12. doi: 10.1007/s11427-012-4411-y. [DOI] [PubMed] [Google Scholar]
- Haselman JT, Olmstead AW, Degitz SJ. Global gene expression during early differentiation of Xenopus (Silurana) tropicalis gonad tissues. Gen Comp Endocrinol. 2015;214:103–113. doi: 10.1016/j.ygcen.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8(3):e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang L, Zhao Y, Gao J, Chen Z. Gonad transcriptome of discus fish (Symphysodon haraldi) and discovery of sex-related genes. Aquac Res. 2017;48:5993–6000. doi: 10.1111/are.13424. [DOI] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal tables of Xenopus laevis (Daudin) 1. Amsterdam: North-Holland; 1956. [Google Scholar]
- Ogunyemi D, Xu J, Mahesan AM, Rad S, Kim E, Yano J, Alexander C, Rotter JI, Chen YD. Differentially expressed genes in adipocytokine signaling pathway of adipose tissue in pregnancy. J Diabetes Mellitus. 2013;3:86–95. doi: 10.4236/jdm.2013.32013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappano WN, Scott IC, Clark TG, Eddy RL, Shows TB, Greenspan DS. Coding sequence and expression patterns of mouse chordin and mapping of the cognate mouse chrd and human CHRD genes. Genomics. 1998;52:236–239. doi: 10.1006/geno.1998.5474. [DOI] [PubMed] [Google Scholar]
- Piprek RP, Pecio A, Kubiak JZ, Szymura JM. Differential effects of busulfan on gonadal development in five divergent anuran species. Reprod Toxicol. 2012;34(3):393–401. doi: 10.1016/j.reprotox.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Piprek RP, Kloc M, Kubiak JZ. Early development of the gonads: origin and differentiation of the somatic cells of the genital ridges. Results Probl Cell Differ. 2016;58:1–22. doi: 10.1007/978-3-319-31973-5_1. [DOI] [PubMed] [Google Scholar]
- Piprek RP, Kloc M, Tassan JP, Kubiak JZ. Development of Xenopus laevis bipotential gonads into testis or ovary is driven by sex-specific cell-cell interactions, proliferation rate, cell migration and deposition of extracellular matrix. Dev Biol. 2017;432:298–310. doi: 10.1016/j.ydbio.2017.10.020. [DOI] [PubMed] [Google Scholar]
- Piprek RP, Damulewicz M, Kloc M, Kubiak JZ. Transcriptome analysis identifies genes involved in sex determination and development of Xenopus laevis gonads. Differentiation. 2018;100:46–56. doi: 10.1016/j.diff.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheider J, Afonso-Grunz F, Hoffmeier K, Horres R, Groher F, Rycak L, Oehlmann J, Winter P. Gene expression of chicken gonads is sex- and side-specific. Sex Dev. 2014;8:178–191. doi: 10.1159/000362259. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A, Orban L. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One. 2008;3:e1791. doi: 10.1371/journal.pone.0001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LX, Teng J, Zhao Y, Li N, Wang H, Ji XS. Gonad transcriptome analysis of high-temperature-treated females and high-temperature-induced sex-reversed neomales in nile tilapia. Int J Mol Sci. 2018;19(3):E689. doi: 10.3390/ijms19030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. The biology and biochemistry of diacylglycerol signalling. EMBO Rep. 2005;6(4):310–314. doi: 10.1038/sj.embor.7400378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Matsuda-Tsukiyama M, Bohgaki M, Terai S, Tanaka S, Hatakeyama S. Ymer acts as a multifunctional regulator in nuclear factor-κB and Fas signaling pathways. Mol Med. 2012;18:587–597. doi: 10.2119/molmed.2011.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shen KN, Fan Z, Huang W, You F, Lou B, Hsiao CD. The testis and ovary transcriptomes of the rock bream (Oplegnathus fasciatus): a bony fish with a unique neo Y chromosome. Genom Data. 2016;7:210–213. doi: 10.1016/j.gdata.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu R, Miyagawa S, Kohno S, Parrott BB, Yamaguchi K, Ogino Y, Miyakawa H, Lowers RH, Shigenobu S, Guillette LJ, Jr, Iguchi T. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genomics. 2016;17:77. doi: 10.1186/s12864-016-2396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Ikeda N, Izutsu Y, Shiba T, Takamatsu N, Ito M. Opposite roles of DMRT1 and its W-linked paralogue, DM-W, in sexual dimorphism of Xenopus laevis: implications of a ZZ/ZW-type sex-determining system. Development. 2010;137:2519–2526. doi: 10.1242/dev.048751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 38 kb)
(XLSX 88 kb)
(XLSX 19 kb)
(XLSX 19 kb)
(XLSX 83 kb)
(XLSX 151 kb)
(XLSX 60 kb)
(XLSX 45 kb)
(XLSX 36 kb)
(XLSX 35 kb)
(XLSX 33 kb)
(XLSX 74 kb)
(XLSX 69 kb)
(XLSX 38 kb)
(XLSX 25 kb)
(XLSX 76 kb)
(XLSX 15 kb)
(XLSX 34 kb)
(XLSX 58 kb)
(XLSX 182 kb)