Abstract
Background
Adjuvant System 03 (AS03) markedly enhances responses to influenza A/H5N1 vaccines, but the mechanisms of this enhancement are incompletely understood.
Methods
Using ribonucleic acid sequencing on peripheral blood mononuclear cells (PBMCs) from AS03-adjuvanted and unadjuvanted inactivated H5N1 vaccine recipients, we identified differentially expressed genes, enriched pathways, and genes that correlated with serologic responses. We compared bulk PBMC findings with our previously published assessments of flow-sorted immune cell types.
Results
AS03-adjuvanted vaccine induced the strongest differential signals on day 1 postvaccination, activating multiple innate immune pathways including interferon and JAK-STAT signaling, Fcγ receptor (FcγR)-mediated phagocytosis, and antigen processing and presentation. Changes in signal transduction and immunoglobulin genes predicted peak hemagglutinin inhibition (HAI) titers. Compared with individual immune cell types, activated PBMC genes and pathways were most similar to innate immune cells. However, several pathways were unique to PBMCs, and several pathways identified in individual cell types were absent in PBMCs.
Conclusions
Transcriptomic analysis of PBMCs after AS03-adjuvanted H5N1 vaccination revealed early activation of innate immune signaling, including a 5- to 8-fold upregulation of FcγR1A/1B/1C genes. Several early gene responses were correlated with HAI titer, indicating links with the adaptive immune response. Although PBMCs and cell-specific results shared key innate immune signals, unique signals were identified by both approaches.
Keywords: adjuvant, AS03, avian influenza, influenza vaccine, systems vaccinology
Transcriptomic analysis of PBMCs after AS03-adjuvanted H5N1 vaccination revealed activation of several innate immune signaling pathways and strong upregulation of Fcγ-receptor genes. Although PBMCs and individual immune cell types shared key innate signals, unique signals were identified by both approaches.
Emerging avian influenza viruses, such as A/H5N1 and A/H7N9, are associated with high mortality and pose a substantial threat for the next influenza pandemic [1–3]. Development of effective vaccines for these strains has been hindered by poor immunogenicity. Multiple dose regimens using higher hemagglutinin content and addition of potent adjuvants have enhanced antibody responses [4]. Oil-in-water adjuvants MF59 [5] and AS03 [6] reduce the antigen dose required to stimulate immune responses to influenza A/H5N1 and other avian influenza vaccines [6–12]. Antigen-sparing approaches are particularly critical in global pandemic preparedness given limitations in production capacity of influenza vaccine antigens.
Conventionally, immune responses to avian influenza vaccines are measured by hemagglutination inhibition (HAI) or neutralizing (Nt) antibody titers 4–6 weeks after the second vaccination. More rapid identification of poor vaccine responders would be an attractive component of pandemic preparedness. Systems vaccinology provides a comprehensive approach to studying vaccine responses by correlating antibody and cell-mediated responses with changes in gene transcript, protein, and metabolite abundance in immune cells [13–16]. This approach may uncover new mechanisms of immune responses to vaccines and adjuvants [17, 18] and identify molecular signatures that correlate with antibody responses as early as 1 day after vaccination [19, 20].
We previously applied a systems vaccinology approach to study an AS03-adjuvanted influenza A/H5N1 vaccine in healthy adults in a Phase I clinical trial evaluating transcriptomic and proteomic responses to vaccination in isolated populations of 6 immune cell types [19–21]. However, this cell separation approach is highly labor and resource intensive. Studying bulk populations of cells, such as peripheral blood mononuclear cells (PBMCs), is less laborious. However, it is unclear whether evaluating bulk PBMCs will provide such detailed immune insights as observed when studying individual immune cell types. In this study, we present our assessment of transcriptomic responses in bulk PBMCs based on future-use samples from our earlier Phase I study [19], identify PBMC gene signatures that predict peak antibody response, and compare PBMC transcriptomic responses to individual immune cell type responses as reported in our earlier study [19].
METHODS
Study Design
We performed a single-center, randomized, double-blinded, controlled, Phase I study enrolling healthy adults 19–39 years old to assess the safety, immunogenicity, and molecular immune responses of intramuscular-inactivated influenza A/H5N1 (3.75 mcg hemagglutinin [HA] A/Indonesia/05/2005) split-virus (SV) vaccine (Sanofi) administered with either AS03 adjuvant (SV-AS03; GlaxoSmithKline) or phosphate-buffered saline (SV-PBS). Detailed methods and results from the trial have been previously reported [19]. In brief, 20 subjects were randomized 1:1 to the SV-AS03 (n = 10) or SV-PBS (n = 10) group. Subjects received two 0.5-mL injections 28 days apart. Subjects and all study personnel were blinded to vaccine assignment.
Laboratory Procedures
Whole blood samples for ribonucleic acid sequencing (RNA-Seq) were collected on study visit days −28, −14, and 0 (prevaccination) and days 1, 3, 7, and 28 (after first vaccination). Sera for HAI and Nt antibody assays were collected at study visit days 0, 1, 3, 7, 28, and 56.
Cell Sorting
Whole blood samples (90 mL) underwent Ficoll separation to yield PBMC and polymorphonuclear cell (PMN) populations. For our earlier, published cell type-specific analyses [19], suspensions of PBMCs or PMNs were subjected to magnetic-activated cell sorting (MACS) then fluorescence-activated cell sorting (FACS) for purification of neutrophils, monocytes, dendritic cells (DCs), natural killer (NK) cells, T lymphocytes, and B lymphocytes [19, 20]. Purified immune cell types and bulk PBMCs were frozen in homogenization buffer and stored at −80°C.
Hemagglutinin Inhibition Antibody Assays
Sera for HAI responses against the homologous A/Indonesia/05/2005 virus were measured as previously described (Southern Research, Birmingham, AL) [22]. Values below the limit of detection (1:10) were imputed as half the limit of detection.
Ribonucleic Acid Sequencing and Data Processing
The RNA-Seq experiments were performed as previously described [19]. Briefly, RNA was extracted from PBMCs from 140 future-use samples from our earlier Phase 1 study (20 subjects at 7 time points), polyadenylated RNAs were isolated, and next-generation sequencing expression libraries were prepared. Paired-end sequencing (25 million, 50 base pairs) was performed on an Illumina HiSeq2000 sequencer. Four samples with insufficient RNA were excluded from downstream processing. Paired-end reads were mapped against the human reference genome (GRCh37, ENSEMBL version 63) using TopHat, version 2.0.0. Gene expression quantification was conducted using Subread, version 1.4.6, counting mapped paired-reads to obtain fragment counts per gene (Supplementary Dataset 1, see Supplementary Text for additional details).
Statistical Analysis
Systematic sample differences in fragment counts were corrected using the trimmed mean of M-value (TMM) normalization method [23]. Negative binomial models, as implemented in edgeR (version 3.2.4) [24], were used to identify genes that were differentially expressed (DE) between vaccine groups (SV-AS03 vs SV-PBS) based on a likelihood ratio test of the group x time interaction adjusting for paired samples (subject effect) and sample percentage guanine-cytosine (GC) content (covariate). Subject-specific prevaccination levels were estimated as the mean of prevaccination samples (days −28, −14, 0) as described in Supplementary Text Section 2.4.1 and [19]. Genes with a false discovery rate (FDR)-adjusted P ≤ .05 (p.adjust R function) and fold change difference between groups of ≥1.5-fold were deemed to be DE. Enrichment analysis for KEGG [25] (version 70.0) and MSigDB (version 4.0) [26] pathways was performed using the goseq R package [27] (version 1.12.0) accounting for gene length bias (FDR-adjusted P ≤ .01). The TMM-normalized log2 fragment counts per million (LCPM) as implemented in edgeR (Supplementary Dataset 2) were used as input for principal component analysis (PCA) and to calculate log2 fold changes used for regularized linear regression analysis (glmnet version 2.0–2 R package) to identify gene responses that best predicted log2 peak HAI titer. Leave-one-out cross-validation was used to select optimal models (see Supplementary Text for additional details).
Ethics Statement
The trial was approved by the Vanderbilt University Human Research Protections Program and is registered on ClinicalTrials.gov (NCT01573312). All subjects provided written informed consent before initiation of study procedures.
RESULTS
Peripheral Blood Mononuclear Cells Showed Strongest Differential Gene Expression Responses on Day 1 After Vaccination
In the SV-AS03 group compared with the SV-PBS group, we identified 474 DE genes for day 1 (Supplementary Table 1); 421 of 474 (89%) were up-regulated, whereas 53 of 474 (11%) were down-regulated from prevaccination. No DE genes between vaccine groups were detected at days 3, 7, or 28. Heatmap analysis of DE genes (Figure 1) confirmed strongest differential responses at day 1, both in fold change magnitude and consistency within the SV-AS03 group. The day 1 signal was primarily driven by up-regulated genes that were strongly increased in most SV-AS03 subjects. The SV-AS03 treatment effect waned quickly, as was evident by reduced within-group clustering and fold changes.
Figure 1.
Heatmaps of baseline log2 fold changes of differentially expressed genes in peripheral blood mononuclear cells for each postvaccination time point. Dendrograms were obtained using complete linkage clustering of uncentered pairwise Pearson correlation distances between log2 fold changes in log2 fragment counts per million. Cells are color-coded by log2 fold change (red, up-regulated from baseline; blue, down-regulated from baseline).
SV-AS03-Responsive Genes Are Known to Play a Role in Innate Immune Signaling and Immune Response to Influenza A Infection
To understand the functional composition of day 1 DE genes, we performed pathway enrichment analysis (Supplementary Text, Tables A13–A19). Enriched KEGG pathways included known innate immune response signaling pathways such as JAK-STAT, tumor necrosis factor (TNF), nuclear factor (NF)-κB, NOD-like receptor, Toll-like receptor (TLR) signaling pathways, and complement and coagulation cascades. Cytokine-cytokine receptor interaction, chemokine signaling pathway, and Reactome pathway-based interferon (IFN) α/β signaling and IFN-γ signaling were also perturbed at day 1. Most DE genes in these pathways were up-regulated in the SV-AS03 relative to the SV-PBS group.
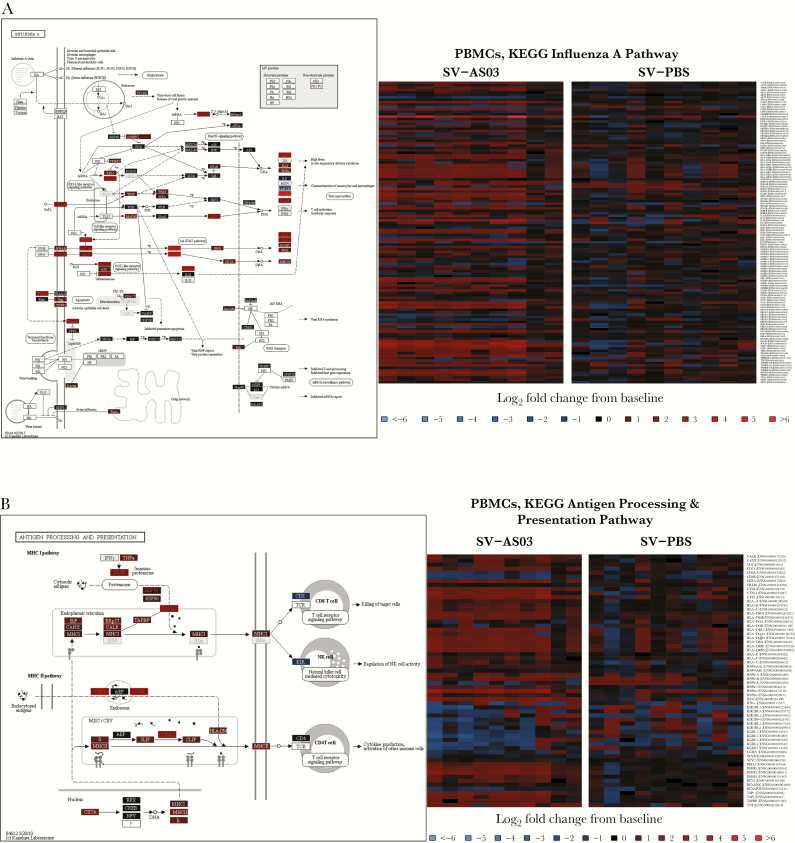
Among KEGG human infectious disease pathways that were overrepresented in SV-AS03-responsive genes, Influenza A had the highest number of overlapping DE genes (20 genes) (Figure 2A). Within this pathway, significant up-regulation of TLR4 was observed in the SV-AS03 group; the corresponding TLR4 receptor is known to recognize viral proteins and induce innate immunity, in part by triggering NF-κB signaling and subsequent innate immune activation and cytokine signaling [28]. This pathway also showed up-regulation of IFN-inducible genes, MXA and OAS, the IFN-γ receptor gene (IFNGR), and the intracellular JAK-STAT signaling pathway, which activates transcription of IFN-inducible genes that trigger antiviral responses. Genes encoding for interferon gamma-induced protein 10 (IP-10) and interleukin-1 were also significantly induced in the SV-AS03 group relative to the SV-PBS group. SOCS3, a negative regulator of the JAK-STAT pathway, was increased, suggesting that some level of negative feedback was triggered as early as day 1.
Figure 2.
Pathway responses for influenza A infection, antigen processing and presentation, and Fc-γ receptor-mediated phagocytosis. (Left) Color-coded KEGG pathway maps for day 1. (Right) Subject-specifc baseline log2 fold changes for pathway genes for day 1 by vaccine group (red, up-regulated; blue, down-regulated). (A) Influenza A. (B) Antigen processing and presentation. (C) FcγR-mediated phagocytosis. Pathway node color gradient encodes log2 fold change difference (LFCD) between vaccine groups (for multigene nodes the median LFCD was used: red, increased log2 fold changes response for the split-virus (SV)-AS03 group relative to the SV-PBS group; blue, decreased log2 fold changes response for the SV-AS03 group relative to the SV-phosphate-buffered saline (PBS) group; black, fold change close to 1; dark gray, genes filtered out due to low overall expression; light gray, gene missing database mapping; white, nonhuman gene). Heatmaps are color coded based on subject-specific log2 fold changes in log2 fragment counts per million.
One of the pathways more strongly associated with the differential response between the SV-AS03 and SV-PBS groups was the KEGG antigen processing and presentation pathway. Several genes associated with major histocompatibility complex (MHC) class I and class II pathways were significantly enriched in the SV-AS03 group at day 1 postvaccination (Figure 2B). For MHC class I, this included immunoproteasome genes (PSME2, PSMB9, PSME2P2) and chaperones for MHC class I loading (HSPA6, HSPA7, and TAP1). The FcγR-mediated phagocytosis pathway exhibited a strong differential response between groups (Figure 2C), with significant up-regulation of FcγRI (FCGRIA, FCGRIB, FCGRIC) and FcγRIIA (FCGRIIA) genes, which encode receptor proteins on phagocytes involved in antigen binding and interacting with immunoglobulin (Ig)G antibodies to regulate immune responses.
Changes in Day 1 Signal Transduction Genes and Day 7 Immunoglobulin Genes Best Correlated With Peak Hemagglutinin Inhibition Antibody Responses
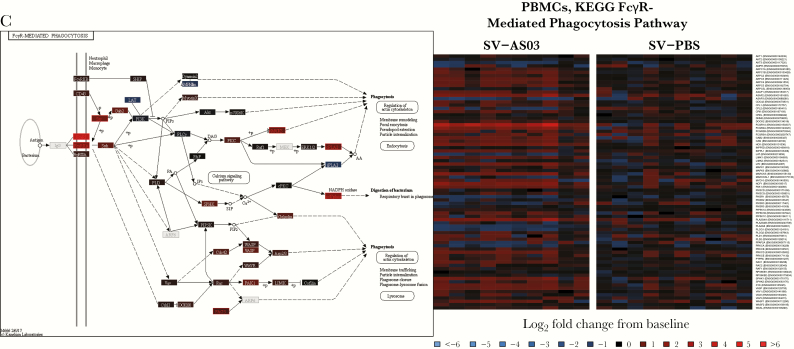
In addition to identifying DE genes between groups, we investigated genes across groups whose changes from prevaccination best predicted peak HAI titer 28 days after the second vaccination using regularized linear regression analysis (Supplementary Table 2; Table A20, Supplementary Text). The day 7 model achieved the best overall fit (17 predictive genes), followed by day 1, day 3, and day 28 (15, 5, and 6 predictive genes, respectively). For all 17 predictive genes in the day 7 model, expression was positively correlated with log2 peak HAI titer (Figure 3). These included several Ig genes (IGKV1−5, IGHG1, IGKV2−24, IGHV1−69, IGHV3−74, IGKV3−15) and MK167, a gene involved in cell proliferation, which had the highest relative impact in predicting HAI titer. At day 1, an increase in expression of several signal transduction genes (STAT1, FCGR1C, FCGR1A, IRF9, PSME2, and SOCS3) was positively correlated with peak titer (Figure 3). Negative correlations were observed for AL138831.1, ADAMTS7, BAIAP3, and HSPG2, indicating that these genes may negatively regulate subsequent antibody response. Scatterplots for all genes are shown in Figures A55–A58, Supplementary Text.
Figure 3.
Combination of gene responses that best predicted peak log2 hemagglutinin inhibition (HAI) titer at day 1 and day 7. The bar plots represent regularized linear regression coefficients of the best model. Scatterplots that summarize individual gene associations between peak HAI titer and gene log2 fold change including locally weighted scatterplot smoothing trend lines are shown for the top and bottom 3 genes for day 1 and the top 6 genes for day 7.
Differential Peripheral Blood Mononuclear Cell Transcriptomics Signals Were Most Similar to Flow-Sorted Monocyte, Dendritic Cell, and Neutrophil Signals
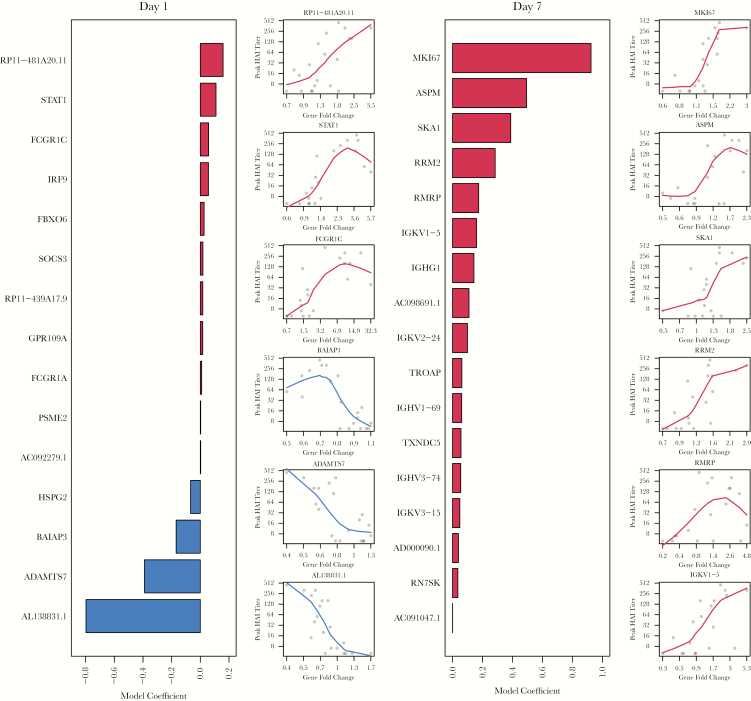
To compare bulk PBMC signals to individual immune cell types, we compared expression profiles using PCA and assessed overlap in DE genes and enriched pathways. Our prior PCA assessment revealed that gene expression profiles based on LCPM clustered according to 3 distinct cell groups: (1) neutrophils, (2) lymphocytes (T, B, and NK cells), and (3) DCs and monocytes [19, 20]. Using PCA on the combined data, PBMC samples formed a unique cluster that was closest to the lymphocyte followed by monocyte/DC clusters (Figure 4). As expected, PBMCs clustered distinctly away from neutrophils. Based on Jaccard’s Similarity Index, the greatest similarity in DE gene expression between PBMCs and individual cell types was for monocytes (Jaccard coefficient 0.21, 251 shared DE genes) followed by DCs (0.18, 106 shared genes) and neutrophils (0.13, 123 shared DE genes) (Figure 5A, Supplementary Table 1). Neutrophils had the most unique DE genes (394) followed by PBMCs, monocytes, and DCs (274, 181, and 70, respectively) (Figure 5B). Five up-regulated genes (IRF1, PARP9, STAT1, GBP1, and EPSTI1) were identified in PBMCs and in each of the 6 immune cell types. In our cell-specific assessments, we identified a core network of 80 genes that were DE in monocytes, neutrophils, and DCs at day 1 postvaccination [19]. Among these 80 genes, 65 (81%) were also identified in PBMCs.
Figure 4.
Principal component analysis of standardized log2 fragment counts per million of peripheral blood mononuclear cells (PBMCs) and individual immune cell subsets. Ninety-nine percent confidence ellipses of the bivariate mean based on a bivariate t-distribution are shown for each cell population.
Figure 5.
Comparison of differentially expressed (DE) genes between peripheral blood mononuclear cells (PBMCs) and individual immune cell subsets. (A) Heatmap summarizing the overlap in DE genes at day 1 in PBMCs and individual immune cell subtypes using the Jaccard Similarity Index. The number of shared DE genes is shown in brackets. (B) Overlapping DE genes at day 1 postvaccination among PBMCs and individual immune cell subtypes. The bar chart indicates the number of unique DE genes or overlapping DE genes between cell populations as indicated by unconnected or connected black circles below the bar chart, respectively. Numbers in round brackets represent the total DE genes at day 1.
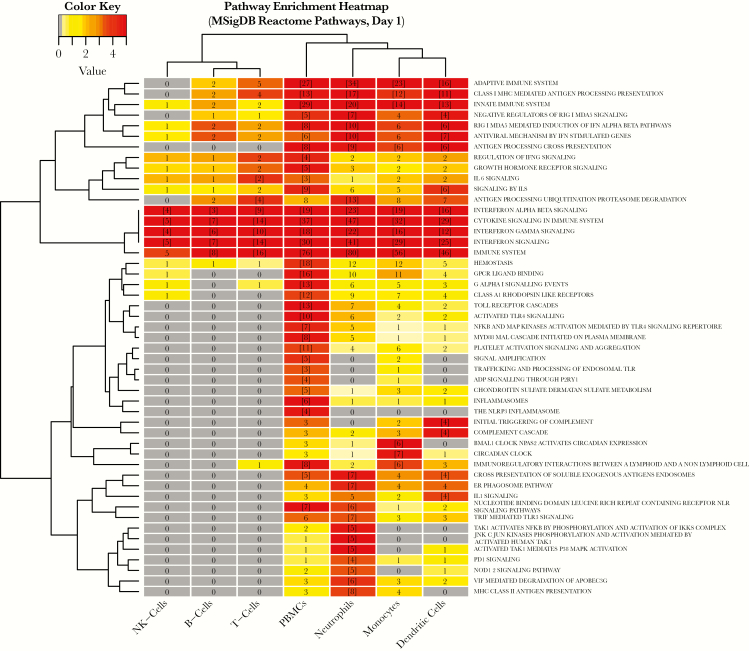
To compare functional composition of DE genes between PBMCs and individual immune cell types, we performed pathway enrichment analysis based on the Reactome database [29] (Supplementary Table 3). By enrichment score, PBMCs Reactome pathway profiles were functionally most similar to neutrophils, monocytes, and DCs (Figure 6, Supplementary Table 3). Of 34 Reactome pathways enriched in PBMCs, 17 were shared by at least 1 cell type, whereas 17 were unique to PBMCs. Most of the shared signals were related to innate immune response signaling, whereas PBMCs showed unique enrichment signals related to hemostasis, Toll-like receptor signaling, platelet activation, inflammasomes, and regulation of IFNG signaling. Fifteen pathways were significantly enriched in at least 1 cell type, but not in PBMCs. Most of these were uniquely identified in neutrophils, including MHC class II antigen presentation. Interferon-γ and IFN α/β signaling were perturbed in all cell populations.
Figure 6.
Comparison of enriched Reactome pathways between peripheral blood mononuclear cells and individual immune cell subsets. Heatmap cell counts represent the number of differentially expressed genes in a pathway. Cells with numbers in brackets indicate significantly enriched sets. Cells are color coded by pathway enrichment score: −1 × log10 (false discovery rate-adjusted P value) (dark red, highly enriched; light yellow, low level of enrichment). Dendrograms were obtained using complete linkage clustering of Euclidean distances between pathway enrichment scores.
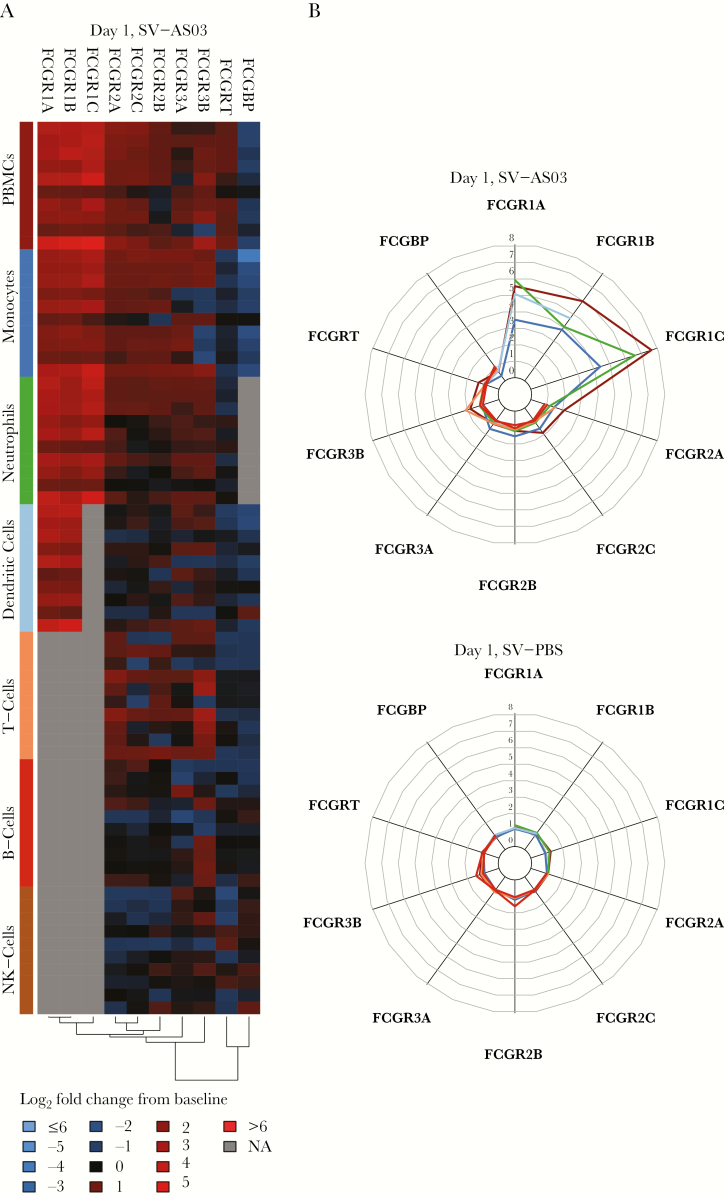
Early Activation of Genes in the Fcγ Receptor Family Was a Key Immune Signal Observed in Gene Expression, Pathway Enrichment, and Predictive Antibody Analyses
We identified (1) significant differential up-regulation of FCGRIA, FCGRIB, FCGRIC, FCGRIIA, and FCGRIIC genes in the SV-AS03 relative to the SV-PBS group, (2) robust coexpression of FCGR1A and FCGR1B at day 1 after vaccination (CPMCD1-008; Figure A18, Supplementary Text), and (3) FCGRIA and FCGR1C as predictors of later peak HAI titer (Figure 3). To further investigate the FcγR signal in PBMCs in the context of our previous cell-specific assessments, we contrasted fold change responses for 10 known FcγR genes using heatmap and radar plot analysis across cell populations (Figure 7). The strongest fold change signals were observed at day 1 for FCGR1A, FCGR1B, and FCGR1C in PBMCs of the SV-AS03 group, which ranged from 5- to 8-fold up-regulation relative to prevaccination. Similar patterns were observed in neutrophils, monocytes, and DCs in our cell-specific assessments, although with lower magnitude of fold-change. FCGR1C was not sufficiently expressed in DCs to be included in the analysis.
Figure 7.
Comparison of day 1 fold change responses for 10 FcγR genes between peripheral blood mononuclear cells (PBMCs) and individual immune cell subsets. (A) Heatmaps of baseline log2 fold changes of FcγR genes in the split-virus (SV)-AS03 group at day 1 with cell types indicated by colored bars in the y-axis. Heatmap cells are color-coded by log2 fold change (red, up-regulated from baseline; blue, down-regulated from baseline; gray, did not meet low expression cutoff). (B) Radar plots contrasting fold changes of FcγR genes at day 1 postvaccination in the SV-AS03 (shown at the top) and SV-phosphate-buffered saline (PBS) groups (shown at the bottom). Colored lines represent the fold change for a particular cell population using the same color key as shown on the y-axis of A.
DISCUSSION
In this assessment of PBMC gene expression signaling after AS03-adjuvanted and unadjuvanted influenza A/H5N1 vaccination, we show that the strongest signals were induced at postvaccination day 1 for the SV-AS03 group. This signal quickly waned, with no further differential gene expression signals between vaccine groups by day 3 postvaccination. Enrichment of several key innate immune signaling pathways was observed, despite innate immune cells representing a minority of the PBMC population relative to adaptive immune cells, affirming previous reports that AS03 enhances immune responses to avian influenza vaccines primarily by triggering induction of innate immune pathways shortly after vaccination [17–19, 21, 30, 31]. Pathway analysis revealed activation of innate immune signaling pathways including IFN, JAK-STAT, NF-κB, NOD-like, Toll-like receptor and TNF signaling, FC receptor-mediated phagocytosis, and enrichment of the antigen presentation and processing and Influenza A immune pathways, with most genes having higher expression in the SV-AS03 group. Robust early IFN-associated innate immune signaling and enhanced antigen processing and presentation have also been associated with milder disease courses in nonhuman primates experimentally infected with H5N1 [32].
When comparing PBMC transcriptomic responses to other human or mouse immunologic transcriptomics datasets in MSigDB [26], day 1 responses were most similar to PBMC responses after live yellow fever virus vaccination, which strongly induced type I IFN signaling [33] or in vitro IFN-γ stimulation of human macrophages [34] (Table A19, Supplementary Text). Overlap with unadjuvanted inactivated seasonal influenza vaccine PBMC responses was statistically significant but was not as pronounced, indicating that the innate immune induction enhanced by AS03-adjuvanted inactivated H5N1 vaccine, mediated by strong IFN signaling, shared more similarity to a live-attenuated viral vaccine than inactivated influenza vaccine [14, 19].
Within the Influenza A pathway, the CXCL10 gene encoding for IP-10 was significantly induced in the SV-AS03 group. Serum IP-10 levels were significantly increased at day 1 and day 3 in the SV-AS03 group in our earlier study, returning to baseline levels by day 7 [19]. In addition, a cluster of 15 coexpressed genes in PBMCs showed a strong increase from prevaccination in the SV-AS03 group at day 1 (CPMCD1D28-012; Figure A22, Supplementary Text). This cluster included CXCL10 among other genes with known functions in JAK-STAT and IFN signaling, further supporting the potential significance of IP-10 in modulating AS03-specific responses after IFN signaling.
For avian influenza vaccines, an HAI titer of ≥1:40 measured 4–6 weeks after the second vaccine dose is considered the standard correlate of protection [35]. However, in influenza pandemic preparedness efforts, early identification of poor vaccine responders is likely to be critical to informing vaccination strategies. Up-regulation of several genes at day 1 or 7 predicted peak HAI titer on day 56, 28 days after the second vaccine dose. In addition to PBMCs, increased day 1 expression of STAT1 (1) was also predictive of higher HAI titers in all 6 individual immune cell types in our earlier study [19], (2) has been strongly correlated with the antibody response to trivalent influenza vaccine [36], and (3) was observed in recipients of an AS03-adjuvanted hepatitis B vaccine [30]. Although these findings require further validation, this suggests a possible role for the use of STAT1 expression in predicting the magnitude of immune responses to an AS03-H5N1 pandemic vaccine 1 day postvaccination, with the potential to inform more personalized vaccine regimens, of particular importance given the need for dose-sparing vaccine strategies in the event of a pandemic. In our previous cell-specific transcriptomics assessments, PSME2, which encodes a protein involved in immunoproteasome formation, was also predictive of seroprotection in monocytes at day 1 [19], and increased expression of the PSME2-associated protein in monocytes at day 3 was predictive of higher HAI titer in our proteomics assessments [21].
We previously reported that most variation in gene expression was attributable to immune cell type [19]. However, when comparing gene expression between individual immune cell subsets and PBMCs, PBMCs clustered distinctly and away from neutrophils (Figure 4), as expected. The closest clusters to PBMCs were lymphocytes followed by monocytes and DCs. Greatest similarity in DE gene expression with PBMCs was seen with innate immune cells (monocytes, DCs, and neutrophils), with many shared genes involved in innate signaling, likely driven by monocytes or DCs. There was substantial overlap in enriched pathways between innate immune cells and PBMCs (Figure 6), although many pathways were unique to PBMCs. Most pathways that were unique to a single cell type were enriched in neutrophils. An important signal in our analysis of NK cells that was not identified in PBMCs was cell proliferation at day 3 in the SV-AS03 group and antigen presentation at day 28 in the SV-PBS group [19]. In addition, at day 1, we observed up-regulation of the MHC class II pathway in neutrophils in our cell-specific analysis, but this was not observed in PBMCs. Despite these differences, and in light of the resource-intensive nature of immune cell type separation, our findings suggest that characterization of bulk PBMCs and neutrophils is a more efficient alternative to postvaccination transcriptomic analysis, at least for influenza vaccine and AS03 adjuvant. Additional targeted cell types or subsets of interest, such as NK cells, could be isolated based on hypotheses pertaining the specific vaccine antigen and/or adjuvant under study. In addition to targeting only certain cell populations of interest, the time points of blood collection could be streamlined for prevaccination and/or postvaccination samples, depending on the hypothesis being tested, which would enable application of this approach to larger study populations.
Development of a universal influenza vaccine that confers protection across diverse strains is a major global public health priority [37]. Influenza HA head antibodies, stimulated by conventional vaccines, only neutralize a few strains, whereas antibodies to the highly conserved HA stalk neutralize a broader array of viruses [38–40]. The Fc-FcγR interactions are pivotal in inducing antistalk antibodies that activate FcγR-expressing NK cells to kill infected cells through antibody-dependent cellular cytotoxicity (ADCC). Strain-specific HA head antibodies do not efficiently interact with FcγRs and thus do not induce NK cell-mediated ADCC [41]. We found that AS03 perturbed the FcγR-mediated phagocytosis pathway (Figure 2C), with strong activation of many FcγR genes, which have important functions in antigen presentation and inducing antibody-mediated effector functions and effector functions of CD8+ and CD4+ T cells or regulatory T cells that recognize peptide-MHC complexes. Thus, the FcγR family serves as a potential link between the innate and adaptive immune systems and may bridge the humoral and cellular adaptive immune responses [42, 43]. We previously identified strong signals for NK cell activation at day 3 postvaccination in the SV-AS03 group [19]. Although we did not see a noticeable increase in FcγR gene expression in NK cells in bulk PBMCs (Figure 7), we hypothesize that AS03 may stimulate low levels of pre-existing antistalk antibodies to activate NK cells to initiate a cytotoxic response to the vaccine antigen. AS03-mediated activation of FcγR genes may have implications for the development of vaccines that promote antistalk antibody responses and thus are more broadly protective across diverse influenza strains.
Although our cell separation procedures [19, 20] isolated highly purified cell populations, this may have filtered out cell phenotypes that were not captured with a single marker. Because gating and choice of CD markers may impact the identity of isolated populations, analyzing bulk PBMCs may capture immune signals from complex cell phenotypes that are not observed when studying highly pure cell populations. For example, plasma cells may not express CD19, the marker used for B-cell selection; thus, these may have been excluded from our B-cell populations. Although individual immune cell types were subjected to MACS and FACS, sorted cells were not activated by the sorting process [20]. Larger studies of AS03-adjuvanted vaccines would be needed to confirm the observed responses, implicated pathways, and predictive ability of the models. A study design that evaluated AS03 administered with or without a vaccine antigen would allow clearer distinction of immune responses to AS03. Small sample size may limit the generalizability of our results. However, our study highlights the strengths of PBMC transcriptomic assessments in identifying potential AS03 mechanisms of action such as the activation of FcγR, IP-10, and antigen processing and presentation-related genes. Direct comparisons to cell-specific transcriptomic results provide critical information that may inform the design of future transcriptomics assessments in vaccine clinical trials.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We acknowledge and thank our study volunteers for their participation in this study. In addition, we acknowledge the contributions of Vanderbilt Vaccine Research Program staff: Gayle Johnson, RN, CCRP; Shanda Phillips, RN, BSN, CCRP, Wendi McDonald, RN, BSN, CCRP; Deborah Myers, CCRP; Roberta Winfrey; Kevin Booth, MBA; Jennifer Nixon, MBA; John Oleis, DPh; and the Vanderbilt Clinical Research Center. We also thank John Mote, Angela Jones, and the Sequencing and Analysis teams of the Genomic Services Laboratory at HudsonAlpha for their support of the genomic studies, and Megan McDonough, MPH, Bernadette Jolles, and Brenda Leung of The Emmes Corporation for their support of clinical data management. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, Tennessee. Flow Cytometry experiments were performed in the VMC Flow Cytometry Shared Resource.
Disclaimer. The Emmes Corporation does not have any patents, products in development, or marketed products to declare.
Financial support. This work was funded by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, under Contract Nos. 272200800007C and 2722001300023I (to K. M. E.) and NIH Grant No. GM64779 (to A. J. L.); the Vanderbilt Clinical and Translational Science Award, NIH Grant RR024975 (UL1TR000445); the Childhood Infections Research Program Grant T32-AI095202-01 (to L. M. H.); the Vanderbilt Department of Pediatrics Turner-Hazinski Award (to L. M. H.); and a VA Merit Award BX001444 (to S. J.). The United States Department of Health and Human Services Biomedical Advanced Research and Development Authority provided the vaccine and adjuvant. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404).
Potential conflict of interest. K. M. E. has received grant funding for group B streptococcal vaccine research from Novartis and has participated as a member on a Novartis Data Safety Monitoring Board on influenza vaccines. C. B. C. has received institutional grant support for staphylococcal vaccine research from Pfizer, Novartis, and GSK and serves as a consultant to GSK (for unrelated staphylococcal vaccine development). L. M. H. has received grant support from Pfizer for pneumococcal vaccine research. J. B. G., T. J. L., and C. E. G. are employed by The Emmes Corporation. The Emmes Corporation is an independent contract research organization and has a contract with NIH-NIAID-Division of Microbiology and Infectious Diseases to collect and analyze data for clinical trials and studies to evaluate the safety and efficacy of vaccines, devices, and therapeutics or infectious diseases. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: IDWeek 2018, October 2018, San Francisco, CA.
References
- 1. Influenza. Avian influenza A(H7N9) virus Available at: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/en/ Accessed 17 August 2017.
- 2. Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO Available at: http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/ Accessed 17 August 2017.
- 3. Russell CA, Fonville JM, Brown AE, et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 2012; 336:1541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med 2006; 354:1343–51. [DOI] [PubMed] [Google Scholar]
- 5. O’Hagan DT, Ott GS, Van Nest G. Recent advances in vaccine adjuvants: the development of MF59 emulsion and polymeric microparticles. Mol Med Today 1997; 3:69–75. [DOI] [PubMed] [Google Scholar]
- 6. Garçon N, Vaughn DW, Didierlaurent AM. Development and evaluation of AS03, an Adjuvant System containing α-tocopherol and squalene in an oil-in-water emulsion. Expert Rev Vaccines 2012; 11:349–66. [DOI] [PubMed] [Google Scholar]
- 7. Chu DW, Hwang SJ, Lim FS, et al. Immunogenicity and tolerability of an AS03(A)-adjuvanted prepandemic influenza vaccine: a phase III study in a large population of Asian adults. Vaccine 2009; 27:7428–35. [DOI] [PubMed] [Google Scholar]
- 8. Gillard P, Chu DW, Hwang SJ, et al. Long-term booster schedules with AS03A-adjuvanted heterologous H5N1 vaccines induces rapid and broad immune responses in Asian adults. BMC Infect Dis 2014; 14:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson LA, Campbell JD, Frey SE, et al. Effect of varying doses of a monovalent H7N9 influenza vaccine with and without AS03 and MF59 adjuvants on immune response: a randomized clinical trial. JAMA 2015; 314:237–46. [DOI] [PubMed] [Google Scholar]
- 10. Langley JM, Frenette L, Ferguson L, et al. Safety and cross-reactive immunogenicity of candidate AS03-adjuvanted prepandemic H5N1 influenza vaccines: a randomized controlled phase ½ trial in adults. J Infect Dis 2010; 201:1644–53. [DOI] [PubMed] [Google Scholar]
- 11. Lasko B, Reich D, Madan A, Roman F, Li P, Vaughn D. Rapid immunization against H5N1: a randomized trial evaluating homologous and cross-reactive immune responses to AS03(A)-adjuvanted vaccination in adults. J Infect Dis 2011; 204:574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moris P, van der Most R, Leroux-Roels I, et al. H5N1 influenza vaccine formulated with AS03 A induces strong cross-reactive and polyfunctional CD4 T-cell responses. J Clin Immunol 2011; 31:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med 2012; 4:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakaya HI, Wrammert J, Lee EK, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 2011; 12:786–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity 2010; 33:516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trautmann L, Sekaly RP. Solving vaccine mysteries: a systems biology perspective. Nat Immunol 2011; 12:729–31. [DOI] [PubMed] [Google Scholar]
- 17. Morel S, Didierlaurent A, Bourguignon P, et al. Adjuvant System AS03 containing α-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine 2011; 29:2461–73. [DOI] [PubMed] [Google Scholar]
- 18. Sobolev O, Binda E, O’Farrell S, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol 2016; 17:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Howard LM, Hoek KL, Goll JB, et al. Cell-based systems biology analysis of human AS03-adjuvanted H5N1 avian influenza vaccine responses: a phase I randomized controlled trial. PLoS One 2017; 12:e0167488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoek KL, Samir P, Howard LM, et al. A cell-based systems biology assessment of human blood to monitor immune responses after influenza vaccination. PLoS One 2015; 10:e0118528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galassie AC, Goll JB, Samir P, et al. Proteomics show antigen presentation processes in human immune cells after AS03-H5N1 vaccination. Proteomics 2017; 17. doi: 10.1002/pmic.201600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noah DL, Hill H, Hines D, White EL, Wolff MC. Qualification of the hemagglutination inhibition assay in support of pandemic influenza vaccine licensure. Clin Vaccine Immunol 2009; 16:558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogata H, Goto S, Fujibuchi W, Kanehisa M. Computation with the KEGG pathway database. Biosystems 1998; 47:119–28. [DOI] [PubMed] [Google Scholar]
- 26. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol 2014; 426:1246–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joshi-Tope G, Gillespie M, Vastrik I, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 2005; 33:D428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burny W, Callegaro A, Bechtold V, et al. Different adjuvants induce common innate pathways that are associated with enhanced adaptive responses against a model antigen in humans. Front Immunol 2017; 8:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Givord C, Welsby I, Detienne S, et al. Activation of the endoplasmic reticulum stress sensor IRE1α by the vaccine adjuvant AS03 contributes to its immunostimulatory properties. NPJ Vaccines 2018; 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muramoto Y, Shoemaker JE, Le MQ, et al. Disease severity is associated with differential gene expression at the early and late phases of infection in nonhuman primates infected with different H5N1 highly pathogenic avian influenza viruses. J Virol 2014; 88:8981–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009; 10:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rock RB, Hu S, Deshpande A, et al. Transcriptional response of human microglial cells to interferon-gamma. Genes Immun 2005; 6:712–9. [DOI] [PubMed] [Google Scholar]
- 35. Administration USDoHaHSFaD. Guidance for industry: clinical data needed to support the licensure of pandemic influenza vaccines Available at: https://www.fda.gov/downloads/ BiologicsBloodVaccines/GuidanceComplianceRegulatory Information/Guidances/Vaccines/ucm091985.pdf. Accessed 1 October 2017.
- 36. Bucasas KL, Franco LM, Shaw CA, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis 2011; 203:921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maron DF. A new push for a universal flu vaccine. Sci Am 2018; 20:143–51. [Google Scholar]
- 38. Khurana S, Loving CL, Manischewitz J, et al. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med 2013; 5:200ra114. [DOI] [PubMed] [Google Scholar]
- 39. Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 2013; 3:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klausberger M, Tscheliessnig R, Neff S, et al. Globular head-displayed conserved influenza H1 hemagglutinin stalk epitopes confer protection against heterologous H1N1 virus. PLoS One 2016; 11:e0153579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 2014; 20:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 2008; 8: 34–47. [DOI] [PubMed] [Google Scholar]
- 43. Nimmerjahn F, Ravetch JV. Fcgamma receptors: old friends and new family members. Immunity 2006; 24:19–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.