Abstract
Background and Objective
Progressive pulmonary fibrosis is the main cause of death in patients with systemic sclerosis (SSc) with interstitial lung disease (ILD) and in those with idiopathic pulmonary fibrosis (IPF). Transforming growth factor-β (TGF-β) and NADPH oxidase- (NOX-) derived reactive oxygen species (ROS) are drivers of lung fibrosis. We aimed to determine the role of the epigenetic readers, bromodomain and extraterminal (BET) proteins in the regulation of redox balance in activated myofibroblasts.
Methods
In TGF-β-stimulated fibroblasts, we investigated the effect of the BET inhibitor JQ1 on the mRNA expression of the prooxidant gene NOX4 and the antioxidant gene superoxide dismutase (SOD2) by quantitative RT-PCR, the antioxidant transcription factor NF-E2-related factor 2 (Nrf2) activity by a reporter assay, and intracellular ROS levels by dichlorofluorescein staining. Myofibroblast activation was determined by α-smooth muscle actin immunocytochemistry. The role of specific BET protein isoforms in NOX4 gene regulation was studied by siRNA silencing and chromatin-immunoprecipitation.
Results and Conclusions
Affymetrix gene array analysis revealed increased NOX4 and reduced SOD2 expression in SSc and IPF fibroblasts. SOD2 silencing in non-ILD control fibroblasts induced a profibrotic phenotype. TGF-β increased NOX4 and inhibited SOD2 expression, while increasing ROS production and myofibroblast differentiation. JQ1 reversed the TGF-β-mediated NOX4/SOD2 imbalance and Nrf2 inactivation and attenuated ROS production and myofibroblast differentiation. The BET proteins Brd3 and Brd4 were shown to bind to the NOX4 promoter and drive TGF-β-induced NOX4 expression. Our data indicate a critical role of BET proteins in promoting redox imbalance and pulmonary myofibroblast activation and support BET bromodomain inhibitors as a potential therapy for fibrotic lung disease.
1. Introduction
Approximately two-thirds of patients with systemic sclerosis (SSc) develop some degree of interstitial lung disease (ILD) [1]. Pulmonary fibrosis, together with pulmonary hypertension, is the leading cause of death in SSc patients [2]. Idiopathic pulmonary fibrosis (IPF) is a progressive and inevitably fatal interstitial lung disease with a median survival since diagnosis of 3-5 years [3]. Both diseases are characterised by accumulation of extracellular matrix by an expanding population of myofibroblasts that show enhanced proliferation, migration, and resistance to apoptosis [4, 5].
Persistent TGF-β signaling is central in driving the myofibroblast phenotype in pulmonary fibrosis [6, 7]. Reactive oxygen species (ROS) are key mediators of TGF-β signaling in pulmonary fibroblasts [8, 9]. Intracellular ROS drive myofibroblast differentiation, and inhibiting ROS production ameliorates lung injury in bleomycin-treated mice [10, 11]. Increased levels of ROS [12, 13] and oxidative DNA damage [14] are observed in patients with SSc, and biomarkers of oxidative stress are elevated in IPF [15, 16], some of which negatively correlate with lung function [17, 18].
The prooxidant enzyme NADPH oxidase (NOX-4) and the antioxidant enzyme Mn-superoxide dismutase (MnSOD or SOD2) are central to intracellular ROS regulation. NOX4 reduces O2 into superoxide anion (O2−) and hydrogen peroxide (H2O2) [19]. NOX4 plays a critical role in TGF-β-induced ROS production, fibroblast-to-myofibroblast differentiation, and pulmonary fibrosis [20, 21]. SOD2 converts O2− to H2O2 in the mitochondria and is an important antioxidant during oxidative stress [22]. The role of SOD2 in myofibroblast differentiation and in pulmonary fibrosis is unknown.
Bromodomain and extraterminal (BET) proteins have emerged as promising therapeutic targets for cancer [23], obesity, inflammation [23], and fibrosis [24, 25]. BET proteins (Brd2, Brd3, Brd4, and BrdT) are epigenetic readers, which bind to acetylated lysine residues on histone and nonhistone proteins via conserved bromodomains and regulate gene expression by recruiting transcriptional activators or repressors [23]. JQ1 is a small molecule inhibitor that inhibits the binding of BET protein bromodomains to acetylated lysines [23]. Recently, BET protein inhibition was shown to reduce lung myofibroblast activation and to have antifibrotic effects in an in vivo model of acute lung injury [24, 25]. However, the role of BET proteins in the regulation of intracellular redox state, in the context of lung fibrosis, has not been addressed.
We hypothesised that BET proteins drive redox imbalance and increased ROS production, contributing to myofibroblast differentiation. We therefore investigated the effect of JQ1 on redox balance, in TGF-β-stimulated pulmonary fibroblasts, by determining the expression of NOX4 and SOD2, the expression and activity of the antioxidant transcription factor NF-E2-related factor 2 (Nrf2), and intracellular ROS levels. The effect of JQ1 on the myofibroblast phenotype was studied by determining αSMA expression and myofibroblast contraction and proliferation. The role of specific BET protein isoforms in NOX4 gene expression was determined using chromatin immunoprecipitation (ChIP) and siRNA knock-down.
2. Materials and Methods
2.1. Reagents
JQ1(+) and its inactive enantiomer JQ1(-) were purchased from Cayman (Cambridge, UK), and TGF-β1 was purchased from R&D systems (Minneapolis, MN).
2.2. Fibroblast Cultures
Primary adult pulmonary fibroblasts were obtained from surgical lung resection samples from non-ILD donors (histologically normal periphery of resected lung cancer) and maintained as previously described [26, 27]. All tissue was obtained with appropriate consent and its use approved by the Ethics Committee of the Royal Brompton and Harefield National Health Service Trust. Subject details, fibroblast cultures, and experimental conditions used in the microarray study, from which data for Table 2 and Figure 1(a) were extracted, have been described previously [27]. Cells were plated for experiments at a density of 7000 cells/cm2 and were at ~90% confluence at the start of the experiment. For the proliferation assay, cells were stimulated at ~50% confluence.
Table 2.
NOX4 and SOD2 gene expression data extracted from microarray analysis.
| Gene | Control (n=10) |
SSc-ILD (n=8) |
Fold change | p value | IPF (n=3) |
Fold change | p value |
|---|---|---|---|---|---|---|---|
| NOX4 | 12.3 | 206.6 | 16.9 | 0.012 | 323.6 | 26.4 | 0.016 |
|
| |||||||
| SOD2 | 2180.4 | 311.0 | -7.0 | <1x10−6 | 29.8 | -73.2 | <1x10−6 |
Basal serum-free gene expression in non-ILD control, SSc-ILD, and IPF lung fibroblasts was determined by Affymetrix microarray analysis [27]. Mean expression levels are given in Affymetrix units. Fold changes for SSc-ILD and IPF fibroblasts, and p values adjusted for multiple comparisons, are relative to the expression levels in non-ILD control cells.
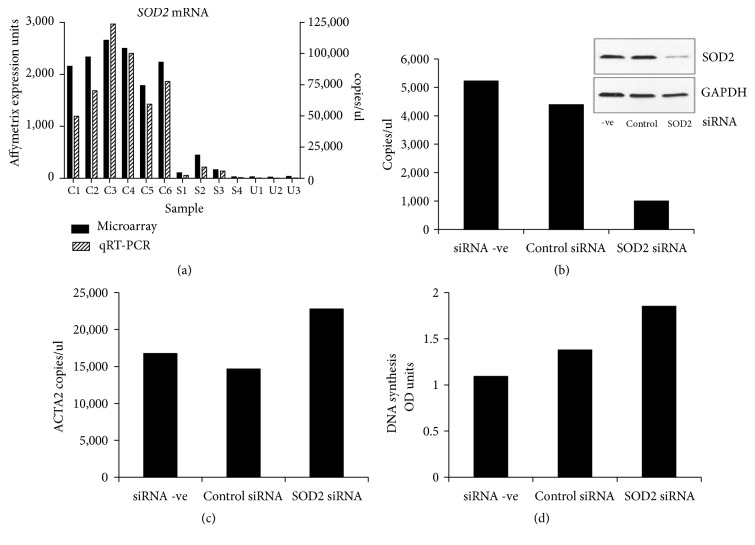
Figure 1.
Effect of SOD2 siRNA knock-down on αSMA expression and cell proliferation. (a) Levels of SOD2 mRNA in non-ILD control (C1-C6), SSc-ILD (S1-S4), and IPF (U1-U3) lung fibroblasts under basal serum-free conditions as determined by Affymetrix microarray analysis (black bars) were confirmed by RT-qPCR (grey bars). (b-d) Non-ILD control lung fibroblasts were either mock-transfected (siRNA –ve) or transfected with nontargeting siRNA (control siRNA), or SOD2-targeting siRNA for 72 h following which SOD2 mRNA and protein (inset) (b) and (c) ACTA2 mRNA expression levels were measured. (d) Proliferation induced by incubation with 3% FBS for 24 h was measured by BrdU incorporation. Data are shown as the mean of three independent experiments performed in two control cell lines ((b) and (c)) or in one control cell line (d), respectively.
2.3. RT-qPCR
Messenger RNA (mRNA) expression was determined using RT-qPCR as previously described [27, 28]. PCR primer details are provided in Table 1.
Table 1.
Primers used for RT-qPCR analysis.
| Gene | Primer sequence |
|---|---|
| NOX4 | forward 5′-CTGCTGACGTTGCATGTTTC-3′ |
|
| |
| reverse 5′-CGGGAGGGTGGGTATCTAA-3′ | |
|
| |
| SOD2 | forward 5′-CACCAGCACTAGCAGCATGT-3′ |
|
| |
| reverse 5′-GAGCCCAGATACCCCAAAAC-3′ | |
|
| |
| ACTA2 | forward 5′-CCCTGAAGTACCCGATAGAACA-3′ |
|
| |
| reverse 5′-GGCAACACGAAGCTCATTG-3′ | |
|
| |
| YWHAZ | forward 5′-ACTTTTGGTACATTGTGGCTTCAA-3′ |
|
| |
| reverse 5′-CCGCCAGGACAAACCAGTAT-3′ | |
|
| |
| HPRT1 | forward 5′-TGACACTGGCAAAACAATGCA-3′ |
|
| |
| reverse 5′-GGTCCTTTTCACCAGCAAGCT-3′ | |
|
| |
| BRD3 | Applied Biosystems TaqMan probe Hs00201284_m1 |
|
| |
| BRD4 | Applied Biosystems TaqMan probe Hs04188087_m1 |
|
| |
| NRF2 (NFE2L2) | QuantiTect Primer Assay QT00027384 |
|
| |
| KEAP1 | QuantiTect Primer Assay QT00080220 |
2.4. siRNA Silencing
Cells were transfected with 3 nM Silencer® Select negative control #1 siRNA or SOD2, Brd3, and Brd4 specific Silencer® Select siRNA (Ambion, Foster City, CA) for 72 h using Lipofectamine® RNAiMAX (Invitrogen) according to the manufacturer's instructions.
2.5. Western Blot Analysis
Whole cell extracts were separated by SDS-PAGE and transferred to a nitrocellulose membrane as previously described [28]. Proteins were detected using anti-SOD2 (Sigma-Aldrich, HPA001814; diluted 1:1000), anti-α-Smooth Muscle Actin (Sigma-Aldrich, A2547; diluted 1:1000), anti-beta Actin (Abcam, Cambridge, UK, ab8227), and mouse anti-GAPDH (Abcam, ab9484; diluted 1:2000) antibodies. Band intensities were quantified using ImageJ [29].
2.6. Proliferation Assay
Proliferation was assessed by measuring BrdU incorporation using the Cell Proliferation ELISA (Roche) according to the manufacturer's instructions.
2.7. Immunocytochemistry
Analysis was performed in 8-well chamber slides as previously described [28]. The primary antibody used was mouse anti-α-Smooth Muscle Actin (Sigma-Aldrich, A2547), used at a dilution of 1:100.
2.8. Collagen Contraction Assay
Collagen matrices were prepared as previously described [18], with some modifications. Briefly, fibroblasts (100,000/well) mixed with neutralised collagen (1.2mg/ml, Invitrogen) supplemented with 2.5μM Mitomycin C (Sigma-Aldrich), 500 nM JQ1(-) or 500 nM JQ1(-), were cast into 24-well plates and allowed to polymerise for 1 h. Additional media (1mL/well) at indicated conditions were added, and the plates were incubated at 37°C. After 60 mins, 1 ng/ml TGF-β1 was added to some of the wells, as indicated, and the plates were incubated for further 24 h. The gels were released and allowed to contract for 4 h before fixing in 4% paraformaldehyde. The gel surface area was measured using ImageJ [29].
2.9. Determination of Intracellular ROS Levels
Intracellular ROS levels were determined by staining cells with the redox-sensitive fluorescent probe 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; Invitrogen) at 5μM for 30 mins and measuring fluorescence using flow cytometry.
2.10. Promoter Reporter Assay
Nrf2 transcriptional activity was assessed using the Cignal Antioxidant Response Element (ARE) reporter assay (Qiagen, Crawley, UK) as described [28], with luminescence detected using the Dual-Luciferase® Reporter Assay System (Promega, Southampton, UK).
2.11. Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the Magna ChIP A/G kit (Merck Millipore, Billerica, MA) and 4 μg of rabbit anti-Brd2 (A302-582A), anti-Brd3 (A302-368A), anti-Brd4 (A301-985A) (Bethyl Laboratories, Cambridge, UK), or Rabbit IgG control antibodies (Millipore, 12-370) as described previously [28]. DNA association was quantified by RT-qPCR (ΔΔ-Ct method) using the following primers: NOX4 forward, 5′-GCTTTAGTTTGGGAGTGGGA-3′, and reverse, 5′-GAAATTTGAGCCGGAAACAG-3′ [30]. DNA used was normalised to input DNA for each sample.
2.12. Statistical Analysis
Data are expressed as mean ± SEM or mean ± SD as indicated. Differences between groups were determined using repeated-measures analysis of variance (ANOVA) followed by a Dunnet's post hoc test. A value of p<0.05 was considered statistically significant. Statistical analysis was performed only in experiments performed on at least three individual cell lines (biological replicates).
3. Results
3.1. Dysregulation of NOX4 and SOD2 Gene Expression in Fibroblasts from Fibrotic Lung
In a previous microarray study, we reported increased NOX4 mRNA expression in pulmonary fibroblasts from patients with SSc-ILD (16.9-fold) and IPF (26.4-fold). Here, we reassessed redox gene expression specifically and found that SOD2 mRNA expression was markedly suppressed in both SSc-ILD (7.0-fold) and IPF (73.2-fold) fibroblasts, compared with non-ILD controls (Table 2, data extracted from [27]). The clinical characteristics of the subjects that donated fibroblasts used for microarray analysis were published previously [27].
3.2. SOD2 Knock-Down Increases αSMA Expression and Inhibits Lung Fibroblast Proliferation
We confirmed the reduction of SOD2 mRNA in fibrotic fibroblasts by RT-qPCR (Figure 1(a), gray bars) and compared it with that seen by microarray (Figure 1(a), black bars). Knock-down (KD) of SOD2 expression with siRNA in lung fibroblasts attenuated SOD2 mRNA levels by approximately 75% (Figure 1(b)), with a corresponding reduction in SOD2 protein levels (Figure 1(b), inset). SOD2 KD also led to an increasing trend in ACTA2 mRNA expression (Figure 1(c)) and cell proliferation (Figure 1(d)).
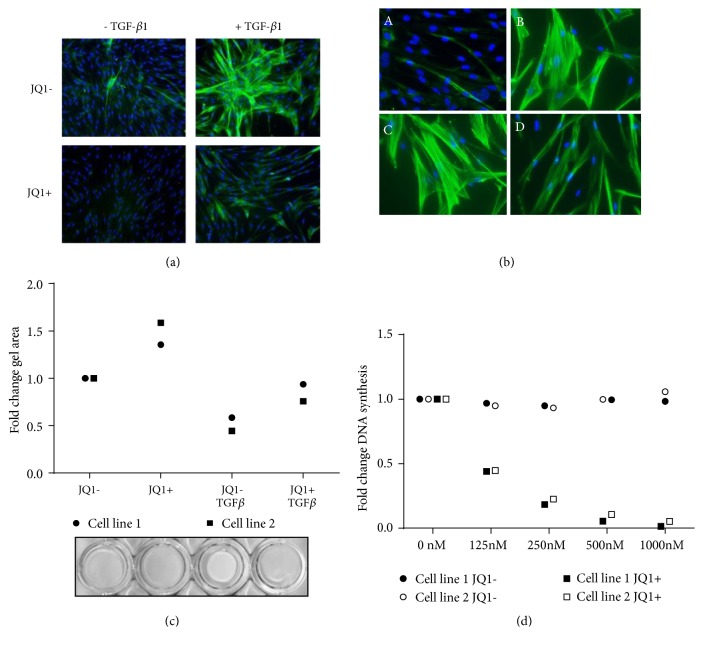
3.3. JQ1 Inhibits TGF-β-Mediated αSMA Fibre Formation, Contractility, and Proliferation of Lung Fibroblasts
TGF-β stimulation of non-ILD control lung fibroblasts for 48 h induced the formation of αSMA-containing stress fibres (Figure 2(a)). Pretreatment with JQ1(+) (500nM) attenuated basal and TGF-β-induced αSMA fibre formation. Importantly, established myofibroblast differentiation induced by 48 h TGFβ stimulation (Figure 2(b), image B) was reversed by treatment with JQ1(+) (Figure 2(b), image D) but not by JQ1(-) (Figure 2(b), image C) for further 48 h.
Figure 2.
Effect of JQ1 on TGF-β-induced myofibroblast phenotypic changes and proliferation. (a) Non-ILD control fibroblasts were serum-starved for 24 h and pretreated with 500 nM JQ1 or JQ1(-) for 2 h before stimulation with TGF-β1 (1ng/ml) for 48 h. Cells were stained for αSMA (green) and nuclear DNA (DAPI/Blue). (b) Cells were cultured in the absence (picture A) or presence (picture B) of TGF-β1 for 48 h before being treated with 500 nM JQ1(-) (picture C) or JQ1(+) (picture D) for further 48 h. (c) Control cells were cast in collagen gels and treated with TGF-β1 and 500 nM JQ1 (+ or -) for 24 h. Gels were released and allowed to contract for 4 h, and the gel area was measured using ImageJ. Data points are mean from experiments in two different cell lines performed in triplicate and expressed as fold change compared to the JQ1- control. (d) Control fibroblasts were serum-starved and pretreated with JQ1 (+ or -) at the indicated concentrations before stimulation with 3% fetal bovine serum (FBS) for 24 h. Proliferation was determined by BrdU incorporation. Data points are mean from experiments in two different cell lines performed in triplicate and expressed as fold change compared to the JQ1- control.
As a functional readout of stress fibre content, we studied the effects of JQ1 on collagen gel contractility. JQ1(+) reduced both baseline and TGF-β-mediated gel contraction, as assessed by increased gel disc area (Figure 2(c)). JQ1(+) also inhibited proliferation induced by fetal bovine serum (FBS; 3%) (Figure 2(d)).
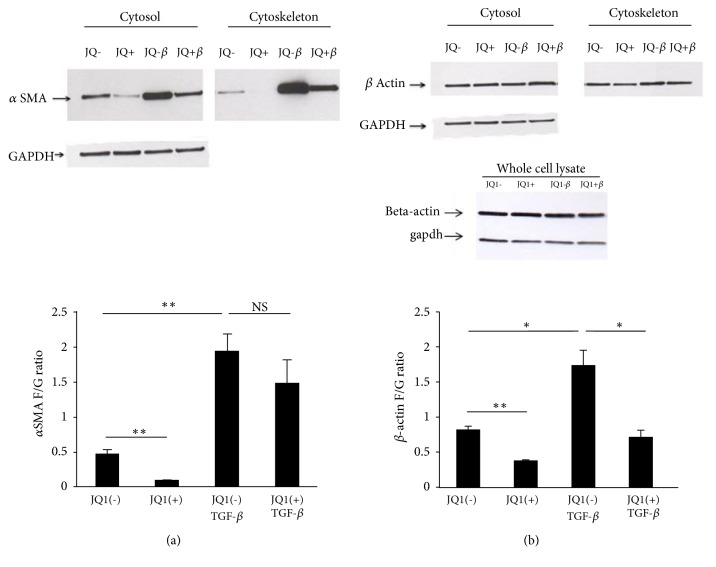
3.4. JQ1 Reduces αSMA Expression and Inhibits β-Actin Intracellular Distribution
TGF-β enhanced both cytosolic and cytoskeletal αSMA protein expression (Figure 3(a), top panel) and caused an approximately 4-fold (p<0.01) shift in αSMA incorporation from globular cytosolic (G-actin) into fibrous cytoskeletal actin (F-actin) (Figure 3(a), graph). JQ1(+) markedly reduced αSMA protein expression in both fractions; however, it did not affect TGF-β-induced αSMA incorporation into fibres. Total β-actin protein expression was not significantly altered by either TGF-β or JQ1(+) (Figure 3(b)). However, the TGF-β-induced redistribution of β-actin from G-actin into F-actin was completely inhibited by JQ1(+) (p<0.05).
Figure 3.
Effect of JQ1 on αSMA and β-actin protein expression and intracellular distribution. Protein expression of αSMA (a) and β-actin (b) was determined by Western blot analysis in non-ILD lung fibroblasts after 48 h in the presence or absence of TGF-β1 and JQ1(+) or JQ1(-). F-actin was separated from G-actin by differential solubility in Triton X-100. Both fractions had the same final volume, and equal volumes were run on SDS-PAGE gels. Total levels of β-actin were also analysed in whole cell extracts. Images are representative of three independent experiments. Signal intensity was measured using ImageJ. Bars represent mean ± SD from three independent non-ILD control cell lines. NS = nonsignificant. ∗p<0.05 and ∗∗ p<0.01.
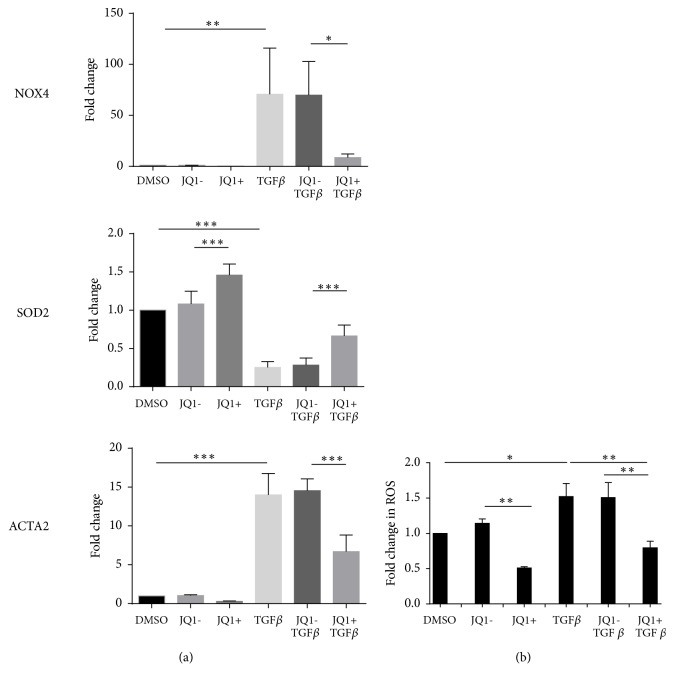
3.5. JQ1 Reverses the TGFβ-Induced Imbalance in NOX4 and SOD2 Expression and Reduces Intracellular ROS Levels and ACTA2 Expression
JQ1(+) led to a reduction in NOX4 and ACTA2 mRNA compared to JQ1(-) after 24 h in non-ILD control pulmonary fibroblasts but did not reach statistical significance. In contrast, the expression of SOD2 mRNA was significantly increased (1.45-fold, p<0.001) (Figure 4(a)). TGF-β increased NOX4 (76-fold, p<0.01) and ACTA2 (14-fold, p<0.001) mRNA levels, while SOD2 mRNA levels were significantly reduced (0.24-fold, p<0.001). JQ1(+) significantly attenuated TGF-β-induced NOX4 (0.09-fold, p<0.05) and ACTA2 (0.31-fold, p<0.001) expression and partially reversed the inhibition of SOD2 mRNA levels by TGF-β (2.9-fold, p<0.01) (Figure 4(a)). In line with these findings, JQ1(+) significantly suppressed baseline (0.51-fold, p<0.01) and TGF-β-induced (0.53-fold, p<0.01) intracellular ROS in pulmonary fibroblasts (Figure 4(b)) There was no effect of JQ1(-) on ROS levels.
Figure 4.
Effect of JQ1 on TGF-β-induced NOX4, SOD2 and ACTA2 mRNA expression, and intracellular ROS. Non-ILD control pulmonary fibroblasts were serum starved and pretreated with 500 nM JQ1(+) or JQ1(-) for 2 h before stimulation with TGF-β1 (1ng/ml) for further 24 h. (a) NOX4, SOD2, and ACTA2 mRNA was determined by RT-qPCR. (b) Intracellular ROS levels were determined by 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate staining. Data are expressed as fold change compared to the DMSO control and bars represent mean ± SD of independent experiments performed in three (a) and four (b) cell lines. ∗p<0.05, ∗∗p<0.01, and ∗∗∗p<0.001.
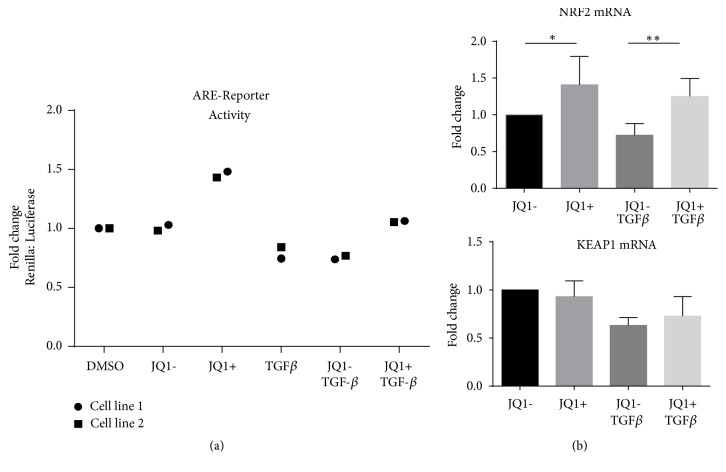
3.6. JQ1 Increases Nrf2 Activity
We have previously shown that BET proteins suppress Nrf2-mediated antioxidant gene transcription in airway smooth muscle cells (ASMCs) [28]. In the current study, JQ1(+) treatment of non-ILD lung fibroblasts led to an increasing trend in Antioxidant Response Element (ARE) promoter-driven gene transcription (Figure 5(a)) and a statistically significant increase in NRF2 mRNA both in absence and presence of TGF-β (Figure 5(b)), while JQ1(-) had no effect. JQ1(+) did not affect the mRNA expression of Keap1, the cytoplasmic inhibitor of Nrf2 (Figure 5(b)).
Figure 5.
Effect of JQ1 on Antioxidant Response Element- (ARE-) dependent transcription and Nrf2 and Keap1 mRNA expression. (a) Non-ILD control fibroblasts were cotransfected for 48 hours with an inducible ARE-responsive Firefly-luciferase reporter construct and a Renilla-luciferase reference construct. Cells were serum-starved for 24 h and pretreated with 500 nM JQ1(+) or JQ1(-) for 2 h and then stimulated with TGF-β1 (1ng/ml) for 24 h. Bars represent fold change compared to DMSO control. Data points shown are two independent experiments performed in triplicate in two individual non-ILD control cell lines expressed as mean fold change compared to DMSO (vehicle control). (b) Cells were serum-starved for 24 h and pretreated with 500 nM JQ1(+) or JQ1(-) for 2 h before being stimulated with TGF-β1 (1ng/ml) for 24 h. NRF2 and KEAP1 mRNA levels were determined by RT-qPCR. Data are expressed as fold change compared to the JQ1- control. Bars represent mean ± SD of independent experiments in three individual non-ILD control lines. ∗p<0.05 and ∗∗p<0.01.
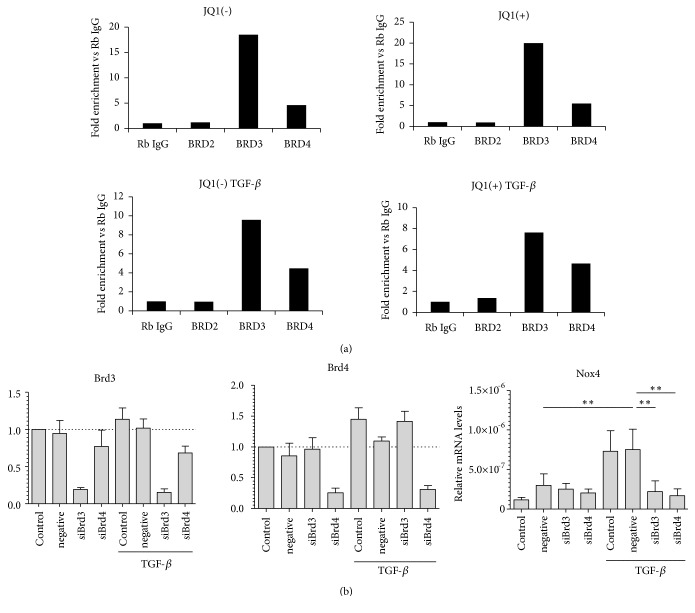
3.7. Brd3 and Brd4 Are Enriched at the NOX4 Promoter and Required for TGFβ-Induced Gene Transcription
ChIP assays in non-ILD fibroblasts from two subjects in two independent experiments demonstrated that Brd3 and Brd4, but not Brd2, bind to the NOX4 promoter (Figure 6(a)). Furthermore, Brd3 or Brd4 siRNA knock-down, which efficiently reduced Brd3 and Brd4 mRNA, respectively, led to inhibition of TGF-β-induced NOX4 mRNA expression, suggesting a role of both these proteins in driving NOX4 gene expression (Figure 6(b)).
Figure 6.
Enrichment of Brd2, Brd3, and Brd4 at the NOX4 promoter and their role in TGFβ-induced Nox4 expression. (a) Non-ILD control fibroblasts were starved for 24 h and pretreated with 500 nM JQ1(+) or JQ1(-) for 2 h before stimulation with TGF-β1 (1ng/ml) for further 4 h. Recruitment of Brd2, Brd3, and Brd4 to the NOX4 promoter was determined by ChIP. Data are expressed as fold enrichment compared to the IgG control. Data shown are the mean of two independent experiments performed in two control cell lines. (b) Non-ILD control fibroblasts were either mock-transfected (Control) or transfected with nonspecific siRNA control (negative) or with specific siRNAs to BRD3 (siBrd3) or BRD4 (siBrd4). Cells were stimulated with TGF-β1 for 24 h and the expressions of BRD3, BRD4, or NOX4 mRNA levels were determined by RT-qPCR. The data is presented as the mean ± SEM of independent experiments on four individual cell lines. ∗∗p<0.01 and ∗∗∗p<0.001.
4. Discussion
We show that the small molecule BET protein inhibitor JQ1 reduces basal and TGF-β-induced NOX4 expression and ROS production. JQ1 also increases the expression of the antioxidant gene SOD2 and the activity of the cytoprotective transcription factor Nrf2 and prevents the inhibition of SOD2 expression by TGF-β. At the same time, JQ1 prevents and reverses TGF-β-mediated myofibroblast differentiation. The suppressive effect of BET protein inhibition on myofibroblast differentiation may be mediated, at least partly, through restoring redox balance via SOD2 regulation, as SOD2 depletion was found to increase αSMA expression and fibroblast proliferation.
The important role of oxidative stress in pulmonary fibrosis has directed therapeutic efforts towards inhibiting ROS generation and enhancing antioxidant capacity [31]. Several therapies involving antioxidant compounds, such as N-acetylcysteine (NAC), have been tested and while promising in preclinical models have ultimately lacked clinical efficacy. Targeting a primary source of intracellular ROS by inhibiting NOX4 activity has lately been a strong focus in antifibrotic drug development. However, there are currently no clinically available, selective NOX4 inhibitors [31].
NOX4 overexpression has been shown to be fundamental in maintaining a myofibroblast phenotype [20]. Here we demonstrate that BET proteins, and specifically Brd3 and Brd4, drive NOX4 expression in pulmonary fibroblasts. Our findings are in line with recent evidence demonstrating a role of Brd4 in TGF-β-induced NOX4 expression in skin and renal fibroblasts [32, 33]. Inhibition of NOX4 expression by JQ1 was accompanied by a reduction in ROS levels, ACTA2 gene expression, stress fibre formation, myofibroblast contraction, and proliferation. NOX4 is one of the early key genes involved in the initiation of a TGF-β-stimulated fibrotic response [8, 20]. Therefore, the antifibrotic effects of JQ1 may be mediated, at least partly, through inhibition of NOX4 expression. Importantly, our finding that Brd3 and Brd4, but not Brd2, are involved in NOX4 regulation by TGF-β suggests that a degree of selectivity may be possible when considering BET inhibitors as possible therapeutic agents in pulmonary fibrosis. However, a study by Tang et al., also performed in human lung fibroblasts, reported that Brd2 and Brd4, but not Brd3, mediate TGF-β-mediated α-SMA expression and myofibroblast activation [24]. This apparent disparity is hard to explain at this point; however, it could suggest that the specificity of BET protein isoforms recruitment to specific gene promoters may be context-specific and dependent on timing and culture conditions. In addition, detailed studies elucidating the roles of specific BET proteins are technically challenging and were relatively limited in both these studies. For example, Tang et al. focused only on Brd4 to show direct binding to promoters of fibrosis-related genes, αSMA (ACTA2), IL6, and PAI-1 (SERPINE1), whereas our study investigated binding of all three BET protein isoforms on the NOX4 promoter. More in-depth investigations will be necessary to delineate the roles of specific BET proteins in progressive fibrosis with a view to targeting them for treatment.
TGF-β-mediated upregulation of NOX4 was accompanied by a reduction in SOD2 expression, in line with our previous findings in ASMC [34]. Attenuated SOD2 expression may contribute to the profibrotic effect of TGF-β, as we show here that knock-down of SOD2 expression results in increased ACTA2 expression and fibroblast proliferation. Reduced SOD2 expression in pulmonary arterial smooth muscle cells of patients with pulmonary arterial hypertension (PAH) has been associated with a phenotype of increased proliferation and lower apoptosis [22]. Downregulation of SOD2 has also been shown to mediate TGF-β-induced epithelial-mesenchymal transition, a profibrotic mechanism, in renal epithelial cells [35]. These findings suggest that the antifibrotic effect of JQ1 may, at least partly, occur through the upregulation of SOD2.
We observed that, during the TGF-β-induced fibrotic state, there was a trend towards reduced expression and activity of Nrf2, a key mediator of cellular antioxidant protection [36]. Furthermore, on reexamination of our previous microarray data, we found that the Nrf2 expression was significantly lower in pulmonary fibroblasts from IPF (fold change -1.77, p=0.0096) and from SSc-ILD (fold change -1.52, p=0.02) lung compared with non-ILD control lung [27]. Reduced Nrf2 gene expression has been reported previously in IPF pulmonary fibroblasts compared to controls, and Nrf2 knock-down has been shown to induce oxidative stress and myofibroblast differentiation [37]. JQ1 increased Nrf2 expression and activity in both absence and presence of TGF-β, indicating that BET protein inhibition can increase antioxidant protection in activated fibroblasts. Our data are consistent with previous findings by our group showing that BET proteins interact with Nrf2 and inhibit ARE-dependent antioxidant gene transcription in ASMC [28]. Studies in WI-38 cells have also reported that BRD4 knock-down leads to increased Nrf2 nuclear accumulation and activity under oxidative stress [38]. A more in-depth investigation, including a study of the interaction of Keap1 with actin fibres, known to be essential for Nrf2 inactivation, is required to better understand the regulation of Nrf2 activity by BET proteins in pulmonary fibroblasts [39].
Our data highlight an important new molecular mechanism regulating redox balance and myofibroblastic differentiation. Future studies will need to confirm this mechanism in fibroblasts from patients with pulmonary fibrosis and in animal models of pulmonary fibrosis to firmly establish a role of BET proteins in regulating redox gene expression and ROS levels during fibrosis.
In summary, our data suggest that BET protein inhibition restores redox balance through its effects on NOX4/SOD2 expression and Nrf2 activity, leading to the reprogramming/dedifferentiation of myofibroblasts. Our findings highlight BET protein inhibitors as potential antifibrotic therapeutic agents. Studies are now urgently needed to assess the suitability of these compounds for pulmonary fibrosis.
Acknowledgments
The research reported in this publication was funded by Raynaud's and Scleroderma Association (RSA) and the Scleroderma Society, by the Dunhill Medical Trust, and by the British Lung Foundation (RG14-10). Charalambos Michaeloudes is supported by the Dunhill Medical Trust (R368/0714). Sharon Mumby is supported by a British Heart Foundation grant (PG/14/27/30679). Ian M. Adcock and Kian Fan Chung are supported by the MRC-ABPI COPD-MAP consortium (G1001367/1). Andrew L. Durham is supported by a grant from the British Lung Foundation. The work was supported by the NIHR Respiratory Disease Biomedical Research Unit at the Royal Brompton and Harefield NHS Foundation Trust and Imperial College London.
Data Availability
All the data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Nihtyanova S. I., Schreiber B. E., Ong V. H., et al. Prediction of pulmonary complications and long term survival in systemic sclerosis. Arthritis & Rheumatology. 2014;66(6):1625–1635. doi: 10.1002/art.38390. [DOI] [PubMed] [Google Scholar]
- 2.Steen V. D., Medsger T. A. Changes in causes of death in systemic sclerosis, 1972–2002. Annals of the Rheumatic Diseases. 2007;66(7):940–944. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gribbin J., Hubbard R. B., Le Jeune I., Smith C. J. P., West J., Tata L. J. Incidence and mortality of idiopathic pulmonary fibrosis and sarcoidosis in the UK. Thorax. 2006;61(11):980–985. doi: 10.1136/thx.2006.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scotton C. J., Chambers R. C. Molecular targets in pulmonary fibrosis: The myofibroblast in focus. CHEST. 2007;132(4):1311–1321. doi: 10.1378/chest.06-2568. [DOI] [PubMed] [Google Scholar]
- 5.Sivakumar P., Ntolios P., Jenkins G., Laurent G. Into the matrix: targeting fibroblasts in pulmonary fibrosis. Current Opinion in Pulmonary Medicine. 2012;18(5):462–469. doi: 10.1097/mcp.0b013e328356800f. [DOI] [PubMed] [Google Scholar]
- 6.Border W. A., Noble N. A. Transforming growth factor beta in tissue fibrosis. The New England Journal of Medicine. 1994;331(19):1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 7.Samarakoon R., Overstreet J. M., Higgins P. J. TGF-beta signaling in tissue fibrosis: redox controls, target genes and therapeutic opportunities. Cellular Signalling. 2013;25(1):264–268. doi: 10.1016/j.cellsig.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain M., Rivera S., Monclus E. A., et al. Mitochondrial reactive oxygen species regulate transforming growth factor-β signaling. The Journal of Biological Chemistry. 2013;288(2):770–777. doi: 10.1074/jbc.m112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thannickal V. J., Fanburg B. L. Activation of an H2O2-generating NADH oxidase in human lung fibroblasts by transforming growth factor beta1. The Journal of Biological Chemistry. 1995;270(51):30334–30338. doi: 10.1074/jbc.270.51.30334. [DOI] [PubMed] [Google Scholar]
- 10.Teixeira K. C., Soares F. S., Rocha L. G., et al. Attenuation of bleomycin-induced lung injury and oxidative stress by N-acetylcysteine plus deferoxamine. Pulmonary Pharmacology and Therapeutics. 2008;21(2):309–316. doi: 10.1016/j.pupt.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Manoury B., Nennan S., Leclerc O., et al. The absence of reactive oxygen species production protects mice against bleomycin-induced pulmonary fibrosis. Respiratory Research. 2005;6:p. 11. doi: 10.1186/1465-9921-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa F., Shimizu K., Muroi E., Hara T., Sato S. Increasing levels of serum antioxidant status, total antioxidant power, in systemic sclerosis. Clinical Rheumatology. 2011;30(7):921–925. doi: 10.1007/s10067-011-1695-4. [DOI] [PubMed] [Google Scholar]
- 13.Sambo P., Baroni S. S., Luchetti M., et al. Oxidative stress in scleroderma: Maintenance of scleroderma fibroblast phenotype by the constitutive up-regulation of reactive oxygen species generation through the NADPH oxidase complex pathway. Arthritis & Rheumatology. 2001;44(11):2653–2664. doi: 10.1002/1529-0131(200111)44:11<2653::AID-ART445>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Avouac J., Borderie D., Ekindjian O. G., Kahan A., Allanore Y. High DNA oxidative damage in systemic sclerosis. The Journal of Rheumatology. 2010;37(12):2540–2547. doi: 10.3899/jrheum.100398. [DOI] [PubMed] [Google Scholar]
- 15.Montuschi P., Ciabattoni G., Pared P., et al. 8-Isoprostane as a biomarker of oxidative stress in interstitial lung diseases. American Journal of Respiratory and Critical Care Medicine. 1998;158(5, part 1):1524–1527. doi: 10.1164/ajrccm.158.5.9803102. [DOI] [PubMed] [Google Scholar]
- 16.Rahman I., Skwarska E., Henry M., et al. Systemic and pulmonary oxidative stress in idiopathic pulmonary fibrosis. Free Radical Biology & Medicine. 1999;27(1-2):60–68. doi: 10.1016/s0891-5849(99)00035-0. [DOI] [PubMed] [Google Scholar]
- 17.Psathakis K., Mermigkis D., Papatheodorou G., et al. Exhaled markers of oxidative stress in idiopathic pulmonary fibrosis. European Journal of Clinical Investigation. 2006;36(5):362–367. doi: 10.1111/j.1365-2362.2006.01636.x. [DOI] [PubMed] [Google Scholar]
- 18.Daniil Z. D., Papageorgiou E., Koutsokera A., et al. Serum levels of oxidative stress as a marker of disease severity in idiopathic pulmonary fibrosis. Pulmonary Pharmacology and Therapeutics. 2008;21(1):26–31. doi: 10.1016/j.pupt.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Martyn K. D., Frederick L. M., von Loehneysen K., Dinauer M. C., Knaus U. G. Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cellular Signalling. 2006;18(1):69–82. doi: 10.1016/j.cellsig.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 20.Hecker L., Vittal R., Jones T., et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nature Medicine. 2009;15(9):1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amara N., Goven D., Prost F., Muloway R., Crestani B., Boczkowski J. NOX4/NADPH oxidase expression is increased in pulmonary fibroblasts from patients with idiopathic pulmonary fibrosis and mediates TGFbeta1-induced fibroblast differentiation into myofibroblasts. Thorax. 2010;65(8):733–738. doi: 10.1136/thx.2009.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer S. L., Marsboom G., Kim G. H., et al. Epigenetic attenuation of mitochondrial superoxide dismutase 2 in pulmonary arterial hypertension: a basis for excessive cell proliferation and a new therapeutic target. Circulation. 2010;121(24):2661–2671. doi: 10.1161/circulationaha.109.916098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller S., Filippakopoulos P., Knapp S. Bromodomains as therapeutic targets. Expert Reviews in Molecular Medicine. 2011;13, article e29 doi: 10.1017/s1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X., Peng R., Ren Y., et al. BET bromodomain proteins mediate downstream signaling events following growth factor stimulation in human lung fibroblasts and are involved in bleomycin-induced pulmonary fibrosis. Molecular Pharmacology. 2013;83(1):283–293. doi: 10.1124/mol.112.081661. [DOI] [PubMed] [Google Scholar]
- 25.Tang X., Peng R., Phillips J. E., et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. The American Journal of Pathology. 2013;183(2):470–479. doi: 10.1016/j.ajpath.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Abraham D. J., Shiwen X., Black C. M., Sa S., Xu Y., Leask A. Tumor necrosis factor alpha suppresses the induction of connective tissue growth factor by transforming growth factor-beta in normal and scleroderma fibroblasts. The Journal of Biological Chemistry. 2000;275(20):15220–15225. doi: 10.1074/jbc.275.20.15220. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl G. E., Stock C. J. W., Shi-Wen X., et al. Microarray profiling reveals suppressed interferon stimulated gene program in fibroblasts from scleroderma-associated interstitial lung disease. Respiratory Research. 2013;14:p. 80. doi: 10.1186/1465-9921-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaeloudes C., Mercado N., Clarke C., et al. Bromodomain and extraterminal proteins suppress NF-E2-related factor 2-mediated antioxidant gene expression. The Journal of Immunology. 2014;192(10):4913–4920. doi: 10.4049/jimmunol.1301984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider C. A., Rasband W. S., Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X., Murphy T. C., Nanes M. S., Hart C. M. PPARγ regulates hypoxia-induced Nox4 expression in human pulmonary artery smooth muscle cells through NF-κB. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2010;299(4):L559–L566. doi: 10.1152/ajplung.00090.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hecker L., Thannickal V. J. Getting to the core of fibrosis: targeting redox imbalance in aging. Annals of Translational Medicine. 2016;4(5):93–93. doi: 10.21037/atm.2015.12.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B., Mu J., Gong Y., et al. Brd4 inhibition attenuates unilateral ureteral obstruction-induced fibrosis by blocking TGF-β-mediated Nox4 expression. Redox Biology. 2017;11:390–402. doi: 10.1016/j.redox.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ijaz T., Jamaluddin M., Zhao Y., et al. Coordinate activities of BRD4 and CDK9 in the transcriptional elongation complex are required for TGFβ-induced Nox4 expression and myofibroblast transdifferentiation. Cell Death & Disease. 2017;8(2) doi: 10.1038/cddis.2016.434.e2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaeloudes C., Sukkar M. B., Khorasani N. M., Bhavsar P. K., Chung K. F. TGF-β regulates Nox4, MnSOD and catalase expression, and IL-6 release in airway smooth muscle cells. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2011;300(2):L295–L304. doi: 10.1152/ajplung.00134.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegel A. J., Fang Y., Liu Y., et al. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor β1: a novel role of miR-382. Nucleic Acids Research. 2010;38(22):8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walters D. M., Cho H.-Y., Kleeberger S. R. Oxidative stress and antioxidants in the pathogenesis of pulmonary fibrosis: A potential role for Nrf2. Antioxidants & Redox Signaling. 2008;10(2):321–332. doi: 10.1089/ars.2007.1901. [DOI] [PubMed] [Google Scholar]
- 37.Artaud-Macari E., Goven D., Brayer S., et al. Nuclear factor erythroid 2-related factor 2 nuclear translocation induces myofibroblastic dedifferentiation in idiopathic pulmonary fibrosis. Antioxidants & Redox Signaling. 2013;18(1):66–79. doi: 10.1089/ars.2011.4240. [DOI] [PubMed] [Google Scholar]
- 38.Hussong M., Börno S. T., Kerick M., et al. The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response. Cell Death & Disease. 2014;5(4) doi: 10.1038/cddis.2014.157.e1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang M.-I., Kobayashi A., Wakabayashi N., Kim S.-G., Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proceedings of the National Acadamy of Sciences of the United States of America. 2004;101(7):2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data used to support the findings of this study are included within the article.