Abstract
We used hierarchical clustering to examine gene expression profiles generated by serial analysis of gene expression (SAGE) in a total of nine normal lung epithelial cells and non-small cell lung cancers. Separation of normal and tumor, as well as histopathological subtypes, was evident by using the 3,921 most abundant transcript tags. This distinction remained when only 115 highly differentially expressed tags were used. Furthermore, these 115 transcript tags clustered into groups suggestive of the unique biological and pathological features of the different tissues examined. Adenocarcinomas were characterized by high-level expression of small airway-associated or immunologically related proteins, whereas squamous cell carcinomas overexpressed genes involved in cellular detoxification or antioxidation. The messages of two p53-regulated genes, p21WAF1/CIP1and 14-3-3σ, were consistently underexpressed in the adenocarcinomas, suggesting that the p53 pathway itself might be compromised in this cancer type. Gene expression patterns observed by SAGE were consistent with results obtained by quantitative real-time PCR or cDNA array analyses by using a total of 43 lung tumor and normal samples. Thus, although derived from only a few tissue libraries, gene expression profiles obtained by using SAGE most likely represent an unbiased yet distinctive molecular signature for the most common forms of human lung cancer.
Lung cancer is the leading cause of cancer death worldwide, and non-small cell lung cancer (NSCLC) accounts for nearly 80% of the disease (1). On the basis of cell morphology, adenocarcinoma and squamous carcinoma are the most common types of NSCLC (2). Although the clinical courses of these tumors are similar, adenocarcinomas are characterized by peripheral location in the lung and often have activating mutations in the K-ras oncogene (3, 4). In contrast, squamous cell carcinomas are usually centrally located and more frequently carry p53 gene mutations (5). Furthermore, the etiology of squamous cell carcinoma is closely associated with tobacco smoking, whereas the cause of adenocarcinoma remains unclear (6, 7). Although many molecular changes associated with NSCLC have been reported (8, 9), the global gene expression pattern associated with these two most common types of lung cancer has not been described. Understanding gene expression patterns in these major tumor types will uncover novel markers for disease detection as well as potential targets for rational therapy of lung cancer.
Several technologies are currently being used for gene expression profiling in human cancer (10). Serial analysis of gene expression (SAGE) (11) is an open system that rapidly identifies any expressed transcript in a tissue of interest, including transcripts that have not yet been identified. This highly quantitative method can accurately identify the degree of expression for each transcript. Comparing SAGE profiles between the tumor and the corresponding normal tissues can readily identify genes differentially expressed in the two samples. By using this method, novel transcripts and molecular pathways have been discovered (12–14). In contrast, cDNA arrays represent a closed system that analyzes relative expression levels of previously known genes or transcripts (15, 16). Because many thousands of genes can be placed on a single membrane or slide for rapid screening, such studies have recently demonstrated molecular profiles of several human cancers (17–20).
Hierarchical clustering is a systematic method widely used in cDNA array data analysis, where the differences between the expression patterns of many genes is generally within a few-fold (21). We reasoned that because SAGE is highly quantitative, hierarchical clustering might be used to organize gene expression profiles generated by SAGE from just a few tissue libraries. To test this, we used SAGE tags that were generated from two of each libraries derived from primary adenocarcinomas, primary squamous cell carcinomas, normal lung small airway epithelial cells (SAEC), or normal bronchial/tracheal epithelial (NHBE) cells, and a lung adenocarcinoma cell line. SAGE tags showing the highest abundance were subjected to clustering analysis. Although each library was derived from a different individual, normal and tumor samples clustered in two separate branches, whereas tissues of different cell types clustered together. Furthermore, SAGE tags clustered into biologically meaningful groups, revealing the important molecular characteristics of these two most common NSCLC subtypes.
Materials and Methods
Tumors and Cell Lines.
Primary lung tumor tissues used for SAGE were microdissected and obtained from Johns Hopkins Hospital after surgery for lung resection because of cancer and as previously described (9). Histologically, the two squamous tumors were moderately differentiated squamous cell carcinomas, whereas the two adenocarcinomas consisted of a well differentiated and a poorly differentiated tumor with a shared common feature of lymphoplasmacytic infiltrations in the adjacent alveolar septa. SAEC and NHBE cells were purchased from Clonetics/BioWhittaker (Walkersville, MD) and propagated following the manufacturer's instructions. We chose these two types of primary cell cultures as normal controls because they represented pure populations of lung epithelial cells from the small and large airways, respectively. An established lung adenocarcinoma cell line, A549, was included in the SAGE analysis to control for potential tissue culture effects on the primary lung epithelial cells. Tumor RNA samples used for quantitative PCR and GeneChip analyses were either purchased from BioChain (Hayward, CA) or obtained in the same manner as samples used for SAGE (9). A549 cells were obtained as a gift from James Herman (Johns Hopkins Oncology Center, Johns Hopkins Medical School).
SAGE Libraries and SAGE Analysis.
Total RNA samples were isolated by RNazol B (Tel-Test, Friendswood, TX) according to the manufacturer's recommendations. Poly(A)+ RNA was extracted by using the Oligotex mRNA Mini Kit (Qiagen, Chatsworth, CA) and the Dynabeads mRNA DIRECT Kit (Dynal, Oslo). SAGE libraries were generated and the tags sequenced as described (21). sage 300 software (http://www.sagenet.org/sage_protocol.htm) was used to identify tag sequences and to quantify the abundance of each tag. The gene identity and UniGene cluster assignment of each SAGE tag was obtained by using the tag-to-gene “reliable” map (updated April 23, 2001) from ftp://ncbi.nlm.nih.gov/pub/sage/map and the table of UniGene clusters (updated May 23, 2001), from http://www.ncbi.nlm.nih.gov/UniGene/.
Normalization and Hierarchical Clustering Analysis.
The cluster 2.11 program (http://rana.lbl.gov) was used for normalization and clustering of the SAGE data. Briefly, the normalization included logarithmic transformation of the data, followed by 10 cycles of centering the data on the median by samples, then by genes, each time scaling the sum of the squares in each sample and each gene to 1. The noncentered Pearson correlation was used for distance calculations and the weighted-average linkage was used for clustering as described (22).
Multidimensional Scaling of Normal Lung and Tumor Samples.
We developed a program based on the classical multidimensional scaling algorithm (23) and used it to determine the relatedness of each library analyzed by SAGE. Each sample was used to generate a unique library. A table of normalized expression levels for each gene in every library was used as a dissimilarity matrix. Normalization was performed by using the cluster 2.11 program, as described above. Multidimensional scaling allows for the calculation of coordinates of objects if the distances between objects are known. The distances between the samples were calculated as 1 − Cnm, where Cnm was the correlation coefficient between libraries n and m. The distance matrix spans an N-dimensional space, where N is the number of libraries in the study. The first three principal coordinates were used to best fit the libraries into a three-dimensional realm for presentation purposes.
Statistical Analysis.
P-chance analysis (available in the sage 300 software and described in ref. 21) was used to select genes most differentially expressed between each tumor and its corresponding normal controls. P-chance uses the Monte Carlo method (24) to calculate the relative probability of detecting an expression difference equal to, or greater than, the observed expression difference between two samples by chance alone. For each tumor type, one of the two tumor libraries was first compared with the two corresponding normal libraries to select genes with a P-chance value of <0.001. At this P-chance, the false positive rate for all selected genes was <0.015. We next selected only those genes with consistent expression patterns in both tumor libraries of the same cell type and combined them with genes selected from the other tumor type by using the same method.
Real-Time Quantitative PCR Analysis.
Five genes identified by SAGE as highly expressed in either adenocarcinomas or squamous cell carcinoma were analyzed by real-time reverse transcription–PCR (RT-PCR) by using 14 RNA samples from lung tumors and controls (25). The real-time RT-PCR probes and primers were designed by using primer express software (PE Biosystems, Foster City, CA). Primer sequences and reaction conditions are published as supporting information on the PNAS web site, www.pnas.org. The relative expression of each gene was calculated as the ratio of the average gene expression levels for tumors of the same cell type compared with its corresponding normal.
Gene Expression Analysis by Using GeneChip.
GeneChip U95A probe arrays were obtained from Affymetrix (Santa Clara, CA). A total of 32 RNA samples were individually prepared, hybridized to the genechip, and scanned by a Hewlett–Packard GeneArray scanner following the protocols provided by the manufacturer. The source and tissue type of each sample used is published as supporting information on the PNAS web site. Six internal genechip standards, β-actin, 18S rRNA, 28S rRNA, glyceraldehyde-3-phosphate dehydrogenase, transferrin receptor, and the transcription factor ISGF-3, were used as controls to ensure the quality of all samples tested.
Results and Discussion
SAGE of NSCLC.
A total of nine independent SAGE libraries were generated from five different normal and tumor samples. A total of 18,300 independent clones were sequenced to generate 374,634 tags that represented 66,502 distinct transcripts (Table 1). Of the 23,056 distinct tags that appeared more than once in all nine libraries combined, 18,595 tags had at least one match to a UniGene cluster, 4,907 tags had multiple matches, 4,319 tags had no match, and 142 tags matched mitochondrial DNA or ribosomal RNA sequences. Accounting for 7% potential sequencing errors (21) in tags that appeared only once in all nine libraries, the total number of distinct transcript tags identified is about 59,000. Although this number exceeds the current estimate of 30,000–40,000 genes predicted in the human genome (26, 27), the discrepancy could be accounted for by alternatively spliced transcripts and polyadenylation usage sites, which can result in multiple SAGE tags for the same gene (26, 28, 29). Alternatively, because our transcript analysis was based on only nine lung samples, it is possible that the current gene estimates are low, because novel tags would be expected when libraries from other tissues are included.
Table 1.
SAGE in NSCLC and normal lung bronchial epithelial cells
| Tissue source | Number of clones | Number of tags |
|---|---|---|
| NHBE-1 | 3,759 | 58,273 |
| NHBE-2 | 4,046 | 59,885 |
| SAEC-1 | 838 | 21,318 |
| SAEC-2 | 1,299 | 26,956 |
| Squamous cell carcinoma-A | 2,259 | 56,817 |
| Squamous cell carcinoma-B | 2,186 | 51,901 |
| Adenocarcinoma-A | 799 | 21,714 |
| Adenocarcinoma-B | 928 | 24,018 |
| Adenocarcinoma cell line A549 | 2,186 | 53,752 |
| Total number | 18,300 | 374,634 |
Summary: Number of unique libraries = 9; number of unique tags = 66,502; number of unique tags that appear >1 = 23,056; number matched to unique UniGene cluster = 18,595.
Hierarchical Clustering of Tumor and Normal Lung Tissues Based on SAGE.
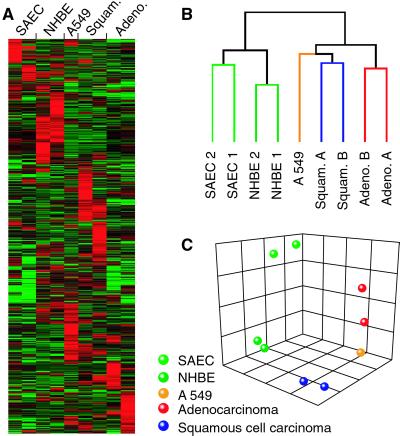
To identify genes that are differentially expressed between the tumors and the normal samples, as well as between the different tumor types, we examined the overall similarities of the libraries derived from each tissue by using hierarchical clustering (22). Because expression differences for more highly expressed genes are less likely to have been observed by chance, a collection of 3,921 SAGE tags appearing at least 10 times in all nine libraries was subjected to the clustering analysis. Although each sample was derived from a different individual and had a unique expression pattern (Fig. 1A), the normal tissues were more similar to each other and the tumor tissues were more alike as a group. Furthermore, the SAEC and NHBE samples each paired together under the normal branch, whereas the adenocarcinomas and the squamous cell tumors clustered together under the tumor branch (Fig. 1B). The adenocarcinoma-derived A549 cell line branched with the NSCLC tumors and demonstrated its relatedness to the two adenocarcinomas in multidimensional scaling (Fig. 1C), which displays the spatial relationship of all nine samples with respect to one another (23).
Figure 1.
Clustering and multidimensional scaling of the SAGE libraries. Only genes with total tag counts of at least 10 are included. (A) Cluster of all nine SAGE libraries. Genes are aligned horizontally, libraries are shown vertically. Red, green, and black indicate genes expressed at high, low, or moderate levels, respectively, in the indicated library. (B) Dendrogram of clustered libraries. (C) Multidimensional scaling indicating the relatedness of the nine libraries.
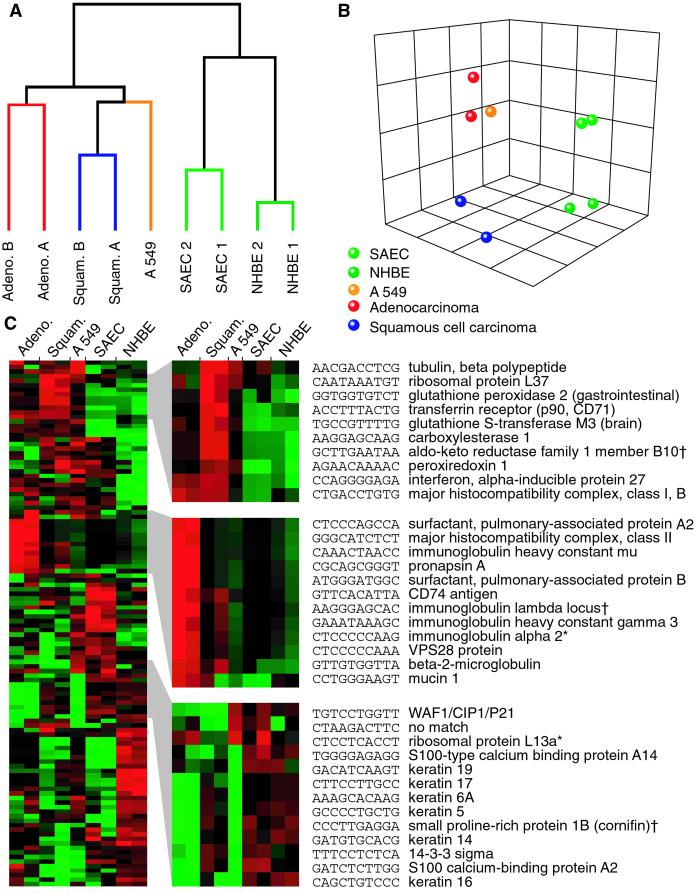
Because gene expression levels were represented by a tag count for each transcript detected in the SAGE libraries, we used Monte Carlo simulation (24) to quantify the significance of gene expression differences between the tumor libraries and the two corresponding normal epithelial cell controls. At a P < 0.001, 58 genes were selected when comparing the two adenocarcinomas to the two SAEC samples, and 71 genes were obtained by comparison of the squamous cell carcinomas to the NHBE cells. Fourteen genes were common to both comparison, and we therefore identified 115 highly differentially expressed transcripts for both tumor types (a list of genes is available as Table 3, which is published as supporting information on the PNAS web site). As expected, when subjected to hierarchical clustering, these 115 genes again separated the nine libraries into the exact same branching patterns (Fig. 2A) as did the nearly 4,000 genes described above. Once again, the A549 cell line branched with the tumor tissues and was located closest to the two adenocarcinomas by multidimensional scaling (Fig. 2B).
Figure 2.
Clustering and multidimensional scaling of the 115 genes highly differentially expressed (P < 0.001) in nine SAGE libraries. (A) Dendrogram of nine clustered libraries by using 115 differentially expressed genes. (B) Multidimensional scaling of the libraries by using 115 differentially expressed genes. (C) Cluster of the 115 genes (Left) with three main clusters (Right) consisting of genes overexpressed in squamous cell carcinoma (Top), overexpressed in adenocarcinoma (Middle), and underexpressed in adenocarcinoma (Bottom), respectively. † indicates that this tag corresponds to more than one gene of the same family. * indicates that the tag corresponds to more than one distinct gene.
Biologically Distinct Clusters of Genes in Different NSCLC Subtypes.
The clustering of the 115 statistically significant genes revealed at least three distinct gene clusters that were highly characteristic of the tumor tissues analyzed (Fig. 2C). Genes most highly expressed in squamous carcinomas of the lung (Fig. 2C Upper) were characterized by transcripts encoding proteins with detoxification and antioxidant properties. These proteins include glutathione peroxidase 2 (GPX2), glutathione S-transferase M3 (GSTM3), carboxylesterase, aldo-keto reductase, and peroxiredoxin 1. Their presence in squamous cell lung cancers most likely represented a cellular response by the bronchial epithelium to environmental carcinogenic insults (30, 31). The clustering of these overexpressed genes highlights the notion that functional variation of these proteins in the population may contribute to lung cancer susceptibility in some patients. Indeed, allelic variations in GSTM3 are susceptibility markers for lung, oral, basal cell carcinoma, and other cancers (32–34). Interferon α-inducible protein 27 is also shown to be overexpressed in 50% of breast cancers (35).
In contrast, the cluster of genes overexpressed in lung adenocarcinoma (Fig. 2C Middle) mostly encoded small airway-associated proteins and immunologically related proteins. The presence of genes for surfactants A2 and B, pronapsin A, and mucin1 in the cluster reflects the origin of tumors derived from small airway epithelial cells, such as type 2 pneumocytes and Clara cells (36, 37). However, high expression of these genes also suggested that these proteins may participate in the tumorigenesis of lung adenocarcinomas. Indeed, mucin1 is also overexpressed in breast cancers and tyrosine phosphorylation of the CT domain of MUC1 mucin leads to activation of a mitogen-activated protein kinase pathway through the Ras-MEK-ERK2 pathway (38, 39). Furthermore, the overexpression of Ig genes in adenocarcinomas may be explained by the extent of B-cell infiltration and the presence of antigen-presenting cells (APC) in the adenocarcinomas used for SAGE analysis. Interestingly, clustering analyses of the SAGE tags revealed that different tumor types preferentially expressed a different set of cell surface markers. Squamous cell cancers appeared to overexpress multihistocompatibility (MHC) class I and CD71 proteins (Fig. 2C Upper), whereas adenocarcinomas had relatively high expression of MHC class II and CD74 antigens. These gene expression differences in tumors indicated that immuno-based cancer therapy might be augmented by exploiting the expression of different tumor surface markers.
Not surprisingly, many of the genes underexpressed in the primary adenocarcinomas and the A549 adenocarcinoma cell line (Fig. 2C Lower) were those that are associated with squamous differentiation. These proteins include S100 proteins, keratins, and the small proline-rich protein 1B (Cornifin). However, two p53-inducible genes, 14-3-3σ (Stratifin) (40) and p21waf1/CIP1 (41, 42), clustered with this group of genes, showing significantly reduced expression in adenocarcinomas. Furthermore, the p21 message was reduced in adeno- as well as squamous tumors. Both p21waf1/CIP1and 14-3-3σ are highly induced, in a p53-dependent manner, in cells treated with ionizing radiation and other DNA-damaging agents (43, 44). Induction of these genes by p53 leads to cell cycle arrest (45). The p53 gene is frequently mutated in squamous carcinomas of the lung, and it is thought that mutations in p53 may contribute to the inability of lung epithelial cells to repair carcinogen-induced damage (46). In contrast, p53 mutations are observed much less frequently in lung adenocarcinomas (5). The reduced expression of both p21waf1/CIP1and 14-3-3σ gene transcripts in adenocarcinomas suggests that inactivation of genes in the p53-pathway plays an important role in this lung tumor type as well. However, reduced expression of the mRNA may not always correlate with a reduction of the gene product. Further studies correlating the molecular status of p53 with the expression of the encoded proteins are needed to assess the involvement of p53 and its downstream genes in the development of lung adenocarcinoma.
Other Genes Differentially Expressed in NSCLC.
It is important to note that the 115 highly differentially expressed genes we have identified represented only a subset of genes whose differential expression could distinguish the molecular characteristics of each cell type as well as the neoplastic condition in the lung. Clearly, additional genes with biological significance to NSCLC could also be identified, depending on the statistical method and the level of significance chosen. For example, when all tags that showed consistent expression within the libraries of the same cell type were compared to identify genes differentially expressed at a 99% confidence interval, a larger number of candidate genes were identified. Specifically, 827 tags showed statistically significant differential expression between the squamous cell carcinomas and the NHBEs, with 71 tags showing at least 10-fold overexpression. A similar comparison of the two adenocarcinoma tumor libraries and the SAECs identified 298 tags showing differential expression, with 20 tags overexpressed at least 10-fold in the tumors. Jointly, 45 tags were differentially expressed in both comparisons, and these genes were either a part of, or further extended, the observations revealed by the 115 genes. For example, small proline-rich protein 3 (SPRR3) was elevated in the squamous tumors but was virtually absent in the adenocarcinomas. SPRR3 is a member of the small proline-rich family of proteins that includes SPRR1 (Cornifin), a gene previously identified as a marker for squamous cell carcinoma (47), and is within the cluster of genes underexpressed in adenocarcinomas (Fig. 2C Lower). SPRR3 is a member of the proteins in the cornified cell envelope that help provide a protective barrier to the epidermal layer of cells (48). Reduced expression of this family of proteins in adenocarcinoma may contribute to the invasive properties of this cancer. Moreover, several members of the tumor necrosis factor (TNF) family of proteins and their receptors have demonstrated increased expression in various cancers including NSCLC (49). Our statistical analysis of the SAGE data revealed that expression of the TNF receptor superfamily member 18 gene was increased in squamous cell tumors in addition to the detoxification and antioxidation genes. TNF promotes T cell-mediated apoptosis (50), and elevated expression of genes in this pathway may provide a mechanism for antiproliferation of the tumor cells. Furthermore, another member of the GST family, GSTM1, was detected at induced levels in the adenocarcinoma tumors. Like GSTM3, GSTM1 is a known susceptibility marker for lung, oral, and other cancers (51–53).
Quantitative PCR and GeneChip cDNA Oligoarray Analyses of Additional NSCLC Tumors.
Because the SAGE libraries were derived from only selected tumor tissues and normal cells, it was essential to determine whether gene expression patterns derived from SAGE could be reproduced in a larger panel of lung tissues by using independent assays. A total of 43 tumor and normal samples were examined by using either quantitative real-time PCR or cDNA array methods. Five genes observed by SAGE as highly overexpressed in either squamous or adenocarcinomas of the lung (listed in Fig. 2C) were examined by real-time RT-PCR by using 10 different NSCLC tumors and four normal controls. As shown in Table 2, real-time RT-PCR indicated that the two squamous-tumor specific genes had consistently high expression ratios in this tumor type compared with its expression in adenocarcinomas. Similarly, the three adenocarcinoma-specific genes had consistently higher expression in this tumor type than in squamous cell cancers, when each was compared with the normal.
Table 2.
Real-time quantitative PCR analysis of SAGE-identified genes
| Spec. | Tag | Accession | Description | Number
of SAGE tags in library*
|
Average
RT-PCR†
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | S1 | S2 | Sq A | Sq B | Ad A | Ad B | Sq/N | Ad/S | ||||
| Sq | GGTGGTGTCT | X53463 | Glutathione peroxidase 2 (GPX2) | 4 | 2 | 0 | 1 | 58 | 41 | 0 | 0 | 11 | 2 |
| Sq | GCCCCCTTCC | AF241229 | Tumor necrosis factor receptor superfamily member 18 | 0 | 1 | 0 | 0 | 11 | 8 | 0 | 0 | 38 | 5 |
| Ad | GAAATAAAGC | Y14737 | Ig heavy constant γ 3 | 0 | 0 | 0 | 0 | 5 | 1 | 293 | 23 | 1 | 17 |
| Ad | GTTCACATTA | AI248864 | CD74 antigen | 0 | 1 | 0 | 1 | 9 | 2 | 86 | 21 | 31 | 93 |
| Ad | GGGCATCTCT | J00196 | Major histocompatibility complex, class II | 0 | 0 | 0 | 0 | 1 | 1 | 51 | 19 | 275 | 1,800 |
Expression of the listed genes was examined in 14 samples, including five squamous cell tumors, four adenocarcinomas, one tumor with adenosquamous morphology, two NHBE cultures, and two SAEC cultures.
The actual number of tag occurrences in the indicated SAGE library is provided.
The average expression of each gene was calculated for the four distinct cell types, and the ratio of differential expression is indicated. Ad, adenocarcinoma; Sq, squamous cell carcinoma; N, NHBE; S, SAEC; Spec., tumor specificity based on SAGE.
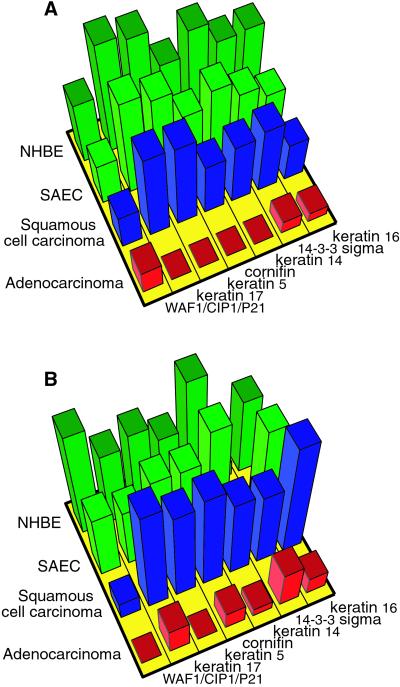
To survey the overall reliability of the molecular clustering obtained from lung SAGE libraries, we used GeneChip cDNA oligoarrays (15, 16) to survey 32 tumor and normal samples (including three samples used in real-time PCR) for relative gene expression. Only 60 of the 115 highly differentially expressed transcript tags were present on the 12,000-element GeneChip (U95A), including 23 of 35 genes from the three main clusters (shown in Fig. 2C). The SAGE tag count and GeneChip values for these 23 genes are shown in Table 5, which is published as supporting information on the PNAS web site. To compare the cDNA array result with SAGE, GeneChip values were averaged among all tumors of the same cell type and compared with that of the corresponding normal samples. Twenty-one of the 23 genes displayed an expression pattern similar to those obtained by SAGE. The expression patterns for the cluster of genes down-regulated in adenocarcinomas are shown (Fig. 3 A and B). These results support the highly reproducible nature of SAGE for most differentially expressed genes. Our data also suggest that hierarchical clustering of the SAGE libraries not only can cluster genes with strong biological significance but also provide precise tissue classification by using just a few tissue samples. Furthermore, because SAGE is independent of the knowledge of the gene sequence or the probe hybridization condition, it allows for an unbiased identification and quantification of gene expression patterns in the tissues of interest.
Figure 3.
Comparison of genes underexpressed in adenocarcinoma by using Affymetrix genechip and SAGE libraries. (A) Histogram of normalized SAGE data shows the average relative expression levels of seven genes that were underexpressed in adenocarcinoma (shown Lower Right in Fig. 2C). (B) Histogram of genechip data shows the normalized average relative expression levels of the same genes as in A. When a genechip expression value was less than 1, it was set to 1 before normalization. Normalization was done in the same manner as for clustering analysis (see Materials and Methods).
In summary, we have used SAGE and hierarchical clustering analyses to identify molecular profiles and clusters of genes specifically associated with two of the most common types of human lung cancer. Although biologically significant and highly reproducible, the gene expression profiles described here may represent only the basic molecular features from which adenocarcinoma and squamous cell carcinoma of the lung can potentially be distinguished. Histological features and clinical behavior of the tumor may depend on less pronounced changes in expression levels for a variety of genes and pathways. Nevertheless, cumulating evidence suggests that gene expression patterns most likely determine the clinical behavior and therapeutic response of the cancer (19, 54). The list of highly differentially expressed genes that we described will likely provide new molecular targets for improved diagnosis, prognosis, and rational therapy. The analyses for the expression of these genes in a larger number of lung tumors with detailed clinical information and outcome will help accomplish this goal.
Supplementary Material
Acknowledgments
We thank Drs. Bert Vogelstein, Kenneth Kinzler, Christoph Lengauer, Scott Kern, Elisabeth Jaffee, and Kent Hunter for critical reading of the manuscript. We thank Drs. Stephen Baylin, Robert Strausberg, and William Travis for advice, Dr. Clarence Wang for assistance with the SAGE data analysis, and Dr. Myung-Soo Lyu and Ms. Jenny Kelly for technical assistance. This work was supported in part by National Cancer Institute (NCI) Lung SPORE CA58184 and Early Detection Research Network Grant CA84986.
Abbreviations
- SAGE
serial analysis of gene expression
- NSCLC
non-small cell lung cancer
- SAEC
normal lung small airway epithelial cell
- NHBE
normal bronchial/tracheal epithelial cells
- GST
glutathione S-transferase
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.American Cancer Society. Cancer Facts and Figures 2001. Atlanta: Am. Chem. Soc.; 2001. [Google Scholar]
- 2.Travis W D, Linder J, Mackay B. In: Lung Cancer Principles and Practice. Pass H I, Mitchell J B, Johnson D H, Turrisi A T, editors. New York: Lippincott–Raven; 1996. pp. 361–395. [Google Scholar]
- 3.Gazdar A F. Anticancer Res. 1994;14:261–267. [PubMed] [Google Scholar]
- 4.Graziano S L, Gamble G P, Newman N B, Abbott L Z, Rooney M, Mookherjee S, Lamb M L, Kohman L J, Poiesz B J. J Clin Oncol. 1999;17:668–675. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 5.Niklinska W, Chyczewski L, Laudanski J, Sawicki B, Niklinski J. Folia Histochem Cytobiol. 2001;39:147–148. [PubMed] [Google Scholar]
- 6.Bennett W P, Hussain S P, Vahakangas K H, Khan M A, Shields P G, Harris C C. J Pathol. 1999;187:8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Hainaut P, Pfeifer G P. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 8.Forgacs E, Zochbauer-Muller S, Olah E, Minna J D. Pathol Oncol Res. 2001;7:6–13. doi: 10.1007/BF03032598. [DOI] [PubMed] [Google Scholar]
- 9.Hibi K, Liu Q, Beaudry G A, Madden S L, Westra W H, Wehage S L, Yang S C, Heitmiller R F, Bertelsen A H, Sidransky D, et al. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- 10.Gray J W, Collins C. Carcinogenesis. 2000;21:443–452. doi: 10.1093/carcin/21.3.443. [DOI] [PubMed] [Google Scholar]
- 11.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 12.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 13.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 14.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett J F, Obaya A J, O'Connell B C, Mateyak M K, Tam W, Kohlhuber F, et al. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. . (First Published February 25, 2000; 10.1073/pnas.050586197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 16.Jordan B R. J Biochem (Tokyo) 1998;124:251–258. doi: 10.1093/oxfordjournals.jbchem.a022104. [DOI] [PubMed] [Google Scholar]
- 17.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 18.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 19.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O P, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 20.Notterman D A, Alon U, Sierk A J, Levine A J. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 21.Zhang L, Zhou W, Velculescu V E, Kern SE, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 22.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox T F, Cox M A. Multidimensional Scaling. New York: Chapman & Hall/CRC; 2001. [Google Scholar]
- 24.Hammersley J M, Handscomb D C. Monte Carlo Methods. New York: Wiley; 1964. [Google Scholar]
- 25.Higuchi R, Fockler C, Dollinger G, Watson R. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 26.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 27.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 28.Mironov A A, Fickett J W, Gelfand M S. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Borka P. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 30.Auerbach O. In: Pulmonary Diseases and Disorders. Fishman A P, editor. New York: McGraw–Hill; 1980. pp. 1388–1396. [Google Scholar]
- 31.Sekido Y, Fong K M, Minna J D. Biochim Biophys Acta. 1998;1378:F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 32.Park L Y, Muscat J E, Kaur T, Schantz S P, Stern J C, Richie J P, Jr, Lazarus P. Pharmacogenetics. 2000;10:123–131. doi: 10.1097/00008571-200003000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Ramsay H M, Harden P N, Reece S, Smith A G, Jones P W, Strange R C, Fryer A A. J Invest Dermatol. 2001;117:251–255. doi: 10.1046/j.0022-202x.2001.01357.x. [DOI] [PubMed] [Google Scholar]
- 34.Reszka E, Wasowicz W. Int J Occup Med Environ Health. 2001;14:99–113. [PubMed] [Google Scholar]
- 35.Rasmussen U B, Wolf C, Mattei M G, Chenard M P, Bellocq J P, Chambon P, Rio M C, Basset P. Cancer Res. 1993;53:4096–4101. [PubMed] [Google Scholar]
- 36.Colby T V, Koss M N, Travis W D. In: Atlas of Tumor Pathology: Tumors of the Lower Respiratory Tract. Rosai J, Sobin L H, editors. Washington, DC: Armed Forces Institute of Pathology; 1995. p. 10. [Google Scholar]
- 37.Chuman Y, Bergman A, Ueno T, Saito S, Sakaguchi K, Alaiya A A, Franzen B, Bergman T, Arnott D, Auer G, et al. FEBS Lett. 1999;462:129–134. doi: 10.1016/s0014-5793(99)01493-3. [DOI] [PubMed] [Google Scholar]
- 38.Taylor-Papadimitriou J, Burchell J, Miles D W, Dalziel M. Biochim Biophys Acta. 1999;1455:301–313. doi: 10.1016/s0925-4439(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 39.Meerzaman D, Shapiro P S, Kim K C. Am J Physiol Lung Cell Mol Physiol. 2001;281:L86–L91. doi: 10.1152/ajplung.2001.281.1.L86. [DOI] [PubMed] [Google Scholar]
- 40.Hermeking H, Lengauer C, Polyak K, He T C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 41.el-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, et al. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 42.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 43.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 44.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 45.Taylor W R, Stark G R. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 46.Therrien J P, Drouin R, Baril C, Drobetsky E A. Proc Natl Acad Sci USA. 1999;96:15038–15043. doi: 10.1073/pnas.96.26.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu R, Wu R, Deng J, Lau D. Lung Cancer. 1998;20:25–30. doi: 10.1016/s0169-5002(97)00097-4. [DOI] [PubMed] [Google Scholar]
- 48.De Heller-Milev M, Huber M, Panizzon R, Hohl D. Br J Dermatol. 2000;143:733–740. doi: 10.1046/j.1365-2133.2000.03768.x. [DOI] [PubMed] [Google Scholar]
- 49.Tran T A, Kallakury B V, Ambros R A, Ross J S. Cancer. 1998;83:276–282. [PubMed] [Google Scholar]
- 50.Holtzman M J, Green J M, Jayaraman S, Arch R H. Apoptosis. 2000;5:459–471. doi: 10.1023/a:1009657321461. [DOI] [PubMed] [Google Scholar]
- 51.Nair U, Bartsch H. IARC Sci Publ. 2001;154:271–290. [PubMed] [Google Scholar]
- 52.Mitrunen K, Jourenkova N, Kataja V, Eskelinen M, Kosma V M, Benhamou S, Vainio H, Uusitupa M, Hirvonen A. Cancer Epidemiol Biomarkers Prev. 2001;10:229–236. [PubMed] [Google Scholar]
- 53.Howells R E, Holland T, Dhar K K, Redman C W, Hand P, Hoban P R, Jones P W, Fryer A A, Strange R C. Int J Gynecol Cancer. 2001;11:107–112. doi: 10.1046/j.1525-1438.2001.011002107.x. [DOI] [PubMed] [Google Scholar]
- 54.Scherf U, Ross D T, Waltham M, Smith L H, Lee J K, Tanabe L, Kohn K W, Reinhold W C, Myers T G, Andrews D T, et al. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.