Abstract
Purpose:
The DEAD-box RNA helicase eIF4A1 carries out the key enzymatic step of cap-dependent translation initiation and is a well-established target for cancer therapy, but no drug against it has entered evaluation in patients. We identified and characterized a natural compound with broad antitumor activities that emerged from the first target-based screen to identify novel eIF4A1 inhibitors.
Experimental Design:
We tested potency and specificity of the marine compound elatol vs. eIF4A1 ATPase activity. We also assessed eIF4A1 helicase inhibition, binding between the compound and the target including binding-site mutagenesis, and extensive mechanistic studies in cells. Finally, we determined maximum tolerated dosing in vivo and assessed activity against xenografted tumors.
Results:
We found elatol is a specific inhibitor of ATP hydrolysis by eIF4A1 in vitro with broad activity against multiple tumor types. The compound inhibits eIF4A1 helicase activity and binds the target with unexpected 2:1 stoichiometry at key sites in its helicase core. Sensitive tumor cells suffer acute loss of translationally regulated proteins, leading to growth arrest and apoptosis. In contrast to other eIF4A1 inhibitors, elatol induces markers of an integrated stress response, likely an off-target effect, but these effects do not mediate its cytotoxic activities. Elatol is less potent in vitro than the well-studied eIF4A1 inhibitor silvestrol but is tolerated in vivo at ~100X relative dosing, leading to significant activity against lymphoma xenografts.
Conclusion:
Elatol’s identification as an eIF4A1 inhibitor with in vivo anti-tumor activities provides proof-of-principle for target-based screening against this highly promising target for cancer therapy.
Keywords: eIF4A1, drug screen, translation initiation, ATF4, cancer therapeutic
Introduction
Cap-dependent translation initiation is the most regulated step of protein production and is activated by multiple oncogenic signaling pathways. Previous studies show targeting this convergence point of signaling promotes strong anti-tumor activities while bypassing resistance mechanisms stemming from the redundancy of upstream pathways (1,2). In 2004, a functional screen by the Pelletier group identified natural compounds that inhibit cap-dependent initiation without shutting down translation as a whole (3). Characterization of hits from the screen – most notably silvestrol, hippuristanol, and pateamine A – determined all have the same cellular target, eIF4A1 (4–6). This founding member of the DEAD-box RNA helicases is the core enzymatic component of the initiation complex, eIF4F, which contains also the scaffolding protein eIF4G and the mRNA cap-binding protein eIF4E. Though binding to 5’ cap by eIF4E, whose availability is tightly regulated by the mTOR Complex 1 (mTORC1), is considered the rate-limiting step of initiation, eIF4A1 has proven a better drug target, being an ATP-dependent catalyst of mRNA unwinding (7).
Anti-eIF4A1 compounds identified through functional screening can work by increasing eIF4A1’s affinity for RNA, which sequesters it out of the eIF4F complex, and in the case of silvestrol promoting particular binding to mRNA species containing polypurine sequences (8). Alternatively, hippuristanol works by blocking the RNA interaction of both free and complexed eIF4A1 (5). We previously reported two compounds that inhibit eIF4A1 ATPase activity leading to detectable activity against tumor cell lines in vitro (9), but by comparison this mode-of-action is minimally explored. Meanwhile, no eIF4A1 inhibitor described to date has led to a drug suitable for evaluation in clinical trials, and thus the preclinical potential of this therapeutic approach remains untested in patients. A proof-of-principle for target-based screening vs. eIF4A1 could help address the need for additional lead compounds suitable for development as cap-dependent translation inhibitors in cancer therapeutics.
The basis of in vivo therapeutic window for inhibiting cap-dependent initiation is thought to be differential dependence between cancer and non-malignant cells for constitutive expression of cap-regulated proteins. Oncoproteins including MYC, MCL1, CYCLIN D3, and others are lost from cancer cells of multiple different cancer types upon eIF4A1 inhibition or knockdown (2,10–12), and several different mRNA structural and sequence elements have been identified as potentially mediating these effects (reviewed in (13)). Regardless, cancer cells are more prone to enter apoptosis than non-malignant cells upon treatment with eIF4A1 inhibitors, and animal studies demonstrate anti-tumor activities in vivo at doses tolerated by the host (2,4). Translationally regulated proteins are mediators of multiple cancer hallmarks, and their loss through eIF4F inhibition is seen as a way to help overcome both upstream signaling redundancies and tumor heterogeneity (14). Moreover, though gene-expression changes at the level of mRNA are often reported as the key outputs of oncogenic signaling and epigenetic reprogramming, protein expression is what actually mediates cellular phenotype. Studies in several systems show poor correlation between mRNA and protein expression, highlighting translation as the step at which the proteome is primarily regulated (15). In cancer, overexpression of components of the translational machinery, including eIF4F subunits, has been reported in a variety of diseases (reviewed in (16)). All of this evidence points to targeting translation directly as a highly promising avenue for cancer therapy for a wide range of cancer types, and a number of different approaches for translational inhibition are under preclinical investigation (16).
Proof-of-principle that inhibiting translation is a tolerable option in humans comes from the drug omacetaxine, which inhibits elongation at the ribosome and is FDA approved for chronic myelogenous leukemia patients resistant to tyrosine kinase inhibitors (17). Inhibition at the more regulated and arguably more promising step of initiation, however, remains untested in patients. eIF4A1 and other DEAD-box helicases are highly conserved enzymes that use the cooperative binding of ATP and RNA to cycle through conformational changes, allowing duplex unwinding (18–20). Inhibition of ATP hydrolysis in these proteins by RNA aptamers or protein interacting partners has been shown to clamp them in place while bound to RNA and prevent their function, but optimized small molecules that work in this way have not yet been discovered (21,22). Here, we report that the marine natural compound elatol is a novel eIF4A1 inhibitor successfully identified through target-based screening. Though off-target effects and synthetic complexities of this particular compound limit its utility in further development, we establish proof-of-principle for target-based screening against eIF4A1 based on inhibition of ATP hydrolysis, yielding a compound with in vivo anti-tumor activity.
Materials and Methods
Cell-free in vitro studies of eIF4A1
Protein purification and assessment of ATPase activity by malachite green were as previously described (9). Helicase assays were performed as per (23). Isothermal titration calorimetry (ITC): eIF4a was dialyzed against buffer A (20 mM MES-KOH, pH 6.0, 10 mM potassium acetate, 2.5 mM MgCl2, 1% glycerol, and 1 mM DTT) for 12 h. eIF4a was supplemented with 2% DMSO to match the ligand solution, degassed, and loaded in the cell of a nano-isothermal titration calorimeter (TA Instruments). A total of 12–20 injections of 0.2 mM elatol in buffer A were made every 200 s over a 3000 s time frame. NanoAnalyze software (TA Instruments) was used to integrate the peaks of the isotherm. The peaks were then integrated from injection start to 75 s post injection and fit to an independent binding model. Replicate experiments were done using 25 and 10 μM eIF4A to add power to the stoichiometric given by the nanoAnalyze software.
ATPase Activity assays
Proteins were prepared as follows: 1 μM eIF4A1 in buffer A (20 mM MES-NaOH, pH 6.0, 100 mM potassium acetate, 2.5 mM MgCl2, 1% glycerol, and 1 mM DTT); 500 nM eIF4A1-K82R in buffer A; and 500 nM eIF4A1-K238E in buffer A. ATP was added to generate samples of each protein containing a gradient of ATP (2 mM, 1 mM, 500 μM, 250 μM, 125 μM, 62.5 μM, 31.25 μM, 0). The assay was carried out at 37°C, and after 1 hour, a 20 μL aliquot of the reactions was added to 40 μL of malachite green solution (9.3 μM malachite green, 53 mM (NH4)2MoO4, 1M HCl, 0.04% Tween 20). After 5 min, the OD660 was read on a GEN5 plate reader (BioTek Synergy 2). This was repeated after 2, 3, and 4 hours. The Michaelis–Menten curves were plotted and the Michaelis-Menten values were calculated (GraphPad Prism Software).
Molecular modeling
Modeling of elatol with eIF4A1 (PDB code: 2ZU6) was performed using Glide docking program (Schrodinger). Initially, the docking grid was created around the binding site and 1 was docked using extra precision (XP) glide docking. Resulting poses were evaluated using docking score and hydrogen bonding interaction with the active site residues.
Cell Lines and Reagents
All cell lines were routinely verified by STR fingerprinting and confirmed mycoplasma negative using the PlasmoTest Kit (Invivogen: REP-PT1). The mouse embryonic fibroblasts eIF2α Serine 51 A/A and corresponding wildtype were a kind gift of Dr. Randal Kaufman at Sanford Burnham Prebys Medical Discovery Institute. The MEFs ATF4 knockout and wildtype were a kind gift of Dr. Peter Johnson at the National Cancer Institute. HBL1, TMD8, U2932, Riva, Toledo, OZ, SU-DHL-4, WSU-DLCL-2, MD901, and SNU-398 cell lines were grown in RPMI culture media (Corning) supplemented with 10% fetal bovine serum (FBS, VWR) and penicillin/streptomycin (P/S, VWR). OCI-Ly2, OCI-Ly3, OCI-Ly10, OCI-Ly19 were grown in IMDM with 20% FBS and P/S. DB, Farage, SU-DHL-10, SU-DHL-6, Karpas-422 were grown in RPMI with 20% FBS and P/S. MDA-MB-468 were grown in DMEM with 10% FBS and P/S (D10). ATF4 wildtype and knockout MEFs were cultured in D10 additionally supplemented with non-essential amino acids (ThermoFisher) and 50 μM β-mercaptoethanol. eIF2α wildtype, and Ser51 A/A mutant MEFs were cultured in D10 media supplemented with NEAA. Silvestrol was purchased from Medchem express (HY-12351). The PERK inhibitor GSK2606414 was purchased from Millipore/Calbiochem (516535). Tunicamycin was purchased from Sigma (T7765). Carboplatin was acquired from the University of Miami Sylvester Comprehensive Cancer Center pharmacy. The retroviral shRNA knockdown vectors were a kind gift from Jerry Pelletier.
Proliferation Assay
Cells were plated at 1 × 105 cells/mL on day 0 and treated with vehicle (DMSO) or indicated concentration of inhibitor and live cells counted every day by trypan blue exclusion. Assessment of elatol effects on cell growth for determination of IC50 in the Harvard/Wellcome cell line collection was carried out as previously described (24).
Western Blotting
As described in (31). Antibodies for CYCLIN D3 (2936), MYC (5605), PIM2 (4730), MCL1 (5453), BCL2 (4223), SURVIVIN (2808), S6 (2217), phospho-S6 Ser240 (5364), 4EBP1 (9452), phospho-4EBP1 Ser65 (9456), eIF2α (5324), ATF4 (11815), PKR (12297), PERK (5683), α-TUBULIN (2144), GAPDH (5147), XBP1 (12782) were purchased from Cell Signaling Technology. Antibodies for HRI (365239), GCN2 were purchased from Santa Cruz Biotechnology. Antibodies for eIF4A1 (31217) and phospo-eIF2α Ser51 (32157) were purchased from Abcam. Puromycin antibody was purchased from Millipore (MABE343). PPIB antibody was purchased from Thermo (PA1–027A).
siRNA knockdown
Control non-targeting siRNA, ATF4 and eIF4A ON-TARGET siRNA pools where purchased from Dharmacon. 5 × 105 cells were plated in D10 media without antibiotics. The following day cells were transfected with 50nM siRNA using Lipofectamine 3000 (Thermo L300015) following manufacturers protocol. Transfection media was replaced with D10 media after 24 hours and cells were collected for analysis at 48 hours. For ATF4 knockdowns cells were treated with DMSO, elatol or tunicamycin at 48 hours and collected for analysis 56 hours following transfection with siRNA.
RT-PCR
As described in (31). Taqman probes were purchased from Thermofisher: 18s rRNA 4319413E-0810041, Gapdh Hs02758991_g1, eif4a1 Hs00426773, eif4a2 Hs00756996_g1, Ccnd3 Hs00236949_m1, Mcl1 Hs01050896_m1, Myc Hs00153408_m1
Viability Assays
Cells were plated at 3–5 × 103 cells/well in serial dilutions of drug ranging two logs with the top concentration for silvestrol 1 μM and the top concentration for elatol 10 μM. Viability was measured after 72 hours using Cell Titer Glo (Promega G7573) following manufacturer’s protocol. Luminescence was detected on the BioTek HT Synergy plate reader and LD50 values calculated using nonlinear regression fit in Graphpad Prism7.
Apoptosis Assays and Annexin Staining
1–3 × 105 cells were plated with indicated drug treatments and washed once with ice cold PBS at indicated time point and stained with PE conjugated Annexin V and 7-aminoactinomycin D (BD Biosciences) according to manufactures protocol. Stained cells were analyzed by flow cytometry on the Attune NxT (Thermo). Data was analyzed using FlowJo v9.9.6 (FlowJo).
Dual-Luciferase Reporter Assay
The Dual-luciferase plasmids were a kind gift from Jerry Pelletier at McGill University. 7 × 105 HEK-293 cells were plated and transfected with 400 ng of plasmid using Lipofectamine 3000 (Thermo L300015). 36 hours after transfection cells were treated as indicated before they were lysed using the dual luciferase reporter assay kit (Promega) and 100 μL added in triplicate to a white bottom 96 well plate for detection of luciferase following the manufacturers protocol. Luciferase units were normalized to the DMSO treated control.
O-propargyl puromycin assay
Performed as in(25), but 1 × 105 cells were plated and treated as indicated and pulsed with 50 μM O-propargyl puromycin (OPP, Jena Biosciences) before being collected and washed two times with PBS. Alexa-488 azide (Thermo A10266) was used for conjugation to C-terminally labeled proteins and samples ran on Attune NxT cytometer (Thermo) and data analyzed using FlowJo v9.9.6 (FlowJo).
Polysome Profiling
20 × 106 cells were treated as indicated and washed with ice-cold PBS with 100 g/mL cycloheximide for 10 minutes prior to lysis. Cells were pelleted at 200xg at 4° C for 10 minutes and lysed in 500 μL of lysis buffer (.3M NaCl, 15 mM MgCl2, 15 mM Tris-HCl pH 7.4, 1% Triton X-100, 100 g/mL cycloheximide, 100 U/mL RNasin). Lysates were cleared and equal A260 units (measured using NanoDrop 2000, Thermo) were loaded onto 10–50% sucrose gradients and centrifuged at 260,343 x g rpm in a SW-41 Ti rotor for 1.5 hours at 4° C. Samples fractionated and 254 nm absorbance recorded using the gradient fractionation system (Brandel).
CLL patient cells
As previously described (25).
Retroviral Complementation Experiments
Generation of the control and eIF4A stable knockdowns in NIH-3T3 cells was performed as described (26).
In Vivo experiments
All in vivo experiments were performed following protocols approved by the relevant Institutional Animal Care and Use Committee. The initial non-tumor bearing maximum tolerated dose studies and SU-DHL-6 xenografts were done with female 8–10 week old SCID mice from the University of Arizona Cancer Center and OCI-Ly3 xenografts done with female 8–10 week old SCID mice purchased from Charles River Laboratories. Complete blood counts were collected from non-tumor bearing mice dosed as indicated and evaluated on a Hemavet950. For SU-DHL-6 xenografts 2×106 cells were washed with ice cold PBS and mixed 1:1 with matrigel (Corning 354234) and injected s.c. into the right flank of SCID mice. Once the tumor reached 60mm3 mice were pair-matched and divided into two treatment groups: vehicle (sterile water with 5.2% tween-80, 5.2% PEG-400) or 20mg/kg elatol i.p. daily for 5 days. Mice were weighed and tumor volume calculated twice weekly. The maximum tolerated dose study of a single i.p. injection of elatol up to 100 mg/kg was done on cohorts of 5 CD1 mice. H&E pathology slides were prepared using standard techniques and analyzed at the University of Miami. For the OCI-Ly3 xenograft study 10 × 106 cells were first washed with ice cold PBS and mixed 1:1 with matrigel and implanted s.c. on the flank of SCID mice. Once tumors reached 500mm3 tumors were dissected and dissociated using the gentleMACs Dissociator (Miltenyi Biotec) and 1 × 106 serially transplanted tumor cells were implanted s.c. into the flank of SCID mice. Once tumors reached 50mm3 mice were pair matched and split into two treatment groups: vehicle or 40mg/kg elatol twice weekly. Mouse weight and tumor volume were monitored twice weekly.
Statistics
All numerical data are based off of biological replicates represented as mean ± SEM. For annexin staining and OPP incorporation unpaired, two-tailed t-tests were performed, with the addition of the two-stage linear setup procedure of Benjamini, Krieger and Yekutieli to compare treatments over time, with p<0.05 as significant. The OCI-Ly3 xenograft experiment was compared using a two-way ANOVA with p<0.05 signficant. All statistical comparisons were performed using GraphPad Prism7. The validity of the malachite green ATPase assay for screening was verified by determining the Z-score with EDTA and DMSO as the positive and negative controls respectively.
Results
Elatol is an eIF4A1 Inhibitor with Broad Anti-Tumor Activity
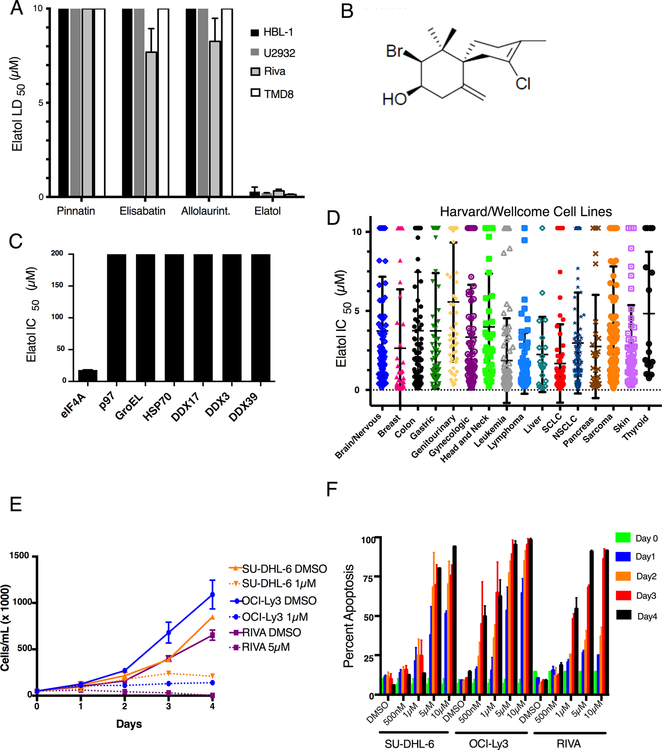
We recently reported a screen for compounds able to inhibit eIF4A1’s ATP hydrolysis and characterized the ATP-competitive inhibitors elisabatin A and allolaurinterol (9). Despite inhibiting eIF4A1 ATP hydrolysis, when we evaluated the initial screen hits against non-Hodgkin lymphoma (NHL) cell lines, where other eIF4A inhibitors show strong anti-tumor activities (2,24,27), elisabatin A and allolaurinterol were inactive up to 10 μM indicating poor cell membrane permeabilization or metabolic liabilities (Fig. 1A). However, the brominated marine terpene elatol (Fig. 1B) identified on the same screening platform was toxic at concentrations less than 1 μM in the preliminary screen of cancer cell lines and warranted further characterization. In the malachite-green assay used to identify eIF4A1 inhibitors in the initial screen, elatol is active against eIF4A1’s ATP hydrolysis at 16.4 μM (Fig. 1C). Counter-screening showed no activity against additional purified ATP-hydrolyzing enzymes, including the bacterial chaperonin GroEL, the human chaperone HSP70 (both considered unlikely to be affected by eIF4A1 modulators, as they have the least similar ATP pockets), the classical Walker A/B protein p97 and three other DEAD-box helicases: DDX3, DDX17, and DDX39A (Fig. 1C). In addition, in a commercial kinome screen, elatol had no activity against 97 oncogenic and related kinases, further validating its specificity (Fig. S1A). To broadly interrogate elatol’s anti-tumor activities, we tested it against the large collection of validated cancer cell lines maintained cooperatively by investigators at Harvard University and the Wellcome Trust (24). In this screen for growth inhibition, we found a wide range of sensitivity to elatol (Fig. 1D) with 37% (344/924) of cell lines having IC50 < 1μM. Leukemia and lymphoma cell lines were the most sensitive tumor type overall, followed by breast and lung, all of which have been previously identified as candidates for eIF4A or eIF4F complex inhibition (12,28). With lymphoma cell lines identified as a sensitive group, we proceeded to test three cell lines derived from diffuse large B-cell lymphoma (DLBCL), an aggressive lymphoma and the most common hematologic malignancy overall. A single elatol treatment completely halted proliferation in these lines (Fig. 1E). Elatol also induced apoptosis in a time- and concentration-dependent manner, with greater than 50% apoptosis by 24 hours in the most sensitive line (Fig. 1F), and the compound was less potent against normal human peripheral-blood mononuclear cells (Fig. S1B). Elatol therefore inhibits eIF4A’s ATP hydrolysis in vitro and is highly toxic in a variety of cancer cell lines.
Figure 1.
Elatol, identified in a novel screen for inhibitors of eIF4A1 ATP hydrolysis, shows broad activity in cancer. A) LD50 of DLBCL cell lines treated with the top 4 eIF4A1 ATP hydrolysis inhibitors based on the malachite green screen. Mean± SEM, n=4. B) Chemical structure of elatol. C) Sensitivity of indicated ATP-hydrolyzing enzymes to elatol determined by malachite green based ATP hydrolysis assay. Mean± SEM, n=3 D) IC50 of cell lines in the Harvard/Wellcome collection to elatol. Grouped as follows: brain/nervous system includes glioma, neuroblastoma and medulloblastoma; breast; colon; gastric includes esophagus and stomach; genitourinary includes prostate, kidney and urinary tract; gynecologic includes cervix, ovary, and uterus; head and neck; leukemia, liver includes liver and biliary tract; lymphoma; SCLC (small cell lung cancer); NSCLC (non-small cell lung cancer); pancreas; sarcoma; skin and thyroid. E) Proliferation of three DLBCL cell lines following a single elatol treatment. Mean ± SEM, n=3 F) Apoptosis in three DLBCL cell lines measured every 24 hours for four days following a single elatol treatment measured by flow cytometry following Annexin V and 7-aminoactinomycin D co-staining. Mean ± SEM, n=3.
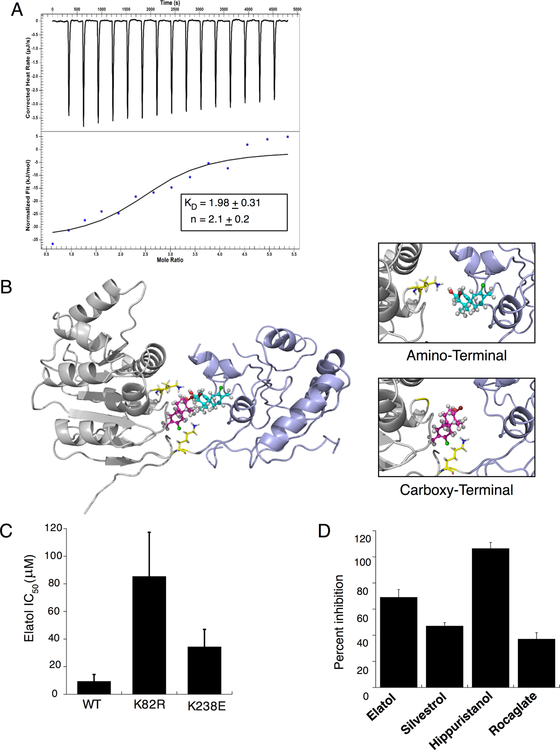
Elatol binds in a 2:1 ratio to eIF4A1 and Disrupts Helicase Activity
To further characterize the interaction between elatol and eIF4A1, we first performed isothermal titration calorimetry. These results confirmed binding (KD 1.98 ± 0.31 μM) and revealed an unexpected 2:1 elatol:eIF4A1 stoichiometry (Fig. 2A). As has been the experience with other eIF4A1-interacting compounds, our attempts to obtain a co-crystal structure with elatol have been unsuccessful. Molecular modeling studies, however, also predicted 2:1 stoichiometry, suggesting binding by two adjacent elatol molecules in the helicase core of eIF4A, with each elatol molecule interacting with a lysine residue from either the amino or carboxy RecA-like domains of eIF4A (Fig. 2B). The in silico model therefore predicts mutation of either lysine, K82 or K238, should lessen drug activity in vitro. K82 is part of the Walker A ATPase motif conserved in all DEAD-box helicases (18,19), and we found the non-conservative substitution K82E resulted in catalytically inactive protein (not shown). More conservative K82R, however, retained full ATPase activity of the wild-type protein but was significantly less sensitive to elatol (IC50 85.5 ± 31.8 μM, Fig. 2C, Fig. S2A). K238E also retained full activity of wild-type and was also less sensitive to elatol, though the effect was less pronounced (IC50 34.2 ± 12.8 μM, Fig. 2C, Fig. S2A). Therefore while elatol’s interactions with both residues in the helicase core likely contribute to inhibition, the interaction with K82 is likely more important. Moreover, these results highlight the ATP binding pocket as a promising drug-binding site for additional development of eIF4A1 inhibitors. Finally, we assessed if elatol affected eIF4A1’s helicase activity in vitro using a fluorescence-based assay in which successful RNA unwinding results in de-quenching (23). Here, we found elatol results in inhibition in a manner similar to the previously characterized eIF4A1 inhibitors hippuristanol, silvestrol, and a silvestrol-derived rocaglate (Fig. 2D and S2B). Elatol’s inhibition of eIF4A1 ATPase therefore depends on binding two distinct pockets in the eIF4A1 core with 2:1 stoichiometry. In addition, elatol inhibits eIF4A1 helicase activity in a cell-free context.
Figure 2.
Elatol binds to the N- and C-termini of eIF4A in a 2:1 stoichiometry. A) Isothermal titration calorimetry of eIF4A1 and elatol. Data fit to an independent binding model using NanoAnalyze software from TA instruments. Mean ± SEM n=3 B) Putative elatol binding with eIF4A1 based on molecular modeling experiments shows two elatol molecules (blue and pink) interacting with key lysines (yellow) in the RNA binding groove between the two helicase domains of eIF4A (gray and purple). C) Malachite green assay for ATP hydrolysis showing IC50 for elatol treatment against wildtype eIF4A1 or proposed lysine mutants in the proposed binding sites. Mean ± SEM n=3 D) eIF4A helicase activity measured following treatment with various known eIF4A inhibitors and elatol. Mean ± SEM n=3.
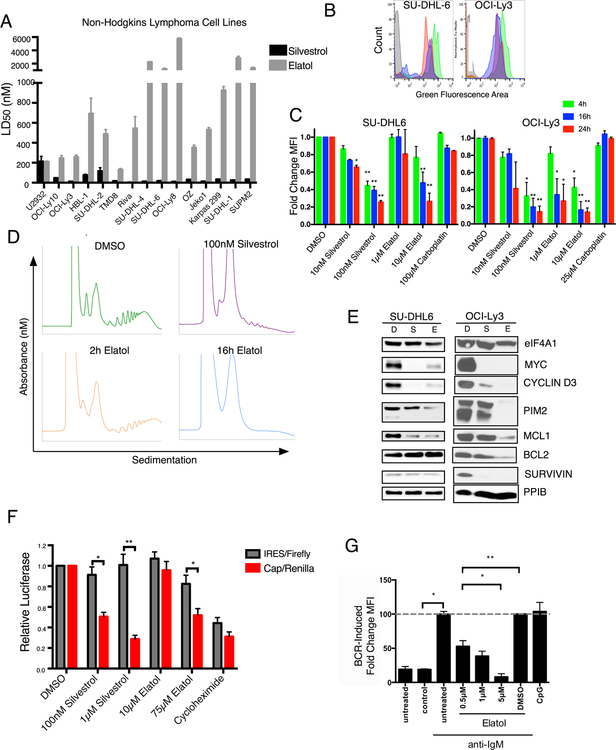
Elatol is a Translation Inhibitor in Cells with Reduced Potency Compared to Silvestrol
Silvestrol’s mechanism as an eIF4A inhibitor and potent activity against a variety of tumor types in vitro are well described (2,4,29). In a panel of lymphoma cell lines, we found elatol was less potent than silvestrol, with LD50 in a metabolic viability assay ranging from 130 to 5,756 nM compared to 2.7 to 213 nM for silvestrol (Fig. 3A). The 2:1 stoichiometry of elatol’s interaction with eIF4A1 would only partially explain this disparity, and additional differences are likely to exist between the compounds. In particular, silvestrol promotes binding by free eIF4A1 to polypurine stretches of mRNAs that contain them (8) while its resulting reduced availability causes ribosome stalling on G-quadruplex structures found in complex 5’ UTRs (29). These unique mechanistic properties of silvestrol would not be expected to apply to elatol, based on its simpler chemical structure and its predicted binding to the target. We next compared elatol and silvestrol in a variety of assays to assess elatol’s effects on protein translation. At concentrations relative to the respective LD50 of either drug in each cell line, elatol like silvestrol causes a global decline of protein synthesis measured by incorporation of OPP, in a concentration- and time-dependent manner (Fig. 3B–C). OPP incorporation is measured on live gated cells, but to ensure these effects on translation are not observed with general cytotoxic agents we compared the DNA-damaging agent carboplatin, again at similar concentrations relative to it’s LD50, and found no significant decline in OPP incorporation (Fig. 3C, Fig. S3A). Elatol also moderately reduced polysome translation at a two-hour treatment compared to silvestrol, but by 16 hours showed complete elimination of polysome translation (Fig. 3D). Western blotting showed elatol, like silvestrol, causes loss of well-established translationally regulated oncoproteins in lymphoma cells (Fig. 3E, S4A). Elatol also down-regulated cap-dependent protein translation more strongly than cap-independent IRES driven expression in a dual-luciferase reporter assay (Fig. 3F). Decreased eIF4A1 function either through knockdown or pharmacologic inhibition results in increased expression of the eIF4A2 transcript (30), a potentially useful marker of drug specificity against eIF4A1. In contrast to silvestrol treatment, which induced eIF4A2 mRNA upregulation in sensitive lymphoma cell lines, we found a mixed response to elatol treatment. eIF4A2 increased in the two more sensitive cell lines OCI-Ly3 and SU-DHL-6 but did not in the less sensitive RIVA cell line (Fig. S3B). Additionally, we observed some decreases in both transcript levels at higher concentrations of elatol, suggesting elatol may have effects on mRNA transcription in addition to translation. We therefore examined mRNA expression of genes whose protein products are lost in response to elatol as in Fig. 3E and again found a mixed response across cell lines, with the most sensitive, OCI-Ly3, showing a decrease in mRNA levels following elatol treatment (Fig. S3C). We therefore interrogated protein and mRNA expression in parallel across a range of elatol concentrations in this cell line and found that while both are affected, protein-expression changes begin as low as 100 nM, below the LD50 of this cell line, while mRNA reductions are not significant until 500–1000 nM, well above the LD50 (Fig. S3D). These results show elatol works more potently as a translation inhibitor but at higher concentrations, well above the LD50, also affects transcription, raising questions about potential cellular off-target effects. Finally, silvestrol has been shown to inhibit deregulated protein translation in B-cell malignancies(31). We found that elatol, like silvestrol (Yeomans, Wilmore, Packham unpublished data) significantly reduced protein translation measured by OPP labeling in primary chronic lymphocytic leukemia (CLL) cells stimulated via the B-cell receptor using anti IgM (25) (Fig. 3G). Addition of caspase inhibitor Q-VD-OPh prevents induction of apoptosis and therefore the loss of translation in the CLL cells is due to inhibition of protein translation. By contrast, elatol did not significantly inhibit OPP labeling in unstimulated primary CLL cells, or non-malignant T-cells which were also present in the patients’ blood samples (Fig. S4B).
Figure 3.
Elatol is toxic to Non-Hodgkin Lymphoma cell lines and inhibits protein translation A) Sensitivity of a collection of Non-Hodgkin Lymphoma cell lines to known eIF4A inhibitor silvestrol or elatol. Viability measured after 72-hour treatment using Promega Cell Titer Glo reagent. LD50 calculated using nonlinear regression fit analysis in Graphpad Prism 7. Mean ± SEM n=4. B) Histogram plot showing OPP labeling in DLBCL cell lines treated with DMSO (green), 100 nM silvestrol (red) or 10 μM elatol (blue) for 24 hours. Unlabeled cells shown in gray. C) Mean fluorescent intensity of live cells labeled with OPP in DLBCL cells treated with indicated concentrations of silvestrol, elatol, or carboplatin for four, 16 or 24 hours. Normalized to DMSO control. Mean ± SEM n=3. * = p< 0.05, ** = p<0.001. D) Polysome profiling of OCI-Ly3 cells treated with DMSO or 100nM silvestrol for 2 hours or 2 μM elatol for 2 or 16 hours. E) Western blot showing protein expression of translationally regulated genes in DLBCL cells treated with DMSO (D), 50 nM silvestrol (S) or 5 μM elatol for 16h. Representative images, n=3. F) Dual luciferase reporter assay measuring cap-dependent versus IRES mediated luciferase expression following an eight-hour treatment with the indicated translational inhibitors. Relative luciferase units normalized to DMSO-treated cells. Mean −/+ SEM, n=3 G) Translation measured by OPP incorporation in CD19+CD5+ CLL patient cells following anti-Ig-M stimulation and elatol treatment. Normalized to anti-Ig-M stimulated but untreated cells. CpG-ODN stimulation used as a control. Mean ± SEM n=4. * = p< 0.05, ** = p<0.001.
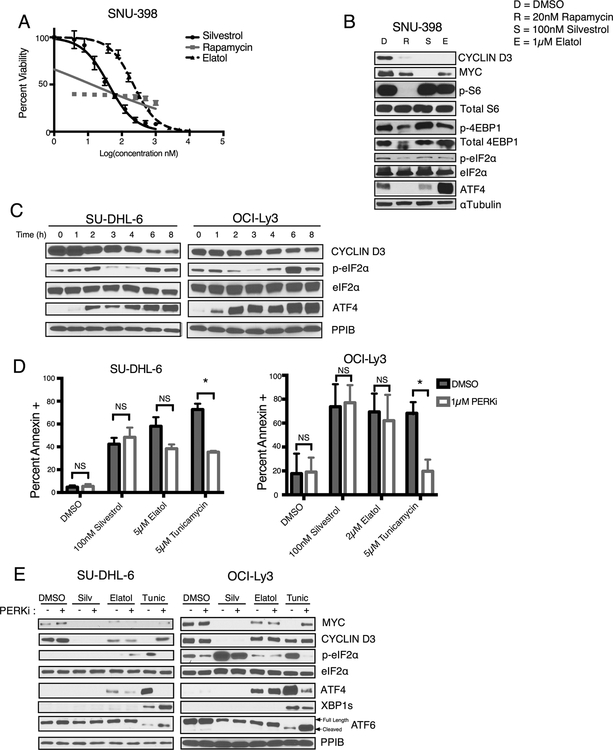
Integrated Stress Mediators Induced by Elatol Do Not Result from an Unfolded Protein Response
Elatol inhibits protein translation, but because of differences compared to silvestrol, we wanted to test for factors in addition to inhibition of eIF4A1 that might mediate elatol’s toxicity in cells. We first employed the hepatocellular carcinoma (HCC) cell line SNU-398, which has constitutive mTORC1 activity due to biallelic loss of its negative regulator TSC2 and therefore high dependence on downstream translational activation (32). In contrast to the cytostatic effect of mTORC1 inhibition by rapamycin, both silvestrol and elatol are highly cytotoxic in SNU-398 (LD50 41 nM and 219 nM respectively, Fig 4A), consistent with previous findings that eIF4A1 inhibition is more apoptotic than the cytostatic effects of decreased eIF4E availability downstream of mTORC1 inhibition (33). Western blotting shows as expected that rapamycin potently inhibits phosphorylation of ribosomal protein S6 and 4EBP1 downstream of mTORC1 (Fig. 4B). Silvestrol and elatol both show no activity against mTORC1 targets, while still strongly down-regulating expression of cap-dependent oncoproteins. We noted, however, that elatol in contrast to silvestrol induced expression of the transcription factor ATF4, which is translationally up-regulated downstream of eIF2α phosphorylation on serine 51 during an integrated stress response (ISR)(34). Though eIF2α S51 phosphorylation was not notably induced by elatol in this experiment, these data point to possible ISR induction by elatol that did not occur in response to silvestrol.
Figure 4.
Elatol treatment induces ATF4 expression, not mediated by the UPR. A) Cell viability of SNU-398 cells measured after 72-hour treatment with translation inhibitors silvestrol, rapamycin or elatol. Mean ± SEM n=4. B) Western blot showing protein expression SNU-398 cells treated with DMSO (D), 200 nM rapamycin (R), 100 nM silvestrol (S) or 1 μM elatol (E) for 16 hours. Representative images. C) Time course showing protein expression in DLBCL cell lines following elatol treatment. Representative images. n=2. D) Cell death measured by flow cytometry following annexin V staining in DLBCL cell lines treated with indicated inhibitors with or without the combination of the PERK inhibitor GSK2606414 for 24 hours. Mean ± SEM n=3. * = p< 0.05, ** = p<0.001. E) Western blot showing protein expression in cells treated for four hours with DMSO, 100nM silvestrol, 5 μM elatol or 5 μM tunicamycin with or without the combination with the PERKi. Representative images. n=3.
In lymphoma cells, as in the TSC2-deficient HCC cells, ATF4 is rapidly and strongly induced by elatol exposure, while eIF2α S51 phosphorylation is variable (Fig. 4C). The breast cancer cell line MDB-MB-468, an elatol sensitive cell line in the Harvard/Wellcome cell line collection, also showed ATF4 induction in response to treatment indicating this is a general effect of elatol treatment, not cell type specific (Fig. S4C). The ISR is a cytoprotective pathway that is activated in response to various cellular insults, and the end result of activation temporarily halts protein synthesis through eIF2α phosphorylation, allowing the cell to attempt to respond to the cellular stress or induce apoptosis if it is too severe (35). The unfolded protein response (UPR) is a type of ISR activated in response to unfolded proteins in the endoplasmic reticulum, leading to eIF2α phosphorylation by the Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK), along with two additional branches indicated by expression of spliced XBP1 (XBP1s) and cleavage of ATF6 respectively. To evaluate if activation of the ISR through the PERK kinase was causing the upregulation of ATF4 expression following elatol treatment we tested the combination of the PERK inhibitor GSK2606414 (PERKi) (36) with drug treatment. PERKi treatment resulted in no significant reversal of elatol’s potency vs. cultured cells, measured at 24 hours by annexin V staining, in contrast to its antagonism of the UPR-inducing compound tunicamycin (Fig. 4D). In a short four-hour treatment where elatol induction of ATF4 was strong, PERKi did not reverse elatol’s activation of ATF4 protein expression, and there was no evidence of overall UPR induction indicated by either XBP1s expression or ATF6 cleavage (Fig. 4E, S4D). Elatol’s induction of ATF4 protein expression therefore is not a result of an unfolded protein response, but its importance to the drug’s activities in cells remains unclear from these results.
ISR Induction by Elatol Does not Mediate its Toxicity to Cells, but eIF4A1 Knockdown Increases Sensitivity to Drug Effects
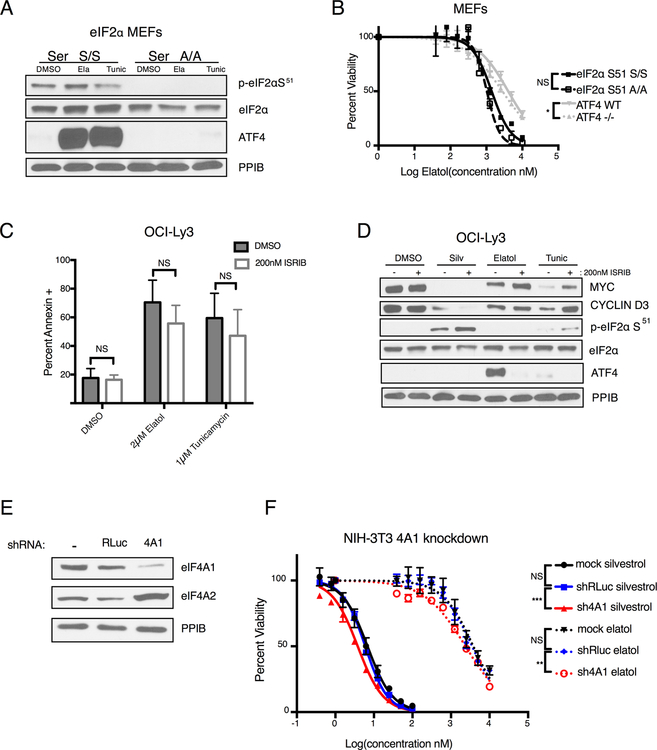
To further assess ISR induction by elatol and determine its role in the drug’s toxicity to cells, we first confirmed that the drug’s induction of ATF4 is downstream of eIF2α S51 phosphorylation. Murine embryonic fibroblasts (MEFs) with both eIF2α alleles mutated to S51A (eIF2α-S51 A/A)(37) show no induction of ATF4 in response to elatol, in contrast to littermate control eIF2α-S51 S/S MEFs (Fig. 5A). There is no significant difference, however, in elatol sensitivity between these cells (Fig. 5B, IC50=1,029 nM (A/A), 1,399 nM (S/S); p=0.9845). We also employed MEFs with biallelic Atf4 deletion(38) in comparison to MEFs from wild-type littermates and found the Atf4-deficient cells are somewhat more sensitive (Fig. 5B, IC50=2,785 nM (Atf4−/−), 4,038 nM (WT); p=0.0138). For confirmation in tumor cells, we used siRNA to knock down ATF4 in elatol-sensitive MDA-MB-468 breast cancer cells and found no change in induction of apoptosis by elatol (Fig. S5A–B). Similarly, the drug ISRIB (39), which ameliorates the short-term translational effects of eIF2α S51 phosphorylation by promoting eIF2B complex assembly (40), also showed no effect on elatol’s toxicity to OCI-Ly3 cells (Fig. 5C), despite strongly limiting its induction of ATF4 (Fig. 5D, S5C). ISR induction by elatol therefore does not mediate its toxic effects and may in fact be a cytoprotective response to either eIF4A1 ATPase inhibition or an off-target effect since MEFs lacking ATF4 were actually more sensitive.
Figure 5.
Elatol induction of ATF4 is mediated by eIF2α phosphorylation, but toxicity is dependent on eIF4A inhibition. A) Western blot showing protein expression in eIF2α wildtype and Ser51 A/A mutant MEFs treated with DMSO, 5 μM elatol or 5 μM tunicamycin for 8 hours. B) Cell viability in MEF cells wildtype or homozygous mutant for alanine at serine 51 of eIF2α and wildtype or homozygous knockout of ATF4 measured using Cell Titer Glo following 72 hour treatment with elatol. Mean ± SEM n=4 * = p< 0.05, ** = p<0.001. C) Western blot showing protein expression in cells treated for four hours with DMSO, 100 nM silvestrol, 5 μM elatol or 5 μM tunicamycin with or without the combination with 200 nM ISRIB. Representative images. n=3. D) Cell death measured by flow cytometry following annexin V staining in DLBCL cell lines treated with indicated inhibitors with or without the combination of the 200 nM ISRIB. Mean ± SEM n=3. * = p< 0.05, ** = p<0.001. E) Western blot showing eIF4A1 and eIF4A2 protein levels in NIH-3T3 cells stably expressing shRLuciferase (shRLuc) control or eIF4A1 shRNA transfected with the indicated proteins. Representative images, n=2. F) Viability of NIH-3T3 cells stably expressing control or eIF4A1 shRNA measured by Cell Titer Glo after being treated with the indicated compounds for 5 days. Mean ± SEM n=4. * = p< 0.05, ** = p<0.001, ***=p<0.0001
In contrast to these results, knockdown of eIf4a1 in NIH/3T3 cells, similar to methods employed previously to establish that eIF4A1 is the key target of silvestrol (26), showed a significant shift in the elatol viability curve, similar to silvestrol (Fig. 5E–F, IC50= 6.0 nM (shRLuc silvestrol), IC50=3.8nM (sh4A1 silvestrol) p<0.0001; IC50= 3,547nM (shRluc elatol), IC50=2,382nM (sh4A1 elatol), p=0.0026). To confirm eIF4A1 loss is not a general sensitizer to cytotoxic agents, we tested carboplatin in the same cells and found no change in drug sensitivity (Fig. S5E). Manipulation of eIF4A1 levels therefore adds to the toxic effects of elatol in a manner similar to the established eIF4A1 inhibitor silvestrol, while multiple manipulations of the ISR pathway do not. Taken together with the in vitro APTase and helicase inhibition, binding and mutational studies, and effects on translationally regulated proteins, these data show elatol functions as an eIF4A1 inhibitor in cells, although with induction of an ISR for unclear reasons and effects on transcription at doses well above IC50, making cellular off targets likely. eIF4A1’s closely homologous but biologically distinct paralogs eIF4A2 and eIF4A3 are high-probability off-targets based on structural similarity. Interestingly, we note in MDB-MB 468 cells that knockdown of either 4A2 or 4A3 results in strong induction of ATF4 (Fig. S5E), but difficulties purifying these proteins from bacteria in a state that preserves their enzymatic activity have prevented cell-free assessment of elatol inhibition (see discussion).
Anti-Tumor Activity and Toxicity of Elatol In Vivo
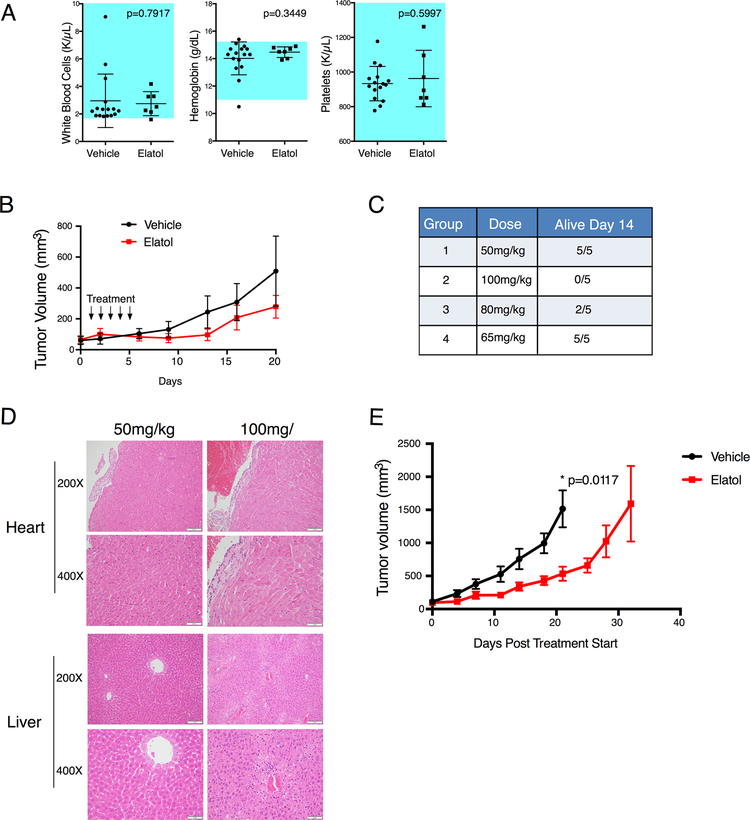
Elatol has been given in vivo in only one prior study, resulting in antitumor activity against engrafted B16 murine melanoma cells, with the compound dosed at 10 mg/kg daily by intraperitoneal (i.p.) or oral (p.o.) administration (41). We tested elatol in vivo in non-tumor bearing SCID mice at 10, 20, and 30 mg/kg daily x 5 days i.p. Dosing at 30 mg/kg had to be stopped after three days due to weight loss and reduced animal activity, but neither 10 mg/kg nor 20 mg/kg resulted in signs of toxicity (Fig S6A). Day 5 complete blood counts of the animals dosed at 20 mg/kg showed no differences from those of vehicle-dosed animals (Fig 6A). We therefore treated SCID mice engrafted with SU-DHL-6 flank tumors at 20 mg/kg. We initially planned multiple five-day cycles, but we found the first cycle resulted in weight loss and overt signs of toxicity, and treatment had to be discontinued (Fig. S6B). The initial cycle halted tumor growth for 12 days from the start of treatment, relative to vehicle-treated animals but afterward tumors resumed enlarging (Fig. 6B). Elatol dosed at 20 mg/kg daily therefore halts tumor growth but was not well tolerated in tumor-bearing animals.
Figure 6.
Elatol is well tolerated in mice up to 65 mg/kg and treatment slows tumor progression in vivo, but high doses show liver and cardiac toxicity. A) Complete blood counts for white blood cells, hemoglobin and platelets of SCID mice following treatment with 20 mg/kg elatol daily for 5 days. Mean ± SEM n=17 and 7 vehicle and elatol respectively. B) SCID mice were engrafted with 2×106 SU-DHL6 cells. Once tumors reached 60mm3 treatment began with 20 mg/kg i.p. daily for 5 days and tumor volume was measured twice per week. Mean ± SEM, n=8. C) Maximum tolerated dose study in normal CD1 mice. Cohorts of 5 mice were given a single indicated dose of elatol and observed daily for signs of morbidity. At time of death due to toxicity or at day 14 if no toxicities were observed mice were sacrificed for organ pathology. D) H&E staining of the heart and liver of mice treated with 50 or 100 mg/kg elatol. Representative images n=5. E) SCID mice were implanted with 1×106 serially transplanted OCI-Ly3 xenograft cells. When tumors reached at least 50 mm3 mice were pair matched and treatment began with either vehicle or 40 mg/kg of elatol i.p. twice weekly. Mean ± SEM, n=8. * = p< 0.05.
To optimize elatol dosing, we performed a maximum-tolerated dosing (MTD) study in female CD-1 mice using a single-dose strategy with five mice per group (Fig. 6C). Group 1 tolerated 50 mg/kg without issue, but all five animals in group 2 died within five days of receiving 100 mg/kg. Dose reduction to 80 mg/kg (group 3) still showed unacceptable toxicity, with 3/5 mice dying or moribund requiring sacrifice within five days, but animals in group 4 tolerated 65 mg/kg without issue. Organ pathology on animals suffering toxicity in group 2 was compared to those that tolerated elatol without issue in group 1 (Fig 6D, S6C). Changes were seen in multiple organs, with damage to heart and/or liver considered most likely cause of morbidity/mortality. Pathology for animals treated at 65 mg/kg was similar to 50 mg/kg (not shown). Elatol’s maximum tolerated dose in non tumor-bearing animals is therefore 65 mg/kg, dramatically higher than for silvestrol, which is typically dosed at 0.2–0.5 mg/kg (4), potentially offsetting the potency difference between the compounds seen in vitro.
We next engrafted OCI-Ly3 cells to SCID mice for additional assessment of elatol’s therapeutic window. Again, initial MTD for non-tumor bearing animals was not tolerated in tumor-bearing animals, with most engrafted animals experiencing morbidity and weight loss after a single dose at 65 mg/kg (not shown). Moving forward, we determined 40 mg/kg dosed twice per week was well-tolerated and led to significant reduction of tumor growth compared to vehicle-treated controls (p=0.0117, Fig. 6E, S6D). Elatol therefore shows proof-of-principle of a pipeline to identify inhibitors of the cap-dependent translation core enzyme eIF4A1 based on target-based cell-free in vitro screening for ATPase activity inhibition, taken through to in vivo therapeutic window.
Discussion
Target-based screening for small-molecule inhibitors of the DEAD-box RNA helicase eIF4A1 is a rational approach for discovery of novel cancer therapeutics for several reasons. First, translational activation is a nearly ubiquitous output of deregulated oncogenic signaling(16). Drugs targeting signaling molecules, typically kinases, have made substantial contributions to cancer therapeutic options, but short-lived responses in most clinical scenarios illustrate the ease with which messenger molecules may be bypassed by parallel or redundant signal (1,2,16). Translation, by contrast, particularly at the level of the eIF4F initiation complex, is a convergence point for these pathways – a bottleneck whose activation is necessary for transcriptional and epigenetic oncogenic outputs to be expressed as cellular phenotype. Both molecular heterogeneity of signaling leading to kinase-inhibitor resistance tracts and the vexing problem of tumor heterogeneity within individual patients may be bypassable with drugs against translation (42). Second, within the eIF4F complex, eIF4A1 has emerged as the most promising pharmacologic target. Innovative functional screening for interrupters of cap-dependent initiation revealed the initial natural compounds with this activity, silvestrol, hippuristanol, and pateamine A (3). Subsequent studies showed all worked by interfering with eIF4A1, the ATP-dependent enzymatic core of the complex. Availability of eIF4E, the cap-binding component of eIF4F, was shown long ago to be rate-limiting in cap-dependent activation (43), but its protein-protein interactions with the eIF4G scaffolding component appear less druggable than eIF4A1’s enzymatic mRNA-unwinding function. Additionally, comparison of translatomes sensitive to either eIF4E (by way of mTORC1) or eIF4A1 inhibition revealed cytotoxicity from eIF4A1 inhibition associated with loss of key pro-survival proteins while eIF4E inhibition was cytostatic (33). Our results in Fig. 4A are highly consistent with these findings, showing cytostasis due to rapamycin in an mTORC1-dependent system but potent cytotoxicity due to the eIF4A1 inhibitors silvestrol and elatol. Overall, we find that cell-free screening for inhibitors of eIF4A1’s RNA-dependent ATPase activity can identify compounds with broad antitumor activities against cultured tumor cells and active in vivo at tolerable doses.
A marine-derived natural compound with previously noted anticancer and antiparasitic properties but no defined mode-of-action (41,44), elatol inhibits eIF4A1 ATPase and helicase activities in vitro. These activities associate with toxicity to a broad range of cancers, with breast, non-small cell lung, and hematapoetic being the most sensitive groups (Fig 1D–F, 3A) consistent with observations with other eIF4A1 inhibitors (31). Like silvestrol, the best characterized previously identified eIF4A1 inhibitor, elatol affects protein synthesis globally and results in rapid loss of translationally regulated oncoproteins like Cyclin D3, MYC, and MCL1 (Fig 3E). The compound also is effective at blocking translation activation by BCR stimulation in primary CLL cells. In vivo, elatol is tolerated at MTD of up to 65 mg/kg in tumor free mice and at 40 mg/kg twice weekly in tumor-bearing animals. These doses are ~100x higher than for silvestrol, typically dosed at 0.2–0.5 mg/kg. Silvestrol’s effects on non-malignant host cells therefore may be significantly higher than elatol’s, suggested also by silvestrol’s >500x increased potency vs. 3T3 fibroblasts in Fig. 5F. Overall, however, elatol is 5–10-fold less potent than silvestrol vs. sensitive tumor cells in vitro, and is impotent against some lines that are silvestrol sensitive. A variety of mechanisms could account for these differences, and an intriguing question is whether elatol’s novel mode of action against the target, being an ATPase inhibitor, is part of the reason. Alternately, elatol’s off-target effects leading to induction of the cytoprotective ISR could be at play. Elatol treatment strongly upregulates ATF4 translation in all cell types we analyzed, an effect dependent, as expected, on eIF2α phosphorylation. We ruled out a role for PERK/UPR induction but otherwise it is not clear from these experiments which eIF2α-phosphorylating kinase may be responsible. Importantly, however, we find elatol toxicity is not dependent on this in MEFs deficient for Atf4 or in breast cancer cells with ATF4 knockdown. Full interrogation this question would be beyond our current scope, particularly since our initial derivatization efforts with elatol have not been successful raising questions about whether or not the compound is a good starting point for further development. We plan intensive additional screening using the approach we have established here, and identification of additional inhibitors with elatol’s mode of action vs. eIF4A1 may not only provide better lead compounds for further development but should clarify the reasons for elatol’s mechanistic differences with silvestrol.
Importantly, using retroviral knockdown we find increased sensitivity to elatol treatment upon loss of eIF4A1 similar to silvestrol and in contrast to the cytotoxic agent carboplatin. These results further establish that elatol is acting on eIF4A1 in a cellular context and that this is the cause of ISR-independent protein translation inhibition we observed. The eIF4A paralogs have increasingly well-characterized divergent biological roles. eIF4A1 is most strongly linked to the eIF4F cap-initiation complex, where it is necessary for efficient translation of most eukaryotic mRNAs (45). eIF4A2 can replace eIF4A1 in cell-free systems but is dispensable for cell survival (30). This factor is implicated instead in microRNA-mediated translational repression (46), although CRISPR deletion of the gene from murine fibroblasts showed it is not necessarily required for this (47). eIF4A3, meanwhile, is a critical component of the exon-junction complex and regulates nonsense mediated mRNA decay (48). Because the three paralogs are so homologous at the amino-acid level, however, especially in their helicase cores, pan-inhibition by elatol would not be surprising, and indeed silvestrol has been reported to interact with both eIF4A1 and eIF4A2 (49), although inhibitory effects on the latter are less well-established. Inhibition of either 4A2 or 4A3 could explain elatol’s effects on ATF4 induction and could also mediate effects on mRNA expression, although as noted we were unable to directly assess them as targets. Regardless, we believe evaluation of the specificity of eIF4A inhibitors for each paralog should become important steps eIF4A1 inhibitor evaluation when technically feasible.
The eIF4A1 inhibitor space has stalled at a preclinical stage, and the only well-defined compounds have complex mechanisms and chemical structures. In over a decade no derivative of silvestrol has made it to clinical evaluation, and its susceptibility to ABCB1 mediated drug efflux poses pharmacologic challenges (50). Elatol’s interesting binding stoichiometry with eIF4A1 suggests immediate ideas for rational derivatization through covalent dimerization that might address some off-target effects, but as mentioned above such efforts were not initially successful. Instead, we see the identification and characterization of elatol as laying the groundwork for a pipeline of novel eIF4A1 inhibitor discovery, including establishment of ATPase measurement as a basis for screening, along with multiple downstream steps for assessment of potency and specificity. Our initial screen was a relatively small one, assessing 500 natural compounds. With much larger-scale efforts now underway, we hope to identify novel eIF4A1 inhibitors suitable for optimization with high anti-tumor potency to go after this promising target for cancer therapy.
Supplementary Material
Translational Significance.
Clinical activity of targeted signaling inhibitors is limited by acquired and de novo resistance, which stems from redundancies in signaling pathways, providing multiple ways for tumor cells to maintain activation of key downstream targets. Cap-dependent translation initiation is a key biologic process downstream from signaling – a convergence point of multiple pathways – that tumor cells depend on more than non-malignant cells to maintain constitutive expression of key oncoproteins. A therapeutic window for targeting the cap-initiation complex eIF4F is well established preclinically, but no drug with this activity has entered evaluation in patients. The DEAD-box RNA helicase eIF4A1 is the enzymatic core of the eIF4F complex and is its most promising avenue for therapeutic targeting. Our study shows a way forward for high-throughput identification and characterization of eIF4A1 inhibitors to bring this highly promising therapeutic avenue to evaluation in patients.
Acknowledgements
The technical assistance of Reynaldo Morales and Loreal del Valle during the isolation and purification of (+)-elatol is gratefully acknowledged. The authors wish to thank Ariel Ramirez Labrada for technical assistance in the OCI-Ly3 xenograft experiment. We thank Francesco Forconi, Isla Henderson, Ian Tracy and Kathy Potter for kind help in collecting, storing and characterising CLL blood samples. Research reported in this publication/press release was supported by the National Cancer Institute of the National Institutes of Health under award number P30 CA023074 through the Experimental Mouse Shared Resource at the University of Arizona.
Financial support: Jonathan H. Schatz- Startup funds from the University of Arizona and University of Miami Sylvester Comprehensive Cancer Center, Eli Chapman-Startup funds University of Arizona, Graham Packham-Cancer Research UK and Southampton Experimental Cancer Medicine Centre. Abimael Rodríguez-NIH-SC1 Program (Grant 1SC1GM086271-01A1). Cyril H. Benes – The Wellcome Trust (grant 10296).
Footnotes
Conflicts of Interest: There are no potential conflicts of interest to disclose
References
- 1.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature [Internet]. Nature Publishing Group; 2014;513:105–9. Available from: 10.1038/nature13572 [DOI] [PubMed] [Google Scholar]
- 2.Schatz JH, Schatz JH, Oricchio E, Oricchio E, Wolfe AL, Wolfe AL, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med [Internet]. 2011;208:1799–9996. Available from: http://www.jem.org/cgi/doi/10.1084/jem.20110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novac O, Guenier A-S, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Research [Internet]. Oxford University Press; 2004;32:902–15. Available from: http://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gkh235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cencic R, Carrier M, Galicia-Vázquez G, Bordeleau M-E, Sukarieh R, Bourdeau A, et al. Antitumor Activity and Mechanism of Action of the Cyclopenta[b]benzofuran, Silvestrol. PLoS ONE. Public Library of Science; 2009;4:e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordeleau M-E, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol [Internet]. 2006;2:213–20. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=16532013&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 6.Bordeleau M-E, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, et al. RNA-Mediated Sequestration of the RNA Helicase eIF4A by Pateamine A Inhibits Translation Initiation. Chemistry & Biology [Internet]. 2006;13:1287–95. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=17185224&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu Rev Biochem [Internet]. 2014;83:779–812. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=24499181&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature [Internet]. 2016;534:558–61. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=27309803&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tillotson J, Kedzior M, Guimarães L, Ross AB, Peters TL, Ambrose AJ, et al. ATP-competitive, marine derived natural products that target the DEAD box helicase, eIF4A. Bioorg Med Chem Lett. 2017;27:4082–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogure T, Kinghorn AD, Yan I, Bolon B, Lucas DM, Grever MR, et al. Therapeutic Potential of the Translation Inhibitor Silvestrol in Hepatocellular Cancer. Vinciguerra M, editor. PLoS ONE [Internet]. 2013;8:e76136 Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=24086701&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinhardt JJ, Peroutka RJ, Mazan-Mamczarz K, Chen Q, Houng S, Robles C, et al. Inhibiting CARD11 translation during BCR activation by targeting the eIF4A RNA helicase. Blood [Internet]. 2014;124:3758–67. Available from: http://www.bloodjournal.org/cgi/doi/10.1182/blood-2014-07-589689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol [Internet]. 2014;15:6814 Available from: http://www.biomedcentral.com/content/pdf/s13059-014-0476-1.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol [Internet]. 2017;4:1 Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=29165424&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu J, Cargnello M, Topisirovic I, Pelletier J. Translation Initiation Factors: Reprogramming Protein Synthesis in Cancer. Trends Cell Biol [Internet]. Elsevier Ltd; 2016;26:918–33. Available from: 10.1016/j.tcb.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 15.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature [Internet]. Nature Publishing Group; 2011;473:337–42. Available from: http://www.nature.com/doifinder/10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 16.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nature Reviews Drug Discovery 2015 14:4 [Internet]. Nature Publishing Group; 2015;14:261–78. Available from: http://www.nature.com/nrd/journal/v14/n4/abs/nrd4505.html [DOI] [PubMed] [Google Scholar]
- 17.Gandhi V, Gandhi V, Plunkett W, Plunkett W, Cortes JE, Cortes JE. Omacetaxine: A Protein Translation Inhibitor for Treatment of Chronic Myelogenous Leukemia. Clinical Cancer Research [Internet]. 2014;20:1735–40. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=24501394&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.PAUSE A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4 A. EMBO J [Internet]. 1992;11:2643–54. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=1378397&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A. RNA Biology [Internet]. 2014;10:19–32. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=22995829&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harms U, Andreou AZ, Gubaev A, Klostermeier D. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Research [Internet]. Oxford University Press; 2014;42:7911–22. Available from: http://nar.oxfordjournals.org/lookup/doi/10.1093/nar/gku440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA [Internet]. Cold Spring Harbor Lab; 2003;9:394–407. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=12649492&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol [Internet]. 2005;12:861–9. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=16170325&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 23.Mendoza O, Gueddouda NM, Boulé J-B, Bourdoncle A, Mergny J-L. A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Research [Internet]. Oxford University Press; 2015;43:e71–1. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=25765657&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. Nature Publishing Group; 2012;483:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeomans A, Yeomans A, Thirdborough SM, Thirdborough SM, Valle-Argos B, Valle-Argos B, et al. Engagement of the B-cell receptor of chronic lymphocytic leukemia cells drives global and MYC-specific mRNA translation. Blood [Internet]. 2016;127:449–57. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=26491071&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 26.Chu J, Galicia-Vázquez G, Cencic R, Mills JR, Katigbak A, Porco JA, et al. CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, eIF4A. Cell Rep. 2016;15:2340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bordeleau M-E, Robert F, Gerard B, Lindqvist L, Chen SMH, Wendel H-G, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest [Internet]. American Society for Clinical Investigation; 2008;118:2651–60. Available from: http://www.jci.org/articles/view/34753#sd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, et al. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell [Internet]. Elsevier Inc; 2015;162:59–71. Available from: http://www.sciencedirect.com/science/article/pii/S009286741500639X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature [Internet]. Nature Publishing Group; 2014;513:65–70. Available from: 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA [Internet]. 2012;18:1373–84. Available from: http://rnajournal.cshlp.org/cgi/doi/10.1261/rna.033209.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters TL, Li L, Tula-Sanchez AA, Pongtornpipat P, Schatz JH. Control of translational activation by PIM kinase in activated B-cell diffuse large B-cell lymphoma confers sensitivity to inhibition by PIM447. Oncotarget. Impact Journals; 2016;7:63362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh H, Huynh H, Hao H-X, Hao HX, Chan SL, Chan SL, et al. Loss of Tuberous Sclerosis Complex 2 (TSC2) Is Frequent in Hepatocellular Carcinoma and Predicts Response to mTORC1 Inhibitor Everolimus. Molecular Cancer Therapeutics [Internet]. 2015;14:1224–35. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=25724664&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 33.Gandin V, Masvidal L, Hulea L, Gravel S-P, Cargnello M, McLaughlan S, et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. Cold Spring Harbor Lab; 2016;26:636–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vattem KM, Vattem KM, Wek RC, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. PNAS [Internet]. 2004;101:11269–74. Available from: http://www.pnas.org/content/101/31/11269.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pakos Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep [Internet]. 2016;17:1374–95. Available from: http://embor.embopress.org/lookup/doi/10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, et al. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}−2,3-dihydro-1 H-indol-5-yl)-7 H-pyrrolo[2,3- d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK). J Med Chem [Internet]. 2012;55:7193–207. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=22827572&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 37.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, et al. Translational Control Is Required for the Unfolded Protein Response and In Vivo Glucose Homeostasis. Molecular Cell [Internet]. Cell Press; 2001;7:1165–76. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=11430820&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 38.Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, et al. C/EBPγ Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF4. Mol Cell Biol [Internet]. 2016;36:693–713. Available from: http://mcb.asm.org/lookup/doi/10.1128/MCB.00911-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. eLife Sciences Publications Limited; 2013;2:e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai JC, Miller-Vedam LE, Anand AA, Jaishankar P, Nguyen HC, Renslo AR, et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science. American Association for the Advancement of Science; 2018;359:eaaq0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campos A, Souza CB, Lhullier C, Falkenberg M, Schenkel EP, Ribeiro-do-Valle RM, et al. Anti-tumour effects of elatol, a marine derivative compound obtained from red algae Laurencia microcladia. Journal of Pharmacy and Pharmacology [Internet]. 2012;64:1146–54. Available from: http://doi.wiley.com/10.1111/j.2042-7158.2012.01493.x [DOI] [PubMed] [Google Scholar]
- 42.Ramon Y Cajal S, Castellvi J, Hümmer S, Peg V, Pelletier J, Sonenberg N. Beyond molecular tumor heterogeneity: protein synthesis takes control. Oncogene. Nature Publishing Group; 2018;:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. American Society for Biochemistry and Molecular Biology; 1985;260:7843–9. [PubMed] [Google Scholar]
- 44.Desoti VC, Lazarin-Bidóia D, Sudatti DB, Pereira RC, Alonso A, Ueda-Nakamura T, et al. Trypanocidal Action of (−)-Elatol Involves an Oxidative Stress Triggered by Mitochondria Dysfunction. Marine Drugs [Internet]. 2012;10:1631–46. Available from: http://www.mdpi.com/1660-3397/10/8/1631/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.PAUSE A, METHOT N, SVITKIN Y, MERRICK WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J [Internet]. 1994;13:1205–15. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=8131750&retmode=ref&cmd=prlinks [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, et al. Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation. Science [Internet]. 2013;340:82–5. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=23559250&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 47.Galicia-Vázquez G, Chu J, Pelletier J. eIF4AII is dispensable for miRNA-mediated gene silencing. RNA [Internet]. 2015;21:1826–33. Available from: http://rnajournal.cshlp.org/lookup/doi/10.1261/rna.052225.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan CC, Dostie J, Diem MD, Feng WQ, Mann M, Rappsilber J, et al. eIF4A3 is a novel component of the exon junction complex. RNA [Internet]. 2004;10:200–9. Available from: http://rnajournal.cshlp.org/content/10/2/200.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers JM, Lindqvist LM, Webb A, Huang DCS, Savage GP, Rizzacasa MA. Synthesis of Biotinylated Episilvestrol: Highly Selective Targeting of the Translation Factors eIF4AI/II. Org Lett [Internet]. 2013;15:1406–9. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=23461621&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 50.Gupta SV, Sass EJ, Davis ME, Edwards RB, Lozanski G, Heerema NA, et al. Resistance to the Translation Initiation Inhibitor Silvestrol is Mediated by ABCB1/P-Glycoprotein Overexpression in Acute Lymphoblastic Leukemia Cells. AAPS J [Internet]. 2011;13:357–64. Available from: http://www.springerlink.com/index/10.1208/s12248-011-9276-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.