Abstract
One of the earliest patterning events in the vertebrate neural plate is the specification of mes/r1, the territory comprising the prospective mesencephalon and the first hindbrain rhombomere. Within mes/r1, an interface of gene expression defines the midbrain-hindbrain boundary (MHB), a lineage restriction that separates the mesencephalon and rhombencephalon. wnt1 is critical to mes/r1 development and functions within the MHB as a component of the MHB gene regulatory network (GRN). Despite its importance to these critical and early steps of vertebrate neurogenesis, little is known about the factors responsible for wnt1 transcriptional regulation. In the zebrafish, wnt1 and its neighboring paralog, wnt10b, are expressed in largely overlapping patterns, suggesting co-regulation. To understand wnt1 and wnt10b transcriptional control, we used a comparative genomics approach to identify relevant enhancers. We show that the wnt1-wnt10b locus contains multiple cis-regulatory elements that likely interact to generate the wnt1 and wnt10b expression patterns. Two of eleven conserved enhancers tested show activity restricted to the midbrain and MHB, an activity that is conserved in the distantly related spotted gar orthologous elements. Three non-conserved elements also play a likely role in wnt1 regulation. The identified enhancers display dynamic modes of chromatin accessibility, suggesting controlled deployment during embryogenesis. Our results suggest that the control of wnt1 and wnt10b expression is under complex regulation involving the interaction of multiple enhancers.
Keywords: wnt1, wnt10b, neural patterning, conserved non-coding element, cis-regulation, vertebrate, zebrafish, spotted gar, ATAC-seq, CAGE-seq
Introduction
Wnt1 is critical to multiple aspects of vertebrate development and human disease, including specification of mouse midbrain dopaminergic neurons (Andersson et al., 2013), stimulation of progenitor proliferation (Falk et al., 2008), dorsal neural tube specification (Ikeya et al., 1997), neural crest development (Dorsky et al., 1998), and bone homeostasis (Ang et al., 2018; Fahiminiya et al., 2013; Keupp et al., 2013; Laine et al., 2013; Makitie et al., 2017; Pyott et al., 2013). Each developmental function of wnt1 requires precise spatial and temporal expression control, and, consequently, disease states can result from errors in wnt1 expression. Thus, understanding the factors behind the dynamic regulation of wnt1 transcription is a crucial step in unraveling the molecular aspects of how it contributes to development and disease.
The highly dynamic wnt1 transcription pattern initiates downstream of early neural posteriorizing signals (Green et al., 2015). The earliest expression of wnt1 is observed in the neural plate of developing vertebrate embryos in the presumptive mesencephalon and anterior rhombencephalon. This pattern evolves such that after neural tube closure wnt1 transcripts are found in the epiphysis, dorsal midline of the midbrain, and at the conserved neural tube constriction known as the midbrain-hindbrain boundary (MHB), or isthmus. Caudal to the isthmus, wnt1 is expressed in the rhombic lip of the hindbrain and in the dorsal spinal cord (McMahon and Bradley, 1990; McMahon et al., 1992; Wilkinson et al., 1987). In some cases, wnt1 expression marks unique functional domains, for example mouse wnt1 homozygous mutants display loss of midbrain and anterior hindbrain, but the dorsal spinal cord appears normal (McMahon and Bradley, 1990; Thomas and Capecchi, 1990). The dispensability of wnt1 to dorsal spinal cord development is likely due to overlapping expression with other Wnt ligands, as wnt1;wnt3a double mutant mice lack dorsal spinal cord fates (Ikeya et al., 1997), and zebrafish wnt1 appears to be functionally redundant with wnt10b and wnt3a (Buckles et al., 2004; Lekven et al., 2003). Nonetheless, in all vertebrates examined to date, wnt1 orthologs share a conserved expression pattern during embryonic stages.
Despite the evolutionary conservation of the vertebrate wnt1 expression pattern, little is known about the molecular basis of its transcriptional regulation. Perhaps the most studied aspect of wnt1 transcription centers on its expression in the early neural plate and midbrain-hindbrain boundary (MHB). In mouse, the earliest wnt1 expression domain marks a region referred to as mes/r1, as the expressing cells comprise the mesencephalon and progenitors for rhombomere 1 (Zervas et al., 2005; Zervas et al., 2004). Within mes/r1, the MHB forms at a position where mesencephalic progenitors that express otx2 abut rhombencephalic progenitors that express gbx orthologs (Hidalgo-Sanchez et al., 1999; Millet et al., 1999; Rhinn et al., 2003; Wassarman et al., 1997). A poorly understood MHB gene regulatory network establishes an interface of wnt1 and fgf8 expressing cells matching the otx2/gbx interface, with otx2/wnt1 expressing midbrain progenitors abutting gbx/fgf8 expressing hindbrain progenitors (Gibbs et al., 2017). Intriguingly, loss of function studies in mouse and zebrafish have shown that this suite of genes comprises an interactive regulatory network required for their combined expression maintenance, but not for their induction (Raible and Brand, 2004; Wurst and Bally-Cuif, 2001). Even though considerable effort has gone into understanding the genetic interactions involved in the mes/r1 and MHB gene regulatory networks, only a small number of direct regulators of wnt1 have been identified, and major mechanistic aspects of the MHB gene regulatory network remain obscure.
Understanding wnt1 transcriptional control requires identifying its full complement of enhancers, and several studies in the mouse made significant progress toward this goal. Expression in mes/r1, as well as in the dorsal spinal cord, requires an enhancer localized to a 5.5 kb region 3’ of the wnt1 transcription unit (Echelard et al., 1994). Mouse knockout and transgene studies established that the 5.5 kb enhancer is necessary and sufficient for wnt1 expression in its normal developmental pattern (Danielian et al., 1997), although some enhancer elements are likely not contained within this region, for example an element required for repression in the ventral telencephalon (Echelard et al., 1994). Within the 5.5 kb enhancer, a 110 bp element conserved between fish and mouse is capable of driving reporter expression in the early midbrain (Rowitch et al., 1998). Importantly, the 110 bp enhancer was found to drive sufficient wnt1 expression to rescue the midbrain and anterior hindbrain deficiency of wnt1 homozygous mutant mice. This element and sites upstream of the wnt1 promoter were discovered to be bound in vivo by Six3, a transcription factor that is essential for repression of wnt1 expression in prospective forebrain (Lagutin et al., 2003). Therefore, cis-regulatory sequences sufficient for much of wnt1 expression have been identified in the mouse, along with a transcription factor that directly represses wnt1 in forebrain domains. However, no direct transcriptional activators of wnt1 have been identified in any system, nor have any direct repressors such as Six3 been verified in systems other than mouse. Considering the degree of vertebrate enhancer functional redundancy revealed in recent studies (Dickel et al., 2018; Osterwalder et al., 2018), it is reasonable to conjecture that additional enhancers of wnt1 transcription have yet to be identified.
To understand the molecular control of wnt1 transcription, we have initiated a comparative genomic and transgenic analysis of wnt1 regulatory sequences in the zebrafish. We present evidence suggesting the identification of a chromosomal interval comprising a wnt1 topological associating domain (TAD), which includes 11 conserved non-coding elements (CNEs), two of which are important regulators of the wnt1 spatial transcription pattern, but all of which show some level of transcriptional regulatory capacity. Additionally, three non-conserved putative regulatory elements are identified as regions of accessible chromatin. Because the neighboring Wnt paralog, wnt10b, is expressed in an almost identical pattern to that of wnt1 in zebrafish, we propose that these regulatory elements interact to drive transcription of both of these neighboring loci.
Materials and Methods.
Identification and cloning of CNEs
Conserved non-coding elements were identified by comparing genomic intervals spanning the wnt1 and wnt10b loci from zebrafish, three-spine stickleback, and spotted gar. The zebrafish genomic interval used comprised coordinates chr23:27,029,742-27,089,756 from genome release Zv7. The stickleback region comprised coordinates chrXII: 11,281,697-11,306,728 from the Feb 2006 genome release (UCSC Genome Browser). The spotted gar region comprised coordinates LG4:18912155-18982547 from the LepOcu1 genome assembly (ensembl.org).
These regions were imported into the zPicture alignment visualization tool in rVista 2.0 (rvista.dcode.org) or the JGI/DOE Vista alignment tool (http://genome.lbl.gov/vista/mvista/submit.shtml). CNE PCR products were amplified from zebrafish or spotted gar genomic DNA (spotted gar tissue sample kindly provided by Dr. Ingo Braasch, Michigan State University, and Dr. John Postlethwait, University of Oregon) and ligated into pT2AW2, a derivative of pT2AL200R150G (Urasaki et al., 2006) in which the EF1α promoter and intron were replaced with the c-fos minimal promoter and an upstream polylinker. PCR primers were designed to amplify each CNE element plus 50 bp of pad sequence with BamHI and HindIII or EcoRI restriction sites on the ends for ligation into corresponding sites in pT2AW2. ATAC-seq, CAGE-seq, ChIP-seq, and RNA-seq datasets presented here are UCSC genome browser tracks derived from published and unpublished datasets that have been curated and in some cases reanalyzed by the DANIO-CODE consortium (https://danio-code.zfin.org).
Zebrafish maintenance, transgenesis and in situ hybridizations
Zebrafish were maintained according to standard procedures (Westerfield, 2000). Wild type strains were either AB, TL, or ABTL hybrids. Transgenic lines were generated by co-injection of 1-3 nl of transgene plasmids (25 ng/μl) with Tol2 transposase mRNA (25 ng/μl) into wild-type 1-cell stage embryos. Embryos were raised to adulthood, then outcrossed to wild-type. Progeny were scored for EGFP fluorescence, and transgenics were raised to establish stable lines. At least two independent stable transgenic lines were examined for each transgene. In cases where position effects caused divergence between expression patterns observed in two lines for a transgene, additional lines were examined. The modal expression pattern was determined to reflect CNE activity. Fluorescence images were obtained on an Olympus BX61 compound microscope or a Nikon SMZ25 fluorescence dissecting microscope. In situ hybridizations were performed as described previously (Buckles et al., 2004).
Results
The Wnt1 locus lies within a highly conserved chromosomal territory.
Current models suggest that enhancers function within the context of chromosomal topological associating domains, or TADs, hypothesized to constrain the interactions of enhancers with their cognate target promoters (Yu and Ren, 2017). The identification of a wnt1 TAD would thus serve to constrain a search for relevant wnt1 enhancer elements. TADs are identified as regions with high degrees of inter-chromosomal association, but microsynteny has also been hypothesized to reflect the conservation of structural TADs (Harmston et al., 2017). We compared genomic intervals spanning wnt1 from diverse vertebrates, as local microsynteny around wnt1 had been previously identified (Gellner and Brenner, 1999). Our comparison revealed that an interval including wnt1, its neighboring paralog wnt10b, and arf3a, is conserved in all vertebrates (Fig. 1). This conserved interval includes the previously noted wnt1 3’ enhancer, orthologs of which are invariably located downstream of the wnt1 transcription unit. Upstream of wnt1, synteny is not observed beyond arf3 in either the ray finned fish (zebrafish, gar) or lobe-finned fish (coelacanth, mouse) lineages. Downstream of wnt1, synteny is observed within the ray-finned fish (lmbr1l-rps26) or within the lobe-finned fish (kmt2d-prkag1), but not between them. Thus, a limited chromosomal region surrounding wnt1 is highly conserved, suggesting an important chromosomal architecture. We hypothesize this interval likely represents a TAD, and we then focused on the zebrafish interval spanning the rps26 and phf8 loci to search for local conserved non-coding elements that might function as enhancers.
Figure 1. Conservation of the wnt1-wnt10b-arf3 genomic interval in vertebrates.
Gene data obtained from the UCSC genome browser, approximate interval size shown is indicated. Genes are indicated with blue arrows that indicate orientation. W1E refers to the conserved wnt1 3’ enhancer identified in mouse and pufferfish. Note the conservation of the interval including W1E through arf3 in all genomes. wnt6 is present between wnt1 and wnt10b in Coelacanth, consistent with a hypothesized ancient Wnt cluster Figure 1. Conservation of the wnt1-wnt10b-arf3 genomic interval in vertebrates. Gene data obtained from the UCSC genome browser, approximate interval size shown is indicated. Genes are indicated with blue arrows that indicate orientation. W1E refers to the conserved wnt1 3’ enhancer identified in mouse and pufferfish. Note the conservation of the interval including W1E through arf3 in all genomes. wnt6 is present between wnt1 and wnt10b in Coelacanth, consistent with a hypothesized ancient Wnt cluster (Nusse, 2001).
Multiple conserved non-coding elements lie within the wnt1-10b chromosomal territory.
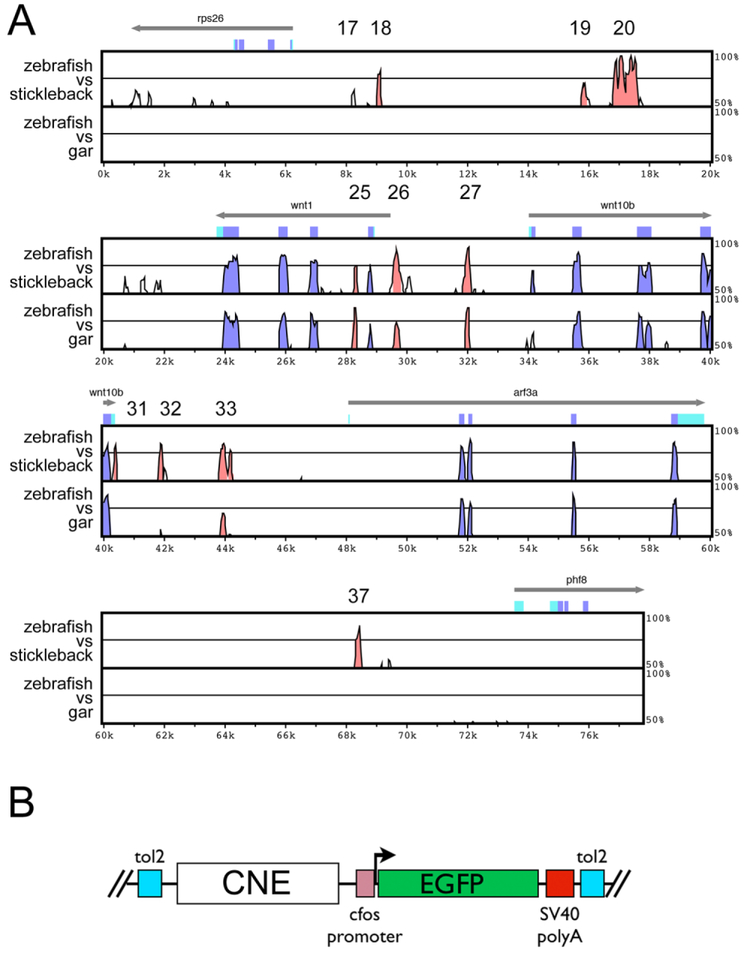
To identify potential wnt1 cis-regulatory elements, we compared wnt1 chromosomal intervals from the zebrafish, three-spine stickleback, and spotted gar genomes (Braasch and Postlethwait, 2017). A Vista comparison of these genomic regions identified several conserved non-coding elements (CNEs) between the rps26 and phf8 loci (Fig. 2A, red peaks) with 11 elements identified from the zebrafish-stickleback comparison and six of these also identified in the zebrafish-gar comparison. Included in the identified CNEs is the ortholog of the previously identified wnt1 3’ enhancer (Fig. 2A, CNE20, identified in a separate spotted gar genomic contig due to incomplete assembly, see supplemental Figure 1).
Figure 2. Analysis of sequence conservation in the wnt1-arf3 genomic interval.
(A) Vista plot comparison of zebrafish to stickleback (top bracket) and zebrafish to spotted gar (bottom bracket). Conserved noncoding elements are represented by red peaks. Numbering is according to vista browser output. The absence of element 20 in the zebrafish-gar comparison is attributable to an incomplete gar contig; element 20 is found on a separate contig. (B) Schematic diagram of the reporter construct used to evaluate transcriptional activity of CNEs. Tol2 elements facilitate transposition via the Tol2 system. Constructs include the cfos minimal promoter.
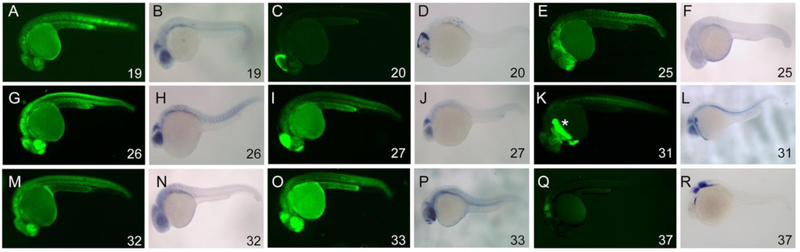
To determine whether these conserved non-coding elements possess transcriptional regulatory activity, we amplified the eleven zebrafish CNE elements for ligation upstream of a c-fos minimal promoter in a Tol2 EGFP reporter plasmid (Fig. 2B; amplification primers and CNE coordinates listed in Supplemental Table 1). All constructs generated some level of reporter fluorescence in transient expression assays (not shown), and we generated stable transgenic lines for nine of the eleven constructs. Expression was determined for at least two independent insertions of each construct, and more were analyzed if discordance was observed between the initial two. We initially screened the EGFP fluorescence patterns at 24-27 hpf and compared them to transcriptional patterns by in situ hybridization (Fig. 3). Most constructs show broad fluorescence at 24-27 hpf. For instance, CNE19 drives widespread low level expression throughout the nervous system and in the posterior notochord, although in situ hybridization at this timepoint detects transcripts most strongly in the posterior midbrain and retina (Fig. 3A,B). CNE25, an element from the first intron of wnt1, drives just detectable levels of expression in somites, telencephalon and putative head mesoderm, but in situ hybridization staining is difficult to detect at this stage (Fig. 3E,F). CNE26 lies upstream of the wnt1 refseq first exon (but see more details below), and drives expression broadly in the nervous system with higher levels in the spinal cord, posterior midbrain, eye and telencephalon. Reporter transcripts at this stage are concentrated in the posterior midbrain and eye, though lower levels are observed in the other expression domains (Fig. 3G,H). CNE31 shows low level expression in the midbrain, retina and posterior notochord, with a very high level of expression in the hatching gland (Fig. 3K,L, asterisk indicates hatching gland). While we observed hatching gland expression somewhat frequently in CNE31 transient expression assays (not shown), not all stable transgenic lines showed this expression domain (not shown). We therefore postulate that this likely represents a positional effect and CNE31 may be more sensitive than other elements to activation in the hatching gland. CNE32 and CNE33 show expression patterns very similar to that of CNE26, though levels are lower and in situ hybridization does not detect expression as readily in the midbrain (Fig. 3M-P). The CNE37 pattern was also variable, but the modal expression pattern appeared to be at very low levels in surface ectoderm over the hindbrain, with additional expression often observed in the hindbrain (Fig. 3Q,R). Of note, CNE37 lies in close proximity to phf8, which encodes a histone demethylase and is expressed in the embryonic brain (Qi et al., 2010). Knockdown of phf8 leads to apoptotic cell death in the hindbrain in a pattern reminiscient of the modal expression we observe driven by CNE37 (Qi et al., 2010), suggesting a potential regulatory interaction with phf8. In contrast to the general expression observed for those elements, we found more specific expression patterns driven by CNE20 and CNE27. CNE20 corresponds to the wnt1 3’ enhancer previously identified in mouse and fugu (Gellner and Brenner, 1999; Rowitch et al., 1998), and expression is localized exclusively to the dorsal midbrain and midbrain-hindbrain boundary (Fig. 3C,D, see also (Gibbs et al., 2017; Gibbs et al., 2014), a pattern strikingly similar to the wnt1/wnt10b expression pattern in the anterior nervous system at this stage. CNE27 expression is observed most strongly in the midbrain, with some expression also in the forebrain (Fig. 3I,J). Curiously, expression detected by in situ hybridization at 27 hpf is limited to a small group of cells in the MHB at very low levels. In sum, these transgenic assays show that multiple conserved noncoding elements function as enhancers, with most showing broad activity spanning known wnt1 expression domains and also ectopic domains. In contrast, two elements show specific activity within known wnt1 expression domains in the midbrain and MHB. One of these was expected (CNE20), but one was not (CNE27), suggesting that more than one enhancer may regulate the wnt1 spatial transcription pattern.
Figure 3. wnt1 CNEs are transcriptionally active.
All images lateral views, 27 hpf embryos.(A,C,E,G,I,K,M,O,Q) EGFP fluorescence. (B,D,F,H,J,L,N,P,R) In situ hybridizations for EGFP transcripts. Note broad GFP fluorescence not matched by transcription pattern for CNE19 (A,B), 25 (E,F), 26 (G,H), 32 (M,N) and 33 (O,P), indicating earlier expression that terminates prior to 27 hpf. Restricted expression is observed for CNE 20 (C,D), 27 (I,J), 31 (K,L) and 37 (Q,R), but the transcription pattern for CNE 27 indicates the cessation of transcription prior to 27 hpf. In contrast, CNE 20 is transcribed robustly in the MHB and dorsal midbrain at 27 hpf. CNE 31expression in the hatching gland (K, asterisk) is likely a positional effect.
Spotted gar wnt1 CNEs are active in the zebrafish midbrain and MHB
To test whether the gar CNEs are similarly active to their zebrafish orthologs, we amplified the elements from gar genomic DNA, ligated them into the same reporter vector used for the zebrafish activity assays and generated stable transgenic lines (primers and CNE coordinates listed in Supplemental Table 1). As shown in Fig. 4, both gar CNE20 and CNE27 are active in 27 hpf zebrafish embryos. Gar CNE20 fluorescence is observed most readily in the MHB and epiphysis, but low level patchy fluorescence is observed in the midbrain and hindbrain (Fig. 4A,B; Fig. 5A). In situ hybridization detects transcripts weakly in the MHB (Fig. 4B, arrow), more strongly in the epiphysis, but not elsewhere. Gar CNE27 fluorescence is also observed in the MHB, with stronger expression than gar CNE20 (Fig. 4C,D), also matched by a relatively robust in situ hybridization signal. These observations point out interesting differences between the gar and zebrafish CNE activities (Fig. 5). The gar CNE20 fluorescence in midbrain and hindbrain likely reflects earlier broader transcriptional activity, with poor maintenance in the MHB (Fig. 4B,5A). Dorsal midbrain expression of zebrafish CNE20 is not recapitulated by the gar ortholog (Fig. 5A,B, arrowheads). The CNE27 orthologs also show differences. While zebrafish CNE27 fluorescence is observed in the midbrain, this must reflect perdurance from earlier transcription as in situ hybridization signal is observed only very weakly in a small number of cells at the MHB (Figs. 3J,5D). In contrast, gar CNE27 fluorescence at the posterior midbrain/MHB closely resembles that of zebrafish CNE20 (cf. 5C to B), and transcriptional activity is robust at the MHB (Fig. 4D). Nonetheless, the gar CNEs are active in midbrain and MHB domains despite 58% sequence identity to the zebrafish orthologs (supplemental Figures 1,2).
Figure 4. Spotted gar CNE 20 and 27 are active in zebrafish.
Expression at 27 hpf, lateral views. EGFP fluorescence (A,C) and corresponding in situ hybridizations for EGFP transcripts (B,D). Note relative differences in expression strength at the MHB (arrows), with 27 being more strongly expressed than 20.
Figure 5. Comparison of zebrafish to gar CNE20 and CNE27 activity.
Lateral views of EGFP fluorescence in 27 hpf zebrafish heads. Note widespread but mottled expression from gar CNE20 (A, arrow) in comparison to the restricted expression of zebrafish CNE20 (B). While the gar 20 element is expressed strongly in the epiphysis (asterisk), zebrafish 20 is expressed at a lower level, but is also observed in the dorsal midbrain (arrowhead). Gar CNE27 is observed strongly in the posterior midbrain and MHB (C, arrow), in comparison to the more diffuse midbrain expression of zebrafish CNE27 (D). This likely reflects different temporal expression dynamics, as zebrafish 27 is not transcribed strongly in the midbrain or MHB at this time (see Fig. 3J).
Overlap between CNEs and open chromatin regions
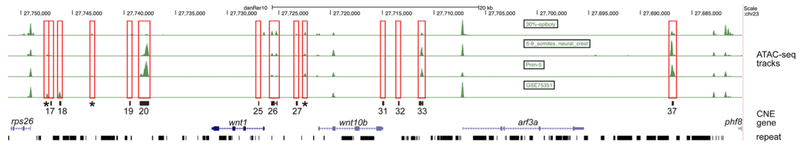
While sequence conservation has been used extensively to identify candidate regulatory elements, the degree to which conserved elements correspond to the suite of regulatory elements for a gene is often unclear (Bulger and Groudine, 2011). Enhancers are frequently sites of specific histone modifications or comprise open chromatin accessible to DNaseI or transposon activity (Calo and Wysocka, 2013). To determine whether the array of conserved sequences we identified might represent the complete set of wnt1 regulatory elements, we took advantage of ATAC-seq datasets recently aggregated through the Danio Code project (danio-code.zfin.org) and available as custom tracks on the UC Santa Cruz genome browser (genome.ucsc.edu). We examined ATAC-seq data from four datasets: 30% epiboly stage (4.7 hpf) whole embryo, 5-9 somite stage (11.7 hpf) neural crest, Prim-5 stage (24 hpf) whole embryo, and a zebrafish melanoma model cell line. As shown in Figure 6, five of the eleven CNEs are also positive for ATAC-seq peaks in embryogenesis stage samples (CNEs 20, 26, 27, 33, and 37), with CNE19 being ambiguous and may overlap with a peak at 30% epiboly and in zebrafish melanoma cell lines. CNE18 is identified as an ATAC-seq peak in a zebrafish melanoma cell line, but not in the embryonic stage samples. In contrast, four CNEs are not represented by ATAC-seq peaks in any of the samples examined (CNEs 17, 25, 31, and 32). Curiously, three additional non-exonic, non-conserved ATAC-seq peaks can be identified in this genomic interval in multiple samples (Fig. 6, marked by asterisks), suggesting these may represent additional enhancers. The most robust ATAC-seq peaks correspond to three CNEs: CNE20, 33, and 37, with one robust peak found between CNE27 and wnt10b exon1. Taken together, these results suggest that some wnt1 CNEs function as enhancers during embryonic development, but other CNEs may function as enhancers in other tissues or stages, for example in bone (Fahiminiya et al., 2013; Keupp et al., 2013; Laine et al., 2013; Pyott et al., 2013). Additionally, wnt1 and wnt10b transcription in zebrafish is likely controlled by additional non-conserved elements.
Figure 6. ATAC-seq peaks identify conserved and non-conserved cis-regulatory elements.
The window spans rps26 through phf8, with CNEs indicated by their numbers below red box outlines. Putative non-conserved enhancers are indicated by asterisks below red boxes. The embryonic sources for each ATAC-seq line are indicated in the black boxes above each line.
CNE26 contains the wnt1 promoter
The Danio-Code project has also aggregated CAGE-seq datasets, which identify the 5’ ends of full-length transcripts. Curiously, these datasets suggest that the 5’ end of the current wnt1 Refseq transcript does not correspond to the transcriptional start site, but rather that the wnt1 transcription start site lies within CNE26 (Fig. 7A). Most transcripts appear to initiate at variable positions within an 86 bp window within the proximal portion of the CNE26 element. In contrast, the majority of wnt10b transcripts initiate within a six bp window 47 bp upstream of the wnt10b Refseq transcript (Fig. 7B). Because our CNE26 reporter transgene has the sequence in the reverse complement orientation relative to the wild type locus but can still drive reporter expression, these results suggest CNE26 likely possesses both promoter and enhancer functions.
Figure 7. CAGE-seq identifies transcription start sites for wnt1 and wnt10b.
Refseq transcripts are shown in blue. (A) wnt1 transcripts originate from a ~86 bp window of CNE 26. (B) wnt10b transcripts originate from a more localized focus at or adjacent to the refseq 5’ end.
Discussion
We have used comparative genomic, transgenesis, and data mining approaches to identify fourteen putative wnt1 regulatory elements, eleven of these being evolutionarily conserved sequences. Of these fourteen putative elements, eleven conserved sequences are confirmed to be transcriptional enhancers, but only five of these eleven are in an open chromatin conformation during embryonic stages. Two elements are in an open conformation in a zebrafish melanoma cell line model, which is interesting considering the derivation of melanocytes from the wnt1-expressing neural crest lineage. Five putative regulatory elements are in an open conformation in the three embryonic stages examined, further suggesting the combinatorial regulation of wnt1. Interestingly, the ultraconserved wnt1 3’ enhancer is in an open conformation only in the post-gastrula stage samples, indicating temporally changing accessibility of this enhancer. And, curiously, one non-conserved sequence is open in the three developmental stages examined. While we did not test that element for transcriptional regulatory activity, its proximity to wnt10b suggests a potential role in its regulation.
The identification of an extensive set of specific enhancers is an important advance in understanding wnt1 and wnt10b transcriptional regulation. While the wnt1 transcription pattern is conserved in vertebrates, wnt10b co-regulation is less clear. Overlapping wnt1-wnt10b expression is observed in zebrafish embryos (Lekven et al., 2003), and evidence suggests a similar pattern in Xenopus (Zhang et al., 2011). In contrast, wnt10b may not be co-expressed with wnt1 during mouse early neural development, as wnt10b is reported to be expressed in the blastocyst, then not again until E8.5, well after wnt1 initiation (Kemp et al., 2005). One possible explanation for the different modes of transcriptional control observed between species may be differential engagement of enhancers with either promoter. Indeed, our reporter transgene constructs used the heterologous c-fos promoter, leaving open the possibility that they may behave differently in cis to the wnt1 or wnt10b promoters. It is notable that conservation of potential regulatory elements has been observed between the mammalian wnt1 and wnt10b loci (Katoh and Katoh, 2005; Qurrat Ul et al., 2011), and ENCODE data suggest the presence of conserved elements in the human wnt1-wnt10b intergenic region that correspond to DNaseI hypersensitive sites suggesting enhancer properties (our observation; genome.ucsc.edu). Because of the conserved architecture but divergent expression of the wnt1-wnt10b gene pair in vertebrates, understanding enhancer engagement with the wnt1 and wnt10b promoters will be an important research avenue.
Enhancers are hypothesized to function within the context of topological associating domains, or TADs, where the TAD structure constrains enhancer-promoter interactions (Remeseiro et al., 2016). Recent studies in the Drosophila blastoderm suggest that a shared enhancer can drive coordinated transcription from two target promoters, i.e. that one enhancer drives transcriptional bursts simultaneously from multiple promoters rather than alternating transcriptional bursts from each promoter (Fukaya et al., 2016). Whether simultaneous or alternating, consistent and prolonged proximity of enhancer and promoter sequences appear to be associated with active transcription (Chen et al., 2018). The identification of critical enhancers for wnt1 and wnt10b in zebrafish will provide the basis for future studies to dissect enhancer-promoter interactions in their TAD architecture in this important vertebrate developmental context, and may provide a model to understand enhancer-promoter dynamics in other complex Wnt loci (Narayanan and Lekven, 2012; Ramel et al., 2004).
wnt1/wnt10b regulation and evolution of brain patterning
Wnt expression in mes/r1 is essential to normal brain development, as loss of function mutations cause the loss of midbrain and cerebellum (Buckles et al., 2004; McMahon and Bradley, 1990; Thomas and Capecchi, 1990). Wnt1 also is a central player in the midbrain-hindbrain boundary gene regulatory network, in which it functions with fgf8, pax2a and engrailed genes in a poorly understood self-maintaining regulatory network (reviewed in Gibbs et al, 2017). Wnt expression must also be limited spatially during neural plate patterning, as ectopic expression can result in abnormal patterning of the forebrain (Lagutin et al., 2003). Thus, positive and negative regulation of Wnt expression in the midbrain and anterior hindbrain is crucial to normal brain patterning. Interestingly, the relative sizes of forebrain subdomains diverge in cichlid fishes that evolve in distinct ecological environments, and the differences in forebrain domain sizes correlate to changes in the Wnt-dependent mechanisms of forebrain patterning (Sylvester et al., 2010). Considering the molecular complexity of the MHB gene regulatory network, such as differential sensitivity of wnt1, pax2 and fgf8 to Sp5 and Irx regulation (Itoh et al., 2002; Tallafuss et al., 2001), deciphering the mechanisms behind their transcriptional regulation is a daunting but intriguing and important goal.
Supplementary Material
Acknowledgements
The authors thank Dr. Jo-Ann Fleming for thoughtful discussions regarding this work and for fish care, and Dr. Laura Beaster-Jones for discussions and assistance during various stages of this project. We are also grateful to Dr. Ingo Braasch and Dr. John Postlethwait for kindly providing spotted gar samples. This work was supported in part by NIH grant 1R01NS088564.
References
- Andersson ER, Salto C, Villaescusa JC, Cajanek L, Yang S, Bryjova L, Nagy II, Vainio SJ, Ramirez C, Bryja V, Arenas E, 2013. Wnt5a cooperates with canonical Wnts to generate midbrain dopaminergic neurons in vivo and in stem cells. Proc Natl Acad Sci U S A 110, E602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang K, Sanchez Rangel E, Yuan Q, Wu D, Carpenter TO, Insogna K, 2018. Skeletal disease in a father and daughter with a novel monoallelic WNT1 mutation. Bone Rep 9, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Postlethwait JH, 2017. The Spotted Gar: Genomic Journeys into a Lost World. J Exp Zool B Mol Dev Evol 328, 593–595. [DOI] [PubMed] [Google Scholar]
- Buckles GR, Thorpe CJ, Ramel MC, Lekven AC, 2004. Combinatorial Wnt control of zebrafish midbrain-hindbrain boundary formation. Mech Dev 121, 437–447. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M, 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J, 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T, 2018. Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet 50, 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Echelard Y, Vassileva G, McMahon AP, 1997. A 5.5-kb enhancer is both necessary and sufficient for regulation of Wnt-1 transcription in vivo. Dev Biol 192, 300–309. [DOI] [PubMed] [Google Scholar]
- Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, Lee EA, Harrington AN, Pham QT, Garvin TH, Kato M, Osterwalder M, Akiyama JA, Afzal V, Rubenstein JLR, Pennacchio LA, Visel A, 2018. Ultraconserved Enhancers Are Required for Normal Development. Cell 172, 491–499 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Moon RT, Raible DW, 1998. Control of neural crest cell fate by the Wnt signalling pathway. Nature 396, 370–373. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP, 1994. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development 120, 2213–2224. [DOI] [PubMed] [Google Scholar]
- Fahiminiya S, Majewski J, Mort J, Moffatt P, Glorieux FH, Rauch F, 2013. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet 50, 345–348. [DOI] [PubMed] [Google Scholar]
- Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid MT, Draganova K, Lang KS, Paratore C, Leveen P, Suter U, Karlsson S, Born W, Ricci R, Gotz M, Sommer L, 2008. Brain area-specific effect of TGF-beta signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell 2, 472–483. [DOI] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M, 2016. Enhancer Control of Transcriptional Bursting. Cell 166, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellner K, Brenner S, 1999. Analysis of 148 kb of genomic DNA around the wnt1 locus of Fugu rubripes. Genome Res 9, 251–258. [PMC free article] [PubMed] [Google Scholar]
- Gibbs HC, Chang-Gonzalez A, Hwang W, Yeh AT, Lekven AC, 2017. Midbrain-Hindbrain Boundary Morphogenesis: At the Intersection of Wnt and Fgf Signaling. Front Neuroanat 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs HC, Dodson CR, Bai Y, Lekven AC, Yeh AT, 2014. Combined lineage mapping and gene expression profiling of embryonic brain patterning using ultrashort pulse microscopy and image registration. J Biomed Opt 19, 126016. [DOI] [PubMed] [Google Scholar]
- Green D, Whitener AE, Mohanty S, Lekven AC, 2015. Vertebrate nervous system posteriorization: Grading the function of Wnt signaling. Dev Dyn 244, 507–512. [DOI] [PubMed] [Google Scholar]
- Harmston N, Ing-Simmons E, Tan G, Perry M, Merkenschlager M, Lenhard B, 2017. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat Commun 8, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Simeone A, Alvarado-Mallart RM, 1999. Fgf8 and Gbx2 induction concomitant with Otx2 repression is correlated with midbrain-hindbrain fate of caudal prosencephalon. Development 126, 3191–3203. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S, 1997. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389, 966–970. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kudoh T, Dedekian M, Kim CH, Chitnis AB, 2002. A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development 129, 2317–2327. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Katoh M, 2005. Identification and characterization of rat Wnt1 and Wnt10b genes in silico. Int J Oncol 26, 841–845. [PubMed] [Google Scholar]
- Kemp C, Willems E, Abdo S, Lambiv L, Leyns L, 2005. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev Dyn 233, 1064–1075. [DOI] [PubMed] [Google Scholar]
- Keupp K, Beleggia F, Kayserili H, Barnes AM, Steiner M, Semler O, Fischer B, Yigit G, Janda CY, Becker J, Breer S, Altunoglu U, Grunhagen J, Krawitz P, Hecht J, Schinke T, Makareeva E, Lausch E, Cankaya T, Caparros-Martin JA, Lapunzina P, Temtamy S, Aglan M, Zabel B, Eysel P, Koerber F, Leikin S, Garcia KC, Netzer C, Schonau E, Ruiz-Perez VL, Mundlos S, Amling M, Kornak U, Marini J, Wollnik B, 2013. Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet 92, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G, 2003. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev 17, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, Lu JT, Pekkinen M, Wessman M, Heino TJ, Nieminen-Pihala V, Aronen M, Laine T, Kroger H, Cole WG, Lehesjoki AE, Nevarez L, Krakow D, Curry CJ, Cohn DH, Gibbs RA, Lee BH, Makitie O, 2013. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med 368, 1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekven AC, Buckles GR, Kostakis N, Moon RT, 2003. Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary. Dev Biol 254, 172–187. [DOI] [PubMed] [Google Scholar]
- Makitie RE, Niinimaki T, Nieminen MT, Schalin-Jantti C, Niinimaki J, Makitie O, 2017. Impaired WNT signaling and the spine-Heterozygous WNT1 mutation causes severe age-related spinal pathology. Bone 101, 3–9. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A, 1990. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62, 1073–1085. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA, 1992. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69, 581–595. [DOI] [PubMed] [Google Scholar]
- Millet S, Campbell K, Epstein DJ, Losos K, Harris E, Joyner AL, 1999. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 401, 161–164. [DOI] [PubMed] [Google Scholar]
- Narayanan A, Lekven AC, 2012. Biphasic wnt8a expression is achieved through interactions of multiple regulatory inputs. Dev Dyn 241, 1062–1075. [DOI] [PubMed] [Google Scholar]
- Nusse R, 2001. An ancient cluster of Wnt paralogues. Trends Genet 17, 443. [DOI] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA, 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott SM, Tran TT, Leistritz DF, Pepin MG, Mendelsohn NJ, Temme RT, Fernandez BA, Elsayed SM, Elsobky E, Verma I, Nair S, Turner EH, Smith JD, Jarvik GP, Byers PH, 2013. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet 92, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y, 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurrat Ul A, Seemab U, Nawaz S, Rashid S, 2011. Integrative analyses of conserved WNT clusters and their co-operative behaviour in human breast cancer. Bioinformation 7, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, Brand M, 2004. Divide et Impera--the midbrain-hindbrain boundary and its organizer. Trends Neurosci 27, 727–734. [DOI] [PubMed] [Google Scholar]
- Ramel MC, Buckles GR, Lekven AC, 2004. Conservation of structure and functional divergence of duplicated Wnt8s in pufferfish. Dev Dyn 231, 441–448. [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Hornblad A, Spitz F, 2016. Gene regulation during development in the light of topologically associating domains. Wiley Interdiscip Rev Dev Biol 5, 169–185. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Lun K, Amores A, Yan YL, Postlethwait JH, Brand M, 2003. Cloning, expression and relationship of zebrafish gbx1 and gbx2 genes to Fgf signaling. Mech Dev 120, 919–936. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Echelard Y, Danielian PS, Gellner K, Brenner S, McMahon AP, 1998. Identification of an evolutionarily conserved 110 base-pair cis-acting regulatory sequence that governs Wnt-1 expression in the murine neural plate. Development 125, 2735–2746. [DOI] [PubMed] [Google Scholar]
- Sylvester JB, Rich CA, Loh YH, van Staaden MJ, Fraser GJ, Streelman JT, 2010. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci U S A 107, 9718–9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallafuss A, Wilm TP, Crozatier M, Pfeffer P, Wassef M, Bally-Cuif L, 2001. The zebrafish buttonhead-like factor Bts1 is an early regulator of pax2.1 expression during mid-hindbrain development. Development 128, 4021–4034. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR, 1990. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346, 847–850. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K, 2006. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Lewandoski M, Campbell K, Joyner AL, Rubenstein JL, Martinez S, Martin GR, 1997. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 124, 2923–2934. [DOI] [PubMed] [Google Scholar]
- Westerfield M, 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed. University of Oregon Press, Eugene. [Google Scholar]
- Wilkinson DG, Bailes JA, McMahon AP, 1987. Expression of the proto-oncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell 50, 79–88. [DOI] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L, 2001. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci 2, 99–108. [DOI] [PubMed] [Google Scholar]
- Yu M, Ren B, 2017. The Three-Dimensional Organization of Mammalian Genomes. Annu Rev Cell Dev Biol 33, 265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas M, Blaess S, Joyner AL, 2005. Classical embryological studies and modern genetic analysis of midbrain and cerebellum development. Curr Top Dev Biol 69, 101–138. [DOI] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL, 2004. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron 43, 345–357. [DOI] [PubMed] [Google Scholar]
- Zhang B, Tran U, Wessely O, 2011. Expression of Wnt signaling components during Xenopus pronephros development. PLoS One 6, e26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.