Abstract
BACKGROUND: Low-grade serous ovarian cancer (LGSOC) is a rare subtype of epithelial ovarian carcinoma. Limited data regarding the molecular-genetic background exist beyond mutations in the RAS signaling pathway. There is a growing need to better characterize these tumors due to chemoresistance and limited therapeutic options in advanced or recurrent disease. METHODS: We performed genome-wide copy number aberration (CNA) profiles and mutation hotspot screening (KRAS, BRAF, NRAS, ERBB2, PIK3CA, TP53) in 38 LGSOC tumor samples. RESULTS: We detected mutations in the RAS-signaling pathway in 36.8% of cases, including seven KRAS, four BRAF, and three NRAS mutations. We identified two mutations in PIK3CA and one mutation in MAP3K1, EGFR, and TP53. CNAs were detected in 86.5% of cases. None of the focal aberrations was correlated with specific clinical characteristics. The most frequently detected CNA was loss of 1p36.33 in 54.1% of cases, with a trend towards lower progression-free survival and overall survival in patients with 1p36.33 loss. CONCLUSIONS: Activating RAS mutations were dominant in our series, with supplementary detection of two PIK3CA mutations which may lead to therapeutic options. Furthermore, we detected 1p36.33 deletions in half of the cases, indicating a role in tumorigenesis, and these deletions may serve as a prognostic marker.
Background
Low-grade serous ovarian cancer (LGSOC) is a rare disease representing only 5%-8% of all ovarian cancers and 6%-10% of all serous ovarian cancers [1], [2], [3]. They can present de novo or as a recurrence from a serous borderline tumor (SBT). LGSOC presents typically in a younger patient group than high-grade serous ovarian cancer (HGSOC) with a median age at diagnosis of 43-55 years and 63 years, respectively [3], [4], [5]. Low grade tumors are more indolent, resulting in a longer overall survival (OS) compared with to HGSOC (81.8-126.2 months vs. 53.8-57 months), although low-grade carcinomas are more resistant to chemotherapy [1], [2], [3], [4], [6]. They have a <5% response rate to first-line chemotherapy compared to the 80% response rate of their high-grade counterpart [1], [7]. The progression-free survival (PFS) of LGSOC is similar compared to that of HGSOC, with a median PFS of 19.5 months [4], although higher PFS rates (25-36 months) have been described for SBT-associated cases [8], [9].
Shih and Kurman introduced a model of two different pathways leading to HGSOC and LGSOC [10]. They describe both serous tumors not only as histologically differentially graded but also as two distinct clinical, molecular, and epidemiological entities. Histological characteristics suggest that low-grade tumors often develop from low–malignant potential tumors in a pathogenic continuum with a 60% presence of SBT in LGSOC, while high-grade tumors arise de novo from the surface epithelium and only have a 2% incidence of concomitant SBT. Hence, the presence of an SBT is a risk factor for developing LGSOC [4], [8], [10], [11]. While nearly all HGSOCs are characterized by genetic loss of TP53 [12], LGSOCs are typically wild-type TP53 and presumably arise in a stepwise fashion from serous cystadenoma, adenofibroma, or serous borderline tumors [13]. LGSOCs harbors a high rate (40%) of activating mutations of the MAPK pathway (Table 1).
Table 1.
Overview of Mutational Analyses in LGSOC Conducted with Either Immunohistochemistry, Polymerase Chain Reaction, Hotspot Genotyping, or Whole Exome/Genome Sequencing
| Author | Journal Publication Year | No. LGSOC |
No. KRAS (%) |
No. BRAF (%) |
No. NRAS (%) |
No. Other (%) |
|---|---|---|---|---|---|---|
| Haas et al. [36] |
Virchows Arch 1999 |
6 | 2 (33%) codon 12 |
|||
| Singer et al. [14] |
J Natl Cancer Inst 2003 |
22 | 8 (36%) codon 12-13 |
7 (32%) codon 599 |
||
| Wong et al. [15] |
Am J Pathol 2010 |
43 | 8 (19%) codon 12-13 |
1 (2%) codon 600 |
||
| Vereczkey et al. [37] | Pathol Oncol Res 2011 | 17 | 4/17 (23.5%) codon 12-13 | 0 (0%) codon 600 |
0 TP53 (0%) | |
| Schlosshauer et al. [38] | Int J Gynecol Pathol 2011 | 4 | 0 (0%) codon 600 |
|||
| Jones et al. [30] |
J Pathol 2012 |
15 | 4 (26.7%) codon 12 |
3 (20%) codon 600 |
1 PIK3CA 345 N/K (6.7%) |
|
| Sundov et al. [39] |
Diagn Pathol 2013 |
11 | 6 (54.5%) codon 12-13 | 0 (0%) codon 600 |
||
| Grisham et al. [21] |
Cancer 2013 |
19 | 3 (15.8%) codon 12-13 | 1 (5.3%) codon 600 | ||
| Farley et al. [17] |
Lancet Oncol 2013 |
34 | 4 (41%) codon 12-13 |
2 (6%) codon 599 |
||
| Emmanuel et al. [9] |
Clin Cancer Res 2014 |
20 | 7 (35%) codon 12 |
2 (10%) codon 600 | 3 (15%) 2*Q61R, Q61K | |
| Gershenson et al. [22] |
Br J Cancer 2015 |
79 | 18 (22.8%) codon 12 | 3 (3.8%) codon 600 | ||
| Hunter et al. [7] |
Oncotarget 2015 |
19 | 4 (21%) codon 12 |
3 (16%) codon 600 |
1 (5.3%) Q61R | 0 HRAS (0%) 0 TP 53 (0%) 15% EIF1AX 11% USP9X |
| Sadlecki et al [40], [41] |
Tumor Biology 2017 |
13, 14 | 2 (15.4%) codon 12 |
0 (0%) codon 599 |
||
| Etemadmoghadam et al. [26] | Cancer Res 2017 | 23 | 5 (22%) codon 12 |
3 (13%) codon 600 |
5 (22%) 3*Q61R 2*Q61K |
2 NF1 (9%) 3 EIF1AX (13%) 3 USP9X (13%) |
| Xing et al. [27] | Human Pathol 2017 | 56 | 2 (3.6%) Q61R | |||
| McIntyre et al. [28] | Histopathology 2017 | 26 | 9 (34.6%) Codon 12 and 61 |
2 (7.7%) Codon 600 |
1 (3.8%) Q61R |
1 MAP2K1 (3.8%) 2 FGFR2 (7.7%) 1 ESR1 (3.8%) |
| TOTAL (%) | 421 | 84/347 (24.2%) | 27/346 (7.8%) | 12/144 (8.3%) | ||
| RANGE | 4-79 | 15.8-54.5% | 0-32% | 3.6-22% |
In 2003, Singer et al. reported the first analysis of KRAS and BRAF mutations in LGSOC and reported a frequency of 36% and 32% positive tumors, respectively [14]. Since then, many studies have confirmed the presence of these mutations in LGSOC, although observed frequencies seem to vary considerably (0%-32% for BRAF and 15.4%-54.5% for KRAS mutations) (Table 1). BRAF mutations are more frequent in SBT and in early-stage LGSOC tumors. As such, BRAF mutated LGSOC tumors are often characterized by a better prognosis [3], [5], [14], [15], [16].
In 2014, Emmanuel et al. broadened the spectrum of mutations in the MAPK pathway by identifying NRAS mutations at a frequency of 15% in 20 LGSOCs with adjacent SBT [9]. Further studies confirmed NRAS as a possible oncogenic driver in LGSOC (Table 1). Hunter et al. also conducted whole exome sequencing in 19 LGSOC cases and identified recurrent mutations in KRAS, BRAF, and NRAS, in addition to somatic mutations in USP9X and EIF1AX [7]. The latter two genes have both been linked to regulation of mTOR, suggesting that mTOR inhibitors may be useful in combination with MEK or RAF inhibitors in LGSOC. In contrast to the latter study, few other genome-wide studies have been performed. As a result, relatively little is known about chromosomal instability in LGSOC. The only more or less consistent finding is that loss of chromosome 1p has been observed in a number of studies (Table 2).
Table 2.
CNA analyses in LGSOC.
| Author | Journal Publication year |
N° LGSOC |
CNA loss/gain | Candidate genes |
|---|---|---|---|---|
| Kuo et al. [33] | Cancer Res 2009 |
12 | Loss: Chr 1p36, Chr 9p21.3 | CHD5, MiR-34a CDKN2A/B |
| Birch et al. [13] | Plos One 2011 |
11 | Loss: chr 1p | - |
| Emmanuel et al. [9] | Clin Cancer Res 2014 |
13 | CNI identical between paired SBT and LGSOC samples Low CNI compared to HGSOC |
- |
| Hunter et al. [7] | Oncotarget 2015 |
13 | Loss: 1p, 9p, 22q Gain: 7, 8 |
CDKN2A/B |
| McIntyre et al. [28] | Histopathol 2017 |
26 | Only report on CNI Low CNI compared to HGSOC |
- |
| Total | 70 |
The most frequent CNAs are indicated. Chr, chromosome; CNI, copy number index.
Since LGSOC is often resistant to chemotherapy, the development of novel targeted therapies has the potential to ameliorate the prognosis of LGSOC patients. The MEK inhibitor selumetinib has shown promising results in LGSOC [17]. However, to select patients for such targeted therapies, it is imperative to upfront define a subset of biomarker-positive patients that will benefit from these therapies.
The purpose of this study is to further analyze the genomic profile of LGSOC using mutational analysis and, to our knowledge, the largest copy number aberration (CNA) analysis of LGSOC.
Methods
Patients and Tumors
We collected fresh-frozen or paraffin-embedded tumor samples and clinical data of 38 patients with LGSOC treated at the University Hospital of Leuven, Belgium. Written consent for the use of tumor tissue and data collection was obtained of each patient, and the study was approved by the local ethics committee (study number s55308). Eligible patients were women diagnosed with LGSOC at our institution between January 1984 and August 2015 of which we had tumor material, either fresh-frozen or paraffin-embedded, and sufficient clinical data. Tumor tissue was collected at the time of surgery being either primary debulking or interval debulking surgery. Clinical data included age at diagnosis, International Federation of Gynecology and Obstetrics (FIGO) stage, laterality, lymph node involvement, ascites, type of surgery, residual tumor after surgery, response to first-line chemotherapy, presence of concomitant borderline ovarian tumor, and recurrence. Staging and grading were performed according to FIGO 2014 classification. All tumor samples were revised by a pathologist, expert in the field of gynecological tumors (P.M.). Response Evaluation Criteria in Solid Tumors (version 1.1) [18] and Gynecologic Cancer InterGroup CA125 response criteria [19] were used to determine response to chemotherapy and to define progressive disease. PFS was calculated as the period between diagnosis and recurrence, progression, or death from any cause, whereas OS was defined as the period between diagnosis and death from any cause and censored at the last date of follow-up if the patient was still alive.
DNA Isolation
Fresh-frozen tumor sections were obtained by cryosectioning biopsies at 10 μm. Three to five 20-μm–thick sections were prepared from the formalin-fixed, paraffin-embedded tissue blocks. A 5-μm hematoxylin and eosin–stained section was used to determine regions with high tumor content. Based on the hematoxylin and eosin staining, only regions containing ≥60% of tumor cells were retained through macrodissection. From these regions, DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Düsseldorf, Germany) for the fresh-frozen samples, and a phenol-chloroform method was used on the formalin-fixed, paraffin-embedded slides.
Hotspot Genotyping
We selected a panel by a query of the Catalogue of Somatic Mutations in Cancer database for the most frequent mutations in ovarian cancer as reported previously and applied this in genotyping of high-grade serous ovarian cancer [20]. For KRAS, BRAF, NRAS, PIK3CA, and PTEN, this resulted in a coverage of greater than 97%, 94%, 97%, 79%, and 7%, respectively. We extended our panel with following genes: AKT2, CDK4, CTNNB1, EGFR, ERBB2, FBXW7, MAP3K1, MAP2K4 and 27, PIK3R1, PDGRA, SMAD4, and TP53.
DNA was aliquoted into 384-well plates and genotyped at the Vesalius Research Center (Leuven, Belgium) using the MassARRAY Compact Analyzer (Sequenom Inc., San Diego, CA). Automated genotyping calls were generated using the MassARRAY RTTM software and were manually reviewed.
Copy Number Variation Analysis
We performed copy number analysis through whole-genome low-coverage (shallow) sequencing on 38 samples. DNA libraries were prepared using KAPA Library Preparation Kits (KK8201, Kapabiosystems, Wilmington, MA) prior to sequencing. Samples were sequenced at low coverage (0.5×) on an Illumina HiSeq 2000 platform. Raw sequencing data were mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA v0.7.10). Using QDNAseq v1.4.2, read counts were generated for bins of size 100 kbp; bins in blacklisted regions were removed, and bin counts were corrected for GC content and mappability applying Loess regression using QDNAseq. ASCAT (v2.0.7) was used to estimate copy number profiles from these bins. Subsequently, GISTIC (v2.0.22) was applied to identify recurrent CNAs in the cohort.
Statistical Analysis
Descriptive statistics were used for patient characteristics. Comparison between groups on clinical parameters was performed by Mann-Whitney U test for continuous variables or Fisher's exact test or Chi-square test for categorical variables. Kaplan-Meier method was used to construct survival curves, and log-rank test was used to calculate the difference in survival curves between groups. Statistical analyses were performed using SAS Software (version 9.4, SAS System for Windows). All tests are two-sided, and we considered a P value of .05 as statistically significant.
Results
Clinical Outcome
We selected 38 patients for which clinical follow-up data and either paraffin-embedded or fresh-frozen tissue was available for mutation analysis. Patient characteristics are detailed in Table 3. Median age at diagnosis was 53.5 years (range 25-76 years). The majority of patients was diagnosed with a FIGO stage III-IV (92.1%), and most of them underwent primary debulking surgery (65.8%) with an 86.8% R0 (no macroscopic residual tumor) resection rate. All patients, except 2 patients with stage I disease and 2 patients treated with an aromatase inhibitor, received platinum-based chemotherapy in either neoadjuvant or adjuvant setting. There were no significant differences in clinical characteristics between wild-type and RAS-mutated patients (Table 3).
Table 3.
Patient Characteristics According to Ras Mutation or Wild-Type
| Ras Mutant N = 14 |
Non-Ras Mutant N = 24 |
P Value | |
|---|---|---|---|
| Median age (years) | 58.5 | 39.5 | .220 |
| Range | (28-68) | (25-76) | |
| Mean age | 53.7 | 47.1 | |
| FIGO stage | |||
| I | 3 (21.4%) | 1 (4.2%) | .105 |
| II | - | - | |
| III | 7 (50%) | 21 (87.5%) | |
| IV | 4 (28.6%) | 2 (8.3%) | |
| Surgery | |||
| Primary debulking | 7 (50%) | 18 (75%) | .163 |
| Interval debulking | 7 (50%) | 6 (25%) | |
| Residual tumor | |||
| R0 | 11 (78.6%) | 22 (91.7%) | .337 |
| R > 1 | 3 (21.4%) | 2 (8.3%) | |
| R0-1 | - | - | |
| Association of borderline component | 6 (42.9%) | 12 (50%) | .745 |
| Initial systemic treatment | |||
| Platinum-based chemo | 13 (92.9%) | 24 (100%) | .368 |
| Hormonal treatment | 1 (7.1%) | - | - |
| Bilateral | 9 (64.3%) | 19 (79.2%) | .217 |
| Unilateral | 5 (35.7%) | 3 (12.5%) | |
| Unknown | - | 2 (8.3%) | |
| Lymph node involvement | |||
| Yes | 6 (42.9%) | 9 (37.5%) | |
| No | 8 (57.1%) | 15 (62.5%) | |
| Ascites | |||
| Yes | 6 (42.9%) | 4 (16.7%) | 1.000 |
| No | 8 (57.1%) | 19 (79.2%) | |
| Unknown | - | 1 (4.2%) |
In 18 patients, a borderline component was also present at the time of diagnosis. The presence of a borderline component was not correlated with a younger age at the time of diagnosis (53.5 vs. 55 years). Patients with a borderline component had a longer PFS and OS rate compared to the pure LGSOC patients: 36 vs. 20.4 months and 130.8 vs. 86.4 months, respectively, but this was not statistically significant (P = .4). SBT-associated tumors were not more likely to be diagnosed in early stage, but the majority of the entire series presented in an advanced stage. Overall, 30/38 patients relapsed (78.9%) with a median PFS of 32 months, and 17/38 succumbed to the disease (45.9%). Median follow-up was 89 months (range 13-372) with two patients lost to follow-up.
Hotspot Genotyping
KRAS and BRAF mutations were mutually exclusive in our series. Overall, mutations in the RAS signaling pathway were identified in 14/38 samples (36.8%). We found seven KRAS mutations (18.4%): six samples had a KRAS G12 V (c.35G > T) and one sample a KRAS G12D (c.35G > A) mutation. One KRAS sample also harbored a MAP3K1 mutation. We found four BRAF mutations (10.5%): three samples had a BRAF V600E (c.1799 T > A) and one sample a BRAF G469A (c.1406G > C) mutation. Activating NRAS mutations were identified in three samples (7.9%) in our series; all were Q61R (c.182 A > G) mutations. These are all known oncogenic mutations. All KRAS, BRAF, and NRAS mutations were mutually exclusive. There were no significant differences regarding age, FIGO stage, and the concomitant presence of borderline between KRAS, BRAF, and NRAS mutated patients.
None of the clinical characteristics—age, FIGO stage, laterality, lymph node involvement, and ascites—correlated significantly with the presence of RAS mutations. Association with borderline component was also equal between the RAS-mutated and the non–RAS-mutated group (Table 3). The median PFS of the RAS-mutated group was 33.6 months (95% CI, 13.2-44.4) compared to 31.2 months (95% CI, 16.8-52.8) in the non–RAS-mutated group (P = .4). Median OS was 130.8 months (95% CI, 51.6-195.6) in the RAS-mutated group versus 170.4 months (95% CI, 54-372) in the non–RAS- mutated group (P = .6). OS of the entire cohort was 137 months.
Furthermore, we found two mutations in the PIK3CA gene: one R93W (c.277C > T) and one H1047RL (c.3140A > GT). One patient also harbored a BRAF mutation besides the PIK3CA mutation, whereas another patient harbored an E746A EGFR mutation (c.2235_2249del15). We found no mutations in ERBB2 in our cohort of 38 LGSOC and two TP53 mutations (c.524G > A and c.817C > T) in one sample.
Copy Number Variation
We conducted a genome-wide CNA analysis on 40 samples: 37 primary LGSOC tumors and 3 paired metastatic samples. CNAs were detected in 86.5% of all the primary tumor samples, whereas the remaining samples (5/37) where completely copy number-stable. Particularly, the chromosome instability index (CIN), which is the fraction of the genome affected by CNAs, was low with a median of 14.4% (IQ range: 6.1%-31.7%). This is substantially lower than in HGSOC. We compared the current dataset to a cohort of 160 stage III/IV HGSOC samples prospectively collected within the OVCAD consortium (FP6 EU-project, www.ovcad.eu) at time of diagnosis (Figure 1A). The median CIN was 53.9% in this group (vs. 14.4%), with an absolute minimum of 8.3% (vs. 0%). Importantly, CIN did also not significantly differ between BRAF, KRAS, and NRAS mutated samples, or between RAS-mutated and RAS wild-type samples (Figure 1B). The CIN was comparable between patients who succumbed to the disease within 60 months of diagnosis and patients who were still alive at 84 months (Figure 1C).
Figure 1.
(A) Fraction of the genome altered in 38 LGSOC versus 160 high-grade serous ovarian cancer OVCAD samples. (B) Percentage of the genome gained (blue), lost (red), or neutral (green) in the Ras-mutated versus the Ras wild-type samples. (C) Percentage of the genome gained (blue), lost (red), or neutral (green) in patients who succumbed to the disease within 60 months of diagnosis (DOD = death of disease) or were still alive after 60 months.
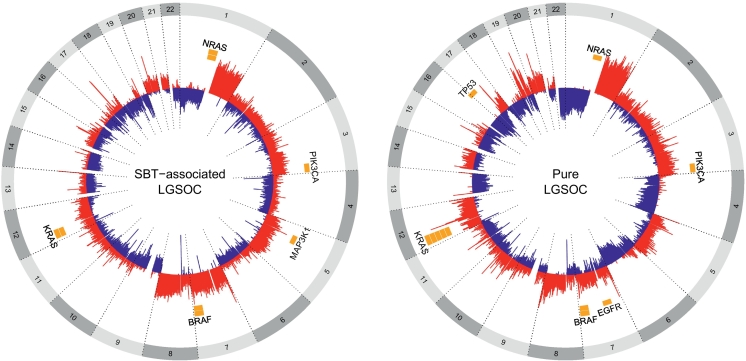
The most frequently reported whole chromosome aberrations were loss of 1p (33%), 6q (24%), 9p (21%), 16p/q (21%), 17p (24%), 18p/q (21%), and 22q (40%) and gain in 1q (40%), 7p/q (26%), and 8q (29%) (Figure 2). Loss of 9p (33%) and gain of 8q (44%) and chromosome 7 (39%) were slightly enriched in the samples with associated borderline component (SBT-LGSOC) compared to pure LGSOC (16%, 21%, and 21%, respectively), but these differences were not statistically significant. None of the other whole arm or focal aberrations correlated with the presence of a borderline component. Nine samples harbored a 17p deletion and 10 samples and 8q gain. Seven in each group (77.8% and 70%, respectively) succumbed to the disease (P = .04 and P = .19, respectively), of which three samples had both CNAs, suggesting a correlation with a poor outcome. Loss of 15q was predominantly found in patients <45 years (43.8 vs. 9.5%, P = .04).
Figure 2.
Circos plot showing oncogene hotspot mutations and the large-scale CNAs in the 38 tumor samples.
We further also analyzed recurrent focal deletions and gains. The most frequent focal alteration was loss of 1p36.33 in 20/37 (54.1%) samples. We could not detect a specific enrichment of 1p36.33 loss in SBT-LGSOC cases, or any significant correlation of loss of 1p36.33 and clinical characteristics, such as age, FIGO stage, bilaterality, lymph node involvement, and the presence of ascites (Table 4). We further looked at the median PFS and OS of patients with LGSOC tumors characterized by 1p36.33 loss. The median PFS of patients with the 1p36.33 alteration was 26.4 months (95% CI, 13.2-50.4) compared to 44.4 months (95% CI, 16.8-67.2) in the group of patients without 1p36.33 loss (P = .40). Similarly, median OS in the 1p36.33 group was 130.8 months (95% CI, 51.6-195.6) compared to 172.8 months (95% CI, 52.8-372) in the group without 1p36.33 loss (P = .52). One sample with loss of 9p harbored a homozygous deletion of the 9p21.3 region which contains CDKN2A/2B.
Table 4.
Patient Characteristics According to Loss of 1p36.33
| 1p36.33 Loss N = 20 |
No 1p36.33 Loss N = 17 |
P Value | |
|---|---|---|---|
| Median age (years) | 47.5 | 55 | .636 |
| Range | (25-76) | (26-76) | |
| Mean age | 48.3 | 51.8 | |
| FIGO stage | |||
| I | 1 (5.0%) | 2 (11.8%) | .630 |
| II | - | - | |
| III | 15 (75%) | 13 (76.4%) | |
| IV | 4 (20%) | 2 (11.8%) | |
| Surgery | |||
| Primary debulking | 14 (70%) | 10 (58.8%) | .512 |
| Interval debulking | 6 (30%) | 7 (41.2%) | |
| Residual tumor | |||
| R0 | 17 (85%) | 15 (88.2%) | 1.000 |
| R > 1 | 3 (15%) | 2 (11.8%) | |
| R0-1 | - | - | |
| Association of borderline component | 9 (45%) | 9 (52.9%) | .745 |
| Initial systemic treatment | |||
| Platinum-based chemo | 20 (100%) | 13 (76.4%) | .036 |
| Hormonal treatment | 0 (0.0%) | 2 (11.8%) | |
| No adjuvant chemotherapy | 0 (0.0%) | 2 (11.8%) | |
| Bilateral | 16 (80%) | 12 (70.6%) | .434 |
| Unilateral | 3 (15%) | 5 (29.4%) | |
| Unknown | 1 (5.0%) | ||
| Lymph node involvement | |||
| Yes | 11 (55%) | 4 (23.5%) | .092 |
| No | 9 (45%) | 13 (76.5%) | |
| Ascites | |||
| Yes | 5 (26.3%) | 5 (29.4%) | 1.000 |
| No | 14 (73.7%) | 12 (70.6%) | |
| Unknown | 1 (5.0%) |
Paired Samples
In three samples, we performed genotyping and copy number analyses on the primary tumor as well as a metastatic site at time of diagnosis (OV0105, OV0185, and OV0771). OV0771 shared the same NRAS mutation in both primary and metastatic sample. For the other two samples, genotyping results were discordant: OV0105 showed a BRAF and PIK3CA mutation in the metastatic sample but not in the primary tumor. OV0185 harbored a PIK3CA mutation in the primary but not in the metastatic sample.
The fraction of copy number alterations was similar in the OV0105 and OV0771 paired samples, while the primary sample of OV0185 showed a higher accumulation of CNAs with respect to the paired metastasis.
Discussion
LGSOCs are genetically stable tumors, primarily characterized by activating mutations in the MAPK pathway and a low number of copy number alterations. We identified a 36.8% mutation rate in the MAPK pathway: 18.4% KRAS, 10.5% BRAF, and 7.9% NRAS mutations, consistent with earlier reports (Table 1). The lower BRAF mutation frequency is in concordance with the finding of Wong et al. who found only one BRAF mutation in 33 advanced LGSOC samples [15]. Our series consists mainly of stage III/IV patients; hence, BRAF mutations are not associated with early stage and a better outcome in advanced LGSOC. However, BRAF mutations are more frequently detected in SBTs, and these BRAF-mutated SBTs are less likely to recur [7], [21].
Wong et al. reported an OS of 77.9 months in 8 LGSOC with either a KRAS or BRAF mutation (7 KRAS, 1 BRAF) versus 47.3 months for 25 LGSOC patients with wild-type KRAS and BRAF (P = .28) [15]. In 79 cases of LGSOC, Gershenson et al. reported a median OS of 106.8 months (95% CI, 50.6-162.9) in KRAS/BRAF mutated patients (21/79) compared to 66.8 months (95% CI, 43.6-90.0) in KRAS/BRAF wild-type patients (P = .018) [22], suggesting a prognostic effect of BRAF/KRAS mutation. In our series, median OS in the Ras-mutated group was 130.8 months compared to 170.4 months in the non–Ras-mutated group (P = .61). Hence, the prognostic effect of BRAF and KRAS mutation cannot be confirmed. In the cohort of Gershenson, however, 69% of KRAS/BRAF wild-type patients and 52.4% of KRAS/BRAF mutated patients had gross residual disease at the time of cytoreductive surgery compared to 13.2% in our series (3 patients with a Ras mutation and 2 wild-type patients). Since residual tumor is the most widely accepted prognostic factor in ovarian cancer [23] and this holds probably even more for LGSOC than for other more rapidly growing ovarian cancers, this could significantly impact OS data. The percentage of patients with stage III/IV disease was comparable between both series (93.8% vs. 92.1%). Hence, our higher R0 rate might explain the longer OS in our cohort. Furthermore, it is not clear how many of the patients in the series of Gershenson were SBT-associated since this might also impact PFS and OS [9]. Additionally, other reports suggest, as we do, a poorer outcome of malignancies with KRAS/BRAF mutations compared with wild-type KRAS/BRAF [24], [25]. We combined all Ras-pathway mutations because the PFS and OS of the patients with an NRAS mutation were comparable to the PFS and OS of patients with KRAS or BRAF mutations.
More recently, NRAS mutations were described in LGSOC. Five studies identified 12 NRAS mutations (either at Q61R or Q61K) in a total of 118 cases, resulting in an average frequency of 9.3% [7], [9], [26], [27], [28]. We found three NRAS mutations in our series. The role of NRAS mutation in the development of LGSOC merits further investigation given new possible targeted therapies against NRAS and its downstream effectors [29].
Furthermore, we observed two mutations in PIK3CA. One patient also harbored a BRAF mutation besides the PIK3CA mutation. Jones et al. previously described a PIK3CA N345K (c1035T > A) mutation in one case out of 15 with pure LGSOC [30]. One patient harbored an EGFR mutation. EGFR mutations are a rarity in SBTs and have not been previously reported in LGSOC. Showeil et al. found a higher nuclear and cytoplasmic staining of EGFR in 61 SBTs and 10 LGSOC tumors compared to benign ovarian tumors or HGSOC tumors but could not detect any mutation [31]. Activating EGFR mutation can upregulate the PI3K pathway. Recurrent mutations in USP9X and EIF1AX have recently been published [7], [26]. Both are linked to regulation of mTOR. All these findings may suggest a role for mTOR inhibitors as an additive to treatment with MEK and RAF inhibitors.
We observed no mutations in ERBB2 in our cohort of 38 LGSOC. We tested ERBB2 given the recent data of ERBB2 mutations in serous borderline tumors [32]. Our study suggests that these mutations are restricted to noninvasive serous pathology.
Unexpectedly, we detected one TP53 mutation. Revision of both the primary surgery specimen and the recurrence specimen 2 years after diagnosis by an expert pathologist in the field of gynecological tumors (P.M.) confirmed the diagnosis of LGSOC. Both the estrogen receptor and progesterone receptor were highly positive. Birch et al. previously described a TP53 mutation in a serous tumor of low malignant potential [13].
The presence of a borderline component is not indicative of Ras-pathway–activated tumors [9]. We did not find a significant difference in association of borderline component between the RAS-mutated group and the non–Ras-mutated group (P = .74). SBT-LGSOC cases did show a longer PFS and OS, as demonstrated by Emmanuel et al., compared to the pure LGSOC patients: 36 vs. 20.4 months and 130.8 vs. 86.4 months, respectively. This difference was not statistically significant (P = .44), but this might be due to the small series. Emmanuel et al. also reported a younger age at time of diagnosis for epithelial tumors associated with a borderline tumor compared to invasive epithelial carcinomas, but 80% of the latter consisted of HGSOC [9]. We did not find a difference in age at onset.
CNAs were detected in 86.5% of all the samples. The majority of cases showed low-level copy number alterations in contradiction to HGSOC that is associated with a very high level of copy number alterations. This is in concordance with previous reports [7], [9], [13], [28], [33]. The fraction of the genome altered is similar for Ras mutated and wild-type tumors (Supplementary data S1). Associating clinical features with somatic mutations or CNAs was difficult because most patients presented with bilateral and/or advanced disease (Table 2).
The most frequently reported whole chromosome aberrations were loss of 1p (33%), 6q (24%), 9p (21%), 16p/q (21%), 17p (24%), 18p/q (21%), and 22q (40%) and gain in 1q (40%), 7p/q (26%), and 8q (29%), and are in concordance with previous studies [7], [9], [13]. The most significant CNA in the series of Hunter et al. [7] was loss of 9p, which was found in 53% of LGSOC cases and in only 2% of SBT cases, indicating a possible role in transition from SBT to an invasive carcinoma. We observed a loss of 9p in 33% of cases of borderline-associated LGSOC samples compared to 16% in pure LGSOC cases (P = .21). 9p contains a candidate gene at locus 9p21.3, CDKN2A/2B. We found one deletion at this locus. Hunter et al. found loss of 1p, 9q, 18q, and X to be common for borderline tumors and LGSOC samples and gain of chromosomes 7, 8, and 22 to be enriched in SBTs but not in LGSOC. In our series, loss of 9p (33%) and gain of 8q (44%) and chromosome 7 (39%) were slightly enriched in the samples with associated borderline component (SBT-LGSOC) vs. 16%, 21%, and 21% in pure LGSOC samples (P = .21, P = .12, P = .26). Overall, we detected similar CNAs. Discrepancy between findings may be explained by the fact that the samples in the series of Hunter et al. were not paired SBT-LGSOC samples and due to the low number of cases analyzed in both series. We analyzed the invasive component of borderline-associated cases. Thus, findings are correlative and caution should be taken to identify these CNAs as markers of transition. We could not withhold an enrichment of other CNAs in patients with an associated borderline component. This suggests that these CNAs can be present in both borderline and LGSOC but are not specific for the transition of borderline to the invasive component given their equal presence in LGSOC without borderline component. This is consistent with Emmanuel et al. who found similar CNAs in LGSOC samples and their associated SBT component [9].
Loss of 1p36.33 was the most frequent focal alteration (54.1%) with no association with a specific clinical characteristic (Table 3). Kuo et al. previously reported loss of 1p36 in LGSOC [33]. They suggested CHD5 and miR-34a as possible targets. Mutation analysis however did not reveal any mutations. However, CHD5 maps to the 1p36.31 region and miR-34a to the 1p36.22 region. We further localized the known loss of 1p36 in LGSOC to the 1p36.33 region. This region contains 70 genes, none of which are known cancer consensus genes (Catalogue of Somatic Mutations in Cancer). However, in a broad range of human cancers, 1p36 is a mutational hotspot which suggests that the loss of tumor suppressor activity maps to this genomic region during tumorigenesis [34]. Genes in the 1p36 region may be affected by haploinsufficiency or promotor methylation since mutations detected in this region are rare. Many tumor suppressor genes do not require classic inactivation by a two-hit manner but contribute to tumorigenesis through reduced dosage of their gene products by mechanisms such as copy number or epigenetic changes, transcriptional repression, or aberrant miRNA regulation. Additionally, 14/37 (33%) of our samples also showed a whole arm loss of 1p, further contributing to the loss of function of these TSGs. In total, 22/37 (59.5%) samples harbored a 1p36 loss.
Finally, loss of 1p36.11-21 was associated with tumor dissemination in microsatellite stable tumors of stage II-IV colon tumors [35]. In our series, PFS and OS tended to be lower in patients with the 1p36.33 alteration compared with those without.
Conclusion
Although mutations in the Ras signaling pathway are the most frequently reported oncogenic events in LGSOC, mutation frequency of the Ras/Raf/MEK/MAPK signaling pathway could be underestimated because of incomplete gene sequencing. It is possible that mutations in genes outside of the exome targeted thus far are responsible or that larger-scale CNA, inversions, or translocations may be important. These possibilities should be tested by whole-genome sequencing of a larger cohort. Nevertheless, a large proportion of LGSOC are “wild type” for the Ras/Raf/MEK/MAPK signaling pathway. The drivers of these wild-type tumors remain undefined.
Loss of the 1p36 region is more frequent in LGSOC than mutations in the RAS signaling pathway. Mutations detected in the 1p36 regions are scarce. Hence, identifying a possible gene or pathway for targeted therapy is not that straightforward since multiple tumor suppressor genes might be affected in this region and they, at their turn, can interact reciprocally or with other pathways. Nevertheless, the focus should shift from trying to target one known mutation towards a broader approach, such as miRNA replacement therapy.
The following are the supplementary data related to this article.
Processed sequencing data on the 38 samples: place of biopsy, hotspot genotyping by Sequenom, altered fraction of segments, and GISTIC focal and whole arm analyses from the shallow sequencing experiment.
Authors' Contributions
Conception and design: E. Van Nieuwenhuysen, I. Vergote.
Acquisition of data: E. Van Nieuwenhuysen.
Analysis and interpretation of data: E. Van Nieuwenhuysen, Pieter Busschaert, Annouschka Laenen, D. Lambrechts, I. Vergote.
Pathological review: P. Moerman.
Writing: E. Van Nieuwenhuysen.
Review and/or revision of the manuscript: all authors.
Funding
This work was supported by the “Vriendtjes tegen Kanker” fund (EVO-FOVTK1-02010).
Disclosure of Potential Conflicts of Interest
There are no potential conflicts of interest to disclose by any of the authors.
Availability of Data and Materials
Research data and data not presented in the manuscript are available as supplementary data (S1). They will also be available upon request to the corresponding author.
Ethical Approval
Written consent for the use of tumor tissue and data collection was obtained of each patient, and the study was approved by the local ethics committee of the University Hospitals Leuven (study number s55308). This study was performed in accordance with the Declaration of Helsinki.
References
- 1.Schmeler KM, Gershenson DM. Low-grade serous ovarian cancer: a unique disease. Curr Oncol Rep. 2008;10(6):519–523. doi: 10.1007/s11912-008-0078-8. [DOI] [PubMed] [Google Scholar]
- 2.Plaxe SC. Epidemiology of low-grade serous ovarian cancer. Am J Obstet Gynecol. 2008;198(4):459. doi: 10.1016/j.ajog.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 3.Gourley C, Farley J, Provencher DM, Pignata S, Mileshkin L, Harter P, Maenpaa J, Kim JW, Pujade-Lauraine E, Glasspool RM. Gynecologic Cancer InterGroup (GCIG) consensus review for ovarian and primary peritoneal low-grade serous carcinomas. Int J Gynecol Cancer. 2014;24(9 Suppl 3):S9–13. doi: 10.1097/IGC.0000000000000257. [DOI] [PubMed] [Google Scholar]
- 4.Gershenson DM, Sun CC, Lu KH, Coleman RL, Sood AK, Malpica A, Deavers MT, Silva EG, Bodurka DC. Clinical behavior of stage II-IV low-grade serous carcinoma of the ovary. Obstet Gynecol. 2006;108(2):361–368. doi: 10.1097/01.AOG.0000227787.24587.d1. [DOI] [PubMed] [Google Scholar]
- 5.Oswald AJ, Gourley C. Low-grade epithelial ovarian cancer: a number of distinct clinical entities? Curr Opin Oncol. 2015;27(5):412–419. doi: 10.1097/CCO.0000000000000216. [DOI] [PubMed] [Google Scholar]
- 6.Bodurka DC, Deavers MT, Tian C, Sun CC, Malpica A, Coleman RL, Lu KH, Sood AK, Birrer MJ, Ozols R. Reclassification of serous ovarian carcinoma by a 2-tier system: a Gynecologic Oncology Group Study. Cancer. 2012;118(12):3087–3094. doi: 10.1002/cncr.26618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter SM, Anglesio MS, Ryland Gl, Sharma R, Chiew YE, Rowley SM, Doyle MA, Li J, Gilks CB, Moss P. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget. 2015 Nov 10;6(35):37663–37677. doi: 10.18632/oncotarget.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shvartsman HS, Sun CC, Bodurka DC, Mahajan V, Crispens M, Lu KH, Deavers MT, Malpica A, Silva EG, Gershenson DM. Comparison of the clinical behavior of newly diagnosed stages II-IV low-grade serous carcinoma of the ovary with that of serous ovarian tumors of low malignant potential that recur as low-grade serous carcinoma. Gynecol Oncol. 2007;105(3):625–629. doi: 10.1016/j.ygyno.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Emmanuel C, Chiew YE, George J, Etemadmoghadam D, Anglesio MS, Sharma R, Russell P, Kennedy C, Fereday S, Hung J. Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver. Clin Cancer Res. 2014;20(24):6618–6630. doi: 10.1158/1078-0432.CCR-14-1292. [DOI] [PubMed] [Google Scholar]
- 10.Shih leM, Kurman RJ. Molecular pathogenesis of ovarian borderline tumors: new insights and old challenges. Clin Cancer Res. 2005;11(20):7273–7279. doi: 10.1158/1078-0432.CCR-05-0755. [DOI] [PubMed] [Google Scholar]
- 11.Medeiros F, Muto MG, Lee Y, Elvin JA, Callahan MJ, Feltmate C, Garber JE, Cramer DW, Crum CP. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am J Surg Pathol. 2006;30(2):230–236. doi: 10.1097/01.pas.0000180854.28831.77. [DOI] [PubMed] [Google Scholar]
- 12.Singer G, Stohr R, Cope L, Dehari R, Hartmann A, Cao DF, Wang TL, Kurman RJ, Shih leM. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29(2):218–224. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 13.Birch AH, Arcand SL, Oros KK, Rahimi K, Watters AK, Provencher D, Greenwood CM, Mes-Masson AM, Tonin PN. Chromosome 3 abnormalities investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer G, Oldt R, III, Cohen Y, Wang BG, Sidransky D, Kurman RJ, Shih leM. Mutations in BRAF and KRAS characterize the development of low-grade serous carcinoma. J Natl Cancer Inst. 2003;95(6):484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 15.Wong KK, Tsang YT, Deavers MT, Mok SC, Zu Z, Sun C, Malpica A, Wolf JK, Lu KH, Gershenson DM. BRAF mutation is rare in advanced-stage low-grade ovarian serous carcinomas. Am J Pathol. 2010;177(4):1611–1617. doi: 10.2353/ajpath.2010.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer G, Kurman RJ, Chang HW, Cho SK, Shih Ie M. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol. 2002;160(4):1223–1228. doi: 10.1016/s0002-9440(10)62549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farley J, Brady WE, Vathipadiekal V, Lankes HA, Coleman R, Morgan MA, Mannel R, Yamada SD, Mutch D, Rodgers WH. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol. 2013;14(2):134–140. doi: 10.1016/S1470-2045(12)70572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Rustin GJ, Vergote I, Eisenhauer E, Pujade-Lauraine E, Quinn M, Thigpen T, du Bois A, Kristensen G, Jakobsen A, Sagae S. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG) Int J Gynecol Cancer. 2011;21(2):419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 20.Despierre E, Yesilyurt BT, Lambrechts S, Johnson N, Verheijen R, van der Burg M, Casado A, Rustin G, Berns E, Leunen K. Epithelial ovarian cancer: rationale for changing the one-fits-all standard treatment regimen to subtype-specific treatment. Int J Gynecol Cancer. 2014;24(3):468–477. doi: 10.1097/IGC.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 21.Grisham RN, Iver G, Garg K, Delair D, Hyman DM, Zhou Q, Iasonos A, Berger MF, Dao F, Spriggs DR. BRAF mutation is associated with early disease and improved outcome in patients with low-grade serous ovarian cancer. Cancer. 2013;119(3):548–554. doi: 10.1002/cncr.27782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gershenson DM, Sun CC, Wong KK. Impact of mutational status on survival in low-grade serous carcinoma of the ovary and peritoneum. Br J Cancer. 2015;113(9):1254–1258. doi: 10.1038/bjc.2015.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergote I, Tropé CG, Amant F, Kristensen GB, Ehlen T, Johnson N, Verheijen RH, van der Burg ME, Lacave AJ, Panici PB. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med. 2010;363(10):943–953. doi: 10.1056/NEJMoa0908806. [DOI] [PubMed] [Google Scholar]
- 24.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the RASCAL II study. Br J Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101(3):465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etemadmoghadam D, Azar WJ, Lei Y, Moujaber T, Garsed DW, Kennedy CJ, Fereday S, Mitchell C, Chiew YE, Hendley J. EIF1AX and NRAS mutations co-occur and cooperate in low-grade serous ovarian carcinomas. Cancer Res. 2017;77(16):4268–4278. doi: 10.1158/0008-5472.CAN-16-2224. [DOI] [PubMed] [Google Scholar]
- 27.Xing D, Survo Rahmanto Y, Zeppernick F, Hannibal CG, Kjaer SK, Vang R, Shih IM, Wang TL. Mutation of NRAS is a rare genetic event in ovarian low-grade serous carcinoma. Hum Pathol. 2017;68:87–91. doi: 10.1016/j.humpath.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIntyre JB, Rambau PF, Chan A, Yap S, Morris D, Nelson GS, Köbel M. Molecular alterations in indolent, aggressive and recurrent ovarian low-grade serous carcinoma. Histopathology. 2017;70(3):347–358. doi: 10.1111/his.13071. [DOI] [PubMed] [Google Scholar]
- 29.Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CM, Queirolo P, Blank CU, Hauschild A, Beck JT, St-Pierre A. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14(3):249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Wang TL, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, Kinzler KW, Papadopoulos N, Shih leM. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226(3):413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Showeil R, Romano C, Valganon M, Lambros M, Trivedi P, Van Noorden S, Sriraksa R, El-Kaffash D, El-Etreby N, Natrajan R. The status of epidermal growth factor receptor in borderline ovarian tumours. Oncotarget. 2016;7(9):10568–10577. doi: 10.18632/oncotarget.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anglesio MS, Arnold JM, George J, Tinker AV, Tothill R, Waddell N, Simms L, Locandro B, Fereday S, Traficante N. Mutation of ERBB2 provides a novel alternative mechanism for the ubiquitous activation of RAS-MAPK in ovarian serous low malignant potential tumors. Mol Cancer Res. 2008;6(11):1678–1690. doi: 10.1158/1541-7786.MCR-08-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo KT, Guan B, Feng Y, Mao TL, Chen X, Jinawath N, Wang Y, Kurman RJ, Shih leM, Wang TL. Analysis of DNA copy number alterations in ovarian serous tumours identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69(9):4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henrich KO, Schwab M, Westermann F. 1p36 Tumor suppression – A matter of dosage? Cancer Res. 2012;72(23):6079–6088. doi: 10.1158/0008-5472.CAN-12-2230. [DOI] [PubMed] [Google Scholar]
- 35.Mayrhofer M, Kultima HG, Birgisson H, Sundström M, Mathot L, Edlund K, Viklund B, Sjöblom T, Botling J, Micke P. 1p36 deletion is a marker for tumour dissemination in microsatellite stable stage II-III colon cancer. BMC Cancer. 2014;14:872. doi: 10.1186/1471-2407-14-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas CJ, Diebold J, Hirschmann A, Rohrbach H, Löhrs U. In serous ovarian neoplasms the frequency of Ki-ras mutations correlates with their malignant potential. Virchows Arch. 1999;434(2):117–120. doi: 10.1007/s004280050314. [DOI] [PubMed] [Google Scholar]
- 37.Vereczkey I, Serester O, Dobos J, Gallai M, Szakács O, Szentirmay Z, Toth E. Molecular characterization of 103 ovarian serous and mucinous tumors. Pathol Oncol Res. 2011;17(3):551–559. doi: 10.1007/s12253-010-9345-8. [DOI] [PubMed] [Google Scholar]
- 38.Schlosshauer PW, Deligdisch L, Penault-Llorca F, Fatemi D, Qiao R, Yao S, Pearl M, Yang Z, Sheng T, Dong J. Loss of p16INK4A expression in low-grade ovarian serous carcinomas. Int J Gynecol Pathol. 2011;30(1):22–29. doi: 10.1097/PGP.0b013e3181ed89b3. [DOI] [PubMed] [Google Scholar]
- 39.Sundov D, Caric A, Mrklic I, Gugic V, Hofman ID, Mise BP, Tomic S. P53, MAPK, topoisomerase II alpha and Ki67 immunohistochemical expression and KRAS/BRAF mutation in ovarian serous carcinomas. Diagn Pathol. 2013;8:21. doi: 10.1186/1746-1596-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadlecki P, Antosik P, Grzanka D, Grabiec M, Walentowicz-Sadlecka M. KRAS mutation testing in borderline ovarian tumors and low-grade ovarian carcinomas with a rapid, fully integrated molecular diagnostic system. Tumour Biol. 2017;39(10) doi: 10.1177/1010428317733984. (1010428317733984) [DOI] [PubMed] [Google Scholar]
- 41.Sadlecki P, Walentowicz P, Bodnar M, Marszalek A, Grabiec M, Walentowicz-Sadlecka M. Determination of BRAF V600E (VE1) protein expression and BRAF gene mutation status in codon 600 in borderline and low-grade ovarian cancers. Tumour Biol. 2017;39(5) doi: 10.1177/1010428317706230. (1010428317706230) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Processed sequencing data on the 38 samples: place of biopsy, hotspot genotyping by Sequenom, altered fraction of segments, and GISTIC focal and whole arm analyses from the shallow sequencing experiment.
Data Availability Statement
Research data and data not presented in the manuscript are available as supplementary data (S1). They will also be available upon request to the corresponding author.