Abstract
Therapies against malignant pleural mesothelioma (MPM) have yielded disappointing results, in part, because pathologic mechanisms remain obscure. In searching for rational molecular targets, we identified metadherin (MTDH), a multifunctional gene associated with several tumor types but previously unrecognized in MPM. Cox proportional hazards regression analysis delineated associations between higher MTDH expression and lower patient survival from three independent MPM cohorts (n = 349 patients). Through in vitro assays with overexpression and downregulation constructs in MPM cells, we characterized the role of MTDH. We confirmed in vivo the phenotype of altered MTDH expression in a murine xenograft model. Transcriptional regulators of MTDH were identified by chromatin immunoprecipitation. Overexpression of both MTDH mRNA (12-fold increased) and protein levels was observed in tumor tissues. MTDH stable overexpression significantly augmented proliferation, invasiveness, colony formation, chemoresistance, and an antiapoptosis phenotype, while its suppression showed opposite effects in MPM cells. Interestingly, NF-κB and c-Myc (in a feed-forward loop motif) contributed to modulating MTDH expression. Knockdown of MTDH expression profoundly retarded xenograft tumor growth. Thus, our findings support the notion that MTDH integrates upstream signals from certain transcription factors and mediates pathogenic interactions contributing to MPM traits. MTDH represents a new MPM-associated gene that can contribute to insights of MPM biology and, as such, suggest other treatment strategies.
Introduction
Malignant pleural mesothelioma (MPM) is an aggressive tumor causally associated with asbestos exposure. Contrary to predictions, the incidence continues to increase worldwide [1]. There are few FDA-approved therapies for MPM, so the dismal median survival time of 12 to 18 months remains unchanged [2]. This therapeutic plateau of conventional chemotherapy contributes to ongoing clinical challenges faced by newer precision medicine-based therapy, particularly as tumor suppressor losses predominate [3]. Clinical trials have failed to identify reliable targeted therapeutic agents [4]. Thus, identification of novel molecular targets is needed to inform about tumor biology and/ or suggest better treatment(s) of MPM.
Metadherin (MTDH) overexpression is observed in diverse malignancies including breast [5], lung [6], esophageal [7], B-cell lymphoma [8], brain tumors [9], gynecologic cancers [10], etc. These observations support the emerging notion that MTDH is a universally important cancer-associated gene. MTDH molecular interactions impact critical signaling pathways that affect common cancer traits like proliferation, evasion of apoptosis, metastasis, angiogenesis, chemoresistance, etc. [11] MTDH fulfills many characteristics to serve as an important molecule regulating multiple events in carcinogenesis. However, this common cancer-associated gene has not previously been implicated with MPM [12], so its role in MPM remains entirely unclear. In contrast to other cancers, gain-of-function somatic mutations have not been consistently identified in MPM. Because of this, identifying genes that are overexpressed and exploring their phenotypic impact could lead to valuable biologic insights.
Among our initial investigations, we confirmed that this gene and its protein product were overexpressed in MPM tissues. Next, we characterized the effects of this gene in cell line models of overexpression and downregulation to demonstrate, overall, that MTDH confers an antiapoptotic phenotype in MPM. This phenotype manifested as an enhanced chemoresistance trait when MTDH was overexpressed above basal conditions and reversed when MDTH was suppressed. Tumor xenograft experiments in mice confirmed that MTDH is important for MPM tumor progression. In further investigations, we uncover a feed-forward regulatory mechanism that conceptually explains the overexpression of MTDH in MPM. Our results underscore the need for ongoing gene discovery to pinpoint relevant target(s) in MPM.
Materials and Methods
Mesothelioma Public Data
We relied on the TCGA-Meso public dataset comprised of 85 (total specimens = 87) MPM tumors with clinical results (gdc.cancer.gov), a genomic profiling (mRNA microarray) of 53 MPM tumors [Memorial Sloan-Kettering Cancer Center (MSKCC)] [13], and a recent sequencing-based transcriptomic analysis of 211 MPM tumors (Genentech, Inc.) [3] as independent validation resources. These RNA datasets (Supplementary Table S1) were derived from analysis of diverse patients undergoing surgical resection of MPM (all three histologic subtypes). Importantly, associated survival outcomes were available among these data.
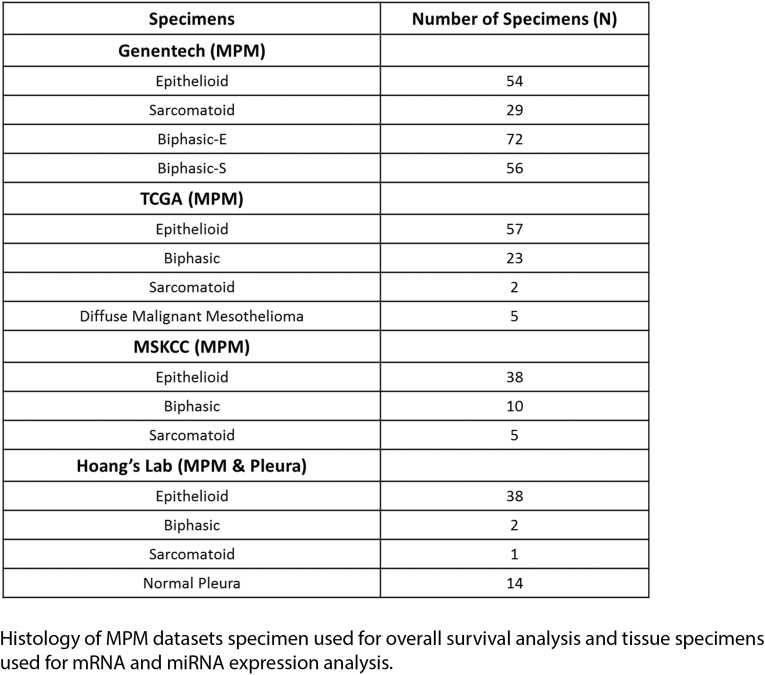
Supplementary Table S1.
Histology of MPM Datasets Specimen Used for Overall Survival Analysis and Tissue Specimens Used for mRNA and miRNA Expression Analysis
Reagents
Cisplatin and pemetrexed chemotherapeutics were used to treat cells (Selleckchem). TNF-α was used as an in vitro stimulatory agent (Sigma-Aldrich). JSH-23 was used as an in vitro inhibitor of p65 activity since it is known to selectively prevent nuclear translocation (Sigma-Aldrich).
Tissues and Cell Culture
Sample collection followed IRB-approved protocols. Deidentified surgical specimens were stored at −°C. We selected 41 MPM tumors of all 3 histologies and 14 unmatched, nonmalignant pleurae obtained from patients undergoing surgery for other diseases not MPM. All these specimens were chosen for our study based on amounts of useable tissue available. Multiple MPM cell lines [14] were tested for native MTDH expression (Supplementary Figure S1). We chose three representative lines (H2452 epithelioid, MSTO-211H biphasic, and H2373 sarcomatoid) to be used for the majority of experiments. The pleural mesothelial cell line MeT-5a was purchased from ATCC, and the peritoneal mesothelial cell line LP-9 was purchased from the Coriell Cell Repository. MPM cells and MeT-5a were cultured and maintained according to ATCC instructions. LP-9 cells were cultured in specific manufacturer media.
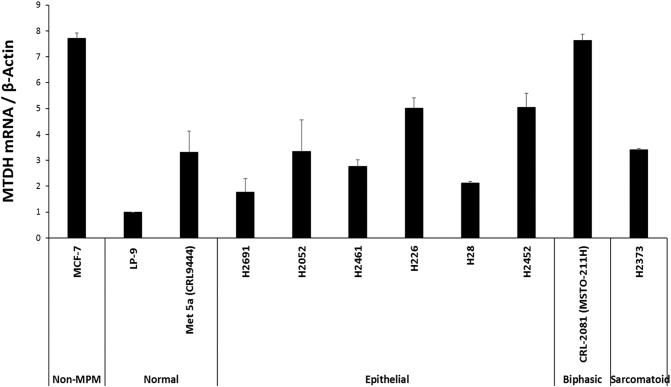
Supplementary Figure S1.
Survey of MTDH basal expression in MPM lines.
Basal expression of MTDH transcript in a panel of NCI MPM cell lines. LP9 is used as the reference normal cell line for MTDH quantitation. MCF-7 breast carcinoma cells are positive control of MTDH expression. qRT-PCR quantitated relative abundance of MTDH in epithelioid MPM lines, a biphasic MPM line, and a sarcomatoid MPM line. The frequency of cell line histologies reflects the pathologic distribution (and availability) of human MPM. H2452, MSTO-211H, and H2373 were chosen as representative cells in subsequent experiments.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from specimens using the TRIzol Plus RNA purification system (Thermo Fisher). Reverse transcription was performed using the Applied Biosystems high-capacity RNA to cDNA synthesis kit. Gene quantitation was determined by TaqMan analysis run on a QuantStudio 6 Flex PCR system (Thermo Fisher) [15]. qRT-PCR primers for gene expression were available from Applied Biosystems (Supplementary Table S2). All independent PCR-based reactions were performed in triplicate.
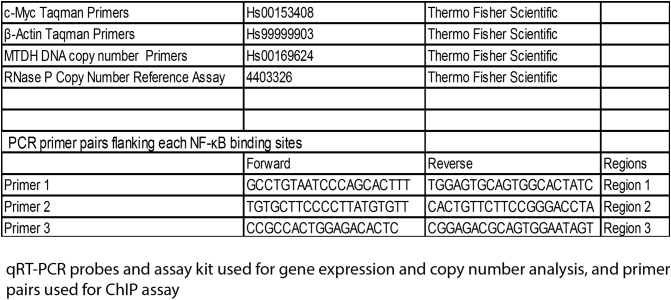
Supplementary Table S2.
qRT-PCR Probes and Assay Kit Used for Gene Expression and Copy Number Analysis, and Primer Pairs Used for ChIP Assay
Western Blotting
Total protein lysates were fractionated on 4%-15% polyacrylamide gels and transferred onto nitrocellulose (Bio-Rad). These primary antibodies were used: anti-MTDH at 1:500 dilution (2F11C3 monoclonal antibody, Thermo Fisher), anti–phospho-p65 at 1:1000 with anti–total p65 at 1:1000 (Cell Signaling Technologies), anti–c-Myc at 1:500 (Abcam), and anti–β-Actin (Sigma-Aldrich). Additionally, antibodies detecting cleaved PARP and caspase-3 (Cell Signaling) were used to assess apoptosis activity. Subsets of samples were randomly selected from the original group of 41 tumors and 14 normal pleurae for this series of experiments. All protein experiments were performed in triplicate. To avoid problems related to incomplete membrane stripping, separate blots were used for each independent experiment.
Proliferation
MPM cells were seeded into 96-well plates (5000 cells/well). Cellular proliferation measurements were carried out by CyQUANT assay kit (Thermo Fisher). Studies were performed in three independent cell preparations.
Cellular Invasion
Cell migration was evaluated by Boyden chamber assays (Millipore) using filter inserts with pores (8 μm). The condition in the top chamber consisted of 0.1×106 cells in serum-free media adjacent to a Matrigel-coated filter. The bottom chamber contained complete media supplemented with 10% FBS as a chemoattractant. After incubation, migratory cells that traversed the filter membrane were fixed and stained with crystal violet. According to manufacturer protocol, invading cells were detached and quantitated using their colorimetric method.
Anchorage-Independent Growth
Colony formation was assessed in soft agarose. Six-well plates were precoated with 0.5% (w/v) agarose in appropriate cell media and allowed to set. Viable cells (5000/well) were resuspended in a heated soft agarose medium consisting of 0.4% (w/v) low-melt agarose. Cultures were incubated for 1-4 weeks. Colonies were stained and counted.
Apoptosis by Flow Cytometry and Specific Proteins
Cell viability of MPM lines was compared against their treated (MTDH overexpression or knockdown) and negative control counterparts. MPM lines (0.5×106 cells/well) were cultured in the presence of a fixed concentration corresponding to the IC50 of cisplatin or pemetrexed in six-well plates for 48 hours. Cells were harvested after enzyme treatment. Apoptotic or dead cells were detected by staining with Annexin V–FITC and propidium iodide during flow cytometry according to the manufacturer's protocol (BD Biosciences). The common caspase-mediated signaling cascade considered a hallmark of apoptosis was assessed by Western blotting of the cleaved forms of PARP and caspase-3 in certain MPM cells.
Copy Number Variation
Quantitation of MTDH gene copy number was determined by qRT-PCR method. Genomic DNA was extracted from a subset of our MPM tumor tissues (n = 25) and normal mesothelial specimens (n = 5) using the Wizard Genomic DNA purification kit (Promega). These subset samples were randomly selected from the original group of 41 tumors and 14 normal pleurae. Primers for MTDH and the endogenous control gene (RNase P) were obtained from Thermo Fisher (Supplementary Table S2).
Immunohistofluorescence
Cryosections (8 μm) were cut from a subset of MPM specimens and processed in a standard fashion for immunolabeling. These primary antibodies were used: lysine-rich carcinoembryonic antigen-related cell-adhesion molecule 1 coisolated (LYRIC, another name for MTDH) (rabbit anti-human; Abcam) and mesothelin (mouse anti-human; Santa Cruz Biotechnology). Fluorochrome-conjugated secondary antibodies were used for detection of LYRIC and mesothelin. Tissue sections were cover slipped with mounting medium containing DAPI stain (Vector Laboratories).
Overexpression and Suppression Constructs
The lentiviral vector pLenti-GIII-UbC containing a MTDH insert of 1749 bp (Applied Biological Materials, cat. #LVP229320) stably overexpressed MTDH in MPM cells. A lentiviral vector pLenti-III-UbC Blank was used as a negative control (Applied Biological Materials, cat. #LV589). The MTDH gene was suppressed using a CRISPR/Cas9 system. MTDH double nickase plasmid (sc-413927-NIC) and a control double nickase plasmid (sc-437281) (Santa Cruz Biotechnology) were transfected. A heterogeneous population of MPM cells was chosen after transfection so MTDH gene is knocked down (not knockout) in all our experiments. c-Myc overexpression relied on a pCMV6 cloning vector carrying a 4707-bp insert (OriGene Technologies). For transient c-Myc gene silencing, a mixture of chemically modified siRNA in the SMARTpool: ON-TARGETplus kit and negative controls (Dharmacon) were used.
Drug Treatment
Cell viability of MPM lines was compared against their treated (MTDH overexpression or suppression) and negative controls. Dose-response was analyzed after 48 hours by CyQUANT assay. Cells were exposed to two-fold increments of cisplatin or pemetrexed, and the IC50 of each drug was calculated in cell lines (GraphPad Prism v7). Three independent triplicate experiments were performed.
Chromatin Immunoprecipitation
To verify NF-κB (RELA) interaction with the promoter region of MTDH, we employed chromatin immunoprecipitation (ChIP). MPM cell lines were prepared using the Magna ChIP A-Chromatin Immunoprecipitation Kit (Millipore) with an anti-p65 antibody. DNA was eluted and purified from complexes, followed by SYBR PCR amplification of three putative binding sites of RELA within the MTDH promoter. Primer pairs flanking these binding sites were used (Supplementary Table S2). Immune complexes formed with nonspecific IgG were a negative control, while IκBα promoter primers (manufacturer kit) were a positive control.
Mice Xenograft Experiments
H2373 parental cells, H2373 cells with MTDH gene overexpression, or H2373 cells with knockdown of MTDH were injected subcutaneously (each group at 2.0×106) on the right flank of NOD.CgPrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice [16] 5-7 weeks old. For each experimental condition, five mice were used. Tumor volume changes were charted during 5 weeks of observation and measured using the formula: V = 1/2 (length × width2) [17]. At the end of observation, mice were sacrificed and tumors were excised. All mice experiments were approved by our Animal Care and Use Committee in accordance with NIH guidelines.
Statistics
Means and standard errors (SEs) were calculated from numerical data. Changes (fold or percentage) indicate the difference between experimental and control samples. Bar graphs depict the mean ± SE for a specific experimental run. SPSS software (IBM Analytics) executed calculations. Two-tailed, unpaired Student t test assessed significance between two conditions. One-way analysis of variance test followed by Tukey's comparison for multiple groups compared the significance of differences between the means of groups. Pearson r correlation coefficient with r > ±0.5 was considered a strong relationship between variables. Spearman's rank correlation coefficient (Rho) was used as a nonparametric measure of rank correlation between variables. Multivariate linear regression was used to model additive contributions of predictor variables (genes) to predict a response variable using the lm function in R (www.r-project.org). The multivariate model was evaluated by variance (adjusted R2) and the significance of each predictor variable's contribution. Cox proportional hazards regression with age as a covariate was used to discover associations between MTDH expression and patient survival. One-tailed P value ≤ .05 was considered significant. The Stouffer's Z score method was used to combine the Cox proportional hazards regression Z scores, and one-tailed P values were computed based on the combined Z score.
Results
Clincal Relevance of Metadherin in Mesothelioma
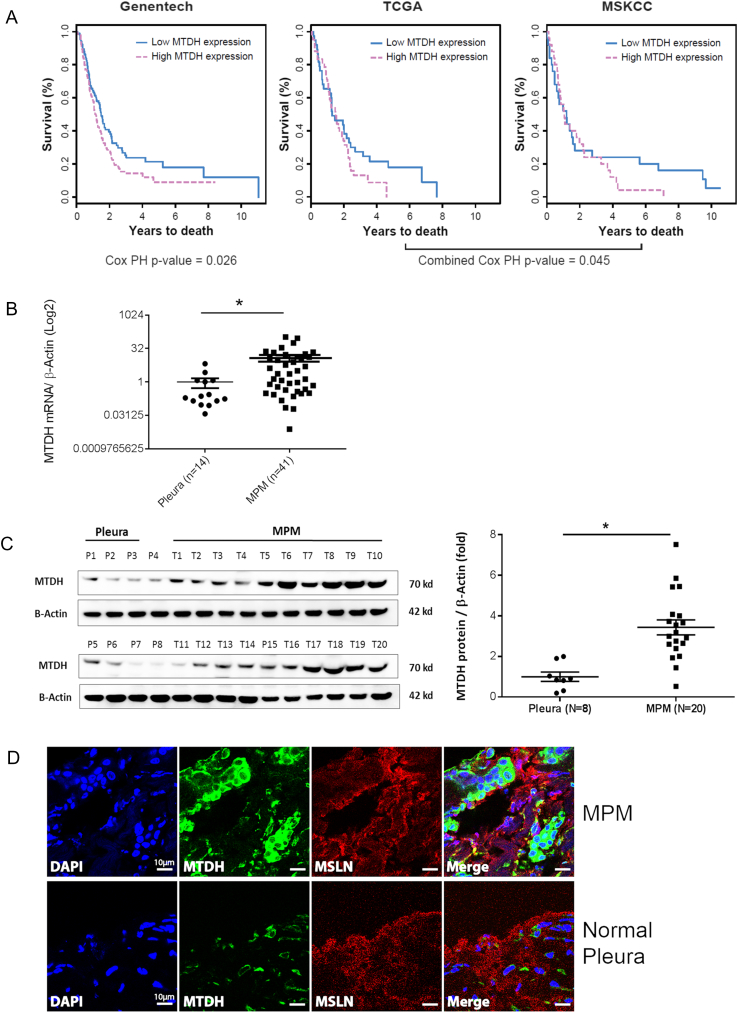
We hypothesized that if MTDH is clinically relevant, the expression of the gene would be related to patient survival. Therefore, we interrogated clinically annotated public MPM transcriptome datasets to assess survival associations with MTDH mRNA expression. In the largest patient cohort (Genentech) [3], we found that a higher expression of MTDH in patient tumors is significantly associated with lower overall survival (Cox proportional hazards P value = .026) (Figure 1A). This survival association was confirmed by observing similar results from combining two other independent datasets (TCGA and MSKCC [13]; combined P value = .045) (Figure 1A). Thus, higher MTDH expression is significantly associated with poor prognosis in MPM.
Figure 1.
Metadherin is a novel prognostic gene overexpressed in MPM.
(A) Kaplan-Meier curves depicting overall survival of MPM patients grouped according to a dichotomized expression of MTDH transcript level. In the Genentech cohort (n = 211), there is a statistically significant reduction in overall survival for those harboring MPM tumors with relatively “high” MTDH expression (Cox proportional hazards P = .026; age is a covariate). This survival association validated in the joint analysis of TCGA (n = 85) and MSKCC (n = 53) cohorts. Their Cox proportional hazards regression Z scores were combined, and P values were computed based on the combined Z score (combined Cox proportional hazards P value = .045). PH is proportional hazards. (B) qRT-PCR verified the mRNA quantity of MTDH in a separate random group of MPM tumors compared to a separate set of unpaired normal pleura tissues. The tumor cohort is comprised of all three MPM histologies (38, epithelioid; 2, biphasic; 1, sarcomatoid). MPM expressed 12-fold more MTDH mRNA compared to normal pleura. (C) Western blotting of the same tissues analyzed by qRT-PCR shows the MTDH protein levels in MPM tumor tissues (T1-T20) and pleural (P1-P8). This subset of epithelioid tumors and pleura specimens was randomly selected from the original tissue cohorts. The associated dot graph (right) quantitates the relative protein abundance of MTDH depicted in the Western blot. (D) Immunohistofluorescence microscopy reveals the cellular location of MTDH protein. Top panel (MPM cells) shows increased staining of MTDH (green), which is predominately expressed in the cytosolic compartment and not at the cell membrane, unlike mesothelin (MSLN, red). Bottom panel is a representative normal pleura specimen showing reduced MTDH staining compared to MPM cells. Nuclei are stained with DAPI (blue). MTDH (green) and MSLN (mesothelin, red). Bars in graphs represent the mean ± SE, *P < .05, **P < .001. Scale bar: 10 μm.
Metadherin Expression in Mesothelioma Tumors
Accordingly, we checked whether MTDH is differentially expressed in randomly selected MPM tumors (n = 41) relative to unmatched normal specimens (n = 14). Most of these available tumor specimens are epithelioid type (similar to the TCGA cohort) because surgical resection is not routinely offered to the other histologic subtypes (www.nccn.org) (Supplementary Table S1). We observed MTDH mRNA overexpressed by 12-fold in MPM tumors relative to normal tissues (P < .05) (Figure 1B). This transcript overabundance was confirmed by observing increased MTDH protein expression by an average of 3.43-fold (P < .05) in a subgroup of available specimens (Figure 1C). Immunohistofluorescence showed dense cytoplasmic MTDH localization in MPM cells (Figure 1D). MTDH also appeared intranuclear, but to a much lesser extent, while it was not observed on the MPM cell surface. This MTDH distribution pattern in MPM cells resembles prostate cancer, where such localization correlated to clinical parameters [18]. Overall, we identified MTDH overexpression at the mRNA and protein levels in MPM tissues.
Metadherin Enhances Chemoresistance via Antiapoptosis
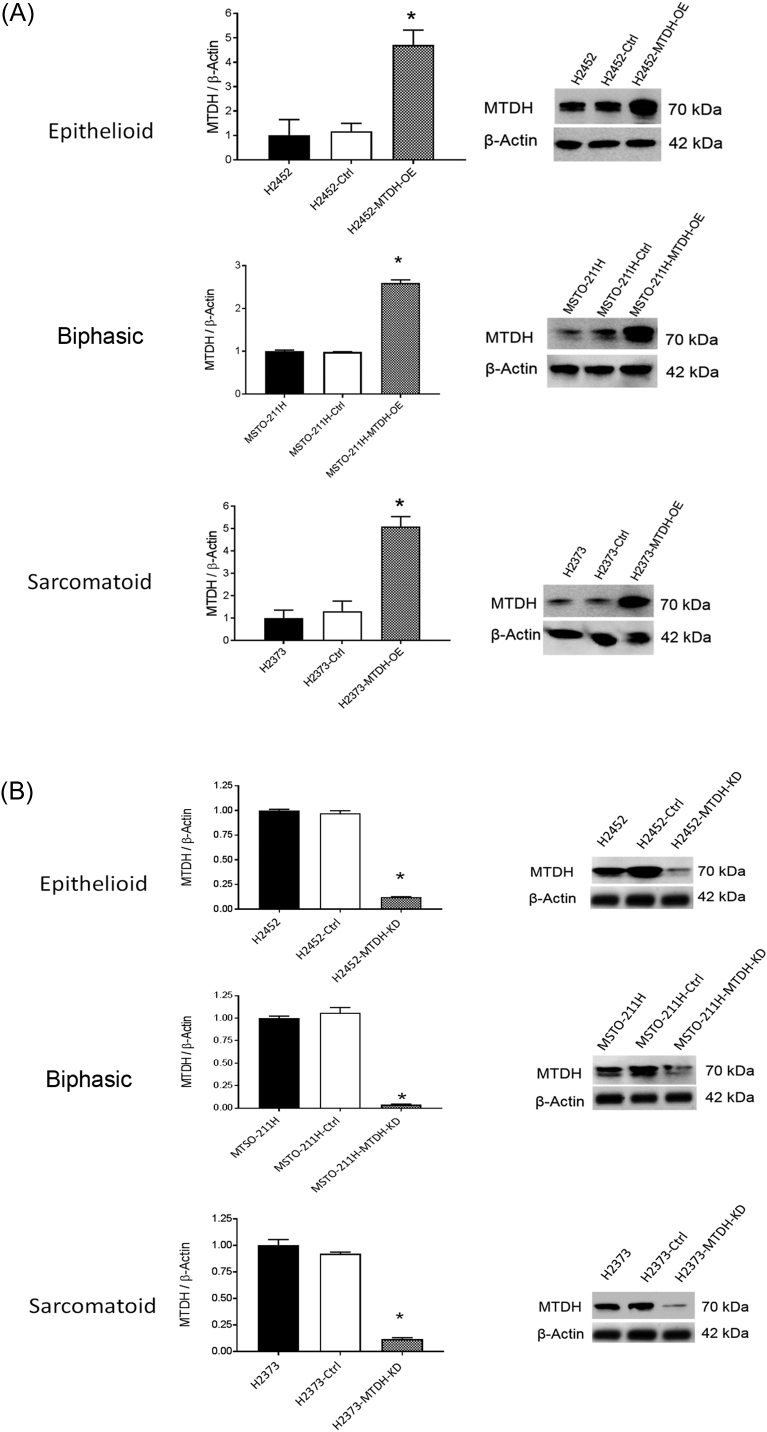
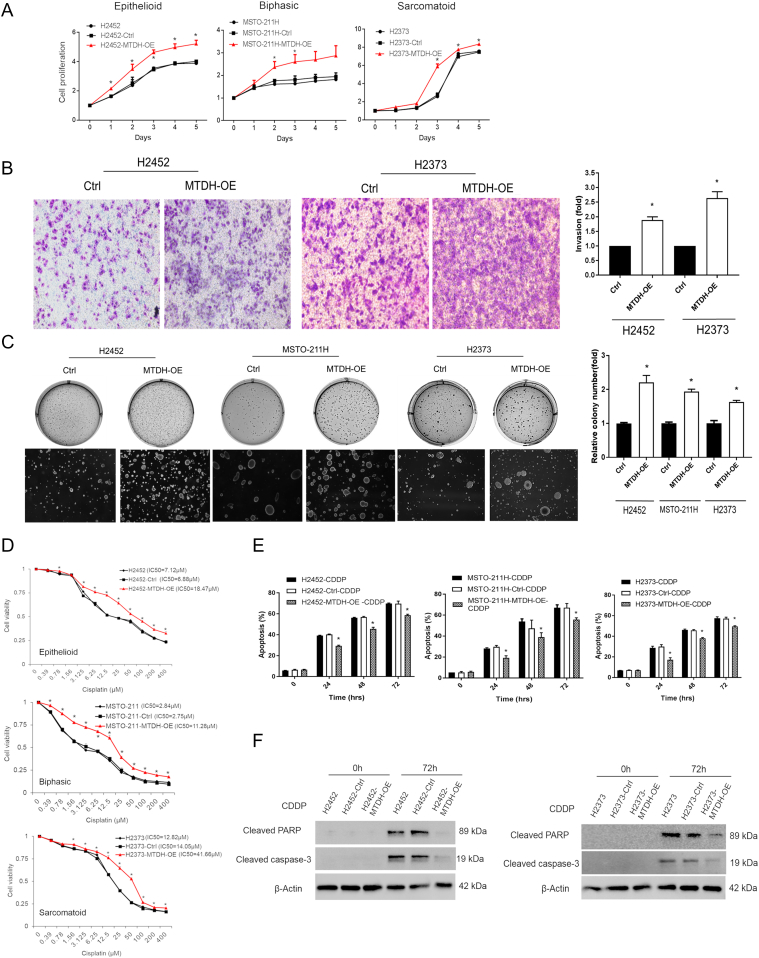
We then characterized the biologic effects of increased MTDH in MPM. The MTDH gene was stably overexpressed in MPM cell lines from all three histotypes (Supplementary Figure S2). Transfected cells exhibited significantly accelerated growth and increased invasiveness (Boyden chamber) (Figure 2, A-B). The number of colonies formed under anchorage-independent conditions was increased compared to parental lines (P < .05) when MTDH was overexpressed (Figure 2C).
Supplementary Figure S2.
Confirmation of MTDH altered expression in transfected MPM lines.
(A) Validation of MTDH overexpression constructs. The MTDH gene was stably overexpressed in three representative MPM lines H2452, MSTO-211H, and H2373 by lentiviral transfection. In all MPM lines, the MTDH protein amount markedly increased above basal levels of parental cells when the MTDH transcript was overexpressed (MTDH-OE). (B) Validation of MTDH knockdown constructs. Although CRISPR/Cas9 gene editing was used, because single clones were not selected, there remained low detection of MTDH transcripts in MPM cell lines. In all MPM lines, the MTDH protein amount markedly decreased below basal levels of parental cells when the MTDH transcript was suppressed (MTDH-KD). MTDH mRNA transcript was assessed by qRT-PCR, and protein amount was assessed by Western blotting. Where applicable, data are presented as mean ± SE. Ctrl is a negative control. * P < .05.
Figure 2.
Biologic effects of metadherin overexpression in MPM.
(A) MTDH overexpression promoted proliferation in MPM cells. MPM cell lines (H2452, MSTO-211H, and H2373) were stably overexpressed with MTDH (red). Data are expressed as relative fold change compared with day 0. On day 3, cell proliferation increased 1.3-fold (H2452), 1.7-fold (MSTO-211H), and 3.0-fold (H2373) (P < .05). (B) Boyden chamber assay showed that MTDH overexpression increases invasive capability of MPM cells by 1.9-fold (H2452) and 2.6-fold (H2373). (C) Soft agar assay in which MTDH overexpression accelerated colony formation. Relative colony numbers increased 2.2-fold (H2452), 1.9-fold (MSTO-211H), and 1.6-fold (H2373) (P < .05). (D) Cell viability was assessed at varying concentrations of cisplatin (CDDP) to calculate IC50 values. MTDH overexpression (red) rendered MPM cells more chemoresistant, requiring 2.6-fold (H2452), 4.0-fold (MSTO-211H), and 3.1-fold (H2373) higher doses of drug to reach IC50 levels. (E) MTDH overexpression significantly decreased apoptosis after 72 hours of treating MTDH-induced cells with a fixed concentration of cisplatin (IC50) by 15.9% (H2452), 16.9% (MSTO-211H), and 14.0% (H2373) compared to parental cells. (F) Western blot showing decreased apoptotic proteins (cleaved PARP and caspase-3) at 72 hours of cisplatin exposure in both H2452 and H2373 MPM cells overexpressing MTDH. Where applicable, data are presented as mean ± SE. Ctrl is overexpression vector control sample. * is P < .05 versus parent cell line and/or negative control specimen. MTDH-OE is stably overexpressed MTDH in MPM cells.
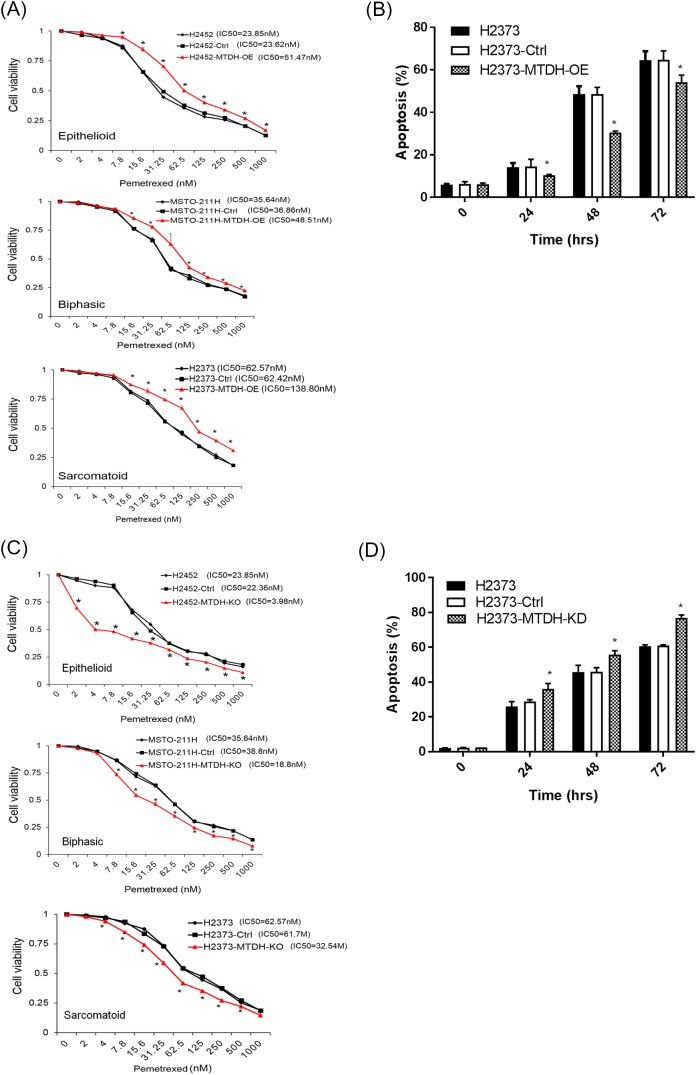
Cell viability was measured under increasing concentrations of cisplatin (Figure 2D) or pemetrexed (Supplementary Figure S3), standard MPM drugs. After 48 hours, cells overexpressing MTDH maintained a greater percentage of viable cells compared to parental cells (P < .05) as drug doses escalated. The IC50 values for each MPM cell line were determined from these cell-killing curves. Our results in MPM are consistent with other solid tumors which implicate MTDH overexpression augmenting chemoresistance [19].
Supplementary Figure S3.
Modulating metadherin expression levels affects pemetrexed sensitivity in MPM cell lines.
(A) Cell viability was assessed at varying concentrations of pemetrexed to calculate IC50 values. MTDH overexpression (red) rendered MPM cells less sensitive to treatment, requiring 2.2-fold (H2452), 1.3-fold (MSTO-211H), and 2.2-fold (H2373) higher doses of drug to reach IC50 levels. (B) Annexin V flow cytometry measured levels of apoptosis when H2373 cells were treated with a fixed dose (IC50) of pemetrexed. Cells overexpressing MTDH (MTDH-OE) had lower levels of apoptosis. (C) Cell viability was assessed at varying concentrations of pemetrexed to calculate IC50 values. MTDH knockdown (red) rendered MPM cells more sensitive to treatment with pemetrexed, requiring 5.8-fold (H2452), 2-fold (MSTO-211H), and 1.9-fold (H2373) lower doses of drug to reach IC50 levels. (D) Annexin V flow cytometry measured levels of apoptosis when H2373 cells were treated with a fixed dose (IC50) of pemetrexed. Cells with reduced expression of MTDH (MTDH-KD) had lower levels of apoptosis. Where applicable, data are presented as mean ± SE. Ctrl is a negative control. OE indicates gene overexpression. KD indicates gene suppression. * P < .05.
To verify the chemoresistance effects, we assessed apoptosis since the chemotherapeutics rely on this cell killing mechanism. MPM cells overexpressing MTDH were exposed to IC50 amounts of cisplatin (Figure 2E) or pemetrexed (Supplementary Figure S3), and the percentage of cells exhibiting apoptosis by flow cytometry at different time points was quantitated. For these cells, the percentage of apoptosis was significantly decreased. Further, cleaved PARP and caspase-3 apoptosis markers were decreased in cells overexpressing MTDH (Figure 2F). These findings demonstrated that MTDH overexpression led to increased proliferation, invasion, and chemoresistance, as well as to decreased apoptosis.
No Common Mechanisms of Metadherin Overexpression in Mesothelioma
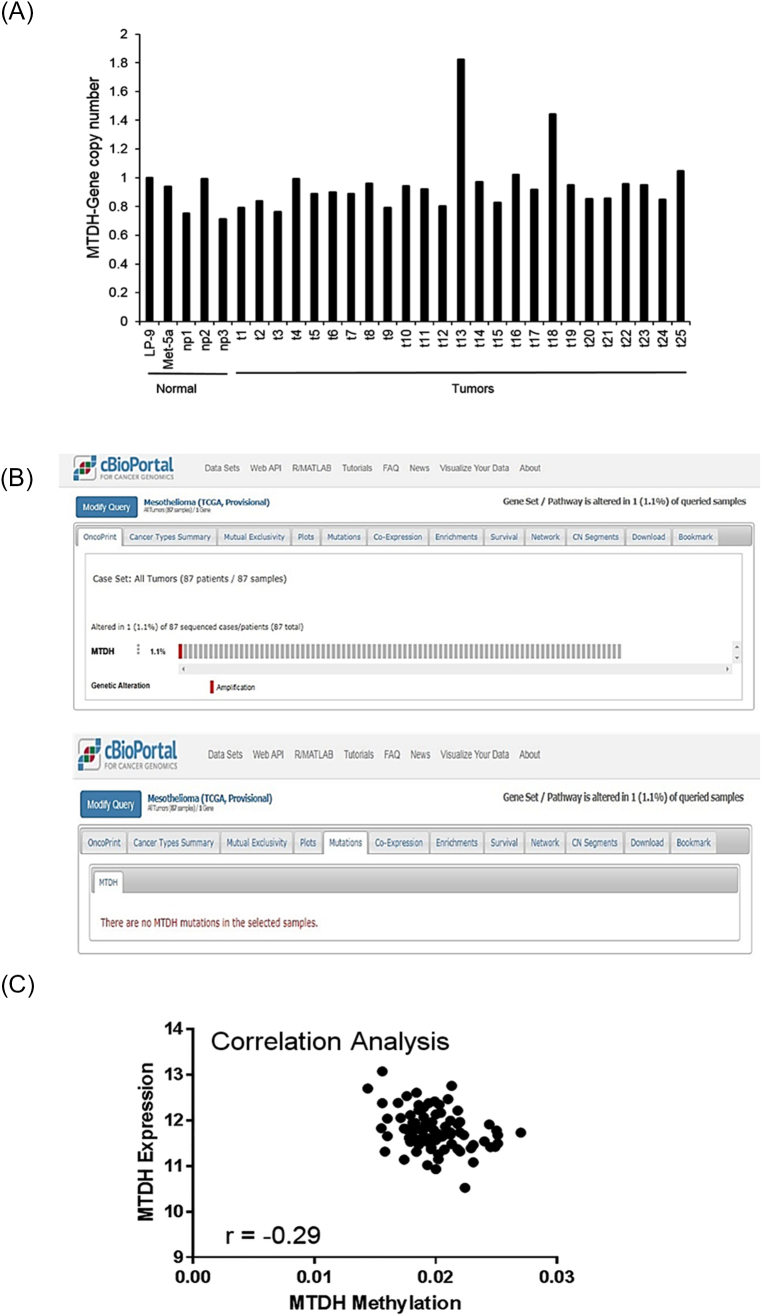
This led us to explore the potential mechanisms driving overabundance of MTDH transcripts. Since chromosome region 8q22.1, containing the MTDH gene locus, is an area of recurrent gain in MPM (18%) [20], this mechanism would be revealed by gene copy number analysis. PCR amplification of MTDH revealed no significant difference in MTDH copies among a subgroup of randomly selected specimens tested (n = 25). Similarly, TCGA showed only a single MPM tumor (1 of 87) with MTDH amplification, no mutations of the MTDH gene, and a weak negative association (Pearson r = −0.29) between the promoter methylation status versus MTDH transcript abundance. Taken together, these negative results (Supplementary Figure S4) implied other phenomena, unlike other solid tumors, were contributing to MTDH overexpression uniquely in MPM.
Supplementary Figure S4.
Common mechanisms of increased gene expression are not responsible for metadherin overexpression in MPM tumors.
(A) MTDH gene copy number was quantitated by qRT-PCR. There was no significant difference in the mean copy number among MPM tumors (t) versus normal pleural tissue (np) as compared to control, normal mesothelial cell lines (MeT-5A, LP9). Two tumor specimens showed increased copy number, but this did not alter statistical results. (B) TCGA data on MPM specimens were queried via cBioPortal. Only a single (1 of 87) MPM specimen showed amplification of MTDH (top). Investigation of MTDH mutational status detected no mutations of MTDH in MPM (bottom). (C) A correlation analysis was performed to assess the methylation status of the MTDH promoter. From the TCGA dataset, there was only a weak correlation (r < ±0.5) between MTDH mRNA expression versus CpG status of the MTDH promoter. Where applicable, data are presented as mean ± SE. Ctrl is overexpression scrambled sample. * is P < .05 versus parent cell line and/or negative control specimen.
Regulation of Metadherin in Mesothelioma: A Feed Forward Loop
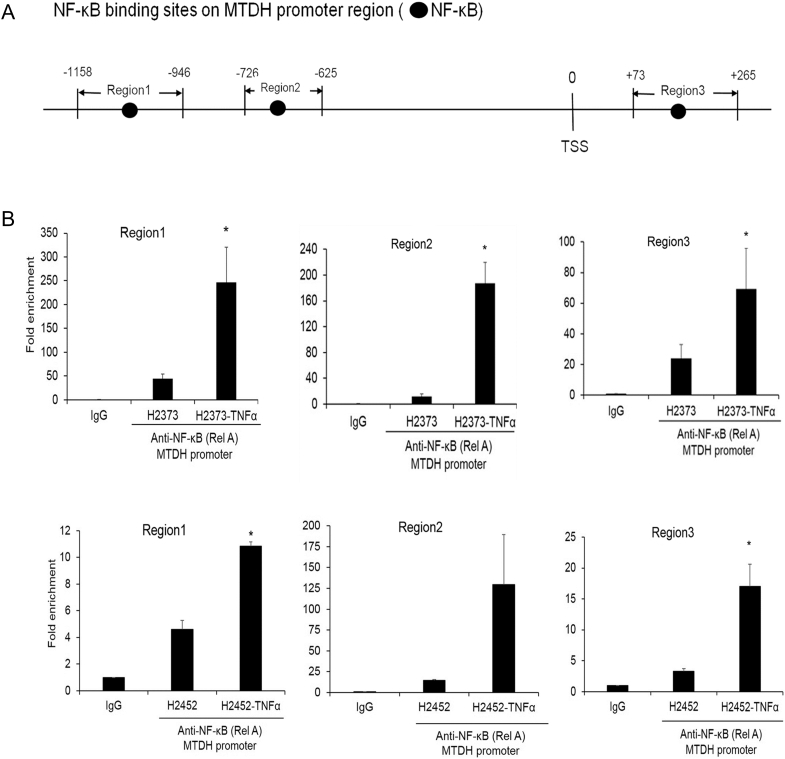
Chronic inflammatory conditions of MPM contribute to maintaining several signaling pathways that could represent plausible mechanism(s) for MTDH overexpression. TNF-α stimulation induces MTDH transcripts in diverse cell types [21], suggesting the necessity of additional intracellular mediators interacting with MTDH. However, these precise biochemical mechanisms remain incompletely defined [22]. Recently, in multiple myeloma [23], the UC Santa Cruz ChIP-sequencing database showed a footprint of p65 (RELA) subunit binding to the MTDH promoter. Since NF-κB activity is prevalent in MPM [24], we queried TRANSFAC 7.0 to identify putative RELA binding sites in the MTDH promoter region (Figure 3A). Using ChIP, we observed under TNF-α–stimulated (10 ng/ml) conditions an increase in RELA binding at three sites (P < .05) (Figure 3B).
Figure 3.
NF-κB directly binds the metadherin promoter and induces metadherin expression.
(A) Schematic of NF-κB binding sites in the promoter region of the MTDH gene. (B) ChIP- qPCR quantification of enrichment of DNA fragments that contain putative NF-κB binding site(s). Where applicable, data are presented as mean ± SE. Ctrl is the parental cell line. * P <. .05 versus parent cell line and/ or negative control specimen (IgG). TSS denotes transcription start site.
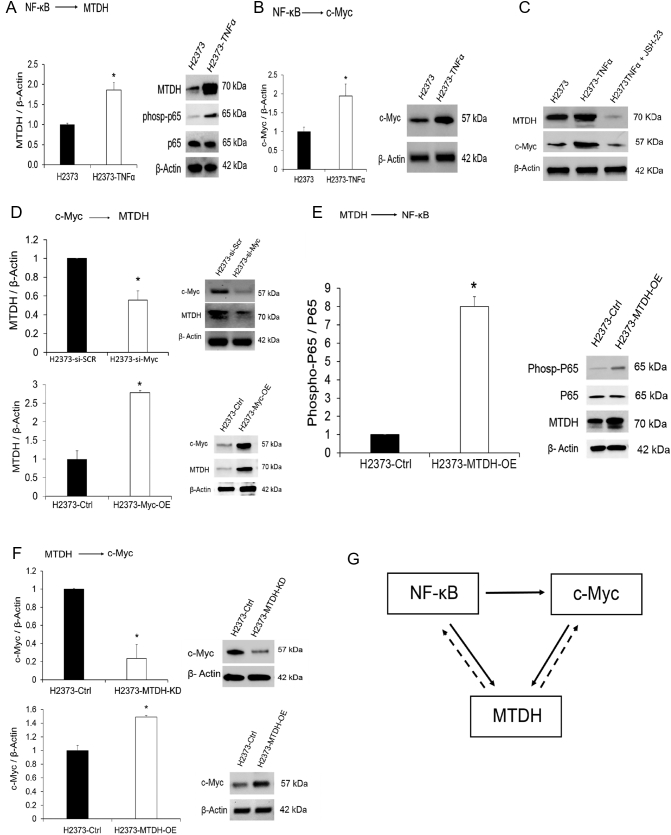
Additionally, NF-κB can directly induce c-Myc [25], a common oncogene. c-Myc regulates MTDH transcription by binding the promoter at two E-boxes [26]. Increasingly, c-Myc is recognized as an important contributor to the MPM malignant phenotype [27]. Taken together, c-Myc should be increased in MPM due to various inducing events and, therefore, may also contribute to MTDH transcript abundance. To verify our deduction, MPM cells were stimulated with TNF-α producing concordant increases in p65 activity, MTDH, and c-Myc abundance (Figure 4, A-B). Conversely, the protein amounts of MTDH and c-Myc reversed with the addition of a p65-specific inhibitor (Figure 4C). When c-Myc was inhibited by siRNA, this induced a decrease in MTDH (Figure 4D). These results experimentally confirm that NF-κB signaling directly regulates MTDH transcript abundance in MPM (for the first time to the best of our knowledge). NF-κB directly regulating MTDH and indirectly inducing MTDH via stimulation of c-Myc suggests a feed-forward loop network motif [28]. Without verifying, as we have done here, that NF-κB directly regulates MTDH, this specific mechanism could not be appreciated. Lastly, we confirmed known, well-documented effects of MTDH overexpression contributing, vice versa, to induction of NF-κB and c-Myc via secondary positive feedback signaling [21] (Figure 4, E-F). The forward path interconnections (Figure 4, A-C) are functionally equivalent to an activated switch once a certain input threshold is sensed and, in MPM, possibly contributes to reinforce and maintain MTDH activity (Figure 4G).
Figure 4.
Metadherin regulation in MPM is influenced by upstream transcription factors.
(A) TNF-α treatment of MPM cells triggers activation of NF-κB signaling as determined by increased phosphorylated p65 protein over a 24-hour duration. MTDH transcript abundance increased (left) as well as protein levels (right). (B) Both c-Myc mRNA (left) and protein (right) expression increased markedly after 24 hours of TNF-α treatment. (C) Treatment of cells with a specific p65 inhibitor (JSH-23) decreased both MTDH and c-Myc protein levels, confirming direct regulation of MTDH and c-Myc by NF-κB. (D) We verified that c-Myc induces MTDH expression. When c-Myc expression was transiently knocked down using siRNA (si-Myc), there was a corresponding decrease observed for both MTDH mRNA (left) and protein (right) levels. Conversely, when c-Myc was overexpressed (Myc-OE), there was a corresponding increase observed for both MTDH mRNA and protein levels. (E) Western blotting confirms that MTDH overexpression results in the activation of NF-κB (p65) in MPM cells, as determined by increased levels of phosphorylated p65 protein. (F) Overexpression and knockdown experiments demonstrate that MTDH positively modulates c-Myc transcript (left) and protein (right) levels in MPM cells. (G) Schematic of a plausible regulatory network controlling MTDH expression in MPM. NF-κB induces feed-forward signaling (solid arrow) that is functionally equivalent to a sensor-coupled switch contributing to maintain activation of MTDH once a threshold of stimulation triggers NF- κB. There are secondary positive feedback loops induced by MTDH itself (dotted arrow). Where applicable, data are presented as mean ± SE. * is P < .05 versus parent cell line and/or negative control specimen. MTDH-OE is stable overexpression of and MTDH-KD is stable gene knockdown of MTDH in MPM cells, respectively.
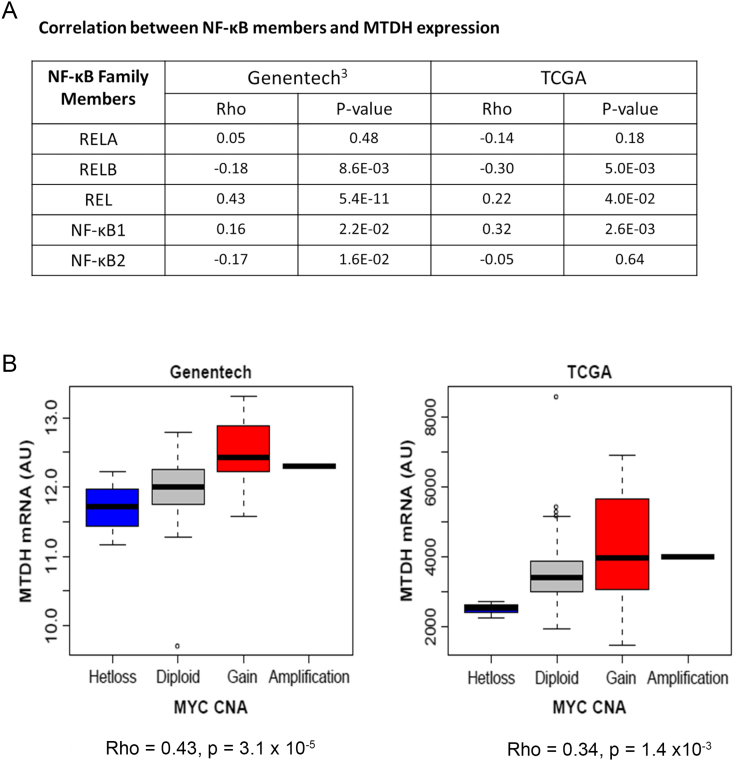
Based on the notion of multiple inflammatory pathways in MPM [29], all NF-κB subunits could plausibly be induced and therefore could bind the MDTH promoter since their DNA-binding domain is very similar [30]. To assess which NF-κB member could likely be involved in regulating MTDH expression, we calculated correlation between the transcript expression of each NF-κB member versus MTDH from TCGA and Genentech [3] MPM datasets. Both REL and NFKB1 had the strongest positive correlations in both independent datasets (Figure 5A). A joint linear model containing all NF-κB members explained 42% of MTDH variation (adjusted R2) in Genentech data [3], and all terms were significant predictors (P < .05) except RELB (P = .075). This result demonstrates that NF-κB members correlate with MTDH expression in human MPM tissue, suggesting they play a role in regulating MTDH in vivo. We performed a similar tissue-level correlation modeling between c-Myc and MTDH using the same external RNA-seq datasets. While c-Myc expression was not significantly associated with MTDH, there was a strong association between c-Myc copy number gains and MTDH mRNA expression consistent with the activator role of c-Myc in MTDH expression (Figure 5B). Overall, this MPM tissue-level analysis supports our in vitro findings of a feed-forward regulatory motif driving MTDH overexpression.
Figure 5.
Tumor tissue analysis of the regulatory network influencing metadherin expression.
(A) Correlation between NF-κB subunits and MTDH expression as derived from tumor tissue datasets. (B) Correlation analysis of c-Myc copy number aberration versus MTDH expression. There was a significant association between c-Myc copy number gains and MTDH mRNA expression (P < .05) among independent patient cohorts. Rho is Spearman's rank correlation coefficient. CNA is copy number aberrations. Genentech RNA-seq data (n = 211). The Cancer Genome Atlas (TCGA)–RNA-seq data (n = 87).
Suppression of Metadherin Exerts an Antitumor Phenotype In Vitro and In Vivo
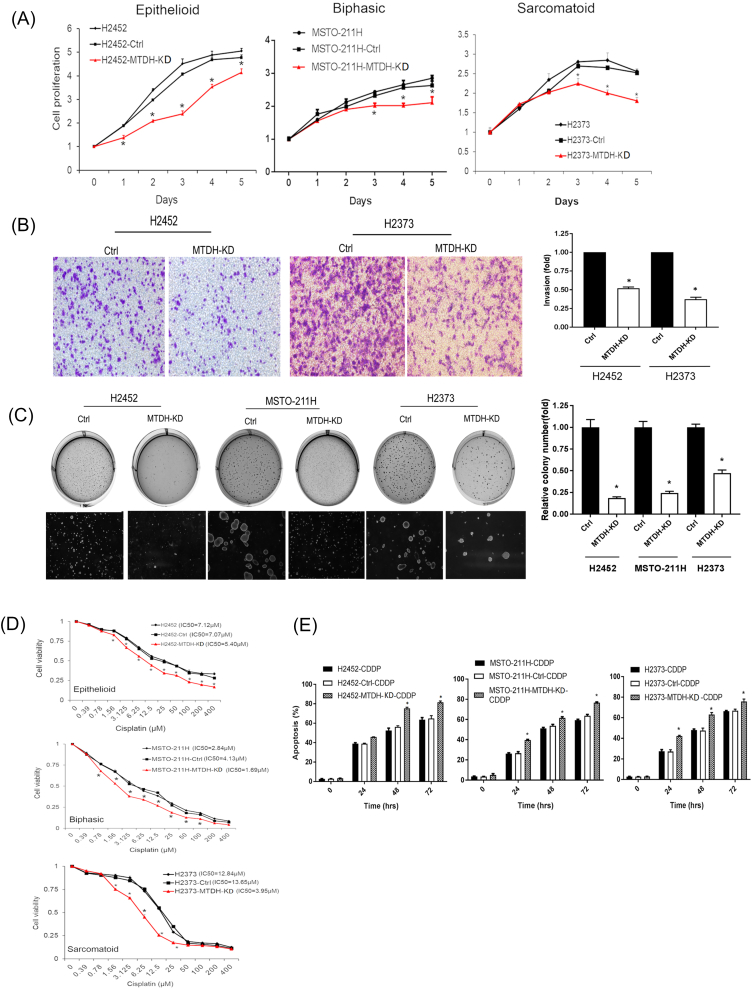
Conversely, we assessed biologic effects associated with knockdown of MTDH expression (Supplementary Figure S5). In a heterogeneous population of stably knockdown MPM cells, we ensured that MTDH transcripts were sufficiently suppressed (Supplementary Figure S2). With MTDH downregulated, cells displayed stunted growth and invasion. MTDH knockdown was also associated with decreased soft agar colony formation by at least 53%. Notably, the IC50 dose of [31] chemotherapy decreased by 3.4-fold in sarcomatoid MPM cells with MTDH knockdown. After 48 hours of chemotherapeutics (IC50) treatment, MTDH knockdown cells were more sensitive to apoptosis by ≥20% (P < .05).
Supplementary Figure S5.
Biologic effects of metadherin suppression in MPM cells.
(A) MTDH knockdown (red) decreased proliferation in MPM cells. On day 3, cell proliferation decreased 47% (H2452), 17% (MSTO-211H), and 20% (H2373) (P < .05). (B) Boyden chamber assay showed that MTDH knockdown decreases invasive capability of MPM cells by 48% (H2452) and 63% (H2373). (C) MTDH knockdown abrogated colony formation. Soft agar assays reveal that relative colony numbers decreased 82% (H2452), 76% (MSTO-211H), and 53% (H2373) (P < .05). (D) Cell viability was assessed at varying concentrations of cisplatin to calculate IC50 values. MTDH knockdown (red) rendered MPM cells more chemosensitive, requiring 24% (H2452), 52% (MSTO-211H), and 70% (H2373) lower doses of drug to reach IC50 levels. (E) Levels of apoptosis were measured after MPM cells were treated with a fixed dose (IC50) of cisplatin. MTDH knockdown significantly augmented apoptosis at 48 hours by 1.5-fold (H2452), 1.2-fold (MSTO-211H), and 1.3-fold (H2373) compared to parental cells. Where applicable, data are presented as mean ± SE. Ctrl is a knockdown scrambled sample. * is P < .05 versus parent cell line and/or negative control specimen. MTDH-KD is stable gene suppression of MTDH in MPM cells.
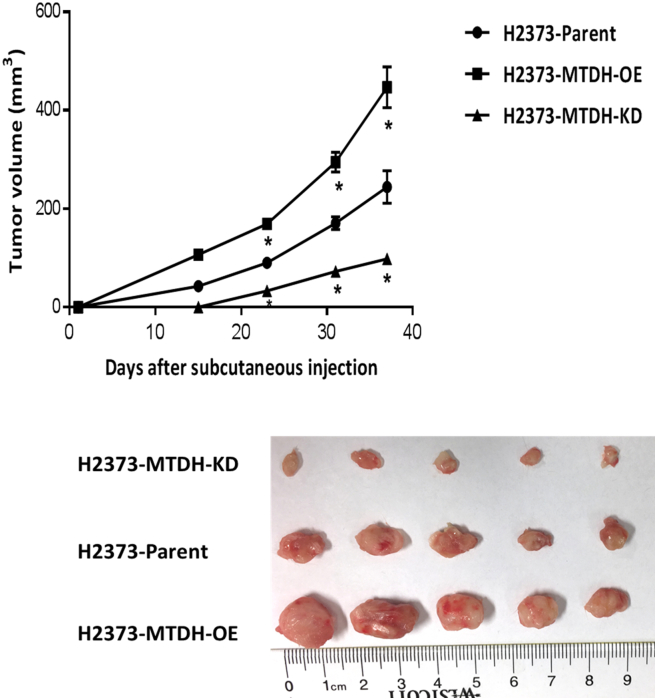
This role of MTDH in MPM is confirmed by in vivo testing. Subcutaneous tumor xenografts were established in NSG mice [16] using H2373 parental cells compared to cells with MTDH overexpressed (OE) and suppressed (KD). During the observation period, xenograft tumor volume was measured at periodic intervals. Notably, MTDH overexpression augmented tumor volume more than two-fold while MTDH suppression inhibited tumor volume more than two-fold as compared to parental MPM cells (P < .05) (Figure 6). The tumor-suppressive effects of targeting MTDH in this xenograft model imply its importance to MPM progression. Collectively, based on its association with prognosis, its overexpression in MPM tissues, and our in vitro/in vivo findings when its expression level is modulated, MTDH appears to play an important maintenance role, augmenting the malignant traits of MPM.
Figure 6.
Modulation of metadherin expression in MPM affects tumor xenograft growth.
Graph depicts tumor growth in a murine xenograft model of subcutaneously injected MPM cells. Three groups of MPM xenografts were assessed. Knockdown of MTDH gene expression resulted in profound tumor inhibition. Photo shows tumor xenografts. * is P < .05 versus parent cell line. MTDH-OE is stable overexpression of and MTDH-KD is stable gene knockdown of MTDH in MPM cells, respectively. Measuring scale is in centimeters.
Discussion
Currently, the normal physiologic role of MTDH remains elusive [32]. MTDH, via a nuclear homing domain, acts as a transcriptional cofactor, but itself does not directly bind DNA or RNA [33]. Accumulating data suggest that MTDH is a critical regulator of malignant traits because of its interactions with a complex network of signaling pathways. In lung cancer, MTDH increases PI3Kp110 and phosphorylation of Akt, all leading to activation of PI3K/Akt signaling while also inhibiting apoptosis by suppressing caspase-3 and enhancing Bcl-2 [34]. In hepatocellular carcinoma, MTDH directly enhanced phosphorylation of ERK and p38 mitogen-activated protein kinases, resulting in hepatocyte transformation and cellular invasiveness [35]. In gastric cancer, MTDH inhibition experiments revealed that it induced coordinated changes in β-catenin, LEF1, and cyclin D1 proteins, thereby establishing its interaction with Wnt signaling to interfere with cell proliferation and augment apoptosis [36]. As a final example, in breast cancer, MTDH knockdown induced upregulation of cell death transcripts TRAIL and BINP3 while inhibiting ALDH3A1 and MET levels, which culminate in enhanced chemosensitivity [37].
Aside from contributing to numerous signaling networks, MTDH expression can be modulated by microRNAs (miRNA or miR), which are short noncoding RNAs implicated in diverse regulatory processes contributing to cancers [38]. In colorectal cancer, ectopic expression of miR-375 exerts tumor suppressive effects by directly targeting MTDH and MAP3K1 to impair cell proliferation and induce apoptosis [39]. Interestingly, in glioma, MTDH itself can induce miR-130b which, in turn, directly regulates levels of PTEN, PPP2CA, and SMAD7 to drive epithelial-to-mesenchymal–like changes and increased invasiveness of tumor cells [40]. Despite an extensive literature regarding the multiple functions of MTDH and its interactions with regulators like miRNA, precise mechanisms as well as the breadth of MTDH involvement across cancers are continuing to be reported.
Here, we identify for the first time a functional role for MTDH in MPM. When MTDH is overexpressed, MPM cells were more resistant to apoptosis, facilitating greater proliferation and invasiveness. When MTDH is suppressed, opposite cellular effects occurred, as confirmed in vivo with murine xenograft results. Our additional unique MPM-specific findings include: a) MTDH expression as a novel, prognostic marker and b) demonstration of NF-κB directly inducing MTDH. The MTDH-driven phenotype we recognized in MPM appears to be, at least in part, increased antiapoptotic mechanism(s) contributing to chemoresistance. When MTDH is forcibly overexpressed, MPM cells demonstrate significantly increased malignant traits like proliferation, invasiveness, and colony formation. Our results suggest that MTDH is central to the overall network of MPM cancer processes.
This novel MPM-associated gene provides insight into how pathogenic transcription factors like NF-κB and c-Myc cooperate. The feed-forward loop is a common regulatory motif that requires an input threshold be reached (inflammatory triggers inducing NF-κB in MPM) before activating downstream elements (c-Myc, MTDH). As the input is ongoing, this regulatory connection serves to reinforce and propagate downstream outputs [28]. Using ChIP analysis, we validate in MPM for the first time that NF-κB directly induces MTDH by binding specific promoter regions. Also, we recognized that NF-κB could regulate c-Myc and confirmed this pathway in MPM cells. Accordingly, the observation that c-Myc overexpression and suppression constructs altered MTDH expression in the same direction suggested to us a feed-forward loop motif (simultaneous direct and indirect pathways between a regulator and target). Furthermore, we found supporting evidence for this regulatory mechanism in MPM tumors (in vivo) via analysis of a large MPM tissue dataset (Genentech [3], n = 211) by modeling correlations resulting in a significantly large adjusted R2 value equivalent to a medium effect size (Cohen's d) [41]. This effect size result is unlikely due to chance. Because NF-κB and MTDH interact with many pathways [21], it is entirely conceivable that there exists alternate regulatory feed-forward loops that could be revealed in systematic analyses, although this is beyond the scope of our current study.
We also observed expected positive feedback loops induced by MTDH. MTDH overexpression in MPM establishes a reinforcing loop between NF-κB and MTDH. This finding is consistent with MTDH interacting with cyclic AMP-responsive element binding protein to induce NF-κB [42]. The other feedback loop occurring with MTDH overexpression is between c-Myc and MTDH. This is consistent with prior studies where MTDH induces c-Myc in a mutually cooperative manner by multiple pathways via Wnt/ β-catenin or promyelocytic leukemia zinc finger [43]. Overall, our in vitro data illustrate that NF-κB inflammatory signaling in MPM is mediated via a regulatory network that could converge on MTDH that, in turn, interacts with other downstream effector pathways [21]. Thus, in MPM, MTDH may represent a novel biomarker of biologic aggressiveness (i.e., chemoresistance driven by increased antiapoptosis).
An inverse relationship between MTDH expression and overall survival has been observed across diverse tumors. A meta-analysis of 827 breast and 651 ovarian cancers demonstrated a higher pooled hazard ratio linking elevated MTDH protein expression with decreased survival [44]. Another meta-analysis of 2999 gastrointestinal cancers (esophageal, colorectal, hepatocellular, etc.) showed MTDH linked to poorer overall and disease-free survivals [45]. Our transcript analysis of MTDH effect on survival in MPM agrees with these other tumors, but we could not assess this effect by protein quantitation due to lack of a large enough MPM cohort clinically annotated.
Conclusions
In summary, we identified a novel mesothelioma-associated gene with an important role in maintaining MPM traits. Tissue analysis validated overexpression of MTDH transcript and protein. MTDH, at least partly, contributes to antiapoptosis and augments chemoresistance as observed in MPM cell assays. Knockdown of MTDH via in vivo experiments underscored the potential of MTDH as a novel MPM therapeutic target. MTDH functions as a network hub that integrates signals from important “undruggable” master transcription factors (NF-κB, c-Myc). This observation supports the notion that MTDH overexpression represents a cancer-specific biomarker [46] since only MPM cells would exhibit upregulation of NF-κB and c-Myc signaling. In turn, MTDH can act as an effector, interacting with myriad pathways such as NF-κB, c-Myc, (shown in MPM), and PI3K-Akt or other yet undiscovered downstream pathways warranting future investigation. Taken together, our results (in vitro and in vivo) coupled with MTDH overexpression in clinical specimens (negative prognostic factor) suggest MTDH as a rational target to explore for better understanding of MPM biology and, in doing so, possibly provide leads towards alternative MPM treatment approaches.
The following are the supplementary data related to this article.
Acknowledgements
We would like to thank Nisan Bhattacharyya for his assistance in lab logistics and James Madigan for critical reading of the final manuscript.
Footnotes
Funding source: This work was supported by the National Institutes of Health Intramural Research Program with funding (ZIA BC 011657) provided to Chuong D. Hoang.
References
- 1.Beckett P, Edwards J, Fennell D, Hubbard R, Woolhouse I, Peake MD. Demographics, management and survival of patients with malignant pleural mesothelioma in the National Lung Cancer Audit in England and Wales. Lung Cancer. 2015;88:344–348. doi: 10.1016/j.lungcan.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Zalcman G, Mazieres J, Margery J, Greillier L, Audigier-Valette C, Moro-Sibilot D, Molinier O, Corre R, Monnet I, Gounant V. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet. 2016;387:1405–1414. doi: 10.1016/S0140-6736(15)01238-6. [DOI] [PubMed] [Google Scholar]
- 3.Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, Gnad F, Nguyen TT, Jaiswal BS, Chirieac LR. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 4.Bononi A, Napolitano A, Pass HI, Yang H, Carbone M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med. 2015;9:633–654. doi: 10.1586/17476348.2015.1081066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Li ZY, Hou XX, Wang X, Luo YH, Ying YP, Chen G. Clinical significance and effect of AEG-1 on the proliferation, invasion, and migration of NSCLC: a study based on immunohistochemistry, TCGA, bioinformatics, in vitro and in vivo verification. Oncotarget. 2017;8:16531–16552. doi: 10.18632/oncotarget.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. doi: 10.1093/carcin/bgp064. [DOI] [PubMed] [Google Scholar]
- 8.Ge X, Lv X, Feng L, Liu X, Gao J, Chen N, Wang X. Metadherin contributes to the pathogenesis of diffuse large B-cell lymphoma. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noch EK, Khalili K. The role of AEG-1/MTDH/LYRIC in the pathogenesis of central nervous system disease. Adv Cancer Res. 2013;120:159–192. doi: 10.1016/B978-0-12-401676-7.00006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Liu J, Lu R, Yu G, Wang X, Zhao Y, Song H, Lin P, Sun X, Yu X. AEG -1 overexpression: a novel indicator for peritoneal dissemination and lymph node metastasis in epithelial ovarian cancers. Int J Gynecol Cancer. 2011;21:602–608. doi: 10.1097/IGC.0b013e3182145561. [DOI] [PubMed] [Google Scholar]
- 11.Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15:5615–5620. doi: 10.1158/1078-0432.CCR-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barone E, Gemignani F, Landi S. Overexpressed genes in malignant pleural mesothelioma: implications in clinical management. J Thorac Dis. 2018;10:S369–S382. doi: 10.21037/jtd.2017.10.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, Linnoila RI, Matthews MJ, Bunn PA, Jr., Carney D. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 17.Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 18.Meng X, Zhu D, Yang S, Wang X, Xiong Z, Zhang Y, Brachova P, Leslie KK. Cytoplasmic metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. J Biol Chem. 2012;287:4485–4491. doi: 10.1074/jbc.C111.291518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X, Thiel KW, Leslie KK. Drug resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res. 2013;120:135–157. doi: 10.1016/B978-0-12-401676-7.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krismann M, Muller KM, Jaworska M, Johnen G. Molecular cytogenetic differences between histological subtypes of malignant mesotheliomas: DNA cytometry and comparative genomic hybridization of 90 cases. J Pathol. 2002;197:363–371. doi: 10.1002/path.1128. [DOI] [PubMed] [Google Scholar]
- 21.Emdad L, Das SK, Dasgupta S, Hu B, Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: signaling pathways, downstream genes, interacting proteins, and regulation of tumor angiogenesis. Adv Cancer Res. 2013;120:75–111. doi: 10.1016/B978-0-12-401676-7.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khuda II, Koide N, Noman AS, Dagvadorj J, Tumurkhuu G, Naiki Y, Komatsu T, Yoshida T, Yokochi T. Astrocyte elevated gene-1 (AEG-1) is induced by lipopolysaccharide as toll-like receptor 4 (TLR4) ligand and regulates TLR4 signalling. Immunology. 2009;128:e700–e706. doi: 10.1111/j.1365-2567.2009.03063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu C, Feng L, Peng H, Yang H, Feng Z, Yang Y. MTDH is an oncogene in multiple myeloma, which is suppressed by bortezomib treatment. Oncotarget. 2016;7:4559–4569. doi: 10.18632/oncotarget.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, Bubici C, Mossman BT, Pass HI, Testa JR. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB–dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci U S A. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Rosa FA, Pierce JW, Sonenshein GE. Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol Cell Biol. 1994;14:1039–1044. doi: 10.1128/mcb.14.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci U S A. 2006;103:17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, Tsao A, Wistuba II. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9:998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossman BT, Shukla A, Heintz NH, Verschraegen CF, Thomas A, Hassan R. New insights into understanding the mechanisms, pathogenesis, and management of malignant mesotheliomas. Am J Pathol. 2013;182:1065–1077. doi: 10.1016/j.ajpath.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 31.Amati M, Tomasetti M, Mariotti L, Tarquini LM, Valentino M, Santarelli L. Assessment of biomarkers in asbestos-exposed workers as indicators of cancer risk. Mutat Res. 2008;655:52–58. doi: 10.1016/j.mrgentox.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Yoo BK, Emdad L, Lee SG, Su ZZ, Santhekadur P, Chen D, Gredler R, Fisher PB, Sarkar D. Astrocyte elevated gene-1 (AEG-1): a multifunctional regulator of normal and abnormal physiology. Pharmacol Ther. 2011;130:1–8. doi: 10.1016/j.pharmthera.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SG, Kang DC, DeSalle R, Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC, the beginning: initial cloning, structure, expression profile, and regulation of expression. Adv Cancer Res. 2013;120:1–38. doi: 10.1016/B978-0-12-401676-7.00001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke ZF, Mao X, Zeng C, He S, Li S, Wang LT. AEG-1 expression characteristics in human non-small cell lung cancer and its relationship with apoptosis. Med Oncol. 2013;30:383. doi: 10.1007/s12032-012-0383-9. [DOI] [PubMed] [Google Scholar]
- 35.Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet JM. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jian-bo X, Hui W, Yu-long H, Chang-hua Z, Long-juan Z, Shi-rong C, Wen-hua Z. Astrocyte-elevated gene-1 overexpression is associated with poor prognosis in gastric cancer. Med Oncol. 2011;28:455–462. doi: 10.1007/s12032-010-9475-6. [DOI] [PubMed] [Google Scholar]
- 37.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 39.Salem SM, Hamed AR, Mosaad RM. MTDH and MAP3K1 are direct targets of apoptosis-regulating miRNAs in colorectal carcinoma. Biomed Pharmacother. 2017;94:767–773. doi: 10.1016/j.biopha.2017.07.153. [DOI] [PubMed] [Google Scholar]
- 40.Tong L, Chu M, Yan B, Zhao W, Liu S, Wei W, Lou H, Zhang S, Ma S, Xu J. MTDH promotes glioma invasion through regulating miR-130b-ceRNAs. Oncotarget. 2017;8:17738–17749. doi: 10.18632/oncotarget.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosco FA, Aguinis H, Singh K, Field JG, Pierce CA. Correlational effect size benchmarks. J Appl Psychol. 2015;100:431–449. doi: 10.1037/a0038047. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava J, Siddiq A, Gredler R, Shen XN, Rajasekaran D, Robertson CL, Subler MA, Windle JJ, Dumur CI, Mukhopadhyay ND. Astrocyte elevated gene-1 and c-Myc cooperate to promote hepatocarcinogenesis in mice. Hepatology. 2015;61:915–929. doi: 10.1002/hep.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou Y, Yu L, Mi Y, Zhang J, Wang K, Hu L. Association of MTDH immunohistochemical expression with metastasis and prognosis in female reproduction malignancies: a systematic review and meta-analysis. Sci Rep. 2016;6 doi: 10.1038/srep38365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Y, Zhang X, Tan Z, Wu P, Xiang X, Dang Y, Chen G. Astrocyte elevated gene-1 as a novel clinicopathological and prognostic biomarker for gastrointestinal cancers: a meta-analysis with 2999 patients. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhatnagar A, Wang Y, Mease RC, Gabrielson M, Sysa P, Minn I, Green G, Simmons B, Gabrielson K, Sarkar S. AEG-1 promoter-mediated imaging of prostate cancer. Cancer Res. 2014;74:5772–5781. doi: 10.1158/0008-5472.CAN-14-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]