Highlights
-
•
The method of building the microgel influencing sugar reduction.
-
•
A single-step way to immobilize crude fungal amylase extracts.
-
•
Microgel as a reusable support of enzymes in starch hydrolysis.
Keywords: Alginate, Chitosan, Beads, Starch, Hydrolysis, Reducing sugars
Abstract
The use of alginate and chitosan polymer in the immobilization of Aspergillus oryzae ATCC 3940 fungal crude enzyme extract (CEE) amylase was presented. The assembly results change in the application of optimal pH and temperature hydrolysis to convert starch to sugar. Bead arrangement in three microgel supports: the internal support phase (IP), the external support phase (EP), and the internal and external support phase (UP). The best results were obtained using IP and EP. Reusing beads evaluated the stability of immobilized enzymes on IP support, remained active and bound during three cycles of reuse. For free and immobilized (IP) activity showed pH ranged from 5.0 to 7.0; optimum thermal enzymatic greater activity at 45 °C. The method of building the microgel influencing sugar reduction, in a single-step way to immobilize crude fungal amylase extracts can be used in industry.
1. Introduction
Enzymes are used for starch hydrolysis (amylase or amylolytic enzymes) have attracted great attention because of biotechnological approaches and economic benefits. There are several types of amylases and the most commonly used in the industry include α-amylases, β-amylases, and glycoamylases [1], [2]. Amylases are found in a wide range of organisms, including plants, animals, and microorganisms, with the latter a suitable source for the production of enzymes in industrial processes because of ease of cultivation [1], [3], [4].
Aspergillus oryzae is known to produce a wide variety of hydrolytic enzymes and has enormous potential to degrade a wide range of compounds [5], [6]. Production of starch hydrolytic enzymes by A. oryzae is induced in presence of starch or maltooligosaccharides and A. oryzae amylases exhibit high efficiency in starch saccharification. [5] A. oryzae is generally recognized as a safe (GRAS) microorganism by the Food and Drug Administration and has been widely used to obtain amylases [7].
In industry, most processes use free enzymes, which are inactivated following completion reaction. The use of immobilized amylases presents advantages in relation to use of free amylases, among which we can mention possibility of reusing is the ability to reuse the biocatalyst for more than one reaction cycle [8] and the use of a continuous process in a bioreactor. In addition, immobilized enzymes increase stability of biocatalyst in relation to variations in pH [9], [10], temperature [11], and the reaction medium, besides facilitating separation and recovery of the biocatalyst and products [12], [13], [14].
Among various options of existing immobilization supports, the advantage of developing a support of alginate-chitosan polymers exists due to gel formed has a greater ability to make connections with the enzymes and synergistically stabilizes polysaccharides chain [15].Thus entrapping the enzymes in the support, so they are not released into the reaction medium during enzymatic hydrolysis [15].
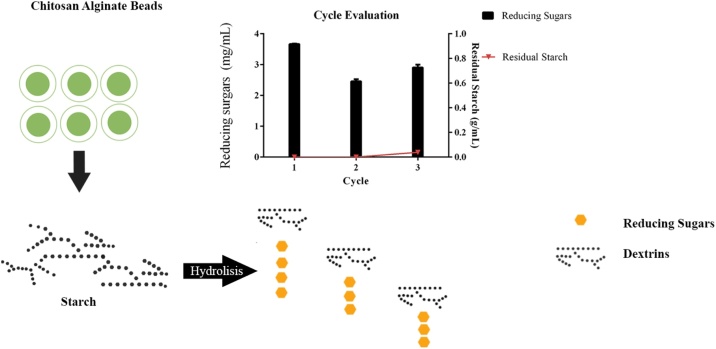
Several types of beads have been described in literature using alginate and chitosan [16], [17]. Many studies have used alginate-chitosan beads to immobilize a wide range of purified enzymes [18], [19]. The present study demonstrates different entrapment procedures dramatically alter enzymatic loading. We report a single-step way to immobilize crude fungal amylase extracts from Aspergillus ATCC 3940 using different preparation procedures of alginate, chitosan, and crude enzymatic extract. These microgels described in this method can be applied as a reusable support of enzymes in starch hydrolysis (Scheme 1).
Scheme 1.
Overall scheme of chitosan alginate bead hydrolysis.
2. Methods
Alginate, chitosan glutaraldehyde 25% and Brilliant Blue G were purchased from Sigma–Aldrich® (USA). Calcium chloride acetic acid and ethanol Vetec® (Brazil). Sabouraud dextrose Prodimol® Biotecnologia S/A (Brazil). Orotophosphoric acid 85% was purchased from Merck®. Rice was purchased from local market.
2.1. Fungal growth and crude enzyme extraction
Aspergillus oryzae ATCC 3940 was used in the present study. The medium usedto obtain A. oryzae spores as Sabouraud dextrose. Inoculum was prepared in 50 mL flasks with 25 mL Sabouraud dextrose medium. After 7 days of incubation at 24 °C, 30 mL of sterile distilled water was added and the spores were suspended. A volume of 10 mL of inoculum (containing 5 × 106 colony forming units/mL) per flask was used.
The cultivation of A. oryzae to obtain crude enzyme extract (CEE) was conducted in autoclaved Erlenmeyer flasks at 121 °C for 20 min containing 25 g of rice and 75 mL of distilled water. After cooling, each flask was inoculated with A. oryzae spores and the inoculated flasks were incubated at a temperature of 24 °C for 7 days. Then, the fungal mass was mixed with 40 mL of 0.1 M acetate buffer (pH 5.0) and homogenized in a blender for 2 min. Following homogenization, obtained mass was centrifuged at 3000 × g for 4 min and the supernatant was filtered using filter paper n°1. Clear supernatant collected was used as CEE containing amylolytic enzymes for evaluation of enzymatic activity [20].
2.2. Preparation of entrapped and covalent binded amylases microgels
Amylase beads were prepared using different order of combination of alginate, chitosan, and crude enzymatic extract (CEE) to evaluate the response of these systems during enzymatic hydrolysis of starch into reducing sugars. The use of glutaraldehyde in immobilization system was also tested to verify the use of this agent in crosslinking the enzymes and supports to increase the stability of the obtained system.
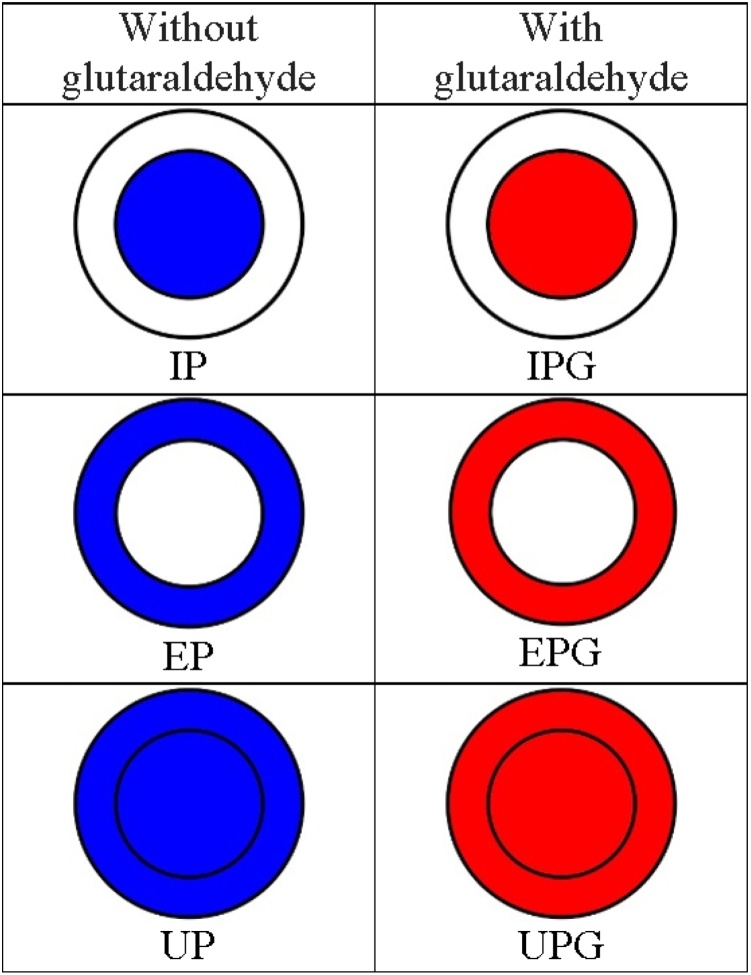
According to order of reactants during preparation of beads, bead received a specific nomenclature to differentiate each model (Fig. 1). In construction of internal phase (IP) support, CEE was mixed with alginate solution (1.89%, w/v) and dripped into a chitosan (0.44%, w/v) and calcium chloride (300 mM) solution. To obtain the external phase (EP) support, the alginate solution (1.89%, w/v) was dripped in CEE, chitosan (0.44%, w/v), and calcium chloride (300 mM) solution. To obtain the unique phase (UP) support, the alginate solution (1.89%, w/v) was dripped in chitosan (0.44%, w/v) and calcium chloride (300 mM) solution. Beads were dried for 7 days at 20 °C. Then, dried beads were immersed in the CEE until the beads swelled. Thus, after swelling of beads, amylases were theoretically present in both internal and external phase of the support (UP). To obtain a chitosan solution for the preparation of all supports, the chitosan was diluted in 2% acetic acid (v/v). Hence, the immobilized amylases assumed IP, EP, and UP conformations in supports in which these conformations assumed by the supports depend on the location of the amylases.
Fig. 1.
Beads produced with different combinations of alginate, chitosan, and amylases (with or without the addition of glutaraldehyde). The colored part of the beads represents the theoretical arrangement of the amylases in the beads. IP, internal phase (amylases were in the internal phase of the support); EP, external phase (amylases were in the external phase of the support); and UP, internal and external phase (amylases were both in the internal and external phase of the support).
The addition of glutaraldehyde crosslinker allowed other conformations of supports: internal phase with glutaraldehyde (IPG), external phase with glutaraldehyde (EPG), and unique phase with glutaraldehyde (UPG). The IPG, EPG, and UPG supports were generated as described above. To obtain IPG and EPG supports, glutaraldehyde 0.5% (v/v) solution was mixed with chitosan (0.44%, w/v) and calcium chloride (300 mM) solution. To obtain the UPG support, glutaraldehyde 1% (v/v) solution was mixed with the CEE.
2.3. Starch hydrolysis assays
Activity was assessed using corn starch (1%, w/v) as substrate and iodometric methods as previously described [21], with modifications. The reaction mix contained 0.5 mL of enzymatic extract and 0.5 mL of soluble starch (1%, w/v) substrate solution. Following incubation at 45 °C for 10 min in a water bath, the reaction was stopped with the addition of 0.5 mL of reactive iodine and 10 mL of distilled water. The absorbance was read at 620 nm in a spectrophotometer.
One unit of activity was defined as the amount of enzyme required to hydrolyze 10 mg of starch in 10 min of reaction under the described assay conditions.
Amylases activity was also assessed using the 3,5-dinitrosalicylic acid (DNS) method [22]. Appropriately diluted enzyme was added to 1 mL of soluble starch (1%, w/v) substrate solution, and following incubation at 45 °C for 10 min in a water bath, reaction was stopped with addition of 2 mL of DNS reagent. Tubes were kept in a boiling water bath for 6 min to develop color and absorbance was read at 540 nm in a spectrophotometer. One unit of activity was defined as mg of reducing sugar liberated in 1 min by 1 mL enzyme under the assay conditions.
The immobilization efficiency (IE, %) was calculated using Eq. (1):
where IE (%) = , where AE is the added enzyme and FE is the free enzyme (1).
The amount of active enzyme in support (active immobilized enzyme, %) was calculated by determining amount of enzyme was available to perform starch hydrolysis, using Eq. (2):
Active immobilized enzyme (%) = , where AIE is the active immobilized enzyme (2).
Overall immobilization yield refers to the percentage of AIE and was calculated using Eq. (3): (%) = AIE/IE where AIE is active immobilized enzyme and IE is the immobilization efficiency (3). In this study, all experiments were done triplicate, and the results are expressed as mean ± SD.
2.4. Protein quantification
Protein concentrations were assessed throughout the process of immobilization and hydrolysis. The supernatant obtained after the process of dripping beads was evaluated to determine the concentration of proteins in beads. Throughout the cycles, the hydrolysis products were evaluated to determine the presence of proteins throughout the bead reuse process [23].
2.5. Reuse of immobilized amylases
To test reuse of immobilized amylases, starch solution (1%, w/v) reactions were conducted in a working volume of 4 mL. Hydrolysis processes were performed at 45 °C for 1 h using 0.22 g of immobilized amylase per assay. Following incubation, beads were removed from the reaction mixture and reused after had been washed three times with distilled water. After each cycle, reducing sugars and the residual starch of amylases were measured.
2.6. pH and temperature of entrapped amylases
Effect of pH and temperature variation in free and immobilized amylases at different pH values were determined measuring residual activity of enzymes incubated for 40 min in water bath at 45 °C in the pH range of 3.0–8.0. After cooling, residual saccharifying amylases activity was measured. Activity of the not incubated enzyme was taken as 100%. The tested buffers were at 0.1 M, acetate (pH 3.0–5.5) and phosphate (pH 6.0–8.0). The thermal stability of both the free and immobilized amylases was determined by measuring the residual activity of the enzymes incubated for 40 min in acetate buffer (0.1 M pH 5.0) in a water bath at temperatures ranging from 10 to 80 °C. After cooling, residual saccharifying activity of the amylases was measured. Also the activity of the not incubated enzyme was taken as 100%.
2.7. FTIR characterization
Infrared spectra of the chitosan-alginate beads were recorded with a FTIR spectrophotometer IR Prestige 21 (Shimadzu®, Japan) and scanned from 4000 to 450 cm−1 at room temperature.
Beads were dried for 72 h at 50 °C. Using same procedure crude enzymatic extract was dripped into KBr. Dried beads were mixed and dried for 24 h. The amount used was 0.2 g of KBr and 0.05 g of each sample
2.8. Scanning electron microscopy (SEM)
SEM was utilized to study the external morphology (size, shape and surface) of the prepared beads. The beads were harvested by filtering it with filter paper, spread on petri dish and dried for 2 days at room temperature and stored in an air-tight container for further use. Samples were also crosslinked with glutaraldehyde. Randomly selected beads were placed on double-sided copper conductive tape fixed on aluminum stubs. The beads were then sputter-coated with a thin layer of gold in a vacuum for 45 s at 20 mA using a coating unit to make it electrically conductive and was analyzed with a SEM instrument (FEI Quanta 250). Beads diameters were measured using a ruler, and their mean value found using scale on the SEM with an Electronscan (Philips Quanta 250) operating at 10–20 kV was used for these measurements in the traditional mode secondary electron detector – SE and Backscattered Electron Detector – BSED detector.
3. Results and discussion
3.1. Immobilization of amylases in microgel
Enzymatic immobilization is widely studied area to replace the use of free enzymes that have a number of disadvantages such as high cost and low stability. The technique of enzymatic immobilization makes it possible to separate enzymes within the products produced through the biochemical process and can be reused in several cycles, because they are retained in matrices or covalently attached to some surface [24].
In this study, different combinations of supports (alginate and chitosan) and crude enzyme extract (amylases) were tested for immobilization. The use of glutaraldehyde in the immobilization system has also been tested to verify the possibility of this agent to perform the crosslinking between the enzymes and supports to increase the stability of the system. Thus, different compositions were evaluated in the preparation of beads: presence or absence of glutaraldehyde and the theoretical arrangement of the amylases in the beads (IP or EP) (Fig. 1).
3.2. Starch hydrolysis using immobilized amylases on different microgel and immobilization efficiency
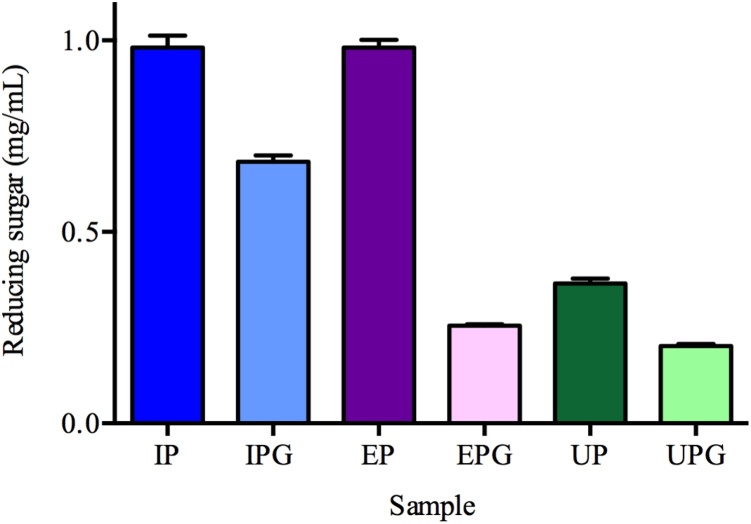
The best results for the conversion of 1% starch solution (w/v) into reducing sugars were obtained using IP and EP supports (Fig. 2). The UP support showed little conversion of starch into reducing sugars, likely due to the low concentration of amylases in this support.
Fig. 2.
Concentrations of reducing sugars obtained in the enzymatic hydrolysis of 1% (w/v) starch solutions using the differentimmobilized amylase beads.
The use of glutaraldehyde decreased the activity of enzymes in the IPG and EPG supports compared with the IP and EP supports, resulting in a lower conversion of starch into reducing sugars. According to Migneault et al. [25], enzymatic activity is inversely proportional to the concentration of glutaraldehyde used because extensive crosslinking may result in a distortion of the enzyme structure (i.e., conformation of the active site). This distortion, the accessibility and accommodation of the substrate may be reduced, thus affecting the retention of biological activity. Although partial enzymatic inactivation due to chemical modification is often unavoidable, in most cases sufficient catalytic activity is retained. Table 1 and Fig. 3 demonstrate the results of the amylase immobilization efficiency and overall immobilization yields on different beads. The maximum immobilization efficiency yield achieved was 97% and 92% on IP and IPG beads, respectively. The higher concentration of amylases in these supports resulted in a greater conversion of starch into reducing sugars (Fig. 2).
Table 1.
Activity and immobilization yields (%) of amylases on different assembled influenced by microgel beads.
| Bead assembly | Immobilization efficiency (%) | Active immobilized enzyme (%) | Overall active immobilization (%) |
|---|---|---|---|
| IP | 97.38 ± 0.03 | 16.72 ± 0.52 | 17.18 ± 0.54 |
| IPG | 92.32 ± 0.76 | 11.49 ± 0.28 | 12.45 ± 0.36 |
| EP | 35.76 ± 1.67 | 2.27 ± 0.04 | 6.38 ± 0.33 |
| EPG | 39.87 ± 8.36 | 0.55 ± 0.00 | 1.43 ± 0.32 |
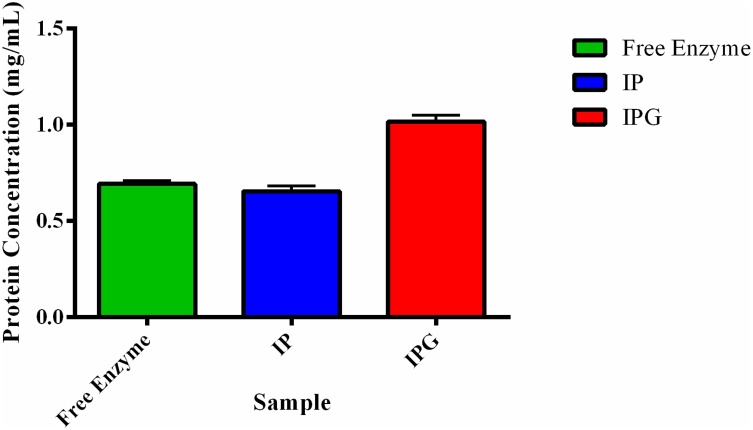
Fig. 3.
Protein concentration in the supernatant after the beads dripping process.
Overall immobilization describes available enzyme during hydrolysis process. Although high immobilization rates are counted, not all enzymes are available for process of starch breaking. The immobilization efficiencies achieved with IP and IPG beads were higher than those of Yazgan et al. [26], which reported 75.7% efficiency of commercial α-amylase Termamyl® immobilized on chitosan beads. Sharma et al. [27] reported α-amylase from A. oryzae was successfully immobilized in calcium agar beads with a high immobilization efficiency of 80%. However, the overall active immobilization in the IP support of 17.18% was lower than the 24% reported by Yazgan et al. [26]. The lower active immobilized enzyme in the support may have resulted in the lower overall immobilization rate, even with a high efficiency of immobilization. Considering the high immobilization efficiency on IP and IPG supports, these beads were chosen for further experiments because amylases arranged in the internal phase of the support (alginate coated externally with chitosan) are more protected from the external environment (e.g., changes in pH and temperature leading to a decrease or loss of enzyme activity) and thus increase the chance of obtaining greater operational stability of the enzymes, which is a requirement for enzyme reusability.
3.3. Protein quantification
The concentrations of IP protein and free enzyme were both the same, with an IP of 0.65 mg/mean and 0.69 mg/mean for CEE, with no statistical difference (p ≤ 0.05) obtained by the Bradford method.
IPG supernantant had a higher concentration of proteins. SEM images show a greater number of fissures, displaying changes made by glutaraldehyde. Due to the changes in macro and micro properties, all these alterations can indeed reduce water absorption, permeability and mechanical and chemical properties [28].
Diffusion experiments show changes in ion permeability. These results indicate that glutaraldehyde chemical modification makes chitosan more hydrophobic [28].
This hydrophobicity reduces protein and support interaction. Chitosan and alginate biocompatibility lessen negative impact on enzyme structure and properties and thus retain high catalytic activity for immobilized proteins.
The presence of reactive functional groups, such as hydroxyl, but also amine and carbonyl functional groups, allows direct reaction/or interaction between the enzyme and the matrix [29].
Glutaraldehyde had altered chitosan and alginate on surface of microgel. This little changes in this polymer, displaying the concentration of protein at 1.05 mg/mL as well.
3.4. Reusability of microgel amylases
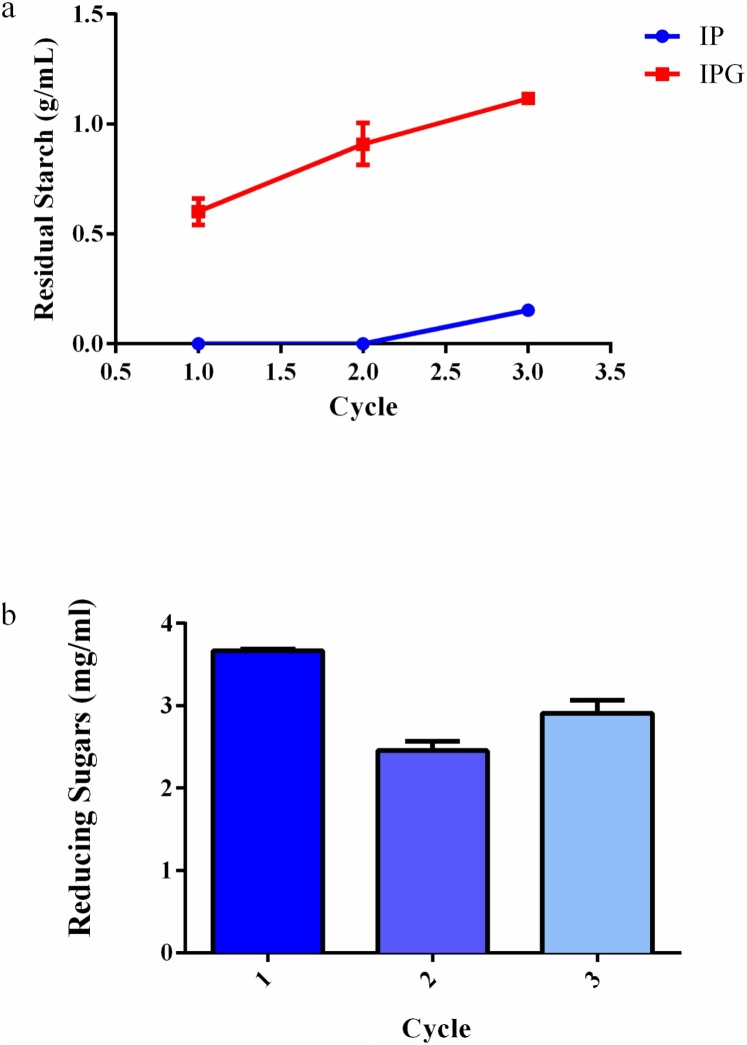
Operational stability of the immobilized amylases in the IP and IPG supports was evaluated reusing beads for three cycles. In Fig. 4a, the dextrinizing activity of immobilized amylases was measured by decreasing the concentration of the starch, the hydrolysis of which results in dextrins. Amylases immobilized on the IP support during two initial cycles converted the entire starch solution into dextrins and in the third cycle, hydrolysis of majority of starch into dextrins was observed. However, with the IPG support, use of glutaraldehyde decreased amylase activity, resulting in reduced hydrolysis of starch solution into dextrins, and in third cycle, no enzymatic activity was observed. Fig. 4b shows the concentration of reducing sugars obtained in enzymatic hydrolysis of 1% starch solution (w/v) during three cycles of reuse using the IP support.
Fig. 4.
Reusability of immobilized amylases. (a) Concentration of residual starch obtained following enzymatic hydrolysis of a 1% (w/v) starch solution during three cycles of reuse with IP and IPG beads. ((b) Concentration of reducing sugars obtained following enzymatic hydrolysis of a 1% (w/v) starch solution during three cycles of reusewith IP beads.
During the cycles of reuse of amylases immobilized on an IP support, the following mean values of reducing sugars were obtained: 3.6, 2.6, and 2.8 mg/mL in first, second, and third cycles, respectively. Therefore, in three cycles of reuse, enzymes remained active and bound to the support, efficiently performing the conversion of starch into reducing sugars. In the work of Yazgan et al. [26], commercial α-amylase Termamyl® immobilized on chitosan beads was reused for 10 cycles and showed a reduction of activity from 100% in the first cycle to 50% in the final cycle. In the work of Sharma et al. [27], immobilized enzymes were reused for seven cycles and the results showed reusability was highly dependent on the use of calcium chloride. Calcium agar beads were more stable and retained approximately 78% activity at the end of six cycles, while the agar beads retained only 20% activity at the end of the sixth cycle. Since the agar beads are porous in nature compared to calcium agar beads, the physical loss of enzyme from carrier is high, which leads to a greater decrease in activity compared to calcium agar beads with repeated use. According to Ertan et al., decreased enzymatic activity during repeated use of immobilized enzymes may be due to enzyme denaturation and/or physical loss of the enzyme from the support [30].
3.5. pH and temperature-optimum of microgel amylases
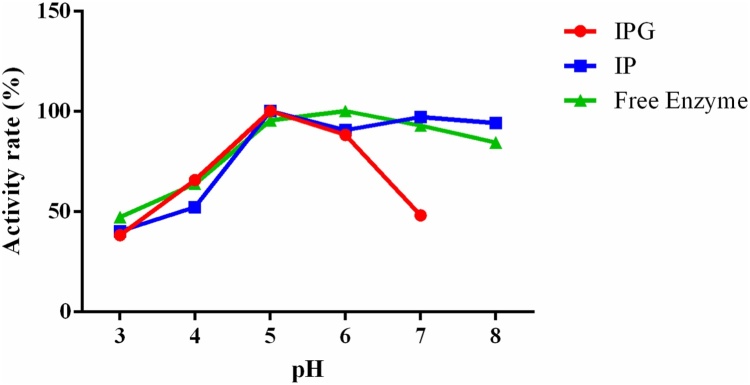
In assays of optimum pH (Fig. 5), both free and immobilized amylases showed greater activity at pH 5.0. For free amylases and amylases immobilized on IP support, a pH range of 5.0–7.0 was obtained. However, beads immobilized on an IPG support showed decreased activity at pH 7.0. Kumar et al. [31] reported similar results and found the optimal pH of free α-amylase from A. oryzae was 5.5 and optimal pH of immobilized enzymes on calcium alginate beads was 6.0. Sharma et al. (2014) [27] reported similar results and found optimal pH of the free and immobilized amylase (on agar beads) from A. oryzae was 6.0.
Fig. 5.
Effect of pH on IP, IPG beads and Free Enzyme (CEE) bead catalyzed reactions.
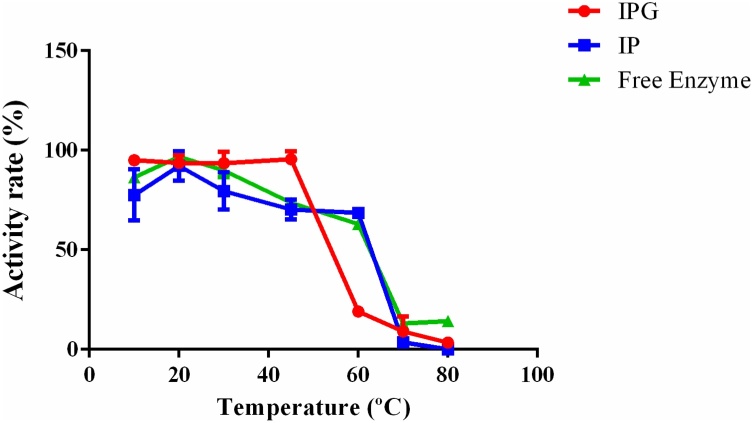
In assays of optimum temperature (Fig. 6), both free and immobilized amylases showed greater activity at 45 °C. At 60 °C, free and immobilized amylases on an IP support still maintained >50% activity, while the IPG support showed a large decrease in activity, maintaining less than 20% of its activity. At 70–80 °C, the free and immobilized enzymes lost their activity.
Fig. 6.
All testes were conduced by hydrolisis of starch at temperatures ranging from 10 to 80 °C.
Kumar et al. [31] described the midpoint of thermal inactivation, where activity is diminished by 50%, for both free and immobilized α-amylase in calcium alginate beads from A. oryzae was between 57 and 63 °C. Sharma et al. [27] reported similar results and described a midpoint of thermal inactivation for both free and immobilized α-amylase on calcium agar beads from A. oryzae was between 57 and 65 °C.
3.6. Fourier-transform infrared spectroscopy (FTIR) analysis of microgel amylase
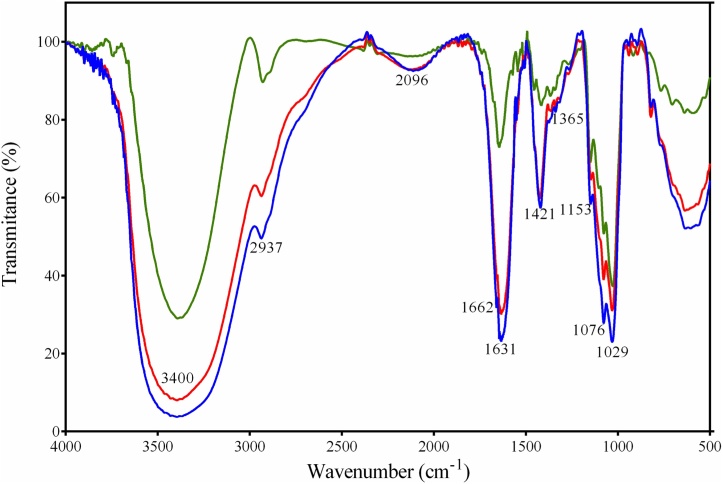
FTIR spectroscopy is typically used for qualitative analysis of organic functional groups. As shown in Fig. 7, the samples IP, (blue) and IPG (red) exhibited a significant broad absorption peak between 3000 and 3700 cm−1, which was assigned to the stretching vibration of the hydroxyl groups of chitosan, alginate, and/or the presence of water and overlapped with the -NH2 stretching vibration peak of chitosan. The peak at approximately 2930 cm−1 was attributed to the sp3 C–H stretching vibration of the chitosan backbone. The efficiency of immobilization procedures in relation to enzyme incorporation on the alginate/chitosan matrix can be confirmed by the presence of characteristics peaks at 1665 cm−1, indicating an amide group (CONH). The peaks at 1631 were attributed to the most typical absorption bands of chitosan, which are related to N–H bending vibrations and C=O stretching, while the band at 1602 cm−1 was ascribed to C=C bending vibrations. The band region of 1345–1421 cm−1 can be ascribed to the bending vibration of C–H methyl groups. According to Simsek-Ege et al. the band observed at 1420 cm−1 in the alginate-chitosan mixture may be attributed to the interaction of –NH + 3 of chitosan with –COO– of alginate [32]. The polysaccharide also exhibited a specific band at 1030–1240 cm−1, which was dominated by ring vibrations overlapped with stretching vibrations of (C–O–H) side groups and the (C–O–C) glycosidic band vibration. The peak at 1082 cm−1 was due to asymmetric C–O–C bridge stretching in the ring of chitosan. As described by Sankalia et al. [33], with the incorporation of enzyme, the spectrum of beads was similar in chitosan–alginate blank beads, except for a shift at specific wavelengths [33]. The shifts in the wavelength at approximately 2929 cm-−1 (C–H stretching), 1642 cm−1 (C=O stretching of secondary amide), and 1080 cm−1 (C–O–C stretching of cyclic ether) may be explained by the interaction of α-amylase with chitosan. Even following serial washing with distilled water, enzymes were maintained, although any signal of a specific bond, indicating binding between support and alginate, or N–N bonds formed, which is the main reaction mediated by glutaraldehyde the main phase in represented conformation enzymes.
Fig. 7.
Internal phase, internal phase and glutaraldehyde, and free enzyme are indicated in blue, red, and green, respectively. Representative FTIR results of the samples demonstrated Free enzyme (Crude enzymatic extract – CEE), shown in green, is retained in the carrier, both in IP (blue) and IPL (red).
3.7. SEM analysis
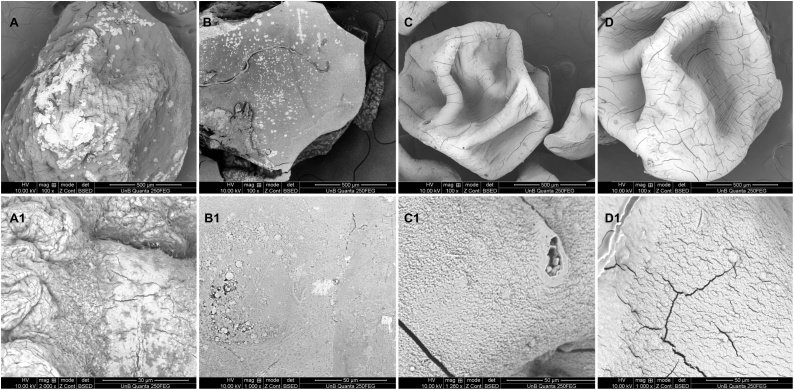
The surface morphology of microgel beads was investigated by SEM analyses as shown in Fig. 8.
Fig. 8.
External morphology of the all formulated beads operating at 10–20 kV, with Back-scattered Electron Detector – BSED. (A) IP-magnification of 100×, (A1) IP-magnification of 1000×; (B) IPG-magnification of 100×, (B1) IPG-magnification of 1000×; (C) EP-magnification of 100×, (C1) EP-magnification of 1000× and (D) EPG-magnification of 100×, (D1) EPG-magnification of 1000×.
Fig. 8 indicated surface morphology of IP, IPG, EP, EPG beads, respectively. As shown in Fig. 8A/A1, the scale bar was 500 μ m and 30 μ m, respectively. The amylase entrapment in IP and IPG produced roughly spherical in shape gel beads. The formation of thick coat on outer surface of the IP and IPG beads could indicate CEE was completely entrapped into interior polymer network.
The surface of the EP and EPG beads were fold and stack due to shrank substantially after drying. During cross-linking process, a large amount of water would be expelled from the cross linked polymer matrices. A portion of water would be expelled into the inner core. The folded surface was resulted from removal of water in the inner core. While, the EP beads had a smoother surface compared to the EPG beads, which was due to the coat of amylase at the surface showed in Fig. 8C1.
4. Conclusions
Distinctive polysaccharide solubilization was performed in this study, leading to assemblies with different enzyme loading, demonstrating that even a simple entrapment method can be explored with different immobilization procedures. Optimal results for the conversion of starch into sugars were obtained using the IP support. The maximum immobilization efficiency yield achieved was 97.38%, with an overall immobilization of 17.18%. During three cycles of reuse of the immobilized amylase, enzymes remained active and bound to the IP support, efficiently converting starch into sugar. Assays of pH and thermal optimization found that immobilized amylase showed greater activity at 45 °C and showed a pH range of 5.0–7.0, with greater activity at pH 5.0. The immobilized system could be improved by increasing concentration of amylase in IP support, which would likely result in higher conversion rates of starch into sugars.
Acknowledgments
The authors are grateful to CNPq, FINATEC, CAPES- Finance Code 001 and FAPDF for financial support of this research.
Contributor Information
Igor A. de Souza, Email: iasphar@gmail.com.
Daniela C. Orsi, Email: danielacastilhoorsi@gmail.com.
Anderson J. Gomes, Email: ajgomes@unb.br.
Claure N. Lunardi, Email: clunardi@unb.br.
References
- 1.Singh S., Singh S., Bali V., Sharma L., Mangla J. Production of fungal amylases using cheap, readily available Agriresidues, for potential application in textile industry. BioMed Res. Int. 2014;2014:1–9. doi: 10.1155/2014/215748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Souza P.M., de Oliveira e Magalhaes P. Application of microbial α-amylase in industry – a review. Braz. J. Microbiol. 2010;41(4):850–861. doi: 10.1590/S1517-83822010000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saleem A., Ebrahim M.K. Production of amylase by fungi isolated from legume seeds collected in Almadinah Almunawwarah, Saudi Arabia. J. Taibah Univ. Sci. 2014;8(2):90–97. [Google Scholar]
- 4.Zaferanloo B., Bhattacharjee S., Ghorbani M.M., Mahon P.J., Palombo E.A. Amylase production by Preussia minima, a fungus of endophytic origin: optimization of fermentation conditions and analysis of fungal secretome by LC–MS. BMC Microbiol. 2014;14(1):55. doi: 10.1186/1471-2180-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machida M., Yamada O., Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 2008;15(4):173–183. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H., Bultema J.B., Dijkhuizen L., van Leeuwen S.S. Reaction kinetics and galactooligosaccharide product profiles of the β-galactosidases from Bacillus circulans, Kluyveromyces lactis and Aspergillus oryzae. Food Chem. 2017;225:230–238. doi: 10.1016/j.foodchem.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki K., Tanaka M., Konno Y., Ichikawa T., Ichinose S., Hasegawa-Shiro S., Shintani T., Gomi K. Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2015;99(4):1805–1815. doi: 10.1007/s00253-014-6264-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X., Hong H., Cheng X., Liu S., Deng T., Guo Z., Wu Z. One-step purification and immobilization of extracellularly expressed sortase A by magnetic particles to develop a robust and recyclable biocatalyst. Sci. Rep. 2017;7(1):6561. doi: 10.1038/s41598-017-06856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virgen-Ortíz J.J., dos Santos J.C.S., Ángel Berenguer-Murcia O., Barbosa R.C., Rodrigues R., Fernandez-Lafuente Polyethylenimine: a very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B. 2017;5(36):7461–7490. doi: 10.1039/c7tb01639e. [DOI] [PubMed] [Google Scholar]
- 10.Luo H., Zhu L., Chang Y., Liu X., Liu Z., Sun H., Li X., Yu H., Shen Z. Microenvironmental pH changes in immobilized cephalosporin C acylase during a proton-producing reaction and regulation by a two-stage catalytic process. Bioresour. Technol. 2017;223:157–165. doi: 10.1016/j.biortech.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 11.Dwevedi A. Springer International Publishing; Cham: 2016. Enzyme Immobilization. [Google Scholar]
- 12.Datta S., Christena L.R., Rajaram Y.R.S. Enzyme immobilization: an overview on techniques and support materials. 3 Biotech. 2013;3(1):1–9. doi: 10.1007/s13205-012-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohamad N.R., Marzuki N.H.C., Buang N.A., Huyop F., Wahab R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015;29(2):205–220. doi: 10.1080/13102818.2015.1008192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirisha V.L., Jain A., Jain A. 2016. Enzyme Immobilization. [Google Scholar]
- 15.Taqieddin E., Amiji M. Enzyme immobilization in novel alginateächitosan core-shell microcapsules. Biomaterials. 2004;25(10):1937–1945. doi: 10.1016/j.biomaterials.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 16.Zusfahair, Ningsih D.R., Kartika D., Fatoni A., Zuliana A.L. Bacillus thuringiensis HCB6 amylase immobilization by chitosan beads. IOP Conf. Ser.: Mater. Sci. Eng. 2017;172:012068. [Google Scholar]
- 17.Gangadharan D., Nampoothiri K.M., Sivaramakrishnan S., Pandey A. Immobilized bacterial α-amylase for effective hydrolysis of raw and soluble starch. Food Res. Int. 2009;42(4):436–442. [Google Scholar]
- 18.Thu T.T.M., Krasaekoopt W. Encapsulation of protease from Aspergillus oryzae and lipase from Thermomyces lanuginoseus using alginate and different copolymer types. Agric. Nat. Resour. 2016;50(3):155–161. [Google Scholar]
- 19.Cubides-Roman D.C., Pérez V.H., de Castro H.F., Orrego C.E., Giraldo O.H., Silveira E.G., David G.F. Ethyl esters (biodiesel) production by Pseudomonas fluorescens lipase immobilized on chitosan with magnetic properties in a bioreactor assisted by electromagnetic field. Fuel. 2017;196:481–487. [Google Scholar]
- 20.Zambare V. Solid state fermentation of Aspergillus oryzae for glucoamylase production on agro residues. Int. J. Life Sci. 2010;4:16–25. [Google Scholar]
- 21.de Oliveira A.P.A., Silvestre M.A., Garcia N.F.L., Alves-Prado H.F., Rodrigues A., da Paz M.F., Fonseca G.G., Leite R.S.R. Production and catalytic properties of amylases from Lichtheimia ramose and Thermoascus aurantiacus by solid-state fermentation. Sci. World J. 2016;2016:1–10. doi: 10.1155/2016/7323875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31 [Google Scholar]
- 23.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Rahim S.N.A., Sulaiman A., Hamzah F., Hamid K.H.K., Rodhi M.N.M., Musa M., Edama N.A. Enzymes encapsulation within calcium alginate-clay beads: characterization and application for cassava slurry saccharification. Proc. Eng. 2013;68:411–417. [Google Scholar]
- 25.Migneault I., Dartiguenave C., Bertrand M.J., Waldron K.C. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques. 2004;37(5) doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 26.Yazgan I., Turner E.G., Cronmiller L.E., Tepe M., Ozturk T.K., Elibol M. Modification of chitosan-bead support materials with l-lysine and l-asparagine for Î-amylase immobilization. Bioprocess Biosyst. Eng. 2018;41(3):423–434. doi: 10.1007/s00449-017-1876-x. [DOI] [PubMed] [Google Scholar]
- 27.Sharma M., Sharma V., Majumdar D.K. Entrapment of -amylase in agar beads for biocatalysis of macromolecular substrate. Int. Scholar. Res. Not. 2014;2014:8. doi: 10.1155/2014/936129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beppu M.M., Vieira R.S., Aimoli C.G., Santana C.C. Crosslinking of chitosan membranes using glutaraldehyde: effect on ion permeability and water absorption. J. Membr. Sci. 2007;301(1-2):126–130. [Google Scholar]
- 29.Zdarta J., Meyer A., Jesionowski T., Pinelo M. A general overview of support materials for enzyme immobilization: characteristics, properties, practical utility. Catalysts. 2018;8(2):92. [Google Scholar]
- 30.Ertan F., Yagar H., Balkan B. Optimization of αâamylase immobilization in calcium alginate beads. Prep. Biochem. Biotechnol. 2007;37(3):195–204. doi: 10.1080/10826060701386679. [DOI] [PubMed] [Google Scholar]
- 31.Kumar R.S.S., Vishwanath K.S., Singh S.A., Rao A.A. Entrapment of α-amylase in alginate beads: single step protocol for purification and thermal stabilization. Process Biochem. 2006;41(11):2282–2288. [Google Scholar]
- 32.Simsek-Ege F.A., Bond G.M., Stringer J. Polyelectrolyte complex formation between alginate and chitosan as a function of pH. J. Appl. Polymer Sci. 2003;88(2):346–351. [Google Scholar]
- 33.Sankalia M.G., Mashru R.C., Sankalia J.M., Sutariya V.B. Reversed chitosanâalginate polyelectrolyte complex for stability improvement of alpha-amylase: optimization and physicochemical characterization. Eur. J. Pharm. Biopharm. 2007;65(2):215–232. doi: 10.1016/j.ejpb.2006.07.014. [DOI] [PubMed] [Google Scholar]