Abstract
Some driver gene mutations, including epidermal growth factor receptor (EGFR), have been reported to be involved in expression regulation of the immunosuppressive checkpoint protein programmed cell death ligand 1 (PD‐L1), but the underlying mechanism remains obscure. We investigated the potential role and precise mechanism of EGFR mutants in PD‐L1 expression regulation in non‐small‐cell lung cancer (NSCLC) cells. Examination of pivotal EGFR signaling effectors in 8 NSCLC cell lines indicated apparent associations between PD‐L1 overexpression and phosphorylation of AKT and ERK, especially with increased protein levels of phospho‐IκBα (p‐IκBα) and hypoxia‐inducible factor‐1α (HIF‐1α). Flow cytometry results showed stronger membrane co‐expression of EGFR and PD‐L1 in NSCLC cells with EGFR mutants compared with cells carrying WT EGFR. Additionally, ectopic expression or depletion of EGFR mutants and treatment with EGFR pathway inhibitors targeting MEK/ERK, PI3K/AKT, mTOR/S6, IκBα, and HIF‐1α indicated strong accordance among protein levels of PD‐L1, p‐IκBα, and HIF‐1α in NSCLC cells. Further treatment with pathway inhibitors significantly inhibited xenograft tumor growth and p‐IκBα, HIF‐1α, and PD‐L1 expression of NSCLC cells carrying EGFR mutant in nude mice. Moreover, immunohistochemical analysis revealed obviously increased protein levels of p‐IκBα, HIF‐1α, and PD‐L1 in NSCLC tissues with EGFR mutants compared with tissues carrying WT EGFR. Non‐small‐cell lung cancer tissues with either p‐IκBα or HIF‐1α positive staining were more likely to possess elevated PD‐L1 expression compared with tissues scored negative for both p‐IκBα and HIF‐1α. Our findings showed important roles of phosphorylation activation of AKT and ERK and potential interplay and cooperation between NF‐κB and HIF‐1α in PD‐L1 expression regulation by EGFR mutants in NSCLC.
Keywords: epidermal growth factor receptor, Hypoxia‐inducible factor‐1α, NF‐κB, non‐small‐cell lung cancer, programmed cell death ligand 1
Abbreviations
- ADC
adenocarcinoma
- APC
allophycocyanin
- EGFR
epidermal growth factor receptor
- HIF‐1α
hypoxia‐inducible factor‐1α
- IKK
IκB kinase
- NSCLC
non‐small‐cell lung cancer
- NF‐κB
nuclear factor‐κB
- p‐
phosphorylated
- PD‐1
programmed cell death 1
- PD‐L1
programmed cell death ligand 1
- PE
phycoerythrin
- SCC
Squamous cell carcinoma
- TKI
tyrosine kinase inhibitor
1. INTRODUCTION
Lung cancer is the leading cause of cancer‐related death around the world. There are approximately 1.30 million new cases and 1.20 million deaths due to lung cancer every year. 1 Non‐small‐cell lung cancer accounts for approximately 80% of lung cancer cases.2 Since the beginning of the 21st century, molecular targeting therapies such as EGFR‐TKIs have shown promising curative effects in NSCLC patients. However, the overall 5‐year survival rate of NSCLC has not apparently been improved due to primary or secondary drug resistance.3, 4 Therefore, new treatments for lung cancer are urgently needed.
Recently, immunotherapies targeting the PD‐1 coinhibitory receptor and its adaptor‐programmed death ligand 1 (PD‐L1) by mAbs have represented a major breakthrough in the treatment of various advanced tumors, including NSCLC.5, 6 Programmed cell death‐1 belongs to the CD28 family and is a type I transmembrane protein mainly expressed on activated T cells. Its ligand PD‐L1 belongs to the B7 family and is widely expressed on dendritic cells, macrophages, activated T and B cells, and non‐immune cells including cancer cells.7, 8 As important immune checkpoint molecules, PD‐1/PD‐L1 interaction suppresses the growth and function of effector T cells by inducing T cell apoptosis, anergy, and exhaustion and regulating the secretion of various cytokines.9, 10 Tumor cells with PD‐L1 overexpression have been reported to be related to aggressive behavior and worse disease control and treatment outcomes.11, 12, 13
A growing amount of evidence has elucidated the intrinsic and extrinsic mechanisms of PD‐L1 expression regulation in cancer cells.14 Inflammatory cytokines, such as IFN‐γ, can induce PD‐L1 expression through the MEK/ERK or JAK/STAT pathway.15, 16 The EML4‐ALK fusion gene and loss of Lkb1 and PTEN have been reported to be involved in intrinsic regulation of PD‐L1 expression in NSCLC.17, 18 Mutated EGFR is the most important driver gene in NSCLC and up to 47.9% of Asian patients harbor EGFR, sensitizing mutations to EGFR‐TKIs.19 In 2013, Akbay et al20 first showed that EGFR mutant in bronchial epithelial cells induced PD‐L1 expression to facilitate immune escape in EGFR‐driven lung tumors. In 2015, D'Incecco et al21 reported that positive PD‐L1 expression was significantly associated with EGFR mutations in a cohort of 125 NSCLC patients. However, the potential role and precise molecular mechanism of PD‐L1 expression regulation by EGFR mutants remain to be explored.
In the present study, we investigated the EGFR status, activation of pivotal EGFR signaling cascades, and PD‐L1 expression in a panel of NSCLC cells and observed apparent associations between PD‐L1 overexpression and phosphorylation activation of ERK and AKT, especially with increased protein levels of p‐IκBα and HIF‐1α. Additionally, we undertook flow cytometry analysis to examine the cell surface expression of EGFR and PD‐L1 in NSCLC cells with different EGFR status. Moreover, ectopic expression or depletion of WT EGFR and EGFR mutants or specific pathway inhibitors was used to elucidate the regulation mechanism of PD‐L1 expression by EGFR in NSCLC cells or xenograft mouse models. The correlations between EGFR status, p‐IκBα, HIF‐1α, and PD‐L1 protein levels were further analyzed in 149 human NSCLC tissue samples.
2. MATERIALS AND METHODS
2.1. Cell lines and cell culture
The human NSCLC cells H522, H661, HCC827, H1299, HCC2935, H1650, H1792, and H1975 were obtained from ATCC (Manassas, VA, USA). Cells were cultured in RPMI‐1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin stock in a humidified atmosphere of 5% CO2 at 37°C.
2.2. Major reagents and Abs
The MEK/ERK inhibitor U0126, PI3K/AKT inhibitor LY294002, NF‐κB inhibitor BAY11‐7082, mTOR inhibitor rapamycin, and HIF‐1α inhibitor PX‐478 were obtained from Selleck Chemicals (Houston, TX, USA. The primary Abs against EGFR [EP38Y] (ab52894), p‐ERK1/2, pT202/pT204) (ab50011), ERK1/2 (ab17942), HIF‐1α (ab51608), PD‐L1 (ab205921), His tag (ab18184), and Actin (ab8226) were purchased from Abcam (Cambridge, UK), and Abs against p‐AKT (Ser473) (#4060), AKT (#4691), p‐S6 (Ser235/236) (#4858), S6 (#2217), and p‐IκBα (Ser32/36) (#9246) were from Cell Signaling Technology (Beverly, MA, USA). Additionally, two primary Abs used for flow cytometry analysis, PE‐PD‐L1 (557924) and APC‐EGFR (563577), and their respective isotype control Abs, PE Mouse IgG1 (κ isotype control, 555749) and APC Mouse IgG2b (κ Isotype control, 557903), were obtained from BD Biosciences (San Jose, CA, USA).
2.3. Expression vectors and siRNA transfection
Expression vectors containing important EGFR mutants were constructed by subcloning the full coding domain sequence of the EGFR gene with e19del, e19del + T790M, L858R, and L858R + T790M, into the pcDNA3.1‐His‐Xpress vector (Invitrogen, Carlsbad, CA, USA). All constructs were restriction mapped and sequenced.
Specific siRNA sequences targeting the EGFR gene (si‐EGFR)22, 23 were synthetized by Beijing Aoke Peak Biotechnology (Beijing, China). Expression vectors and siRNA transfections were carried out as described.24 Briefly, exponentially growing NSCLC cells were seeded into 6‐well plates (2 × 105 cells/well). The next day, 2 μg of each expression vector or siRNA sequence was mixed with 6 μL Lipofectamine 2000 (Invitrogen) plus 250 μL Opti‐MEM medium (Invitrogen) for 20 minutes and then added to cells. The empty vector and mismatched siRNA transfections were used as controls. At 48 hours post‐transfection, cells were harvested for further analysis.
2.4. Flow cytometry
The NSCLC cells were collected and washed twice in cold flow cytometry staining buffer (PBS containing 0.2% [w/v] BSA), then resuspended with cold staining buffer to a final concentration of 1 × 106 cells/100 μL. Cell suspension was aliquoted into 100 μL to each tube, and the primary Abs, PE‐PD‐L1 and APC‐EGFR, were added and incubated for 30 minutes on ice in the dark. The respective isotype control Abs, PE Mouse IgG1 and APC Mouse IgG2b, were used according to the manufacturer's instructions. The cells were washed twice with staining buffer to remove unbound Abs and then analyzed on a flow cytometer (Accuri C6; BD Biosciences). Side‐scatter and forward‐scatter profiles were used to eliminate cell doublets. Cells were routinely sorted twice and data were analyzed with BD Accuri C6 software.
2.5. Pathway inhibition experiment
H1975 cells carrying EGFR (L858R + T790M) were selected for EGFR pathway inhibition experiments. H1975 cells received 2 dose treatments (1× and 3×) of each pathway inhibitor (3 and 9 μmol/L U0126, 15 and 45 μmol/L LY294002, 4 and 12 nmol/L rapamycin, 2.5 and 7.5 μmol/L BAY11‐7082, and 25 and 75 μmol/L PX‐478). We used DMSO‐treated cells as control. After treatment for 48 hours, total cellular proteins were extracted for further analysis.
2.6. Western blot analysis
The treated cells were lysed on ice for 25 minutes using RIPA Lysing Buffer supplemented with protease inhibitors, and the solution was cleared by centrifugation at 14 000 g for 20 minutes. The total protein concentration was quantified utilizing a BCA protein assay kit, and equal amounts of proteins (20‐40 μg/lane) were separated on 10%‐12% SDS‐PAGE gels and electrotransferred to PVDF membranes. The membranes were then blocked with 5% nonfat milk and probed with specific primary Abs: anti‐EGFR, anti‐p‐ERK1/2 (pT202/pT204), anti‐ERK1/2, anti‐HIF‐1α, anti‐PD‐L1, anti‐p‐AKT (Ser473), anti‐AKT, anti‐p‐IκBα (Ser32/36), anti‐S6, anti‐p‐S6 (Ser235/236), anti‐His, and anti‐Actin (1:1000‐1:5000 dilution). After washing with 0.2% Tween 20/PBS buffer 4 times, membranes were incubated with HRP‐conjugated secondary Ab and visualized using the ECL system (GE Healthcare, Little Chalfont, UK).
2.7. In vivo animal model experiment
Female BALB/c nude mice (5‐6 weeks old) were obtained from Beijing Vital River Laboratory Animal Company (Beijing, China) and maintained under specific pathogen‐free conditions in the Laboratory Animal Department of Beijing Cancer Hospital and Institute (Beijing, China). Care of experimental animals was in accordance with institutional animal care and use committee guidelines.
Non‐small‐cell lung cancer cells H1975 were injected s.c. into the lateral root of posterior limb of nude mouse (3 × 106 cells/mouse). The day of cell injection was designated as day 0. The tumor size of H1975 xenograft mouse models was monitored every 4 days using digital calipers, and tumor volumes were calculated according to the following formula: tumor volume (mm3) = (short axis in mm)2 × (long axis in mm) × 0.52. When the tumor size reached 100‐150 mm3, mice were randomly divided into 5 experimental groups (5 mice per group) and treated with U0126 (i.p., at 20 mg/kg), LY294002 (i.p., at 15 mg/kg), BAY11‐7082 (i.p., at 5 mg/kg), PX‐478 (p.o. gavage at 20 mg/kg), or an equivalent volume of DMSO (used as control). All 5 inhibitors were given once every 3 days within 16 days. Mice were killed and tumors were photographed on indicated days. Total proteins of tumors of each group were extracted, and western blot analysis was used to test the expression of p‐IκBα, HIF‐1α, and PD‐L1. The remaining part of each tumor was routinely formalin‐fixed, paraffin‐embedded, serially sectioned, and subjected to immunohistochemistry to examine the expression of PD‐L1 protein, described below.
2.8. Immunohistochemical analysis
For immunohistochemistry analysis of the expression of p‐IκBα, HIF‐1α, and PD‐L1, tumor samples were obtained from 149 NSCLC patients who had undergone surgical resection at Beijing Cancer Hospital. Tumor tissues obtained from xenotransplanted tumor models were also included. All the specimens had been routinely formalin‐fixed, paraffin‐embedded, and serially sectioned at 5 μm in thickness.
All tissues were stained using the streptavidin‐peroxidase immunohistochemical method. Briefly, the slides were deparaffinized in xylene, rehydrated in graded ethanol, and then treated with PBS containing 3% hydrogen dioxide to block endogenous peroxidase. Slides were preincubated in 10% goat serum to block nonspecific binding and then incubated with specific primary Abs against PD‐L1 (ab205921; Abcam), HIF‐1α (ab51608; Abcam), and p‐IκBα (#9246; Cell Signaling Technology) separately at 4°C overnight. Sections were subsequently rinsed and incubated with biotin‐conjugated IgG from Santa Cruz Biotechnology (Santa Cruz, CA, USA) with 1:10 000 dilution for 15 minutes and then with streptavidin‐peroxidase conjugate for 15 minutes. The signals were developed with DAB‐H2O2 solution. The slides were counterstained with 5% hematoxylin and then examined by light microscopy. Sections without primary Ab treatment were used as negative control. Immunohistochemical evaluation of PD‐L1, p‐IκBα, and HIF‐1α in NSCLC specimens was based on the intensity and extent of tumor cell staining. Moderate to strong cell surface PD‐L1 expression in ≥5%, p‐IκBα cytoplasm staining in ≥10%, and HIF‐1α nuclear staining in ≥10% of tumor cells were defined as positive results.25 All NSCLC sections were histopathologically reviewed by 2 trained pathologists.
2.9. DNA extraction and EGFR genotyping
The cancer tissues from all the 149 NSCLC samples were separated using manual microdissection and incubated overnight at 56°C in 50 μL digestion buffer containing 10 mg/mL proteinase K, 0.5% Tween‐20, 1 mmol/L EDTA, pH 8.0, and 50 mmol/L Tris, pH 8.5. The next day, proteinase K was inactivated by incubation of the samples at 100°C for 10 minutes. DNA samples were stored at −80°C until analysis. The EGFR status was also determined by PCR direct sequencing using the primers as previously reported.26 Polymerase chain reaction was carried out using 50 ng each sample DNA as template and negative controls (extracted slices of paraffin blocks containing no tissue) were included. The EGFR status was identified by direct sequencing using the ABI 3700 DNA sequencer (PE Applied Biosystems).
2.10. Statistical analysis
SPSS 16.0 software (SPSS, Chicago, IL, USA) was used in determining statistical significance. The continuous variables from different groups are shown as mean ± SD and were compared using t tests. The correlations among EGFR status, p‐IκBα, HIF‐1α, and PD‐L1 protein levels in NSCLC specimens were analyzed using χ2 tests. Corresponding P values < .05 were considered statistically significant.
3. RESULTS
3.1. Activation of EGFR signaling pathway and PD‐L1 expression in NSCLC cells
To investigate the correlation between the activation of the EGFR pathway and PD‐L1 expression in NSCLC cells, we examined the EGFR status and expression levels of EGFR, its pivotal downstream effectors (p‐ERK1/2, ERK1/2, p‐AKT, AKT, p‐IκBα, and HIF‐1α), and PD‐L1 in a total of 8 NSCLC cells. Sequencing results showed that 4 NSCLC cells possessed EGFR mutation (HCC827, HCC2935, and H1650 with EGFR [e19del] and H1975 with EGFR [L858R + T790M], respectively). As shown in Figure 1A, western blot results revealed obviously elevated phosphorylation levels of ERK1/2, AKT, and IκBα and increased expression levels of HIF‐1α and PD‐L1 in 3 NSCLC cells with EGFR mutants and the highest or a moderate expression of EGFR (HCC827, HCC2935, and H1975), in comparison with the other NSCLC cells carrying WT EGFR (H522, H661, H1792, and H1299) or cells with EGFR mutant but relatively low EGFR expression (H1650). Notably, apparent associations of PD‐L1 overexpression with increased p‐IκBα and HIF‐1α protein levels were observed in these 3 NSCLC cell lines with EGFR mutants. Due to the important roles of cell surface expression of EGFR and PD‐L1, further flow cytometry analysis was carried out using specific Abs labeled with different fluorescent proteins. The percentages of cells with both EGFR and PD‐L1 expression on cell surface were 88.9%, 55.8%, and 62.7% in 3 NSCLC cell lines with mutated EGFR (HCC827, HCC2935, and H1975, respectively), which were obviously higher than the remaining NSCLC cell lines carrying WT EGFR (Figure 1B).
Figure 1.
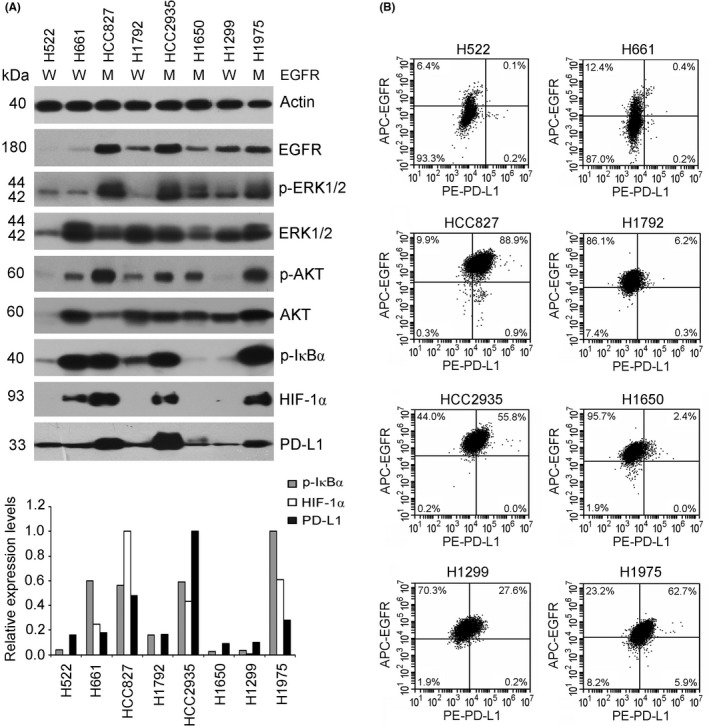
Activation of the epidermal growth factor receptor (EGFR) signaling pathway and programmed cell death ligand 1 (PD‐L1) expression in non‐small‐cell lung cancer (NSCLC) cells. A, Eight human NSCLC cell lines were cultured, and cellular proteins were extracted for western blot analysis of the protein levels of EGFR, phosphorylated (p‐)ERK1/2, ERK1/2, p‐AKT, AKT, p‐IκBα, hypoxia‐inducible factor‐1α (HIF‐1α), and PD‐L1 (top panel). Protein bands of p‐IκBα, HIF‐1α, and PD‐L1 were quantified and normalized to internal control Actin (bottom panel). M, EGFR mutant; W, WT EGFR. B, Flow cytometry analysis using specific Abs labeled with different fluorescent proteins (APC‐EGFR and PE‐PD‐L1) was used to examine the cell surface expression of EGFR and PD‐L1 of 8 NSCLC cell lines. APC, allophycocyanin; PE, phycoerythrin. The respective isotype control Abs, PE Mouse IgG1 and APC Mouse IgG2b, were used as controls
3.2. Effects of activation or inhibition of EGFR signaling pathway on PD‐L1 expression regulation in NSCLC cells
To compare the effects of WT EGFR and different EGFR mutants on PD‐L1 expression, H661 cells were transfected with WT EGFR or EGFR mutant expression vectors (e19del, e19del + T790M, L858R, and L858R + T790M). As shown in Figure 2A, ectopic expression of WT EGFR significantly elevated phosphorylation levels of ERK, AKT, S6, and IκBα and expression levels of HIF‐1α and PD‐L1 in H661 cells. In comparison with WT EGFR, transfection of all 4 EGFR mutants showed stronger capability of activating downstream signaling pathway effectors and upregulating PD‐L1 protein expression. Notably, transfection of EGFR (e19del + T790M) or EGFR (L858R + T790M) mutant showed stronger upregulation effects than EGFR (e19del) or EGFR (L858R), respectively. Subsequent specific siRNA‐induced depletion of EGFR mutant and WT EGFR obviously suppressed signaling pathway activation and PD‐L1 expression in H1975 and H1299 cells, respectively (Figure 2B,C). To elucidate the precise roles of signaling effectors of the EGFR pathway in PD‐L1 expression regulation, 5 kinds of pathway inhibitor (U0126, LY294002, Rapamycin, BAY11‐7082, and PX‐478) were used to treat H1975 cells separately. As shown in Figure 2D, treatment with the 5 inhibitors induced obviously decreased protein levels of p‐IκBα, HIF‐1α, and PD‐L1 and displayed significant dose‐dependent relationship in H1975 cells, with BAY11‐7082 and PX‐478 treatments playing the strongest roles.
Figure 2.
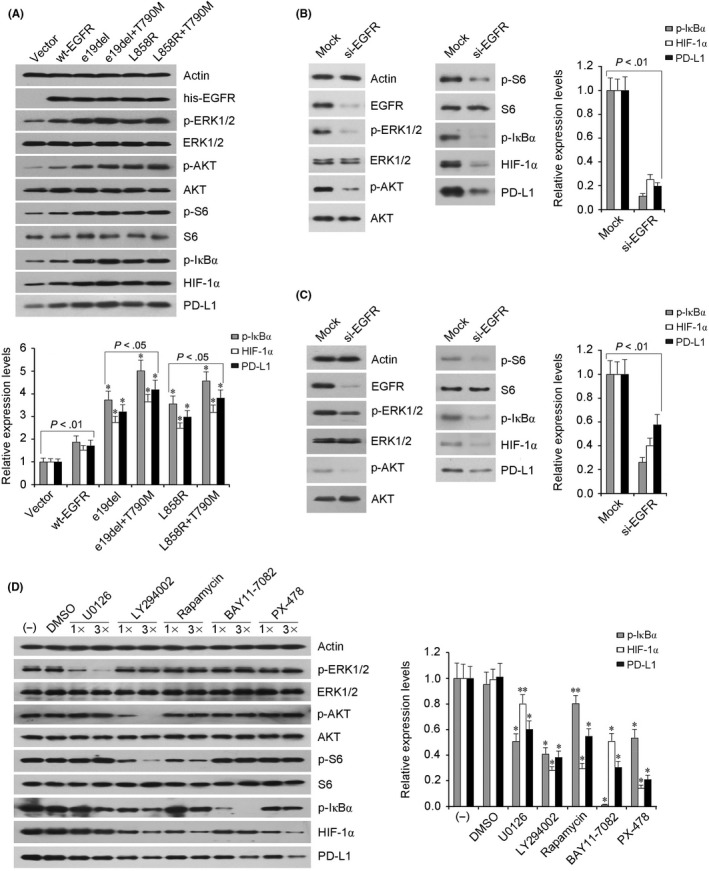
Effects of activation or inhibition of the epidermal growth factor receptor (EGFR) signaling pathway on programmed cell death ligand 1 (PD‐L1) expression in non‐small‐cell lung cancer cells. A, H661 cells transfected with WT EGFR (wt‐EGFR) or EGFR mutant expression vectors (e19del, e19del + T790M, L858R, and L858R + T790M) showed apparently elevated protein levels of phosphorylated (p‐)ERK, p‐AKT, p‐S6, p‐IκBα, hypoxia‐inducible factor‐1α (HIF‐1α), and PD‐L1. *P < .01 vs wt‐EGFR transfection. B,C Specific siRNA (si‐EGFR) transfection induced significantly downregulated EGFR expression, followed by obviously decreased protein levels of p‐ERK, p‐AKT, p‐S6, p‐IκBα, HIF‐1α, and PD‐L1 in H1975 (B) and H1299 (C) cells, respectively. D, H1975 cells received 2 dose treatments (1× and 3×) of 5 kinds of pathway inhibitor, with DMSO‐treated cells used as control. Actin was used as internal control and the graph indicates the relative protein levels of p‐IκBα, HIF‐1α, and PD‐L1. Mean ± SD values for continuous variables of 3 experiments. *P < .01 and **P < .05 vs DMSO group
3.3. Treatment with EGFR pathway inhibitors suppressed tumor growth and PD‐L1 expression in xenograft mouse model
To further testify the roles of signaling effectors (p‐ERK, p‐AKT, p‐IκBα, and HIF‐1ɑ) in PD‐L1 expression regulation by EGFR mutant in NSCLC in vivo, a xenotransplanted tumor mouse model bearing H1975 cells was established and received treatments of corresponding pathway inhibitors. As shown in Figure 3A, the tumor burdens were significantly decreased by treatments with U0126, LY294002, BAY11‐7082, and PX‐478 compared with control group (DMSO). The greatest inhibition effects on tumor growth were observed in PX‐478 and BAY11‐7082 groups.
Figure 3.
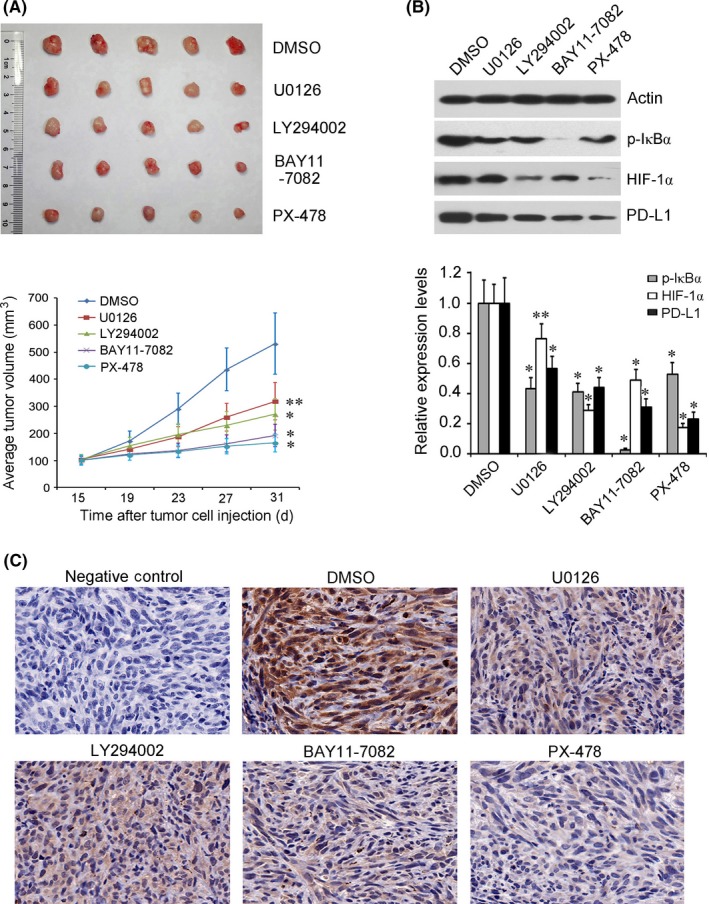
Inhibitors of the epidermal growth factor receptor (EGFR) signaling pathway suppressed tumor growth and programmed cell death ligand 1 (PD‐L1) expression in a xenograft mouse model of non‐small‐cell lung cancer. Xenotransplanted tumor mouse model bearing H1975 cells was established and received treatments of 4 pathway inhibitors (U0126, LY294002, BAY11‐7082, and PX‐478, separately). A, All 4 experimental groups showed significantly decreased tumor burden compared with control group (DMSO). B,C. Western blot analysis (B) and immunohistochemistry analysis (C) showed that the protein levels of phosphorylated (p‐)IκBα, hypoxia‐inducible factor‐1α (HIF‐1α), and PD‐L1 were significantly downregulated to varying degrees in all inhibitor groups, with PX‐478 and BAY11‐7082 displaying the strongest inhibition action. Original magnification, 400×. *P < .01 and **P < .05 vs DMSO group
Western blot analysis showed that the protein levels of HIF‐1α, p‐IκBα, and PD‐L1 were significantly downregulated to varying degrees in transplanted tumor tissues of all 4 pathway inhibitor groups compared with the control group. Further immunohistochemistry analysis confirmed the change in PD‐L1 protein level in each experimental group. Notably, PX‐478 and BAY11‐7082 groups showed the strongest inhibition effects on PD‐L1 expression (Figure 3B,C).
3.4. Correlations among EGFR status, p‐IκBα, HIF‐1α, and PD‐L1 protein levels in NSCLC tissues
The representative examples of p‐IκBα, HIF‐1α, and PD‐L1 staining are shown in Figure 4. The expression patterns of PD‐L1 according to clinical characteristics, EGFR status, p‐IκBα protein level, and HIF‐1α expression are summarized in Table 1. A total of 54 (36.2%), 64 (43.0%), and 53 (35.6%) of 149 NSCLC specimens were scored p‐IκBα, HIF‐1α, and PD‐L1 staining positive, respectively. The expression of PD‐L1 protein was not significantly associated with clinical characteristics. The proportion of tissues with positive PD‐L1 staining was much higher in NSCLC tissues with either p‐IκBα or HIF‐1α positive staining than in tissues with both p‐IκBα and HIF‐1α negative staining (44.4% vs 22.0%; P = .005). As for EGFR genotyping, PCR amplification was successful in 142 of 149 NSCLC samples and EGFR mutants were detected in 46 (32.4%) of 142 NSCLC specimens. The NSCLC tissues harboring EGFR mutants presented significantly increased positive rate of PD‐L1 expression in comparison with tissues with WT EGFR (47.8% vs 30.2%, P = .041).
Figure 4.
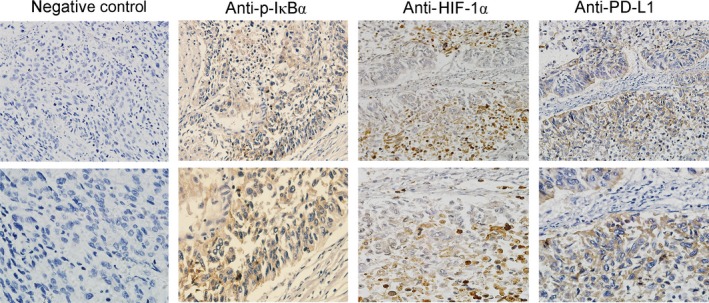
Immunohistochemistry staining of phosphorylated (p‐)IκBα, hypoxia‐inducible factor‐1α (HIF‐1α), and programmed cell death ligand 1 (PD‐L1) proteins in non‐small‐cell lung cancer tissues. Human non‐small‐cell lung cancer tissues (n = 149) were included for immunohistochemistry analysis of the expression of p‐IκBα, HIF‐1α, and PD‐L1 proteins. Positive expression of p‐IκBα (Anti‐p‐IκBα), HIF‐1α (Anti‐HIF‐1α), and PD‐L1 (Anti‐PD‐L1) is shown as brown staining in cytoplasm, cell nucleus, and cell membrane, respectively. Section without Ab treatment was used as negative control. Original magnification, 200 × (top panels) and 400 × (bottom panels)
Table 1.
Associations of clinical and genetic factors with programmed cell death ligand 1 (PD‐L1) expression in patients with non‐small‐cell lung cancer (n = 149)
| Variable | Total | PD‐L1, n (%) | P‐valuea | |
|---|---|---|---|---|
| Positive | Negative | |||
| Age, years | ||||
| ≤60 | 61 | 24 (39.3) | 37 (60.7) | 0.423 |
| >60 | 88 | 29 (33.0) | 59 (67.0) | |
| Gender | ||||
| Male | 96 | 35 (36.5) | 61 (63.5) | 0.761 |
| Female | 53 | 18 (34.0) | 35 (66.0) | |
| Smoking status | ||||
| Smoker | 54 | 23 (42.5) | 31 (57.5) | 0.177 |
| Nonsmoker | 95 | 30 (31.6) | 65 (68.4) | |
| Pathology | ||||
| ADC | 76 | 25 (32.9) | 51 (67.1) | |
| SCC | 73 | 28 (38.4) | 45 (61.6) | 0.486 |
| TNM stage | ||||
| I | 14 | 3 (21.4) | 11 (78.6) | 0.529 |
| IIa | 63 | 21 (33.3) | 42 (66.7) | |
| IIb | 59 | 23 (39.0) | 36 (61) | |
| III | 13 | 6 (46.1) | 7 (53.9) | |
| Lymph node metastasis | ||||
| Positive | 72 | 29 (40.3) | 43 (59.7) | 0.246 |
| Negative | 77 | 24 (31.2) | 53 (68.8) | |
| EGFR statusb | ||||
| Wild‐type | 96 | 29 (30.2) | 67 (69.8) | 0.041 |
| Mutated | 46 | 22 (47.8) | 24 (52.2) | |
| p‐IκBα expression | ||||
| Positive | 54 | 23 (42.6) | 31 (57.4) | 0.177 |
| Negative | 95 | 30 (31.6) | 65 (68.4) | |
| HIF‐1α expression | ||||
| Positive | 64 | 27 (42.2) | 37 (57.8) | 0.143 |
| Negative | 85 | 26 (30.6) | 59 (69.4) | |
| p‐IκBα and HIF‐1α expression | ||||
| Positivec | 90 | 40 (44.4) | 50 (55.6) | 0.005 |
| Negatived | 59 | 13 (22.0) | 46 (78.0) | |
Two‐sided χ2 test.
No information for some patients.
Either phosphorylated (p‐)IκBα or hypoxia‐inducible factor‐1α (HIF‐1α) positive.
Both p‐IκBα and HIF‐1α negative.
ADC, adenocarcinoma; EGFR, epidermal growth factor receptor; SCC, squamous cell carcinoma.
Elevated protein levels of p‐IκBα and HIF‐1α were also detected in tissues carrying EGFR mutants compared to tissues with the WT EGFR gene (56.0% vs 44.0%, P = .029 and 56.4% vs 43.6%, P = .012, respectively; Table 2). Additionally, the proportion of tissues with positive p‐IκBα staining was significantly higher in NSCLC tissues with HIF‐1α positive staining than HIF‐1α negative staining (59.2% vs 40.8%; P = .002), as shown in Table 3.
Table 2.
Association of epidermal growth factor receptor (EGFR) status with phosphorylated (p‐)IκBα or hypoxia‐inducible factor‐1α (HIF‐1α) expression in patients with non‐small‐cell lung cancer (n = 142)
| Variables | Total | EGFR statusa (%) | P‐valueb | |
|---|---|---|---|---|
| WT | Mutated | |||
| p‐IκBα expression | ||||
| Positive | 50 | 28 (56.0) | 22 (44.0) | |
| Negative | 92 | 68 (73.9) | 24 (26.1) | 0.029 |
| HIF‐1α expression | ||||
| Positive | 62 | 35 (56.4) | 27 (43.6) | |
| Negative | 80 | 61 (76.2) | 19 (23.8) | 0.012 |
No information for some patients.
Two‐sided χ2 test.
Table 3.
Association of phosphorylated (p‐)IκBα expression with hypoxia‐inducible factor 1α (HIF‐1α) expression in patients with non‐small‐cell lung cancer (n = 149)
| Variable | Total | HIF‐1α expression (%) | P‐valuea | |
|---|---|---|---|---|
| Positive | Negative | |||
| p‐IκBα expression | ||||
| Positive | 54 | 32 (59.2) | 22 (40.8) | |
| Negative | 95 | 32 (33.6) | 63 (66.4) | 0.002 |
Two‐sided χ2 test.
4. DISCUSSION
Overexpression of PD‐L1 has been detected in various kinds of human tumors, including NSCLC, and has been reported to be a poor prognostic indicator of overall survival and a predictive marker of good response to new immunotherapy drugs.27, 28 Some driver gene mutations including EGFR have been reported to be involved in intrinsic regulation of PD‐L1 expression in various tumors, including NSCLC.29 Epidermal growth factor receptor is a member of the ErbB family of receptor tyrosine kinases. EGFR mutants promote tumorigenesis of NSCLC by constitutively activating downstream signaling effectors, including PI3K/AKT, RAS/ERK, and others.30, 31 Activated AKT, as a key downstream effector of the EGFR pathway, has been reported to activate NF‐κB and thereby regulate PD‐L1 expression.32 As an important transcription factor, NF‐κB transactivates PD‐L1 expression by binding directly to the PD‐L1 promoter.33, 34 NF‐κB is bound and inhibited by IκBs in the cytoplasm of unstimulated cells. Upon stimulation, IκBs are phosphorylated by upstream IKK kinases and NF‐κB is released and translocates into nucleus to transcript target genes, including PD‐L1. By phosphorylation of IKK kinases, which in turn phosphorylate IκBα, activation of PI3K/AKT can further activate NF‐κB.35, 36, 37
In the present study, we investigated the association of EGFR status and EGFR signaling pathway activation with PD‐L1 expression in 8 human NSCLC cell lines. Genotyping and western blot results showed that the EGFR status and EGFR expression together affected phosphorylation activation of ERK1/2, AKT, and IκBα and expression of HIF‐1α and PD‐L1 in NSCLC cells. Notably, strong accordance between PD‐L1 expression and protein levels of p‐IκBα and HIF‐1α was observed in the majority of NSCLC cells examined. Further flow cytometry analysis indicated that NSCLC cells with mutated EGFR showed clearly higher percentages of cells that were both EGFR and PD‐L1 positive on the cell surface than the remaining NSCLC cell lines carrying WT EGFR. Another important transcription factor, HIF‐1α, increases the expression of PD‐L1 in hypoxic cancer cells by binding directly to the hypoxia response element in the PD‐L1 promoter.38, 39 Upregulated HIF‐1α expression by a reduction in oxygen‐dependent degradation in hypoxia leads to the expression of genes involved in tumor angiogenesis, metastasis/migration, glucose metabolism, cell proliferation, chemoresistance, and immune adaptation.40, 41, 42 Apart from hypoxia, genetic changes and abnormal activation of important signaling pathways in tumors can induce HIF‐1α expression under normoxia conditions. VHL, PTEN, BRAF, and SDH gene mutations have been reported to regulate HIF‐1α expression under normoxia.40, 43 Activation of AKT upregulates HIF‐1α protein translation through downstream component mTOR/S6.44
Our sequencing results revealed EGFR (L858R + T790M) mutant in H1975 cells and WT EGFR in H661 and H1299 cells, and these 3 cell lines were used to investigate the precise molecular mechanism of PD‐L1 expression regulation by EGFR in NSCLC cells. H661 cells transfected with 4 kinds of common EGFR mutant or H1975 cells with si‐EGFR showed obviously up‐ or downregulated protein levels of p‐ERK, p‐AKT, p‐IκBα, HIF‐1α, and PD‐L1, respectively. In contrast, H661 cells transfected with WT EGFR or H1299 cells with si‐EGFR showed relatively mild changes in the protein levels mentioned above. We further treated H1975 cells with 5 kinds of pathway inhibitor (U0126, LY294002, Rapamycin, BAY11‐7082, and PX‐478) to explore the potential roles of pivotal signaling effectors of the EGFR pathway in regulation of PD‐L1 expression in NSCLC cells. Western blot analysis revealed significantly suppressed protein levels of p‐IκBα, HIF‐1α, and PD‐L1 in H1975 cells treated with all 5 inhibitors, with BAY11‐7082 and PX‐478 showing the strongest inhibitory effects. These results suggested important roles of p‐AKT and p‐ERK and potential interplay and cooperation between NF‐κB and HIF‐1α in regulation of PD‐L1 expression by EGFR mutants in NSCLC cells. It has been reported that activated NF‐κB can directly bind to the promoter of HIF‐1α and promote HIF‐1α transcription under hypoxia or normoxia conditions in multiple cells, including cancer cells.45, 46 Notably, some studies have revealed a positive feedback loop between HIF‐1α and NF‐κB pathways. Hypoxia‐inducible factor‐1α can induce transcription of IKKβ through the hypoxia response element present in the promoter of the IKKβ gene and mediate consequent nuclear translocation and activation of NF‐κB.47 Hypoxia‐inducible factor‐1α has also been reported to directly transcript NF‐κB expression under hypoxia.45, 48 Based on previous research reports and our existing research findings, we developed a possible model for PD‐L1 expression regulation by EGFR mutants (Figure 5).
Figure 5.
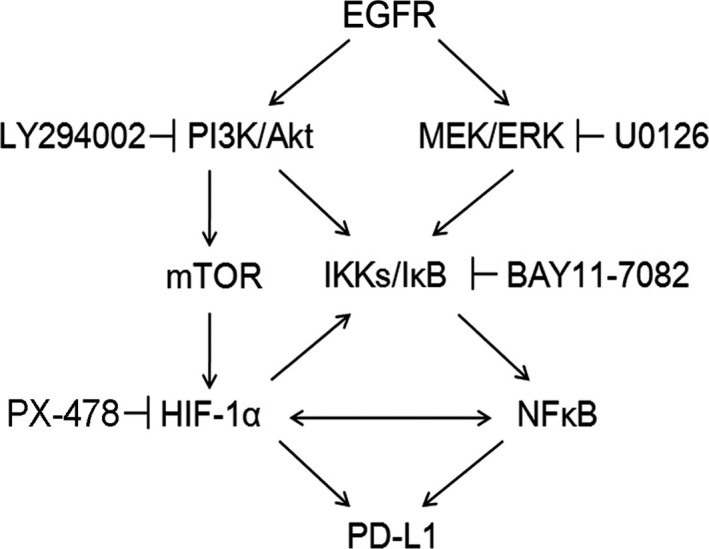
Flowchart of programmed cell death ligand 1 (PD‐L1) expression regulation by epidermal growth factor receptor (EGFR) mutants through downstream signaling effectors phosphorylated (p‐)AKT, p‐ERK, nuclear factor‐κB (NF‐κB), and hypoxia‐inducible factor‐1α (HIF‐1α). IKK, IκB kinase
We next determined the roles of pivotal EGFR signaling effectors in regulation of PD‐L1 expression in NSCLC in vivo. A xenotransplanted tumor mouse model bearing H1975 cells was established and treated with corresponding pathway inhibitors. Subsequent western blot and immunohistochemistry analyses showed that the protein levels of p‐IκBα, HIF‐1α, and PD‐L1 were significantly downregulated to varying degrees in all inhibitor groups, with PX‐478 and BAY11‐7082 treatments displaying the strongest inhibition effects on PD‐L1 expression. These results were consistent with our research findings in vitro and further suggested the important roles of HIF‐1α and p‐IκBα in PD‐L1 expression regulation by EGFR mutants in NSCLC.
To investigate the correlations among EGFR mutants, pivotal EGFR signaling effectors, and PD‐L1 expression, we further examined EGFR status and protein levels of p‐IκBα, HIF‐1α, and PD‐L1 in 149 human NSCLC tissues. The mutation rate of the EGFR gene was 32.4% in this NSCLC patient group, and NSCLC tissues carrying EGFR mutants showed elevated protein levels of PD‐L1, p‐IκBα, and HIF‐1α, compared to tissues with WT EGFR (P = .041, .029, and .012, respectively), which was supported by various previous reports that PD‐L1 expression was upregulated in NSCLC tissues harboring EGFR mutations.21, 49, 50 We next undertook an association analysis of HIF‐1α and p‐IκBα positive staining with PD‐L1 expression and revealed that NSCLC tissues with either HIF‐1α or p‐IκBα positive staining presented significantly increased positive rates of PD‐L1 expression compared with tissues that were stained negative for both HIF‐1α and p‐IκBα (P = .005). Moreover, a high degree of correlation was observed between protein expression of HIF‐1α and phosphorylation level of IκBα (P = .002). These statistical analysis results showed significant correlations among EGFR mutants, p‐IκBα, HIF‐1α, and PD‐L1 protein levels in NSCLC tissues. In contrast, some other retrospective studies have reported that PD‐L1 positivity was more frequent in NSCLC tissues carrying WT EGFR and other studies have shown no association between PD‐L1 expression and EGFR mutations.51, 52, 53 The discrepancies among these studies might be caused by the heterogeneous study population and variable definitions of PD‐L1 expression. Additionally, our present findings indicated the necessity to explore the effects of genetic and environmental factors on NF‐κB and HIF‐1α in the studying of PD‐L1 expression regulation by EGFR mutants. Notably, several recent clinical trials and retrospective studies have revealed no efficacy of the PD‐1/PD‐L1 inhibition strategy in EGFR mutated NSCLC compared with WT EGFR.54, 55 This lack of response might be partially attributed to the complexity of regulatory effects of EGFR mutants on PD‐L1 expression and the suppression of tumor‐infiltrating lymphocytes caused by EGFR pathway activation, which is likely responsible for the uninflamed tumor microenvironment and immunosuppression.56, 57
Taken together, our data indicated important roles of p‐AKT, p‐ERK, NF‐κB, and HIF‐1α and potential interplay and cooperation between NF‐κB and HIF‐1α in PD‐L1 expression regulation by EGFR mutants in NSCLC cells. Considering the widespread existence of hypoxia in solid tumors and its induction effect on HIF‐1α expression, the influence of driver mutants combined with hypoxic conditions on PD‐L1 expression regulation and tumor immune escape merits further investigation in expanded NSCLC cases.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
ACKNOWLEDGMENTS
This work was supported by: the National Natural Science Foundation Key Program (Grant Nos. 81630071 and 81330062); National Key Research and Development Project Precision Medicine Special Research (Grant No. 2016YFC0902300); National High Technology Research and Development Program 863 (Grant No. SS2015AA020403); Beijing Municipal Natural Science Foundation (Grant Nos. 7152029 and 7172045); Collaborative Innovation Center for Cancer Medicine; Peking University‐Tsinghua University Joint Center for Life Sciences Clinical Investigator; CAMS Innovation Fund for Medical Sciences (Grant No. CIFMS 2016‐I2M‐3‐008); and National Natural Science Foundation (Grant Nos. 81101778; 81472206). This study was approved by both the Ethics and the Academic committees of Beijing Cancer Hospital, and informed consent was obtained from each subject.
Guo R, Li Y, Wang Z, et al. Hypoxia‐inducible factor‐1α and nuclear factor‐κB play important roles in regulating programmed cell death ligand 1 expression by epidermal growth factor receptor mutants in non‐small‐cell lung cancer cells. Cancer Sci. 2019;110:1665–1675. 10.1111/cas.13989
Funding information
Beijing Municipal Natural Science Foundation, Grant/Award Number 7152029, 7172045; National Key Research and Development Project Precision Medicine Special Research, Grant/Award Number 2016YFC0902300; National Natural Science Foundation of China, Grant/Award Number 81101778, 81472206; CAMS Innovation Fund for Medical Sciences, Grant/Award Number CIFMS 2016‐I2M‐3‐008; National Natural Science Foundation Key Program, Grant/Award Number 81330062, 81630071; National High Technology Research and Development Program 863, Grant/Award Number SS2015AA020403
REFERENCES
- 1. Siegel R, De Santis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220‐241. [DOI] [PubMed] [Google Scholar]
- 2. Kocher F, Hilbe W, Seeber A, et al. Longitudinal analysis of 2293 NSCLC patients: a comprehensive study from the TYROL registry. Lung Cancer. 2015;87:193‐200. [DOI] [PubMed] [Google Scholar]
- 3. Korpanty GJ, Graham DM, Vincent MD, et al. Biomarkers that currently affect clinical practice in lung cancer: EGFR, ALK, MET, ROS‐1, and KRAS. Front Oncol. 2014;4:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dienstmann R, Rodon J, Prat A, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25:552‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti‐PD‐L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti‐PD‐1 antibody in cancer. N Engl J Med. 2012;366:2443‐2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okazaki T, Honjo T. PD‐1 and PD‐1 ligands: from discovery to clinical application. Int Immunol. 2007;19:813‐824. [DOI] [PubMed] [Google Scholar]
- 8. Zou W, Wolchok JD, Chen L. PD‐L1 (B7‐H1) and PD‐1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi L, Chen S, Yang L, et al. The role of PD‐1 and PD‐L1 in T‐cell immune suppression in patients with hematological malignancies. J Hematol Oncol. 2013;6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanmamed MF, Chen L. Inducible expression of B7‐H1 (PD‐L1) and its selective role in tumor site immune modulation. Cancer J. 2014;20:256‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan Y, Ma K, Wang C, et al. Prognostic value of PD‐L1 and PD‐1 expression in pulmonary neuroendocrine tumors. Onco Targets Ther. 2016;9:6075‐6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takada K, Okamoto T, Shoji F, et al. Clinical significance of PD‐L1 protein expression in surgically resected primary lung adenocarcinoma. J Thorac Oncol. 2016;11:1879‐1890. [DOI] [PubMed] [Google Scholar]
- 13. Wu P, Wu D, Li L, et al. PD‐L1 and survival in solid tumors: a meta‐analysis. PLoS One. 2015;10:e0131403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD‐L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51:221‐228. [DOI] [PubMed] [Google Scholar]
- 15. Liu J, Hamrouni A, Wolowiec D, et al. Plasma cells from multiple myeloma patients express B7‐H1 (PD‐L1) and increase expression after stimulation with IFN‐{gamma} and TLR ligands via a MyD88‐, TRAF6‐, and MEK‐dependent pathway. Blood. 2007;110:296‐304. [DOI] [PubMed] [Google Scholar]
- 16. Chen J, Feng Y, Lu L, et al. Interferon‐gamma‐induced PD‐L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217:385‐393. [DOI] [PubMed] [Google Scholar]
- 17. Marzec M, Zhang Q, Goradia A, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD‐L1, B7‐H1). Proc Natl Acad Sci USA. 2008;105:20852‐20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu C, Fillmore CM, Koyama S, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD‐L1 expression. Cancer Cell. 2014;25:590‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small‐cell lung cancer: meta‐analyses by ethnicity and histology (mutMap). Ann Oncol. 2013;24:2371‐2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akbay EA, Koyama S, Carretero J, et al. Activation of the PD‐1 pathway contributes to immune escape in EGFR‐driven lung tumors. Cancer Discov. 2013;3:1355‐1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Incecco A, Andreozzi M, Ludovini V, et al. PD‐1 and PD‐L1 expression in molecularly selected non‐small‐cell lung cancer patients. Br J Cancer. 2015;112:95‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wichmann H, Güttler A, Bache M, et al. Targeting of EGFR and HER2 with therapeutic antibodies and siRNA: a comparative study in glioblastoma cells. Strahlenther Onkol. 2015;2:180‐191. [DOI] [PubMed] [Google Scholar]
- 23. Chen N, Fang W, Lin Z, et al. KRAS mutation‐induced upregulation of PD‐L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;9:1175‐1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kakudo N, Morimoto N, Ogawa T, et al. Hypoxia enhances proliferation of human adipose‐derived stem cells via HIF‐1ɑ activation. PLoS One. 2015;10:e0139890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang YL, Yang CY, Lin MW, et al. High co‐expression of PD‐L1 and HIF‐1α correlates with tumour necrosis in pulmonary pleomorphic carcinoma. Eur J Cancer. 2016;60:125‐135. [DOI] [PubMed] [Google Scholar]
- 26. Giaccone G, Gallegos Ruiz M, Le Chevalier T, et al. Erlotinib for frontline treatment of advanced non‐small cell lung cancer: a phase II study. Clin Cancer Res. 2006;12:6049‐6055. [DOI] [PubMed] [Google Scholar]
- 27. Ma W, Gilligan BM, Yuan J, et al. Current status and perspectives in translational biomarker research for PD‐1/PD‐L1 immune checkpoint blockade therapy. J Hematol Oncol. 2016;9:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goel G, Sun W. Advances in the management of gastrointestinal cancers‐an upcoming role of immune checkpoint blockade. J Hematol Oncol. 2015;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang W, Pang Q, Zhang X, et al. Programmed death‐ligand 1 is prognostic factor in esophageal squamous cell carcinoma and is associated with epidermal growth factor receptor. Cancer Sci. 2017;108:590‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akca H, Tani M, Hishida T, et al. Activation of the AKT and STAT3 pathways and prolonged survival by a mutant EGFR in human lung cancer cells. Lung Cancer. 2006;54:25‐33. [DOI] [PubMed] [Google Scholar]
- 31. Sordella R, Bell DW, Haber DA, et al. Gefitinib‐sensitizing EGFR mutations in lung cancer activate anti‐apoptotic pathways. Science. 2004;305:1163‐1167. [DOI] [PubMed] [Google Scholar]
- 32. Lin K, Cheng J, Yang T, et al. EGFR‐TKI down‐regulates PD‐L1 in EGFR mutant NSCLC through inhibiting NF‐κB. Biochem Biophys Res Commun. 2015;463:95‐101. [DOI] [PubMed] [Google Scholar]
- 33. Xiong HY, Ma TT, Wu BT, et al. IL‐12 regulates B7‐H1 expression in ovarian cancer‐associated macrophages by effects on NF‐κB signalling. Asian Pac J Cancer Prev. 2014;15:5767‐5772. [DOI] [PubMed] [Google Scholar]
- 34. Huang G, Wen Q, Zhao Y, et al. NF‐κB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS One. 2013;8:e61602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghosh S, Dass JF. Study of pathway cross‐talk interactions with NF‐κB leading to its activation via ubiquitination or phosphorylation: a brief review. Gene. 2016;584:97‐109. [DOI] [PubMed] [Google Scholar]
- 36. Vallabhapurapu S, Karin M. Regulation and function of NF‐kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693‐733. [DOI] [PubMed] [Google Scholar]
- 37. Aggarwal BB, Takada Y, Oommen OV. From chemoprevention to chemotherapy: common targets and common goals. Expert Opin Investig Drugs. 2004;13:1327‐1338. [DOI] [PubMed] [Google Scholar]
- 38. Barsoum IB, Smallwood CA, Siemens DR, et al. A mechanism of hypoxia‐mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665‐674. [DOI] [PubMed] [Google Scholar]
- 39. Noman MZ, Desantis G, Janji B, et al. PD‐L1 is a novel direct target of HIF‐1α, and its blockade under hypoxia enhanced MDSC‐mediated T cell activation. J Exp Med. 2014;211:781‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Semenza GL. HIF‐1 inhibitors for cancer therapy: from gene expression to drug discovery. Curr Pharm Des. 2009;15:3839‐3843. [DOI] [PubMed] [Google Scholar]
- 41. Lee YH, Bae HC, Noh KH, et al. Gain of HIF‐1α under normoxia in cancer mediates immune adaptation through the AKT/ERK and VEGFA axes. Clin Cancer Res. 2015;21:1438‐1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zou J, Li P, Lu F, et al. Notch1 is required for hypoxia‐induced proliferation, invasion and chemoresistance of T‐cell acute lymphoblastic leukemia cells. J Hematol Oncol. 2013;6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Masoud GN, Li W. HIF‐1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang BH, Jiang G, Zheng JZ, et al. Phosphatidylinositol 3‐kinase signaling controls levels of hypoxia‐inducible factor 1. Cell Growth Differ. 2001;12:363‐369. [PubMed] [Google Scholar]
- 45. Jiang Y, Zhu Y, Wang X, et al. Temporal regulation of HIF‐1 and NF‐κB in hypoxic hepatocarcinoma cells. Oncotarget. 2015;6:9409‐9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yao G, Zhang Q, Doeppner TR, et al. LDL suppresses angiogenesis through disruption of the HIF pathway via NF‐κB inhibition which is reversed by the proteasome inhibitor BSc2118. Oncotarget. 2015;6:30251‐30262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mak P, Li J, Samanta S, et al. ERβ regulation of NF‐κB activation in prostate cancer is mediated by HIF‐1. Oncotarget. 2015;6:40247‐40254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walmsley SR, Print C, Farahi N, et al. Hypoxia‐induced neutrophil survival is mediated by HIF‐1alpha‐dependent NF‐kappaB activity. J Exp Med. 2005;201:105‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Azuma K, Ota K, Kawahara A, et al. Association of PD‐L1 overexpression with activating EGFR mutations in surgically resected nonsmall‐cell lung cancer. Ann Oncol. 2014;25:1935‐1940. [DOI] [PubMed] [Google Scholar]
- 50. Song Z, Yu X, Cheng G, et al. Programmed death‐ligand 1 expression associated with molecular characteristics in surgically resected lung adenocarcinoma. J Transl Med. 2016;14:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cha YJ, Kim HR, Lee CY, et al. Clinicopathological and prognostic significance of programmed cell death ligand‐1 expression in lung adenocarcinoma and its relationship with p53 status. Lung Cancer. 2016;97:73‐80. [DOI] [PubMed] [Google Scholar]
- 52. Yoneshima Y, Ijichi K, Anai S, et al. PD‐L1 expression in lung adenocarcinoma harboring EGFR mutations or ALK rearrangements. Lung Cancer. 2018;118:36‐40. [DOI] [PubMed] [Google Scholar]
- 53. Cooper WA, Tran T, Vilain RE, et al. PD‐L1 expression is a favorable prognostic factor in early stage non‐small cell carcinoma. Lung Cancer. 2015;89:181‐188. [DOI] [PubMed] [Google Scholar]
- 54. Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD‐1 pathway blockade in Non‐Small Cell Lung Cancer: a retrospective analysis. Clin Cancer Res. 2016;22:4585‐4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non‐small‐cell lung cancer (OAK): a phase 3, open‐label, multicentre randomised controlled trial. Lancet. 2017;389:255‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang S, Zhang Y, Wang Y, et al. Amphiregulin confers regulatory T cell suppressive function and tumor invasion via the EGFR/GSK‐3β/Foxp3 axis. J Biol Chem. 2016;291:21085‐21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu S, Liu D, Shen B, et al. Immunotherapy strategy of EGFR mutant lung cancer. Am J Cancer Res. 2018;8:2106‐2115. [PMC free article] [PubMed] [Google Scholar]


