Abstract
The use of immune checkpoint inhibitors targeting PD‐1 and PD‐L1 in advanced non‐small cell lung cancer (NSCLC) has been one of the most significant improvements in recent years. However the resistance mechanisms of immune checkpoint inhibitors require further investigation. Herein we attempted to determine the possible resistance mechanism of nivolumab in a male smoker with advanced adenosquamous carcinoma. After experiencing disease progression on systematic chemotherapy, he was administered nivolumab as a result of high PD‐L1 expression. Larger panel gene detection was performed after the failure of nivolumab treatment to investigate the possible resistance mechanism and a new EGFR exon 21 L858R mutation was detected. After a achieving a response with gefitinib, the patient suffered a rapid relapse and died of tumor progression. This case represents the first time EGFR exon 21 L858R has been detected as an acquired resistance mutation to nivolumab. Patients with high PD‐L1 expression may exhibit a poor response to EGFR‐tyrosine kinase inhibitors. Large panel gene detection remains the optimal choice when confronted with drug resistance.
Keywords: Acquired resistance mechanism, EGFR exon 21 L858R, gefitinib, lung cancer, nivolumab
Introduction
Lung cancer is one of the most aggressive malignant tumors and the most common cause of cancer‐related death in the world.1 The identification of molecular events underlying pathogenesis has improved the clinical treatment and outcome of lung cancers.2, 3, 4, 5 Among all of the new treatment strategies, immunotherapy is one of the most promising. Studies have proven that agents targeting immune checkpoint receptors (ICIs) are effective and well tolerated as second‐line treatment in non‐small cell lung cancer (NSCLC) with PD‐L1 overexpression or without.6, 7 However, like all antitumor treatments, the effect of immunotherapy is largely limited by its drug resistance. Few studies have reported the resistance mechanism to ICIs. Herein, we present a case of a patient who developed resistance to nivolumab. Next generation sequencing (NGS) was used to investigate the drug resistance mechanism and therefore provide further treatment strategies.
Case presentation
A 48‐year‐old smoking man came to the hospital complaining of three months of back pain. Computed tomography (CT) showed a tumor in the right upper lobe of the lung and pleural disseminations; whole‐body bone scintigraphy showed bone metastasis of thoracic vertebra. The right upper lobe of the lung was resected by video‐assisted thoracic surgery for diagnosis. Histological examination showed adenosquamous carcinoma with CK, CK7, and CK5/6 expression on tumor cells (Fig 1b). A large NSG panel (Burning Rock, Guang Zhou, China), including 168 somatic driver genes related to lung cancer (supplementary material 1) was taken of both tumor tissue and plasma, and mutations in the tested genes were not detected (Fig 1a). The depth of gene detection was 1000X for tumor tissues and 10 000X for plasma. After palliative surgery, the patient commenced chemotherapy of pemetrexed combined with cisplatin. A CT scan taken after four cycles of chemotherapy revealed pulmonary metastasis and involvement of the right sacroiliac joint (Fig 1b) and the patient was evaluated with progressive disease (PD). A biopsy of the bone confirmed lung cancer metastasis by pathological examination. NGS of both metastatic bone tissue and plasma was conducted again with the same panel and still no mutation was detected (Fig 1a). In addition, immunohistochemistry (IHC) of the bone metastatic tissue showed PD‐L1 expression (SP142, Roche, Basel, Switzerland) in 65% of tumor cells (Fig 1b) and as a result chemotherapy was ceased and nivolumab administered. After three cycles of nivolumab treatment (3 mg/kg every two weeks), the patient was evaluated via CT as immunity unconfirmed PD (iUPD) (Fig 2a). Nivolumab treatment was not suspended until the patient was evaluated as immunity confirmed PD (iCPD), with dramatically increasing metastasis after another three cycles of nivolumab (Fig 2a). NGS of the plasma was implemented again to explore the possible resistance mechanism and a new EGFR exon 21 L858R mutation was detected (Figs 1a, 2c). The treatment was altered to gefitinib (250mg, orally, once a day) and after a month, CT revealed a partial response (Fig 2a). However the patient experienced a rapid relapse and died of tumor progression two months later. The tumor biomarkers CEA, CA12‐5, and CA19‐9 accordingly presented dynamic changes, along with different treatments for this patient (Fig 2b).
Figure 1.
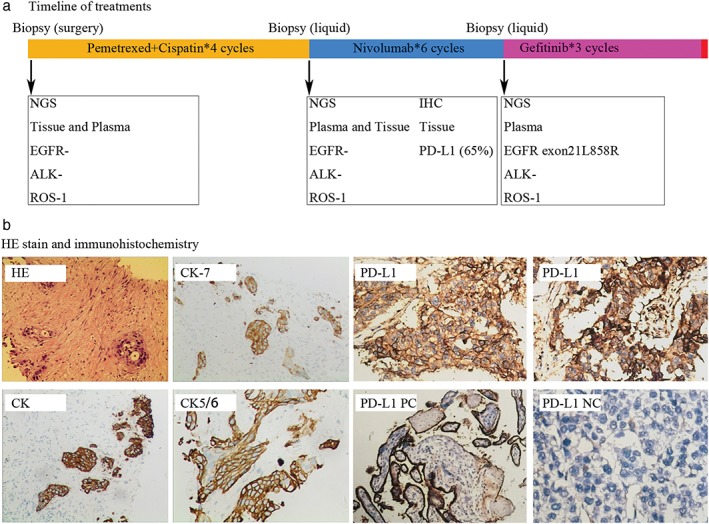
(a) The timeline of next generation sequencing (NGS) and the various treatments administered. (b) Hematoxylin and eosin (HE) stain and immunohistochemistry (IHC) of CK, CK7, CK5/6, and PD‐L1.
Figure 2.
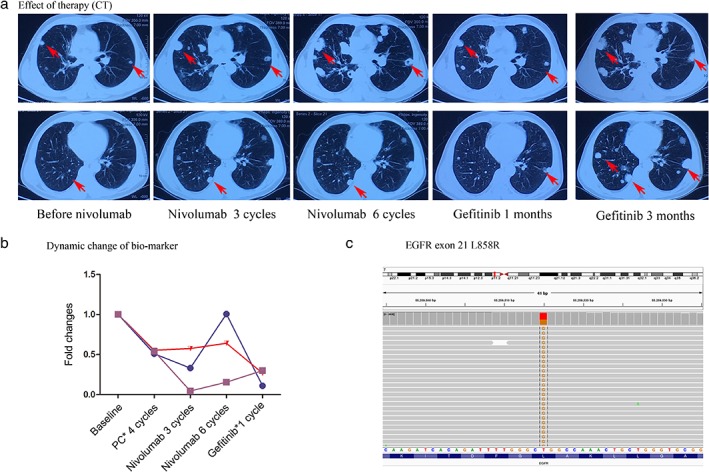
(a) Computed tomography (CT) images of the patient. Red arrows show diffusion metastasis of the lung. (b) The dynamic change of biomarkers with CA19‐9, CEA, and CA12‐5 after each treatment ( ) CA19‐9, (
) CA19‐9, ( ) CEA, and (
) CEA, and ( ) CA12‐5. (c) Mutation of EGFR exon 21 L858R detected by next generation sequencing.
) CA12‐5. (c) Mutation of EGFR exon 21 L858R detected by next generation sequencing.
The patient signed and provided written informed consent for the use of his data in publication.
Discussion
Although PD‐1 inhibitors are becoming the hotspot of anti‐cancer treatment and research, data is relatively limited regarding drug resistance mechanisms. The tumor interferon signaling pathway and JAK1/2 mutations have been suggested as potential resistance mechanisms to PD‐1 inhibitors.8, 9 Herein, for the first time, we detected an EGFR exon 21 L858R mutation as a potential acquired resistance mechanism of nivolumab in an NSCLC patient that was treated with gefitinib.
Two meta‐analyses reported that ICIs are less effective in pretreated EGFR mutated lung cancer patients than in those with wild type EGFR, which is probably related to a lower tumor mutation burden10 and the uninflamed and immunosuppressive tumor microenvironment of the latter subgroup of patients.11 However EGFR mutation has never been reported as an acquired resistance mechanism of ICIs. In our case, the patient showed a poor response to nivolumab with PD‐L1 expression in over 50% of tumor cells (although the predictive nature of tumor PD‐L1 expression in PD‐L1 inhibitor application is supported by KEYNOTE‐010 and CheckMate 057 trials). A new EGFR mutation was detected after icPD, which strongly suggests that EGFR exon 21 L858R is an acquired resistance mutation of nivolumab. Furthermore, this patient's original driver gene status was confirmed twice as negative by NGS of both tissue and blood. The second panel was taken before nivolumab was administered and the EGFR exon 21 L858R was detected right after acquired resistance to nivolumab, which largely reduces the odds that the exon 21 L858R mutation was the result of other drugs in the serial therapy. There is also the possibility that a portion of the tumors driven by exon 21 L858R existed below measurable limits in the beginning but became detectable after serial treatments, with the increase occurring during nivolumab application. In our opinion this supports the theory that an exon 21 L858R mutation is a resistance mechanism of nivolumab. However, more basic experiments and clinical research are warranted to prove this conclusion.
Few studies have investigated the effect of PD‐L1 expression on EGFR‐TKI efficacy and have reported controversial conclusions. Lin et al. found that when treating an EGFR mutant NSCLC patient with EGFR‐TKIs, PD‐L1‐positive patients had longer progression‐free and overall survival than PD‐L1 negative patients.12 However, several other studies drew the opposite conclusion.13, 14, 15 The patient in our report experienced a rapid relapse after gefitinib treatment and thus exhibited significantly short progression‐free and overall survival. More preclinical and clinical research is warranted to elucidate the underlying biology mechanism and other appropriate predictive biomarkers are required when treating patients with both PD‐L1 overexpression and EGFR mutations.
In conclusion, our results suggest that EGFR exon 21 L858R is an acquired resistance mechanism of nivolumab. Current information concerning cross‐talk between EGFR signaling pathways and PD‐1/PD‐L1 remains complicated and contentious, thus no consensus has been established regarding the optimal treatment approach for EGFR mutant/PD‐L1 positive NSCLC patients. EGFR exon 21 L858R mutant NSCLC patients may be less responsive to nivolumab. Gene detection should be conducted to identify the accurate resistance mechanism to provide precision treatment for patients.
Disclosure
No authors report any conflict of interest.
Supporting information
Supplementary material. A large gene detection panel including 168 lung cancer related gene mutations.
Acknowledgment
This work was partially supported by Natural Science Foundation of Hunan Province (2018RS3106, 2017SK2134 and 2018JJ2238).
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Klaeger, S. , Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., Schoof M., Canevari G., Casale E., Depaolini S.R., Feuchtinger A., Wu Z., Schmidt T., Rueckert L., Becker W., Huenges J., Garz A.K., Gohlke B.O., Zolg D.P., Kayser G., Vooder T., Preissner R., Hahne H., Tõnisson N., Kramer K., Götze K., Bassermann F., Schlegl J., Ehrlich H.C., Aiche S., Walch A., Greif P.A., Schneider S., Felder E.R., Ruland J., Médard G., Jeremias I., Spiekermann K., Kuster B., The target landscape of clinical kinase drugs. Science, 2017. 358: pii: eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim HS, Mendiratta S, Kim J et al. Systematic identification of molecular subtype‐selective vulnerabilities in non‐small‐cell lung cancer. Cell 2013; 155: 552–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet 2013; 382: 720–31. [DOI] [PubMed] [Google Scholar]
- 5. Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non‐small‐cell lung cancer. Lancet 2016; 388: 1012–24. [DOI] [PubMed] [Google Scholar]
- 6. Borghaei H, Paz‐Ares L, Horn L et al. Nivolumab versus docetaxel in advanced nonsquamous non‐small‐cell lung cancer. N Engl J Med 2015; 373: 1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herbst RS, Baas P, Kim DW et al. Pembrolizumab versus docetaxel for previously treated, PD‐L1‐positive, advanced non‐small‐cell lung cancer (KEYNOTE‐010): A randomised controlled trial. Lancet 2016; 387: 1540–50. [DOI] [PubMed] [Google Scholar]
- 8. Benci JL, Xu B, Qiu Y et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 2016; 167: 1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shin DS, Zaretsky JM, Escuin‐Ordinas H et al. Primary resistance to PD‐1 blockade mediated by JAK1/2 mutations. Cancer Discov 2017; 7: 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rizvi NA, Hellmann MD, Snyder A et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015; 348: 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rangachari D, VanderLaan PA, Shea M et al. Correlation between classic driver oncogene mutations in EGFR, ALK, or ROS1 and 22C3‐PD‐L1 ≥50% expression in lung adenocarcinoma. J Thorac Oncol 2017; 12: 878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soo RA, Kim HR, Asuncion BR et al. Significance of immune checkpoint proteins in EGFR‐mutant non‐small cell lung cancer. Lung Cancer 2017; 105: 17–22. [DOI] [PubMed] [Google Scholar]
- 13. Soo RA, Lim SM, Syn NL et al. Immune checkpoint inhibitors in epidermal growth factor receptor mutant non‐small cell lung cancer: Current controversies and future directions. Lung Cancer 2018; 115: 12–20. [DOI] [PubMed] [Google Scholar]
- 14. Su S, Dong ZY, Xie Z et al. Strong programmed death ligand 1 expression predicts poor response and De novo resistance to EGFR tyrosine kinase inhibitors among NSCLC patients with EGFR mutation. J Thorac Oncol 2018; 13: 1668–75. [DOI] [PubMed] [Google Scholar]
- 15. Haratani K, Hayashi H, Tanaka T et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation‐positive non‐small‐cell lung cancer based on T790M status after disease progression during EGFR‐TKI treatment. Ann Oncol 2017; 28: 1532–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. A large gene detection panel including 168 lung cancer related gene mutations.


