Abstract
Metabolic engineering requires fine-tuned gene expression for most pathway optimization applications. To develop a suitable suite of promoters, traditional bacterial promoter engineering efforts have focused on modifications to the core region, especially the −10 and −35 regions, of native promoters. Here, we demonstrate an alternate, unexplored route of promoter engineering through randomization of the UP element of the promoter—a region that contacts the alpha subunit carboxy-terminal domain instead of the sigma subunit of the RNA polymerase holoenzyme. Through this work, we identify five novel UP element sequences through library-based searches in Escherichia coli. The resulting elements were used to activate the E. coli core promoter, rrnD promoter, to levels on par and higher than the prevalent strong bacterial promoter, OXB15. These relative levels of expression activation were transferrable when applied upstream of alternate core promoter sequences, including rrnA and rrnH. This work thus presents and validates a novel strategy for bacterial promoter engineering with transferability across varying core promoters and potential for transferability across bacterial species.
Keywords: UP element, Promoter engineering, Expression modulation
1. Introduction
Metabolic engineering efforts can rewire microbial organisms into cellular factories capable of producing industrial chemicals [1], therapeutics [2], food supplements [[3], [4], [5]], and alternative fuels [6,7]. In this regard, these microbes can replace traditional chemical synthesis, reduce the use of harsh solvent reagents, provide precise stereochemistry, and reduce costs. To effectively engineer these pathways and introduce novel, synthetic circuitry into cells, gene expression must be modified [8,9]. At first approximation, rate control of gene expression is exerted by transcriptional initiation, which implicates the promoter sequence as a valuable synthetic part.
Historically, gene expression control in bacterial systems like Escherichia coli was achieved through a subset of isolated, non-native promoters including the T5 bacteriophage promoter [10] and the T7 phage promoter and polymerase [11]. While such promoter elements are effective at strong heterologous overexpression, they often consume too many cellular resources, leading to reduced cell growth and subsequently decreased overall product yields [12]. As an alternative strategy, native promoter structures can be engineered through both rational and evolutionary approaches. In this regard, these modified promoters function as tunable knobs for precise control of gene expression and, in the case of metabolic pathways, regulated metabolic flux [13].
A variety of approaches exist to alter the strength of a promoter, including random selection strategies based on error-prone PCR of a functional promoter [14,15]. Upon gaining an understanding of molecular interactions at the promoter, it is possible to view the complete bacterial RNA polymerase holoenzyme complex as consisting of two major parts: the core enzyme and a sigma subunit [16]. Within this architecture, the sigma subunit is responsible for transcription initiation aided by the recognition of two specific sequences within bacterial promoters, the −35 and the −10 (Pribnow Box) regions [17]. This binding event subsequently results in recruitment of the core enzyme [16,18]. Modifications to these −35 and −10 regions, including altering the spacer region, can substantially alter (and even abolish) promoter activity [19,20]. As a further complication in this process, the vast majority of bacterial species contain multiple different sigma subunits that vary in usage depending on cell state and environmental stress [21]. In many cases, the conserved −10 and −35 regions and accompanying spacer sequence requirements differs for each sigma factor within a single organism [22], and thereby complicate promoter engineering efforts relying solely on this region.
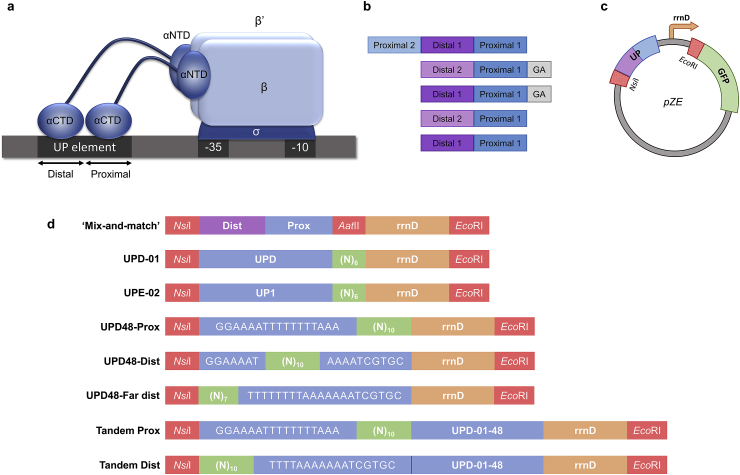
Recent methods for promoter engineering in fungal hosts have demonstrated that upstream regions can exert significant control on promoter activity in a manner that is independent of the core promoter region [23,24]. An analogous upstream interaction occurs in bacterial promoters through interactions with the RNA polymerase holoenzyme [25]. Specifically, at a region upstream of the −35 element, at approximately the −40 to −60 positions, exist so-called UP elements that are capable of activating transcription through contact with the alpha subunit carboxy-terminal domain (αCTD) of the RNA polymerase core enzyme [[26], [27], [28]]. The sequence motif of UP elements is typically rich in adenine and thymine dinucleotide tracts and is generally divided into two parts: the proximal and distal regions. It is believed that each respective region interacts with a single αCTD monomer [28] (Fig. 1a), thus serving an analogous role as upstream activating sequence elements for yeast promoters. Thus far, modifying promoter activity through the engineering of UP elements has received limited attention in the field [29].
Fig. 1.
UP element architecture and library design. a The UP element lies at the −40 to −60 positions in canonical bacterial promoters. The UP element can be divided into distal and proximal regions, each thought to interact with precisely one of two total αCTD monomers. b Conceptual design of the 7 UP elements comprising the initial mix-and-match style library. Sequences corresponding to each distal and proximal region can be found in the Appendix (Table SV). Grey boxes present on the end of sequences UPB and UPC indicate presence of two additional base pairs, ‘GA,’ downstream of the proximal region. Choice of designs based on previous publication [28]. c UP elements are cloned into a pZE plasmid upstream of the E. coli core promoter, rrnD, and reporter GFP gene. d Initial cloning scheme utilizes adjacent restriction enzyme sites, NsiI and AatII, for insertion of rationally designed UP element sequences. Libraries UPD-01 and UPE-02 randomize the hexameric AatII site of the two highest performing variants from the initial cloning scheme. The three UPD48 libraries are based on the sequence of the highest performing variant (UPD-01-48) across both libraries, UPD-01 and UPE-02, and randomize the distal, far distal, and proximal regions of this basis sequence, respectively.
Here, we seek to establish both semi-rational and library-based design strategies to alter bacterial promoter activity through modifications only to the UP element by using the E. coli rRNA promoter, rrnD, as the core promoter element. To do so, we established a small mix-and-match style initial library based on distal and proximal elements. Subsequent library designs were built in a hierarchal manner (Fig. 1d), resulting in the characterization of five novel UP elements able to activate expression of the rrnD core promoter by up to 9-fold. The performance of these promoters is benchmarked by comparing transcriptional capacity to a commonly used strong, synthetic constitutive promoter (OXB 15). Finally, we demonstrate the rather generic activation potential of these UP elements by placing them upstream of additional rRNA core promoters, rrnA and rrnH, and achieve a proportion expression actuation. As such, this work thus develops a unique strategy for bacterial promoter engineering through altering simply the UP elements alone in a manner that is independent of the core promoter itself.
2. Materials and methods
2.1. Media and strain propagation
The expression vectors were cloned and propagated in Escherichia coli dH10β. Cultures were cultivated in Luria-Bertani (LB) medium [30] (Teknova) at 37 °C with 225 r.p.m. orbital shaking. LB was supplemented with 25 mg/ml kanamycin (Gold Biotechnology) for plasmid maintenance and propagation. Electrotransformations were completed by adding 2 μL of ligation mixtures to 50 μL E. coli dH10β competent cells. This mixture was placed in a 2 mm electroporation cuvette (Bioexpress) and electroporated with the Biorad Genepulser Xcell at 2.5 kV. Transformants were recovered with 950 μL SOC medium (Cellgro) for 1 h at 37 °C and plated on LB agar (1.5%) containing 25 μg/ml kanamycin. Plates were incubated overnight and at 37 °C. To isolate plasmids, single colonies were picked from plates and grown overnight in 5 ml LB. Plasmids were isolated (QIAprep Spin Miniprep Kit, Qiagen) from resulting cultures and confirmed by sequencing.
2.2. Plasmid construction
All pZE plasmid constructs (Table SI) were assembled using restriction digest cloning. Oligonucleotides listed (Table SII) were purchased from Integrated DNA Technologies. Plasmid inserts were assembled through double stranding reactions with Phusion DNA polymerase (NEB) per manufacturer instructions. All digests were performed using restriction enzymes (NEB) per manufacturer instructions. Assembled inserts and digests were purified using the QiaQUICK PCR purification kit (Qiagen). Prior to ligation, digested plasmids were phosphatase with Antarctic Phosphatase (NEB) for 3–18 h and heat inactivated for 20 min at 65 °C. Ligations (T4 DNA Ligase, NEB) were performed for 3–18 h in a 10:1 insert to backbone ratio at 16 °C followed by heat inactivation at 65 °C for 10 min. Insertion of UP element-core promoter-reporter gene sequences into pACYC plasmid constructs (Table SI), reporter gene substitutions, and rrnA and rrnH core promoter substitutions were performed via Gibson assembly.
2.3. Fluorescence measurements with flow cytometry and FACS
All cultures were inoculated from cell stocks stored at – 80 °C in 15% glycerol into 3 ml LB in biological triplicate. Cultures were grown overnight at 37 °C in a 225 RPM orbital shaker. Each culture was restarted by inoculation of 20 μL culture to 3 ml M9 minimal media containing 5 g/L d-glucose and supplemented with 0.1% casamino acids. Samples tested in exponential phase were grown for 6 h after inoculation. Samples tested in stationary phase were grown for 16 h after inoculation. Samples were set on ice prior to measurements to stop growth. GFP fluorescence was measured (BD Accuri C6 cytometer) at an excitation wavelength of 488 nm and emission wavelength of 530 ± 15 nm. 10,000 events per sample were collected at a flow rate of 14 μL/min. Average fluorescence and standard deviation was calculated from the geometric mean fluorescence values of technical triplicates. Collected data was analyzed using FlowJo software. β-lactamase expression levels were measured via fluorescence-coupled cleavage activity through the Fluorocillin™ Green 345/530 β-lactamase substrate kit (Invitrogen).
2.4. qPCR
E. coli cultures were grown in analogous conditions to those used in Flow cytometry and FACS, grown to exponential phase. 1 ml aliquots of each culture were set on ice in preparation for flow cytometry. One of each of the biological triplicate cultures was taken for each strain variant, and an OD measurement was made. Aliquots of this culture normalized by OD were then pelleted via centrifugation and used in RNA extraction per manufacturer instructions (Trizol® Max™ Bacterial RNA Isolation kit). Extracted RNA was reverse transcribed (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems). qPCR was performed by normalizing the total RNA concentrations added to each reaction (6.67 ng/μL), then quantification in technical triplicate (Power SYBR Green Master Mix, ThermoFisher Scientific). GFP transcript levels were measured relative to that of the housekeeping gene hcaT [31] (Viia 7 Real Time PCR Instrument, Life Technologies). Primers used for quantification listed in Table SII.
3. Results
3.1. A semi-rational library of proximal and distal elements can alter promoter activity
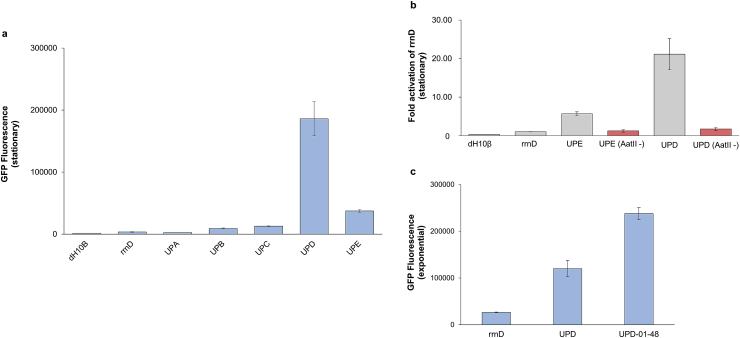
Initially, we sought to evaluate whether a mix-and-match strategy (i.e. combinatorial, separate assembly of proximal and distal elements) could reconstitute a functional and strong UP element. To do so, sequences for three proximal and two distal regions, comprised of 9 and 14 base pairs respectively, were chosen from a previous study [28]. We then constructed 5 distinct combinations of these elements for testing promoter performance and UP element function (Fig. 1b) (Table SIII). In this effort, we sought to identify the strongest UP element based on the ability to amplify expression of the base rrnD promoter, an E. coli rRNA promoter [32,33]. To do so, each variant was cloned upstream of the 37 base pair rRNA promoter, rrnD, and a GFP reporter gene in a pZE plasmid (Table SI) (Fig. 1c) between AatII (between the core promoter and UP element) and NsiI (adjacent to the upstream region of the UP element) restriction sites. Through flow cytometry, we demonstrate that two of our selected combinations (named UPD and UPE), activate expression of GFP with respect to the core promoter (Fig. 2a).
Fig. 2.
Results of initial libraries. a Stationary phase mean fluorescence measurements of mix-and-match UP elements UPE and UPA-UPD. b Stationary phase fold activation over rrnD of initial cloning scheme UP element constructs UPD and UPE, with and without hexameric AatII restriction site. In both cases, loss of AatII severely decreases UP element performance. c Exponential phase comparison of fluorescence levels of UPD-01-48, the highest expressing variant from Library UPD-01 (which randomized the hexameric AatII sequence of UPD) to UPD and core promoter rrnD, positive and negative controls, respectively. All error bars represent standard deviation from technical triplicates.
3.2. Removing the impact of restriction sites on UP element performance
While the UP elements established above demonstrate function in this system and demonstrate the ability to perform combinatorial assembly of UP elements, the constructs contain restriction site cloning scars that can be strong affecters of gene expression [34,35]. Moreover, these scars can impact the transferability of these UP elements to other core promoter constructs. To evaluate the impact of these scars (particularly from AatII), we simply removed these sites and indeed saw a significant decrease in expression activation (Fig. 2b). In an effort to ameliorate the library and thus impact from this site, two new UP element libraries were designed by randomizing the hexameric AatII sites of UPD and UPE (thus keeping the number of base pairs the same) in an effort to seamlessly clone the construct while maintaining function. These libraries, termed Library UPD-01 and Library UPE-02, were established, and highly functional members of this library were isolated using FACS (aimed at isolating the top 10% of fluorescent cells). From these libraries, a total of 48 fluorescent colonies were chosen for further testing through individual culture flow cytometry. Overall, it was found that members of the UPD-based library (Library UPD-01) showed greater fluorescence activation than members of the UPE-based library (Library UPE-02) (Fig. S1). The best performing UP element from this library, UPD-01-48, activated GFP expression 2.0-fold compared to UPD, and 8.9-fold compared to rrnD in a final exponential phase screen (Fig. 2c).
3.3. Libraries based on regional randomization of UPD yield variants stronger than UPD, but not stronger than UPD-01-48
Our highest expression-activating candidate from initial library testing, UPD-01-48, was used as a starting point in design of three additional libraries. Prior to this endeavor, a variety of additional strategies were undertaken to explore sequence variations outside of the UPD-01-48 base sequence (Fig. S2, Fig. S3). Unfortunately, large potential library size made these designs challenging to effectively screen and most failed to produce activating UP elements. To avoid future screening of extraordinarily large library sizes, we aimed to limit potential sequence diversity to approximately 1 million potential variants to balance between transformation efficiency of E. coli [36] and the ability to sample exhaustively.
In three new libraries, UPD48-Dist, UPD48-Far dist, and UPD48-Prox, the distal, far distal, and proximal regions of UPD-01-48 were randomized, respectively (Fig. 1d). The far distal region was selected according to previous works suggesting additional interactions contributing to αCTD binding in this region [37]. These libraries were evaluated using a similar protocol to libraries UPD-01 and UPE-02, however, sorts and screens were conducted in exponential phase rather than stationary phase, as initial screens of libraries in stationary phase suffered from high standard deviations and low percentages of fluorescent cells (Fig. S4) due to known growth phase dependence of rrn core promoters [38].
Though many variants of these libraries were identified that were stronger than UPD (in fact, more than 20% of candidates screened from each library had fluorescence levels greater than or equal to that of UPD), very few variants reached fluorescence levels comparable to that of UPD-01-48 (Fig. S5). As a result, these finding suggest a possible route to identify new UP elements, but performance was maximized by the UPD-01-48 element isolated as described above.
3.4. Sequence homology analysis reveals GC-rich preference
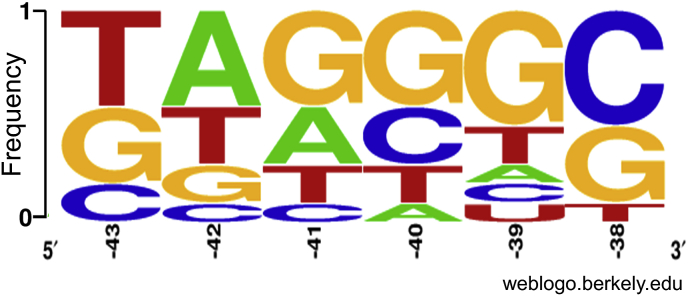
To determine whether activating UP elements identified in Library UPD48-Prox shared any sequence homology with activating elements identified in Library UPD-01, we sequenced a small group of UP elements emerging in the top 10% fluorescence values for each respective library sort. We then performed sequence alignment on this group and found GC rich sequences in the −38 to −40 positions (Fig. 3). This was unexpected, as previously synthesized UP elements were reported to be rich in AT tracts to facilitate interaction with the αCTD (Fig. S6) [28]. These findings, in addition to the limited success of libraries as described above, prompted us to evaluate an alternative method to further improve performance.
Fig. 3.
Sequence alignment between Libraries UPD-01 and UPD48-Prox. WebLogo representation of sequence homology between libraries UPD-01 and UPD48-Prox, the best performing library from the three different UPD-01-48-based libraries. The following UP elements were included in the webLogo: UPD-01-1, UPD-01-6, UPD-01-12, UPD-01-18, 29–47, UPD-01-48, UPD48-Prox-18, UPD48-Prox-30, UPD48-Prox −53, UPD48-Prox −80, UPD48-Prox-84 and UPD. WebLogo y-axis represents relative nucleotide frequencies instead of bits.
3.5. Tandem repeat libraries activate expression of UP element UPD-01-48
It has been previously shown that sequences up to 100 base pairs upstream of the transcription start site can further contribute to RNAP association [26,33,39]. Additionally, endogenous promoters such as the Fis promoter have upstream tandem RNAP binding sites that facilitate Fis-controlled gene expression [40]. Furthermore, our prior work in fungal [23,24,41] and mammalian [42] promoters with tandem upstream elements suggest that promoters can be enhanced by tandem upstream repeats. As a result, it is possible that placing UP element sequences in tandem could further activate gene expression by providing an extended interaction region for αCTD to bind.
The initial strategy selected to improve UP element function was to place tandem repeats of UP element sequences upstream of the core promoter. Using a small library of four variations comprised of combining previously identified UP element sequences (Table SV), we were unable to achieve designs that activated expression compared to that of UPD (Fig. S7). Based on the inability to semi-rationally create a tandem UP element, we then created two library-based searchers to further explore the sequence space of tandem UP elements. To do so, we established one library that randomized solely a proximal region and another that randomized solely a distal region in two tandemly placed UPD-01-48 sequences (based on its strong performance in our prior library selections). These libraries were termed tandem-proximal (TP) and tandem-distal (TD), respectively.
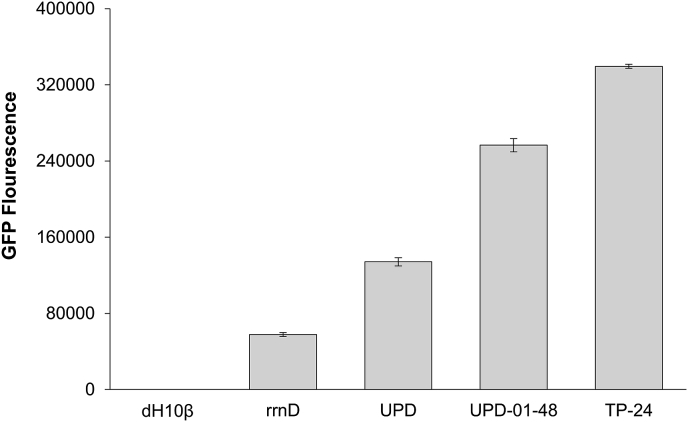
In contrast to results above, the approach of using a tandem UP element library enabled us to establish and identify highly functional elements and stronger promoters. Specifically, this approach was the most fruitful of the attempted design strategies so far with over 87% of the members of both proximal and distal libraries having a fluorescence greater than the core promoter (Fig. S8). A similar FACS based sorting strategy was used for these promoters as described above with many variants selected and analyzed. Among the isolated variants tested and characterized, one element from the tandem proximal library, TP-24, had a fluorescence value greater than that of UPD-01-48, and was the highest-activating UP element identified in this study (Fig. 4). Ultimately, the wide range of expression (from low to high) seen in this library suggest that the DNA upstream of the even a single UP element of the promoter can have a significant effect on the function of the downstream promoter performance.
Fig. 4.
Tandem UP element library performance. Comparison of fluorescence levels between highest expressing tandem-proximal variant, TP-24, and positive controls, 29–48 and UPD, and negative controls, rrnD and untransformed dH10β. Fluorescence measured in exponential phase. Error bars represent standard deviation from technical triplicates.
3.6. Final collection of promoters through UP element engineering and transcriptional profiling compare favorably with other strong bacterial promoters
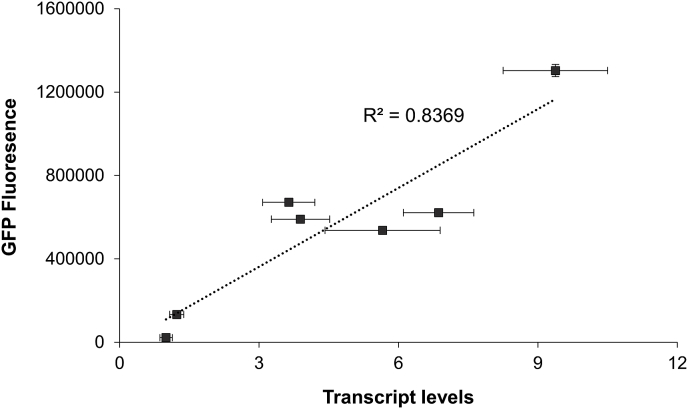
To fully characterize the range of potential expression enabled by an UP element engineering approach, five UP elements spanning low to high activation of the rrnD core promoter were chosen. These elements included UP-01-48, TP24, and one element from each of the UPD-01-48-based libraries (UPD48-Dist, UPD48-Prox, UPD48-Far dist). These specific elements were chosen in order to encompass a range of medium to high expression levels, thus demonstrating the capacity of UP element engineering to achieve modular expression control. Using this final UP element set evaluated here, the strongest performing sequence, TP-24, activates rrnD expression 9.4-fold, and the weakest sequence, UPD48-Prox-18, activates rrnD expression 4.4-fold (Table 1). To demonstrate that these effects seen at the fluorescence level were indeed due to expression level changes, we conducted qPCR measurements. Collectively, this data (Fig. 5) demonstrates a strong linear relationship between GFP transcript and GFP fluorescence, thus implicating the performance of these UP elements at the transcriptional level.
Table 1.
Performance of select UP elements compared to commonly used promoters. Fold-activation over rrnD calculated from fluorescence data pictured in Fig. 6
| Variant | Fold-improvement in expression over rrnD |
|---|---|
| UPD48-Prox-18 | 4.41 |
| UPD48-Dist-66 | 4.65 |
| UPD48-Far Dist-62 | 5.36 |
| UPD-01-48 | 6.49 |
| TP24 | 9.38 |
| OXB15 | 6.84 |
Fig. 5.
qPCR confirmation of expression effects. Fluorescence measurements vs transcript levels of selected UP elements. Transcript levels normalized to levels of no-promoter no-UP element pZE control. Fluorescence measurements made in exponential phase. Error bars represent standard deviation of technical triplicates.
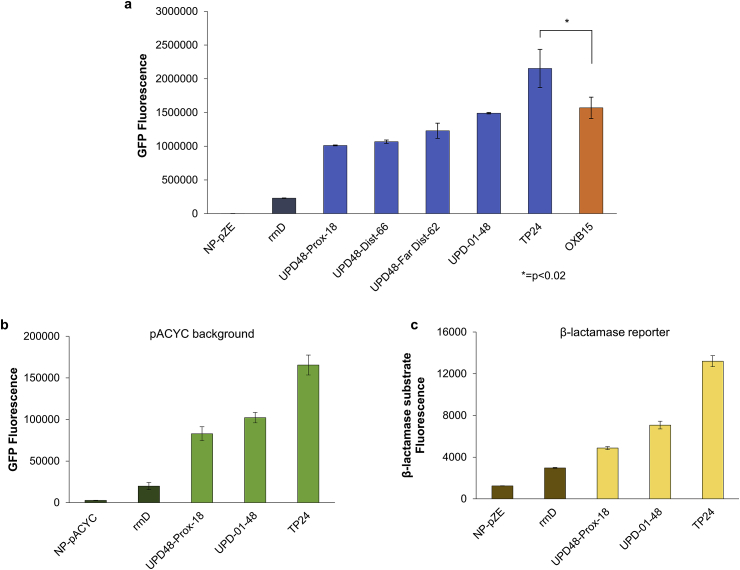
We sought to benchmark the performance of these UP element engineered promoters with another strong, commonly used promoter. To do so, we chose OXB15, a high-level expression promoter created from mutagenesis of the bacterial recA promoter [43] as a point of reference due to its free availability and due to its signature level of expression in bacterial systems. The OXB15 promoter, while not the highest expression-level promoter from the suite of 20 constitutive OXB promoters, was chosen for benchmarking our UP elements due to the unfavorable effects we exhibited on cell growth that stemmed from the reportedly higher-expressing promoters such as OXB 20. Thus, direct comparisons could be made with OXB15 due to similar growth kinetics. Following plasmid assembly, fluorescence levels were measured using flow cytometry in the exponential growth phase. This comparative analysis demonstrates that the strongest-activating UP element, TP-24, activates rrnD expression to a level higher than the strongest used common promoter, OXB15 (Fig. 6a). Specifically, while OXB15 has a 6.8-fold higher fluorescence compared with rrnD, UP element TP-24 is 9.4-fold higher than this same rrnD baseline. Based on these results, it is evident that the use of UP element engineering is a novel promoter engineering strategy in E. coli, with the ability to generate promoters that are on-par and higher than commonly used promoters in the same expression activation range.
Fig. 6.
Capacity of UP elements to modulate expression levels. a Fluorescence levels of final 5 UP elements in pZE background, encompassing a range of medium to high activation, compared to other strong bacterial promoter OXB15. Fluorescence measurements of selected UP elements shown in blue, commercial/benchmarking OXB15 promoter shown in orange. b Fluorescence levels of select UP elements in pACYC background. c Fluorescence levels resulting from β-lactamase activity assay. In each plot, negative controls rrnD and no-promoter no-UP element plasmid shown in darker color. Error bars represent standard deviation of biological triplicates. All samples in each plot were cultured and measured in exponential phase same-day for accuracy of comparison.
Additionally, relative activation levels of rrnD are conserved when UP element, core promoter, and reporting gene are used in a differing plasmid background (pACYC184), showing the relative activation of UP elements is independent of plasmid copy number (Fig. 6b and Fig. S9). Finally, upon replacing the GFP reporter gene with TEM-1 β-lactamase, fluorescence levels from a β-lactamase activity assay show similar fold activation across select UP elements (Fig. 6c). These results suggest UP element activation may be independent of the transcribed gene.
3.7. Engineered UP elements are functional and transferrable across multiple core promoter elements
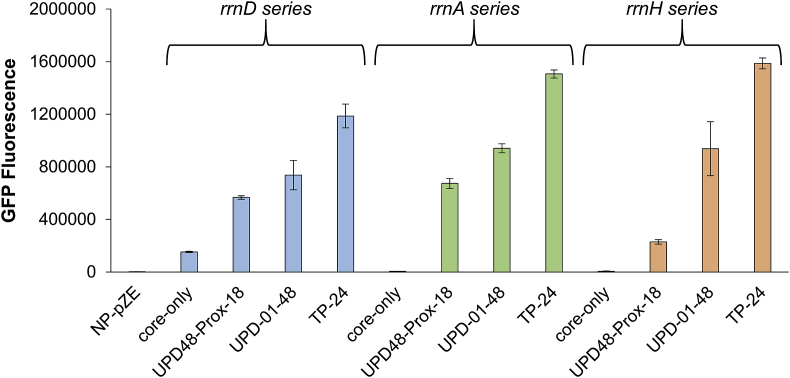
Finally, we sought to investigate whether the expression modulation enabled by these engineered UP elements was transferrable to other alternative core promoters. To this end, we selected a medium-low, medium, and high expressing UP element variant (specifically, the UPD48-Prox, UP-01-48, and TP24 elements) and cloned these upstream of two alternative core promoters (rrnA or rrnH) to drive GFP expression. Fluorescence measurements of these resulting strains via flow cytometry demonstrate a conservation of expression modulation by these UP elements across each of these core promoters (Fig. 7). Indeed, strong linear relationships were seen between GFP transcript levels and GFP fluorescence across each set of these core promoters once again suggesting that these UP elements function through altered transcriptional rates (Fig. S10). The highest-activating UP element, TP-24, was able to activate expression of the rrnA and rrnH minimal core promoters to approximately 360 and 390-fold, respectively (Table 2).
Fig. 7.
Capacity of UP elements to modulate expression levels across multiple rRNA core promoters. Fluorescence levels of medium, medium-high, and high expression-modulating UP elements upstream of variable rRNA core promoters. rrnD, rrnA, and rrnH core promoter strains shown in blue, green, and orange, respectively. Negative control, no-promoter no-UP element pZE, shown in grey. Error bars represent standard deviation of biological triplicates. All samples cultured and measured in exponential phase same-day for accuracy of comparison.
Table 2.
Fold-activation of select UP elements with respect to various downstream rRNA cores. Quantities calculated from fluorescence data pictured in Fig. 7.
| rRNA core promoter |
|||
|---|---|---|---|
| rrnD | rrnA | rrnH | |
| UPD48-Prox-18 | 3.7 | 161 | 42 |
| UPD-01-48 | 4.8 | 224 | 174 |
| TP-24 | 7.8 | 359 | 391 |
4. Discussion
Through this work, we present the development and validation of a set of unique UP element sequences capable of modulating the activity of a core promoter in E. coli. This effort was accomplished through the development of several sequential rational and semi-rational library design strategies. The highest activating UP elements discovered through these library approaches were evaluated on a sequence basis. Surprisingly, GC-rich tracts were favored in the −38 to −40 positions of the UP element. This finding contradicts previous studies that suggest UP elements are characterized by AT tracts thought to facilitate binding of the RNA polymerase αCTD [34]. Based on this discovery, it is possible that these regions actually serve as a spacer sequence that optimizes αCTD interaction with the UP element by positioning it such as to force interactions with higher AT content upstream regions.
While several libraries were considered in this work, the highest performing libraries were the tandem libraries which demonstrates a function of far-upstream DNA on influence promoter function in bacteria. The randomization of the tandem proximal region was particularly fruitful, with 87% of candidates activating expression when compared to UPD. From this library, we identified the highest activating UP element in this study, TP-24, which increased expression of rrnD by 9.4-fold. The wide range of activations seen in the tandem-proximal library suggests that DNA upstream of the UP element has a significant effect on promoter function. While follow-up studies would be required to further elucidate the mechanism of tandemly repeated UP element effects, we conjecture that this could be due to recruitment of additional transcriptional regulators or changes in local DNA topology.
After screening our hierarchal library designs, we sought to evaluate five different UP elements spanning medium to high activation of core promoter expression with respect to commercially available bacterial promoters. We find that expression activation due to UP element engineering compared favorably to the OXB15 promoter, a high expression-level promoter previously reported and commonly used. We also show UP element expression activation is independent of plasmid copy number and can be realized through varying reporters. Finally, we show that engineered UP elements are portable and modular. Specifically, these regions can be placed upstream of variable core promoters to yield similar relative expression modulations. These results echo the results of hybrid promoter engineering efforts in other host organisms. The results from this study show that promoter engineering through UP element manipulation alone is competitive with other strategies of promoter engineering and can be applied to fine-tune performance of multiple core promoters. As such, we postulate that these UP element sequences could activate core promoters pertaining to various other sigma factors to predictable degrees. Finally, it appears that our proposed workflow of UP element design and the general strategy of hybrid promoter engineering could serve as a model to be used in other bacterial species for creating unique promoters.
5. Conclusion
Through this work, we have demonstrated the capacity to engineer UP elements for varying the expression of E. coli promoters. Prior to this work, most promoter engineering efforts attempted to modulate expression level through manipulation of the spacer sequence between the −10 and −35 promoter regions and other core promoter elements. In contrast, the αCTD is conserved across a single bacterial species and more independent of host factors like sigma-factors, making these elements reliable for activating expression across different species. In this work, we characterize a group of 5 UP elements capable of moderate to high activation of a core rrnD promoter. Additionally, we demonstrate that these UP elements can activate additional varying rRNA core promoters to predictable levels of expression. These promoters and accompanying strategy have potential for fine-tuned expression in bacterial species.
Conflicts of interest
All authors declare no conflict of interest.
Funding
This study was funded by the Air Force Office of Scientific Research under Award No. FA9550-14-1-0089.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2019.04.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Curran K.A., Leavitt J.M., Karim A.S., Alper H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Ro D.K., Paradise E.M., Quellet M., Fisher K.J., Newman K.L., Ndungu J.M. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 3.Becker J., Klopprogge C., Herold A., Zelder O., Bolten C.J., Wittmann C. Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum-over expression and modification of G6P dehydrogenase. J Biotechnol. 2007;132:99–109. doi: 10.1016/j.jbiotec.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.W., Keasling J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng. 2001;72:408–415. doi: 10.1002/1097-0290(20000220)72:4<408::aid-bit1003>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 5.Sun T., Miao L., Li Q., Dai G., Lu F., Liu T. Production of lycopene by metabolically-engineered Escherichia coli. Biotechnol Lett. 2014;36:1515–1522. doi: 10.1007/s10529-014-1543-0. [DOI] [PubMed] [Google Scholar]
- 6.Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 7.Cordova L.T., Alper H.S. Central metabolic nodes for diverse biochemical production. Curr Opin Chem Biol. 2016;35:37–42. doi: 10.1016/j.cbpa.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H., Lin M., Shi H., Ji W., Huang L., Zhang X. Programming a Pavlovian-like conditioning circuit in Escherichia coli. Nat Commun. 2014;5:3102. doi: 10.1038/ncomms4102. [DOI] [PubMed] [Google Scholar]
- 9.Hussain F., Gupta C., Hirning A.J., Ott W., Matthews K.S., Josic K. Engineered temperature compensation in a synthetic genetic clock. Proc Natl Acad Sci. 2014;111:972–977. doi: 10.1073/pnas.1316298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rommens J., Macknight D., Pomeroy-aoney L., Jay E. Gene expression: chemical synthesis and molecular cloning of a bacteriophage T5 (T5P25) early promoter. Nucleic Acids Res. 1983;11:5921–5940. doi: 10.1093/nar/11.17.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakley J.I., Strothkamp R.E., Sarris A.H., Coleman J.E. T7 RNA polymerase: promoter structure and polymerase binding. Biochemistry. 1979;18:528–537. doi: 10.1021/bi00570a023. [DOI] [PubMed] [Google Scholar]
- 12.Del Vecchio D., Murray R.M. Synthetic biology. In: Baillieul J., Samad T., editors. Encycl. Syst. Control. Springer; 2015. [Google Scholar]
- 13.Leavitt J.M., Alper H.S. Advances and current limitations in transcript-level control of gene expression. Curr Opin Biotechnol. 2015;34:98–104. doi: 10.1016/j.copbio.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Alper H., Fischer C., Nevoigt E., Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazeck J., Alper H.S. Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol J. 2013;8:46–58. doi: 10.1002/biot.201200120. [DOI] [PubMed] [Google Scholar]
- 16.Weaver R. McGraw-Hill Higher Education; Fifth: 2011. Molecular biology. [Google Scholar]
- 17.Murakami K.S., Masuda S., Darst S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 18.Hinkle D.C., Chamberlin M.J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972;70:157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- 19.Rud I., Jensen P.R., Naterstad K., Axelsson L. A synthetic promoter library for constitutive gene expression in Lactobacillus plantarum. Microbiology. 2006;152:1011–1019. doi: 10.1099/mic.0.28599-0. [DOI] [PubMed] [Google Scholar]
- 20.Jensen P.R., Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton-Foot M., Gey van Pittius N.C. The complex architecture of mycobacterial promoters. Tuberculosis. 2013;93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Ruff E.F., Thomas Record M., Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–1062. doi: 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazeck J., Garg R., Reed B., Alper H.S. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng. 2012;109:2884–2895. doi: 10.1002/bit.24552. [DOI] [PubMed] [Google Scholar]
- 24.Blazeck J., Liu L., Redden H., Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77:7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hook-Barnard I.G., Hinton D.M. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Biol. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 26.Ross W., Gourse R.L. Sequence-independent upstream DNA- αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc Natl Acad Sci. 2005;102:291–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H., Tang H., Ebright R.H. Functional interaction between RNA polymerase α subunit C-terminal domain and σ70 in UP-element and activator-dependent transcription. Mol Cell. 2003;11:1621–1633. doi: 10.1016/s1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 28.Estrem S.T., Ross W., Gaal T., Chen Z.W.S., Niu W., Ebright R.H. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Q., Fong S.S. Study of in vitro transcriptional binding effects and noise using constitutive promoters combined with UP element sequences in Escherichia coli. J Biol Eng. 2017;11:33. doi: 10.1186/s13036-017-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J., Russell D. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- 31.Zhou K., Zhou L., Lim Q., Zou R., Stephanopoulos G., Too H.P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao L., Ross W., Appleman J.A., Gaal T., Leirmo S., Schlax P.J. Factor independent activation of rrnB P1. An “Extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 33.Aiyar S.E., Gourse R.L., Ross W. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase α subunit. Proc Nat Acad Sci USA. 1998;95:14652–14657. doi: 10.1073/pnas.95.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirzadeh K., Martínez V., Toddo S., Guntur S., Herrgård M.J., Elofsson A. Enhanced protein production in Escherichia coli by optimization of cloning scars at the vector-coding sequence junction. ACS Synth Biol. 2015;4:959–965. doi: 10.1021/acssynbio.5b00033. [DOI] [PubMed] [Google Scholar]
- 35.Crook N.C., Freeman E.S., Alper H.S. Re-engineering multicloning sites for function and convenience. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr346. e92–e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dower W.J., Miller J.F., Ragsdale C.W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhodius V.A., Mutalik V.K., Gross C.A. Predicting the strength of UP-elements and full-length E. coli σE promoters. Nucleic Acids Res. 2012;40:2907–2924. doi: 10.1093/nar/gkr1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda M., Shimada T., Akira I. Strength and regulation of seven rRNA promoters in Escherichia coli. PLoS One. 2015 doi: 10.1371/journal.pone.0144697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis C.A., Capp M.W., Record M.T., Saecker R.M. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:285–290. doi: 10.1073/pnas.0405779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerganova V., Maurer S., Stoliar L., Japaridze A., Dietler G., Nasser W. Upstream binding of idling RNA polymerase modulates transcription initiation from a nearby promoter. J Biol Chem. 2015;290:8095–8109. doi: 10.1074/jbc.M114.628131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blazeck J., Miller J., Pan A., Gengler J., Holden C., Jamoussi M. Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Appl Microbiol Biotechnol. 2014;98:8155–8164. doi: 10.1007/s00253-014-5895-0. [DOI] [PubMed] [Google Scholar]
- 42.Cheng J.K., Alper H.S. Transcriptomics-guided design of synthetic promoters for a mammalian system. ACS Synth Biol. 2016;5:1455–1465. doi: 10.1021/acssynbio.6b00075. [DOI] [PubMed] [Google Scholar]
- 43.Oxford Genetics . 2016. pSF-OXB15 (OG558) Strong promoter E. coli vector.https://www.oxfordgenetics.com/Products/Plasmids/Details/Bacterial/pSF-OXB15/OG558 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.