Genome sequencing in potato plants regenerated from protoplasts reveals widespread changes in chromosome number and structure.
Abstract
Nontransgenic genome editing in regenerable protoplasts, plant cells free of their cell wall, could revolutionize crop improvement because it reduces regulatory and technical complexity. However, plant tissue culture is known to engender frequent unwanted variation, termed somaclonal variation. To evaluate the contribution of large-scale genome instability to this phenomenon, we analyzed potatoes (Solanum tuberosum) regenerated from either protoplasts or stem explants for copy number changes by comparison of Illumina read depth. Whereas a control set of eight plants that had been propagated by cuttings displayed no changes, all 15 protoplast regenerants tested were affected by aneuploidy or structural chromosomal changes. Certain chromosomes displayed segmental deletions and duplications ranging from one to many. Resampling different leaves of the same plant found differences in three regenerants, indicating frequent persistence of instability. By comparison, 33 regenerants from stem explants used for Agrobacterium-mediated transformation displayed less frequent but still considerable (18%) large-scale copy number changes. Repetition of certain instability patterns suggested greater susceptibility in specific genomic sites. These results indicate that tissue culture, depending on the protocol used, can induce genomic instability resulting in large-scale changes likely to compromise final plant phenotype.
Protoplast modification via nucleoprotein complexes results in high efficiency editing and transgene-free genomes (Woo et al., 2015; Andersson et al., 2018). When regeneration is possible, at least some of the resulting plants should display only the targeted changes. In contrast, regulatory compliance of genome-edited plants when using stably integrated transgenes requires, at a minimum, elimination of any transgene by meiotic segregation. This, however, is not a viable approach in clonally propagated, highly heterozygous crops because the optimal parental genotype is unlikely to be frequent in the progeny. Somaclonal variation, i.e. genotypic and phenotypic differences from the source plant (Landsmann and Uhrig, 1985; Lee and Phillips, 1988; Bao et al., 1996), has been associated with epigenetic changes (Stroud et al., 2013; Ong-Abdullah et al., 2015; Han et al., 2018), single nucleotide mutations (Miyao et al., 2012), chromosomal structure changes, and aneuploidy (Lee and Phillips, 1988). Notwithstanding the high likelihood of large phenotypic impact, chromosomal mutation, such as aneuploidy and structural changes, remain poorly documented. Potato (Solanum tuberosum) is a major caloric source in both industrialized and developing countries. Its breeding is complicated by its autotetraploid and highly heterozygous genome, which also hinders the use of seed-based propagation and breeding (Potato Genome Sequencing Consortium et al., 2011; Hardigan et al., 2017). In the 1980’s, potato emerged as a model for somaclonal variation, even spurring commercial interest as an alternative to breeding (Evans, 1989; Karp et al., 1989). Multiple varieties were shown to be amenable to protoplasting followed by regeneration. Notwithstanding the interest, production of relevant commercial varieties through this method has been limited (Krishna et al., 2016). The possibility of chromosomal instability was highlighted in cytological studies, but structural changes were detected infrequently (Karp et al., 1982; Creissen and Karp, 1985; Sree Ramulu et al., 1986). Interestingly, the outcome of the regeneration process was thought to result in a binary outcome: either somaclonal variants, or normal plants that resembled in all characteristics to the protoplast-derived variety. Implicitly, the assumption was that the latter plants had dodged the “somaclonal bullet.” Lacking a mechanistic understanding of what causes somaclonal variation, this assumption is often extended to explant regenerants used for Agrobacterium or biolistic transformation. In other words, off-types commonly encountered among plants produced by dedifferentiation and proliferation of somatic cells may be the product of mutagenic mechanisms whose frequency can be minimized, and phenotypic screens are adequate to address the problem. The need to explore these assumptions is underscored by the unexplained yield drag connected with transgenic modifications (Elmore et al., 2001). To enhance our molecular understanding of somaclonal variation, we studied copy number changes in potatoes regenerated from either protoplasts or from explants used for Agrobacterium transformation. We found an unexpected degree of chromosomal and segmental instability in the genomes of protoplast regenerants, whereas plants regenerated from stems displayed less genome instability, but still at a significant frequency.
RESULTS AND DISCUSSION
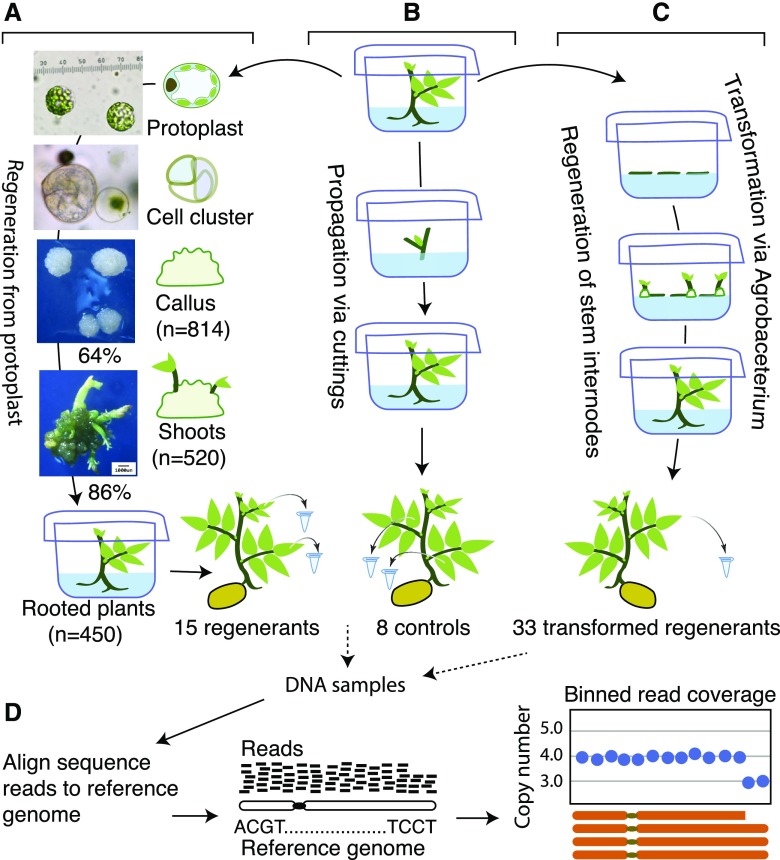
We produced protoplasts from autotetraploid potato variety Desiree and induced regeneration through callus and shoot formation (Fig. 1A). In preparation for genome editing, we wanted to measure the rate of sterility and abnormalities among regenerated plants. In two experiments we collected ∼400 plants, some derived from the same callus and thus the same protoplast (Supplemental Table S1). Overall, the phenotype of 100 plants grown in the greenhouse resembled that of the starting clone. Most displayed the expected nuclear content (Supplemental Table S2). A minority displayed changes from slight to obvious (Fig. 2; Supplemental Table S3). To investigate whether chromosomal alterations were associated with these abnormalities, we used whole genome sequencing to analyze the karyotypes of five phenotypically abnormal regenerants, ten phenotypically normal regenerants, and eight control samples that were not regenerated from protoplasts. For each individual, an average of 7.58 million Illumina sequence reads were generated and mapped to the S. tuberosum Group Phureja DM v4.04 reference genome (Hardigan et al., 2016). Read counts (mean = 1155; std = 268) binned in 0.25-Mb consecutive, nonoverlapping genomic bins were standardized for chromosome copy dosage using the counts from a single control plant (Fig. 1C; see “Materials and Methods”). S. tuberosum cv ‘Desiree’ has 12 chromosomes, with each present in four copies (tetrasomy). Eight control plants propagated by nodal cutting without protoplasting or regeneration and the replicate samplings in five of them displayed regular genomes (Fig. 3A).
Figure 1.
Plant production and analysis. A to D, Schematic representation of experimental workflow. Autotetraploid potato var Desiree was cultured axenically and either protoplasted and regenerated (A), propagated from nodal buds without callus formation or regeneration (B), or regenerated from stem explants after Agrobacterium transformation (C). Cumulative numbers or process efficiencies for two experiments are shown in A. Derivation of dosage plots (D) was used to detect copy number variation for chromosomes.
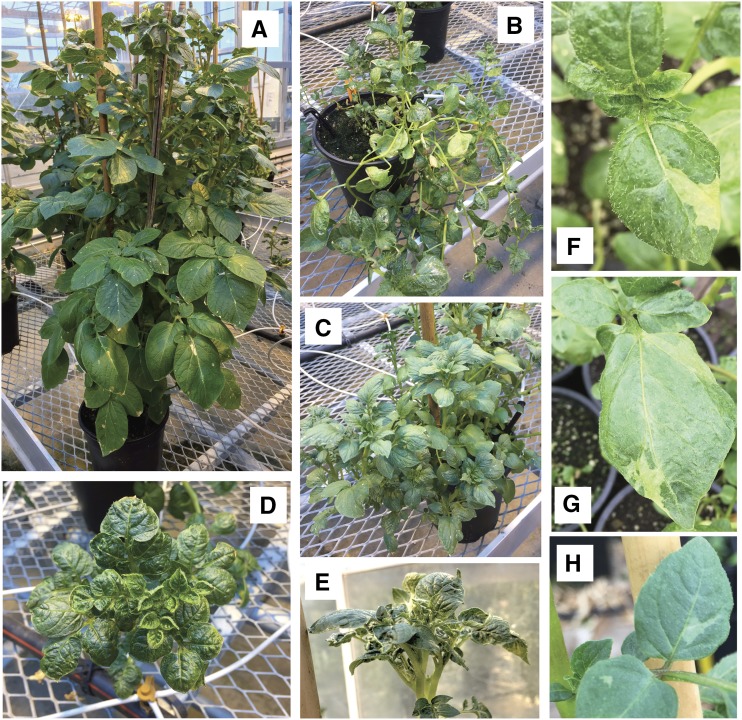
Figure 2.
Phenotype of potato plants regenerated from protoplasts. A, Normal phenotype. B to E, Abnormal phenotypes. F to H, Leaf variegation displayed by tuber-propagated clones of original regenerant 86 (F, G) and 63 (H). Chimerism of phenotype matches genomic chimerism. See Figures 3 and 4 for genomic details illustrating persistent instability.
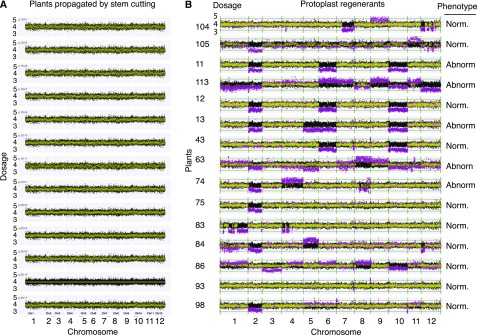
Figure 3.
Frequent genome dosage changes in plants regenerated from protoplasts. Each horizontal track represents genomic dosage values of one individual. Dosage on y axis is plotted versus 250-kb chromosomal bins on the x axis, arrayed consecutively for the 12 chromosomes of potato. To provide the range variation expected from normal (Norm) genomes, the control dataset of 8 propagated plants is plotted in black for each plot track. Individual sample data points are yellow if not statistically different from controls and magenta if they display significant divergence according to the Z-score statistics with 5% false discovery rate. Four genomic copies are expected from autotetraploidy. Bins with high variability were dropped (see “Materials and Methods”). A, Dosage plots from controls consist of 8 plants propagated using stem cuttings. Five controls were sampled twice, and each preparation is plotted independently. The next to last control plant (p.2D-10) was used for standardization of all others read counts. B, Dosage plots for 15 individuals regenerated from protoplasts. Two to four independent samples are plotted together for each plant, except for plant 105 (See Supplemental Fig. S1 for separate plots of all biological replicates). Because calli could be resampled, it is possible that some plants may derive from the same protoplast. Abnorm, abnormal.
Standardized read coverage of regenerated plants was compared with that in controls in order to identify outliers. In each plant, multiple bin measurements differed from the expectation of four copies (Supplemental Fig. S1). They encompassed segments ranging from a few bins to whole chromosomes (Fig. 1C). Reliability of each structural variant identification increases with the number of contiguous outliers enabling robust conclusions.
Cloning by culturing stem cuttings on artificial media, a common horticultural practice, did not cause genome instability. The process of protoplast regeneration, however, engendered high instability, which affected both normal and abnormal regenerated plant types. Among the 15 regenerated plants (Fig. 3B), those phenotypically abnormal had more changes affecting whole chromosomes (2.83 vs 1.13; P = .032, 2-tailed t test). However, neither the mean occurrence of any change, small or large, nor mean number of outlier bins differed between phenotypically normal and abnormal regenerated cohorts, suggesting that screening for normal individuals may not select for plants with intact genomes (Supplemental Tables S4 and S5).
After scoring the large multiple-bin changes we concluded that all chromosomes could be affected, but with varying frequency (7% to 73%). In plants 74, 83, 104, and 105, a single chromosome (1, 4, 8, and 12) displayed multiple deletions or duplications (Fig. 3B). This degree of fragmentation resembles chromothripsis, as described in human cancer (Leibowitz et al., 2015) and in plants (Tan et al., 2015). The full extent of rearrangements cannot be inferred accurately from our data; at least some of the apparent chromosomal discontinuities could be accounted for if the genome of S. tuberosum var Desiree differs from that of the DM1-3 reference used here for sequence alignment (Supplemental Fig. 2), as demonstrated for the pericentromeric region of chromosome 5 (de Boer et al., 2015). Even if multiple deletions have occurred, our analysis does not reveal whether the shattered chromosome fragments form a single reshuffled, but syntenic chromosome as in classical chromothripsis (Leibowitz et al., 2015; Tan et al., 2015), or a diaspora of translocations to different chromosomes. In any case, our results indicate that genome instability results both in aneuploidy and in double-stranded DNA breaks, likely leading to rearrangements upon repair.
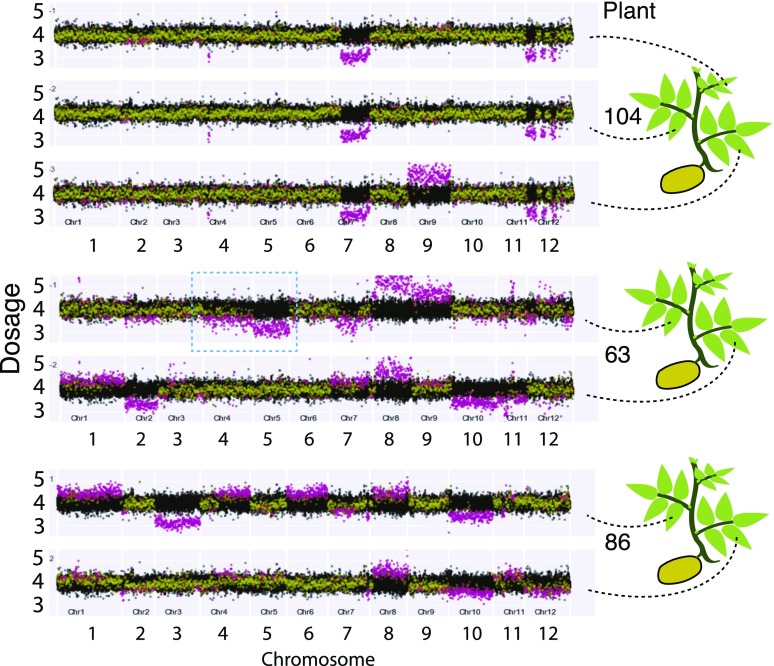
Genome instability may be catastrophic or chronic. Three plants displayed variations between samples originating from different leaves of the same plant (Fig. 4; Supplemental Fig. 2), such as pentasomy for chromosome 9 in one of three samples of plant 104. Consistent with the karyotypic chimerism displayed by these plants, incomplete dosage shifts were found in several plants (63, 84, 86, 98, 113; Figs. 3B and 4). Uniquely among the 96 plants tested for DNA content by flow cytometry, plant 86 displayed peaks consistent with both 4X and 8X DNA content (Supplemental Table S2). Chimerism for ploidy type may thus help explain the smaller dosage shift for that particular plant. However, none of the other plants exhibited DNA content exceeding tetraploidy. Thus, the intermediate dosage shifts must originate from chimerism in aneuploidy in the sampled tissue. Consistent with this hypothesis, shoots from tubers produced by the primary regenerants exhibited variegation for leaf characteristics (Fig. 2, F–H). We concluded that instability could persist through development, generating new variation.
Figure 4.
Persistent instability in protoplast regenerants. Each horizontal track represents genomic dosage values in one leaf. Samples from the same individuals are grouped as indicated on the right. Dosage on y axis is plotted versus 250-kb chromosomal bins on the x axis, arrayed consecutively for the 12 chromosomes of potato. To provide the range variation expected from normal genomes, the dataset of 8 controls (13 samples) is plotted in black for each plot track. Individual sample data points are yellow if not statistically different from controls and magenta if they display significant divergence (false discovery rate = 0.05). Four genomic copies are expected from autotetraploidy. Bins with high variability were dropped (see “Materials and Methods”). See Figure 2 for variegation phenotype of plant 63 and 86. Chr, chromosome.
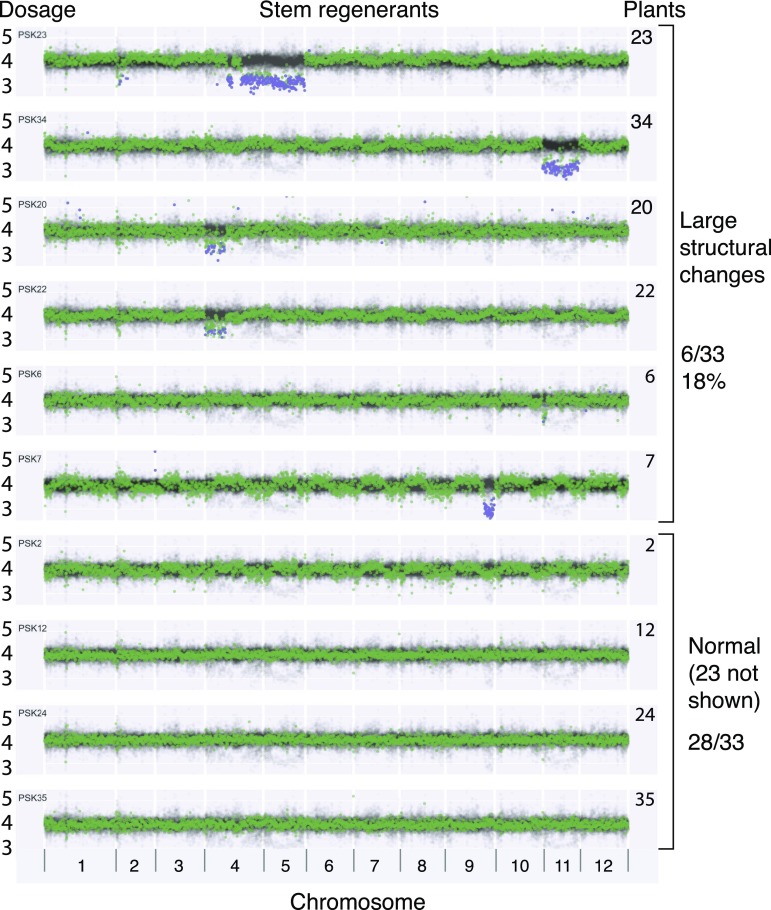
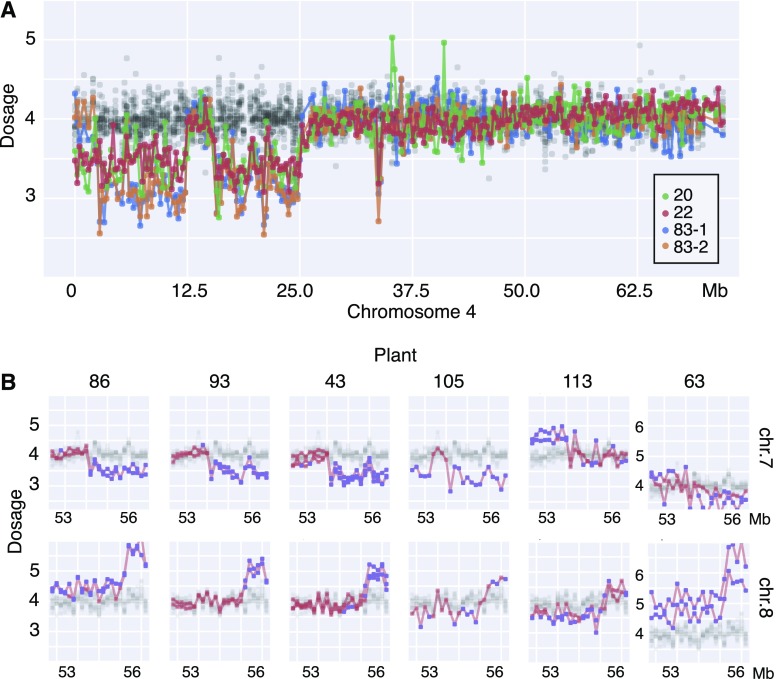
Next, we wondered how much genomic instability occurred in plants regenerated in the process of explant regeneration, such as used in Agrobacterium transformation (Van Eck et al., 2007). This method for potato transformation uses a short period on callus inducing medium followed by regeneration medium. Short callus induction is thought to help avoid polyploidy and other chromosome alterations (Beaujean et al., 1998). By the same genomic method, we examined 33 independent Desiree potato plants regenerated from Agrobacterium co-cultivated stem internode sections. Because these plants were propagated axenically, their morphology could not be phenotyped. Six of these displayed altered genomes involving two trisomies and five large scale deletions (Fig. 5). Two plants produced by independent explants displayed the same compound deletion on chromosome 4. Notably, a very similar compound deletion was displayed in chromosome 4 of protoplast regenerant 83 (Fig. 6A). Another surprising pattern was visible in three, possibly six, plants from protoplasts and consisted of terminal arm changes, a deletion and a duplication, respectively, on chromosomes 7 and 8 (Fig. 6B).
Figure 5.
Genome dosage changes in plants regenerated from stem internodes during Agrobacterium transformation. Dosage on y axis is plotted versus 250-kb chromosomal bins on the x axis, arrayed consecutively for the 12 chromosomes of potato. The green points on each horizontal track represent genomic dosage values of one individual. To represent variation, the same cumulative dataset of all 33 plants is plotted in low opacity black for each track. Only four of the plants with normal genome profiles are shown. The Z-score statistics was calculated using the whole dataset of 33 plants because controls specific to these conditions were not available. The false discovery rate alpha parameter was set at 20%, and the positive points were marked blue. Four genomic copies are expected from autotetraploidy.
Figure 6.
Conserved chromosomal changes across independent individuals. Each horizontal track represents genomic dosage values plotted in 250-kb bins along chromosomes. Control measurements from propagated individuals are shown in black at low opacity. A, Similar deletion events in chromosome (chr.) 4 of plant 83 (protoplast regenerant, two biological replicates, blue and orange) and plant 20 and 22 (stem internode regenerants, green and brick red). See Figures 3 and 4 and “Materials and Methods” for details of dosage plots method. B, Dosage plots for terminal left regions of chr. 7 and chr. 8. Close up of the “deletion-7, duplication-8” paired pattern for three very clear examples (left) and three additional likely case (right). All except 105 have two biological replicates (independent samples from the same plant). The y axis is shifted for the rightward most plot set to display high dosage points. Orange data points are not significantly different from controls, while blue data points use false discovery rate = 0.05.
Taken together, our analysis provides compelling proof of large-scale, frequent genomic instability as a consequence of regeneration from protoplasts in potato. The high penetrance of this instability syndrome in protoplast regenerants may depend on cell type and treatment. Instability in potato is probably aided by polyploidy buffering (Comai, 2005). Notably, the high frequency of polyploids among regenerants of diploid potato (Sree Ramulu et al., 1986) could result from selection for lower imbalance. In diploid species, monosomy is highly deleterious and trisomy can be fairly deleterious (Henry et al., 2010). Most highly rearranged chromosomes, such as those produced by chromothripsis, are similarly deleterious (Tan et al., 2015). We expect that a corresponding analysis in a diploid would find fewer large-scale abnormalities because gross dosage and structural variants will be subject to negative selection during growth and regeneration.
Comparison of instability in protoplast and stem internode regenerants was very informative. First, sampling genomic integrity in stem regenerants, a regeneration procedure aimed at minimizing tissue culture-derived variation (Beaujean et al., 1998), indicated that this method resulted in significantly less instability (χ2 test, P < 0.001). Whereas our investigation is limited to two conditions, it is consistent with the long-standing proposal that experimental conditions such as time on callus induction medium will affect instability (Müller et al., 1990; Bairu et al., 2011). Second, the occurrence of similar deletions in independent individuals, such as in one plant regenerated from protoplasts and two plants from stems (Fig. 6), strengthened the hypothesis that DNA breaks are not random, but occur at specific fragile sites.
Our data provide sequence-based evidence of multiple, common genomic changes, whose frequency reflects the original cell type and the applied tissue culture environment (Lee and Phillips, 1988). Protoplasting followed by regeneration results in the most severe outcome, but considerable genomic risk occurs in regeneration from stem internodes. Experimental variables may affect outcome in multiple ways (Bairu et al., 2011). The observed syndrome is consistent with failure of one or more of the major mechanisms contributing to genome stability (Lee and Phillips, 1988; Lee et al., 2016; Lee and Seo, 2018). Frequent aneuploidy could be triggered by mitotic malfunction, such as a defective spindle, resulting in missegregation, chromosome loss, and perhaps rescue, possibly through restitution of micronuclei. Collapse of genome maintenance in the defective micronuclear environment is thought to result in chromosome-specific shattering (Crasta et al., 2012; Zhang et al., 2015; Ly et al., 2017). The remarkably conserved chromosomal breaks suggest locus-specific epigenetic upheaval, the presence of fragile sites (Durkin and Glover, 2007; dela Paz et al., 2012), and the disruptive effects of transposon activation (Hirochika et al., 1996; Stroud et al., 2013; Ong-Abdullah et al., 2015; Han et al., 2018).
Whatever the causes, the observed genomic dosage changes are likely to impact phenotype, and this type of change could well underlie the somaclonal syndrome. Confirmation of this syndrome in other species will affect the prospects for protoplast utilization. In vegetatively propagated crops, the load of genomic changes is likely to affect agronomic performance. In sexually propagated crops, recombination during backcrossing to an agronomically fit parent should offset the negative effect of most genomic changes, with the possible exception of those tightly linked to any locus of interest. Lastly, if genomic instability is pervasive and frequent during commonly used procedures that involve dedifferentiation of specialized cells and regeneration, measures to understand its causes and ameliorate the consequences should be undertaken. For example, if fragile sites are responsible for instability, their deletion by genome editing could improve tissue culture performance of varieties targeted for manipulation.
MATERIALS AND METHODS
Plant Regeneration from Protoplast and Growth
A single axenic cutting of Solanum tuberosum group Tuberosum cv Desiree (PI 310467) was obtained in 2015 from the US Potato Genebank. Cuttings propagated from this original source were aseptically maintained in vitro at 16-h light/8-h dark, 25°C and 40 µmol m-2s-1 light intensity. Cuttings were propagated in magenta cubes on medium containing Murashige and Skoog salts (Murashige and Skoog, 1962) and Gamborg vitamins (Gamborg et al., 1968), supplemented with 1.5 mg/ml (6 µM) silver thiosulfate (Perl et al., 1988), 3% (w/v) Suc, 590 mg/L MES, and 0.7% (w/v) agar, pH 5.8. Potato protoplasts were prepared from 1 g of leaves harvested from in vitro grown plants following procedures of Tan, with modifications (Tan et al., 1987). Protoplasts were resuspended at 0.4–0.6 × 106 protoplasts/mL and embedded in an alginate solution as in Perales and Schieder, 1993, with some medium modifications, including substitution of liquid Murashige and Skoog basal medium with liquid 8P medium (Kao and Michayluk, 1975). Callus and shoot regeneration was achieved as described (Shahin, 1985). After 7 to 14 d, multicellular structures still embedded within the alginate disks were transferred to callus stimulating medium, TM-3. Microcalli were formed in the dark at 28°C. Microcalli 1 to 2 mm in size were transferred to TM-4 medium, 16-h light/8-h dark, 25°C and 40 µmol m−2s−1 light intensity for shoot regeneration. Shoots began to form after several weeks on TM-4. Shoots several millimeters in size with a good meristem were excised from the callus clump and placed on the propagation medium for rooting. Calli were kept for continued regeneration, and the removed shoot was transferred for rooting. One to three shoots per callus clump were excised and rooted on propagation medium. The callus of origin was not recorded, and it is possible that plants with close identification numbers may have originated from the same callus. In two experiments, we regenerated about 400 plantlets, 101 of which were transplanted to the greenhouse. From these, we selected 10 regenerated plants that appeared normal and 5 plants that looked stunted or otherwise abnormal. At the same time, through axenic nodal cutting propagation in tissue culture, we produced 8 rooted plants. These were transferred to the same greenhouse to be used as controls that had not experienced regeneration. All sampled plants were transferred to the greenhouse at the same time and grown in the same environment. Plants were acclimated to greenhouse conditions (16-h light/8-h dark) in flats under a plastic dome for 1 week before transplanting to 1-Gal pots. Plants were allowed to flower and tuber; however, greenhouse conditions favored tuber formation over flowering.
Regenerated Plant Ploidy Detection by Flow Cytometry
The youngest fully expanded leaf was selected from each greenhouse-grown plant for ploidy assessment by flow cytometry. Leaves were sliced with a sharp razor blade, and nuclei were stained with 4’,6-diamidino-2-phenylindole), processed with Sysmex’s (formerly Partec) CyFlow Space ploidy analyzer and analyzed with CyPad software.
Regenerated Plant Pollen Viability Testing
Flowers collected for viability testing were stained with fluorescein diacetate (FDA). Working solution was prepared freshly for each experiment from 2 stock solutions combining 5 mL of solution 1 (100 mg/L boric acid, 700 mg/L CaCl22H2O, and 200 g/L Suc) with 2–4 drops of solution 2 (2 mg FDA/mL acetone), until working solution becomes just slightly cloudy. These were kept on ice. Anthers were submerged in a drop of the FDA stain solution, incubated for 30 min in the dark, and then examined with a fluorescent microscope.
Plant Regeneration from Stem Cuttings
Stem cuttings regenerated plants of the same variety Desiree were obtained from the potato transformation center at Cornell University (J. Van Eck laboratory). Thirty-three plants were regenerated according to a protocol that minimizes the callus induction phase to 48 h (Van Eck et al., 2007). The Agrobacterium used in the co-cultivation experiment contained one of two vectors: either a vector expressing a CRISPR-Cas9 system targeted against the CENH3 gene of potato located on chromosome 1, or a vector carrying a variant CENH3 for complementation. Internode segments of 1 cm length were cut from 6-week-old plants grown under axenic conditions. The explants were incubated for 10 min with Agrobacterium suspension, blotted on sterile filter paper to remove excess media, and then transferred onto callus inducing media without any antibiotics. The plates were incubated at 19°C in the dark for 48 h. The explants were then transferred onto selective plant regeneration medium containing appropriate antibiotics, kanamycin for selection of vector T-DNA carrying a variant CENH3, and hygromycin for CAS9. The explants were subcultured on selective plant regeneration medium on a weekly basis for the first months and once every 2 weeks for the second month. Large calli were transferred onto magenta boxes containing selective plant regeneration medium with appropriate antibiotics. After ∼8 weeks, regenerated shoots were excised and transferred onto shoot propagation/rooting medium containing 300 mg/L timentin. One month after incubation on rooting media with timentin, the plants were transferred onto tubes containing rooting media without timentin. Agrobacterium-free plants were then transferred onto soil and acclimatized in growth chamber and moved to greenhouse for further analysis. An additional nontransformed control plant provided by the Van Eck laboratory was added to the analysis. The control tissue was incubated as described above, except that Agrobacterium was omitted and regeneration was induced on a medium without antibiotics.
DNA Extraction
For the protoplast regenerant analysis, DNA was extracted from young, fully expanded leaves of 23 plants: 8 controls and 15 regenerated individuals. At a minimum, two independent samples were collected from each selected plant. For the stem regenerant analysis, DNA was extracted from a single, fully expanded leaf of each individual. Two 4-mm hole punches were taken from leaf tissue and grinded in 500 µL buffer with a tungsten bead at 20.0 frequency 1/s for 1 min in the QIAGEN TissueLyserII. The plate was rotated and then grinded for an additional 1 min. Two methods were then used to extract DNA from the two independent samples collected from each plant. In one method, DNA was extracted using a QIAGEN DNeasy plant mini kit, following the manufacturer’s instruction, and, in the other method, DNA was extracted using a CTAB DNA extraction protocol (Henry et al., 2015). The two DNA extraction techniques yielded DNA of comparable quantity and quality.
DNA Quantification and Library Preparation
Concentration of all extracted DNA samples was measured using SYBR Green (Thermo Fisher Scientific) and processed on an automated plate reader (Tsai et al., 2011). Samples were adjusted to 20 ng/µL before 50 µL of each DNA sample was sheared to an average size of 300 bp with a Covaris E220 ultrasonicator. The KAPA Bioscience Hyper Prep Kit (KAPA Biosiences KK8504; www.kapabiosystems.com) was used for library construction following manufacturer methods. Libraries used custom 8-bp dual-indexed adapters. Libraries constructed from protoplast-regenerated material were sequenced on an Illumina HiSeq 4000 in 100-nt single-end mode at the QB3 core facility (http://qb3.berkeley.edu) at the University of California, Berkeley, and libraries constructed from explant-regenerated material were sequenced on an Illumina NovaSeq 6000 in 150-nt paired-end mode at the University of California San Francisco Center for Advanced Technology. Library construction and sequencing details are provided in Supplemental Table S6.
Dosage Analysis
Raw reads were trimmed to remove adapter and low-quality (trimmed when 5-nt 5′-3′ sliding window average base quality < Q20) sequence using custom Python scripts available on our lab Web site (https://github.com/Comai-Lab/allprep). The DM1-3 v4.04 genome assembly (Hardigan et al., 2016) as well as DM1-3 chloroplast and mitochondrion sequences were retrieved from http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml, concatenated, and used as the reference for sequencing read alignment by Burrows-Wheeler Aligner (BWA)-mem (Li, 2014) with default parameters. PCR duplicates were removed from the resulting Sequence Alignment Map (SAM) files using a custom Python script available from our lab (overamp.py; http://comailab.genomecenter.ucdavis.edu/index.php/Bwa-doall). Only reads with mapping quality ≥10 were retained for further analysis. Standardized coverage values were derived as described previously (Henry et al., 2010, 2015). Due to differences in experimental design and sequencing type between the protoplast and stem regeneration experiments, reads were processed slightly differently in the two experiments.
In the protoplast experiment, standardized coverage values for each sample were derived in fixed-size, nonoverlapping bins by computing the fraction of all reads with an alignment start position within the bin at hand, standardizing this fraction by the corresponding fraction of a nonregenerated control sample, and then multiplying the standardized value by four to represent the expected tetraploid state. Outlier bins were removed as follows: for each sample, the per-bin fractions of mapped reads to total reads for the sample at hand were used to compute a within-sample mean and sd. If in that sample, a bin exhibited a fraction greater than two standard deviations from the within-sample mean, the bin was withheld from analysis in all protoplast-derived samples and controls. Statistically significant deviation between control and test samples was then assessed for individual bins. Because karyotype variation was detected between resampling of the same protoplast-regenerated plant, each sequencing library generated from protoplast-derived samples was analyzed independently. For each bin, the distribution of the 13 control values was compared with the dosage value of each single sample from the regenerated plants, and a Z score was calculated for each value. A two-tailed P value of Z statistics was derived and corrected for multiple testing using the statsmodels’ multipletests ‘fdr_tsbh’ method, yielding Benjamini-Hochberg corrected P and the connected false discovery rate (α = 0.05; Benjamini and Hochberg, 1995; Seabold and Perktold, 2010). Because a nonregenerated control sequenced in the same machine run was not available for the stem internode regeneration experiment, each sample was instead standardized to the corresponding mean bin fractions of all 34 explant-regenerated samples. Outlier bins were not removed and significant per-sample deviations in relative coverage were assessed using the entire explant-regenerated dataset with false discovery rate of 20%.
Accession Numbers
Sequencing data reported in this article have been deposited at National Center for Biotechnology Information Sequence Read Archive under project ID PRJNA510212. SRA run IDs are listed in Supplemental Table 6.
Supplemental Data
The following supplemental materials are available:
Supplemental Figure S1. Dosage profiles from all individual DNA preparations from all plants that had two or more DNA samples.
Supplemental Figure S2. Interpretation of Chr. 1 dosage plot.
Supplemental Table S1. Average regeneration rates in Desiree protoplasts.
Supplemental Table S2. Flow cytometry analysis of nuclear genome content.
Supplemental Table S3. Pollen viability.
Supplemental Table S4. Relation of phenotype to genome instability.
Supplemental Table S5. Normal and dosage outlier 250kb bins in protoplast regenerated plants.
Supplemental Table S6. Plant sequencing information.
Acknowledgments
We acknowledge Benny Ordonez and Meric Lieberman for help and assistance during this work, and Isabelle M. Henry for advice and editing suggestions. We thank Joyce Van Eck and Michelle Tjahjadi at Cornell University for potato transformation.
Footnotes
Articles can be viewed without a subscription.
This work was supported by the National Science Foundation (NSF) (Plant Genome IOS Grant 1444612: Rapid and Targeted Introgression of Traits via Genome Elimination; to L.C.); Innovative Genomics Institute, UC Berkeley (“Segmental editing of the potato genome” award); and H.M. Clause, Inc. (a graduate student fellowship to M.F.).
References
- Andersson M, Turesson H, Olsson N, Fält A-S, Ohlsson P, Gonzalez MN, Samuelsson M, Hofvander P (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164: 378–384 [DOI] [PubMed] [Google Scholar]
- Bairu MW, Aremu AO, Van Staden J (2011) Somaclonal variation in plants: Causes and detection methods. Plant Growth Regul 63: 147–173 [Google Scholar]
- Bao PH, Granata S, Castiglione S, Wang G, Giordani C, Cuzzoni E, Damiani G, Bandi C, Datta SK, Datta K, et al. (1996) Evidence for genomic changes in transgenic rice (Oryza sativa L.) recovered from protoplasts. Transgenic Res 5: 97–103 [DOI] [PubMed] [Google Scholar]
- Beaujean A, Sangwan RS, Lecardonnel A, Sangwan-Norreel BS (1998) Agrobacterium-mediated transformation of three economically important potato cultivars using sliced internodal explants: An efficient protocol of transformation. J Exp Bot 49: 1589–1595 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Comai L. (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creissen GP, Karp A (1985) Karyotypic changes in potato plants regenerated from protoplasts. Plant Cell Tissue Organ Cult 4: 171–182 [Google Scholar]
- de Boer JM, Datema E, Tang X, Borm TJA, Bakker EH, van Eck HJ, van Ham RCHJ, de Jong H, Visser RGF, Bachem CWB (2015) Homologues of potato chromosome 5 show variable collinearity in the euchromatin, but dramatic absence of sequence similarity in the pericentromeric heterochromatin. BMC Genomics 16: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dela Paz JS, Stronghill PE, Douglas SJ, Saravia S, Hasenkampf CA, Riggs CD (2012) Chromosome fragile sites in Arabidopsis harbor matrix attachment regions that may be associated with ancestral chromosome rearrangement events. PLoS Genet 8: e1003136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Elmore RW, Roeth FW, Nelson LA, Shapiro CA, Klein RN, Knezevic SZ, Martin A (2001) Glyphosate-resistant soybean cultivar yields compared with sister lines. Agron J 93: 408–412 [Google Scholar]
- Evans DA. (1989) Somaclonal variation--genetic basis and breeding applications. Trends Genet 5: 46–50 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Han Z, Crisp PA, Stelpflug S, Kaeppler SM, Li Q, Springer NM (2018) Heritable epigenomic changes to the maize methylome resulting from tissue culture. Genetics 209: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan MA, Crisovan E, Hamilton JP, Kim J, Laimbeer P, Leisner CP, Manrique-Carpintero NC, Newton L, Pham GM, Vaillancourt B, et al. (2016) Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 28: 388–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardigan M.A., Laimbeer F.P.E., Newton L., Crisovan E., Hamilton J.P., Vaillancourt B., Wiegert-Rininger K., Wood J.C., Douches D.S., Farré E.M., et al. (2017). Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc Natl Acad Sci USA 114: E9999–E10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L (2010) Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Zinkgraf MS, Groover AT, Comai L (2015) A system for dosage-based functional genomics in poplar. Plant Cell 27: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93: 7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KN, Michayluk MR (1975) Nutritional requirements for growth of Vicia hajastana cells and protoplasts at a very low population density in liquid media. Planta 126: 105–110 [DOI] [PubMed] [Google Scholar]
- Karp A, Nelson RS, Thomas E, Bright SW (1982) Chromosome variation in protoplast-derived potato plants. Theor Appl Genet 63: 265–272 [DOI] [PubMed] [Google Scholar]
- Karp A, Jones MGK, Foulger D, Fish N, Bright SWJ (1989) Variability in potato tissue culture. Am Potato J 66: 669–684 [Google Scholar]
- Krishna H., Alizadeh M., Singh D., Singh U., Chauhan N., Eftekhari M., Sadh R.K. (2016). Somaclonal variations and their applications in horticultural crops improvement. Biotech 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsmann J, Uhrig H (1985) Somaclonal variation in Solanum tuberosum detected at the molecular level. Theor Appl Genet 71: 500–505 [DOI] [PubMed] [Google Scholar]
- Lee K, Seo PJ (2018) Dynamic epigenetic changes during plant regeneration. Trends Plant Sci 23: 235–247 [DOI] [PubMed] [Google Scholar]
- Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol Plant Mol Biol 39: 413–437 [Google Scholar]
- Lee J-K, Choi YL, Kwon M, Park PJ (2016) Mechanisms and consequences of cancer genome instability: Lessons from genome sequencing studies. Annu Rev Pathol 11: 283–312 [DOI] [PubMed] [Google Scholar]
- Leibowitz ML, Zhang C-Z, Pellman D (2015) Chromothripsis: A new mechanism for rapid karyotype evolution. Annu Rev Genet 49: 183–211 [DOI] [PubMed] [Google Scholar]
- Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30: 2843–2851 10.1093/bioinformatics/btu356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, Cleveland DW (2017) Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 19: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Nakagome M, Ohnuma T, Yamagata H, Kanamori H, Katayose Y, Takahashi A, Matsumoto T, Hirochika H (2012) Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol 53: 256–264 [DOI] [PubMed] [Google Scholar]
- Müller E, Brown PT, Hartke S, Lörz H (1990) DNA variation in tissue-culture-derived rice plants. Theor Appl Genet 80: 673–679 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ong-Abdullah M, et al. (2015) Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales EH, Schieder O (1993) Plant regeneration from leaf protoplasts of apple. Plant Cell Tissue Organ Cult 34: 71–76 [Google Scholar]
- Perl A, Aviv D, Galun E (1988) Ethylene and in vitro culture of potato: Suppression of ethylene generation vastly improves protoplast yield, plating efficiency and transient expression of an alien gene. Plant Cell Rep 7: 403–406 [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium; Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, Orjeda G, Guzman F, et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Seabold S, Perktold J (2010). Statsmodels: Econometric and statistical modeling with python. In Proceedings of the ninth Python in Science Conference (SciPy society Austin), p. 61. [Google Scholar]
- Shahin EA. (1985) Totipotency of tomato protoplasts. Theor Appl Genet 69: 235–240 [DOI] [PubMed] [Google Scholar]
- Sree Ramulu K, Dijkhuis P, Roest S, Bokelmann GS, Groot B (1986) Variation in phenotype and chromosome number of plants regenerated from protoplasts of dihaploid and tetraploid potato. Plant Breed 97: 119–128 [Google Scholar]
- Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE (2013) Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Korf I, Lysak MA, Comai L, Chan SW (2015) Catastrophic chromosomal restructuring during genome elimination in plants. eLife 4: e06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M-MC, Boerrigter HS, Kool AJ (1987) A rapid procedure for plant regeneration from protoplasts isolated from suspension cultures and leaf mesophyll cells of wild Solanum species and Lycopersicon pennellii. Plant Sci 49: 63–72 [Google Scholar]
- Tsai H, et al. (2011) Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol 156: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eck J, Conlin B, Garvin DF, Mason H, Navarre DA, Brown CR (2007) Enhancing beta-carotene content in potato by rnai-mediated silencing of the beta-carotene hydroxylase gene. Am J Potato Res 84: 331 [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Zhang C-Z, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D (2015) Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]