ESA1 is involved in embryo sac abortion in hybrid backcross progeny derived from common wild rice and cultivated rice.
Abstract
The emergence of sterile individuals in the hybrid backcross progeny of wild and cultivated rice limits the use of wild rice alleles for improving cultivated rice, but the molecular mechanisms underlying this sterility remain unclear. Here, we identified the semisterile introgression line YIL42, derived from a cross between the indica rice variety Teqing (Oryza sativa) and Oryza rufipogon accession YJCWR (Yuanjiang common wild rice), which exhibits semisterility. Using positional cloning, we isolated EMBRYO SAC ABORTION 1 (ESA1), which encodes a nuclear-membrane localized protein containing an armadillo repeat domain. A mutation in ESA1 at position 1819 (T1819C) converts a stop codon into an Arg (R) codon, causing delayed termination of protein translation. Analysis of transgenic lines indicated that the difference in ESA1 protein structure between O. rufipogon–derived ESA1 and Teqing-derived esa1 affects female gamete abortion during early mitosis. Fertility investigation and expression analysis indicated that the interaction between ESA1T1819 and unknown gene(s) of Teqing affects spikelet fertility of the hybrid backcross progeny. The ESA1T1819 allele is present in O. rufipogon but absent in O. sativa, suggesting that variation in ESA1 may be associated with interspecific hybrid incompatibility between wild and cultivated rice. Our findings provide insight into the molecular mechanism underlying female sterility, which is useful for improving the panicle seed setting rate of rice and for developing a strategy to overcome interspecific hybrid sterility between cultivated rice and wild rice.
The panicle seed setting rate is a major determinant of grain yield in rice (Oryza sativa). Both male and female sterility can compromise rice spikelet fertility. Recently, many genes causing male sterility have been reported, but few genes involved in female sterility have been identified. Histological analysis has shown that genes causing female sterility affect the developing embryo sac and the process of fertilization, inducing abnormal megaspore development after meiosis (Chen et al., 2008; Kubo et al., 2016; Koide et al., 2018); leading to abnormal polar nucleus position (Yu et al., 2016); and blocking pollen tube growth in the style transmission tract (Li et al., 2013; Xu et al., 2017). However, the mechanism underlying female gamete abortion remains unclear. So far, a lot of work in rice fertility has focused on researching hybrid sterility. Several genes for intraspecific hybrid sterility in rice have been reported (Chen et al., 2008; Long et al., 2008; Mizuta et al., 2010; Yang et al., 2012; Kubo et al., 2016; Yu et al., 2016; Shen et al., 2017), but few interspecific hybrid sterility genes have been detected.
Known genes underlying interspecific hybrid sterility include O. sativa/Oryza glumaepatula S27/S28 (Yamagata et al., 2010); O. sativa/Oryza nivara DUPLICATED GAMETOPHYTIC STERILITY 1 (DGS1)/DGS2 (Nguyen et al., 2017); O. sativa/Oryza glaberrima S1 (Xie et al., 2017b; Koide et al., 2018); and O. sativa/Oryza meridionalis qHMS7 (Yu et al., 2018). The identification of these fertility-related genes established the foundation for elucidating the mechanism behind interspecific hybrid sterility and speciation in rice. However, except for the nonallelic Hwi1/Hwi2 interactions that result in hybrid weakness between Oryza rufipogon and O. sativa (Chen et al., 2014), no genes involved in causing hybrid backcross progeny sterility from common wild rice and Asian cultivated rice have been identified.
YJCWR (Yuanjiang common wild rice), an O. rufipogon accession from Yuanjiang county, Yunnan province, China, grows on hillsides at 780 m above sea level, far from cultivated rice fields (Morishima, 1997). The rate of seed fertility was ∼54.53% in a cross between YJCWR and indica cultivated rice (Yuan et al., 1998), implying that hybridization between YJCWR and cultivated rice might result in hybrid sterility. In the current study, we identified a gene, EMBRYO SAC ABORTION 1 (ESA1), whose alleles differ between Teqing (O. sativa) and YJCWR (O. rufipogon). This difference leads to female sterility due to an abnormality after meiosis in the near-isogenic line derived from an interspecific cross between Teqing and YJCWR. ESA1 encodes a nuclear-membrane localized armadillo (ARM) repeat protein. Silencing of ESA1 using an RNA interference (RNAi)-ESA1 construct transformed into the semisterile introgression line YIL42 (a progeny of an interspecific cross between Teqing and YJCWR) restores its fertility. Our findings provide insight into the molecular mechanism underlying female sterility, which is helpful for improving the panicle seed setting rate of rice and could be used to develop a strategy to overcome the barrier of hybridization between cultivated rice and wild rice.
RESULTS
Development of a Near-Isogenic Line with a Low Seed Set Ratio
To identify the loci for spikelet fertility in the hybridization between O. rufipogon and O. sativa, we investigated the seed set ratio in a set of introgression lines derived from a cross between the indica rice variety Teqing and O. rufipogon accession YJCWR. We identified the YJCWR introgression line (YIL42), which was semisterile (Supplemental Figure S1). The seed set ratio was ∼87.73% in the female parent Teqing and 80.30% in the male parent YJCWR (Fig. 1), whereas that in YIL42 was 20.67%, indicating the semisterility of the hybrid backcross progeny (Supplemental Figure S2). Genotype analysis showed that YIL42 carried six O. rufipogon–derived chromosomal segments on chromosomes 1, 3, 5, 8, and 9 (Supplemental Figure S2). We identified a major quantitative trait loci (QTL) for the reduced seed set ratio near marker rice microsatellite (RM)24 on chromosome 1, which explained ∼42% of the phenotypic variance in the F2 population derived from a backcross between the introgression line YIL42 and the recipient parent Teqing.
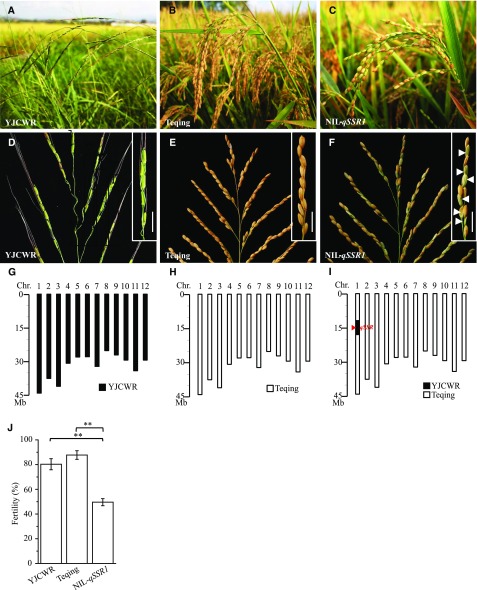
Figure 1.
Comparison of the main panicle and fertility of YJCWR, Teqing, and NIL-qSSR1. A–C, Plant: YJCWR (A), Teqing (B), and NIL-qSSR1 (C). D–F, Main panicle: YJCWR (D), Teqing (E), and NIL-qSSR1 (F). The fertility of the branch is shown in the top right corner; the white arrow indicates normal seeds. Scale bars = 2 cm. G–I, introgression genetics: YJCWR (G), Teqing (H), and NIL-qSSR1(I). Black boxes represent the chromosome (Chr.) segments from YJCWR; white boxes represent the segments from Teqing. The red triangle is the location of the ESA1 gene. J, Fertility of YJCWR, Teqing, and NIL-qSSR1. Data are means ± sd (n = 20). The double asterisks represent a significant difference determined by Student’s t test at P < 0.01.
Based on the results of QTL mapping, we developed a near-isogenic line of QTL for seed setting rate, NIL-qSSR1, with only one O. rufipogon introgressed segment harboring qSSR1 (Fig. 1, C, F, and I). Analysis of spikelet fertility showed that the seed set ratio was 49.59% in NIL-qSSR1, and 51.66% in F1 plants derived from a cross between NIL-qSSR1 and Teqing (Fig. 1J; Supplemental Figure S3B). The similar seed set of F1 plants and NIL-qSSR1 indicated that the O. rufipogon–derived allele at qSSR1 was a dominant allele. In addition, phenotype observation showed that, except for the seed set ratio, other yield-related traits did not significantly differ between NIL-qSSR1 and Teqing (Supplemental Figure S4).
Embryo Sac Abortion Causes Semisterility in NIL-qSSR1
We carried out a cross to explain the low seed set ratio in NIL-qSSR1. When NIL-qSSR1 was crossed as the male parent and the photo-thermo sensitive male sterile line Guangzhan 63s was used as the female parent, Guangzhan 63s had a normal seed setting rate, which is similar to the results of the control cross in which Teqing was used as the male parent (Supplemental Figure S5, A, B, and E). However, when Teqing was used as the male parent, NIL-qSSR1 as the female parent displayed semisterility, which is consistent with the results of NIL-qSSR1 as the female parent using saturated pollination by NIL-qSSR1 (Supplemental Figure S5, C–E). We infer from these results that the spikelet semisterility in NIL-qSSR1 is caused by female abortion.
To investigate the fertility of the stamen, we compared the panicles, spikelets, anthers, and mature pollen grains of NIL-qSSR1 and Teqing. Panicles at the heading stage and spikelets were normal, and there was no defect in anther dehiscence (Supplemental Figure S6, A–H). Meanwhile, mature pollen grains observed by scanning electron microscopy and transmission electron microscopy were normal (Supplemental Figure S6, I–P). We also plumped pollen grains from Teqing and NIL-qSSR1 and stained them with I2-KI and 4′-6-diamidino-2-phenylindole (DAPI; Supplemental Figure S7, A–D) and found that pollen viability in both Teqing and NIL-qSSR1 was normal. Furthermore, no difference in pollen tube elongation was observed between Teqing and NIL-qSSR1 (Supplemental Figure S7, E–J).
Further observations of embryo sac development in NIL-qSSR1 revealed no apparent defects in megasporogenesis before the tetrad stage (Fig. 2, A–F). However, the megaspore near the chalaza degenerated after the tetrad stage, resulting in an inability to form mature embryo sacs during female gamete development in NIL-qSSR1 (Fig. 2, G–N). Observations of embryo sac development in NIL-qSSR1 showed that the viability of mature embryo sacs was 54.62% (Supplemental Figure S3B). Taken together, these results confirm that the partial abortion of female gametes causes semisterility in NIL-qSSR1. We designated the gene conferring female sterility as ESA1.
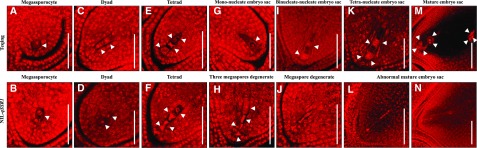
Figure 2.
Comparison of embryo sac development of Teqing and NIL-qSSR1 plants. A and B, Megasporocyte stage of Teqing and NIL-qSSR1. The white arrow indicates megasporocyte. C and D, Dyad stage of Teqing and NIL-qSSR1. The white arrow indicates two cells in the dyad. E and F, Tetrad stage of Teqing and NIL-qSSR1. The white arrow indicates four cells in the tetrad. G, Mononucleate embryo sac stage of Teqing. The white arrow indicates mononucleate embryo sac. H, Three megaspores near the micropyle degenerate in succession of NIL-qSSR1. The white arrow indicates three degenerate megaspores near the micropyle, and one megaspore near the chalaza. I, Binucleate-nucleate embryo sac of Teqing. The white arrow indicates two nuclei. J, Megaspore degenerate of NIL-qSSR1. K, Tetra-nucleate embryo sac of Teqing. The white arrow indicates four nuclei. M, Mature embryo sac stage of Teqing. The white arrow indicates eight nuclei. L and N, Abnormal mature embryo sac of NIL-qSSR1. Scale bars = 50 μm.
Fine-Mapping of ESA1
To further map ESA1, we selected individuals that were heterozygous at the qSSR1 locus (Z10 ∼ RM446), containing one chromosomal segment from Teqing and one from YJCWR by self-crossing to construct a NIL-qSSR1 segregation population. Due to the location of the qSSR1 locus near the centromere of chromosome 1, we developed 22,992 plants, genotyped them, and selected the recombinant plants to investigate the seed setting rate. Initially, we were only able to use BH recombinant plants (“B” represents the Teqing homozygous genotype; “H” represents the heterozygous genotype) for preliminary positioning interval analysis. Some heterozygous recombinant plants cannot be used directly for location analysis (e.g. AH recombinant plants. “A” represents the NIL-qSSR1 homozygous genotype; “H” represents the heterozygous genotype, but all the plants were semisterile) because ESA1 is a dominant allele.
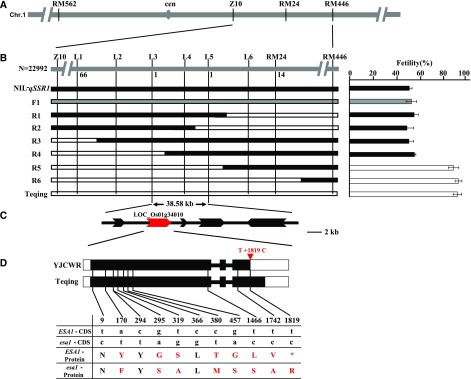
We obtained homozygous recombinant plants from all types of key heterozygous individuals at the qSSR1 locus by self-crossing, and investigated the phenotypes including the seed setting rate and embryo sac viability. Finally, according the genotypes and phenotypes of homozygous recombinant plants, we delimited ESA1 to a 38.58-kb region between markers L3 and L5, which is predicted to contain five genes based on the rice genome annotated project (http://rice.plantbiology.msu.edu/), designated open reading frames (ORFs) 1–5 (Fig. 3, A–C). According to the annotation, ORFs 3–5 encode retrotransposon proteins, ORF 1 encodes a hypothetical protein, and ORF 2 encodes an expressed protein. RT-quantitative PCR (RT-qPCR) analysis showed that ORF 1 was expressed at low levels in different tissues including leaves, roots, and culms at the mature stage and developing panicles (Supplemental Figure S8A), whereas ORF 2 was highly expressed in the different developing panicle stages (Fig. 4A).
Figure 3.
Fine-mapping and gene candidates for ESA1. A, The target gene primarily mapped between Z10 and RM446 near the centromere of chromosome (Chr) 1. B, ESA1 was further delimited to a 38.58-kb region between the L3 and L5 markers. C, Five genes in this region based on the annotation of Nipponbare. D, The gene structures of LOC_Os01g34010 in Teqing and NIL-qSSR1. White boxes, 5′-and 3′-untranslated region; black boxes, coding sequences; lines between boxes, introns; red triangle, single nucleotide change, T1819C, that elongates the peptide chain; black lowercase, 15 SNPs; black uppercase, the same aa; red uppercase, the different aa.
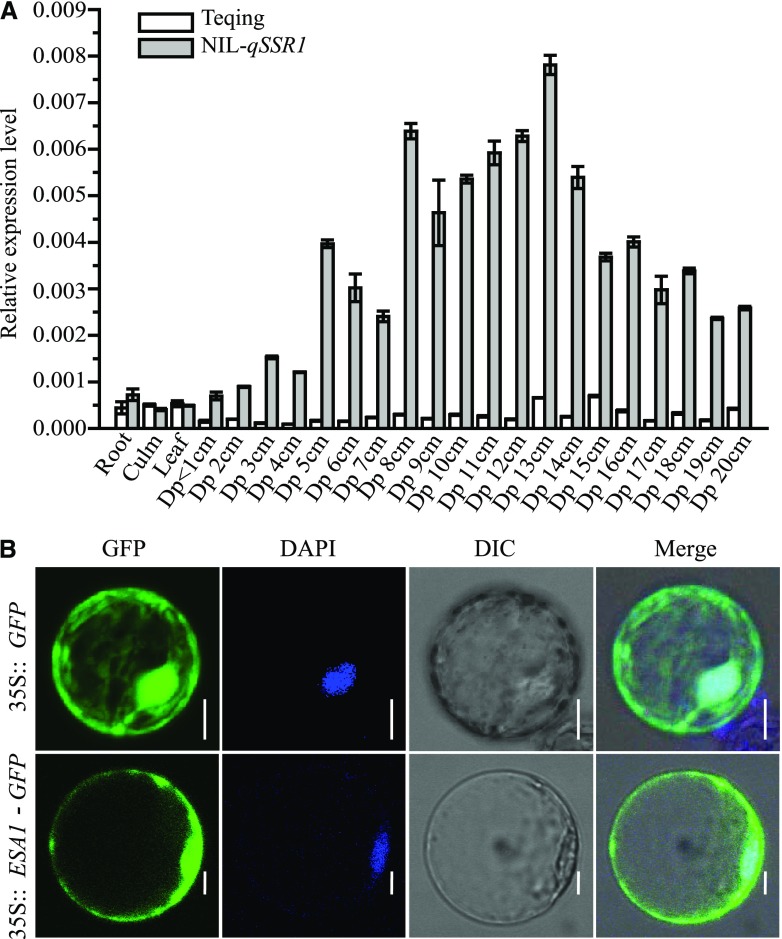
Figure 4.
Transcriptional characterization of ESA1. A, Expression pattern of ESA1 in leaves, roots, and culms at the mature stage and different developing panicle stages among Teqing and NIL-qSSR1. Dp: Developing panicle. Data are means ± sd (n = 3). B, Subcellular localization of ESA1 protein. 35S::GFP (top) and 35S::ESA1-GFP (bottom) constructs were transiently expressed in rice protoplast cells. Scale bars = 5 μm. DAPI, 4′-6-diamidino-2-phenylindole; DIC, differential interference contrast.
We also compared the sequences of ORF 2 in NIL-qSSR1 and Teqing and identified a single nucleotide change, T to C, at position 1819 in exon 3 of ORF 2. This single-base substitution led to the elongation of the peptide chain in Teqing, which altered the structure of the protein. This protein also had seven amino acid differences compared with the wild rice version (Fig. 3D). Therefore, we focused on ORF 2 as a candidate for ESA1.
Functional Analysis of ESA1
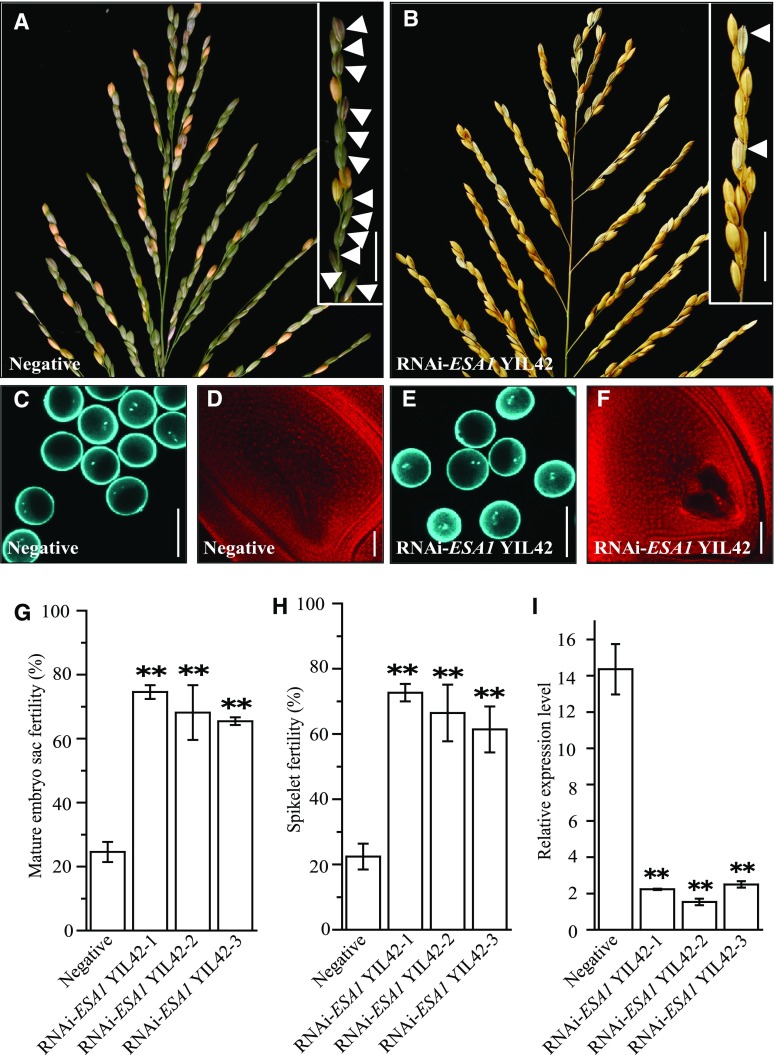
To examine whether ESA1 causes the female sterility phenotype of NIL-qSSR1, we performed transgenic analysis by introducing an RNAi-ESA1 construct into the YIL42 background (Fig. 5, A and B). Analysis of 11 independent T2 RNAi-ESA1 YIL42 transgenic lines showed that the mature embryo sac and spikelet fertility was much higher in RNAi-ESA1 YIL42 positive transgenic plants than in the control nontransgenic plants (Fig. 5, G and H). Most of the pollen grains and embryo sacs in RNAi-ESA1 YIL42 transgenic plants showed no obvious abortion (Fig. 5, C–F). RT-qPCR analysis showed that the expression of ESA1 in RNAi-ESA1 YIL42 transgenic plants was significantly lower than in control plants (Fig. 5I). These results indicate that ORF 2 (LOC_Os01g34010) is ESA1.
Figure 5.
Functional analysis of ESA1. A and B, Main panicle comparison of negative plant and RNAi-ESA1 YIL42 plants. The fertility of the branch is shown in the top right corner. The white arrow indicates normal seeds. Scale bars = 2 cm. C and E, DAPI staining of mature pollen of negative plants and RNAi-ESA1 YIL42 plants. D and F, Mature embryo sac of negative plants and RNAi-ESA1 YIL42 plants. G, Mature embryo sac fertility comparison of negative plants and RNAi-ESA1 YIL42 transgenic lines. Data are means ± sd (n = 3). H, Spikelet fertility comparison of negative plants and RNAi-ESA1 YIL42 transgenic lines. Data are means ± sd (n = 20). I, The relative expression levels of ESA1 in negative plants and RNAi-ESA1 YIL42 transgenic representative plants. Data are means ± sd (n = 3). The double asterisks in (G–I) represent a significant difference determined by Student’s t test at P < 0.01.
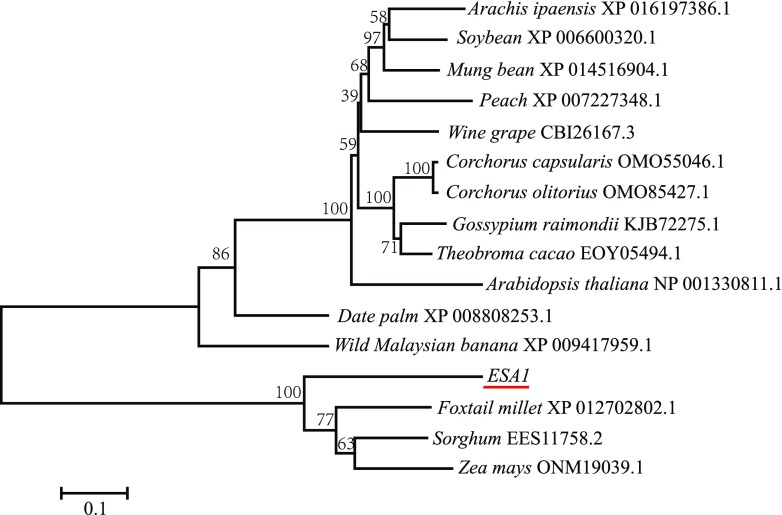
Sequence analysis of genomic DNA, complementary DNA (cDNA), and rapid amplification of cDNA ends (RACE) cDNA products revealed that ESA1 cDNA in O. rufipogon is 2417 bp long, with an ORF of 1821 bp (Supplemental Figure S9), an 82 bp 5′-untranslated region, and a 514 bp 3′-untranslated region, containing three exons and two introns and encoding a 606 amino acid protein of unknown function (Supplemental Figure S10). InterPro analysis revealed that the deduced protein contains an armadillo repeat domain at position 211–525 and shares homology with deduced proteins in other plant species, such as maize (Zea mays), sorghum (Sorghum bicolor), and foxtail millet (Setaria italica; Fig. 6). Transient expression analysis in rice protoplast cells revealed that the ESA1-GFP fusion protein is localized to both the nucleolus and cell membrane (Fig. 4B).
Figure 6.
Phylogenetic tree of ESA1 homologs. Numbers on branches represent bootstrap values (based on 1000 replications). The scale bar (0.1) = the number of amino acid substitutions per site.
Expression Pattern of ESA1
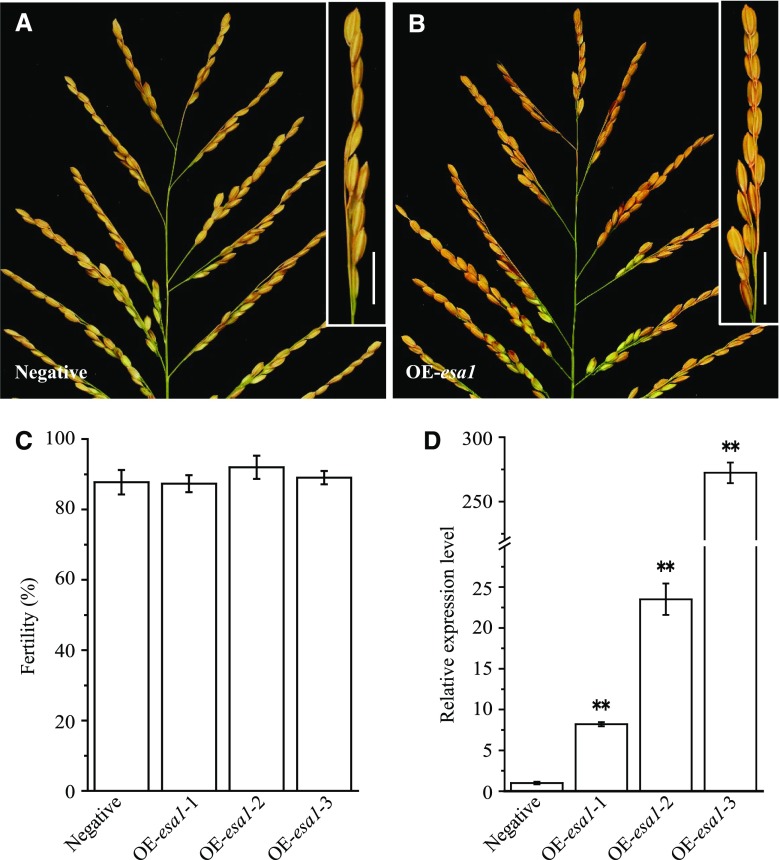
We analyzed the expression profile of ESA1 in different organs using RT-qPCR. ESA1 was expressed at low levels in leaves, roots, and culms at the mature stage, but at higher levels in developing panicles, specifically after the meiotic stages (Fig. 4A). ESA1 was highly expressed in developing panicles in NIL-qSSR1, but esa1, the Teqing version of the gene, was barely expressed in Teqing. Meanwhile, we also analyzed the expression profile of ESA1 in developing panicles of YJCWR. The expression of ESA1 in YJCWR was similar to that in NIL-qSSR1 (Supplemental Figure S8B). ESA1 caused sterility in the Teqing background, but not in the YJCWR background, which indicates that female sterility is not caused by differential expression levels of ESA1 in different genetic backgrounds. Instead, sterility might be due to a deleterious interaction between ESA1 and epistatic gene(s) in Teqing. To further investigate whether female semisterility was caused by differential expression or the change in the protein structure of ESA1, we performed transgenic analysis. We introduced a construct overexpressing esa1 driven by the maize Ubiquitin promoter into Teqing plants (Fig. 7, A and B). Twelve overexpressing transgenic lines were obtained. For both transgenic positive and negative plants, no significant difference in fertility was observed from the T1 to the T2 generation (Fig. 7C). RT-qPCR analysis showed that esa1 was expressed at higher levels in the positive plants than the negative plants (Fig. 7D). These results indicate that female semisterility is caused by a change in ESA1 protein structure.
Figure 7.
Overexpression analysis of esa1. A and B, Main panicle comparison of negative plant and esa1 overexpression (OE-esa1) plants. The fertility of the branch is shown in the top right corner. The white arrow indicates normal seeds. Scale bars = 2 cm. C, Fertility comparison of negative lines and OE-esa1 transgenic lines. Data are means ± sd (n = 20). D, Relative expression levels of esa1 in negative and OE-esa1 transgenic plant panicles. Data are means ± sd (n = 3). The double asterisks represent a significant difference determined by Student’s t test at P < 0.01.
We detected differentially expressed genes in YIL42 and RNAi-ESA1 YIL42 via RNA-sequencing (RNA-seq)and found that 1155 and 705 genes were down-regulated and up-regulated, respectively, in RNAi-ESA1 YIL42 compared with YIL42. Further analysis found that genes associated with cell wall degradation, like xyloglucan endotransglucosylase and polygalacturonase, were down-regulated in RNAi-ESA1 YIL42, but no related genes were up-regulated in RNAi-ESA1 YIL42 (Supplemental Figure S11). These findings suggest that ESA1 may affect female gametophyte abortion by regulating cell degradation pathways.
Nucleotide Diversity Analysis of ESA1
We sequenced the 2615-bp genome region of ESA1 in 36 wild rice varieties (Supplemental Table S1) and 113 cultivated rice varieties, including 61 indica and 52 japonica cultivars (Supplemental Table S2). Sequence variation analysis revealed eight haplotypes (H2–H9) in wild rice, but only three haplotypes (H1, H2, and H3) in cultivated rice. A sequence comparison between Teqing and NIL-qSSR1 revealed 15 single nucleotide polymorphisms (SNPs) in the coding region of ESA1. Among these SNPs between cultivated and wild rice, we found the key site, T1819C, which alters the length of the peptide chain. Wild rice varieties possessed both C and T at this site, whereas in cultivated rice, all cultivars examined had C at the 1819 site (Supplemental Figure S12). These results suggest that variation in ESA1 may be associated with interspecific hybrid incompatibility between wild and cultivated rice. The consistency in the ESA1 genotype among all cultivated rice makes it easier to transfer genes between cultivated rice varieties, and the divergence at ESA1 among cultivated and wild rice introduces a reproductive barrier, which may affect the formation of new species.
DISCUSSION
In this study, we identified ESA1 as a gene affecting interspecific hybrid progeny female sterility in rice. InterPro analysis showed that the encoded protein contains an armadillo (ARM) repeat at position 211–525. ARM repeats fold into the right-handed super helical structure of an alpha helix, which is involved in protein-protein interactions (Peifer et al., 1994; Huber et al., 1997). Several ARM repeat proteins regulate various biological processes, including embryogenesis, signal transduction, ubiquitination pathways, transcriptional regulation, reproductive development, mitosis, and protein degradation (Samuel et al., 2006; Tewari et al., 2010). In rice, SPOTTED LEAF 11, a U-box/ARM repeat protein with E3 ubiquitin ligase activity, negatively regulates programmed cell death (Zeng et al., 2004; Vega-Sánchez et al., 2008). According to our RNA-seq results, genes involved in cell wall degradation are down-regulated in RNAi-ESA1 YIL42 plants, suggesting that ESA1 may affect embryonic sac development by regulating cell degradation pathways. Intriguingly, at the S5 locus, ORF5 encodes an aspartic protease associated with programmed cell death in reproductive organs (Chen et al., 2008). In addition, both OgTPR1 harboring trypsin-like peptidase and ribosome biogenesis regulatory domains and SSP-gla containing specific peptidase domain at the S1 locus deveried from O. glaberrima are involved in protein degradation (Xie et al., 2017b; Koide et al., 2018). These findings suggest that the molecular mechanism of female gametophyte abortion may be related to degradation pathways.
In general, postzygotic reproductive isolation mechanisms are referred to as hybrid incompatibility in the F1 generation or hybrid breakdown in the F2 or later generations (Kubo, 2013). To date, some F1 sterility causal genes at a single locus have been cloned (Long et al., 2008; Yang et al., 2012; Yu et al., 2016, 2018; Shen et al., 2017; Xie et al., 2017b; Koide et al., 2018). The Bateson-Dobzhansky-Muller model proposes that deleterious epistasis in hybrids between two divergent populations or species leads to hybrid failure and prevents gene flow, providing a genetic explanation for hybrid incompatibility (Bateson, 1909; Dobzhansky, 1937; Muller, 1942). So far, several causal genes for hybrid sterility at two or more loci have been identified. The locus pairs DOPPELGANGER1 (DPL1)/DPL2 (Mizuta et al., 2010), S27/S28 (Yamagata et al., 2010), and DGS1/DGS2 (Nguyen et al., 2017) cause hybrid male sterility; the double recessive genotype results in the abortion of gametes. The nonallelic interaction of Hwi1/Hwi2 at two segregating loci has been shown to result in hybrid weakness between common wild rice (O. rufipogon) and the indica variety Teqing (O. sativa). Hwi1 derives from common wild rice, and Hwi2 from Teqing (Chen et al., 2014). Chromosome segment substitution lines (CSSLs) cannot be developed only to effectively discover genes/ QTLs, but also to evaluate the multiple-gene interactions. If a sterility phenotype is masked in a certain genetic background, additional interacting genes can be postulated (Kubo, 2013). Multiple sets of epistatic networks, including has1-hahashsa3, EFS-S24-S35, and Hwi1-Hwi2, have already been identified using CSSLs (Kubo and Yoshimura, 2005; Kubo et al., 2011; Chen et al., 2014). Among the three unlinked epistatic loci, hsa1-has2-hsa3, only the hsa1 locus has been cloned (Kubo et al., 2016). In our study, the rates of spikelet fertility were normal in YJCWR and Teqing, but NIL-qSSR1, a rice CSSL derived from a cross between YJCWR and Teqing, was semisterile. The expression profiles of ESA1 in developing panicles show that the expression of ESA1 in YJCWR is similar to that in NIL-qSSR1 (in the Teqing background). Complementary nonallelic interaction caused by ESA1 and an epistatic gene(s) in Teqing leads to progeny semisterility between YJCWR and Teqing. Nucleotide diversity analysis showed that the SNP site, T1819C, changes the peptide chain length, and whereas only 1819C exists in cultivated rice, the site was polymorphic in wild rice. The variable ESA1 protein structure led to the interaction with nonallelic gene(s) in cultivated rice, which might cause the interspecific hybrid incompatibility between wild rice and cultivated rice.
Using marker L1, we analyzed the genotypic segregation ratio of F2 populations constructed by YIL42 and Teqing and the segregation population from later generations of NIL-qSSR1 heterozygous individuals by self-crossing. We found the segregation ratio of qSSR1/qSSR1 (AA): qSSR1/qssr1 (Aa): qssr1/qssr1 (aa) was 2: 3: 1 (i.e. the gamete ratio of A:a was 7:5), which indicates that the A and a gametes have different transmission proportions. As shown in Supplemental Figure S3, observations of embryo development suggested that the viability of the mature embryo sac in NIL-qSSR1 was 54.62%, and 56.10% in F1 plants derived from a cross between NIL-qSSR1 and Teqing. Meanwhile, pollen fertility of F1 plants was 77.38%, although the male gamete fertility of NIL-qSSR1 and Teqing was normal. According to these results, we could infer two conclusions: (1) About half of the “A” female gametes and half of the “a” female gametes are sterile, i.e. female gamete fertility is controlled by the genotype of the sporophyte (Aa). If it was gametophyticly controlled, the embryo sac sterility would be less in the F1 (∼ 50%) plants. (2) About half of the “a” male gametes were killed, i.e. there might exist hybrid male sterility gene(s) at the qSSR1 locus. Whether ESA1 directly or indirectly causes hybrid male sterility at the qSSR1 locus need further research.
To overcome hybrid sterility, rice breeders have identified several neutral alleles or wide-compatibility varieties harboring compatible alleles that do not cause sterility in the heterozygous state. The natural variation in wide compatibility genes S5-n and S7-n allows highly fertile hybrids to be produced by intersubspecies crosses (Chen et al., 2008; Yang et al., 2012; Yu et al., 2016). The neutral allele, S1mut, was recently created by mutagenesis. Heterozygous hybrids harboring S1mut/S1g and S1mut/S1s did not exhibit sterility (Koide et al., 2018). The introduction of both Sc-n and the Sa-compatible (neutral) allele (Sa-n) by CRISPR/Cas9 plant genome editing can help overcome hybrid male sterility (Shen et al., 2017; Xie et al., 2017a). In the current study, silencing of ESA1 in YIL42 restored spikelet fertility and would improve the fertility of hybrid backcross progeny between O. rufipogon and O. sativa. Therefore, ESA1, which is related to the development of the embryo sac, will help to overcome the hybrid backcross progeny sterility between common wild rice and cultivated rice and promote the application of beneficial genes in wild rice.
CONCLUSION
In this study, we identified a gene near the centromere of chromosome 1, ESA1, that is involved in female gamete abortion during early mitosis. We identified ESA1 in hybrid backcross progeny from a rice CSSL derived from a cross between cultivar Teqing and Yuanjiang common wild rice. ESA1 encodes a nuclear-membrane localized protein containing an ARM repeat domain. Transgenic analysis indicated that changes in protein structure between ESA1 and esa1 affect female sterility. Sequence analysis found that the SNP site, T1819C, which changes the peptide chain length, was polymorphic in wild rice, but only 1819C exists in cultivated rice, suggesting that the variation in ESA1 might be associated with interspecific hybrid incompatibility between wild and cultivated rice. ESA1 knockdown in YIL42 transgenic lines restored fertility, and may be useful for overcoming the barrier of distant hybridization between cultivated rice and wild rice.
MATERIALS AND METHODS
Plant Materials
An introgression line with semisterility, YIL42, was identified from a set of Orzya rufipogon and Orzya sativa introgression lines derived from a cross between an accession of Yuanjiang common wild rice (O. rufipogon, YJCWR) and the high-yielding indica cultivar (O. sativa) Teqing (Tan et al., 2007). The other materials used in this study are listed in Supplemental Tables S1 and S2, including 61 indica varieties, 52 japonica varieties, and 36 wild rice varieties.
Evaluation of Pollen and Embryo Sac Viability
Ten individuals from Teqing and NIL-qSSR1 were examined to determine pollen fertility. Six florets from three panicles of each plant were collected 1 to 2 d before flowering. The pollen from one anther per floret from each plant was mixed and stained with 1% (w/v) iodine potassium iodide (I2-KI) solution, and four views were obtained by light microscopy with at least 100 pollen grains in each view. Nuclei were stained with DAPI, and the pollen was observed under an FV1000 fluorescence microscope (Olympus).
The affinity between the pollen and stigma was examined by observing the behavior of the pollen grains on the stigma after pollination. Twenty florets were collected at 30 min after flowering and used to examine the adherence of pollen on stigmata, pollen germination, and elongation of the pollen tube under an FV1000 fluorescence microscope (Olympus). In vivo pollen germination and elongation assays were conducted as described by Chhun et al. (2007).
To observe embryo sac development in Teqing and NIL-qSSR1, panicles at various developmental stages were excised and fixed in formaldehydea–cetic acid solution (18:1:1 [v/v/v] mixture of formalin, 70% [v/v] ethanol, and acetic acid). The samples were placed under a vacuum for 30 min, incubated for 24 h at room temperature, and stored in 70% (v/v) methanol at 4°C. Before staining, the samples were transferred to 70% (v/v) ethanol, and the lemma and palea were removed to expose the ovaries. The tissue was processed through an ethanol series (50, 30, and 15% [v/v]) and distilled water (20 min per incubation). Each whole ovary was incubated in 1 mol/L hydrochloric acid for 15 min, incubated in 1% (w/v) eosin-Y for 8 h, and washed in distilled water until colorless. The ovaries were incubated in citric acid-disodium hydrogen phosphate buffer (0.1 mol/L, pH 5.0) for an additional 8 h and dyed with 20 μg/mL Hoechst stain at 25°C in the dark for 24 h. The samples were washed three or four times with distilled water and passed through an ethanol series (30%, 50%, 70%, and 90% [v/v]; 20 min per incubation) and absolute ethanol (three times; 2 h each), incubated in 1:1 ethanol and methyl salicylate for 1 h, and cleared three times in methyl salicylate (2 h each the first two times and > 15 h the last time; Dai et al., 2006). To examine embryo sac development of the parents, about 30-80 florets per plant from panicles were sampled at various developmental stages. To investigate mature embryo sac viability of the key recombinant plants and transgenic plants, about 30-50 florets per plant from the middle and top parts of the panicles were sampled during flowering time. The ovaries were observed under an Olympus FV1000 fluorescence microscope (Olympus).
Primers
The primers used in this study are listed in Supplemental Table S3.
Vector Construction and Plant Transformation
An RNAi construct contained the 304 bp at the cDNA sequence position 849-1152, and the reverse complementary sequence 436 bp at the cDNA sequence position 849-1284 driven by the maize Ubiquitin promoter. The esa1 overexpression construct (pOE-esa1) harbored the sequence of the esa1 coding sequence, also under the control of the maize Ubiquitin promoter. The two constructs were introduced into Agrobacterium tumefaciens strain EHA105. Subsequently, the RNAi-ESA1 construct was transferred into YIL42, and the esa1 overexpression constructwas introduced into Teqing. Phenotypic measurements were conducted in transgenic plants from the T1 and T2 generations.
RT-qPCR and 5′-and 3′-RACE
Total RNA was extracted from various samples using TRIzol (Invitrogen) and purified using an RNeasy Kit (Qiagen) following the manufacturer’s instructions. For RT-qPCR, first-strand cDNA synthesis was performed with the GoScript Reverse Transcription System (Promega). The expression levels of ESA1 and other genes were analyzed on a CFX96 real-time system (Bio-Rad). Diluted cDNA was amplified using SYBR Green Master Mix (Bio-Rad). The amplification of ACTIN was used as an internal control to normalize transcript levels. Each set of experiments was repeated three times, and the relative quantification method (2−ΔΔCT) was used to evaluate quantitative variation. 5′-and 3′-RACE were performed using a 5′-Full RACE Kit and 3′-Full RACE Core Set (Takara), respectively, according to the manufacturer’s instructions.
Subcellular Localization
The p35S:: ESA1-GFP construct containing the ESA1 coding sequence fused with GFP driven by the CaMV35S promoter was produced and introduced into rice protoplasts. Confocal imaging analysis was performed under an Olympus FV1000 fluorescence microscope (Tokyo, Japan).
Analysis of Differentially Expressed Genes from RNA-seq
The differentially expressed genes were identified using Differentially Dxpressed Genes seq package with the random sampling model based on the read count for each gene in different libraries (Wang et al., 2010). The false discovery rate method (Benjamini and Yekutieli, 2001) was used to determine the threshold of the P value in multiple tests. The false discovery rate adjusted P value ≤ 0.001 and the absolute value of log2Ratio ≥ 1 were taken as the threshold to judge the significance of gene expression difference.
Sequencing and Data Analysis
QTL analysis was performed using MapManager QTX (Manly et al., 2001). Multiple sequence alignment was performed using CLUSTAL_X (Thompson et al., 1997). A phylogenetic tree was constructed using MEGA 5 (Tamura et al., 2011).
Accession Numbers
Sequence data from this article can be found in the Michigan State University Rice Genome Annotation Project and National Center for Biotechnology Information databases under the following locus names and accession numbers: HSA1a, LOC_Os12g39880; HSA1b, LOC_Os12g39920; PTB1, LOC_Os05g05280; OsCNGC13, LOC_Os06g10580; Sa (SaM), LOC_Os01g39680; Sa (SaF), LOC_Os01g39670; qHMS7 (ORF3), LOC_Os07g45194; Sc, LOC_Os03g14310; S5 (ORF3), LOC_Os06g10990; SPOTTED LEAF 11, LOC_Os12g38210; ESA1, MK510182; Hwi1-25L1, KJ410135; Hwi1-25L2, KJ410136; Hwi2, KJ410137; S5 (ORF5), EU889294; OgTPR1, KY457222; qHMS7 (ORF2), XM_026027159.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Development of the ESA1 locus-containing near isogenic line NIL-qSSR1.
Supplemental Figure S2. Comparison of main panicles and fertility between Teqing and YIL42.
Supplemental Figure S3. Fertility of mature embryo sac, pollen and spikelet, genotypic segregation distortion and the female and male gametes transmission proportions.
Supplemental Figure S4. Comparison of agronomic traits between Teqing and NIL-qSSR1.
Supplemental Figure S5. Seed set ratio after self-crossing and hybridization.
Supplemental Figure S6. Comparison of panicle, spikelet, anthers and mature pollen grains between Teqing and NIL-qSSR1.
Supplemental Figure S7. Characterization of pollen fertility of Teqing and NIL-qSSR1.
Supplemental Figure S8. Expression analysis of ORF 1 and ORF 2 (ESA1).
Supplemental Figure S9. Coding sequences of ESA1 and esa1 (related to Figure 3D).
Supplemental Figure S10. Amino acid variations between ESA1 and esa1.
Supplemental Figure S11. Differentially expressed genes in YIL42 and RNAi-ESA1 YIL42 via RNA-seq analysis.
Supplemental Figure S12. Haplotype analysis of 15 SNPs in the coding region of ESA1.
Supplemental Table S1. Wild rice accessions used in the haplotype and nucleotide diversity analyses.
Supplemental Table S2. Rice cultivars used in the haplotype and nucleotide diversity analyses.
Supplemental Table S3. Primers used in this study.
Footnotes
This work was supported by the National Transgenic Major Project of China (grant no. 2016ZX08001-004) and National Natural Science Foundation of China (NSFC) (grant no. 30771319).
References
- Bateson W. (1909) Heredity and variation in modern lights. In Seward AC. (ed.) Darwin and Modern Science, Cambridge University Press, Cambridge, 85–101 [Google Scholar]
- Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Chen C, Chen H, Lin YS, Shen JB, Shan JX, Qi P, Shi M, Zhu MZ, Huang XH, Feng Q, et al. (2014) A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun 5: 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K, Zhao J, Qiu S, Zhang X, Yao J, et al. (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of Indica–Japonica hybrids in rice. Proc Natl Acad Sci USA 105: 11436–11441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, Kitano H, Ashikari M, Matsuoka M, Ueguchi-Tanaka M (2007) Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19: 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Huang QC, Qin GY, Li GP (2006) Observation on particular embryo sac of rice using confocal microscopy. North China Agriculture 21: 26–29 [Google Scholar]
- Dobzhansky T. (1937) Genetics and the origin of species. Columbia University Press, New York [Google Scholar]
- Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90: 871–882 [DOI] [PubMed] [Google Scholar]
- Koide Y, Ogino A, Yoshikawa T, Kitashima Y, Saito N, Kanaoka Y, Onishi K, Yoshitake Y, Tsukiyama T, Saito H, et al. (2018) Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc Natl Acad Sci USA 115: E1955–E1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T. (2013) Genetic mechanisms of postzygotic reproductive isolation: An epistatic network in rice. Breed Sci 63: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yoshimura A (2005) Epistasis underlying female sterility detected in hybrid breakdown in a Japonica–Indica cross of rice (Oryza sativa L.). Theor Appl Genet 110: 346–355 [DOI] [PubMed] [Google Scholar]
- Kubo T, Yoshimura A, Kurata N (2011) Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189: 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Takashi T, Ashikari M, Yoshimura A, Kurata N (2016) Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol Plant 9: 221–232 [DOI] [PubMed] [Google Scholar]
- Li S, Li W, Huang B, Cao X, Zhou X, Ye S, Li C, Gao F, Zou T, Xie K, et al. (2013) Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat Commun 4: 2793. [DOI] [PubMed] [Google Scholar]
- Long Y, Zhao L, Niu B, Su J, Wu H, Chen Y, Zhang Q, Guo J, Zhuang C, Mei M, et al. (2008) Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci USA 105: 18871–18876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH Jr., Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Mizuta Y, Harushima Y, Kurata N (2010) Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc Natl Acad Sci USA 107: 20417–20422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima H. 1997. An observation of wild rice population in Hainan and Yuanjiang China. Report of National Institute of Genetics, pp. 358–375, https://shigen.nig.ac.jp/rice/oryzabase/asset/ricereport/Part_II.pdf
- Muller HJ. (1942) Isolating mechanisms, evolution and temperature. Biol Symp 6: 71-125 [Google Scholar]
- Nguyen GN, Yamagata Y, Shigematsu Y, Watanabe M, Miyazaki Y, Doi K, Tashiro K, Kuhara S, Kanamori H, Wu J, et al. (2017) Duplication and loss of function of genes encoding RNA polymerase III subunit C4 causes hybrid incompatibility in rice. G3 (Bethesda) 7: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76: 789–791 [DOI] [PubMed] [Google Scholar]
- Samuel MA, Salt JN, Shiu SH, Goring DR (2006) Multifunctional arm repeat domains in plants. Int Rev Cytol 253: 1–26 [DOI] [PubMed] [Google Scholar]
- Shen R, Wang L, Liu X, Wu J, Jin W, Zhao X, Xie X, Zhu Q, Tang H, Li Q, et al. (2017) Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat Commun 8: 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LB, Liu FX, Xue W, Wang GJ, Ye S, Zhu ZF, Fu YC, Wang XK, Sun CQ (2007) Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J Integr Plant Biol 49: 871–884 [Google Scholar]
- Tewari R, Bailes E, Bunting KA, Coates JC (2010) Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol 20: 470–481 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Zeng L, Chen S, Leung H, Wang GL (2008) SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Xie Y, Niu B, Long Y, Li G, Tang J, Zhang Y, Ren D, Liu YG, Chen L (2017a) Suppression or knockout of SaF/SaM overcomes the Sa-mediated hybrid male sterility in rice. J Integr Plant Biol 59: 669–679 [DOI] [PubMed] [Google Scholar]
- Xie Y, Xu P, Huang J, Ma S, Xie X, Tao D, Chen L, Liu YG (2017b) Interspecific hybrid sterility in rice is mediated by OgTPR1 at the S1 locus encoding a peptidase-like protein. Mol Plant 10: 1137–1140 [DOI] [PubMed] [Google Scholar]
- Xu Y, Yang J, Wang Y, Wang J, Yu Y, Long Y, Wang Y, Zhang H, Ren Y, Chen J, et al. (2017) OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genet 13: e1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y, Yamamoto E, Aya K, Win KT, Doi K, Sobrizal, Ito T, Kanamori H, Wu J, Matsumoto T, et al. (2010) Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc Natl Acad Sci USA 107: 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao X, Cheng K, Du H, Ouyang Y, Chen J, Qiu S, Huang J, Jiang Y, Jiang L, et al. (2012) A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Yu X, Zhao Z, Zheng X, Zhou J, Kong W, Wang P, Bai W, Zheng H, Zhang H, Li J, et al. (2018) A selfish genetic element confers non-Mendelian inheritance in rice. Science 360: 1130–1132 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhao Z, Shi Y, Tian H, Liu L, Bian X, Xu Y, Zheng X, Gan L, Shen Y, et al. (2016) Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 203: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan PR, Yang CD, Zhou N, He QR (1998) Studies on differentiation of Yuan-jiang common wild rice in Yunnan, China. II, Preliminary study on compatibility between common wild rice and indica and japonica cultivated rice. Agricultural Archaeology 1: 38–40 [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]