Screening of an annotated library of biologically active molecules for inhibitory effects on auxin-induced Ca2+ entry in BY-2 cells yielded several new inhibitors for investigating Ca2+ signaling.
Abstract
Many signal perception mechanisms are connected to Ca2+-based second messenger signaling to modulate specific cellular responses. The well-characterized plant hormone auxin elicits a very rapid Ca2+ signal. However, the cellular targets of auxin-induced Ca2+ are largely unknown. Here, we screened a biologically annotated chemical library for inhibitors of auxin-induced Ca2+ entry in plant cell suspensions to better understand the molecular mechanism of auxin-induced Ca2+ and to explore the physiological relevance of Ca2+ in auxin signal transduction. Using this approach, we defined a set of diverse, small molecules that interfere with auxin-induced Ca2+ entry. Based on annotated biological activities of the hit molecules, we found that auxin-induced Ca2+ signaling is, among others, highly sensitive to disruption of membrane proton gradients and the mammalian Ca2+ channel inhibitor bepridil. Whereas protonophores nonselectively inhibited auxin-induced and osmotic stress–induced Ca2+ signals, bepridil specifically inhibited auxin-induced Ca2+. We found evidence that bepridil severely alters vacuolar morphology and antagonized auxin-induced vacuolar remodeling. Further exploration of this plant-tailored collection of inhibitors will lead to a better understanding of auxin-induced Ca2+ entry and its relevance for auxin responses.
The plant hormone auxin is a potent regulator of a diverse set of developmental processes, ranging from embryogenesis, postembryonic organogenesis, and regeneration to tropic growth responses (Vanneste and Friml, 2009). These pluripotent effects in plant development make auxin a key player in the plant’s developmental plasticity. Moreover, auxin is subject to extensive cross-talk with many other signaling pathways for flexible integration in auxin-regulated development (Chaiwanon et al., 2016; Liu et al., 2017). Decades of extensive research have led to the formulation of a canonical auxin signaling pathway. In short, the perception of auxin occurs via the auxin-induced stabilization of a coreceptor complex constituted by TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB) and Aux/indole-3-acetic acid (IAA) proteins, resulting in the ubiquitination and proteolysis of the latter. Consequently, Aux/IAA-interacting AUXIN RESPONSE FACTORs (ARFs) can become active (Lavy and Estelle, 2016; Weijers and Wagner, 2016). This auxin signaling mechanism can explain many of the plant’s responses to auxin. In addition, a nontranscriptional branch of TIR1/AFB-based auxin perception was recently connected to the nontranscriptional inhibition of elongation (Fendrych et al., 2018), vacuolar remodeling (Löfke et al., 2015), and activation of Ca2+ signaling (Dindas et al., 2018).
A large body of literature describes the role of Ca2+ in a variety of cellular processes in plants in the context of responses to light, and biotic and abiotic stress (for review, see Tuteja and Mahajan, 2007; Kudla et al., 2010, 2018). However, little is known about the role of Ca2+ signaling downstream of auxin. Interestingly, a few reports connect Ca2+ to auxin transport regulation (Dela Fuente and Leopold, 1973; Benjamins et al., 2003; Zhang et al., 2011; Rigó et al., 2013). More recently, auxin-induced cytosolic Ca2+ increase was proposed to contribute to auxin’s inhibitory effect on root growth and auxin-regulated root hair growth via the nonselective cation channel CNGC14 (Shih et al., 2015; Dindas et al., 2018). Jointly, these reports illustrate the importance of Ca2+ in auxin physiology. Despite this recent progress, it is clear that much remains to be uncovered about the underlying signaling mechanism and its cellular targets.
Several types of plant Ca2+ channel types exist in relatively large gene families, as illustrated in a few examples in Arabidopsis (Arabidopsis thaliana): 20 CYCLIC NUCLEOTIDE-GATED CHANNELs (CNGCs; Ma and Berkowitz, 2011), 20 GLU RECEPTOR-LIKE CHANNELs (GLRs; Forde and Roberts, 2014), and 16 OSCA/TMEM63 channels (Murthy et al., 2018; Zhang et al., 2018). This genetic complexity greatly hinders pinpointing the Ca2+ channels that are involved in a given Ca2+ regulated process. Therefore, the involvement of Ca2+ in any process in plants is often deduced from using Ca2+ chelators such as EGTA and 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acidor very nonselective Ca2+ channel blockers such as La3+ and Gd3+. However, these treatments do not reveal anything about the molecular nature of the Ca2+ channel involved. Moreover, some important side effects need to be considered when using these treatments to manipulate Ca2+, for example, acidification caused by EGTA and1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid releasing four H+ when binding two Ca2+ and the efficient precipitation of phosphates by La3+ (for review, see De Vriese et al., 2018).
The importance of Ca2+ in human physiology and neurology led to the development of an extensive pharmacological toolbox to manipulate specific groups of Ca2+ channels. Simple drug treatments thus allow manipulation of specific Ca2+ channels and evaluation of its effect on any process of interest. Unfortunately, the Ca2+ signaling machinery in plants has diverged significantly from the one in the animal kingdom, with many of the Ca2+ signaling components of animals being absent in plants and vice versa (Nagata et al., 2004; Edel et al., 2017). For instance, channels associated with muscle and nerve Ca2+ signal transduction in animals, such as l-type voltage-dependent Ca2+ channels (Zuccotti et al., 2011), inositol 1,4,5-triphosphate receptors (Nixon et al., 1994), and ryanodine receptors (Lanner et al., 2010), are either missing or significantly different in plants. Hence, many Ca2+ channel inhibitors established in animal systems are arguably of limited use in plants. Only a few ionotropic GLR inhibitors (DNQX, CNQX, MNQX, and AP5) were also shown to have inhibitory effects on specific plant GLR-based Ca2+ channeling activities (for review, see De Vriese et al., 2018), but the molecular nature and specificity of these inhibitory effects remain to be characterized. These examples illustrate that there is an important need to develop Ca2+ channel inhibitors directly in plant systems to stimulate plant Ca2+ research.
Here, we set out to identify small molecules that modify the shape and amplitude of auxin-induced cytosolic Ca2+ dynamics and use them to further explore the role of Ca2+ signaling in cellular auxin responses. We screened a collection of biologically active and structurally diverse compounds for inhibitors of auxin-induced Ca2+ signaling in transgenic tobacco (Nicotiana tabacum) BY-2 cell lines that express yellow fluorescent protien (YFP)-apoaequorin as a reporter of Ca2+ signaling. Our primary screen identified 80 potential inhibitors, of which 67 were reconfirmed in a confirmation screen. Based on annotated biological functions, we found that protonophores nonselectively interfered with auxin-induced Ca2+. Moreover, we found that auxin-induced Ca2+ signals were much more sensitive to bepridil than hyperosmotic-stress–induced Ca2+ signals. This differential drug sensitivity is consistent with distinct Ca2+ channel types being involved in the Ca2+ response (Yuan et al., 2014; Shih et al., 2015; Dindas et al., 2018). Interestingly, we found that bepridil had a severe impact on vacuolar morphology, to the extent that it antagonized auxin-induced vacuolar remodeling. The resulting set of inhibitors thus represents a valuable expansion of the toolbox with nonspecific and semiselective inhibitors for exploring the role of Ca2+ signaling in plants.
RESULTS
Assay Development and Chemical Screen
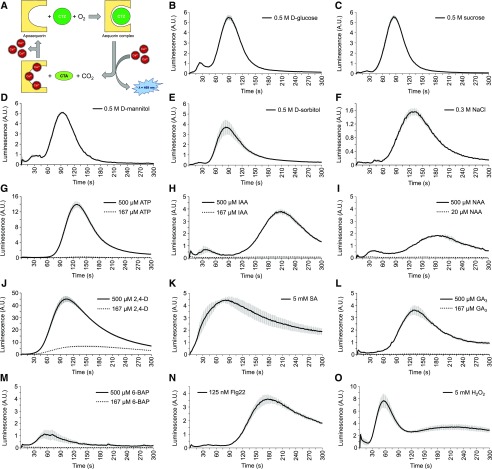
Unlike whole plants, which are composed of complex mixtures of cell types with different stimulus sensitivities, tobacco BY-2 cell suspension cultures are highly homogenous in terms of cell type and developmental stage, thus making them highly suited for high-throughput screening in a multiwell format. A BY-2 cell suspension line expressing YFP-fused apoaequorin (YA; Mehlmer et al., 2012) was established for luminescence-based quantification of Ca2+ signals (Shimomura et al., 1962; Knight et al., 1991). Aequorin is a bioluminescent photoprotein that emits blue light upon Ca2+ binding and has been used extensively as a Ca2+ sensor (Shimomura et al., 1962; Shimomura, 2005; Fig. 1A). The YFP fused to the aequorin was used to visualize expression of the probe in the BY-2 cells but did not contribute to the sensor activity. The assay was miniaturized to 96-well plate format and validated by assessing the effect of 14 known elicitors of Ca2+ signaling. The known elicitors induced distinct signals in this BY-2 cell culture (Fig. 1, B–O), corroborating the suitability of the cells to assess the effect on Ca2+ signaling.
Figure 1.
Fourteen elicitors induce a distinct Ca2+ signal in aequorin-expressing BY-2 cells. A, Schematic representation of aequorin complex formation and bioluminescent reaction. A functional aequorin complex is formed upon binding of apoaequorin with its substrate coelenterazine (CTZ) in the presence of O2. The binding of three Ca2+ ions leads to the conversion of CTZ into coelenteramide (CTA) and CO2, upon which blue light (λ = 469 nm) is emitted. B–O, Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with various potential elicitors: 0.5 M d-Glucose (B), 0.5 M Sucrose (C), 0.5 M d-mannitol (D), 0.5 M d-sorbitol (E), 0.3 M NaCl (F), 167 µM and 500 µM ATP (G), 167 µM and 500 µM IAA (H), 20 µM and 500 µM NAA (I), 167 µM and 500 µM 2,4-d (J), 5 mM salicylic acid (SA; K), 167 µM and 500 µM gibberellin (GA3; L), 167 µM and 500 µM 6-benzylaminopurine (6-BAP; M), 125 nM flg22 (N), and 5 mM H2O2 (O). The data represent average luminescence values of 3 individual measurements in the same multiwell plate. Error bars represent ± sem. A.U., arbitrary units.
The BY-2 cell suspensions were poorly responsive to exogenous application of auxins (Fig. 1, H–J). This probably reflects an auxin insensitivity that is associated with prolonged culturing of the cells in relatively high 2,4-d concentrations (0.2 mg/L ≈ 1 µM in the growth medium). Therefore, the 2,4-d concentration was increased to 500 µM, which resulted in a robust auxin-induced Ca2+ response. Immediately after 2,4-d addition, the luminescent signal rapidly increased and reached a maximum after ∼90 s, which attenuated to close to baseline levels around 300 s after addition (Fig. 1J). By using the viability stains propidium iodide (PI) and fluorescein diacetate (FDA), no obvious difference in cellular integrity was observed after 1-h treatment with 2,4-d, suggesting that the immediate Ca2+ response to 2,4-d (within minutes) is unlikely the result of a defect in cellular viability (Supplemental Fig. S1).
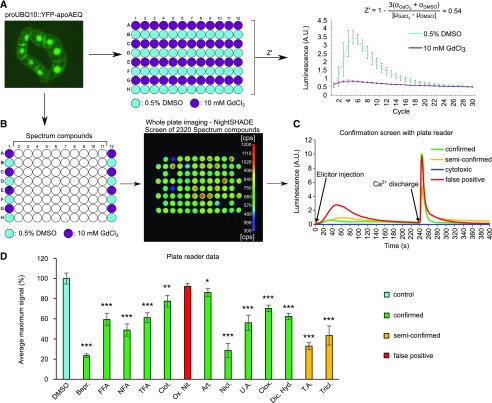
Next, we aimed to evaluate the quality of our assay by calculating its Z-prime (Z′) factor, which is a commonly used statistical indicator of quality for high-throughput assays (Zhang et al., 1999). It allows estimation of the quality of the bioassay in discriminating the effect of hit molecules from read-out variation based on two parameters: (1) the ‘separation band’ of the assay: the difference between the mean of the negative control plus three times the standard deviations of the negative control and the mean of positive control minus three times the standard deviations of the positive controls and (2) the dynamic range of the assay: the difference between the mean of the positive and negative controls. In a good assay, the means of the positive and negative controls differ strongly from each other, whereas the standard deviations are very low (Fig. 2A). An ideal assay has a Z′ factor close to 1, and assays with a Z′ factor between 0.5 and 1 are generally considered to be excellent (Zhang et al., 1999). Here, we calculated the Z′ value of our assay based on the means and standard deviations of the peak intensities of mock-treated cells (dimethyl sulfoxide [DMSO]) and cells treated with a nonspecific Ca2+ channel inhibitor (GdCl3; Fig. 2A). The Z′ factor was 0.54, suggesting that our assay is robust enough to reliably distinguish potential inhibitors from well-to-well variation. Subsequently, we used this experimental setup to screen the Spectrum collection (MicroSource Discovery Systems) of 2,320 annotated compounds (Fig. 2B). This resulted in the identification of 80 molecules that reduced the maximal response induced by 500 µM 2,4-d to less than 55% of that of the respective DMSO controls. Next, we used a more sensitive well-per-well reconfirmation assay via a plate reader, which also allowed assay of the viability of the cells at the end of the assay by adding a discharge solution. This treatment causes disruption of the membrane integrity and flooding of the cytoplasm with Ca2+, which binds the remnant reconstituted aequorin, and thus generates a sharp, strong luminescence peak (discharge peak; Fig. 2C). Out of the 80 primary hits, 67 were reconfirmed for reducing the auxin-responsive Ca2+ signal by more than 45% (Supplemental Table S1). Of these 67 confirmed hits, 39 showed no obvious short-term cytotoxicity, as evidenced by the presence of a large Ca2+ discharge peak (Fig. 2C, green; Supplemental Table S1, confirmed). However, 28 confirmed hits caused a reduction (Fig. 2C, yellow; Supplemental Table S1, semiconfirmed) or even complete absence of the Ca2+ discharge peak, indicating a defect in Ca2+ compartmentalization mechanisms and/or cell viability (Fig. 2C, blue; Supplemental Table S1, cytotoxic). Based on structural features, the 67 hits retained after the confirmation screen could be organized into 43 clusters of structurally similar compounds (Supplemental Table S1).
Figure 2.
Schematic representation of primary screen for Ca2+ signaling inhibitors. A and B, Multiwell setup for screening inhibitors of auxin-induced Ca2+ responses via YFP-apoaequorin-expressing BY-2 cells. Based on the means and standard deviations of both the positive (10 mM GdCl3, purple) and negative (0.5% [v/v] DMSO, cyan) controls of a test run, a Z′ score of 0.54 could be calculated (A), supporting the robustness of the assay. With use of this setup, the Spectrum library of 2320 compounds was screened for inhibitors of 2,4-d-induced Ca2+ signaling (B). The outer columns contained positive (10 mM GdCl3, purple) and negative (0.5% DMSO, cyan) controls, with the assay compounds in the 10 inner columns (50 µM; white). Addition of auxin induced a rapid luminescence-based signal that was detected with the NightSHADE luminescence imaging system. From this library, 80 hit compounds were retained that caused a maximum signal less than 55% of that of the DMSO-treated control cells. C, Confirmation screen of the 80 hit compounds in a multiwell plate reader. After 240 s, 50 µL discharge solution (0.1 M CaCl2 and 20% [v/v] ethanol) was added and luminescence was measured for the remaining 160 s. Based on their Ca2+ response and burst patterns, the tested compounds could be further categorized into 4 groups: confirmed (green), semiconfirmed (yellow), cytotoxic (blue), and false-positive (red) compounds. D, Maximum 2,4-d-induced Ca2+ response signal of YFP-apoaequorin-expressing BY-2 cells pretreated with 50 µM of 13 selected hit compounds. The data represent average maximum luminescence values of 4–8 individual measurements in comparison with the average of 4–8 DMSO controls in the same multiwell plate. Error bars represent ± sem. Bars are color-coded based on the underlying Ca2+ response and burst patterns. Student’s t test p-values: *P < 0.05, **P < 0.01, and ***P < 0.001. A.U., arbitrary units; Bepr., bepridil; FFA, flufenamic acid; NFA, niflumic acid; TFA, tolfenamic acid; Clot., clotrimazole; Ox. Nit., oxiconazole nitrate; Art., artemether; Nicl., niclosamide; U.A., (+)-usnic acid; Clox., cloxyquin; Dic. Hyd., dicyclomine hydrochloride; T.A., tannic acid; Tricl., triclosan.
Given that both the primary screen and confirmation screen represented single-well analyses, we aimed to further validate a part of our dataset using multiple biological repeats. Therefore, we selected 13 commercially available hit molecules representing a large chemical diversity for further validation (Table 1). The auxin-induced Ca2+ responses were analyzed in 4–8 replicates on YFP-apoaequorin-expressing BY-2 cells (Mehlmer et al., 2012). Out of the 13 tested chemicals, 10 could be confirmed to strongly modify the 2,4-d induced Ca2+ signature, while maintaining a robust discharge peak (Fig. 2D). Together, these data highlight that our set of 67 hits after the confirmation screen is rich in potent modifiers of auxin-induced Ca2+ signaling.
Table 1. Thirteen compounds selected for further validation experiments.
| Name | Reported Bioactivities | Reference |
|---|---|---|
| Artemether | Antimalarial agent, HMGCoA inhibitor | Korade et al. (2016) |
| Bepridil hydrochloride | Calcium-blocking agent, antiarrhythmic, antihypertensive, calmodulin antagonist | Narahara et al. (1992) |
| Clotrimazole | Antifungal, antibacterial, sterol biosynthesis inhibitor | Qiu et al. (2017) |
| Cloxyquin | Antibacterial, antifungal | Hongmanee et al. (2007) |
| Dicyclomine hydrochloride | Anticholinergic | Ali et al., 2018 |
| Flufenamic acid | Anti-inflammatory, analgesic, antipyretic | Habjan and Vandenberg (2009) |
| Niclosamide | Anthelmintic, teniacide | Monin et al. (2016) |
| Niflumic acid | Analgesic, anti-inflammatory | Hogg et al. (1994) |
| Oxiconazole nitrate | Antifungal, sterol biosynthesis inhibitor | Jegasothy and Pakes (1991) |
| Tannic acid | Nonspecific enzyme/receptor blocker | Isenburg et al. (2005) |
| Tolfenamic acid | Anti-inflammatory, analgesia | Pentikäinen et al. (1981) |
| Triclosan | Anti-infective, antibacterial, antifungal | Heath et al. (1999) |
| (+)-Usnic acid | Antibacterial | Latkowska et al. (2006) |
Fenamates Alter the Shape of Auxin-Induced Ca2+
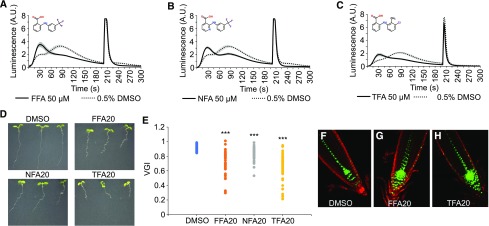
Among the 67 confirmed hit compounds, we found four highly related fenamate-type chemicals: (1) flufenamic acid (FFA), (2) niflumic acid (NFA), (3) tolfenamic acid (TFA), and (4) flunixin meglumine (structures of all compounds described in the manuscript are shown in Supplemental Fig. S2). Unlike any other tested hit compounds, which simply reduced the amplitude of the Ca2+ response, these fenamates had a dramatic effect on the shape of the Ca2+ response. Treatment with FFA, NFA, or TFA reduced the maximum of the Ca2+ signal, but also revealed a novel Ca2+ signal that preceded the maximum (Fig. 3, A–C). Such an effect on the Ca2+ signal shape was not observed for any of the other hit compounds that were selected for validation.
Figure 3.
Fenamates alter the shape of auxin-induced Ca2+. A–C, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM FFA (A), NFA (B), or TFA (C). Discharge solution was added after 200 s. The data represent average luminescence values of 4–8 individual measurements (solid lines) in comparison with the average of 4–8 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. D, Phenotype of wild-type Col-0 seedlings grown for 7 d in presence of 0.1% DMSO or 20 µM FFA, NFA, or TFA. E, Vertical Growth Index (VGI) values for the roots from (D). For each treatment, 42 to 47 roots were measured. Student’s t test p-values: ***P < 0.001. F–H, Confocal microscopy images of 5-d-old DR5rev::VENUS-N7 seedlings grown on 0.1% (v/v) DMSO (F), 20 µM FFA (G), and 20 µM TFA (H). Green: DR5rev::VENUS-N7 signal; red: propidium iodide staining. A.U., arbitrary units.
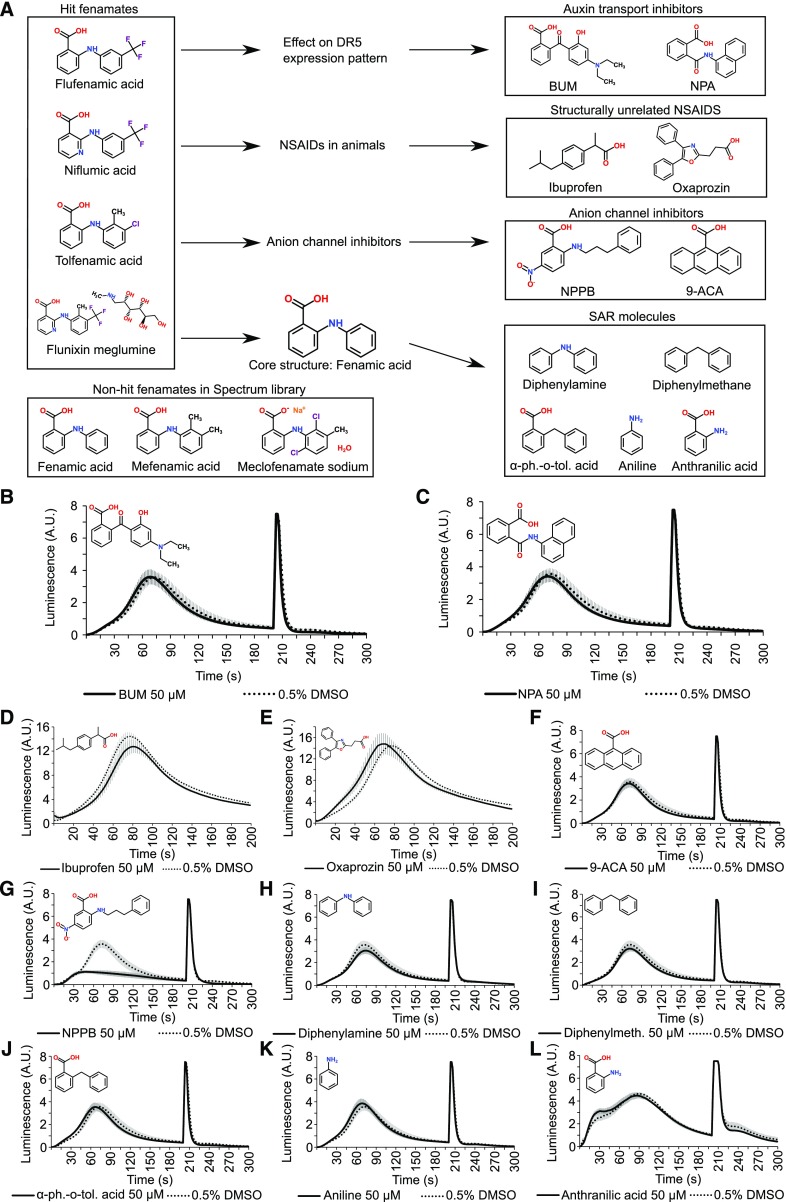
Seedlings grown for 7 d in the presence of 20 µM FFA, NFA, or TFA had significantly shorter roots than seedlings grown on control plates and displayed a reduced gravitropic root growth, as indicated by a reduced vertical growth index (Fig. 3, D and E; Supplemental Fig. S3). Consistently, we observed spreading of the expression of the synthetic auxin response reporter DR5rev::VENUS-N7 in the columella and stem cell niche (Fig. 3, F–H), reminiscent of an inhibitory effect on auxin transport. However, neither of the two known auxin transport inhibitors, 2-[4-(diethylamino)-2-hydroxybenzoyl]benzoic acid (BUM) and 1-N-naphtylphtalamic acid (NPA), had an obvious effect on auxin-induced Ca2+ (Fig. 4, A–C), suggesting that a block in auxin transport does not explain the effect of fenamates on Ca2+ signaling.
Figure 4.
Functional and structural fenamate analogs do not alter the shape of auxin-induced Ca2+. A, Overview of a small-scale structure-activity relationship analysis of the fenamates. Based on known and observed functions and structures of the hit fenamates, a set of functional and structural analogs was investigated. B and C, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM of the auxin transport inhibitors BUM (B) and NPA (C). Discharge solution was added after 200 s. The data represent average luminescence values of 4 individual measurements (solid lines) in comparison with the average of 4 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. D and E, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM of the NSAIDs ibuprofen (D) and oxaprozin (E; note: measurements were done with a new plate reader [GloMax Navigator – Promega] because the old one was defective). The data represent average luminescence values of 8 individual measurements (solid lines) in comparison with the average of 8 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. F–L, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM of 2 anion channel inhibitors (9-ACA and NPPB; F and G) and various compounds structurally similar to fenamates (H–L). Discharge solution was added after 200 s. The data represent average luminescence values of 4 individual measurements (solid lines) in comparison with the average of 4 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. BUM, 2-[4-(diethylamino)-2-hydroxybenzoyl]benzoic acid; NPA, 1-N-naphtylphtalamic acid; 9-ACA, 9-anthracenecarboxylic acid; NPPB, 5-nitro-2-(3-phenylpropylamino) benzoic acid; α-ph.-o-tol. acid, α-phenyl-o-toluic acid; A.U., arbitrary units.
The inhibition of cyclooxygenase activity by fenamates renders them suitable for use as nonsteroidal anti-inflammatory drugs (NSAIDs; Graham, 2016), and NFA has reported anion channel inhibitory activities (Diatloff et al., 2004; Gilliham and Tester, 2005). Based on these reported bioactivities, we investigated whether other NSAIDs and anion channel inhibitors have a similar effect on Ca2+ signaling (Fig. 4A). Of two structurally unrelated NSAIDs (ibuprofen and oxaprozin) and two unrelated anion channel inhibitors, 5-nitro-2-(3-phenylpropylamino) benzoic acid (NPPB) and 9-anthracenecarboxylic acid (9-ACA), only NPPB inhibited auxin-induced Ca2+ (Fig. 4, D–G). However, the effect of NPPB was clearly distinct from the effect of fenamates on Ca2+ response dynamics (Fig. 4G), suggesting that the observed effects of fenamates on Ca2+ signaling are independent of any additional effects on cyclooxygenases or anion channels.
These analyses suggest that the effect of fenamates on Ca2+ signaling is not related to any of their known (cyclooxygenase inhibition and anion channel inhibition) and observed (auxin transport inhibition) biological activities but is rather a distinct feature associated with the chemical structure. Therefore, we also tested the effect of a range of commercially available structurally similar compounds to see whether the effect of the fenamates could be explained by the presence of a specific substructure (Fig. 4A). However, every tested molecular variant (diphenylamine, diphenylmethane, α-phenyl-o-toluic acid, aniline, and anthranilic acid) failed to inhibit/modify auxin-induced Ca2+ signals at 50 µM (Fig. 4H-L), suggesting that the fenamate-core structure represents the biological activity on Ca2+. Moreover, when revisiting the primary screen data of all fenamates that were included in the Spectrum library, we found that fenamic acid and mefenamic acid showed a tendency to reduce the Ca2+ response changes, albeit with a lower potency than the validated fenamates, whereas the effect of meclofenamate sodium was negligible (Fig. 4A).
When we evaluated the 2,4-d-sensitive root growth, no obvious resistance or hypersensitivity could be observed, suggesting that fenamates do not affect auxin perception or 2,4-d uptake (Supplemental Fig. S4A). Similarly, no obvious resistance or hypersensitivity to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) could be observed in seedlings grown in the presence of these fenamates (Supplemental Fig. S4A). In addition, we also evaluated the effects of NPPB, 9-ACA, BUM, NPA, and the fenamate analogs on root growth and their sensitivity to 2,4-d and ACC (Supplemental Fig. S4B). Interestingly, unlike FFA, NFA, and TFA, diphenylamine, aniline, and anthranilic acid caused a significant 2,4-d resistance, which further illustrates a completely different mode of action between the effective fenamates and diphenylamine, aniline, and anthranilic acid.
Protonophores Impair the Ca2+ Response to Distinct Stimuli and Render Roots Insensitive to 2,4-d
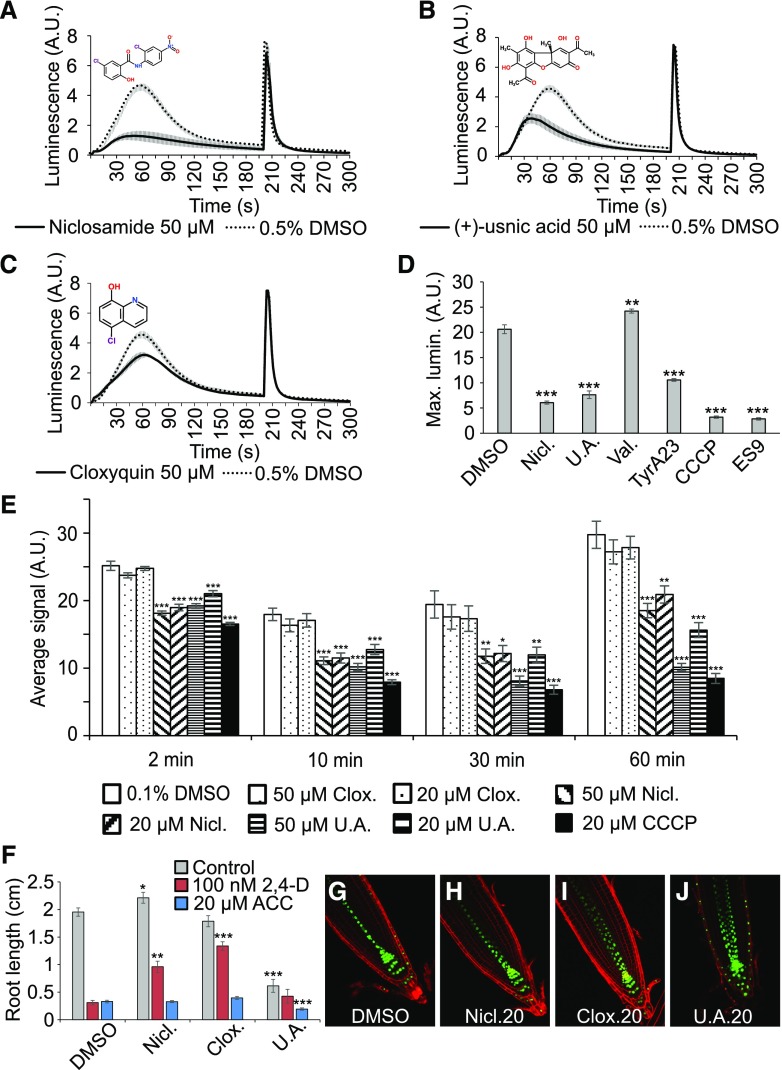
Inspection of the 67 confirmed hits revealed a total of at least 13 molecules with reported protonophore activities in different organisms (Supplemental Table S1), suggesting an important contribution of H+ gradients to auxin-induced Ca2+. We selected niclosamide, (+)-usnic acid, and cloxyquin as structurally diverse representatives of this group of hits. Both niclosamide and (+)-usnic acid could be confirmed as potent inhibitors of auxin-induced Ca2+ signals (Fig. 5, A and B), whereas cloxyquin caused only a modest reduction in the peak of the Ca2+ signal (Fig. 5C). To test whether the suspected protonophore activities of niclosamide and (+)-usnic acid are the underlying cause of their observed inhibitory effect on Ca2+ signals, we also analyzed three well-characterized, but structurally unrelated protonophores: carbonyl cyanide m-chlorophenyl hydrazone (CCCP), tyrphostin A23 (TyrA23), and endosidin9 (ES9; Dejonghe et al., 2016). In addition, we included the K+ selective ionophore valinomycin (25) to account for general disruption of gradients of monovalent cations. Similar to niclosamide and (+)-usnic acid, the three tested protonophores were potent inhibitors of 2,4-d-induced Ca2+ signaling (Fig. 5D). Importantly, protonophore treatment did not cause obvious defects in Ca2+ discharge profiles in the BY-2 cells, suggesting that an impaired Ca2+ compartmentalization cannot explain the inhibition of 2,4-d-induced Ca2+. Unlike the protonophores, valinomycin did not reduce the maximal Ca2+ responses (Fig. 5D), suggesting that 2,4-d-induced Ca2+ signaling specifically requires proton-based gradients, rather than K+ gradients. These findings are entirely consistent with the recently reported very tight interdependence between H+ and Ca2+ dynamics (Behera et al., 2018).
Figure 5.
Protonophores impair the 2,4-d-induced Ca2+ response and render roots insensitive to 2,4-d. A–C, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM niclosamide (A), (+)-usnic acid (B), or cloxyquin (C). Discharge solution was added after 200 s. The data represent average luminescence values of 4 individual measurements (solid lines) in comparison with the average of 4 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. D, Maximum 2,4-d-induced luminescence in YFP-apoaequorin-expressing BY-2 cells treated with 50 µM of the hit protonophores niclosamide and (+)-usnic acid, the ionophore valinomycin, or the nonhit protonophores tyrphostin A23, CCCP, and ES9. Four individual measurements were performed for each treatment and compared with the average of 8 DMSO controls in the same multiwell plate. Error bars represent ± sem. Student’s t test p-values: **P < 0.01 and ***P < 0.001. E, ATP measurements of BY-2 cells pretreated with 0.5% (v/v) DMSO (white), 20 µM CCCP (black), or 20 or 50 µM cloxyquin (dots), niclosamide (diagonal stripes), or (+)-usnic acid (horizontal stripes). ATP was measured 2, 10, 30, and 60 min after compound treatment. Error bars represent ± sem. Student’s t test p-values: *P < 0.05, **P < 0.01, and ***P < 0.001. F, Average primary root length of wild type Col-0 seedlings grown for 7 d on 1/2 MS medium in presence of 0.1% (v/v) DMSO or 20 µM niclosamide, cloxyquin, or (+)-usnic acid (gray bars) and supplemented with 100 nM 2,4-d (red bars) or 20 µM ACC (blue bars). Root lengths represent average of 12–21 roots. Error bars represent ± sem. Student’s t test p-values: *P < 0.05, **P < 0.01, and ***P < 0.001. G-J, Confocal microscopy images of 5-d-old DR5rev::VENUS-N7 seedlings grown on 0.1% (v/v) DMSO (G), 20 µM niclosamide (H), 20 µM cloxyquin (I), or 20 µM (+)-usnic acid (J). Green: DR5rev::VENUS-N7 signal; red: propidium iodide staining. The image in (G) is identical to Figure 3F. A.U., arbitrary units.
Because protonophores dissipate H+-gradients across membranes, including the mitochondrial membranes, they are expected to interfere with ATP production due to mitochondrial uncoupling. Therefore, we evaluated the evolution of ATP content in BY-2 cells during 60-min treatments with 20 or 50 µM niclosamide, (+)-usnic acid, or cloxyquin (Fig. 5E). As a positive control we included the well-described protonophore CCCP (20 µM). Within 2 min, niclosamide, (+)-usnic acid, and CCCP caused a significant reduction of cellular ATP levels compared with DMSO-treated cells. Niclosamide had a milder effect on the ATP levels than (+)-usnic acid and CCCP. In contrast, cloxyquin did not interfere with ATP production, even at prolonged incubation times. This suggests that niclosamide and (+)-usnic acid have protonophore activities in plants, which could explain how they inhibit 2,4-d-induced Ca2+ signals.
Next, we evaluated the effects of these molecules on root growth (Fig. 5F). Although cloxyquin did not cause any noticeable defects, plants treated with niclosamide had on average slightly longer primary roots, whereas (+)-usnic acid had a strong inhibitory effect on the primary root length, suggesting that they have distinct cellular targets. Moreover, (+)-usnic acid induced DR5rev::VENUS-N7 expression in the lateral root cap, which was not observed with niclosamide and cloxyquin (Fig. 5, G–J). When cotreated with 100 nM 2,4-d and niclosamide or cloxyquin [but not (+)-usnic acid], the seedlings had significantly longer roots than the controls (Fig. 5F). However, because cloxyquin caused only a slight inhibition of 2,4-d-induced Ca2+ signaling and did not disturb ATP production in BY-2 cells, the observed resistance to 2,4-d is likely unrelated to impairment of Ca2+ signaling or protonophore activity. Importantly, the protonophores did not render root growth resistant to ACC (Fig. 5F), as seen for 2,4-d uptake-defective aux1 mutants (Swarup et al., 2001), suggesting that the observed 2,4-d resistance is not due to impaired AUX1-mediated auxin uptake.
Bepridil Is a Potent Inhibitor of Auxin-Induced Ca2+ Signaling and Modifies Vacuolar Morphology
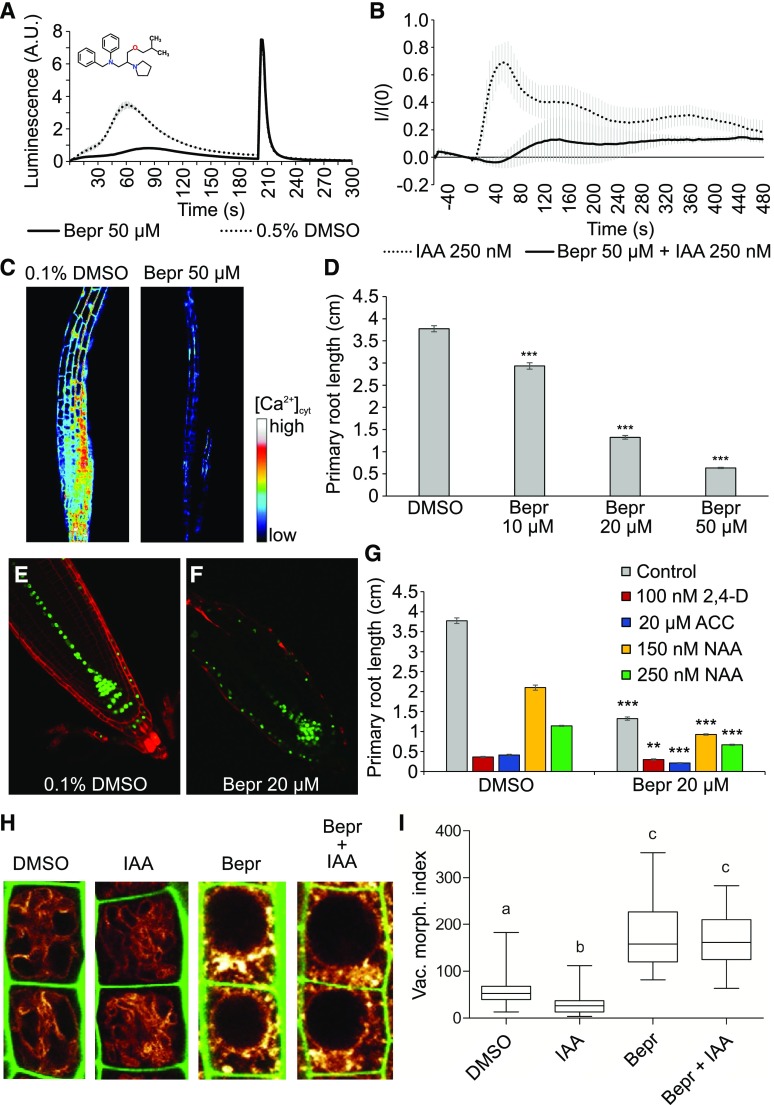
Bepridil is the only molecule inhibitor with reported Ca2+ channel inhibitory effects that we identified in our chemical screen (Yatani et al., 1986; Sarajärvi et al., 2012; Lipsanen et al., 2013). Also, in the validation, bepridil robustly interfered with the auxin-induced Ca2+ response compared with mock-treated cells, while maintaining a strong response to the discharge solution (Fig. 6A). This suggests that bepridil is a potent inhibitor of 2,4-d-induced Ca2+ signaling in BY-2 cells. Moreover, bepridil potently inhibited the rapid IAA-induced Ca2+ response in roots of Arabidopsis seedlings expressing the intensiometric Ca2+ sensor red-fluorescent GECO1 (R-GECO1; Fig. 6, B and C; Supplemental Video S1).
Figure 6.
Bepridil is a potent inhibitor of auxin-induced Ca2+ signaling. A, 2,4-d-Induced Ca2+ response of YFP-apoaequorin-expressing BY-2 cells treated with 50 µM bepridil. Discharge solution was added after 200 s. The data represent average luminescence values of 4 individual measurements (solid lines) in comparison with the average of 4 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. B, Fluorescent intensity in R-GECO1-expressing Arabidopsis seedlings after IAA treatment. Seedlings were pretreated for 30 min with 50 µM bepridil (solid lines) or 0.1% (v/v) DMSO (dotted lines). IAA (250 nM) was added at time point 0, and R-GECO1 fluorescence intensity was monitored for 8 min. The data represent average fluorescence values of 4 individual measurements in comparison with the average of 3 DMSO controls. Error bars represent ± sem. C, Snapshot of the peak Ca2+ signal elicited in R-GECO1 seedlings by 250 nm IAA after a 30-min pretreatment with 0.1% (v/v) DMSO (left) or 50 µM bepridil (right). The snapshots were taken from Supplemental Video S1 30 s after IAA addition. D, Average primary root length of wild type Col-0 seedlings grown for 7 d on 1/2 MS medium in presence of 0.1% (v/v) DMSO or 10, 20, or 50 µM bepridil. Root lengths represent the average of 54–60 roots. Error bars represent ± sem. Student’s t test p-value: ***P < 0.001. E and F, Confocal microscopy images of 5-d-old DR5rev::VENUS-N7 seedlings grown on 0.1% (v/v) DMSO (E) and 20 µM bepridil (F). Green: DR5rev::VENUS-N7 signal; red: propidium iodide staining. G, Average primary root length of wild type Arabidopsis seedlings grown for 7 d on 1/2 MS medium in presence of 20 µM bepridil or 0.1% (v/v) DMSO (gray) and supplemented with 100 nM 2,4-d (red), 20 µM ACC (blue), 150 nM NAA (yellow), or 250 nM NAA (green). Root lengths represent the average of 51–61 roots. Error bars represent ± sem. Student’s t test p-values in comparison with DMSO: **P < 0.01 and ***P < 0.001. H and I, The effect of bepridil on vacuolar morphology. pUBQ10::VAMP711-YFP seedlings (6 d old) were pretreated with 50 µM bepridil or solvent control for 5 H, followed by 3-h treatments with (DMSO), 250 nM IAA (IAA), 50 µM bepridil (Bepr), or 50 µM bepridil and 250 nM IAA (Bepr + IAA). Tonoplast-localized VAMP711-YFP (orange) as vacuolar marker and propidium iodide stain (green) for decorating the cell wall were used for confocal imaging of atrichoblast cells (H). The quantification of the vacuolar morphology index was performed with 4 vacuoles of late meristematic atrichoblasts per root, with 10–14 roots used for each treatment (I). Statistical analysis was performed using one-way ANOVA (Kruskal-Wallis test) followed by Dunn´s multiple comparison test, b: P < 0.05, c: P < 0.001. A.U., arbitrary units.
Bepridil treatment on Arabidopsis seedlings caused a dose-dependent reduction in primary root length (Fig. 6D), which was associated with altered DR5rev::VENUS-N7 expression in the lateral root cap (Fig. 6, E and F). Cotreatment of seedlings with 20 µM bepridil and either 100 nM 2,4-d, 20 µM ACC, 150 nM NAA, or 250 nM NAA did not result in noticeable growth resistance to any of the hormone treatments (Fig. 6G). Because vacuolar remodeling was proposed to be part of the mechanism by which auxin inhibits root growth (Löfke et al., 2015) and because bepridil interferes with auxin-induced Ca2+ signaling, we next investigated the effect of bepridil on vacuoles by analyzing the localization of the tonoplast marker VAMP711-YFP (Fig. 6, H and I). Bepridil dramatically altered vacuolar morphology, inducing swollen and roundish central vacuoles in Arabidopsis roots (Fig. 6H). Notably, auxin treatments induce smaller luminal vacuoles, which consequently affect cellular elongation rates (Löfke et al., 2015; Scheuring et al., 2016). However, in the presence of bepridil, the auxin effect on vacuolar morphology was abolished (Fig. 6, H and I), tentatively suggesting that auxin-regulated vacuolar remodeling could depend on a bepridil-sensitive step.
During the vacuolar morphology experiments, the tonoplast marker VAMP711-YFP seemed to display some ectopic subcellular pattern. Therefore, we also analyzed several fluorescent, late endosomal markers. Each of these markers had an aberrant localization pattern after bepridil treatment (Supplemental Fig. S5). These observations suggest that bepridil has pleiotropic effects on late endosomal trafficking, which could complicate the interpretation of the auxin resistance of vacuolar remodeling after bepridil treatment. When lowering the bepridil concentration, we found that vacuolar morphology was still aberrant at 10 µM, but was no longer obviously impaired at 5 µM (Supplemental Fig. S6).
Suc-Induced Ca2+ Signals Are Highly Sensitive to Fenamates and Sterol Biosynthesis Inhibitors, But Not to Bepridil
Next, we sought to evaluate the specificities of the identified inhibitors. The auxin response requires CNGC14 for eliciting Ca2+ (Shih et al., 2015; Dindas et al., 2018), whereas hyperosmotic stress is predicted to activate OSCA/TMEM63-type mechanosensitive Ca2+-permeable channels (Yuan et al., 2014; Murthy et al., 2018; Zhang et al., 2018). Thus, auxin and hyperosmotic stress activate two distinct Ca2+ entry mechanisms.
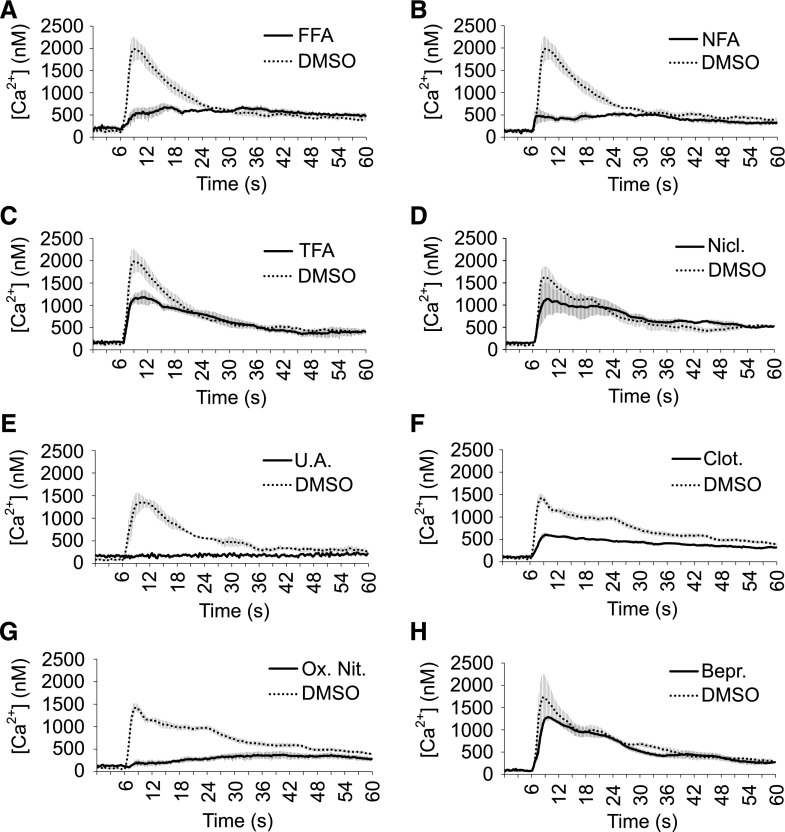
We used 0.5 M Suc as a hyperosmotic stimulus. When eliciting YFP-apoaequorin-expressing Arabidopsis seedlings, we observed a very fast and transient rise in [Ca2+]cyt (Fig. 7; Supplemental Fig. S7), as was previously described for such hyperosmotic treatments (Furuichi et al., 2001; Stephan et al., 2016). The Suc-induced Ca2+ signal was characterized by an initial large peak in free [Ca2+]cyt that reached a maximum value (1.5–2 µM range) within seconds. After reaching the maximum peak value, the Ca2+ signal quickly decreased until it reached an elevated steady-state concentration 30–40 s after elicitor addition (Fig. 7; Supplemental Fig. S7).
Figure 7.
Suc-induced Ca2+ signals are highly sensitive to fenamates, protonophores, and imidazoles. A-H, Suc-induced Ca2+ responses of YFP-apoaequorin-expressing Arabidopsis seedlings treated with fenamates (A–C), protonophores (D and E), imidazoles (F and G), or bepridil (H). The data represent average luminescence values of 4 individual measurements (solid lines) in comparison with the average of 4 DMSO controls (dotted lines) in the same multiwell plate. Error bars represent ± sem. FFA, flufenamic acid; NFA, niflumic acid; TFA, tolfenamic acid; Nicl., niclosamide; U.A., (+)-usnic acid; Clot., clotrimazole; Ox. Nit., oxiconazole nitrate; Bepr., bepridil.
We evaluated the fenamates (FFA, NFA, and TFA), the protonophores [niclosamide, (+)-usnic acid, ES9, and CCCP], and bepridil for their ability to inhibit hyperosmotic-stress–induced Ca2+. Additionally, we also tested two imidazole-type fungicides (clotrimazole and oxiconazole nitrate), which were initially identified as inhibitors of 2,4-d-induced Ca2+ signals but were not further pursued in the context of auxin responses due to a poor reproducibility in follow-up experiments (Supplemental Fig. S8). All tested fenamates, three protonophores [(+)-usnic acid, ES9, and CCCP] and, surprisingly, both imidazoles potently interfered with Suc-induced Ca2+ (Fig. 7; Supplemental Fig. S7). This suggests that hyperosmotic stress Ca2+ entry is much more sensitive to inhibition of sterol biosynthesis than 2,4-d-induced Ca2+ responses. On the other hand, although bepridil and niclosamide were potent inhibitors of auxin-induced Ca2+, they did not inhibit the Suc-induced Ca2+ response (Fig. 7, D and H). Niclosamide was the only one of the tested protonophores that could not inhibit Suc-induced Ca2+, suggesting that its inhibitory effect on auxin-induced Ca2+ may not be related solely to its protonophore activity. Together, these data illustrate that auxin and hyperosmotic stress Ca2+ responses show different pharmacological sensitivities.
DISCUSSION
The molecular mechanism by which auxin regulates transcriptional changes is largely captured by the very well-characterized TIR1/AFB-based degradation of Aux/IAA transcriptional corepressors. This pathway accounts for much of the auxin-regulated transcriptional changes and, thus, the cellular response. Recently, TIR1/AFB-based auxin perception was also found to be required for nontranscriptional cellular responses, such as rapid and reversible inhibition of root growth (Fendrych et al., 2018). The inhibitory effect of auxin root elongation is correlated with alkalinization of the apoplast (Barbez et al., 2017), activation of Ca2+ signals through CNCG14 (Shih et al., 2015), and remodeling of the vacuole (Löfke et al., 2015). Alkalinization of the apoplast is possibly the result of the coordinated inhibition of plasma membrane H+-ATPase activity and AUX1-mediated H+/IAA− uptake from the apoplast (Dindas et al., 2018). Moreover, auxin-induced alkalinization of the apoplast is coupled to CNGC14-dependent auxin-induced Ca2+ signaling in the epidermis (Shih et al., 2015). Importantly, root elongation of cngc14 mutants is mildly insensitive to inhibitory auxin levels (Shih et al., 2015), suggesting that auxin-induced cytosolic Ca2+ increase is part of the root growth inhibitory auxin signaling pathway. We identified bepridil as a potent inhibitor of auxin-induced cytosolic Ca2+ increase and found that bepridil interferes strongly with auxin-induced vacuolar remodeling. This makes it tempting to speculate that auxin-induced cytosolic Ca2+ increase controls vacuolar morphology. However, the strong pleiotropic effects on late endosomal compartments preclude drawing such strong conclusions. Therefore, it will be interesting to identify the molecular targets of bepridil and characterize its function in auxin-regulated vacuolar remodeling.
The identification of bepridil as a potent inhibitor of auxin-induced Ca2+ is consistent with its reported Ca2+ channel blocker function in animals (Yatani et al., 1986; Sarajärvi et al., 2012; Lipsanen et al., 2013) and suggests, in line with cngc14’s defects in auxin-induced cytosolic Ca2+ increase (Shih et al., 2015), that CNGC14 might be a bepridil target. However, electrophysiological experiments demonstrated that bepridil inhibits outward rectified K+ currents in plant protoplasts (Thomine et al., 1994). This hints at a functional coupling of auxin-induced Ca2+ entry with outward rectified K+ currents as described for Ca2+ spiking during nodulation (Ané et al., 2004; Charpentier et al., 2008, 2016). However, auxin activates inward rectified K+ currents in Arabidopsis and maize (Thiel and Weise, 1999; Philippar et al., 2004), and we found that the K+ selective ionophore valinomycin had no effect on auxin-induced Ca2+ signals. Instead, it seems more likely that bepridil targets CNGC14 and/or other CNGCs via structural features that they share with outward rectifying K+ channels.
Recently, the protonophore carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone was shown to completely inhibit the ATP-induced Ca2+ response, whereas pretreatment with the ionophore nigericin had only marginal effects on the Ca2+ and pH transients (Behera et al., 2018). These observations are perfectly in line with our findings showing that protonophores are potent inhibitors of auxin- and osmotic-stress–induced Ca2+ signaling and strengthen previous notions that Ca2+ and pH are functionally coupled in several cellular processes in plants, including root hair and pollen tube growth (Herrmann and Felle, 1995; Monshausen et al., 2008; Michard et al., 2017), cold stress response (Gao et al., 2004), and touch response (Monshausen et al., 2009). Also, disruption of the transmembrane pH gradient via alkalinization of the apoplast interfered with auxin-induced Ca2+ entry in root hairs (Dindas et al., 2018). This was explained by a need for a proton gradient to drive AUX1-mediated H+/IAA− symport into the cell. In this model, the inability of IAA to enter the cell in aux1 prevents TIR1-mediated CNGC14 activation (Dindas et al., 2018). In turn, cngc14 has a defect in AUX1 activity, providing a mechanistic model for the coupling of Ca2+ and H+ dynamics during auxin response. However, NAA-induced cytosolic Ca2+ increase was recently also coupled to a cytoplasmic acidification (Behera et al., 2018), which cannot be explained by AUX1-mediated H+ uptake, because NAA is not a good substrate for AUX1 (Yang et al., 2006).
In summary, by exploring a small subset of hits, we readily identified several new inhibitors of auxin-induced Ca2+. Further exploration of these inhibitors will thus lead to novel insights in the mechanism (e.g. protonophores) and the physiological relevance of auxin-induced Ca2+ (e.g. vacuolar remodeling). Importantly, the use of a library of annotated molecules has the added advantage that many of the compounds are commercially available and thus are easily accessible to researchers for further characterization and analysis of structural derivatives with a higher inhibitory potency and specificity. Identification of the molecular targets will be key for further refining the inhibitors in terms of specificity and affinity.
MATERIALS AND METHODS
BY-2 Cell Lines and Arabidopsis (Arabidopsis thaliana) Plant Lines
We stably transformed wild-type tobacco BY-2 (Nicotiana tabacum cv Bright Yellow 2) cell suspensions with a kanamycin-resistant construct for constitutive expression of YFP-fused apoaequorin (AEQ) driven by an ubiquitin (UBQ10) promoter as previously described (Mehlmer et al., 2012). The transgenic BY-2 cell lines were maintained by weekly dilution (1:40) in modified Linsmaier and Skoog medium. The cell cultures were agitated on a rotary shaker at 130 rpm at 25°C in the dark and used in experiments 5 d after subculture.
Transgenic Col-0 Arabidopsis seedlings that carry a proUBQ10::YFP-apoAEQ cassette were used in our experiments (Mehlmer et al., 2012). These lines were generated using Agrobacterium-mediated transformation via the floral-dip method (Clough and Bent, 1998). Transformants were selected based on phosphinothricin resistance and YFP expression levels and were made homozygous in subsequent generations.
The other used plant lines expressing R-GECO1 (Keinath et al., 2015), DR5rev::VENUS-N7 (Heisler et al., 2005), Ara7-mRFP (Jia et al., 2013), VAMP727-YFP (Ebine et al., 2008), 2xFYVE-YFP (Vermeer et al., 2006), VAMP711-YFP (Geldner et al., 2009), and VAMP711-mCherry (Geldner et al., 2009) have been described previously.
Compounds
The compounds used for the primary screen and confirmation screen belong to the Spectrum compound library (MicroSource Discovery Systems) and were dissolved in DMSO. The compounds used in follow-up experiments [FFA, NFA, TFA, diphenylamine, diphenylmethane, α-phenyl-o-toluic acid, aniline, anthranilic acid, NPPB, 9-ACA, NPA, BUM, clotrimazole, oxiconazole nitrate, artemether, niclosamide, (+)-usnic acid, cloxyquin, bepridil, TyrA23, CCCP, and valinomycin] were obtained from Sigma-Aldrich and dissolved in DMSO. Coelenterazine-h was obtained from Promega and dissolved in methanol.
Primary Screen Setup
The Spectrum library of 2320 compounds with a wide range of reported biological activities and structural diversity (MicroSource Discovery Systems) was screened for inhibitors of 2,4-d-induced Ca2+ signaling. The individual compounds and controls were added to 100 µL YFP-apoaequorin-expressing BY-2 cells in white 96-well plates, with a final concentration of 50 µM per well. Negative (0.5% [v/v] DMSO) and positive (10 mM GdCl3) controls were added to both outer columns of each multiwell plate. After a 30-min incubation period in the dark to reduce background signals, 100 µL 2,4-d (final concentration 500 µM) was added to each well with a liquid handling robot (Tecan Freedom EVO200 with 96-channel head) and immediately transferred to the luminescence imaging system (NightSHADE LB 985 in vivo Plant Imaging System, Berthold Technologies) to ensure capturing the peak signal in each well. The induced luminescent signal was measured for 30 cycles with a 10-s exposure time per cycle. The maximum signal in each well was calculated and normalized to the average of the negative control of the corresponding multiwell plate.
Luminescence Measurements of Ca2+ Responses in BY-2 Cells during Confirmation Screen and Follow-Up Experiments
After subculturing 5 d, YFP-apoAEQ-expressing BY-2 cells were collected by centrifugation, washed, and resuspended in fresh BY-2 medium. Aequorin was reconstituted in the BY-2 cells by adding 2.5 μm of coelenterazine-h (Promega) for 3 h under agitation in the dark. Afterward, 100 µL of reconstituted BY-2 cells was added to each well of a white 96-well plate (PerkinElmer), and the cells were preincubated with the hits identified in the primary screen for 30 min to 1 h in the dark. For measuring elicitor-induced Ca2+ responses per well, 100 µL elicitor solution was added to the cells, after which the aequorin-induced light emission was measured every 1.5 s for 200 s (every 2 s for 240 s during the confirmation screen) with a luminescence plate reader (all measurements were done in a LUMIstar Galaxy, BMG LABTECH, unless mentioned otherwise). Immediately after this measurement, the remaining reconstituted aequorin was discharged by the addition of 50 µL of discharge solution (0.1 M CaCl2 and 20% [v/v] ethanol), and luminescence was measured every 1.5 s for an additional 100 s (every 2 s for 160 s during the confirmation screen). The bioluminescence signal of our transgenic BY-2 cell lines could not be converted to absolute [Ca2+]cyt values because the total luminescent signal after aequorin discharge could not be completely detected in situ due to saturation of the plate reader detector. Therefore, all treatments were always evaluated relative to controls within each corresponding multiwell plate. This normalization also accounted for day-to-day variation in amplitude and shape of the Ca2+ signals.
Luminescence Measurements of Ca2+ Responses in Arabidopsis Seedlings
Gas-sterilized seeds were grown on plates containing half-strength Murashige and Skoog (MS) medium for 3 d until germination. Freshly germinated seedlings were transferred individually to wells of sterile, white, 96-well microplates (PerkinElmer) containing 130 µL of medium composed of 1/2 MS salts (2.2 g/L), 0.5 g/L MES, 1% (w/v) Suc, and 0.08% (w/v) phyto agar. This low phyto agar concentration provided modest support for the growing seedlings while still allowing rapid mixing of injected pretreatment compounds and elicitor solutions. The transparent lids of the 96-well plates were sealed with Parafilm to prevent medium evaporation. The seedlings were grown in the plates for 2 to 3 d in a growth chamber under continuous light conditions at 21°C. The evening before measurement, 130 µL of water containing 2.5 µM coelenterazine-h (Ctz-h; Promega) was added to each well to allow overnight reconstitution of apoaequorin into functional aequorin. During this incubation, the plate was covered in aluminum foil to prevent light-induced degradation of Ctz-h. The following day, 130 µL of the medium was removed and replaced with 130 µL of a solution containing 2× the final concentrations of the compounds in water. After a 1-h preincubation period, 130 µL medium was removed just before the luminescence measurements, to allow the addition of elicitor solutions.
The seedling-containing 96-well plates were analyzed in a luminescence plate reader (LUMIstar Galaxy, BMG LABTECH). Luminescence was first measured every 0.3 s for 6 s to establish a baseline reading, after which 100 µL of a 2× elicitor solution was automatically added and luminescence was further measured every 0.3 s for 54 s. Subsequently, the plate was removed from the plate reader and 100 µL of solution was removed from each well. The plate was then placed into the plate reader again and luminescence was measured every 0.3 s for 6 s to establish a baseline signal. Subsequently, 100 µL of discharge solution (2 M CaCl2 and 50% [v/v] ethanol) was automatically added and luminescence was further measured every 0.3 s for 54 s to determine the remaining aequorin in the seedlings.
Quantification of Ca2+ Response Data in Transgenic YFP-Apoaequorin-Expressing Arabidopsis Seedlings
Raw light data measured by the LUMIstar Galaxy plate reader were converted to calcium concentration by applying the empirically determined formula pCa = 0.332588(-logk) + 5.5593 (Knight et al., 1996). The rate constant k in this formula equals the elicitor-induced luminescence counts per second divided by the total remaining counts. The total remaining counts were determined after adding discharge solution (2 M CaCl2 + 50% [v/v] ethanol).
Phenotyping
Wild-type gas-sterilized Arabidopsis seeds were plated on 1/2 MS medium supplemented with the appropriate compounds and/or hormones (3 rows/plate, 0.5 cm between seeds). For the primary root length experiments, the plated seeds were first stratified for 3 d in the dark at 4°C and subsequently transferred to a growth chamber under continuous light conditions at 21°C. After 7 d of growth, the plates were scanned and the primary root lengths of the seedlings were measured with ImageJ. For each treatment 10–61 individual roots were counted.
Late Endosomal Marker Localization
Seedlings (3 to 4 d old) from the endomembrane marker lines (2xFYVE-YFP, Ara7-mRFP, and VAMP727-YFP) were pretreated with control medium or 50 µM bepridil for 5 h. The control medium (CaPLUS) consists of the following components dissolved in MilliQ (for 0.5 L): 25 mL MS basal salt micronutrient solution, 5 g Suc, 0.05 g myoinositol, 0.25 g MES, 0.413 g NH4NO3, 0.045 g MgSO4, 0.475 g KNO3, 0.043 g H2KO4P-, and 0.083 g CaCl2, with pH set to 5.7.
For imaging of the endomembrane markers, the confocal laser scanning microscopes Leica SP2 (Leica Microsystems) and Zeiss 710 (Zeiss) were used. Fluorescence emission of mRFP (excitation [ex] 561 nm/emission [em] 570-630 nm) and YFP (ex 514 nm/em 520-565 nm) was detected using a 63× water objective (NA 1.2, digital zoom 1,2×). Images were analyzed using Fiji (Schindelin et al., 2012).
R-GECO1 Visualization
Seedlings from the R-GECO1 line were pretreated for 30 min with 50 µM bepridil or 0.1% DMSO and mounted in a specialized imaging chamber as previously described (Himschoot et al., 2018). The imaging chamber was mounted on the stage of an Ultra View Vox spinning disc microscope (PerkinElmer), and 250 nm IAA was added to elicit a Ca2+ response in the samples. R-GECO1 fluorescence intensity was monitored for 8 min after elicitor addition and processed as previously described (Himschoot et al., 2018). Per treatment, 3 to 4 individual measurements were performed.
DR5rev::VENUS-N7 Visualization
DR5rev::VENUS-N7 seedlings were grown for 5 d on 1/2 MS plates containing the appropriate compounds or 0.1% (v/v) DMSO. For each treatment, 5 seedlings were stained with freshly prepared PI solution (15 μM in distilled water) for 2 min, rinsed twice in water, and subsequently spread on a glass microscope slide. Fluorescence emission of DR5rev::VENUS-N7 (ex 514 nm/em 535-590 nm) and PI (ex 514 nm/em 570-630 nm) was visualized and imaged using a Leica SP2 confocal microscope (Leica Microsystems).
Analysis of Vacuolar Morphology
Analysis of vacuolar morphology was carried out on 6-d-old seedlings of a tonoplast marker line (pUBQ10::VAMP711-YFP) that were grown on solid 1/2 MS medium. The samples were pretreated for 5 h with 50 µM bepridil or solvent control (DMSO). Subsequently, the seedlings were transferred to plates containing either DMSO, 50 µM bepridil, 250 nM IAA, or bepridil and IAA (50 µM and 250 nM, respectively). Afterward, seedlings were grown for another 3 h before image acquisition. Roots were mounted in PI solution (0.02 mg/mL) to counterstain cell walls and report viability. YFP was excited at 514 nm (fluorescence emission: 525–555 nm) and PI at 561 nm (fluorescence emission: 644–753 nm) using a Leica TCS SP5 confocal laser scanning microscope equipped with a Leica HCX PL APO CS 63 × 1.20 water-immersion objective. Confocal images were analyzed using ImageJ. To calculate the vacuolar morphology index, the longest and widest distance of the biggest luminal structure was measured and multiplied (Löfke et al., 2015). The atrichoblast cells were quantified before the onset of elongation (late meristematic). To depict this region, the first cell being twice as long as wide was considered as the onset of elongation. Starting from this cell, the next cell toward the meristem was excluded (as it usually shows either partial elongation and/or already substantial vacuolar expansion), and vacuoles of the subsequent 4 cells were quantified as described previously (Dünser et al., 2017).
BY-2 Viability Assay
BY-2 cells (5 d old) were treated with H2O, 1% (v/v) DMSO, 500 µM 2,4-d, or a combination of 1% (v/v) DMSO and 500 µM 2,4-d. After 1 h of treatment, the cells were stained with freshly prepared FDA-PI staining solution (15 µM FDA and 15 µM PI in BY-2 medium) for 5 min in the dark. Afterward, the cells were washed with fresh medium and 100 µL of cells was spread on a glass microscope slide per treatment. For each treatment, ∼100 cells were counted using an Ultra View Vox spinning disc microscope (PerkinElmer) and classified as alive or dead based on their individual uptake of FDA and PI. For each treatment, 2–4 such individual measurements were performed.
ATP Content Determination
Wild-type tobacco BY-2 cell cultures were used and maintained as described earlier. After subculturing 5 d, the BY-2 cell suspension was diluted 5 times in modified Linsmaier and Skoog medium and preconditioned in the dark for 1 h on a shaker before use. The cells were subsequently distributed in white 96-well plates (95 µL/well) and 5 µL of the appropriate compounds [DMSO, cloxyquin (50 and 20 µM final concentration), niclosamide (50 and 20 µM final concentration), (+)-usnic acid (50 and 20 µM final concentration), and CCCP (20 µM final concentration)] was added with a Freedom EVO robot (Tecan). All compounds were dissolved in DMSO with the final DMSO concentration for each treatment being 0.66% (v/v). There were 8 repeats per treatment. The ATP levels were detected by adding 80 µL of the ATPlite 1step Luminescence Assay System (PerkinElmer) after incubation of the cells in the presence of the compounds for the indicated time. Luminescence was measured with an EnVision 2104 Multilabel Reader (PerkinElmer).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers E14214.1 (apoaequorin), JN258411.1 (R-GECO1), NM_115290.5 (VAMP727-At3g54300), and NM_119367.3 (VAMP711- AT4G32150).
Supplemental Data
The following supplemental materials are available:
Supplemental Figure S1. DMSO and 2,4-d have no significant impact on BY-2 cell viability.
Supplemental Figure S2. Overview of chemical structures.
Supplemental Figure S3. Higher magnification of root phenotypes of fenamate-treated plants.
Supplemental Figure S4. Diphenylamine, aniline, and anthranilic acid make roots resistant to 2,4-d.
Supplemental Figure S5. Bepridil has a profound impact on endomembrane trafficking.
Supplemental Figure S6. Bepridil has a profound impact on vacuolar morphology.
Supplemental Figure S7. The protonophores ES9 and CCCP alter Suc-induced Ca2+ signals.
Supplemental Figure S8. Imidazoles are not robust inhibitors of 2,4-d induced Ca2+.
Supplemental References. References for Supplemental Table S1.
Supplemental Table S1. List of confirmed hit compounds (excel file).
Supplemental Video S1. Bepridil inhibits the IAA-induced Ca2+ response in Arabidopsis roots expressing R-GECO1 (.avi file).
Acknowledgments
We thank Dr. Simon Stael for critical reading of the manuscript.
Footnotes
This work was supported by the Special Research Fund of Universiteit Gent (Ghent University) (to K.D.V. and E.H.); Ministero dell’Istruzione, dell’Università e della Ricerca (Ministry of Education, Research and Universities), Fondo per gli Investimenti della Ricerca di Base (FIRB) 2010 RBFR10S1LJ_001; University of Milan Transition Grant - Horizon 2020, Fondo di ricerca Linea 1A Progetto “Unimi Partenariati H2020” and Piano di Sviluppo di Ateneo 2016, 2017 (to A.C.); Austrian Academy of Sciences (Österreichischen Akademie der Wissenschaften) (DOC fellowship to K.D.V.); Vienna Research Group (VRG) program of the Vienna Science and Technology Fund (Wiener Wissenschafts-, Forschungs- und Technologiefonds); Austrian Science Fund (FWF) (Projects P26568-B16 and P26591-B16); and EC | European Research Council (ERC) (starting grant 639478-AuxinER to J.K.-V.) and the ERC StG PROCELLDEATH (project no. 639234 to M.K.N.).
Articles can be viewed without a subscription.
References
- Ali A, Jadhav A, Jangid P, Patil R, Shelar A, Karuppayil SM (2018) The human muscarinic acetylcholine receptor antagonist, Dicyclomine targets signal transduction genes and inhibits the virulence factors in the human pathogen, Candida albicans. J Antibiot (Tokyo) 71: 456–466 [DOI] [PubMed] [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Barbez E, Dünser K, Gaidora A, Lendl T, Busch W (2017) Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: E4884–E4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Xu Z, Luoni L, Bonza MC, Doccula FG, De Michelis MI, Morris RJ, Schwarzländer M, Costa A (2018) Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Ampudia CSG, Hooykaas PJJ, Offringa R (2003) PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol 132: 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang W, Zhu JY, Oh E, Wang ZY (2016) Information integration and communication in plant growth regulation. Cell 164: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, et al. (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dejonghe W, Kuenen S, Mylle E, Vasileva M, Keech O, Viotti C, Swerts J, Fendrych M, Ortiz-Morea FA, Mishev K, et al. (2016) Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat Commun 7: 11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Fuente RK, Leopold AC (1973) A role for calcium in auxin transport. Plant Physiol 51: 845–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vriese K, Costa A, Beeckman T, Vanneste S (2018) Pharmacological strategies for manipulating plant Ca2+ signalling. Int J Mol Sci 19: 1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatloff E, Roberts M, Sanders D, Roberts SK (2004) Characterization of anion channels in the plasma membrane of Arabidopsis epidermal root cells and the identification of a citrate-permeable channel induced by phosphate starvation. Plant Physiol 136: 4136–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Müller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. (2018) AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünser K, Gupta S, Ringli C, Kleine-Vehn J (2017) LRX- and FER-dependent extracellular sensing coordinates vacuolar size for cytosol homeostasis. bioRxiv, doi: 10.1101/231043 [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel KH, Marchadier E, Brownlee C, Kudla J, Hetherington AM (2017) The evolution of calcium-based signalling in plants. Curr Biol 27: R667–R679 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J (2018) Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Roberts MR (2014) Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T, Mori IC, Takahashi K, Muto S (2001) Sugar-induced increase in cytosolic Ca2+ in Arabidopsis thaliana whole plants. Plant Cell Physiol 42: 1149–1155 [DOI] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliham M, Tester M (2005) The regulation of anion loading to the maize root xylem. Plant Physiol 137: 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GG. (2016) Fenamates. In Parnham MJ, ed, Compendium of Inflammatory Diseases. Springer International Publishing AG, Birkhauser, Basel [Google Scholar]
- Habjan S, Vandenberg RJ (2009) Modulation of glutamate and glycine transporters by niflumic, flufenamic and mefenamic acids. Neurochem Res 34: 1738–1747 [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO (1999) Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274: 11110–11114 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Herrmann A, Felle HH (1995) Tip growth in root hair-cells of Sinapis-Alba L - significance of internal and external Ca2+ and pH. New Phytol 129: 523–533 [Google Scholar]
- Himschoot E, Krebs M, Costa A, Beeckman T, Vanneste S (2018) Calcium ion dynamics in roots: Imaging and analysis. Methods Mol Biol 1761: 115–130 [DOI] [PubMed] [Google Scholar]
- Hogg RC, Wang Q, Large WA (1994) Action of niflumic acid on evoked and spontaneous calcium-activated chloride and potassium currents in smooth muscle cells from rabbit portal vein. Br J Pharmacol 112: 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongmanee P, Rukseree K, Buabut B, Somsri B, Palittapongarnpim P (2007) In vitro activities of cloxyquin (5-chloroquinolin-8-ol) against Mycobacterium tuberculosis. Antimicrob Agents Chemother 51: 1105–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenburg JC, Simionescu DT, Vyavahare NR (2005) Tannic acid treatment enhances biostability and reduces calcification of glutaraldehyde fixed aortic wall. Biomaterials 26: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Jegasothy BV, Pakes GE (1991) Oxiconazole nitrate: Pharmacology, efficacy, and safety of a new imidazole antifungal agent. Clin Ther 13: 126–141 [PubMed] [Google Scholar]
- Jia T, Gao C, Cui Y, Wang J, Ding Y, Cai Y, Ueda T, Nakano A, Jiang L (2013) ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J Exp Bot 64: 2817–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Waadt R, Brugman R, Schroeder JI, Grossmann G, Schumacher K, Krebs M (2015) Live cell imaging with R-GECO1 sheds light on flg22- and chitin-induced transient [Ca(2+)]cyt patterns in Arabidopsis. Mol Plant 8: 1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Korade Z, Kim HY, Tallman KA, Liu W, Koczok K, Balogh I, Xu L, Mirnics K, Porter NA (2016) The effect of small molecules on sterol homeostasis: Measuring 7-dehydrocholesterol in Dhcr7-deficient Neuro2a cells and human fibroblasts. J Med Chem 59: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218: 414–431 [DOI] [PubMed] [Google Scholar]
- Lanner JT, Georgiou DK, Joshi AD, Hamilton SL (2010) Ryanodine receptors: Structure, expression, molecular details, and function in calcium release. Cold Spring Harb Perspect Biol 2: a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latkowska E, Lechowski Z, Bialczyk J, Pilarski J (2006) Photosynthesis and water relations in tomato plants cultivated long-term in media containing (+)-usnic acid. J Chem Ecol 32: 2053–2066 [DOI] [PubMed] [Google Scholar]
- Lavy M, Estelle M (2016) Mechanisms of auxin signaling. Development 143: 3226–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsanen A, Flunkert S, Kuptsova K, Hiltunen M, Windisch M, Hutter-Paier B, Jolkkonen J (2013) Non-selective calcium channel blocker bepridil decreases secondary pathology in mice after photothrombotic cortical lesion. PLoS One 8: e60235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K (2017) Crosstalk complexities between auxin, cytokinin, and ethylene in arabidopsis root development: From experiments to systems modeling, and back again. Mol Plant 10: 1480–1496 [DOI] [PubMed] [Google Scholar]
- Löfke C, Dünser K, Scheuring D, Kleine-Vehn J (2015) Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4: e05868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA (2011) Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol 190: 566–572 [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Parvin N, Hurst CH, Knight MR, Teige M, Vothknecht UC (2012) A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J Exp Bot 63: 1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Simon AA, Tavares B, Wudick MM, Feijó JA (2017) Signaling with ions: The keystone for apical cell growth and morphogenesis in pollen tubes. Plant Physiol 173: 91–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monin MB, Krause P, Stelling R, Bocuk D, Niebert S, Klemm F, Pukrop T, Koenig S (2016) The anthelmintic niclosamide inhibits colorectal cancer cell lines via modulation of the canonical and noncanonical Wnt signaling pathway. J Surg Res 203: 193–205 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Messerli MA, Gilroy S (2008) Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol 147: 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S (2009) Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SE, Dubin AE, Whitwam T, Jojoa-Cruz S, Cahalan SM, Mousavi SAR, Ward AB, Patapoutian A (2018) OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7: e41844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Iizumi S, Satoh K, Ooka H, Kawai J, Carninci P, Hayashizaki Y, Otomo Y, Murakami K, Matsubara K, et al. (2004) Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol Biol Evol 21: 1855–1870 [DOI] [PubMed] [Google Scholar]
- Narahara KA, Singh BN, Karliner JS, Corday SR, Hossack KF (1992) Bepridil hydrochloride compared with placebo in patients with stable angina pectoris. Am J Cardiol 69: 37D–42D [DOI] [PubMed] [Google Scholar]
- Nixon GF, Mignery GA, Somlyo AV (1994) Immunogold localization of inositol 1,4,5-trisphosphate receptors and characterization of ultrastructural features of the sarcoplasmic reticulum in phasic and tonic smooth muscle. J Muscle Res Cell Motil 15: 682–700 [DOI] [PubMed] [Google Scholar]
- Pentikäinen PJ, Neuvonen PJ, Backman C (1981) Human pharmacokinetics of tolfenamic acid, a new anti-inflammatory agent. Eur J Clin Pharmacol 19: 359–365 [DOI] [PubMed] [Google Scholar]
- Philippar K, Ivashikina N, Ache P, Christian M, Lüthen H, Palme K, Hedrich R (2004) Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J 37: 815–827 [DOI] [PubMed] [Google Scholar]
- Qiu W, Ren B, Dai H, Zhang L, Zhang Q, Zhou X, Li Y (2017) Clotrimazole and econazole inhibit Streptococcus mutans biofilm and virulence in vitro. Arch Oral Biol 73: 113–120 [DOI] [PubMed] [Google Scholar]
- Rigó G, Ayaydin F, Tietz O, Zsigmond L, Kovács H, Páy A, Salchert K, Darula Z, Medzihradszky KF, Szabados L, et al. (2013) Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 25: 1592–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarajärvi T, Lipsanen A, Mäkinen P, Peräniemi S, Soininen H, Haapasalo A, Jolkkonen J, Hiltunen M (2012) Bepridil decreases Aβ and calcium levels in the thalamus after middle cerebral artery occlusion in rats. J Cell Mol Med 16: 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Löfke C, Krüger F, Kittelmann M, Eisa A, Hughes L, Smith RS, Hawes C, Schumacher K, Kleine-Vehn J (2016) Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc Natl Acad Sci USA 113: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HW, DePew CL, Miller ND, Monshausen GB (2015) The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr Biol 25: 3119–3125 [DOI] [PubMed] [Google Scholar]
- Shimomura O. (2005) The discovery of aequorin and green fluorescent protein. J Microsc 217: 1–15 [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59: 223–239 [DOI] [PubMed] [Google Scholar]
- Stephan AB, Kunz HH, Yang E, Schroeder JI (2016) Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc Natl Acad Sci USA 113: E5242–E5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Weise R (1999) Auxin augments conductance of K+ inward rectifier in maize coleoptile protoplasts. Planta 208: 38–45 [Google Scholar]
- Thomine S, Zimmerman S, Van Duijn B, Barbier-Brygoo H, Guern J (1994) Calcium channel antagonists induce direct inhibition of the outward rectifying potassium channel in tobacco protoplasts. FEBS Lett 340: 45–50 [DOI] [PubMed] [Google Scholar]
- Tuteja N, Mahajan S (2007) Calcium signaling network in plants: An overview. Plant Signal Behav 2: 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobena-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr., Munnik T (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67: 539–574 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Yatani A, Brown AM, Schwartz A (1986) Bepridil block of cardiac calcium and sodium channels. J Pharmacol Exp Ther 237: 9–17 [PubMed] [Google Scholar]
- Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B, et al. (2014) OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514: 367–371 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73 [DOI] [PubMed] [Google Scholar]
- Zhang J, Vanneste S, Brewer PB, Michniewicz M, Grones P, Kleine-Vehn J, Löfke C, Teichmann T, Bielach A, Cannoot B, et al. (2011) Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev Cell 20: 855–866 [DOI] [PubMed] [Google Scholar]
- Zhang M, Wang D, Kang Y, Wu JX, Yao F, Pan C, Yan Z, Song C, Chen L (2018) Structure of the mechanosensitive OSCA channels. Nat Struct Mol Biol 25: 850–858 [DOI] [PubMed] [Google Scholar]
- Zuccotti A, Clementi S, Reinbothe T, Torrente A, Vandael DH, Pirone A (2011) Structural and functional differences between L-type calcium channels: Crucial issues for future selective targeting. Trends Pharmacol Sci 32: 366–375 [DOI] [PubMed] [Google Scholar]