AtMDN1 plays roles in embryogenesis and root meristem maintenance, and its mutation leads to defects in nuclear export of the pre-60S ribosome and in pre-rRNA processing.
Abstract
Ribosome biogenesis is an orchestrated process that relies on many assembly factors. The AAA-ATPase Midasin 1 (Mdn1) functions as a ribosome assembly factor in yeast (Saccharomyces cerevisiae), but the roles of MDN1 in Arabidopsis (Arabidopsis thaliana) are poorly understood. Here, we showed that the Arabidopsis null mutant of MDN1 is embryo-lethal. Using the weak mutant mdn1-1, which maintains viability, we found that MDN1 is critical for the regular pattern of auxin maxima in the globular embryo and functions in root meristem maintenance. By detecting the subcellular distribution of ribosome proteins, we noted that mdn1-1 impairs nuclear export of the pre-60S ribosomal particle. The processing of ribosomal precusor RNAs, including 35S, 27SB, and 20S, is also affected in this mutant. MDN1 physically interacts with PESCADILLO2 (PES2), an essential assembly factor of the 60S ribosome, and the observed mislocalization of PES2 in mdn1-1 further implied that MDN1 plays an indispensable role in 60S ribosome biogenesis. Therefore, the observed hypersensitivity of mdn1-1 to a eukaryotic translation inhibitor and high-sugar conditions might be associated with the defect in ribosome biogenesis. Overall, this work establishes a role of Arabidopsis MDN1 in ribosome biogenesis, which agrees with its roles in embryogenesis and root development.
The ribosome is a ribonucleoprotein complex that is responsible for protein synthesis and therefore plays a major role in cell division, growth, and development. The eukaryotic 80S ribosome consists of the 60S large subunit and 40S small subunit. The 60S subunit contains three distinct ribosomal RNAs (rRNAs, 25S/28S, 5.8S, and 5S) and more than 40 ribosomal proteins (RPs), and is responsible for peptidyl transfer and peptide synthesis (Cech, 2000; Yusupova and Yusupov, 2014). The 40S subunit comprises the 18S rRNA and more than 30 RPs, and plays a role in mRNA decoding (Yusupova and Yusupov, 2014; Weis et al., 2015a). Distinctively, plants such as Arabidopsis (Arabidopsis thaliana) possess up to 81 different types of RPs, which each have two to seven members (Carroll et al., 2008).
Ribosome biogenesis is a highly orchestrated process that starts in the cell nucleolus with transcription of a large precursor rRNA (prerRNA) by RNA polymerase I (Woolford and Baserga, 2013). This precursor contains the mature 18S, 5.8S, and 25S/28S rRNA, 5′ and 3′ external transcribed spacers, and internal transcribed spacers 1 and 2 (Lafontaine, 2015). Each transcribed spacer has two to five endonucleolytic sites (Henras et al., 2015; Tomecki et al., 2017). At the beginning of ribosome biogenesis, a large 90S preribosomal particle is formed with the earliest 35S (in yeast), 45S (in plants), or 47S (in humans) prerRNA and a number of RPs and ribosomal biogenesis factors (RBFs, also termed “assembly factors,” “nonribosomal factors,” or “transacting factors”; Kornprobst et al., 2016). Subsequently, the 90S is split by an endonucleolytic cleavage at the A2 site in internal transcribed spacer 1 to form the pre-60S and pre-40S particles (Henras et al., 2008; Woolford and Baserga, 2013).
Each particle follows a separate processing pathway for maturation (Zemp and Kutay, 2007; Kressler et al., 2010). Compared with rapid nuclear export of the pre-40S particle, the pre-60S particle requires a series of more complicated maturation events in the nucleolus, nucleoplasm, and cytoplasm (Lafontaine, 2015; Barrio-Garcia et al., 2016). Nonetheless, at later maturation steps of the pre-40S particle that occur in the cytoplasm, an 80S-like complex is formed with pre-40S and pre-60S particles and the translation initiation factor eIF5B (in yeast). The complex is critical for the 3′ end cleavage at site D of the 20S prerRNA to mature to 18S rRNA, which is a quality control step (also called “test drive”) to ensure proper maturation of cytoplasmic preribosomal particles (Lebaron et al., 2012; Strunk et al., 2012; García-Gómez et al., 2014). In all of these processes, ∼250 RBFs play essential roles in coordinated assembly of rRNAs and RPs through processing prerRNA, stabilizing preribosomal particles, or controlling nuclear export of the preribosomes (Kressler et al., 2012; Woolford and Baserga, 2013).
In both eukaryotes and bacteria, many of the RBFs are identified as energy-consuming enzymes (ATP-dependent RNA helicases, protein kinases, ATPases, and GTPases) and WD40-domain (beta-propellers)–containing proteins (Kaczanowska and Rydén-Aulin, 2007; Strunk and Karbstein, 2009; Kressler et al., 2012; Matsuo et al., 2014; Kornprobst et al., 2016). In yeast, three ATPases associated with diverse cellular activities (AAA)-type ATPases, namely ribosome export7, Mdn1/Ribosome export associated1, and diazaborine resistance gene1, contribute to distinct steps during the 60S ribosomal subunit biogenesis (Kressler et al., 2012). Ribosome export7 is the earliest acting ATPase in establishing the 60S ribosome and functions in removing Nop-7-associated1 from late nucleolar pre-60S particles (Kressler et al., 2008). Diazaborine resistance gene1 plays roles in cytoplasmic pre-60S ribosome maturation relying on interaction with Ribosomal-like Protein24 (Lo et al., 2010). Mdn1, a large dynein motor protein (∼550 kD), drives nuclear export of preribosomal particles through removal of biogenesis factors at different critical checkpoints of 60S ribosome assembly (Ulbrich et al., 2009; Bassler et al., 2010). Mdn1 is conserved in different species and consists of a ring-shaped structure built up by six tandem AAA protomers and a long tail protruding from the preribosome with a metal ion-dependent adhesion site (MIDAS)-domain at its terminus (Garbarino and Gibbons, 2002; Barrio-Garcia et al., 2016). The MIDAS-domain plays roles in Mdn1 contact with the substrates located at a second distinct position of pre-60S particles (Ulbrich et al., 2009). In yeast, a WD40 protein, Ytm1, is a pre-60S ribosome maturation factor that can directly interact with MIDAS through its N-terminal MIDAS interacting domain (Bassler et al., 2010). Ytm1 is enriched in the nucleolar pre-60S particles (Miles et al., 2005; Bassler et al., 2010). The interacting reaction is necessary for release of the Ytm1 complex (Ytm1-Erb1-Nop7), which is essential for the maturation and nuclear export of the ribosome.
In yeast, Mdn1 depletion is lethal (Galani et al., 2004). In HeLa cells, the small interfering RNA-mediated knockdown of MDN1 results in a severe growth defect (Raman et al., 2016). These observations suggest that eukaryotic cell viability is sensitive to a defect in MDN1 function. In Arabidopsis, the loss of MDN1 function leads to delayed development of the female gametophyte (Chantha et al., 2010). In maize (Zea mays), the reduced function of RIBOSOME EXPORT ASSOCIATED1 (the counterpart of MDN1) results in delayed embryo, endosperm, and seedling development (Qi et al., 2016). Furthermore, we previously characterized an Arabidopsis mutant, dwarf and short root (dsr1; renamed mdn1-1 in this study), which contained a missense mutation in MDN1 and displayed phenotypes such as a short root and low seed set under normal growth conditions (Li et al., 2016b). These results imply that MDN1 is also vital for normal plant growth and development. Through transcript profile analyses, we found that many biological processes were affected in mdn1-1 (Li et al., 2016b). Nevertheless, the molecular function of MDN1 in Arabidopsis remains poorly understood.
In this study, through phenotypic analyses of the viable homozygous mdn1-1 mutant, we further demonstrate that MDN1 is critical for root meristem cell proliferation and auxin-mediated early embryo development. Through detection of the distribution of RPs in cells and prerRNA levels, we shed light on the roles of MDN1 in ribosome biogenesis. Our results suggest that the molecular function of MDN1 is tightly associated with its roles in plant development.
RESULTS
MDN1 Is Essential for Embryo Development
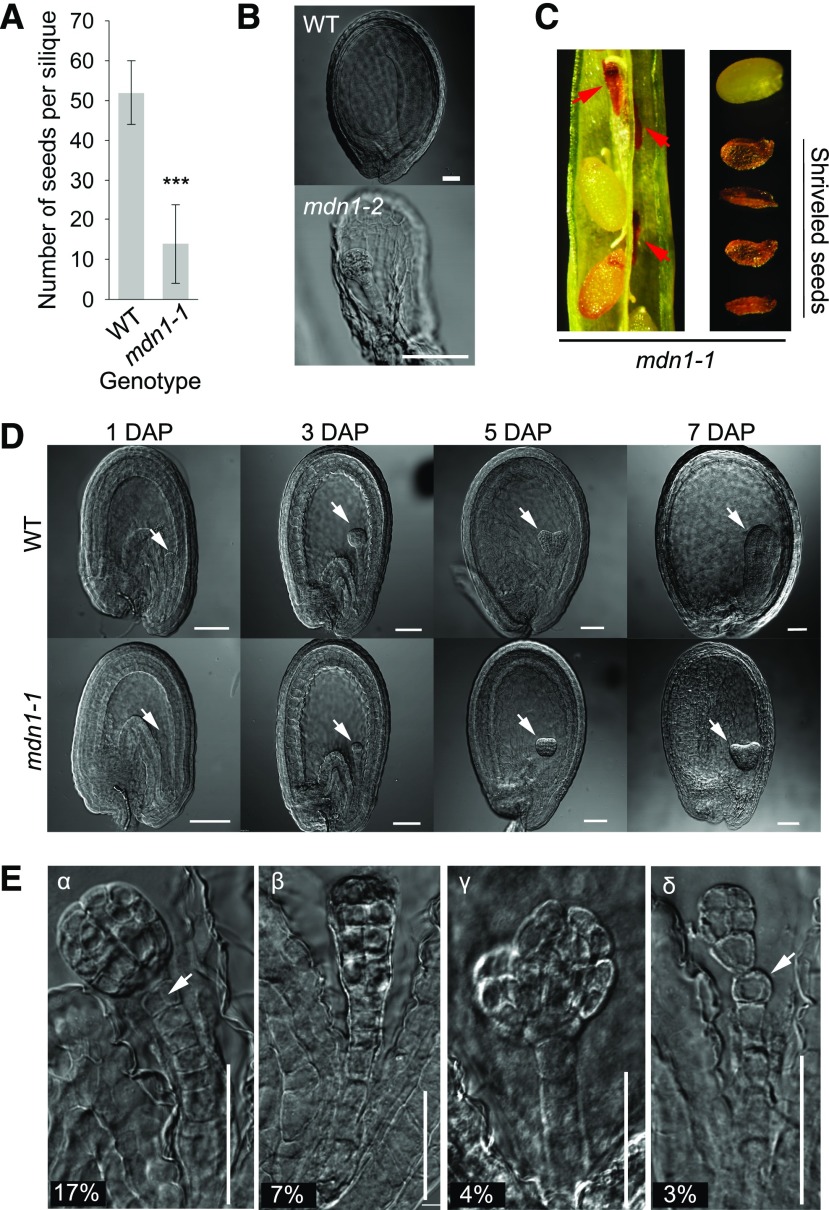
We focused on the yield traits of mdn1-1. The silique was shorter, and the number of plump seeds per silique was significantly less than that of the wild type (Fig. 1A). The phenotype of the mdn1-1 silique has been presented in our previous study (Li et al., 2016b). As described by Chantha et al. (2010), a heterozygous T-DNA insertion mutant allele of MDN1 (Salk_057010), which we call “mdn1-2/+” in this study, also exhibits the phenotype of reduced seed set per silique. Chantha et al. (2010) further found that the mdn1-2 female gametophyte development is strongly retarded but can progress to maturity, and its seed set is increased by delayed pollination (Chantha et al., 2010). However, no progeny of mdn1-2/+ is homozygous, implying that the loss of MDN1 function leads to embryonic lethality (Chantha et al., 2010). In this study, to confirm this result, the aborted ovules in the mdn1-2/+ siliques were detected with differential interference contrast (DIC) microscopy. We found that, in addition to the unfertilized ovules, most embryos were arrested at the early-globular stage (Fig. 1B), suggesting that MDN1 is required for maintaining embryo development. This result is consistent with the high expression of the MDN1 gene in embryos (Li et al., 2016b). Furthermore, several brown but shriveled seeds were observed in immature siliques of mdn1-1 (Fig. 1C). The above observations impelled us to check the embryo developmental phenotype of mdn1-1.
Figure 1.
Embryo developmental phenotypes of the mdn1 mutants. A, The number of plump seeds per silique of the wild type and mdn1-1. Error bars represent sd (n = 15). Student’s t test was applied (***P < 0.01). B, Embryos of the aborted seeds from the mdn1-2 siliques at 12 DAP observed with DIC optics. Scale bars = 50 μm. C, Shriveled seeds from the mdn1-1 siliques at 12 DAP. Red arrowheads indicate the abnormal seeds. D, Embryos of wild type (WT) and mdn1-1 at different developmental stages observed with DIC optics. White arrowheads indicate the embryos. Scale bars = 50 μm. E, Phenotypes of four types of malformed globular embryos of mdn1-1 at 4 DAP. The percentage of each type is indicated below. For the α-type, the white arrowhead indicates the abnormal cell division pattern. For the δ-type, the white arrowhead indicates the misshaped suspensor cell. Scale bars = 50 μm.
We found that at 1 d after pollination (DAP), most embryos of both wild type and mdn1-1 were categorized within the 1- to 2-cell stages (Fig. 1D). At 3 DAP, ∼55% of wild-type embryos were at the globular stage, whereas most of the mdn1-1 embryos were categorized within the 4- to 8-cell stages (Fig. 1D; Supplemental Fig. S1A). At 5 DAP, ∼89% of mdn1-1 embryos were at the globular stage. By contrast, 45% of wild-type embryos had developed into the heart stage (Fig. 1D; Supplemental Fig. S1A). Consistently, from 6 to 11 DAP, most embryos of mdn1-1 showed a delayed developmental phenotype (Fig. 1D; Supplemental Fig. S1A). At 10 DAP, most embryos of the wild type reached the cotyledonary stage (Supplemental Fig. S1A). At 11 DAP, 6% and 17% of mdn1-1 embryos remained at the early-torpedo and torpedo stages, respectively (Supplemental Fig. S1A). These results indicated that mdn1-1 embryo development was clearly retarded compared with that of the wild type. More importantly, a portion of the mdn1-1 embryos was noted that displayed abnormal morphology. Four types (α-, β-, γ-, and δ-type) of malformed embryos were observed at the globular stage (Fig. 1E). The suspensor of the wild type comprises a single file of 7–9 cells (ten Hove et al., 2015). Despite the embryo proper of both α- and β-types apparently developing normally, some of the suspensor cells were divided aberrantly (Fig. 1E). The α-type accounted for 17% of the detected embryos (n = 582), and the cell adjacent to the hypophysis underwent a transverse division (Fig. 1E). By contrast, several suspensor cells of the β-type were divided aberrantly (Fig. 1E; Supplemental Fig. S1B). The proportion of β-type was 7%. The γ-type (4%) suspensor developed normally; however, the embryo proper appeared split (Fig. 1E). Approximately 3% of embryos were δ-type, which displayed misshapen cells in the suspensor (Fig. 1E). These observations further demonstrated that MDN1 is critical for early embryo development.
Auxin Distribution Is Altered in the Globular Embryo of mdn1-1
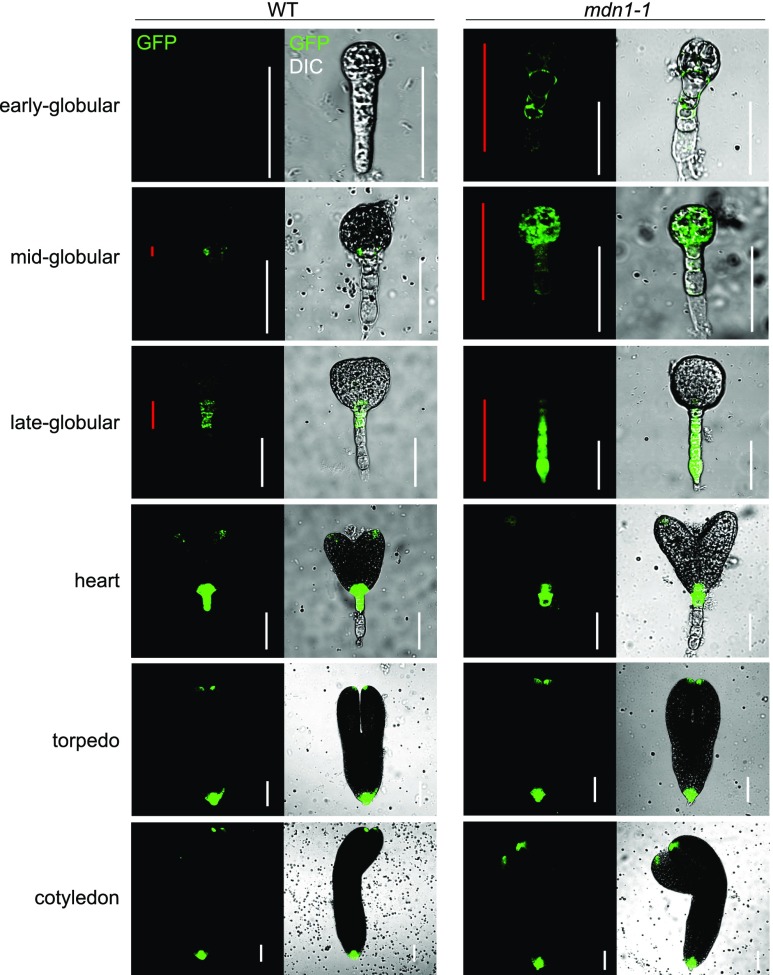
The plant hormone auxin plays an essential role in almost all aspects of plant development, including embryogenesis (Zhao, 2010). Auxin gradients are vital for the establishment of the apical-basal axis of the Arabidopsis embryo (Friml et al., 2003). The abnormal developmental pattern of the mdn1-1 embryo led us to examine whether the auxin homeostasis in embryos was affected. To determine this, we assessed the activity of the auxin-responsive DR5 reporter in embryos. As shown in Figure 2, at the early globular stage, an extremely weak level of auxin was observed in the wild-type proembryo proper. By contrast, a strong ectopic signal of the DR5 reporter was observed in the proembryo proper and suspensor. In the midglobular embryo of the wild type, an auxin response was observed around the hypophysis. However, the signal of DR5 could be clearly observed in the entire embryo of mdn1-1. At the late-globular stage, auxin was primarily localized in the hypophysis and the upper area of the suspensor in the wild-type embryo, whereas a strong auxin response was observed not only in the hypophysis but also in the entire suspensor of mdn1-1. Notably, at the heart stage and later, the auxin distribution appeared to be normal in the mdn1-1 embryo. These results revealed that the mutation of MDN1 in mdn1-1 influenced the steady state of auxin predominantly in the globular stage, which was consistent with the abnormal cell division patterns in globular embryos.
Figure 2.
mdn1-1 embryos display altered auxin distribution. DR5rev::GFP expression in wild-type (WT) and mdn1-1 embryos at the indicated stages. Red lines indicate the distribution range of GFP signals in embryo. White lines are scale bars (= 50 μm). Images were acquired using the same optical setting.
MDN1 Functions in Root Meristem Growth
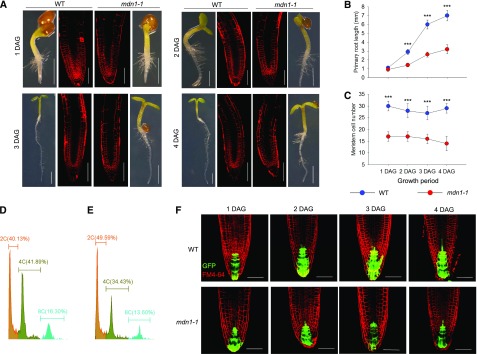
In our previous study, we presented a rough phenotype of the mdn1-1 root, which was approximately threefold shorter compared with that of the wild type at 7 d after germination (DAG) under normal growth conditions (Li et al., 2016b). To dissect the mechanism that resulted in this phenotype, kinematic analyses were performed on the primary root from 1 to 4 DAG. MDN1 was highly expressed in the region of the root apical meristem (RAM) as indicated by the GUS reporter (Li et al., 2016b), and therefore, we assessed both primary root length and RAM size (estimated by counting the cortical cells from the quiescent center to the basal cell of the elongation region). The results showed that at 1 DAG, no significant difference was detected in the root length between wild type and mdn1-1 (Fig. 3, A and B), but the number of meristem cells in mdn1-1 was clearly reduced (17 ± 2 cells [sd, n = 20]) compared with that of the wild type (30 ± 2 cells [sd, n = 20]; Fig. 3, A and C). However, at 2 DAG, the root length of the wild type was 2.9 ± 0.3 mm (sd, n = 20), whereas that of mdn1-1 was only 1.4 ± 0.2 mm (sd, n = 20; Fig. 3B). With the extension of culture time, the root length difference between wild type and mdn1-1 increased (Fig. 3B). Consistently, the number of meristem cells of mdn1-1 remained significantly less than that of the wild type (Fig. 3C). These results implied that the retarded root growth of mdn1-1 was tightly associated with the reduced number of meristem cells.
Figure 3.
Root development phenotypes of mdn1-1. A, Seedling and RAM phenotypes at the indicated stages grown under normal growth conditions. The images of seedlings were obtained with an optical microscope (scale bars = 1 mm). The images of FM4-64–stained RAM were obtained using a CLSM (scale bars = 20 μm). B, Kinematic analyses of primary root length of wild type and mdn1-1 from 1 to 4 DAG. C, Kinematic comparison of the root meristem cell number between wild type and mdn1-1 from 1 to 4 DAG. B and C, Error bars represent sd (n = 10), and Student’s t test was applied (***P < 0.01). D and E, Ploidy analysis of nuclei isolated from root cells of wild type and mdn1-1, respectively, using flow cytometry. Five independent experiments were performed, and representative results are presented. F, The expression pattern of DR5rev::GFP in root tips of wild type and mdn1-1 at the indicated stages. Scale bars = 20 μm. WT, wild type.
The reduced number of cortical cells in the RAM of mdn1-1 suggested the progression of cell division was defective. To test this hypothesis, the ploidy level of 4-DAG seedling roots was examined with flow cytometry. We found that ∼40.13%, 41.89%, and 16.30% of the nuclei had 2C, 4C, and 8C DNA content, respectively, in the wild type (Fig. 3D). In contrast to that in the wild type, mdn1-1 had more nuclei (49.59%) with 2C DNA content and fewer nuclei with higher DNA contents (Fig. 3E). The results suggested that more cells of the mdn1-1 root were at the G0/G1 stage than those of the wild type, which further implied that the mutation of MDN1 affects cell division and proliferation.
Additionally, through detecting DR5rev::GFP (green fluorescent protein) reporter expression in the root tip during the first 4 DAG, no significant difference in auxin distribution was observed between wild type and mdn1-1 (Fig. 3F). However, it was noteworthy that the auxin maxima in the root tip appeared to be decreased in mdn1-1 compared with that in wild type from day three(Fig. 3F). On account of the number of cortical cells in the RAM of mdn1-1 was less than that of wild type at the beginning of germination, we believed that the stunted phenotype of the mdn1-1 root was not associated with the auxin level in the root tip.
MDN1 Plays a Role in Nuclear Export of the Pre-60S Particle
The yeast counterpart of MDN1 is localized in the nucleus and is required for nuclear export of the pre-60S ribosomal subunit (Galani et al., 2004). In human cells, the MDN1 counterpart can be detected in both the nucleus and cytoplasm (Romes et al., 2016) and also plays a role in 60S ribosome biogenesis (Raman et al., 2016). In Arabidopsis, the results of proteomic analyses have revealed the presence of MDN1 in both the nucleus and cytoplasm (Palm et al., 2016). To further investigate MDN1 subcellular distribution, the nuclear and cytoplasmic protein components were successively isolated and screened with the anti-MDN1 antibody. Because of the excessively large Mr of MDN1 (∼611 kD), regular western-blot analysis was unfeasible. Consequently, a dot-immunoblot assay was performed. The results showed that in both wild type and mdn1-1, MDN1 was detected clearly in the nuclear fractions (Supplemental Fig. S2), whereas low signals were observed in the cytoplasmic fractions (Supplemental Fig. S2). In this experiment, BRASSINAZOLE INSENSITIVE PALE GREEN2 (BPG2, a chloroplast stroma-localized protein) and Histone3 were employed as the cytoplasmic and nuclear controls, respectively. The results revealed that MDN1 was predominantly localized in the nucleus, and that the mutation of MDN1 in mdn1-1 did not affect its protein localization. These results impelled us to further check whether the molecular function of MDN1 in ribosome biogenesis was conserved in Arabidopsis and affected in the mdn1-1 mutant.
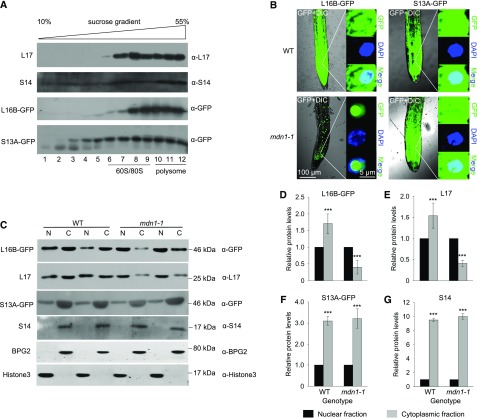
Total cellular proteins were fractionated on a 10% to 55% sucrose (Suc) density gradient using ultracentrifugation and then tested with immunoblot analysis (Fig. 4A). The distribution of small and large ribosomal subunits, 80S monosomes, and polysomes in the Suc gradient were indicated with the L17 (a 60S-associated RP) and S14 (a 40S-associated RP) blotting bands (Fig. 4A). We found that the distribution of L16B (another 60S associated RP)-GFP fusion protein in the Suc gradient was consistent with that of L17 and that of S13A (another 40S-associated RP)-GFP in the Suc gradient was similar to that of S14 (Fig. 4A). The results suggested that the GFP-tag did not affect the L16B and S13A embedding in the ribosomal particles. Therefore, to determine whether MDN1 played a role in nuclear export of preribosomal subunits, L16B-GFP and S13A-GFP were employed as the markers of large and small ribosomal subunits, respectively. We found that in the seedling root of the wild-type background, both L16B-GFP and S13A-GFP proteins were scattered throughout the cytoplasm and nuclei (Fig. 4B). However, in the root of mdn1-1, most of the L16B-GFP proteins were apparently blocked in nuclei, particularly in the nucleoli, based on the low GFP signals in the cytoplasm (Fig. 4B). By contrast, the distribution of S13A-GFP in mdn1-1 was similar to that in the wild type (Fig. 4B). These results suggested that nuclear export of the large ribosomal subunit was impaired, but no clear effect on the export of the 40S ribosome was observed in the mdn1-1 mutant.
Figure 4.
Nuclear export of pre-60S ribosomal particles is impaired in mdn1-1. A, Cofractionation analyses of L16B-GFP and S13A-GFP with 60S and 40S ribosomal subunits, respectively, using Suc gradient sedimentation and immunoblotting. The factions of 60S/80S and polysomes are indicated. L17 and S14 were employed as markers of 60S and 40S ribosomal subunits, respectively. B, Subcellular localization analyses of L16B-GFP and S13A-GFP in wild-type and mdn1-1 background. Nuclei were indicated by 4′,6-diamino-phenylindole staining. Images were acquired using the same optical setting. Scale bars = 100 µm (left images) and 5 µm (right images). C, Western-blot analyses of L16B-GFP, L17, S13A-GFP, and S14 in nuclear (N) and cytoplasmic (C) fractions extracted from 5-d-old wild-type (WT) and mdn1-1 seedlings. Each lane was loaded with 10-μg protein. Six biological replicates were performed, and representative results are presented. D–G, The protein levels of L16B-GFP (D), L17 (E), S13A-GFP (F), and S14 (G) in the cytoplasmic fraction relative to that in the nuclear fractions of wild type (WT) and mdn1-1. The protein levels were according to gray values of the western-blot bands. Error bars represent sd (n = 6), and Student’s t test was applied (***P < 0.01).
To further confirm this result, the cytoplasmic and nuclear fractions were separately isolated from 5-DAG seedlings, and immunoblot assays were performed (Fig. 4C). In the wild-type background, L16B-GFP was detected in both cytoplasmic and nuclear fractions, but the level was higher in the cytoplasmic fraction (Fig. 4, C and D). By contrast, in the mdn1-1 background, the level of L16B-GFP was higher in the nuclear fraction than that in the cytoplasmic fraction (Fig. 4, C and D). Furthermore, consistent results were observed after detection of the level of L17 (Fig. 4, C and E). By contrast, the homeostasis of S13A-GFP and S14 in the nuclear and cytoplasmic fractions was not clearly altered in mdn1-1 (Fig. 4, C, F, and G). Likewise, BPG2 and Histone3 were employed as the loading controls of cytoplasmic and nuclear fractions, respectively (Fig. 4C). These results implied that the maturation of the pre-60S ribosomal subunit might be affected in mdn1-1 because this mutation of MDN1 impaired the conserved function of the protein in nuclear export of the pre-60S ribosome.
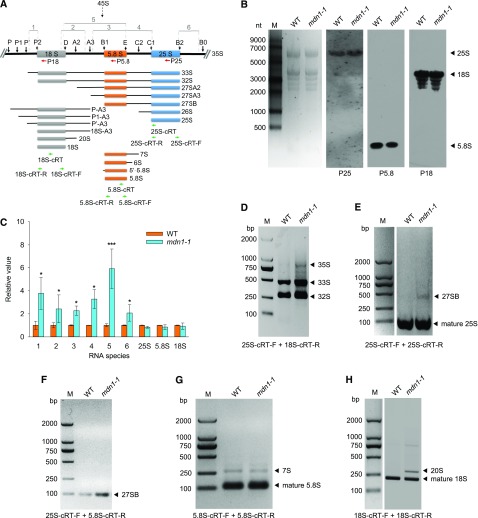
The prerRNA Processing Is Affected in the mdn1-1 Mutant
To further investigate whether rRNA levels were affected in mdn1-1, total RNA was isolated from 5-DAG seedlings of the wild type and mdn1-1, and northern-blot assays were performed. The levels of mature 25S, 5.8S, and 18S rRNAs were detected with the biotin-labeled specific oligoprobes (Fig. 5A). The results showed that the abundance of these mature rRNAs was not clearly altered in mdn1-1 (Fig. 5B). We further detected the levels of prerRNA intermediates using reverse transcription-quantitative PCR (RT-qPCR). The templates for PCR amplification are indicated with lines and the numbers 1–6 in Figure 5A. We found that the levels of all detected intermediates were significantly higher in mdn1-1 than those in the wild type (Fig. 5C). Notably, the most upregulated fragment was 5 (Fig. 5C), which suggested the level(s) of 35S, 33S, and/or 32S prerRNA(s) was upregulated in mdn1-1. The high abundance of 35S in mdn1-1 was further suggested by the upregulation of 6 (Fig. 5C). To further confirm this result, circular RT-PCR (cRT-PCR) was carried out. The results showed that, indeed, the 35S, 33S, and 32S prerRNAs were all accumulated at higher levels in mdn1-1 than in wild type (Fig. 5D). We also detected the transcript levels of four different variants (VARs) of the 45S prerRNA using RT-PCR according to a previous study (Pontvianne et al., 2010). The results showed that three variants (VAR2, VAR3, and VAR4) could be clearly detected in both wild type and mdn1-1 at the seedling stage, and no significant differences in their expression levels were observed between the two detected lines, suggesting that the rRNA transcription from rDNA was not affected in mdn1-1 (Supplemental Fig. S3) Therefore, the accumulation of prerRNAs was likely on account of defects in rRNA processing in mdn1-1. Moreover, the mature rRNA levels were further detected by RT-qPCR (Fig. 5C), and the results were consistent with those of northern-blot analysis (Fig. 5B).
Figure 5.
PrerRNA processing in mdn1-1. A, Diagram of the rRNAs and pathways of prerRNA processing in Arabidopsis. The major processing intermediates are illustrated. The black vertical arrows indicate the processing sites. Regions 1–6 indicate the fragments that are employed for RT-qPCR amplification. The regions used for oligonucleotide probes are indicated with red horizontal arrows. The regions of the primers used for reverse transcription and cRT-PCR amplification are indicated with green horizontal arrows. B, Northern-blot analyses of the mature rRNA levels in 5-d-old seedlings of wild type and mdn1-1 with specific probes indicated in (A). M, RNA size marker. Left, the ethidium-bromide–stained gel image, which is shown as a loading control. C, Levels of the intermediates indicated in (A) and three mature rRNAs in mdn1-1 relative to that in wild type. Arabidopsis Actin7 was employed as the internal control to normalize the values. Error bars represent sd (n = 4). Student’s t test was applied (***P < 0.01, and *P < 0.05). D, Increased accumulation of the 35S, 33S, and 32S prerRNAs in mdn1-1 determined by PCR with 25S-cRT-F and 18S-cRT-R primers. E and F, Increase of the 27SB intermediate in mdn1-1 validated by PCR with 25S-cRT-F and 25S-cRT-R primers (E) and 25S-cRT-F and 5.8S-cRT-R primers (F). G, Unchanged levels of the 7S prerRNA and 5.8S rRNA in mdn1-1 checked by PCR with 5.8S-cRT-F and 5.8S-cRT-R primers. H, Increased accumulation of the 20S intermediate in mdn1-1 determined by PCR with 18S-cRT-F and 18S-cRT-R primers. D–H, Bands indicated by arrowheads were cloned and sequenced. M, DNA markers; WT, wild type; bp, basepair.
Because nuclear export of the pre-60S ribosome was affected in mdn1-1 (Fig. 4), the processing of 25S and 5.8S rRNA precursors might be affected as well. To test this hypothesis, the precise 3′ and 5′ ends of the pre-25S and pre-5.8S rRNA intermediates were determined by cRT-PCR. Through PCR amplification with specific primers, notably, a band signal of unprocessed 27SB was observed in mdn1-1 (Fig. 5E), suggesting the accumulation of this 25S rRNA precursor. The upregulation of 27SB in mdn1-1 was further confirmed by PCR amplification with another pair of primers (Fig. 5F). Additionally, mature 5.8S and unprocessed 7S prerRNA bands were detected; however, no clear differences in these band signals were observed between wild type and mdn1-1 (Fig. 5G). These results implied that MDN1 was involved in the 25S rRNA maturation, and further suggested its roles in 60S ribosome biogenesis.
A previous study suggested that the final steps for maturing the pre-40S ribosomal subunit is 60S ribosome-dependent in plant (Weis et al., 2014). Considering the pre-60S maturation was impaired in mdn1-1, the maturation of pre-40S might likewise be affected. Using cRT-PCR, indeed, a strong band signal of a PCR product originated from the 20S prerRNA was observed in mdn1-1 (Fig. 5H), suggesting the accumulation of this 18S rRNA precursor. Collectively, the above results suggest that in mdn1-1, although the levels of mature rRNAs are not clearly altered, the prerRNA processing is affected, which is tightly associated with the defect in nuclear transport of the pre-60S ribosome.
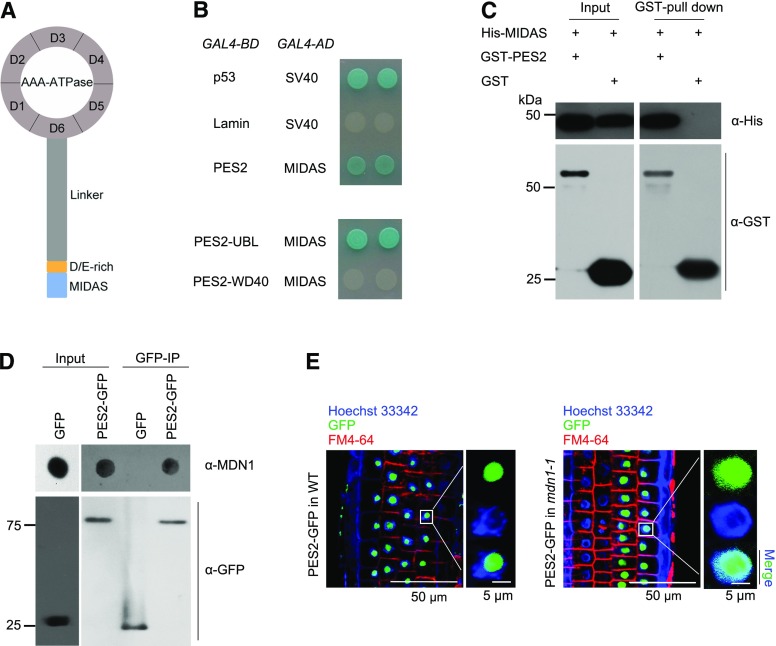
MDN1 Is Associated with PESCADILLO2
The MDN1 protein consists of several domains (Fig. 6A). The C-terminal MIDAS domain is well conserved among all eukaryotes (Garbarino and Gibbons, 2002; Li et al., 2016b). In yeast, this domain plays a critical role in Mdn1 interaction with its substrate Ytm1 (Ulbrich et al., 2009; Bassler et al., 2010). The Arabidopsis PESCADILLO2 (PES2) is the homolog of yeast Ytm1 (Supplemental Fig. S4A). Furthermore, two previous studies demonstrate that PES2 is localized in the nucleolus and plays critical roles in 60S ribosome biogenesis (Cho et al., 2013; Ahn et al., 2016). As a consequence, we questioned whether the MIDAS domain of Arabidopsis MDN1 directly interacted with PES2. To address this issue, the yeast-2-hybrid assay was performed, and a positive result was observed (Fig. 6B, upper). Next, the proteins of His-6-tagged MIDAS and glutathione S-transferase (GST)-tagged PES2 were expressed in bacteria and further purified. Through a GST-tag pull-down assay, we confirmed the physical association between MIDAS and PES2 (Fig. 6C). We further obtained the homozygous transgenic lines of 35S::PES2-GFP. To investigate whether PES2 interacted with the MDN1 protein in vivo, anti-GFP immunoprecipitated (IP) assays were performed in homozygous transgenic plants of 35S::PES2-GFP and those of 35S::GFP. The precipitated substrates were further tested by dot-immunoblot with the anti-MDN1 antibody. We found that the GFP-fused PES2 captured MDN1 (Fig. 6D). By contrast, no reaction between the GFP-tag and MDN1 was observed (Fig. 6D).
Figure 6.
The interaction between MDN1 and PES2 in vitro and in vivo. A, Illustration of the MDN1 protein domains. Ring-shaped AAA-ATPase consists of six tandem AAA protomers (D1–D6), a long flexible linker, a D/E-rich domain, and the MIDAS domain at the tip. B, Yeast-2-hybrid assays on protein interaction between MIDAS and PES2, the UBL-domain of PES2 (PES2-UBL), or the WD40-domain of PES2 (PES2-WD40). Plasmids expressing the indicated GAL4-BD and GAL4-AD were transformed into the yeast strain Y2HGold. The interaction of p53 and the SV40 large T-antigen was used as the positive control, while that of lamin and SV40 was used as the negative control. Solutions of transformed yeast were spotted onto the SD/-Ade/-His/-Leu/-Trp/X-α-Gal/AbA medium. C, GST-tagged PES2 interacts with the His-6-tagged MIDAS in a GST pull-down assay. D, IP analysis of PES2-GFP with the MDN1 protein. Protein extracts from the homozygous transgenic seedlings of 35S::PES2-GFP and 35S::GFP were subjected to IP with the monoclonal anti-GFP antibody. The MDN1 protein was detected by dot-immunoblot with the anti-MDN1 antibody. E, Subcellular localization of PES2-GFP in RAM cells of 4-d-old seedling of the wild-type (WT) or mdn1-1 background. The nucleoplasm was indicated with the Hoechst 33342 staining, and the cellular outline was indicated with the FM4-64 dye. Scale bars = 50 µm (left images) and 5 µm (right images).
The PES2 protein contains a N-terminal ubiquitin-like (UBL) domain (also called “Notchless-like” domain) and a WD40 domain (Supplemental Fig. S4B). In yeast, this UBL-domain is referred to as the “MIDAS interacting domain” because it interacts with the MIDAS-domain of Mdn1 (Bassler et al., 2010). To determine whether the UBL-domain was critical for interacting with MIDAS, both the UBL and WD40 of PES2 were employed, and the yeast-2-hybrid assays were performed. The results showed that the UBL domain could directly interact with MIDAS, whereas a negative result was observed in the interaction assay between WD40 and MIDAS (Fig. 6B, bottom). Collectively, the above results suggest that MDN1 is physically associated with PES2 through the MIDAS-UBL interaction in Arabidopsis.
The PES2 Localization Is Affected in the mdn1-1 Mutant
A previous study has revealed that the PES2 protein lacking the UBL domain can be detected not only in nucleolus but also in nucleoplasm (Ahn et al., 2016), suggesting that this domain is critical for maintaining the nucleolar localization of its protein. Because MDN1 could interact with the UBL domain of PES2, we questioned whether the subcellular localization of PES2 was affected in mdn1-1. To address this issue, we analyzed the distribution of PES2-GFP fusion protein in the root meristem cells of wild type or mdn1-1. The results showed that the PES2-GFP signal was predominantly observed in nucleolus of the wild-type background (Fig. 6E), which was consistent with the previous study (Ahn et al., 2016). In contrast, in the mdn1-1 background, the GFP signal was clearly observed throughout the nucleus (Fig. 6E), indicating the mislocalization of PES2. Because PES2 is associated with the 60S ribosome (Ahn et al., 2016), the result further supports a role of MDN1 in 60S ribosome biogenesis.
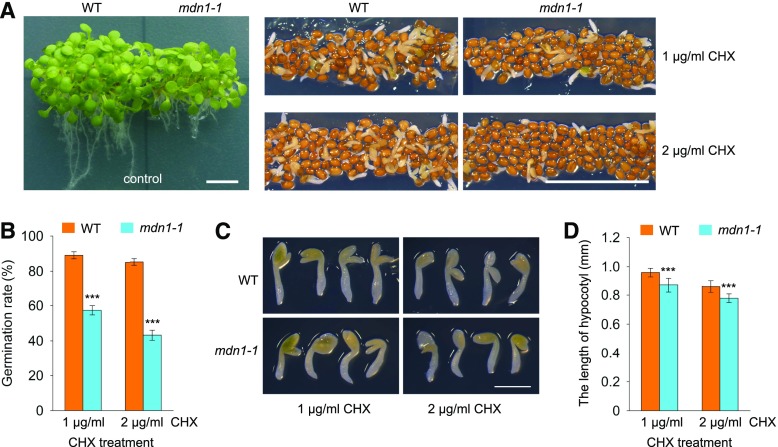
The mdn1-1 Mutant Shows Hypersensitivity to a Protein Synthesis Inhibitor
A previous study revealed that seed germination (radicle protrusion) is sensitive to a defect in de novo protein synthesis (Rajjou et al., 2004). Under normal growth conditions, we previously found that the germination rate of mdn1-1 was significantly lower than that of the wild type (Li et al., 2016b). Furthermore, in this study, we found that ribosome biogenesis was affected in mdn1-1. These results implied that the translational efficiency might be reduced in mdn1-1. To verify this hypothesis, cycloheximide (CHX), a eukaryotic protein synthesis inhibitor, was employed to test its influence on seed germination. At 2 weeks after sowing, the germination rate of the wild type grown on medium containing 1 μg/mL CHX reached 89% (Fig. 7, A and B). By contrast, only ∼57.3% of mdn1-1 seeds showed radicle emergence (Fig. 7, A and B). Under the 2 μg/mL CHX condition, the seeds of the wild type showed 85% radicle emergence, whereas the germination rate of mdn1-1 decreased to 43.2% (Fig. 7, A and B). As a control, both wild type and mdn1-1 grown in the CHX-free medium developed into mature seedlings (Fig. 7A). Additionally, we assessed the total length of hypocotyl with radicle from the germinated seeds grown under the CHX condition. The results showed that the length of hypocotyl of mdn1-1 was slightly but significantly less than that of the wild type (Fig. 7, C and D). Thus, these observations suggest a more severe defect in protein synthesis in mdn1-1 than that in the wild type, which might result from impaired ribosome biogenesis in the former.
Figure 7.
Phenotypes of mdn1-1 grown under CHX-treatment conditions. A, Phenotypes of wild type and mdn1-1 grown on solid 1/2 MS medium containing 0 (control), 1 μg/mL, or 2 μg/mL CHX for 2 weeks. Scale bars = 50 mm. B, Germination rate analyses of wild type and mdn1-1 grown under the indicated CHX-treatment conditions. Data are mean values of three biological replicates ± sd. Student’s t test was applied (***P < 0.01). C, Seedling phenotypes of wild type and mdn1-1 grown under CHX-treatment conditions for 2 weeks. Scale bar = 1 mm. D, Comparison analyses of the length of hypocotyl with radicle between wild type and mdn1-1 grown under CHX-treatment conditions for 2 weeks. Error bars = sd (n = 10). Student’s t test was applied (***P < 0.01). WT, wild type.
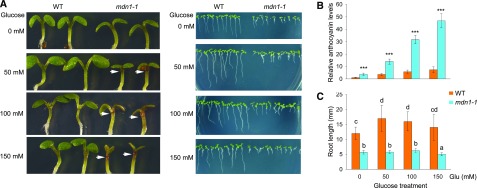
The mdn1-1 Mutant Is Sensitive to High Concentrations of Sugar
Ribosome biogenesis is an extremely energy-consuming anabolic process in eukaryotic cells. Considering that ribosome biogenesis is impaired in mdn1-1, we were interested in its phenotype under different energy conditions. Furthermore, under normal growth conditions, in addition to the stunted phenotype, the seedlings of mdn1-1 also showed an anthocyanin accumulation phenotype in the upper hypocotyl area (Fig. 3A). Numerous genes involved in anthocyanin biosynthesis are sugar-induced (Solfanelli et al., 2006), and Arabidopsis seedlings grown under high levels of sugar (Suc or glucose [Glc]) always show the accumulation of anthocyanins (Martin et al., 2002). Therefore, we proposed that the overaccumulation of anthocyanin in mdn1-1 might be associated with sugar in the growth medium. To test this assumption, we assessed the anthocyanin levels in 5-d-old seedlings grown on medium containing 0, 50, 100, or 150 mm Glc (Fig. 8). Notably, under the Glc-free condition, although no obvious accumulation of anthocyanins was observed in the upper hypocotyl area by the naked eye, the levels of anthocyanins in the whole seedling of mdn1-1 were significantly higher than those in the wild type (Fig. 8, A and B). Under the 50- or 100-mm Glc condition, mdn1-1 respectively possessed ∼3.8-/5.6-fold higher levels of anthocyanins than those of the wild type (Fig. 8, A and B). Under the 150-mm Glc treatment, the accumulation of anthocyanins in mdn1-1 was ∼6.4-fold higher than that in wild type, and a repressed cotyledon expansion phenotype was also observed in mdn1-1 (Fig. 8, A and B). By contrast, the cotyledon expansion of the wild type was normal (Fig. 8A). As an osmotic control, mannitol had no clear effect on the induction of anthocyanin biosynthesis in either wild type or mdn1-1 (Supplemental Fig. S5). Additionally, when the wild type was grown under the 50- or 100-mm Glc condition, the root length was significantly longer than that grown under Glc-free medium (Fig. 8, A and C). However, the root growth of mdn1-1 displayed no response to these sugar levels (Fig. 8, A and C). Moreover, with a further increase in level of Glc (150 mM), the root growth of mdn1-1 was stunted further (Fig. 8, A and C). These results revealed that the growth of mdn1-1 is sensitive to a high-sugar condition.
Figure 8.
Phenotypes of mdn1-1 grown under Glc-treatment conditions. A, Phenotypes of wild type and mdn1-1 grown on solid MS medium containing the indicated concentration Glc for 5 d. The white arrowheads indicate the accumulated anthocyanins. B, Relative anthocyanin levels in the seedlings shown in (A). The anthocyanin content of wild type grown on Glc-free medium was set to 1. Error bars = SD (n = 3). Student’s t test was applied (***P < 0.01). C, Comparison analyses of root length of seedlings shown in (A). Data are mean values of 10 replicates ± sd. Statistically significant differences are indicated by different lowercase letters (P < 0.01, Duncan's multiple range test). WT, wild type.
DISCUSSION
We demonstrate here that MDN1, as a probable RBF in Arabidopsis, plays a role in 60S ribosome maturation with involvement in its nuclear export. A large-scale in silico analysis identified 174 genes encoding orthologous proteins of the yeast RBFs in the Arabidopsis genome (Simm et al., 2015). However, to date, only a very small proportion of these members have been characterized in detail (Weis et al., 2015a). Nevertheless, some common phenotypes are observed for mutants of these RBFs, including defects in gametophyte development, embryonic lethality, impaired root growth, and altered leaf shape (Weis et al., 2015a). These phenotypes are constantly observed for RP mutants (Byrne, 2009). The coincidence might result from the ribosome acting as the exclusive machine for protein synthesis in cells. As shown in previous studies (Chantha et al., 2010; Li et al., 2016b; Qi et al., 2016) and in this paper, the common phenotypes were also observed for the mutants of MDN1, which were tightly associated with the role of MDN1 in ribosome biogenesis.
mdn1-1 is a weak MDN1 mutant allele that maintains its viability. Nevertheless, embryo development was delayed in mdn1-1 (Fig. 1D), which was most likely linked with defects in ribosome biogenesis based on impaired nuclear export of the pre-60S ribosome particles (Fig. 4). Moreover, embryo malformation resulting from an altered cell division pattern was observed in mdn1-1 (Fig. 1E), which was tightly associated with perturbed auxin distribution (Fig. 2). An altered auxin distribution in embryos is observed for other RBF mutants. APUM24 (ARABIDOPSIS PUMILIO PROTEIN24) is a nucleolar-localized Pumilio protein, which is responsible for proper prerRNA processing (Shanmugam et al., 2017; Maekawa et al., 2018). The embryo of the apum24 mutant exhibits an abnormal cell division pattern likely resulting from the disrupted auxin distribution and transport (Shanmugam et al., 2017). Furthermore, in some RP mutants, such as rpl27ac-1d, the mislocalization of auxin in the embryo is also observed (Szakonyi and Byrne, 2011). These observations indicate that the auxin homeostasis is sensitive to defects in ribosome biogenesis (also called “ribosomal stress”). Additionally, embryos are arrested at the globular stage in mdn1-2 (Fig. 1B) and in mutants of essential RBFs or RPs (Byrne, 2009; Weis et al., 2015a), suggesting the cell proliferation in the proembryo is independent of newly synthesized ribosomes. However, the maternally inherited ribosomes might be insufficient for the postglobular stage development. Therefore, we propose that the effect of ribosome stress on embryo development might be through various pathways, including those that are auxin-involved and ribosome-dose–dependent.
In addition to the defects in embryo development, we also demonstrated that root growth was retarded in mdn1-1, which was tightly associated with the significant reduction in meristem cells (Fig. 3, A–C). We also found that the cell cycle was affected in the mdn1-1 root (Fig. 3E). During the first 2 DAG, the reprogramming in root cells attributed to the MDN1 mutation was apparently auxin-independent because the auxin maxima and distribution in the root tip were not clearly altered (Fig. 3F). Later, however, the auxin maxima in the root tip of mdn1-1 were reduced (Fig. 3F). Whether the mdn1-1 root development at the later stage is associated with auxin transport and/or responsiveness requires further identification.
In many RP and RBP mutants, auxin is suggested as a prominent signal in the pathways of ribosome-mediated defects in plant development (Degenhardt and Bonham-Smith, 2008; Abbasi et al., 2010). However, how auxin responds to ribosome stress remains elusive. A recent report revealed that ANAC082, a NAC transcription factor, mediates a set of stress responses triggered by perturbation of ribosome biogenesis in Arabidopsis, similar to the roles of p53 in mammalian cells (Ohbayashi et al., 2017). Thus, establishing clearly the relation between auxin and ANAC082 would contribute to our understanding about the roles of ribosomes in plant development. The transcriptional level of ANAC082 is upregulated inroot initiation defective2 and upregulated inroot initiation defective3 (two mutants with impaired prerRNA processing; Ohbayashi et al., 2017). However, based on the results of RNA-sequencing published in our previous study (Li et al., 2016b), no significant change in the ANAC082 transcriptional level was observed in the seedlings of mdn1-1 compared with that in the wild type, suggesting that other factors act downstream of ribosome stress because of the complexity of ribosome biogenesis pathway.
We also found that the rRNA processing was affected in mdn1-1 (Fig. 5). The increased accumulation of the 35S and 27SB prerRNAs elicited by the loss of MDN1 function was observed in both yeast and Arabidopsis (Galani et al., 2004), suggesting the role of MDN1 in these rRNA processing might be conserved. In yeast, the depletion of Mdn1 leads to reduced synthesis of the mature 5.8S rRNA because of inhibited processing of the 7S prerRNA (Galani et al., 2004). However, the steady state of the mature 5.8S rRNA was not clearly affected in the Arabidopsis mutant mdn1-1 (Fig. 5, B and G). The difference suggests that MDN1 may evolve distinct functions in Arabidopsis. Furthermore, it is noteworthy that the 20S prerRNA was clearly accumulated in mdn1-1 (Fig. 5H). Akin to mdn1-1, the Arabidopsis mutants of the LARGE SUBUNIT GTPASE1-2 gene also exhibit a defect in 20S prerRNA processing (Weis et al., 2014). LARGE SUBUNIT GTPASE1-2 is a 60S/pre-60S-associated GTPase that functions in ribosome biogenesis (Weis et al., 2014). Together, these results exemplify that in Arabidopsis, the 60S ribosomal subunit is vital for the 20S prerRNA processing. Consistently, in yeast, the 60S subunit is revealed as a chaperon of the pre-40S particle in the translation-like cycle, which is a licensing step in the maturation of the 40S subunit (Strunk et al., 2012).
In yeast, the molecular function of Ribosome export associated1/Mdn1 is well studied. The progression in exploring the roles of Mdn1 fills a vacancy in the nuclear export pathway of the pre-60S ribosomal particle. As a mechanoenzyme, Mdn1 strips off RBFs likely by physical force at critical transition points of ribosome maturation (Barrio-Garcia et al., 2016). Ytm1 is determined to be a substrate of Mdn1 in the nucleolus (Bassler et al., 2010). Typically, Ytm1, Erb1, and Nop7 interact with one another and form a Nop7-subcomplex of preribosome (Tang et al., 2008). This complex also exists in mammalian cells, which is called the “PeBoW complex” (Hölzel et al., 2005). Moreover, the interaction patterns among the Arabidopsis counterparts of these three proteins, PES (Nop7), BLOCK OF CELL PROLIFERATION1 (Erb1), and PES2 (Ytm1), have been detected in tobacco (Nicotiana benthamiana), and positive results were observed (Cho et al., 2013; Ahn et al., 2016). These observations imply that the Nop7-subcomplex is conserved in eukaryotes. More importantly, in yeast, every member of the Nop7-subcomplex is necessary for its association with the preribosome (Tang et al., 2008). As a consequence, the interaction between Mdn1 and Ytm1 leads to disassociation of the Nop7-subcomplex from the preribosome, which is an essential step for ribosome biogenesis (Kressler et al., 2012). In the nucleoplasm, Mdn1 targets to another substrate, Rsa4 (Ribosome Assembly4; Ulbrich et al., 2009). Rsa4 is proposed to be a relay-like factor that transmits the mechano-chemical energy of Mdn1 into the preribosome for relocation of the premature rRNA helix 89 (Bassler et al., 2014). Notably, both of the substrates of Mdn1 are WD40-containing proteins, although the interacting sites are not located at the WD40 domain (Bassler et al., 2010). The substrates are targeted by Mdn1 through its C-terminal MIDAS-domain, which interacts with the N-terminal UBL domain of the substrates (Ulbrich et al., 2009; Bassler et al., 2010). The MIDAS-UBL interaction pattern is also observed in human cells (Romes et al., 2016). In this study, we determined that PES2 was physically associated with MDN1 through the MIDAS-UBL interaction pattern (Fig. 6). Additionally, the MIDAS from potato (Solanum chacoense), ScMDN1, interacts with the UBL-domain of ScNOTCHLESS (a homolog of Rsa4) in the yeast-2-hybrid assay (Chantha and Matton, 2007). Thus, the MIDAS-UBL interaction pattern is conserved in eukaryotes. Interestingly, two recent studies have identified a dock for the MIDAS domain that is located at the AAA domain of Mdn1 in yeast (Chen et al., 2018; Sosnowski et al., 2018). This intramolecular binding reaction is required for the Mdn1 function and likely helps the MIDAS domain interact with its substrate (Chen et al., 2018; Sosnowski et al., 2018).
PES2 plays a critical role in plant development (Ahn et al., 2016). The inducible RNA interference mutant of PES2 shows severe growth arrest (Ahn et al., 2016). Similar phenotypes are observed in RNA interference mutants of the other two members of the “PeBoW” complex (Cho et al., 2013; Ahn et al., 2016). Further analyses reveal that PES2 is associated with the ribosome, and the association is C-terminal WD40-domain–dependent (Ahn et al., 2016). In particular, lack of the N-terminal UBL domain leads to the PES2 protein distribution throughout the nucleus (Ahn et al., 2016). This result is consistent with the PES2 mislocalization in the mdn1-1 background (Fig. 6E). Together with the UBL-MIDAS interaction pattern of PES2 with MDN1, these observations reveal that MDN1 is required for the proper subcellular localization of PES2, and this process might be vital for ribosome biogenesis in Arabidopsis.
Accounting for the roles of MDN1 in ribosome biogenesis, the translation inhibitor sensitivity of its mutant was understandable (Fig. 7). Moreover, a sugar-sensitive phenotype indicated by the accumulation of anthocyanins was observed in mdn1-1 (Fig. 8). This phenotype is also reported in mutants of APUM24 and BIOGENESIS OF RIBOSOMES IN XENOPUS1-2 (Maekawa et al., 2018). Similar to APUM24, BRX1-2 is a conserved RBF that is involved in prerRNA processing (Weis et al., 2015b). Furthermore, the expression of several genes encoding RPs is sugar-inducible (Kojima et al., 2007). As a consequence, a close connection between ribosome biogenesis and energy status in plant cells is suggested, although the mechanism is still unknown. In yeast and mammalian cells, this connection involves epigenetic regulation of rRNA synthesis and mechanistic targeting of rapamycin (mTOR) signaling pathways (Oakes et al., 2006; Murayama et al., 2008; Pelletier et al., 2018). Uncovering the mechanism of ribosome biogenesis responding to energy status in plant cells is the next challenge.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. mdn1-1 was a site-changed mutant that was identified in our previous study (Li et al., 2016b). The T-DNA insertion mutant of MDN1 (mdn1-2, SALK_057010) was obtained from the Arabidopsis Biological Resource Center. Seeds were surface-sterilized successively using 70% (v/v) ethanol and 10% (v/v) H2O2 and plated on solid 1/2 Murashige and Skoog (MS) medium containing 1% (w/v) Suc (pH 5.7). After a 3-d stratification at 4°C, plates were moved to a phytotron (22°C to 23°C) with long-day (16-h light (75 µmol·m−2·s−1)/8-h dark cycles) conditions. At 10 DAG, plants were transferred into soil. For the CHX treatment, sterilized seeds were plated on the 1/2 MS medium with the addition of 1 or 2 μg/mL CHX. For the Glc treatment, Suc in the MS medium was replaced with Glc. The anthocyanins quantitation was performed as described in Solfanelli et al. (2006).
Construction of Transgenic Plants
For the 35S::GFP construct, the GFP sequence was inserted into the pCAMBIA2300-35S vector. For the 35S::L16B-GFP, 35S::S13A-GFP, and 35S::PES2-GFP constructs, the coding sequences of L16B, S13A, and PES2 were amplified by PCR and then, respectively, cloned into the above 35S::GFP construct. These final constructs were then introduced into wild type or mdn1-1 by Agrobacterium tumefaciens (GV3101)-mediated floral dip method. For selection, 50 μg/mL Kanamycin was added to the 1/2 MS medium. All of the primers were listed in Supplemental Table S1.
Embryo Observation
For observation of the entire ovule and its embryo, the pistils at different stages after pollination from wild type or the mdn1 mutants were collected and immersed in the fixative solution of ethanol/acetic acid (9:1, v/v) for at least 2 h. The samples were then successively incubated in 90%, 70% (v/v) ethanol, and clearing solution (chloral hydrate/glycerol/water = 8:1:2, w/v/v) for 1 h, respectively. The cleared pistils were placed on the labeled glass slide. The embryo in ovule was observed and photographed using DIC optics of a confocal laser scanning microscope (CLSM, FV1200; Olympus).
Antibodies and Immunoblotting
For the MDN1 antibody production, the MIDAS-domain (5089–5393 of MDN1) was employed as the antigen. The polyclonal antibody of MDN1 was produced in rabbit (ABMART). The antibody specificity was tested with the total proteins extracted from wild type and 35S::MIDAS-GFP transgenic seedlings. The anti-GFP, anti-GST, and anti-His mouse monoclonal antibodies were purchased from EarthOx Life Sciences. The polyclonal antibody of BPG2 was introduced in our previous study (Li et al., 2016a). The polyclonal antibody of Histone3 and S14 were purchased from Agrisera (AS10710 and AS122111, respectively). The L17 monoclonal antibody was purchased from Santa Cruz (sc-517047). For extraction of the nuclear and cytoplasmic fractions, a Minute Cytoplasmic & Nuclear Extraction Kit (Invent Biotechnologies, SC-003) was used following the user instruction. Extracted proteins were measured using a Pierce BCA Protein Assay Kit (P23225; Thermo Fisher Scientific). For the dot-immunoblotting analysis, 2-μg nuclear or cytoplasmic proteins extracted from 5-DAG seedlings were dropped onto the nitrocellulose membrane. Then the air-dried membrane was blocked with 5% (w/v) nonfat milk/TBS-T and successively incubated with the primary (anti-MDN1, 1:100) and secondary antibodies (1:20,000) at room temperature. For western-blot analysis, 10-μg nuclear or cytoplasmic proteins were separated in SDS-PAGE gels. The separated proteins were then transferred to a polyvinylidene difluoride membrane (0.45 μm; Millipore) and successively incubated with the primary (anti-GFP, 1:500; anti-S14, 1:500; anti-L17, 1:1,000; anti-BPG2, 1:500; anti-Histone3, 1:200) and secondary antibodies (1:20,000). The dot or band signals were detected with a Super ECL Assay Kit (EarthOx). The band gray analysis was performed with the software ImageJ (National Institutes of Health).
Suc Gradient Sedimentation
Total proteins were extracted from 500-mg seedlings with a Minute Total Protein Extraction Kit for Plant Tissues (SN-009; Invent Biotechnologies) according to the user manual. Five-hundred microliters of the extraction were loaded onto a 4.5-mL 10% to 55% (w/v) Suc gradient, and then centrifuged at 45,000 rpm for 2 h at 4°C using a SW 55Ti rotor (Beckman). Fractions of 400 μL each were collected and 20 μL of each selected fraction was analyzed by SDS-PAGE and western blot.
Detection of Fluorescence Signals
For detecting the expression of DR5rev::GFP in embryo, the embryos at different stages were isolated from ovules with a syringe needle on a labeled glass slide under a microscope (SZX16; Olympus). The GFP signals in embryo and RAM were detected by a CLSM (excitation wavelength: 488 nm, emission wavelength: 507 nm, FV1200; Olympus) under the same setting parameters. The nucleus was stained with 1 μg/mL 4′,6-diamino-phenylindole for 10 min, and then scanned using a CLSM (excitation wavelength: 345 nm, emission wavelength: 455 nm, FV1200; Olympus). The DNA double-stranded localized in nucleoplasm was stained with 10 μg/mL Hoechst 33342 for 5 min, and then scanned using a CLSM (excitation wavelength: 345 nm, emission wavelength: 455 nm, FV1200; Olympus). For indicating the membrane of RAM cells, roots were immersed in FM4-64 solution (10 μg/mL in Hanks’ Balanced Salt Solution) and incubated on ice for 5 min. Images were then obtained by excitation with the 596-nm laser line and emission with a 680–750-nm bandpass filter (FV1200; Olympus).
Nuclear Ploidy Analysis
For extraction of nuclei, 50-mg seedling roots were quickly, finely chopped with a sharp razor blade in 1-mL buffer-1 (100 mm citric acid, 0.5% [v/v] polysorbate-20, pH 2–3). The resulting slurry was filtered through a 70-μm nylon filter to eliminate cell debris. After an incubation on ice for 5–10 min, the filtered slurry was centrifuged at 1,000 revolutions per min for 5 min at 4°C. After centrifugation, the supernatant was discarded. The pellets were then resuspended with 600-μL buffer-1:buffer-2 (400 mm Na2HPO4·12H2O, 1:2) and filtered through a 40-μm nylon filter. RNase and propidium iodide were added to the suspension and incubated for 30 min at room temperature. After the incubation, the suspension containing nuclei was measured with a flow cytometer (CytoFlex A00-1-1102; Beckman Counter) at a wavelength of 488 nm. For each sample, at least 10,000 nuclei were collected.
RT-qPCR
To detect the relative levels of prerRNA intermediates, RNA was prepared from 5-DAG seedlings using RNAiso Plus reagent (Takara) and a DNase Kit (Takara), and RT was performed with PrimeScript II First Strand cDNA Synthesis Kit (Takara) with random primers. The RT-qPCR was performed with a TB Green Premix EX Taq II (Tli RNaseH Plus) Kit (Takara). Relative gene expression levels were calculated using the △△CT method and normalized relative to the expression levels of Actin7. Four independent biological replicates were performed. The primer sequences were listed in Supplemental Table S1.
Northern-Blot Analysis
For RNA gel blot analysis, total RNA was isolated using RNAiso Plus reagent (Takara) from the 5-DAG seedlings and digested with a DNase Kit (Takara). Thirty nanograms of total RNA was separated on 1% (w/v) agarose gels and transferred to a positively charged nylon membrane (Hybond N+; Amersham Bioscience). Biotin-labeling of oligonucleotide probes, used for detecting mature 18S, 5.8S, and 25S rRNA, were performed using a Biotin 3′ End DNA Labeling Kit (Beyotime) following the manufacturer’s instructions. RNA hybridization was performed using a NorthernMax-Gly Kit (complete Northern-Blotting Kit; Invitrogen) following the manufacturer’s instructions. The detection was performed using a Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Beyotime) following the manufacturer’s instructions. Probe sequences are listed in Supplemental Table S1.
cRT-PCR
cRT-PCR analysis was performed according to the method described in Abbasi et al. (2010). Total RNA (5 ng) was prepared from 5-DAG seedlings grown on 1/2 MS solid medium using RNAiso Plus reagent (Takara) and a DNase Kit (Takara) and circularized with T4 RNA ligase (Takara). As indicated in Figure 5A, to detect RNA containing the 35S, 33S, or 32S prerRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 25S-cRT primer, followed by PCR using 25S-cRT-F and 18S-cRT-R PCR primers. To detect RNA containing the 25S rRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 25S-cRT primer, followed by PCR using 25S-cRT-F and 25S-cRT-R PCR primers. To detect RNA containing the 27SB prerRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 25S-cRT primer, followed by PCR using 25S-cRT-F and 5.8S-cRT-R PCR primers. To detect RNA containing the 18S rRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 18S-cRT primer, followed by PCR using 18S-cRT-F and 18S-cRT-R PCR primers. To detect RNA containing the 5.8S rRNA sequence, the first-strand cDNA was synthesized with circularized RNA and the 5.8S-cRT primer, followed by PCR using 5.8S-cRT-F and 5.8S-cRT-R PCR primers. The PCR was performed with a Premix Taq (EX Taq Version 2.0; Takara). After a 40-cycle amplification, the cRT-PCR products were analyzed by 2% (w/v) agarose gels and the DNA fragments were collected and cloned into the pMD19-simple vector (Takara) and confirmed by sequencing. Three independent biological replicates were performed. The primer sequences are listed in Supplemental Table S1.
IP Assay
For detecting the PES2 substrates, the IP assay was performed in the 35S::PES2-GFP transgenic seedlings. Seedlings of 35S::PES2-GFP were ground to fine powder in liquid N. Total proteins were extracted with a Minute Total Protein Extraction Kit for Plant Tissues (SN-009; Invent Biotechnologies,) according to the user manual. The tissue powder was solubilized with native lysis buffer and then Complete Protease Inhibitor Cocktail (Roche) was added. The solution was incubated on ice for 5 min with periodic mixing, and then centrifuged at 13,000 g for 5 min at 4°C. The supernatant was incubated with anti-GFP mAb Agarose (D153-8; MBL) for 30–120 min at 4°C. The agarose was then washed 3–5 times with the cold lysis buffer (P87788; Thermo Fisher Scientific). The IP proteins of anti-GFP were detected with the MDN1 antibody by dot-immunoblot.
Yeast-2-Hybrid Assay
The GAL4 yeast-2-hybrid assay was performed according to the manufacturer’s instructions (Clontech Laboratories). The DNA fragment encoding the MDN1 C-terminal MIDAS domain was cloned into the pGADT7 prey vector. The DNA fragments encoding the full PES2 protein, the PES2 N-terminal domain (PES2-UBL), and the PES2 C-terminal domain (PES2-WD40), were cloned into the pGBKT7 bait vector. The bait and prey constructs were cotransformed into the yeast strain Y2HGold via the PEG/LiAc transformation procedure. After transformation, the yeast cells were spread on the SD/-Leu/-Trp medium and the stringent SD/-Leu/-Trp/-His/-Ade selective medium and grown under dark condition at 30°C for 3 d.
Protein Purification and Pull-Down Assay
The MIDAS-domain was fused N-terminally with a His-6-tag using the pET28a vector (Novagen). PES2 was fused N-terminally with a GST-tag using the pGEX6P-1 vector. The fusion constructs were transformed into Escherichia coli strain Rosetta DE3. Transformants (500 mL) were grown to an A600 nm value of 0.6 at 37°C and then incubated with 0.1 mm isopropyl-β-d-thiogalactopyranoside for 5–6 h at 20°C. The cells were lysed in phosphate-buffered saline (at pH 7.4; FG701-01; Transgen Biotechnology) containing protease inhibitor cocktail and lysozyme on ice by ultrasonication. After centrifugation at 13,000 g for 10 min at 4°C, the supernatants were collected. For purification of His-6-tagged MIDAS, the Ni-NTA Purification System was used according to the user instruction (K95001; Invitrogen). For GST-pull down assay, the Pierce GST protein Interaction Pull-Down Kit (P21516; Thermo Fisher Scientific) was used. The supernatants were incubated with pre-equilibrated Glutathione Agarose resin for 2 h at 4°C. Then the resin was incubated with the purified His-MIDAS protein for 2 h at 4°C. The bait-prey proteins were eluted with a Glutathione Elution buffer and then prepared for SDS-PAGE and western-blot analysis.
Bioinformatic Analysis
The amino acid sequences of MDN1 and PES2 were employed to search for counterparts in the available sequence database (http://blast.ncbi.nlm.nih.gov/). Multiple sequence alignment was performed using ClustalX (Plate-Forme de Bio-Informatique). The protein structure predictions were made using the on-line software SWISS-MODEL (https://www.swissmodel.expasy.org/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: MDN1, At1g67120; L16B, At4g18730; S13A, At4g00100; PES2, At5g15550; Actin7, At5g09810.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Embryo development is impaired in mdn1-1.
Supplemental Figure S2. Subcellular localization analysis of MDN1.
Supplemental Figure S3. 45S prerRNA VAR expression in wild type and mdn1-1.
Supplemental Figure S4. Sequence alignment analysis and predicted structure of PES2.
Supplemental Figure S5. Phenotypes of wild type and mdn1-1 grown under mannitol-treatment conditions.
Supplemental Table S1. Sequences of primers and probes used in this study.
Acknowledgments
We thank Prof. Zhao-Jun Ding (Shandong University) for generously sharing the seeds of DR5rev::GFP.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant 31500257) and the Shandong Academy of Agricultural Sciences (Agricultural Scientific and Technological Innovation Project no. CXGC2018E13).
References
- Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB (2010) APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J 64: 960–976 [DOI] [PubMed] [Google Scholar]
- Ahn CS, Cho HK, Lee DH, Sim HJ, Kim SG, Pai HS (2016) Functional characterization of the ribosome biogenesis factors PES, BOP1, and WDR12 (PeBoW), and mechanisms of defective cell growth and proliferation caused by PeBoW deficiency in Arabidopsis. J Exp Bot 67: 5217–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Bassler J, Beckmann R, Hurt E (2016) Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat Struct Mol Biol 23: 37–44 [DOI] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010) The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38: 712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Lee W, Stier G, Clark SA, et al. (2014) A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 207: 481–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Heazlewood JL, Ito J, Millar AH (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 7: 347–369 [DOI] [PubMed] [Google Scholar]
- Cech TR. (2000) Structural biology. The ribosome is a ribozyme. Science 289: 878–879 [DOI] [PubMed] [Google Scholar]
- Chantha SC, Matton DP (2007) Underexpression of the plant NOTCHLESS gene, encoding a WD-repeat protein, causes pleitropic phenotype during plant development. Planta 225: 1107–1120 [DOI] [PubMed] [Google Scholar]
- Chantha SC, Gray-Mitsumune M, Houde J, Matton DP (2010) The MIDASIN and NOTCHLESS genes are essential for female gametophyte development in Arabidopsis thaliana. Physiol Mol Biol Plants 16: 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Suzuki H, Kobayashi Y, Wang AC, DiMaio F, Kawashima SA, Walz T, Kapoor TM (2018) Structural insights into Mdn1, an essential AAA protein required for ribosome biogenesis. Cell 175: 822–834 e818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HK, Ahn CS, Lee HS, Kim JK, Pai HS (2013) Pescadillo plays an essential role in plant cell growth and survival by modulating ribosome biogenesis. Plant J 76: 393–405 [DOI] [PubMed] [Google Scholar]
- Degenhardt RF, Bonham-Smith PC (2008) Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol 147: 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E (2004) Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem 279: 55411–55418 [DOI] [PubMed] [Google Scholar]
- Garbarino JE, Gibbons IR (2002) Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez JJ, Fernández-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J (2014) Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet 10: e1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue MF, Chakraborty A, Gleizes PE (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, et al. (2005) Mammalian WDR12 is a novel member of the Pes1–Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol 170: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M (2007) Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Sáez-Vasquez J, Echeverría M, Nakagawa T, et al. (2007) Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J 49: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Koš-Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R, et al. (2016) Architecture of the 90S pre-ribosome: A structural view on the birth of the eukaryotic ribosome. Cell 166: 380–393 [DOI] [PubMed] [Google Scholar]
- Kressler D, Roser D, Pertschy B, Hurt E (2008) The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J Cell Biol 181: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, Bassler J (2012) The power of AAA-ATPases on the road of pre-60S ribosome maturation—molecular machines that strip pre-ribosomal particles. Biochim Biophys Acta 1823: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL. (2015) Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22: 11–19 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D (2012) Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol 19: 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Huang JG, Yu SW, Li YY, Sun P, Wu CA, Zheng CC (2016a) Arabidopsis YL1/BPG2 is involved in seedling shoot response to salt stress through ABI4. Sci Rep 6: 30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Yu SW, Li K, Huang JG, Wang XJ, Zheng CC (2016b) The mutation of Glu at amino acid 3838 of AtMDN1 provokes pleiotropic developmental phenotypes in Arabidopsis. Sci Rep 6: 36446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Ishida T, Yanagisawa S (2018) Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30: 209–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, Hurt E (2014) Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL Jr (2005) Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol Cell Biol 25: 10419–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. (2008) Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133: 627–639 [DOI] [PubMed] [Google Scholar]
- Oakes ML, Siddiqi I, French SL, Vu L, Sato M, Aris JP, Beyer AL, Nomura M (2006) Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol Cell Biol 26: 3889–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I, Lin CY, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M (2017) Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29: 2644–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm D, Simm S, Darm K, Weis BL, Ruprecht M, Schleiff E, Scharf C (2016) Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol 13: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, Volarević S (2018) Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat Rev Cancer 18: 51–63 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. (2010) Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Zhu J, Wu Q, Wang Q, Li X, Yao D, Jin Y, Wang G, Wang G, Song R (2016) Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol 170: 971–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman N, Weir E, Müller S (2016) The AAA ATPase MDN1 acts as a SUMO-targeted regulator in mammalian pre-ribosome remodeling. Mol Cell 64: 607–615 [DOI] [PubMed] [Google Scholar]
- Romes EM, Sobhany M, Stanley RE (2016) The crystal structure of the ubiquitin-like domain of ribosome assembly factor Ytm1 and characterization of its interaction with the AAA-ATPase midasin. J Biol Chem 291: 882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam T, Abbasi N, Kim HS, Kim HB, Park NI, Park GT, Oh SA, Park SK, Muench DG, Choi Y, et al. (2017) An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J 92: 1092–1105 [DOI] [PubMed] [Google Scholar]
- Simm S, Fragkostefanakis S, Paul P, Keller M, Einloft J, Scharf KD, Schleiff E (2015) Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum. Bioinform Biol Insights 9: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski P, Urnavicius L, Boland A, Fagiewicz R, Busselez J, Papai G, Schmidt H (2018) The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1. eLife 7: e39163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Karbstein K (2009) Powering through ribosome assembly. RNA 15: 2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K (2012) A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi D, Byrne ME (2011) Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65: 269–281 [DOI] [PubMed] [Google Scholar]
- Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL Jr (2008) Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol Biol Cell 19: 2844–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D (2015) Building a plant: Cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska-Placzek M (2017) Comparison of preribosomal RNA processing pathways in yeast, plant and human cells—focus on coordinated action of endo- and exoribonucleases. FEBS Lett 591: 1801–1850 [DOI] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E (2009) Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138: 911–922 [DOI] [PubMed] [Google Scholar]
- Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E (2014) The 60S associated ribosome biogenesis factor LSG1-2 is required for 40S maturation in Arabidopsis thaliana. Plant J 80: 1043–1056 [DOI] [PubMed] [Google Scholar]
- Weis BL, Kovacevic J, Missbach S, Schleiff E (2015a) Plant-specific features of ribosome biogenesis. Trends Plant Sci 20: 729–740 [DOI] [PubMed] [Google Scholar]
- Weis BL, Palm D, Missbach S, Bohnsack MT, Schleiff E (2015b) atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr., Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G, Yusupov M (2014) High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem 83: 467–486 [DOI] [PubMed] [Google Scholar]
- Zemp I, Kutay U (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]