A BES/BZR-type transcription factor, TaBZR2, activates TaGST1 to scavenge reactive oxygen species and mediates crosstalk between brassinosteroids and drought signaling pathways.
Abstract
BRI1-EMS suppressor (BES)/brassinazole-resistant (BZR) family transcription factors are involved in a variety of physiological processes, but the biological functions of some BES/BZR transcription factors remain unknown; moreover, it is not clear if any of these proteins function in the regulation of plant stress responses. Here, wheat (Triticum aestivum) brassinazole-resistant 2 (TaBZR2)-overexpressing plants exhibited drought tolerant phenotypes, whereas downregulation of TaBZR2 in wheat by RNA interference resulted in elevated drought sensitivity. electrophoretic mobility shift assay and luciferase reporter analysis illustrate that TaBZR2 directly interacts with the gene promoter to activate the expression of T. aestivum glutathione s-transferase-1 (TaGST1), which functions positively in scavenging drought-induced superoxide anions (O2−). Moreover, TaBZR2 acts as a positive regulator in brassinosteroid (BR) signaling. Exogenous BR treatment enhanced TaBZR2-mediated O2− scavenging and antioxidant enzyme gene expression. Taken together, we demonstrate that a BES/BZR family transcription factor, TaBZR2, functions positively in drought responses by activating TaGST1 and mediates the crosstalk between BR and drought signaling pathways. Our results thus provide new insights into the mechanisms underlying how BES/BZR family transcription factors contribute to drought tolerance in wheat.
As sessile organisms, plants encounter various environmental stresses, such as drought and salt stresses, that severely affect growth and productivity (Jeong et al., 2010; Takasaki et al., 2010; Yu et al., 2013; Zhang et al., 2017; Qi et al., 2018). Plants have developed elaborate mechanisms to cope with such challenges via changes at the physiological and biochemical levels as well as at the cellular and molecular levels (Yamaguchi-Shinozaki and Shinozaki, 2006; Zhang et al., 2012b; Yu et al., 2013; Liu et al., 2018; Qi et al., 2018). These adaptive strategies are highly sophisticated processes regulated by an intricate signaling network and by orchestrating expression of stress-responsive genes (Ramegowda et al., 2015; Liu et al., 2018; Wu et al., 2018). Stress-responsive genes can be classified into two groups: effector genes and regulatory genes (Huang et al., 2013; Liu et al., 2014; Kidokoro et al., 2015).
Effector genes encode enzymes required for osmoprotectants, late embryogenesis abundant proteins, aquaporin proteins, chaperones, and detoxification enzymes, which protect cell membrane integrity, control ion balances, and scavenge reactive oxygen species (ROS; Huang et al., 2013; Liu et al., 2014). Regulatory genes encode protein kinases and transcription factors, which function in signal perception, signal transduction, and transcriptional regulation of gene expression (Huang et al., 2013; Liu et al., 2014). Transcription factors, such as the dehydration responsive element-binding (DREB)/C-repeat binding factor (CBF) family (Liu et al., 2013b, 2018; Kidokoro et al., 2015), APETALA2/ethylene responsive factor family (Seo et al., 2010; Rong et al., 2014), myeloblastosis family (Li et al., 2009; Seo et al., 2009, 2011), NAC (NAM, ATAF, and CUC) family (Hao et al., 2011; Mao et al., 2015; Wang et al., 2018), WRKY family (Zhou et al., 2008; Wang et al., 2015), and basic Leu zipper family (Tang et al., 2012; Song et al., 2013; Ma et al., 2018), can bind to cis-regulatory elements to modulate the expression of various downstream genes, ultimately regulating adaptive responses to unfavorable environmental conditions.
BRI1-EMS suppressor (BES)/brassinazole-resistant (BZR) transcription factors form a small plant-specific gene family (Wang et al., 2002; Yin et al., 2005; Bai et al., 2007). Members of the BES/BZR family of transcription factors, which function redundantly in BR response, are key components of the BR signaling pathway (Wang et al., 2002; Yin et al., 2002, 2005; Li et al., 2010). BES1 and BZR1 are two well-known BES/BZR family members that function as positive regulators in Arabidopsis (Arabidopsis thaliana) BR signaling. Gain-of-function mutants bes1-D and bzr1-1D can partially suppress the dwarf phenotypes of brassinosteroid insensitive1 (bri1) and are resistant to the BR biosynthesis inhibitor brassinazole (BRZ; Wang et al., 2002; Yin et al., 2002). OsBZR1 functions as a positive regulator in the rice (Oryza sativa) BR signaling pathway, and 14-3-3 proteins inhibit OsBZR1 nuclear accumulation to negatively regulate BR signaling (Bai et al., 2007). GmBEHL1 mediates the crosstalk between BR signaling and nodulation signaling pathways that negatively regulates symbiotic nodulation in soybean (Glycine max; Yan et al., 2018).
In addition to their essential roles in BR signaling, BES/BZR family members have been shown to function in Arabidopsis responses to drought and to stress from both high and low temperatures (Oh et al., 2012; Li et al., 2017; Ye et al., 2017). The drought-induced transcription factor RD26 mediates crosstalk between BR and drought pathways via reciprocal inhibition between RD26 and BES1 transcriptional activities (Ye et al., 2017). BZR1-PIF4 interaction integrates BR signaling and environmental signals (Oh et al., 2012). BZR1 positively regulates Arabidopsis freezing tolerance via DREB/CBF-dependent and DREB/CBF-independent pathways (Li et al., 2017).
Bread wheat (Triticum aestivum) is a cereal crop that is widely grown throughout the world. Drought profoundly affects wheat growth and productivity worldwide. Although a few BES/BZR family members have been characterized in model plants, the biological functions of wheat BES/BZR family transcription factors remain largely unknown. In this study, both drought and exogenous BR treatments induced expression of a BES/BZR family transcription factor gene, TaBZR2. We then analyzed the function of TaBZR2 through generating overexpression and RNA interference (RNAi) transgenic wheat plants. Moreover, electrophoretic mobility shift assay (EMSA) and luciferase (LUC) reporter analysis illustrated that TaBZR2 functions positively in drought tolerance by directly upregulating the transcriptional activity of T. aestivum glutathione s-transferase-1 (TaGST1). Furthermore, TaBZR2 acts as a link between BR and drought signaling pathways.
RESULTS
Identification of Stress-Responsive BES/BZR Transcription Factors in Wheat
In previous whole-transcriptome analyses of drought and BR on wheat, the transcript of TraesCS3D02G139300.1 was induced by both drought and exogenous BR treatments and exhibited the greatest stress-inducible gene response (Supplemental Table S1). Sequence alignment analysis revealed that this transcript encodes a protein that shows high sequence similarity with rice BES/BZR transcription factor OsBZR2 (∼87%; https://blast.ncbi.nlm.nih.gov/Blast.cgi). We thus named this transcript TaBZR2 and selected it for further analysis of its role in drought responses.
Protein structure analysis illustrated that the TaBZR2 amino acid sequence contained an N-terminal DNA binding domain and 29 putative glycogen synthase kinase3 (GSK3)-like kinase phosphorylation sites (S/TXXXS/T) but no putative PEST domain (a region rich in Pro, Glu, Ser, and Thr; http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind) or potential 14-3-3 binding site (RXXXpSXP, where X is any amino acid, R is Arg, pS is phospho-Ser, and P is Pro) was identified (Rechsteiner and Rogers, 1996; Wang et al., 2002; Yin et al., 2002; Bai et al., 2007; Supplemental Fig. S1A). To explore the relationships among wheat BZRs and their orthologs from other plant species, a phylogenetic tree was constructed by amino acid sequence alignment. TaBZR2 was classified into subgroup V, and the BES/BZRs derived from monocots clustered separately from those of dicots, suggesting a potential functional diversity between dicot and monocot plants (Supplemental Fig. S1B).
Drought and Exogenous BR Induced TaBZR2 Expression and the Nuclear Accumulation of TaBZR2 Protein
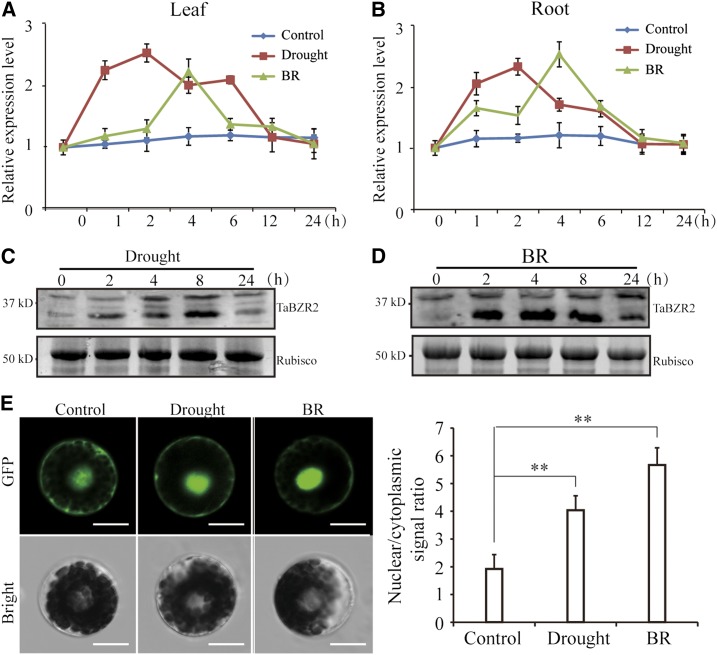
We confirmed the expression patterns of TaBZR2 in drought and BR responses by reverse transcription quantitative PCR (RT-qPCR) and immunoblot assays. Drought induced TaBZR2 expression in both shoots and roots, reaching a peak at 2 h (Fig. 1, A and B). TaBZR2 expression increased after treatment with exogenous BR and peaked at 4 h in BR-treated leaves and roots (Fig. 1, A and B). Furthermore, drought and exogenous BR treatments increased the abundance of TaBZR2 protein (Fig. 1, C and D). To better understand the biological functions of TaBZR2, we investigated the subcellular localization of TaBZR2 protein in response to drought and exogenous BR treatments. The TaBZR2- GFP fluorescence signal was observed in both the cytoplasm and nucleus under unstressed conditions (Fig. 1E). Upon drought and exogenous BR treatments, TaBZR2 proteins translocated from the cytoplasm to the nucleus as shown by the nuclear/cytoplasmic signal ratio (Fig. 1E).
Figure 1.
Expression and localization of TaBZR2 in wheat under BR and drought conditions. A and B, The expression profile of TaBZR2 in 2-week–old wheat seedling leaf and root tissue under drought and BR treatments for the indicated time. Transcript levels were quantified by RT-qPCR assays. The expression of β-actin was analyzed as internal control. Each data point is the mean (±se) of three experiments. C and D, Protein level of TaBZR2 in 2-week–old wheat seedlings after drought and BR treatments for the indicated time. Total proteins were extracted and subjected to immunoblot analysis with anti-TaBZR2 antibodies. Rubisco was used as a loading control. E, Localization of TaBZR2 protein under drought and BR conditions. The nuclear/cytoplasmic signal ratio represents nuclear-accumulated TaBZR2 versus cytoplasmic-accumulated TaBZR2. Images were observed under a laser scanning confocal microscope. Scale bar = 12 μm. Each data point is the mean (±se) of 10 biological replicates (**P < 0.01; Student’s t test).
Overexpression of TaBZR2 Significantly Improves Drought Tolerance in Transgenic Wheat
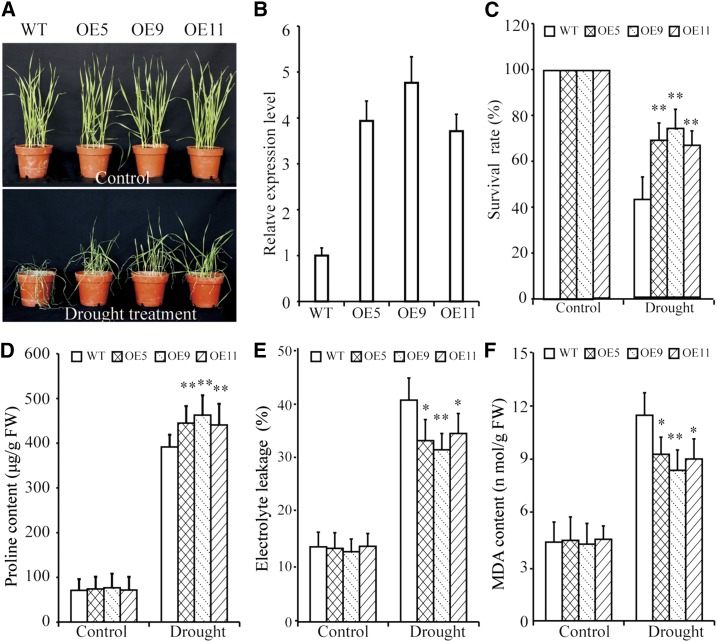
To investigate the drought tolerance associated with TaBZR2, we generated transgenic bread wheat plants on the wheat cv ‘Fielder’ background in which TaBZR2, driven by the maize (Zea mays) Ubiquitin promoter, was overexpressed. Three independent transgenic lines that exhibited high TaBZR2 expression level based on RT-qPCR assays were chosen for further analysis (Fig. 2B). No differences were observed between the TaBZR2-overexpressing (OE5, OE9, and OE11) and wild-type plants under normal growth conditions (Fig. 2A). Drought treatment caused obvious differences in growth and physiology of both TaBZR2-overexpressing and wild-type plants. Upon drought treatment, compared with control plants, TaBZR2-overexpressing plants had significantly delayed leaf rolling and higher survival rates (Fig. 2, A and C). Moreover, the Pro contents were significantly higher in TaBZR2-overexpressing plants than in wild-type plants under drought conditions (Fig. 2D). The TaBZR2-overexpressing plants had significantly lower electrolyte leakage levels and malondialdehyde (MDA) contents compared to wild-type plants under drought conditions (Fig. 2, E and F). Thus, TaBZR2 regulated physiological processes that improve the drought tolerance of transgenic wheat plants.
Figure 2.
TaBZR2-overexpressing wheat plants exhibit improved drought tolerance. A, Phenotypes of TaBZR2-overexpressing (OE5, OE9, and OE11) and wild-type wheat plants under well-watered and drought conditions. B, RT-qPCR analysis of TaBZR2 gene expression in TaBZR2-overexpressing and wild-type plants. The expression of β-actin was analyzed as an internal control. Each data point is the mean (±se) of three experiments. C, Survival rate of the control and water-stressed plants (without irrigation for 21 d). D, Pro content in seedlings under normal and drought conditions. E, Electrolyte leakage in seedlings under normal and drought conditions. F, MDA content in seedlings under normal and drought conditions. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). The asterisks indicate significant differences between TaBZR2-overexpressing and wild-type wheat plants (Student’s t test, *P < 0.05 and **P < 0.01). WT, wild type.
Suppression of TaBZR2 Enhances Drought Sensitivity in Wheat
To further explore the function of TaBZR2 in drought responses, we produced two independent TaBZR2-RNAi lines (Ri3 and Ri7) and determined the expression of TaBZR2 using RT-qPCR assays. The expression level of TaBZR2 decreased in the two lines (Fig. 3B; Supplemental Fig. S2), implying that TaBZR2 was successfully suppressed. There were no obvious differences in the growth performance and physiology between TaBZR2-RNAi and wild-type plants under normal growth conditions (Fig. 3A). However, upon drought treatment, the survival rates were significantly lower in TaBZR2-RNAi plants than in wild-type plants under drought conditions (Fig. 3C). Moreover, drought-treated TaBZR2-RNAi lines had significantly lower Pro contents, higher electrolyte leakage levels, and higher MDA contents compared to wild-type plants under drought conditions (Fig. 3, D–F).
Figure 3.
TaBZR2-RNAi wheat plants show enhanced drought sensitivity. A, Phenotypes of TaBZR2-RNAi (Ri3 and Ri7) and wild-type wheat plants under well-watered and drought conditions. B, RT-qPCR analysis of TaBZR2 gene expression in TaBZR2-RNAi and wild-type plants. The expression of β-actin was analyzed as an internal control. Each data point is the mean (±se) of three experiments. C, Survival rate of the control and water-stressed plants (without irrigation for 18 d). D, Pro content in seedlings under normal and drought conditions. E, Electrolyte leakage in seedlings under normal and drought conditions. F, MDA content in seedlings under normal and drought conditions. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). The asterisks indicate significant differences between TaBZR2-RNAi and wild-type wheat plants (Student’s t test, *P < 0.05 and **P < 0.01). WT, wild type.
TaBZR2 Positively Regulates the Expression of Multiple Stress-Related Genes
To explore how TaBZR2 contributes to drought tolerance, we performed RNA sequencing (RNA-Seq) assays to evaluate the differential gene expression between TaBZR2-overexpressing and wild-type wheat plants under both normal and drought conditions. As shown in Figure 4A, using a threshold of a 2-fold change and a Student’s t test significance cutoff of P < 0.05, a comparison of the RNA-Seq data from TaBZR2-overexpressing and wild-type plants under normal conditions identified 1,399 upregulated and 1,064 downregulated genes in TaBZR2-overexpressing plants (TaBZR2-OEN [overexpressing normal]) compared with those in wild-type plants (wild-type normal [WTN]). Upon drought treatment, the expression of 728 and 1,496 genes in the TaBZR2-overexpressing plants (TaBZR2-OED [overexpressing drought] ) was up- or downregulated, respectively, compared with that in wild-type plants (wild-type drought [WTD]). In total, 20,224 differentially expressed genes (DEGs) were identified between drought-treated and normal growth wild-type plants (WTD/WTN). Cluster and Venn diagram analyses revealed that the expression patterns of all DEGs in TaBZR2-OED compared with that in WTD (TaBZR2-OED/WTD) did not significantly overlap with TaBZR2-OEN compared with WTN (TaBZR2-OEN/WTN), or WTD compared with WTN (WTD/WTN). These results demonstrated that TaBZR2 significantly affects the global gene expression profile in wheat, indicating that unknown mechanisms may underlie the drought tolerance of transgenic wheat.
Figure 4.
Analysis of the expression levels of TaBZR2 downstream genes. A, Venn diagrams comparing the up- and downregulated genes between wild-type plants under normal and drought conditions (WTN and WTD), and TaBZR2-overexpressing and wild-type plants under normal (TaBZR2-OEN/WTN) and drought conditions (TaBZR2-OED/WTD). B, Functional categorization analysis of candidate TaBZR2 target genes in biological process under drought conditions. C, The expression levels of drought-responsive genes in TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants. Two-week–old wheat seedlings treated with 15% (w/v) PEG 6000 for 6 h were used for RNA isolation. Transcript levels were quantified by RT-qPCR assays, and the expression of β-actin was used as an internal control. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). WT, wild type.
Gene ontology (GO) analysis revealed that the DEGs between the drought-treated TaBZR2-overexpressing and wild-type plants were significantly enriched in biological process categories including “response to abiotic stimulus,” “response to water stress,” and “regulation of metabolic and biosynthetic processes” (Fig. 4B). Interestingly, we found that the expressions of a range of well-known stress-related genes were among the upregulated DEGs for the drought-treated TaBZR2-overexpressing plants (Supplemental Table S2). Note that we also used RT-qPCR assays to successfully verify the upregulated expression trends for the genes identified from the RNA-Seq data, including TaGST1, TaLEA3 (late embryogenesis abundant), TaDHN3 (dehydration-stress inducible protein), T. aestivum Δ-1-pyrroline-5-carboxylate synthetase, TaPOD21 (peroxidase), and T. aestivum sucrose non-fermenting1-type Ser/Thr protein kinase (Fig. 4C). Consistent with a direct functional impact of TaBZR2 on the expression of these known stress-related genes, we also used RT-qPCR to examine the expression of these genes in the aforementioned drought-treated TaBZR2-RNAi plants and found that their expression was significantly reduced compared to both drought-treated TaBZR2-RNAi and wild-type plants (Fig. 4C). GST genes, encoding ROS-scavenging enzymes, function in protecting plants against oxidative damage under stress conditions (Jha et al., 2011; Rong et al., 2014). Δ-1-pyrroline-5-carboxylate synthetase is the key enzyme for Pro synthesis (Yoshiba et al., 1995; Zhuo et al., 2018). Dehydrins are responsive to various environmental stresses and exhibit multiple biochemical activities, such as buffering water, sequestering ions, stabilizing membranes, or acting as chaperones (Kovacs et al., 2008; Tang et al., 2012; Rong et al., 2014; Zhuo et al., 2018). Suc non-fermenting-1-related protein kinase2 is implicated in stress signaling transduction via abscisic acid-dependent and -independent pathways (Yoshida et al., 2002; Zhang et al., 2011), TaBZR2 could modulate the expression of numerous stress-responsive genes under drought conditions, contributing to the drought tolerance of the transgenic wheat.
TaBZR2 Functions Positively in Scavenging Drought-Induced Superoxide Anions
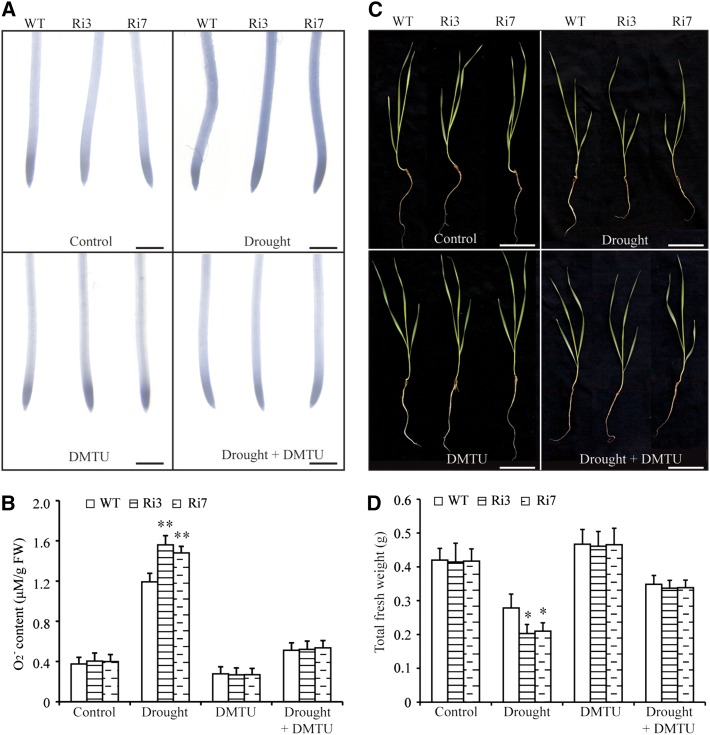
Environmental stimuli, including drought, salt, and high and low temperatures, induce the accumulation of toxic ROS, including H2O2 and superoxide anions (O2−), which, if not controlled, can eventually lead to oxidative damage (Dat et al., 2000; Wang et al., 2017). TaBZR2 has a role in activating antioxidant enzyme gene expression. To investigate whether TaBZR2 participates in scavenging ROS, we analyzed the ROS contents between TaBZR2-RNAi and wild-type wheat lines under normal and drought conditions. There was no significant difference in H2O2 accumulation between TaBZR2-RNAi and wild-type wheat lines under unstressed and drought conditions (Supplemental Fig. S3). The O2− contents of TaBZR2-RNAi and wild-type wheat lines were similar under unstressed conditions (Fig. 5, A and B). Nevertheless, the O2− contents were significantly higher in the TaBZR2-RNAi plants than in the wild-type plants under drought conditions (Fig. 5, A and B). 1,3-dimethyl-2-thiourea (DMTU) acts as a O2− scavenging reagent (Lv et al., 2018). When DMTU was added to the growth medium to reduce O2−, the O2− contents of TaBZR2-RNAi wheat lines recovered to a level similar to that of the wild-type plants (Fig. 5, A and B).
Figure 5.
TaBZR2 functions positively in scavenging drought-induced O2−. A, NBT staining in primary root tips of TaBZR2-RNAi and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 1 mm of DMTU, or medium containing 15% (w/v) PEG 6000 + 1 mm of DMTU for 72 h. The strength of color shows the concentration of O2− in the root tips. Scale bar = 1 mm. B, Measurements of the O2− contents of TaBZR2-RNAi and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 1 mm of DMTU, or medium containing 15% (w/v) PEG 6000 + 1 mm of DMTU for 72 h. Each data point is the mean (±se) of six biological replicates. The asterisks indicate significant differences between TaBZR2-RNAi and wild-type wheat plants (Student’s t test, **P < 0.01). C, Phenotypes of TaBZR2-RNAi and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% PEG 6000, medium containing 1 mm of DMTU, or medium containing 15% (w/v) PEG 6000 + 1 mm of DMTU. Scale bar = 2 cm. D, Measurement of the total fresh weight of TaBZR2-RNAi and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 1 mm of DMTU, or medium containing 15% (w/v) PEG 6000 + 1 mm of DMTU. Each data point is the mean (±se) of six biological replicates. The asterisks indicate significant differences between TaBZR2-RNAi and wild-type wheat plants (Student’s t test, *P < 0.05). WT, wild type.
To investigate whether the TaBZR2-mediated O2− scavenging was associated with the positive role of TaBZR2 in drought responses, we compared the biomass of TaBZR2-RNAi wheat plants with that of the wild-type wheat plants grown on half-strength Hoagland’s nutrient solution supplemented with different concentrations of polyethylene glycol (PEG) 6000 and DMTU (0, 15% [w/v] PEG 6000, 1 mm of DMTU, and 15% [w/v] PEG 6000 + 1 mm of DMTU). Biomass was similar for the wild-type and TaBZR2-RNAi plants grown on half-strength Hoagland’s nutrient solution containing 0 and 1 mm of DMTU (Fig. 5, C and D). However, biomass was significantly larger in the wild-type plants than in the TaBZR2-RNAi plants under drought conditions (Fig. 5, C and D). DMTU treatment can partially suppress drought (15% [w/v] PEG 6000)-induced biomass reduction. Importantly, the biomass of TaBZR2-RNAi plants was comparable with that of the wild-type plants grown on half-strength Hoagland’s nutrient solution containing 15% (w/v) PEG 6000 and 1 mm of DMTU (Fig. 5, C and D). These results indicate that TaBZR2 has a role in scavenging O2− to alleviate drought stress.
TaBZR2 Functions as a Positive Regulator of TaGST1 Expression by Binding to Its Promoter and Activating Transcription
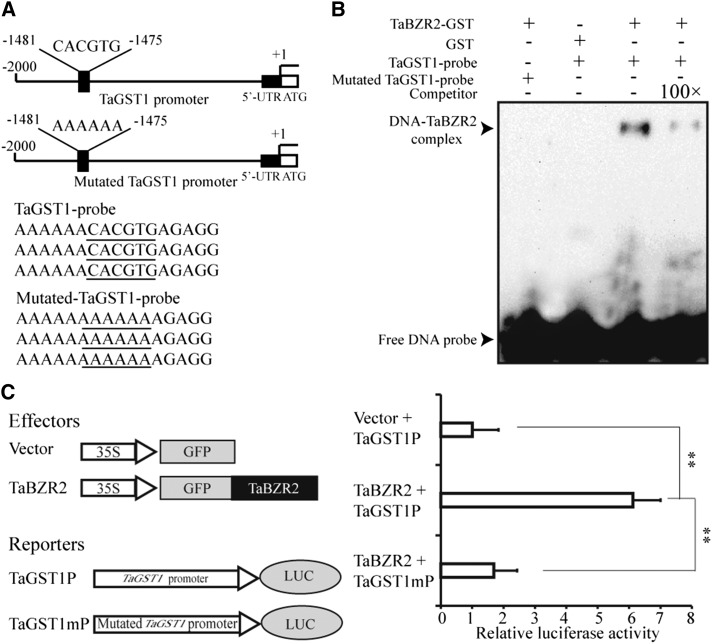
RNA-Seq and RT-qPCR analyses both indicated that the expression of TaGST1 is upregulated by TaBZR2 overexpression, so the transcription of this gene may be activated by TaBZR2. Previous studies have revealed that BES/BZR family transcription factors can bind to E-box (5′-CANNTG-3′) cis-elements to regulate the expression of target genes; we detected 10 E-box cis-elements in the TaGST1 promoter. We thus used EMSAs to investigate whether TaBZR2 can directly bind to the TaGST1 promoter in vitro. The EMSAs showed that the TaBZR2-GST fusion protein was able to bind to the TaGST1 promoter; no such binding was observed for the control GST protein (Fig. 6, A and B). Further, the observed binding to the biotin-labeled target sequences was dramatically reduced when unlabeled competitor target DNA sequences were added, and no binding was detected when adding the mutated biotin-labeled TaGST1 probes (Fig. 6, A and B). Having determined that TaBZR2 can bind the TaGST1 promoter in vitro, we next used a wheat protoplast transient expression system to assess whether this binding can drive TaGST1 gene expression in vivo. A pGreen II 0800 vector harboring a LUC reporter gene driven by the TaGST1 promoter (∼2,000 bp) was cotransformed into wheat protoplasts transfected with an empty pJIT16318 vector or a pJIT16318-TaBZR2 vector. Compared with the empty-vector control samples, the protoplasts expressing TaBZR2 exhibited significantly increased expression of the reporter (Fig. 6C). To further test if the activation effect of TaBZR2 on the TaGST1 was through binding to the E-box (CACGTG, −1,475 to −1,481), the TaGST1 promoter containing the mutated E-box was inserted into the pGreen II 0800 vector and coexpressed with the pJIT16318-TaBZR2 vector in wheat protoplasts. The results demonstrated that the E-box mutation (AAAAAA, −1,475 to −1,481) disrupted TaBZR2-mediated activation of the TaGST1 promoter (Fig. 6C), indicating that the TaBZR2 transcription factor is a positive regulator of TaGST1 expression.
Figure 6.
TaBZR2 directly regulates the expression of TaGST1. A, The diagram shows the structure of the TaGST1 promoter. The sequences represent TaGST1 probe sequences. The underlined sequences indicated the core elements or mutated core elements in the TaGST1 probe. B, EMSA of TaBZR2 binding to the promoter of TaGST1. Biotin-labeled TaGST1 probes (normal and mutated) were incubated with GST or GST-TaBZR2 protein. 100× competitor fragments were added to analyze the specificity of binding. C, TaBZR2 increases TaGST1 promoter activity in wheat protoplasts. TaBZR2 was cotransfected with either TaGST1 promoter or mutated TaGST1 promoter. The LUC to REN ratio is shown and indicates the activity of the transcription factors on the expression level of the promoters. Each data point is the mean (±se) of 10 biological replicates (**P < 0.01; Student’s t test).
Overexpression of TaGST1 Significantly Improved Drought Tolerance in Transgenic Wheat by Reducing O2− Contents
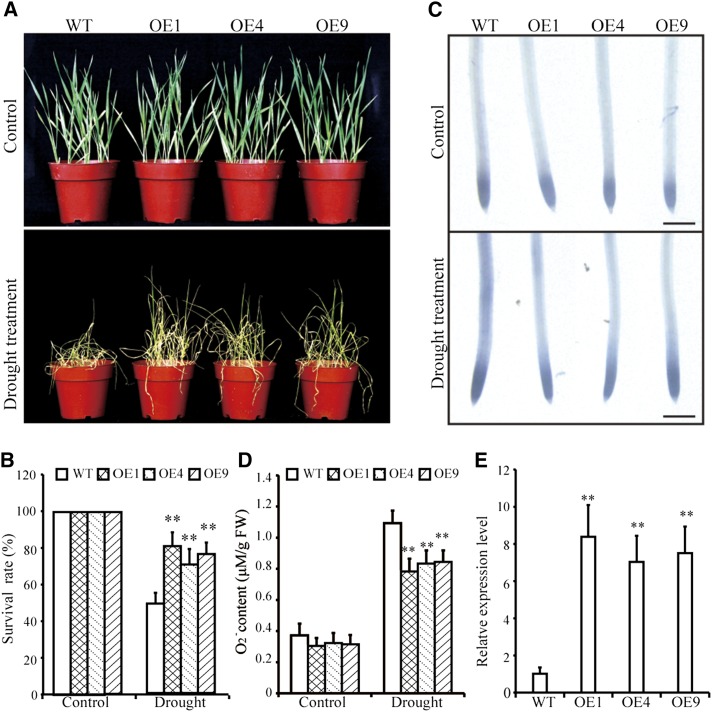
To investigate the function of TaGST1 in the drought response, we generated transgenic wheat plants that overexpressed TaGST1 under the control of the maize Ubiquitin promoter. Three independent homozygous T3 transgenic lines with relatively high expression of TaGST1 were selected for additional phenotypic analyses (Fig. 7E). Under normal growth conditions, there were no notable differences in plant growth or physiology between TaGST1-overexpressing (OE1, OE4, and OE9) and wild-type plants. However, upon drought treatment, the survival rate of TaGST1-overexpressing plants was significantly higher than that of wild-type plants (Fig. 7, A and B). Moreover, the drought-treated TaGST1-overexpressing plants had significantly lower O2− content compared to wild-type plants (Fig. 7, C and D).
Figure 7.
TaGST1 overexpression promotes drought tolerance in transgenic wheat. A, Phenotypes of TaGST1-overexpressing and wild-type plants under normal and drought conditions. B, Survival rate of control and water-stressed plants (15% [w/v] PEG 6000 treatment for 14 d). Each data point is the mean (±se) of three experiments (10 seedlings per experiment). C, NBT staining in primary root tip of TaGST1-overexpressing and wild-type seedlings with 0 or 15% (w/v) PEG 6000 treatment for 72 h. The strength of color shows the concentration of O2− in the root tips. Scale bar = 1 mm. D, Measurements of the O2− contents of TaGST1-overexpressing and wild-type plants under normal and drought conditions. Each data point is the mean (±se) of six biological replicates. E, RT-qPCR analysis of TaGST1 gene expression in TaGST1-overexpressing and wild-type wheat seedlings. The expression of β-actin was used as an internal control. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). The asterisks indicate significant differences between TaGST1-overexpressing and wild-type plants (Student’s t test, **P < 0.01). WT, wild type.
TaBZR2 Is a Positive Regulator in the BR Signaling Pathway
To obtain more detailed evidence for the role of TaBZR2 in BR responses, we transformed the BR-insensitive mutant bri1-5 with a TaBZR2 overexpression construct under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Two independent homozygous T3 transgenic lines (35S:TaBZR2/bri1-5-B3 and -B7) that strongly expressed TaBZR2 were selected for further phenotypic analysis. Overexpression of TaBZR2 partially suppressed the dwarf phenotypes of bri1-5 mutant plants (Supplemental Fig. S4, A and B). Compared with bri1-5 mutant plants, 35S:TaBZR2/bri1-5 transgenic plants had enhanced tolerance to the BR biosynthetic inhibitor BRZ (Supplemental Fig. S4B). In addition, compared with bri1-5 mutant plants, 35S:TaBZR2/bri1-5 transgenic plants showed reduced expression of the BR biosynthesis genes Constitutive Photomorphogenesis and Dwarfism and DWARF4 and increased expression of the BR signaling gene the small auxin up RNAs (SAUR)-AC (Supplemental Fig. S4C).
To obtain further insights into the role of TaBZR2 in the wheat BR signaling pathway, we investigated the BR sensitivity of TaBZR2 transgenic wheat plants. TaBZR2-overexpressing and TaBZR2-RNAi wheat plants exhibited altered BR sensitivity as indicated by their root length in the absence or presence of BR. In the absence of BR, there was no significant difference in root lengths between TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants (Fig. 8A). However, in the presence of BR, the root lengths of TaBZR2-overexpressing lines were shorter than that of wild-type plants. Moreover, compared with wild-type plants, TaBZR2-RNAi plants exhibited BR-insensitive phenotypes with longer roots (Fig. 8A). Previous studies have shown that BES/BZR family transcription factors can bind to the BR-response element (BRRE) in the promoters of feedback-regulated BR biosynthetic genes to repress their expression (He et al., 2005). EMSAs demonstrated that TaBZR2 can bind to the BRRE cis-regulatory elements in the promoter of the BR biosynthetic gene T. avestivum CYP90D2 (TaD2; Fig. 8B). Furthermore, RT-qPCR assays revealed that, compared with wild-type plants, TaBZR2-overexpressing plants showed reduced expression of the BR biosynthetic genes TaD2 and TaDWARF, whereas TaBZR2-RNAi plants showed enhanced expression of the BR biosynthetic genes TaD2 and TaDWARF in the absence and presence of BR (Fig. 8C). The TaBZR2-modulated inhibition of TaD2 and TaDWARF expressions were larger in the presence of BR than in the absence of BR (Fig. 8C). Our data suggest that TaBZR2 functions as a positive regulator in BR signaling.
Figure 8.
TaBZR2 is a positive regulator in the BR signaling pathway. A, Phenotypes of TaBZR2-overexpressing (OE5, OE9, and OE11), TaBZR2-RNAi (Ri3 and Ri7), and wild-type wheat plants grown in half-strength Hoagland’s liquid medium or medium containing 1 μm of BR. Scale bar = 2 cm. Root length of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants grown on half-strength Hoagland’s medium that contained different concentrations of BR (0, 0.25, or 1 μm) in the light for 7 d. Each data point is the mean (±se) of three experiments (20 seedlings per experiment). The asterisks indicate significant differences between TaBZR2 transgenic (TaBZR2-overexpressing lines and TaBZR2-RNAi lines) and wild-type plants (Student’s t test, *P < 0.05). B, EMSA of TaBZR2 binding to the BRREs in the promoter of TaD2. Biotin-labeled BRRE probes (normal and mutated) were incubated with GST or GST-TaBZR2 protein. 100× competitor fragments were added to analyze the specificity of binding. C, The expression levels of BR biosynthetic genes in TaBZR2 transgenic (TaBZR2-overexpressing lines and TaBZR2-RNAi lines) and wild-type plants. The expression of β-actin was used as an internal control. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). WT, wild type.
TaBZR2 Is Involved in BR-mediated Drought Responses
To investigate whether TaBZR2 has a role in BR-mediated drought responses, we investigated the expression of stress-responsive genes encoding antioxidant enzymes, including TaGST1, TaPOD21, and TaDHN3, in response to BR treatment by RT-qPCR analyses. Upon exogenous BR treatment, the expression of these genes in TaBZR2-overexpression plants was enhanced compared to wild-type plants, whereas their expression in TaBZR2-RNAi plants was reduced under normal and drought conditions (Fig. 9A). In addition, drought and BR treatments enhanced the abundance of dephosphorylated TaBZR2 proteins in the TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type plants (Fig. 9B). The amounts of dephosphorylated TaBZR2 proteins were larger in TaBZR2-overexpression plants than in wild-type plants. Nevertheless, compared to wild-type plants, the amount of dephosphorylated TaBZR2 proteins in TaBZR2-RNAi plants was smaller upon drought and BR treatments (Fig. 9B). The phosphorylation status of the BES/BZR family transcription factors is usually used as the biochemical maker for BR signaling outputs (Zhang et al., 2009). Treatment of immunoprecipitated protein with protein phosphatase eliminated the slowly migrating band (Supplemental Fig. S5), strongly suggesting that the fast band is unphosphorylated and the slow band is phosphorylated TaBZR2. In addition, the O2− accumulation of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type plants was similar under normal conditions. When exposed to induced drought conditions, O2− accumulation increased in the roots of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type plants. The O2− contents of TaBZR2-overexpressing plants under drought conditions was significantly lower than that of wild-type plants, whereas the O2− accumulation was significantly higher in TaBZR2-RNAi plants than in wild-type plants (Fig. 9, C and D). Exogenous BR treatment repressed the O2− accumulation in wheat plants under normal and drought conditions. Compared to wild-type plants, BR-mediated O2− scavenging was enhanced in TaBZR2-overexpressing plants, whereas BR-mediated O2− scavenging was reduced in TaBZR2-RNAi plants under drought conditions (Fig. 9, C and D). These results indicated that TaBZR2 participates in BR-mediated O2− scavenging.
Figure 9.
TaBZR2 regulates wheat drought tolerance through the BR-dependent pathway. A, The expression levels of stress-responsive genes in TaBZR2 transgenic (TaBZR2-overexpressing lines and TaBZR2-RNAi lines) and wild-type plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 10 nm of BR, or medium containing 15% (w/v) PEG 6000 + 10 nm of BR for 6 h. Each data point is the mean (±se) of three experiments (10 seedlings per experiment). B, Protein level of TaBZR2 in TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants upon drought and BR treatments for 6 h. Total proteins were extracted and subjected to immunoblot analysis with anti-TaBZR2 antibodies. Rubisco was used as a loading control. C, NBT staining in primary root tip of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 10 nm of BR, or medium containing 15% (w/v) PEG 6000 + 10 nm of BR for 72 h. The strength of color shows the concentration of O2− in the root tips. Scale bar = 1 mm. D, Measurements of the O2− contents of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants grown in half-strength Hoagland’s liquid medium, medium containing 15% (w/v) PEG 6000, medium containing 10 nm of BR, or medium containing 15% (w/v) PEG 6000 + 10 nm of BR for 72 h. Each data point is the mean (±se) of six biological replicates. The asterisks indicate significant differences between TaBZR2 transgenic (TaBZR2-overexpressing lines and TaBZR2-RNAi lines) and wild-type plants (Student’s t test, **P < 0.01). WT, wild type.
DISCUSSION
Plant genomes have many kinds of transcription factors that function importantly in plant adaption to extreme environmental conditions (Zhang et al., 2017; Liu et al., 2018; Ma et al., 2018). BES/BZR proteins constitute another important family of plant-specific transcription factors (Wang et al., 2002; Yin et al., 2002, 2005), but comparatively little is known about their biological functions in drought responses. In this study, a drought-inducible BES/BZR-type transcription factor gene TaBZR2 was identified from wheat drought transcriptome data, and follow-up work illustrated that overexpression of TaBZR2 enhanced drought tolerance in transgenic wheat plants with larger accumulation of osmoprotectant metabolites, higher membrane stability, and lower ROS contents compared with the control plants under drought conditions, whereas TaBZR2-RNAi wheat lines exhibited the opposite trend. These results suggest that TaBZR2 functions positively in regulating drought responses in wheat.
BES/BZR family members regulate the expression of target genes by interacting with BRRE and/or or E-box cis-elements in their promoters (Goda et al., 2004; Nemhauser et al., 2004; He et al., 2005; Wang et al., 2006; Walcher and Nemhauser, 2012; Li et al., 2017). For example, BZR1 binds to BRRE and/or E-box elements in the promoters of the genes encoding CBF1/DREB1A, CBF2/DREB1B, and WRKY6 to modulate their expression, contributing to freezing tolerance in Arabidopsis (Li et al., 2017), and BES1 directly binds to the E-box element of the SAUR-AC15 promoter to enhance auxin signaling in Arabidopsis (Goda et al., 2004; Walcher and Nemhauser, 2012). Our EMSA and LUC reporter analyses demonstrated that TaBZR2 directly binds to the promoter of TaGST1 to activate its transcription. GST genes encode detoxification enzymes that function in maintaining cell redox homeostasis and protecting organisms against oxidative stress under stress conditions (Jha et al., 2011; Rong et al., 2014; Qi et al., 2018). Our data illustrated that, compared with the wild-type plants, the TaGST1-overexpressing wheat lines exhibited drought tolerance phenotypes with lower O2− contents under drought conditions, which was consistent with the positive role of TaBZR2 in scavenging drought-induced O2−. These results indicate that TaBZR2 has a role in activating TaGST1 for scavenging O2−, and consequently alleviate drought stress.
Genetic and molecular studies have greatly increased our understanding of the BR signaling pathway in model plants (Bai et al., 2007; Yu et al., 2008; Ye et al., 2011; Chen et al., 2017). BRs are perceived by a Leucine-rich repeat-receptor kinase, BRI1, and transduces the signal to activate the BES/BZR family transcription factor, which regulates the expression of a large number of genes (Wang et al., 2002; Yin et al., 2002, 2005; Bai et al., 2007; Oh et al., 2012; Jiang et al., 2013; Shimada et al., 2015; Yan et al., 2018). Consistent with the positive role of BES/BZR family members in the BR signaling pathway (Wang et al., 2002; Yin et al., 2002; Yan et al., 2018), TaBZR2 positively regulates BR signaling in wheat. Exogenous application of BR protects plants from drought stress (Kagale et al., 2007; Xia et al., 2009; Divi et al., 2010, 2016; Nawaz et al., 2017). Previous studies have shown that some components of the BR signaling pathway are involved in drought responses (Koh et al., 2007; Sahni et al., 2016). Overexpression of the Arabidopsis BR biosynthetic gene DWARF4 confers drought tolerance in Brassica napus (Sahni et al., 2016). OsGSK1 is a negative regulator of rice BR signaling: its T-DNA knockout mutants display enhanced tolerance to drought and other abiotic stresses (Koh et al., 2007). Considering that TaBZR2 functions as a positive regulator in drought responses, it is possible that TaBZR2 has a role in mediating the crosstalk between BR and drought responses. A recent study has shown that BR is involved in regulation of the accumulation of O2− (Lv et al., 2018). For example, the BR-deficient mutant det2-9 accumulates more O2− in roots (Lv et al., 2018). Our data demonstrated that the expression of some TaBZR2 target genes encoding antioxidant enzymes, including TaGST1, TaPOD21, and TaDHN3, was upregulated upon exogenous BR treatment. Furthermore, exogenous application of BR can enhance TaBZR2-mediated activation of antioxidant enzymes and scavenging of O2− under drought conditions. Our data indicated that TaBZR2 participates in BR-mediated drought response partially by reducing the accumulation of O2−.
It is worth noting that recent studies also illustrated that several components of the BR signaling pathway negatively regulate drought responses (Chen et al., 2017; Ye et al., 2017). For example, in contrast to a positive role of TaBZR2 in drought responses, AtBES1 negatively regulates plant drought tolerance (Ye et al., 2017). BES/BZR family transcription factor genes derived from monocots clustered separately from those of dicots by phylogenetic analysis. Protein structure analyses illustrated that the amino acid sequence of TaBZR2 has an N-terminal binding domain and GSK3-like kinase phosphorylation sites, but no 14-3-3 binding domain and PEST motif were identified, which was different from the well-known BES/BZR family members like AtBES1, AtBZR1, and OsBZR1 (Wang et al., 2002; Yin et al., 2002; Bai et al., 2007). Furthermore, TaBZR2 exhibited a different BR regulated mobility shift pattern with AtBZR1 and AtBES1. Previously, studies revealed that all of phosphorylated AtBZR1 and AtBES1 were dephosphorylated upon BR treatment (He et al., 2002; Yin et al., 2002), whereas BR treatment caused partially phosphorylated TaBZR2 to convert to the dephosphorylated form. These results indicate that although BES/BZR family members function positively in BR signaling, protein structural differences and the different mechanisms of action may lead to functional differences in environmental stress responses. Our study expands the known functional scope of the BES/BZR family members, and its basic insights should inform the work of both plant abiotic stress researchers and wheat breeders and biotechnologists.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wheat (Triticum aestivum) plants used for molecular analysis were grown in a greenhouse at 70% relative humidity, 25°C/23°C day/night temperatures, and long-day conditions (16-h light/8-h dark photoperiod) with a light intensity of ∼300 μmol m−2 s−1. The wheat cv ‘KeNong 199’ was used to amplify complementary DNA (cDNA) sequences of TaBZR2 and TaGST1. The wheat cv ‘Fielder’ was used as the receptor material to generate transgenic plants. To analyze the expression of TaBZR2 under abiotic stress conditions, wheat cv ‘KeNong 199’ seedlings were grown in half-strength Hoagland’s liquid medium in a greenhouse with 70% relative humidity, 25°C/23°C day/night temperatures, long-day conditions (16-h light/8-h dark photoperiod), with a light intensity of ∼400 μmol m−2 s−1 for 2 weeks. For BR and drought stress treatments, the roots of wheat seedlings were immersed in half-strength Hoagland’s solution containing 1 μm of 24-epi-brassinolide (EBL) solution (Sigma-Aldrich) and 15% (w/v) PEG 6000. Leaves and roots were sampled at 0, 1, 2, 4, 8, 12, and 24 h, and then immediately frozen in liquid N and stored at −80°C before RNA extraction. The Arabidopsis (Arabidopsis thaliana) plants were subsequently grown in a greenhouse at 23°C under long-day conditions (16-h light/8-h dark photoperiod) and a light intensity of ∼100 μmol m−2 s−1. For Arabidopsis, the seeds were germinated on half-strength Murashige and Skoog (Caisson Labs) media supplemented with 2% (w/v) Suc and grown for a week, after which the seedlings were transplanted into soil. The plants were subsequently grown in a greenhouse with 70% relative humidity, 23°C, and long-day conditions (16-h light/8-h dark photoperiod) with a light intensity of ∼100 μmol m−2 s−1 for 3 weeks. The Arabidopsis BR-insensitive mutant bri1-5 was used for transformation.
Generation of Transgenic Arabidopsis and Wheat
To generate TaBZR2 transgenic wheat plants, the coding regions (coding sequences, CDS) of TaBZR2D were cloned into the plant transformation vector pWMB110 driven by the maize (Zea mays) Ubiquitin promoter. The 198-bp TaBZR2 specific fragment was synthesized by Beijing AuGCT, which was then fused in both sense and anti-sense orientations to flank the 508-bp rice (Oryza sativa) zinc finger type family protein gene intron 6. This recombinant DNA was then inserted into the pWMB110 vector to generate the pWMB110-TaBZR2-RNAi construct. To generate TaGST1-overexpression wheat plants, the TaGST1 CDS were also inserted into the pMWB110 vector, driven by the maize Ubiquitin promoter. Genetic transformations were performed using an Agrobacterium tumefaciens-mediated transformation system. To isolate positive transgenic wheat lines, leaves of 10-d–old transgenic wheat seedlings grown in half-strength Hoagland’s nutrient solution were used for RNA isolation, and then RT- and RT-qPCR analyzes were performed. For Arabidopsis, the CDS of TaBZR2 was introduced into the plant transformation vector pBI121 under the control of the CaMV 35S promoter. The resultant constructs were confirmed by sequencing and then transformed into BR-insensitive mutant bri1-5 plants via the vacuum infiltration method (Bechtold and Pelletier, 1998). Homozygous T3 seeds of the transgenic lines were used for phenotypic analyses. Primers used in these studies are in Supplemental Table S3.
Drought Stress Treatment
For drought tolerance assays, TaBZR2 transgenic and wild-type wheat seedlings were planted in pots containing mixed soil (1:1 vermiculite/humus) and cultured normally in the greenhouse for 3 weeks (until seedlings were at the 3-leaf stage), after which these seedlings were deprived of water until significant differences in wilting were observed between transgenic and wild-type wheat plants. Three independent experiments were performed. TaGST1 transgenic and wild-type wheat seedlings were planted in pots containing mixed soil (1:1 vermiculite/humus) and cultured normally in the greenhouse for 3 weeks (until seedlings were at the 3-leaf stage), after which 15% (w/v) PEG 6000 solution was applied to the bottom of the plates for ∼14 d until significant differences in wilting were observed between transgenic and wild-type wheat plants. Three independent experiments were performed.
BR Sensitivity Assays
For BR sensitivity assays, sterilized seeds of TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants were maintained at 4°C for 1 week, after which the germinated seeds were transplanted into half-strength Hoagland’s solution containing different concentrations of EBL (0, 0.25, and 1 μm). After 7 d of growth at 23°C under long-day conditions (16-h light/8-h dark photoperiod), images were taken, and the primary root length for each seedling was evaluated using an Expression 11000XL Root System Scanning Analyzer (Epson). To perform hypocotyl elongation assays, the sterilized seeds of wild-type plants, 35S:TaBZR2/bri1-5 transgenic Arabidopsis plants, and bri1-5 plants were sown on half-strength Murashige and Skoog growth media supplemented with various concentrations (0, 0.25, and 0.5 μm) of BRZ and then kept at 4°C in the dark for 3 d. After 7 d of growth at 22°C under dark conditions, images were taken, and the lengths of hypocotyls were measured.
RNA Extraction and RT-qPCR Assays
The total RNA from Arabidopsis and wheat seedlings was extracted using Trizol reagent (TaKaRa), and their DNA was digested using RNase-free DNaseI (TaKaRa). First-strand cDNA was synthesized using a PrimeScript First-Strand cDNA Synthesis Kit (TaKaRa). RT-qPCR was performed with an ABI 7500 Real-Time PCR system (Thermo Fisher Scientific) in conjunction with SYBR (Thermo Fisher Scientific) to monitor double stranded DNA products. The reaction was conducted at 95°C for 5 min, then 42 cycles of 95°C for 15 s, 58°C to 60°C for 25 s, and 72°C for 30 s. A quantitative analysis using the 2-ΔΔCT method was subsequently performed (Le et al., 2011). Each experiment was performed with at least three independent biological replicates. For each primer pair, the amplification efficiency was checked using a melting-curve analysis. For wheat, β-actin was used as the internal control and actin was used an internal control for Arabidopsis (Liu et al., 2013a). The specific primers used for RT-qPCR are listed in Supplemental Table S3.
Immunoblot Assay
Wheat cv ‘KeNong 199’ seedlings were grown in half-strength Hoagland’s liquid medium in a greenhouse with 70% relative humidity, 25°C/23°C day/night temperatures, and long-day conditions (16-h light/8-h dark photoperiod) with a light intensity of ∼400 μmol m−2 s−1 for 2 weeks. For BR and drought stress treatments, the roots of wheat seedlings were immersed in half-strength Hoagland’s solution containing 1 μm of EBL solution and 15% (w/v) PEG 6000. Leaves were sampled at 0, 2, 4, 8, and 12 h and then used to extract total protein. Plant protein was isolated with lysis buffer (50 mm of Tris at pH 7.5, 1 mm of EDTA, 150 mm of NaCl, 10 mm of MgCl2, 10% [v/v] glycerol, 1 mm of phenylmethanesulfonyl fluoride, 5 mm of dithiothreitol, protease inhibitor cocktail Complete Minitablets [Roche], and 0.2% [v/v] Nonidet P-40). For phosphatase treatment, the extracted plant proteins were treated with the Lambda protein phosphatase (P0753S; New England BioLabs) according to the manufacturer’s instructions. The dephosphorylation reaction took place at 30°C for 30 min in a thermal cycler (Bio-Rad). TaBZR2 proteins were subsequently detected by immunoblotting using Anti-TaBZR2 antibodies at a 1:1,000 dilution. IRDye 800CW anti-rabbit IG (H + L) at a 1:10,000 dilution (LI-COR) was used as a second antibody. The immunoblots were developed via an Odyssey CLx Infrared Imaging System (LI-COR).
Subcellular Localization
Transient expression assays were conducted as described in Liu et al. (2013a). TaBZR2 was inserted into the subcellular localization vector pJIT16318, which contains a CaMV 35S promoter and a C-terminal GFP. Approximately 4 × 104 mesophyll protoplasts were isolated from 10-d–old wheat seedlings and then transfected with pJIT16318-TaBZR2 plasmids by PEG-mediated transformation. The transfected protoplasts were then incubated at 23°C for 12 h. GFP fluorescence in the transformed protoplasts was imaged using a confocal laser-scanning microscope (LSM 700; Zeiss).
Measurements of Pro Content, Electrolyte Leakage Level, and MDA Content
For assays of physiological traits, 3-week–old wheat seedlings at the 3-leaf stage were treated with drought conditions for ∼12 d. Approximately 0.2 g of wheat leaf leaves were harvested for measurements of physiological parameters. Absorbance values were measured with a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific). The Pro concentration was determined as described in Zhang et al. (2012a). The electrolyte leakage was examined in accordance with methods described in Cao et al. (2007), and the MDA content was assayed as described in Zhang et al. (2012a). All of the measurements were repeated three times.
Measurements of O2− Content and H2O2 Content
To investigate the contents of O2− and H2O2, 2-week–old wheat seedlings grown on half-strength Hoagland’s nutrient solution supplemented with different concentrations of PEG 6000 and BR (0, 15% [w/v] PEG 6000, 10 nm of BR, 15% [w/v] PEG 6000 + 10 nm of BR, 1 mm of DMTU, and 15% [w/v] PEG 6000 + 1 mm of DMTU) for 72 h. The O2− contents were measured following the protocol of the Superoxide Anion Content Detection Kit (BC1295; Solarbio Life Science). The H2O2 contents were measured following the protocol of the H2O2 Content Detection Kit (BC3595; Solarbio Life Science). For Nitro-blue tetrazolium (NBT) staining, the wheat roots were immersed in NBT stain solution for 30 min and the dark blue color appeared following the protocol of the Alkaline Phosphatase Activity Detection Kit (Amersco). The staining reaction was stopped by the addition of an excess of 95% ethanol. Images were observed and photographed under a stereomicroscope (Leica).
RNA-Seq Assays
TaBZR2-overexpressing (OE9) and wild-type (‘Fielder’) plants were grown in half-strength Hoagland’s liquid medium in a greenhouse with 70% relative humidity, 25°C/23°C day/night temperatures, and long-day conditions (16-h light/8-h dark photoperiod) with a light intensity of ∼400 μmol m−2 s−1 for 2 weeks. Then, the wheat seedlings were transferred to fresh half-strength Hoagland’s solution that contained 15% (w/v) PEG 6000. Leaves were sampled at 0 and 6 h for transcriptome sequencing experiments, and three biological replicates were used. The RNA-Seq analysis was performed by the Allwegene Company. Total RNA was extracted from the samples using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and RNA sequencing was conducted on an Illumina HiSeq platform. RNA-Seq data were analyzed as described in Mortazavi et al. (2008). DEGs were selected using DESeq (1.10.1) with a relative change threshold of 2-fold (P < 0.05, false discovery rate < 0.01; Anders and Huber, 2010). GO categories were identified using the GOseq R package (Young et al., 2010). The genome annotation and functional categorization are based on the National Center for Biotechnology Information nonredundant protein sequences (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/).
EMSA
The CDS of TaBZR2 was inserted into the pGEX-4T-1 vector. The GST and GST-TaBZR2 fusion proteins were expressed in Escherichia coli (BL21) and purified by glutathione-Sepharose TM 4B (GE Healthcare) according to the manufacturer’s protocol. The biotin-labeled probes used in this assay were synthesized (Beijing AuGCT), and the sequences are listed in Supplemental Table S3. Double stranded DNA was obtained by heating oligonucleotides at 95°C for 10 min and annealing at room temperature. The EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, 2 mg of purified fusion protein GST-TaBZR2 or GST protein was added to the binding reaction. The binding reaction took place at 25°C for 30 min in a thermal cycler (Bio-Rad). The mixture was separated on a 6% polyacrylamide mini gel, and then the DNA was transferred to a nylon membrane (Millipore). The signal was visualized with an EasySee Western Blot Kit (TransGen).
Transcriptional Activation Assays in Wheat Protoplasts
For the transcriptional activation assay, the promoter fragment of TaGST1 was inserted into LUC reporter plasmid pGreen II 0800, which contained a Renilla luciferase (REN) gene under the control of the CaMV 35S promoter used as an internal control. The effector plasmids and the reporter plasmids were cotransformed into protoplasts by PEG-mediated transformation. After culturing for 16 h at 23°C, the activities of LUC and REN were separately determined using a Dual-Luciferase Reporter Assay System (E1910; Promega).
Antibody Preparation
Anti-TaBZR2 was generated by Wuhan Abclonal Biotechnology. TaBZR2 CDs (453–999 bp) were inserted into the pET32a vector. Purified His-TaBZR2 (151–333 amino acids) fusion protein was injected into rabbits to produce TaBZR2 polyclonal antibodies. Immunoblots were performed using antiserum against TaBZR2 and visualized with an EasySee Western Blot Kit (TransGen).
Accession Number
RNA-Seq data described in this study can be found in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP071191.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence and phylogenetic analyses of TaBZR2.
Supplemental Figure S2. The expression level of BES/BZR family transcription factor genes in TaBZR2-RNAi and wild-type wheat plants.
Supplemental Figure S3. Measurements of H2O2 contents in TaBZR2-overexpressing, TaBZR2-RNAi, and wild-type wheat plants under normal and drought conditions.
Supplemental Figure S4. TaBZR2 overexpression partially rescued the dwarf phenotypes of bri1-5 plants.
Supplemental Figure S5. Immunoblot analysis of TaBZR2 protein.
Supplemental Table S1. Wheat BZRs responsive to drought and BR treatments.
Supplemental Table S2. Analysis of stress-related genes in TaBZR2-overexpressing and wild-type wheat plants under drought conditions.
Supplemental Table S3. Primers and probes used in this study.
Acknowledgments
We are grateful to Drs. Rui-Lian Jing and Yong-Fu Fu (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing wheat seeds and for the BiFC system, respectively. We also thank Dr. Dongying Gao (Department of Plant Sciences, University of Georgia, Athens, GA, USA) for suggestions on the manuscript.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0100600), the National Transgenic Key Project of the Ministry of Agriculture of China (grant no. 2018ZX0800909B), the National Natural Science Foundation of China (grant no. 31871624), and the Technological Innovation Projects of Modern Agriculture of Hebei Province.
Articles can be viewed without a subscription.
References
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y (2017) Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29: 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Rahman T, Krishna P (2010) Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol 10: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Rahman T, Krishna P (2016) Gene expression and functional analyses in brassinosteroid-mediated stress tolerance. Plant Biotechnol J 14: 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, et al. (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68: 302–313 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XS, Wang W, Zhang Q, Liu JH (2013) A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162: 1178–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha B, Sharma A, Mishra A (2011) Expression of SbGSTU (tau class glutathione s-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol Biol Rep 38: 4823–4832 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang C, Wang X (2013) Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. J Integr Plant Biol 55: 1198–1211 [DOI] [PubMed] [Google Scholar]
- Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P (2007) Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225: 353–364 [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K, Mizoi J, Myint Phyu Sin Htwe N, Fujita Y, Sekita S, Shinozaki K, et al. (2015) Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J 81: 505–518 [DOI] [PubMed] [Google Scholar]
- Koh S, Lee SC, Kim MK, Koh JH, Lee S, An G, Choe S, Kim SR (2007) T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65: 453–466 [DOI] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol 147: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res 18: 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S (2017) BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant 10: 545–559 [DOI] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y (2010) Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shi Y, Yang S (2018) Insights into the regulation of C-repeat binding factors in plant cold signaling. J Integr Plant Biol 60: 780–795 [DOI] [PubMed] [Google Scholar]
- Liu P, Xu ZS, Pan-Pan L, Hu D, Chen M, Li LC, Ma YZ (2013a) A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot 64: 2915–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang X, Wang H, Xin H, Yang X, Yan J, Li J, Tran LSP, Shinozaki K, Yamaguchi-Shinozaki K, et al. (2013b) Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9: e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX (2014) bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201: 1192–1204 [DOI] [PubMed] [Google Scholar]
- Lv B, Tian H, Zhang F, Liu J, Lu S, Bai M, Li C, Ding Z (2018) Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet 14: e1007144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Liu C, Li Z, Ran Q, Xie G, Wang B, Fang S, Chu J, Zhang J (2018) ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol 178: 753–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LSP, Qin F (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6: 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nawaz F, Naeem M, Zulfiqar B, Akram A, Ashraf MY, Raheel M, Shabbir RN, Hussain RA, Anwar I, Aurangzaib M (2017) Understanding brassinosteroid-regulated mechanisms to improve stress tolerance in plants: A critical review. Environ Sci Pollut Res Int 24: 15959–15975 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Song CP, Wang B, Zhou J, Kangasjärvi J, Zhu JK, Gong Z (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 60: 805–826 [DOI] [PubMed] [Google Scholar]
- Ramegowda V, Basu S, Gupta C, Pereira A (2015) Regulation of grain yield in rice under well-watered and drought stress conditions by GUDK. Plant Signal Behav 10: e1034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Rong W, Qi L, Wang A, Ye X, Du L, Liang H, Xin Z, Zhang Z (2014) The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol J 12: 468–479 [DOI] [PubMed] [Google Scholar]
- Sahni S, Prasad BD, Liu Q, Grbic V, Sharpe A, Singh SP, Krishna P (2016) Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci Rep 6: 28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YJ, Park JB, Cho YJ, Jung C, Seo HS, Park SK, Nahm BH, Song JT (2010) Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol Cells 30: 271–277 [DOI] [PubMed] [Google Scholar]
- Shimada S, Komatsu T, Yamagami A, Nakazawa M, Matsui M, Kawaide H, Natsume M, Osada H, Asami T, Nakano T (2015) Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 27: 375–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Li QT, Liu YF, Zhang FX, Ma B, Zhang WK, Man WQ, Du WG, Wang GD, Chen SY, et al. (2013) Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J Exp Bot 64: 4329–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284: 173–183 [DOI] [PubMed] [Google Scholar]
- Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher CL, Nemhauser JL (2012) Bipartite promoter element required for auxin response. Plant Physiol 158: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wei J, Song N, Wang N, Zhao J, Kang Z (2018) A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J Integr Plant Biol 60: 432–443 [DOI] [PubMed] [Google Scholar]
- Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, et al. (2015) GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J 83: 224–236 [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang W, Qin M, Li S, Qiao M, Liu Z, Xiang F (2017) Drought tolerance conferred in soybean (Glycine max L.) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol 58: 1764–1776 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Wang Q, Chong K, Wang F, Wang L, Bai M, Jia C (2006) The brassinosteroid signal transduction pathway. Cell Res 16: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wang M, Shen J, Chen D, Zheng Y, Zhang W (2018) ZmOST1 mediates abscisic acid regulation of guard cell ion channels and drought stress responses. J Integr Plant Biol doi:10.1111/jipb.12714 [DOI] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ (2009) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150: 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yan Q, Wang L, Li X (2018) GmBEHL1, a BES1/BZR1 family protein, negatively regulates soybean nodulation. Sci Rep 8: 7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Li L, Yin Y (2011) Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol 53: 455–468 [DOI] [PubMed] [Google Scholar]
- Ye H, Liu S, Tang B, Chen J, Xie Z, Nolan TM, Jiang H, Guo H, Lin HY, Li L, et al. (2017) RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat Commun 8: 14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7: 751–760 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C (2013) Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol 162: 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mao X, Jing R, Chang X, Xie H (2011) Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot 62: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Xia C, Jia J, Liu X, Kong X (2012a) A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J Exp Bot 63: 5873–5885 [DOI] [PubMed] [Google Scholar]
- Zhang N, Yin Y, Liu X, Tong S, Xing J, Zhang Y, Pudake RN, Izquierdo EM, Peng H, Xin M, et al. (2017) The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol 175: 1878–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA 106: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu X, Wang X, Zhou M, Zhou X, Ye X, Wei X (2012b) An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol 196: 1155–1170 [DOI] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6: 486–503 [DOI] [PubMed] [Google Scholar]
- Zhuo C, Liang L, Zhao Y, Guo Z, Lu S (2018) A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by upregulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ 41: 2021–2032 [DOI] [PubMed] [Google Scholar]