The sultr3 quintuple mutant unequivocally demonstrates that sulfate transporter subfamily 3 is responsible for more than half of the chloroplast sulfate uptake and influences downstream sulfate assimilation and ABA biosynthesis as well as sulfate-induced stomatal closure.
Abstract
Plants are major sulfur reducers in the global sulfur cycle. Sulfate, the major natural sulfur source in soil, is absorbed by plant roots and transported into plastids, where it is reduced and assimilated into Cys for further metabolic processes. Despite its importance, how sulfate is transported into plastids is poorly understood. We previously demonstrated using single Arabidopsis (Arabidopsis thaliana) genetic mutants that each member of the sulfate transporter (SULTR) subfamily 3 was able to transport sulfate across the chloroplast envelope membrane. To resolve the function of SULTR3s, we constructed a sultr3 quintuple mutant completely knocking out all five members of the subfamily. Here we report that all members of the SULTR3 subfamily show chloroplast membrane localization. Sulfate uptake by chloroplasts of the quintuple mutant is reduced by more than 50% compared with the wild type. Consequently, Cys and abscisic acid (ABA) content are reduced to ∼67 and ∼20% of the wild-type level, respectively, and strong positive correlations are found among sulfate, Cys, and ABA content. The sultr3 quintuple mutant shows obvious growth retardation with smaller rosettes and shorter roots. Seed germination of the sultr3 quintuple mutant is hypersensitive to exogenous ABA and salt stress, but is rescued by sulfide supplementation. Furthermore, sulfate-induced stomatal closure is abolished in the quintuple mutant, strongly suggesting that chloroplast sulfate is required for stomatal closure. Our genetic analyses unequivocally demonstrate that sulfate transporter subfamily 3 is responsible for more than half of the chloroplast sulfate uptake and influences downstream sulfate assimilation and ABA biosynthesis.
Sulfur is an essential macronutrient for plants as it participates in many biological processes, including the biosynthesis of Cys and Met, the resistance against diseases and pests, and the detoxification of reactive oxygen species, xenobiotics, and heavy metals (Leustek et al., 2000; Saito, 2000; Xiang et al., 2001; Takahashi et al., 2011; Álvarez et al., 2012). Sulfate is the main form of inorganic sulfur in the natural environment, and the oxidized sulfur in sulfate must be reduced and assimilated to Cys before entering other metabolic processes (Leustek, 2002). Incorporation of sulfur into plant metabolism requires uptake from the soil and coordinated transport of sulfate through dedicated sulfate transporters (Takahashi et al., 2000; Yoshimoto et al., 2002). Then, sulfate is reduced to sulfide in plastids by 5'-adenylylsulfate (APS) reductase and sulfite reductase and finally fixed by O-acetyl-Ser (thiol) lyase into Cys (Khan et al., 2010; Hell and Wirtz, 2011).
Sulfate transporters are anion transporters dedicated to sulfate uptake and transport in plants. The typical sulfate transporters are pH-dependent proton/sulfate cotransporters with 10–12 membrane-spanning helices and a STAS (Sulfate Transporter and AntiSigma factor antigonist) domain (Smith et al., 1995; Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). The STAS domain is thought to be critical for the function of sulfate transport since MOT1 and MOT2, molybdate transporters previously named sulfate transporter (SULTR)5;2 and SULTR5;1, lack the STAS domain (Shibagaki and Grossman, 2004; Tomatsu et al., 2007; Gasber et al., 2011). In Arabidopsis (Arabidopsis thaliana), 12 sulfate transporters were initially identified and subdivided into four groups based on their phylogenic relationships and kinetic properties (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). Two root-specific, high-affinity transporters of group 1, SULTR1;1 and SULTR1;2, are involved in sulfate uptake from soil (Yoshimoto et al., 2002). SULTR2;1 and SULTR2;2, two low-affinity transporters of group 2, transport sulfate to xylem for root-to-shoot transport (Takahashi et al., 2000), whereas SULTR1;3 mediates shoot-to-root sulfate translocation through phloem (Yoshimoto et al., 2003). SULTR4;1 and SULTR4;2 of group 4 are both tonoplast-localized transporters and coordinately facilitate the efflux of sulfate from the vacuole, the main site for cellular sulfate storage (Kataoka et al., 2004b).
However, the function of group 3 transporters remains fragmentary. SULTR3;5 was reported to reinforce SULTR2;1’s function in root-to-shoot sulfate translocation, with more sulfate accumulating in roots of the sultr3;5 mutant under low sulfur conditions (Kataoka et al., 2004). Other work also reported increased sulfate and decreased free Cys content in Arabidopsis seeds of the single defective mutant of group 3 sulfate transporters, with total sulfur supply unaffected, indicating a decline in sulfur reduction and assimilation in these defective mutants (Zuber et al., 2010). Our previous work demonstrated that SULTR3;1 is chloroplast-localized and involved in sulfate uptake across the chloroplast envelope membrane (Cao et al., 2013). Single knockout mutants of group 3 sulfate transporters show decreased chloroplast sulfate uptake, indicating that these sulfate transporters may also be involved in chloroplast sulfate transport (Cao et al., 2013). Because chloroplasts are the main site for sulfate reduction in plants (Hell and Wirtz, 2011; Takahashi et al., 2011), Cys levels also decreased in the sultr3;1 mutant due to a decline in sulfur assimilation (Cao et al., 2013).
Sulfate was reported to be a signal under drought stress that reinforces the effect of abscisic acid (ABA) in stomatal closure (Goodger et al., 2005; Ernst et al., 2010). Sulfate can keep the R-Type anion channel Quickly Activating Anion Channel1 open, which regulates stomata movement (Meyer et al., 2010) and induces the expression of 9-cis-Epoxycarotenoid Dioxygenase 3, a rate-limiting enzyme for ABA synthesis, in guard cell through unknown pathways (Malcheska et al., 2017). Besides sulfate, sulfide was also reported to act as a signaling molecule to induce stomatal closure (Lisjak et al., 2010; Jin et al., 2013; Honda et al., 2015). ABA biosynthesis is connected with the availability of Cys because the activity of Abscisic Aldehyde Oxidase 3, a key enzyme in ABA biosynthesis, relies on Cys as the sulfur donor for its molybdenum cofactor sulfuration catalyzed by sulfurase ABA3 (Bittner et al., 2001; Xiong et al., 2001; Mendel and Hänsch, 2002; Llamas et al., 2006). AAO3 activity is reduced in sultr3;1 and can be restored by exogenous application of Cys (Cao et al., 2014). In addition, SULTR3 single mutants were reported to show reduced ABA levels under normal and salt stress conditions and were hypersensitive to exogenous ABA and salt during the germination stage (Cao et al., 2014). Two other mutants in sulfur assimilation, sir1-1 and apr2, also showed delayed germination under ABA and salt stress conditions (Cao et al., 2014).
In a companion study, we identified Cys as a trigger of ABA biosynthesis in Arabidopsis, which explains the drought-sensitive phenotype of Cys-synthesis depleted mutants and provides a mechanism for sulfate-induced stomatal closure (Batool et al., 2018). Here, we address the biological relevance of the remaining SULTR3 group members for sulfate allocation in plant cells and its potential impact for production of the phytohormone ABA. We demonstrate that the four remaining sulfate transporters of SULTR3 subfamily, SULTR3;2 (AT4G02700), SULTR3;3 (AT1G23090), SULTR3;4 (AT3G15990), and SULTR3;5 (AT5G19600), are localized in plastids. Thus, we generated double, triple, quadruple, and quintuple mutants of the SULTR3 subfamily and provided direct evidence for the redundant function of all SULTR3s in sulfate uptake into the chloroplast. All SULTR3s contributed almost equally to the sulfate uptake rate in isolated chloroplasts. The decreased sulfate allocation into chloroplasts significantly impacted steady-state levels of Cys and ABA in double, triple, quadruple, and quintuple mutant of SULTR3. The quintuple mutant displayed the lowest chloroplast sulfate uptake rate, and significantly reduced Cys and ABA content, but was viable. These findings highlight the biological relevance of SULTR3s for subcellular partitioning of sulfate in plant cells and uncover the existence of a yet unidentified back-up system for sulfate uptake into plastids. The remarkably high correlation between the sulfate uptake rate into plastids and the syntheses of Cys and ABA prompted us to test the germination rates of mutant seeds on high ABA and salt stress. As expected, the higher-order sultr3 mutants displayed progressively increasing sensitivity to both stresses compared with the wild type, and the sultr3 quintuple mutant was most strongly affected. Furthermore, the sensitive germination phenotype of the mutants can be rescued by feeding sulfide. As a result of its 50% decreased sulfate uptake rate, the sultr3 quintuple mutant failed to close stomata upon sulfate administration. Our findings demonstrate a crucial role of SULTR3s for dynamic transport of sulfate into the chloroplasts to promote stress-induced synthesis of Cys, which in turn triggers biosynthesis of the phytohormone ABA to coordinate rapid adaptive responses such as stomatal closure.
RESULTS
Subcellular Localization of SULTR3 in Plants
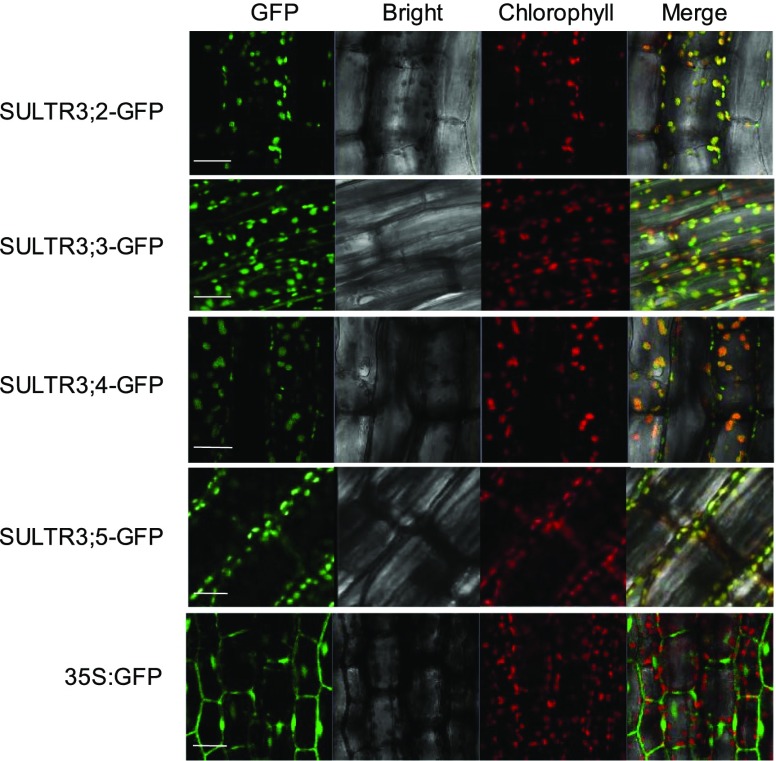
The initial characterization of group 3 SULTRs revealed that SULTR3;1 is localized in the chloroplast envelope and that loss-of-function mutants for most SULTR3 family members suffer from decreased sulfate uptake into isolated chloroplast (Cao et al., 2013). To provide direct evidence for the action of the five SULTR3s in the chloroplast envelope, we fused the remaining SULTR3;2, SULTR3;3, SULTR3;4, and SULTR3;5 to the N terminus of GFP (Supplemental Figure S1), and determined the subcellular localization of the fusion proteins in hypocotyls of T2 transgenic lines (Fig. 1). The GFP-specific signal of all tested SULTR3-GFP fusion proteins colocalized with the autofluorescence of chlorophyll. We did not observe any significant signal at the plasma membrane or the tonoplast. These results suggested that SULTR3;2 to SULTR3;5 were also integrated in the chloroplast envelope as shown previously for SULTR3;1.
Figure 1.
Group 3 sulfate transporters are all localized on plastid envelop membrane. Confocal images of hypocotyl cells of SULTR3-GFP transgenic plants. The panels from left to right represent GFP fluorescence of the SULTR3-GFP fusion protein, bright field, autofluorescence of chlorophyll, and the merged images, respectively. Scale bars = 100 μm.
sultr3 Quintuple Mutant of Subfamily 3 Sulfate Transporters
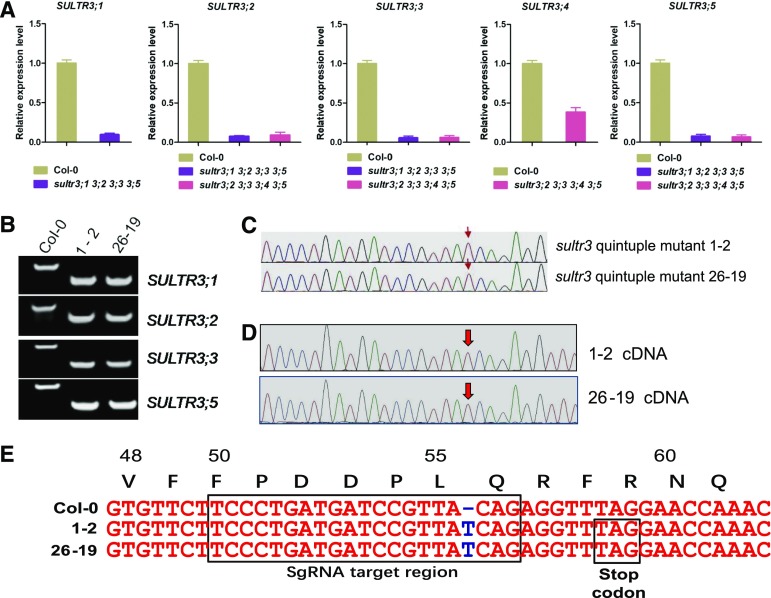
To address the biological function of the SULTR3 subfamily, we aimed to construct double, triple, quadruple, and quintuple mutants of group 3 SULTRs by genetic crossing of established transfer DNA (T-DNA) insertion lines of SULTR3;1, SULTR3;2, SULTR3;3, SULTR3;4, and SULTR3;5 ordered from Arabidopsis Biological Resource Center (ABRC). We obtained four double, three triple, and two quadruple mutant lines as described in Supplemental Table S1. The T-DNA insertion in the 5′ untranslated region of SULTR3;4 in the sultr3;4 line did not entirely abolish expression of SULTR3;4. Thus, the steady-state SULTR3;4 transcript level was decreased to 33% of wild-type level in the first quadruple mutant (sultr3;2 sultr3;3 sultr3;4 sultr3;5). Because transcription of SULTR3;1, SULTR3;2, SULTR3;3, and SULTR3;5 in the second quadruple mutant (sultr3;1 sultr3;2 sultr3;3 sultr3;5) was absent (Fig. 2A), we decided to eliminate SULTR3;4 activity by the CRISPR/Cas9 technology in this quadruple mutant. This strategy resulted in creation of two independent SULTR3 quintuple mutant lines lacking activities of all SULTR3 members. The identities of the SULTR3;1, SULTR3;2, SULTR3;3, and SULTR3;5 loci in the quintuple mutants were confirmed by PCR amplification of the T-DNA insertion sites in the respective loci from genomic DNA (Fig. 2B). The CRISPR-Cas9-mediated genomic editing of the SULTR3;4 locus in both sultr3 quintuple lines, 1-2 and 16-29, was verified by sequencing of the genomic region (Fig. 2C) and the transcribed mRNA (Fig. 2D). As expected, a frameshift mutation was created by a thymidine (T) insertion in the single guide RNA (sgRNA)-targeted region, resulting in early termination at the 59th codon of SULTR3;4 mRNA translation (Fig. 2E).
Figure 2.
Creation and identification of quintuple mutant of group 3 sulfate transporters. A, Relative expression level of SULTR3;1, SULTR3;2, SULTR3;3, SULTR3;4, and SULTR3;5 in Col-0 wild type, sultr3;1 sultr3;2 sultr3;3 sultr3;5 and sultr3;2 sultr3;3 sultr3;4 sultr3;5 T-DNA insertion quadruple mutants. The expression of SULTR3;4 in sultr3;2 sultr3;3 sultr3;4 sultr3;5 quadruple mutant was knockdown rather than knockout as for the other SULTR3 genes. B, T-DNA insertion identification of SULTR3;1, SULTR3;2, SULTR3;3, and SULTR3;5 in two quintuple mutant lines 1-2 and 26-19. The results confirm that 1-2 and 26-19 are homozygote lines of SULTR3;1, SULTR3;2, SULTR3;3, and SULTR3;5. C and D, Sequencing results of genomic DNA (C) and complementary DNA (D) of SULTR3;4 in two quintuple-mutant lines 1-2 and 26-19. Both results demonstrate that 1-2 and 26-19 are homozygote lines with a single thymidine (T) nucleotide insertion in the SULTR3;4 coding region. E, Schematic illustration of early termination of a 58-amino acid SULTR3;4 translation fragment in two quintuple mutant lines 1-2 and 26-19 due to the frameshift mutation by a T nucleotide insertion in the sgRNA-targeted region.
Knockout of Sulfate Transporter Subfamily 3 Results in Growth Retardation
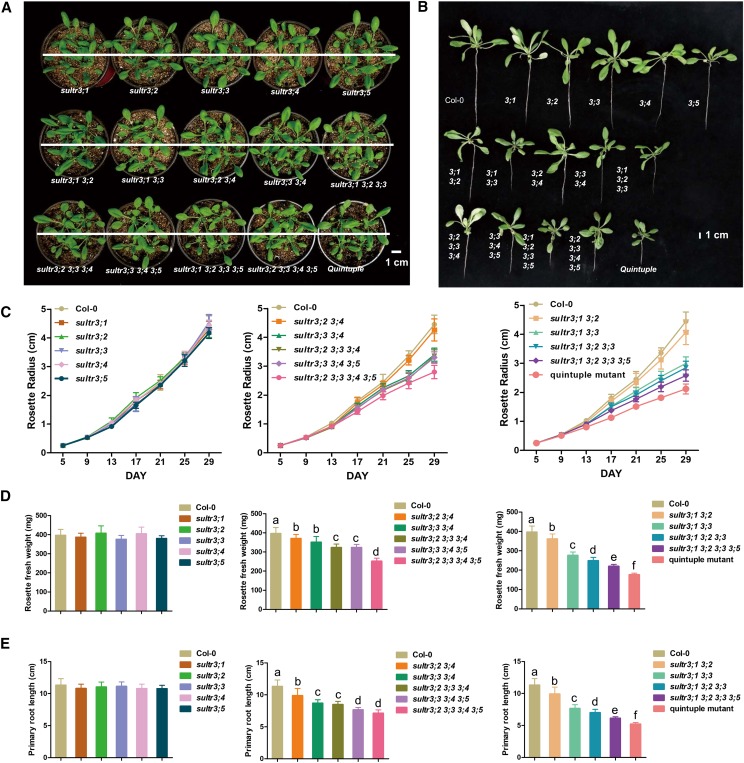
Whereas single knockout mutants of the SULTR3s grew like the wild type, as reported previously (Cao et al., 2013), the higher-order mutants of SULTR3 showed various degrees of growth retardation resulting in smaller rosettes and shorter roots when compared with wild-type plants (Fig. 3, A and B). A progressive reduction in fresh weight, rosette diameter, and primary root length from double mutants to the SULTR3 quintuple mutant was observed. Comparison of the various higher-order SULTR3 mutants did not reveal a dominant isoform of SULTR3 responsible for the observed growth retardation. As expected, the SULTR3 quintuple mutant suffered the most severe growth retardation (Fig. 3, C–E). However, organ development in the vegetative and generative phases of growth was normal in all SULTR3 mutants when compared with wild type, explaining the fertility of the SULTR3 quintuple mutant. These results showed that the depletion of SULTR3 activity located at the chloroplast envelope strongly affects plant growth. Surprisingly, loss of all SULTR3 isoforms did not cause lethality, suggesting that sulfate import into the chloroplast was not entirely abolished in the SULTR3 quintuple mutant or that sulfate is reduced and incorporated into Cys in another subcellular compartment.
Figure 3.
Multigene defective mutants of SULTR3 show various degrees of growth retardation. A and B, Phenotype of aerial part (A) and whole plant (B) of SULTR3 knockout mutants and wild-type plants (Col-0). Plants were photographed 29 d after germination. Scale bars = 1 cm. C, Time-course change of rosette diameters. Values are means ± sd (n = 20 plants for each line). D, Fresh weight of rosette leaves. Values are means ± sd (n = 20 plants for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001). E, Length of primary roots. Values are means ± sd (n = 20 plants for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001).
Group 3 Sulfate Transporters Show Redundancy in Chloroplast Sulfate Uptake, Cys Level, and ABA Content
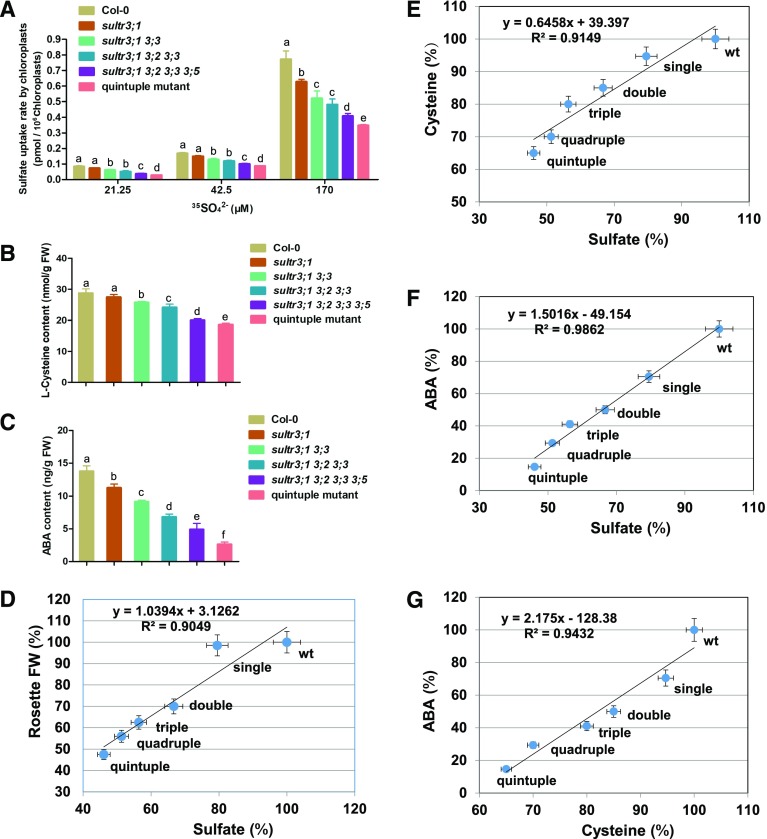
To address the contribution of group 3 sulfate transporters in transporting sulfate into chloroplast, we determined incorporation of radioactive 35S-labeled sulfate into freshly isolated chloroplasts from the five higher-order SULTR3 loss-of-function mutants and the wild type. The Hill reaction was applied to ensure that the isolated chloroplasts were intact and photosynthetically active (Supplemental Figure S2). The number of chloroplasts was adjusted to the same concentration for each line before the 35S-uptake assay. The in organello sulfate uptake was assayed at three exogenous sulfate concentrations: 21.25, 42.5, and 170 µM. Under all tested external sulfate regimes, the sulfate uptake rates of the isolated chloroplasts from single, double, triple, quadruple, and quintuple mutants were decreased when compared with the wild-type control (Col-0). 35S-sulfate uptake rates were negatively correlated with the number of disrupted SULTR3 genes, with a progressive reduction from the single mutant to the sultr3 quintuple mutant, the latter displaying the lowest sulfate uptake rate (Fig. 4A). We also measured the sulfate levels in isolated chloroplasts (Supplemental Figure S3A). Despite the technical difficulty of accurately measuring chloroplast sulfate levels, a negative correlation of sulfate level in chloroplasts was observed with the number of disrupted SULTR3 genes. These results demonstrated the functional redundancy of group 3 sulfate transporters in chloroplast sulfate uptake, indicating that each of the SULTR3 members contributes to sulfate transport across the chloroplast envelop membrane. This finding directly linked the decreased sulfate uptake rate into chloroplasts with the observed growth retardation of the allelic series of SULTR3 mutants. Under 170 µM, sulfate uptake of chloroplasts from the sultr3 quintuple mutant decreased to less than half (∼45%) of the wild-type level, which indicates the existence of another yet unidentified sulfate transporter system in chloroplasts that safeguards sulfate import in the absence of SULTR3.
Figure 4.
Loss of SULTR3 genes decreases sulfate uptake rate of chloroplasts, Cys levels, and ABA content. A, Sulfate uptake by isolated chloroplasts of the SULTR3 knockout mutants and wild-type plants (Col-0) under three different sulfate conditions. Values are means ± sd (n = three experiments for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001). B, Cys content in SULTR3 knockout mutants and wild-type plants (Col-0) under normal conditions. Values are means ± sd (n = three experiments for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001). C, ABA content in 8-d-old SULTR3 knockout mutants and wild-type plants (Col-0). Values are means ± sd (n = three experiments for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001). D, Correlation between chloroplast sulfate uptake and growth. A simple regression was made with the chloroplast sulfate uptake (as a percentage of the wild type) at 170 µM in (A) and rosette fresh weight (FW, as a percentage of the wild type) in Fig. 3D. E, Correlation between chloroplast sulfate uptake and Cys content. A simple regression was made with the chloroplast sulfate uptake (as a percentage of the wild type) at 170 µM in (A) and Cys content (as a percentage of the wild type) in (B). Correlation between chloroplast sulfate uptake and ABA content is also shown. A simple regression was made with the chloroplast sulfate uptake (as a percentage of the wild type) at 170 µM in (A) and ABA content (as a percentage of the wild type) in (C). F, Correlation between Cys and ABA content. A simple regression was made with Cys content in (B) and ABA content (as a percentage of the wild type).
Because plastidic sulfate reduction provides the precursor for Cys biosynthesis in other subcellular compartments, we determined the foliar Cys concentration in the SULTR3 double, triple, quadruple, and quintuple mutants and found it decreased to 90%, 84%, 70%, and 65% of that of wild-type plant, respectively (Fig. 4B). These results strongly support the hypothesis that growth retardation in the higher-order SULTR3 mutants was a direct consequence of depleted sulfur fixation into Cys, due to decreased sulfate reduction capability in plastids. In agreement, we found a very stringent correlation of sulfate import rate into chloroplasts with the growth retardation of plants (R2 = 0.9049; Fig. 4D) and the foliar Cys steady-state level (R2 = 0.9149; Fig. 4E). To validate sulfate assimilation into Cys, we measured the more robust metabolite glutathione (Supplemental Figure S3). Similar to Cys levels, reduced glutathione (GSH) levels were negatively correlated with the number of disrupted SULTR3 genes. The quintuple mutant had the lowest GSH level, as expected.
Our previous work showed that steady-state ABA levels in SULTR3 single mutants decreased (Cao et al., 2014), which could be explained by the stimulating effect of Cys on ABA biosynthesis. In support of this hypothesis, the ABA content of the sultr3 double, triple, quadruple, and quintuple mutants decreased to 68%, 50%, 36%, and 20% of that of wild-type plants, respectively (Fig. 4C). Remarkably, the foliar steady-state ABA level correlated well with the sulfate uptake rate into chloroplast (R2 = 0.9862; Fig. 4F) and the Cys supply of the cells (R2 = 0.9432; Fig. 4G).
Loss of SULTR3 Transporters Alters Responses to ABA and Salt Stress during Seed Germination
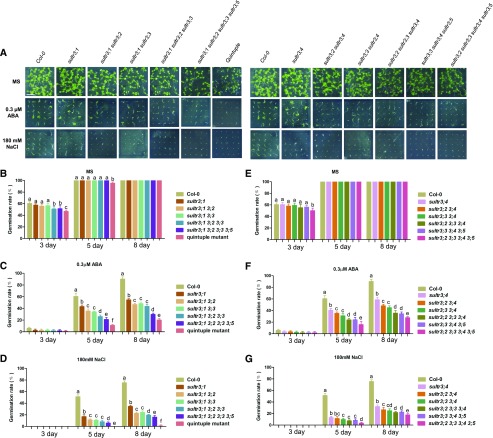
It is known that loss of single SULTR3 transporters can alter seed germination response to exogenous ABA and salt conditions. We assayed seed germination of the multigene defective mutants under ABA and salt conditions (Fig. 5A). Again, we observed a progressive reduction in germination rate from single to SULTR3 quintuple mutants, with the latter again having the lowest rate under both ABA and NaCl treatment (Fig. 5, B and D–G). At 5 d after sowing, seed germination rate of the SULTR3 quintuple mutant was only 10% under 0.3 µM ABA, whereas more than 60% of wild-type seeds germinated. At 8 d after sowing, only 20% of the SULTR3 quintuple mutant seeds germinated, whereas more than 90% of wild-type seeds germinated (Fig. 5C). Seed germination under salt stress showed similar patterns. Compared with 0.3 µM ABA, seed germination of all sultr3 mutants was more sensitive to 180 mM NaCl than that of the wild type. The germination rate of sultr3;1 knockout mutant seed was only 33% of that of the wild type, and no radical emergence of SULTR3 quintuple mutant seeds was found even 5 d after sowing (Fig. 5D). At 8 d after sowing, only 5% of SULTR3 quintuple mutant seeds germinated, whereas more than 75% of the wild-type seeds germinated. Consistent with alterations in Cys and ABA content, seed germination inhibition upon treatment with exogenous ABA and salt was directly proportional to the number of mutated SULTR3 genes, demonstrating the correlation of Cys and ABA content with the plant abiotic stress response during germination stages.
Figure 5.
Loss of SULTR3 genes alters seed germination response to ABA and salt stress. A, SULTR3 knockout mutants are hypersensitive to exogenous ABA and salt stress during germination. Plants were photographed 8 d after germination. Scale bar = 1 cm. B–G, Germination rates of SULTR3 knockout mutants and wild-type seeds (Col-0) under normal, ABA, and salt conditions in (A). Seeds were evaluated at 5 d and 8 d after sowing and were considered germinated when the radicles penetrated the seed coats. Values are means ± sd (n = three experiments with 45 seeds for each line). Letters indicate statistically significant differences between groups determined with one-way repeated measure ANOVA (P < 0.001).
Seed germination of SULTR3 quadruple and quintuple mutants was also delayed under normal conditions compared with that of the wild type. For triple mutants, sultr3;1 sultr3;2 sultr3;3, but not sultr3;2 sultr3;3 sultr3;4 or sultr3;3 sultr3;4 sultr3;5, showed a germination delay on d 3 after sowing (Fig. 5B), which indicates that SULTR3;1 may have a more critical role than other SULTR3 genes in the early germination stage.
If disruption of SULTR3 genes caused the ABA- and salt-sensitive germination phenotypes of the mutants, one would predict that the observed defects could be partially rescued by feeding sulfide. Indeed, 1 mM NaHS could rescue the ABA- and salt-sensitive germination phenotypes of the mutants (Supplemental Figure S4), which further confirms the role of SULTR3 transporters in the seed germination response to ABA and salt stress.
SULTR3 Transporters Are Essential for Sulfate-Induced Stomatal Closure
In our companion manuscript we demonstrated that sulfate triggers ABA production after incorporation into Cys (Batool et al., 2018). These findings prompted us to test whether the sultr3 quintuple mutant is defective in dynamic induction of stomatal closure upon short-term sulfate application. The wild type rapidly closed stomata upon addition of sulfate (15 mM MgSO4, P < 0.001), Cys (0.5 mM, P < 0.001), or ABA (50 µM, P < 0.001). As expected, addition of Gly (0.5 mM) as a control had only marginal influence on stomatal aperture of the wild type or the sultr3 quintuple mutant. In contrast with the wild type, application of sulfate to the sultr3 quintuple mutant failed to induce stomatal closure, strongly suggesting that the sultr3 mutant was unable to incorporate the sulfate into Cys for promotion of ABA biosynthesis (Fig. 6). In order to rule out a potential impact of sultr3 quintuple mutations on ABA sensing or promotion of ABA biosynthesis by Cys, we also applied Cys and ABA to the sultr3 quintuple mutant. ABA and Cys were able to induce stomatal closure proving that ABA sensing and promotion of ABA biosynthesis by Cys was functional in the sultr3 quintuple mutant. Taken together these findings demonstrate that loss of all SULTR3 isoforms strongly impairs dynamic and efficient transport of sulfate into the chloroplast for stress-induced Cys biosynthesis.
Figure 6.
Sulfate-induced stomatal closure requires sulfate import into chloroplast mediated by SULTR3. A, Stomatal images for each treatment. Impact of sulfate (white, 15 mM MgSO4), Cys (yellow, 500 µM), Gly (green, 500 µM), and ABA (gray, 50 µM) on stomatal aperture of the wild type and the SULTR3 quintuple mutant suffering from decreased sulfate uptake into chloroplasts. Control refers to water adjusted to pH 5.5 (black). Epidermal peels were obtained from 5-week-old wild type and SULTR3 quintuple plants grown on soil under short-day conditions and treated for 180 min with the effectors dissolved in water at pH 5.5. Scale bar = 10 μm. B, Stomatal aperture measured for the treatments in (A). Values are means ± se (n ≥ 60 stomata, derived from ≥ 6 individual plants). Letters indicate statistically significant differences between groups determined with the one-way repeated measure ANOVA (P < 0.001).
DISCUSSION
Sulfate Transporters of Group 3 Control Sulfate Import into Chloroplasts
Plants rely on a series of pH-dependent proton/sulfate cotransporters, simply referred to as sulfate transporters, for transport of the charged oxyanion sulfate across plasma membranes. In Arabidopsis, 12 known sulfate transporters are subdivided into four groups based on their phylogenic relationships. The biological function of the five plasma membrane-localized transporters of group 1 and 2 in the uptake and partitioning of sulfate at the whole plant level is well established, whereas tonoplast-embedded SULTR4;1 and SULTR4;2 facilitate intracellular remobilization of vacuolar stored sulfate (for review, see Takahashi et al., 2011).
In contrast, the role of group 3 SULTRs is poorly understood. Our previous work demonstrated that SULTR3;1 localized to the chloroplast envelop membrane and was involved in sulfate influx into chloroplasts. Here, we demonstrate that the remaining four SULTR3 isoforms also localize to plastids (Fig. 1). This localization provides a molecular explanation for the decreased sulfate uptake rate of chloroplasts isolated from single loss-of-function mutants for the other four SULTR3 subfamily members (Cao et al., 2013). We applied CRISPR-Cas9-based gene editing and genetic crossing of established SULTR3 loss-of-function mutants to successively eliminate all group 3 SULTRs to explore the relevance of each isoform for sulfate import into chloroplasts during vegetative growth under nonstressed conditions. The sulfate influx rate continually decreased upon elimination of additional SULTR3 isoforms, revealing the functional redundancy of SULTR3 proteins (Fig. 3). These results strengthen the conclusion that not only SULTR3;1, but all group 3 SULTRs, contribute to sulfate transport across the chloroplast membrane. Remarkably, the comparatively large number of group 3 SULTRs found in Arabidopsis is conserved in the reference grass, Brachypodium distachyon (SULTR3;1 to SULTR3;5), whereas redundant SULTRs in group 2 and group 4 are absent in this species (Tombuloglu et al., 2017). In Medicago truncalata, the number of group 3 sulfate transporter is even higher (Gallardo et al., 2014). Gene duplication often allows specification of function, e.g. via modulation of transcriptional responses. Indeed, SULTR3 transporters respond differentially to diverse stresses (Gallardo et al., 2014). Notably, the transcriptional regulation of SULTR1 subfamily is the hallmark of the sulfate-deficiency response and the key to surviving amid a limited sulfur supply, strongly suggesting that stress-induced transcriptional regulation of SULTRs is biologically relevant and controlled by highly specific signal transduction pathways (Maruyama-Nakashita et al., 2004; Rouached et al., 2008). Spatial separation of SULTR3 transcription has been reported in vegetative and generative organs and will add to the specification of SULTR3 transporter activity in vascular plants (Kataoka et al., 2004a; Zuber et al., 2010).
Plastids, especially chloroplasts, are the exclusive site for de novo reduction of APS-bound sulfur to sulfide in plants. Thus, transport of sulfate into chloroplasts is essential for plants. Chloroplasts isolated from the SULTR3 quintuple mutant displayed only 45% of wild-type sulfate uptake, revealing that SULTR3 contributed significantly to chloroplast sulfate influx. Impaired plastid sulfate uptake capacity caused a significant decrease in plant growth (Fig. 4). However, the viability of the sultr3 quintuple mutant provides evidence for the existence of a yet unidentified backup system for chloroplast sulfate influx, which safeguards sulfate incorporation into plastids in the absence of SULTR3s. The contribution of this system to sulfate import into plastids in the presence of the five SULTR3s remains elusive because it fails to facilitate fast transport of sulfate required for sulfate-induced stomatal closure in the sultr3 quintuple mutant (Fig. 6). Candidates for this backup system are plastid localized ATP-binding cassette (ABC)-type transporters or the triose-P/P-translocator (TPT). Evidence for the involvement of chloroplast-localized ABC-type transporter complexes evolved from the analysis of sulfate uptake into chloroplasts from green algae (Chen et al., 2003, 2005; Chen and Melis, 2004). However, no homologs of these ABC-type transporters were identified in vascular plants (Buchner et al., 2004).
Exchange of sulfate specifically with triose-P and not Glc-6-P was shown previously in isolated plastids from wild-type Spinacia oleracea (Hampp and Ziegler, 1977). Furthermore, isolated spinach chloroplasts are capable of exchanging sulfate with phosphate. This strict counter-exchange followed a saturation kinetic with a Km of 2.5 mM and a rate of 25 µmol sulfate mg−1 chlorophyll h−1 and external phosphate acted as a competitive inhibitor (KI: 0.7 mM) of sulfate import by chloroplasts (Mourioux and Douce, 1979). Both results suggest that the TPT is involved in the loading of sulfate into plastids. In Arabidopsis, the tpt-1 mutant, which is defective in the chloroplast TPT (AT5g46110), displays less than 5% of wild-type triose P–specific transport but shows no apparent growth retardation under nonstressed conditions (Schneider et al., 2002). Together with our findings, these independent studies point toward a significant contribution of plastid membrane-localized TPT1 for sulfate import into plastids. The established SULTR3 quintuple mutant will open new avenues to address the function of the potential plastidic sulfate-import systems in planta mentioned above.
Uptake of Sulfate into Chloroplasts Impacts Cys and ABA Biosynthesis
Restriction of sulfate reduction in plastids strongly limits Cys synthesis and downstream processes like glutathione biosynthesis and translation (Khan et al., 2010; Dong et al., 2017; Speiser et al., 2018). On the one hand, sulfate reduction is controlled by regulation of APS Reductase and APS kinase to effectively control the flux of sulfur into primary sulfur metabolism or secondary sulfur metabolism (Loudet et al., 2007; Mugford et al., 2009; Ravilious et al., 2012). On the other hand, it might be restricted by sulfate transport from the cytosol into chloroplasts. The strict correlation between the steady-state Cys level and the sulfate-uptake capacity into chloroplasts (Fig. 3) strongly suggests that such limitation is biologically relevant and underpins the importance of tight control of sulfate transport over the chloroplast membrane via SULTRs of group 3.
Very recently, Cys biosynthesis has been established as a relevant trigger for ABA production and stomatal closure (Batool et al., 2018). The decrease of ABA levels in all sultr3 mutants suggests that sulfate uptake into chloroplasts does also limit ABA production via modulation of Cys supply. As a consequence of perturbed ABA biosynthesis, germination of the higher-order sultr3 mutants (double, triple, quadruple, and quintuple) is strongly impaired amid ABA and high salt stress (Fig. 5). In concordance with the hypothesis that sulfate reduction perturbs ABA biosynthesis during germination, the sir1-1 and apr2 mutants are also hypersensitive to ABA and high salt stress (Cao et al., 2014).
Drought stress strongly impairs sulfur metabolism in an organ-specific manner and is highly connected at multiple levels with sulfate transport, sulfate assimilation, and sulfate use in sulfation reactions (Estavillo et al., 2011; Chan et al., 2013; Ahmad et al., 2016). Remarkably, sulfate has been identified as an early accumulating signal in the xylem sap of drought-stressed maize, common hop, and poplar (Ernst et al., 2010; Korovetska et al., 2014; Malcheska et al., 2017). Because petiole feeding of sulfate is sufficient to close stomata in poplar and Arabidopsis, when sulfate reduction and Cys biosynthesis are functional, we tested whether active sulfate transport into the plastids is mandatory for sulfate-induced stomatal closure. Indeed, loss-of all SULTR3 isoforms in the SULTR3 quintuple mutant abolishes sulfate-induced stomatal closure (Fig. 6). These results demonstrate that active transport of sulfate into the plastids by SULTR3-type transporters is a prerequisite for sulfate-induced ABA biosynthesis. Indeed transcription of SULTR3;1, SULTR3;2, SULTR3;3, and SULTR3;5 is strongly enriched in guard cells (Bauer et al., 2013), supporting the hypothesis that active sulfate metabolism in the guard cells contributes to ABA production in response to petiole-fed sulfate or drought-induced root-to-shoot transport in the xylem (Ernst et al., 2010; Korovetska et al., 2014; Malcheska et al., 2017). Indeed drought stress and high salt stress differentially affect transcription of SULTR3 members in leaves (Gallardo et al., 2014), supporting a role of the SULTR3s during abiotic stresses that affects water management of the plant.
In conclusion, our findings in this study using the multigene defective mutants of SULTR3 subfamily members unambiguously show their functional redundancy in chloroplast sulfate uptake and consequent influence on Cys, glutathione, and ABA biosynthesis, which upon mutation result in growth retardation and altered stress responses. We also demonstrate a critical role of SULTR3s for dynamic transport of sulfate into the chloroplasts to promote stress-induced synthesis of Cys, which in turn triggers biosynthesis of the phytohormone ABA to regulate stomatal closure. The highly positive correlations between chloroplast sulfate availability and Cys and ABA levels implies that they are highly coordinated and tightly regulated.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Columbia, Col-0, and mutants with Col-0 background) was grown on half-strength Murashige and Skoog (MS) solid medium (Sigma) that contained 1% (w/v) Suc at 22°C under 16-h-light/8-h-dark cycles. The plants for isolation of chloroplasts and protoplasts were grown on soil for 4 weeks under the same light regime.
Identification of the Multigene Knockout Mutants
The single knockout mutants sultr3;1, sultr3;2, sultr3;3, sultr3;4, and sultr3;5 were T-DNA insertion lines (SALK_023190, SALK_023980, SALK_000822C, CS859766, and SALK_127024, respectively) ordered from the ABRC. The single mutants were crossed to obtain the double, triple, and quadruple mutants. Quintuple mutants were created by knocking out SULTR3;4 expression in sultr3;1 sultr3;2 sultr3;3 sultr3;5 quadruple mutant using the pYAO-based CRISPR/Cas9 system (Yan et al., 2015). The SULTR3;4 genomic region targeted by sgRNA was TCCCTGATGATCCGTTACAG with AGG as the protospacer adjacent motif sequence, following the recommendation from Massachusetts Institute of Technology online software (http://crispr.mit.edu). Double-stranded DNA of the targeted region was created by annealing sgRNA-P1 and sgRNA-P2, and introduced into the pAtU6-26-sgRNA-SK cassette before fusion into the pCAMBIA1300::pYAO-Cas9 plasmid.
The homozygote of each SULTR3 gene was identified by PCR using a common primer LBb1.3 and gene-specific primer pairs 3;1-F/R for SULTR3;1, 3;2-F/R for SULTR3;2, 3;3-F/R for SULTR3;3, 3;4-F/R for SULTR3;4, and 3;5-F/R for SULTR3;5, respectively. Homozygote lines were further confirmed byreal time-quantitative PCR (RT-qPCR), with primer pairs 3;1-qF/qR for SULTR3;1, 3;2-qF/qR for SULTR3;2, 3;3-qF/qR for SULTR3;3, 3;4-qF/qR for SULTR3;4, and 3;5-qF/qR for SULTR3;5, respectively. Two homozygotes of SULTR3;4 in the quintuple mutants were screened and confirmed by sequencing using primer pairs seq-LP1/RP1 for genome and seq-LP2/RP2 for complementary DNA amplification. All primer sequences are listed in Supplemental Table S2.
Localization of SULTR3;1-GFP Fusion Protein
The full-length coding regions of SULTR3;2 to SULTR3;5 were amplified by PCR and fused into GFP fusion binary vector pGWB5 (Invitrogen). The SULTR3-GFP construct was transferred into the wild type, and the T2 generation was used for fluorescence imaging. Fluorescence in hypocotyl of SULTR3-GFP transgenic plants was observed by using a confocal microscope (Carl Zeiss LSM510) under 488 nm excitation. The emission wavelength was restricted to 530 nm for green fluorescence and 650 nm for red autofluorescence of chlorophyll.
RT-qPCR
Total RNA was extracted from primary root material of 4-d-old wild-type seedlings using TRIzol reagent (Invitrogen) and reverse transcribed with the Takara RT kit (Invitrogen) in accordance with the manufacturer’s instructions. All RT-qPCR assays were performed using a SYBR kit (Transgene) in a One Step real-time PCR system (Applied Biosystem) as described in the manufacturer’s protocol. Each assay consisted of three biological replicates and was performed twice. Ubiquitin5 was used as the control in RT-qPCR assays with primer pairs Ubiquitin5-qF/qR (Supplemental Table S2).
Isolation of Intact Chloroplasts
Plants were grown under day-neutral conditions at 22°C and sampled in the early morning to avoid starch accumulation in chloroplasts. Crude chloroplasts were obtained from the rosette leaves of 4-week-old plants using isolation buffer (pH 8.0) and purified using Percoll gradients and activated with high light as described (Kunst, 1998). Hill reaction was carried out to determine integrity and photosynthetic activity of chloroplasts using- 2,6-dichloroindophenol dye as described (Bregman, 1990).
Sulfate Uptake by Purified Chloroplasts and Measurement of Chloroplast Sulfate Levels
Chloroplast sulfate uptake assay was carried out as described previously (Cao et al., 2013). Three exogenous sulfate concentrations (21.25, 42.5, and 170 μM) were used in the uptake assay. The reaction buffer (0.33 M sorbitol, 50 mM HEPES, 10 mM NaCl, 2 mM MgCl2, 2 mM EDTA, 0.5 mM KH2PO4, pH 7.0) containing 42.5, 85, or 340 μM Na235SO4 and 1 mM ATP was mixed with same volume of chloroplast suspension to initiate the uptake assay.
To measure chloroplast sulfate levels, chloroplasts were purified as described above and dried in a 70°C oven, then ground to powder. Sulfate concentrations were measured using a turbidimetry method described previously (Tabatabai, 1974). Briefly, the samples were boiled in hydrochloric acid and filtered. The filtrate was reacted with barium chloride as a turbidifier. The absorbance was measured using a spectrophotometer at 410 nm. A series of potassium sulfate dilutions were used to generate the standard curve. The sulfate content in each sample was determined using the standard curve.
Determination of ABA, Cys, and GSH Content
Wild-type and SULTR3 knockout mutants were germinated and grown on half-strength MS medium for 8 d before being sampled. ABA was extracted and quantified by ELISA (Sigma) as described previously (Yang et al., 2001). Cys and GSH were quantified as described elsewhere (Xiang and Oliver, 1998).
Germination Response to Exogenous ABA and Salt Stress and Rescue of the Mutant Phenotypes with Sulfide
Sterilized seeds were vernalized for 3 d and then plated on half-strength MS medium that contained 1% Suc and 0 or 3 μM ABA or 180 mM NaCl in the presence or absence of 1 mM NaHS. Three replicate plates (45 seeds per plate) were used for each wild-type and mutant line. The plates were kept at 22°C under long-day conditions. Seed germination was evaluated from day to day, and seeds were considered germinated when the radicles broke the seed coat.
Stomatal Aperture Bioassay
The assay was conducted as recently described (Batool et al., 2018). Epidermal strips were peeled off the abaxial side of wild-type and quintuple mutant leaves and floated on distilled water for 2 h under constant light. The strips were then transferred to distilled water (pH 5.5) supplemented without (control) or with effectors (15 mM MgSO4, 0.5 mM Cys, 0.5 mM Gly, and 50 μM ABA) for 3 h. Stomata were imaged before and after treatment with a conventional wide-angle microscope (Leica DMIRB). Each experiment was performed at least in triplicate and showed the same results.
Statistical Analysis
Statistically significant differences were determined with one-way repeated measure ANOVA. Lowercase alphabetical letters indicate significant difference (P < 0.05) between two neighboring letters.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SULTR3;1 (AT3G51895), SULTR3;2 (AT4G02700), SULTR3;3 (AT1G23090), SULTR3;4 (AT3G15990), and SULTR3;5 (AT5G19600).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Schematic of pGWB5::SULTR3-GFP fusion construct.
Supplemental Figure S2. Activity determination of isolated chloroplasts by Hill Reaction.
Supplemental Figure S3. Sulfate and GSH levels.
Supplemental Figure S4. Rescuing mutant phenotype by sulfide.
Supplemental Table S1. Mutants constructed and used in the experiments.
Supplemental Table S2. Primer pairs used in the experiments.
Acknowledgments
The authors thank Dr. Qi Xie from Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for providing the CRISPR-Cas9 gene editing system and the ABRC for SULTR3 T-DNA insertional lines.
Footnotes
This work was supported by National Natural Science Foundation of China (grant nos. 31572183 and 31770273) and the Ministry of Science and Technology of China (grant nos. 2018ZX08009-11B, 2016ZX08005-004-003, and 2016ZX08001003).
References
- Ahmad N, Malagoli M, Wirtz M, Hell R (2016) Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots. BMC Plant Biol 16: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, Bermúdez MA, Romero LC, Gotor C, García I (2012) Cysteine homeostasis plays an essential role in plant immunity. New Phytol 193: 165–177 [DOI] [PubMed] [Google Scholar]
- Batool S, Uslu VV, Rajab H, Ahmad N, Waadt R, Geiger D, Malagoli M, Xiang CB, Hedrich R, Rennenberg H, et al. (2018) Sulfate is incorporated into Cysteine to trigger ABA production and stomatal closure. Plant Cell 30: 2973–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Bregman A. (1990) Laboratory Investigations in Cell and Molecular Biology, 3rd ed. John Wiley and Sons, New York [Google Scholar]
- Buchner P, Stuiver CE, Westerman S, Wirtz M, Hell R, Hawkesford MJ, De Kok LJ (2004) Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol 136: 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Wirtz M, Hell R, Oliver DJ, Xiang CB (2013) SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J 73: 607–616 [DOI] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, Xiang CB (2014) Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J 77: 604–615 [DOI] [PubMed] [Google Scholar]
- Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ (2013) Balancing metabolites in drought: The sulfur assimilation conundrum. Trends Plant Sci 18: 18–29 [DOI] [PubMed] [Google Scholar]
- Chen HC, Melis A (2004) Localization and function of SulP, a nuclear-encoded chloroplast sulfate permease in Chlamydomonas reinhardtii. Planta 220: 198–210 [DOI] [PubMed] [Google Scholar]
- Chen HC, Yokthongwattana K, Newton AJ, Melis A (2003) SulP, a nuclear gene encoding a putative chloroplast-targeted sulfate permease in Chlamydomonas reinhardtii. Planta 218: 98–106 [DOI] [PubMed] [Google Scholar]
- Chen HC, Newton AJ, Melis A (2005) Role of SulP, a nuclear-encoded chloroplast sulfate permease, in sulfate transport and H2 evolution in Chlamydomonas reinhardtii. Photosynth Res 84: 289–296 [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ernst L, Goodger JQD, Alvarez S, Marsh EL, Berla B, Lockhart E, Jung J, Li P, Bohnert HJ, Schachtman DP (2010) Sulphate as a xylem-borne chemical signal precedes the expression of ABA biosynthetic genes in maize roots. J Exp Bot 61: 3395–3405 [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Courty P-E, Le Signor C, Wipf D, Vernoud V (2014) Sulfate transporters in the plant’s response to drought and salinity: Regulation and possible functions. Front Plant Sci 5: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasber A, Klaumann S, Trentmann O, Trampczynska A, Clemens S, Schneider S, Sauer N, Feifer I, Bittner F, Mendel RR, Neuhaus HE (2011) Identification of an Arabidopsis solute carrier critical for intracellular transport and inter-organ allocation of molybdate. Plant Biol (Stuttg) 13: 710–718 [DOI] [PubMed] [Google Scholar]
- Goodger JQ, Sharp RE, Marsh EL, Schachtman DP (2005) Relationships between xylem sap constituents and leaf conductance of well-watered and water-stressed maize across three xylem sap sampling techniques. J Exp Bot 56: 2389–2400 [DOI] [PubMed] [Google Scholar]
- Hampp R, Ziegler I (1977) Sulfate and sulfite translocation via the phosphate translocator of the inner envelope membrane of chloroplasts. Planta 137: 309–312 [DOI] [PubMed] [Google Scholar]
- Hell R, Wirtz M (2011) Molecular biology, biochemistry and cellular physiology of Cysteine metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yamada N, Yoshida R, Ihara H, Sawa T, Akaike T, Iwai S (2015) 8-Mercapto-cyclic GMP mediates hydrogen sulfide-induced stomatal closure in Arabidopsis. Plant Cell Physiol 56: 1481–1489 [DOI] [PubMed] [Google Scholar]
- Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y (2013) Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol Biochem 62: 41–46 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004a) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Haas FH, Samami AA, Gholami AM, Bauer A, Fellenberg K, Reichelt M, Hänsch R, Mendel RR, Meyer AJ, et al. (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22: 1216–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korovetska H, Novák O, Jůza O, Gloser V (2014) Signalling mechanisms involved in the response of two varieties of Humulus lupulus L. to soil drying: I. changes in xylem sap pH and the concentrations of abscisic acid and anions. Plant Soil 380: 375–387 [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Leustek T. (2002) Sulfate metabolism. The Arabidopsis Book 1: e0017, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Lisjak M, Srivastava N, Teklic T, Civale L, Lewandowski K, Wilson I, Wood ME, Whiteman M, Hancock JT (2010) A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol Biochem 48: 931–935 [DOI] [PubMed] [Google Scholar]
- Llamas A, Otte T, Multhaup G, Mendel RR, Schwarz G (2006) The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J Biol Chem 281: 18343–18350 [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Malcheska F, Ahmad A, Batool S, Müller HM, Ludwig-Müller J, Kreuzwieser J, Randewig D, Hänsch R, Mendel RR, Hell R, et al. (2017) Drought-enhanced xylem sap sulfate closes stomata by affecting ALMT12 and guard cell ABA synthesis. Plant Physiol 174: 798–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Yamaya T, Takahashi H (2004) Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol 45: 340–345 [DOI] [PubMed] [Google Scholar]
- Mendel RR, Hänsch R (2002) Molybdoenzymes and molybdenum cofactor in plants. J Exp Bot 53: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid KA, Geiger D, Marten I, Martinoia E, Hedrich R (2010) AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Mourioux G, Douce R (1979) [Sulfate transport across the limiting double membrane or envelope, of spinach chloroplasts]. Biochimie 61: 1283–1292 [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, Nakazato Y, Noji M, Takahashi H, Kramell R, et al. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious GE, Nguyen A, Francois JA, Jez JM (2012) Structural basis and evolution of redox regulation in plant adenosine-5′-phosphosulfate kinase. Proc Natl Acad Sci USA 109: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Wirtz M, Alary R, Hell R, Arpat AB, Davidian J-CE, Fourcroy P, Berthomieu P (2008) Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol 147: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3: 188–195 [PubMed] [Google Scholar]
- Schneider A, Häusler RE, Kolukisaoglu U, Kunze R, van der Graaff E, Schwacke R, Catoni E, Desimone M, Flügge U-I (2002) An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J 32: 685–699 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Grossman AR (2004) Probing the function of STAS domains of the Arabidopsis sulfate transporters. J Biol Chem 279: 30791–30799 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser A, Silbermann M, Dong Y, Haberland S, Uslu VV, Wang S, Bangash SAK, Reichelt M, Meyer AJ, Wirtz M, Hell R (2018) Sulfur partitioning between glutathione and protein synthesis determines plant growth. Plant Physiol 177: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabai MA. (1974) A rapid method for determination of sulfate in water samples. Environ Lett 7: 237–243 [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Tomatsu H, Takano J, Takahashi H, Watanabe-Takahashi A, Shibagaki N, Fujiwara T (2007) An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc Natl Acad Sci USA 104: 18807–18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombuloglu H, Filiz E, Aydın M, Koc I (2017) Genome-wide identification and expression analysis of sulphate transporter (SULTR) genes under sulfur deficiency in Brachypodium distachyon. J Plant Biochem Biotechnol 26: 263–273 [Google Scholar]
- Xiang C, Oliver DJ (1998) Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10: 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Werner BL, Christensen EM, Oliver DJ (2001) The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol 126: 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H (2003) Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131: 1511–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber H, Davidian JC, Aubert G, Aimé D, Belghazi M, Lugan R, Heintz D, Wirtz M, Hell R, Thompson R, Gallardo K (2010) The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol 154: 913–926 [DOI] [PMC free article] [PubMed] [Google Scholar]