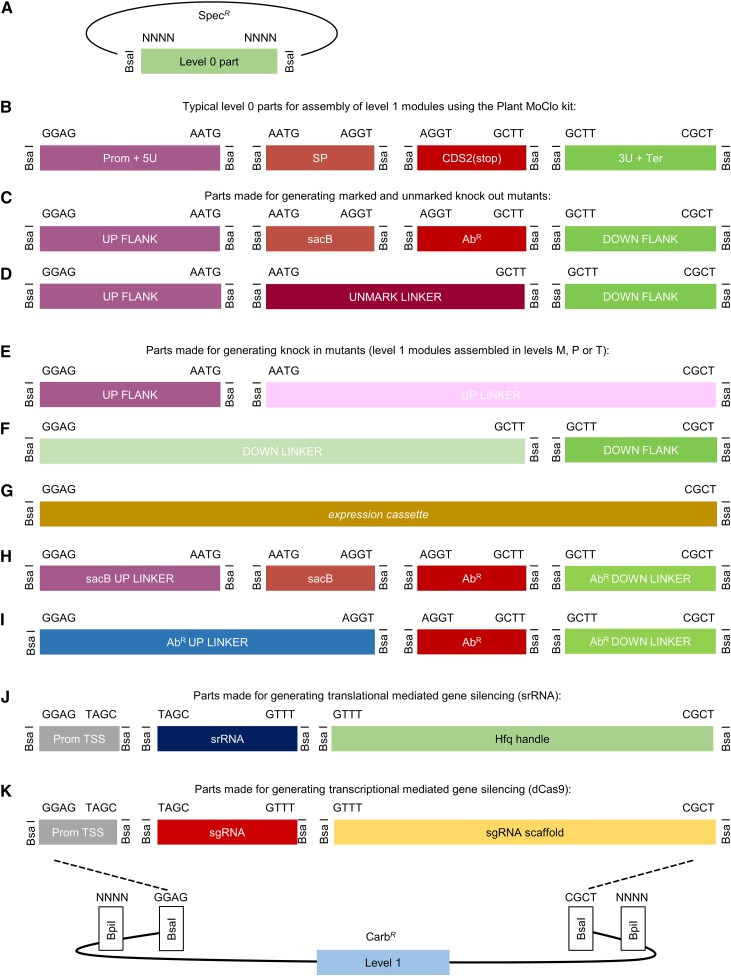
A Golden Gate-based assembly standard developed for cloning and transformation in cyanobacteria is compatible with, and builds on, the broadly established plant modular cloning syntax.
Abstract
Recent advances in synthetic biology research have been underpinned by an exponential increase in available genomic information and a proliferation of advanced DNA assembly tools. The adoption of plasmid vector assembly standards and parts libraries has greatly enhanced the reproducibility of research and the exchange of parts between different labs and biological systems. However, a standardized modular cloning (MoClo) system is not yet available for cyanobacteria, which lag behind other prokaryotes in synthetic biology despite their huge potential regarding biotechnological applications. By building on the assembly library and syntax of the Plant Golden Gate MoClo kit, we have developed a versatile system called CyanoGate that unites cyanobacteria with plant and algal systems. Here, we describe the generation of a suite of parts and acceptor vectors for making (1) marked/unmarked knock-outs or integrations using an integrative acceptor vector, and (2) transient multigene expression and repression systems using known and previously undescribed replicative vectors. We tested and compared the CyanoGate system in the established model cyanobacterium Synechocystis sp. PCC 6803 and the more recently described fast-growing strain Synechococcus elongatus UTEX 2973. The UTEX 2973 fast-growth phenotype was only evident under specific growth conditions; however, UTEX 2973 accumulated high levels of proteins with strong native or synthetic promoters. The system is publicly available and can be readily expanded to accommodate other standardized MoClo parts to accelerate the development of reliable synthetic biology tools for the cyanobacterial community.
Much work is focused on expanding synthetic biology approaches to engineer photosynthetic organisms, including cyanobacteria. Cyanobacteria are an evolutionarily ancient and diverse phylum of photosynthetic prokaryotic organisms that are ecologically important and are thought to contribute ∼25% of the total oceanic net primary productivity (Castenholz, 2001; Flombaum et al., 2013). The chloroplasts of all photosynthetic eukaryotes, including plants, resulted from the endosymbiotic uptake of a cyanobacterium by a eukaryotic ancestor (Keeling, 2004). Therefore, cyanobacteria have proved useful as model organisms for the study of photosynthesis, electron transport, and associated biochemical pathways, many of which are conserved in eukaryotic algae and higher plants. Several unique aspects of cyanobacterial photosynthesis, such as the biophysical carbon concentrating mechanism, also show promise as a means for enhancing productivity in crop plants (Rae et al., 2017). Furthermore, cyanobacteria are increasingly recognized as valuable platforms for industrial biotechnology to convert CO2 and water into valuable products using solar energy (Ducat et al., 2011; Tan et al., 2011; Ramey et al., 2015). They are metabolically diverse and encode many components (e.g. P450 cytochromes) necessary for generating high-value pharmaceutical products that can be challenging to produce in other systems (Nielsen et al., 2016; Wlodarczyk et al., 2016; Pye et al., 2017; Stensjö et al., 2018). Furthermore, cyanobacteria show significant promise in biophotovoltaic devices for generating electrical energy (McCormick et al., 2015; Saar et al., 2018).
Based on morphological complexity, cyanobacteria are classified into five subsections (I to V; Castenholz, 2001). Several members of the five subsections reportedly have been transformed (Vioque, 2007; Stucken et al., 2012), suggesting that many cyanobacterial species are amenable to genetic manipulation. Exogenous DNA can be integrated into or removed from the genome through homologous recombination-based approaches using natural transformation, conjugation (triparental mating), or electroporation (Heidorn et al., 2011). Exogenous DNA can also be propagated by replicative vectors, although the latter are currently restricted to a single vector type based on the broad-host range RSF1010 origin (Mermet-Bouvier et al., 1993; Huang et al., 2010; Taton et al., 2014). Transformation tools have been developed for generating “unmarked” mutant strains (lacking an antibiotic resistance marker cassette) in several model species, such as Synechocystis sp. PCC 6803 (Synechocystis hereafter; Lea-Smith et al., 2016). More recently, markerless genome editing using clustered regularly interspaced short palindromic (CRISPR)-based approaches has been demonstrated to function in both unicellular and filamentous strains (Ungerer and Pakrasi, 2016; Wendt et al., 2016).
Although exciting progress is being made in developing effective transformation systems, cyanobacteria still lag behind in the field of synthetic biology compared to bacterial (heterotrophic), yeast, and mammalian systems. Relatively few broad host-range genetic parts have been characterized, but many libraries of parts for constructing regulatory modules and circuits are starting to become available, albeit using different standards, which makes them difficult to combine (Huang and Lindblad, 2013; Camsund et al., 2014; Albers et al., 2015; Markley et al., 2015; Englund et al., 2016; Immethun et al., 2017; Kim et al., 2017; Taton et al., 2017; Ferreira et al., 2018; Li et al., 2018; Liu and Pakrasi, 2018; Wang et al., 2018). One key challenge is clear: parts that are widely used in Escherichia coli behave very differently in model cyanobacterial species such as Synechocystis (Heidorn et al., 2011). Furthermore, different cyanobacterial strains generally show a wide variation regarding functionality and performance of different genetic parts (e.g. promoters, reporter genes, and antibiotic resistance markers; Taton et al., 2014, 2017; Englund et al., 2016; Kim et al., 2017). This suggests that parts need to be validated, calibrated, and perhaps modified for individual strains, including model species and strains that may be more commercially relevant. Rapid cloning and assembly methods are essential for accelerating the “design, build, test, and learn” cycle, which is a central tenet of synthetic biology (Nielsen and Keasling, 2016).
The adoption of new cloning and vector assembly methods (e.g. Isothermal [Gibson] Assembly and Modular Cloning [MoClo]), assembly standards, and part libraries has greatly enhanced the scalability of synthetic biology-based approaches in a range of biological systems (Moore et al., 2016; Vazquez-Vilar et al., 2018). Recent advances in synthetic biology have led to the development of standards for Type IIS restriction endonuclease-mediated assembly (commonly known as Golden Gate cloning) for several model systems, including plants (Sarrion-Perdigones et al., 2013; Engler et al., 2014; Andreou and Nakayama, 2018). Based on a common Golden Gate MoClo syntax, large libraries are now available for fusion of different genetic parts to assemble complex vectors cheaply and easily without proprietary tools and reagents (Patron et al., 2015). High-throughput and automated assembly are projected to be widely available soon through DNA synthesis and construction facilities, such as the United Kingdom DNA Synthesis Foundries, where MoClo is seen as the most suitable assembly standard (Chambers et al., 2016).
Here, we describe the development of an easy-to-use system called CyanoGate that unites cyanobacteria with plant and algal systems. This system builds on the established Golden Gate MoClo syntax and assembly library for plants (Engler et al., 2014) that has been adopted by the OpenPlant consortium (www.openplant.org), iGEM competitions as “Phytobricks,” and the MoClo kit for the microalga Chlamydomonas reinhardtii (Crozet et al., 2018). First, we constructed and characterized a suite of known and new genetic parts (level 0) for use in cyanobacterial research, including promoters, terminators, antibiotic resistant markers, neutral sites (NS), and gene repression systems (Na et al., 2013; Yao et al., 2016; Sun et al., 2018). Second, we designed an additional level of acceptor vectors (level T) to facilitate integrative or replicative transformation. We characterized assembled level T vectors in Synechocystis and in Synechococcus elongatus UTEX 2973 (UTEX 2973 hereafter), which has a reported doubling time similar to that of Saccharomyces cerevisiae under specific growth conditions (Yu et al., 2015; Ungerer et al., 2018a, 2018b). Lastly, we developed an online tool for assembly of CyanoGate and Plant MoClo vectors to assist with the adoption of the CyanoGate system.
RESULTS AND DISCUSSION
Construction of the CyanoGate System
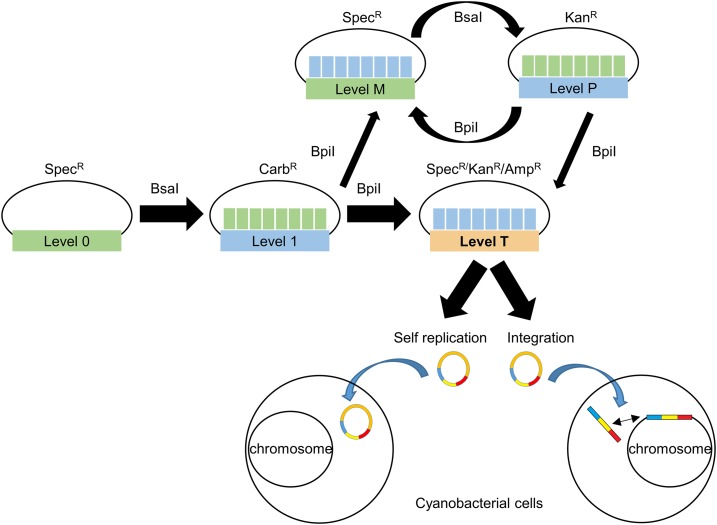
The CyanoGate system integrates with the two-part Golden Gate MoClo Plant Tool Kit, which can be acquired from Addgene (standardized parts [Kit no. 1000000047] and backbone acceptor vectors [Kit no. 1000000044]; www.addgene.org; Engler et al., 2014). A comparison of the benefits of MoClo- and Gibson assembly-based cloning strategies is shown in Supplemental Information S1. The syntax for level 0 parts was adapted for prokaryotic cyanobacteria to address typical cloning requirements for cyanobacterial research (Fig. 1). New level 0 parts were assembled from a variety of sources (Supplemental Table S1). Level 1, M, and P acceptor vectors were adopted from the MoClo Plant Tool Kit, which facilitates assembly of level 0 parts in a level 1 vector, and subsequently up to seven level 1 modules in level M. Level M assemblies can be combined further into level P and cycled back into level M to produce larger multimodule vectors if required (Supplemental Information S2). Vectors >50 kb in size assembled by MoClo have been reported (Werner et al., 2012). Modules from level 1 or level P can be assembled in new level T vectors designed for cyanobacterial transformation (Fig. 2). We found that both UTEX 2973 and Synechocystis produced recombinants following electroporation or conjugation methods with level T vectors. For the majority of the work outlined below, we relied on the conjugation approach.
Figure 1.
Adaptation of the Plant Golden Gate MoClo level 0 syntax for generating level 1 assemblies for transfer to level T. A, The format for a level 0 MoClo acceptor vector with the part bordered by two BsaI sites. B, Typical level 0 parts from the Plant MoClo kit (Engler et al., 2014), where parts of the same type are bordered by the same pair of fusion sites (for each fusion site, only the sequence of the top strand is shown). Note that the parts are not drawn to scale. C and D, The syntax of the Plant MoClo kit was adapted to generate level 0 parts for engineering marked and unmarked cyanobacterial mutant strains (Lea-Smith et al., 2016). E to I, To generate knock-in mutants, short linker parts (30 bp) were constructed to allow assembly of individual flanking sequences, or marker cassettes (AbR or sacB), in level 1 vectors for subsequent assembly in level T. J and K, Parts required for generating synthetic srRNA or CRISPRi level 1 constructs. See Supplemental Information S2 and S3 for workflows. 3U+Ter, 3′UTR and terminator; AbR, antibiotic resistance cassette; AbR DOWN LINKER, short sequence (∼30 bp) to provide CGCT overhang; AbR UP LINKER, short sequence (∼30 bp) to provide GAGG overhang; CDS2(stop), coding sequence with a stop codon; DOWN FLANK, flanking sequence downstream of target site; DOWN FLANK LINKER, short sequence (∼30 bp) to provide GGAG overhang; Prom+5U, promoter and 5′ UTR; Prom TSS, promoter transcription start site; sacB, levansucrase expression cassette; sacB UP LINKER, short sequence (∼30 bp) to provide GAGG overhang; sgRNA, single guide RNA; SP, signal peptide; srRNA, small regulatory RNA; UP FLANK, flanking sequence upstream of target site; UP FLANK LINKER, short sequence (∼30 bp) to provide CGCT overhang; UNMARK LINKER, short sequence to bridge UP FLANK and DOWN FLANK.
Figure 2.
Extension of the Plant Golden Gate MoClo Assembly Standard for cyanobacterial transformation. Assembly relies on one of two Type IIS restriction endonuclease enzymes (BsaI or BpiI). Domesticated level 0 parts are assembled into level 1 vectors. Up to seven level 1 modules can be assembled directly into a level T cyanobacterial transformation vector, which consists of two subtypes (either a replicative or an integrative vector). Alternatively, larger vectors with more modules can be built by assembling level 1 modules into level M, then cycling assembly between level M and level P, and finally transferring from level P to level T. Antibiotic selection markers are shown for each level. Level T vectors are supplied with internal antibiotic selection markers (shown), but additional selection markers could be included from level 1 modules as required. See Supplemental Table S1 and Supplemental Information S4 for the full list and maps of level T acceptor vectors.
Integration—Generating Marked and Unmarked Knock-Out Mutants
A common method for engineering stable genomic knock-out and knock-in mutants in several cyanobacteria relies on homologous recombination via integrative (suicide) vectors using a two-step marked-unmarked strategy (Supplemental Information S3; Lea-Smith et al., 2016). Saar et al. (2018) recently used this approach to introduce up to five genomic alterations into a single Synechocystis strain. First, marked mutants are generated with an integrative vector carrying two sequences (∼1 kb each) identical to the regions of the cyanobacterial chromosome flanking the deletion/insertion site. Two gene cassettes are inserted between these flanking sequences: a levansucrase expression cassette (sacB) that confers sensitivity to transgenic colonies grown on Suc and an antibiotic resistance cassette (AbR) of choice. Second, unmarked mutants (carrying no selection markers) are generated from fully segregated marked lines using a separate integrative vector carrying only the flanking sequences and selection on plates containing Suc.
We adapted this approach for the CyanoGate system (Fig. 1). To generate level 1 vectors for making knock-out mutants, sequences flanking the upstream (UP FLANK) and downstream (DOWN FLANK) sites of recombination were ligated into the plant MoClo Prom+5U (with overhangs GGAG-AATG) and 3U+Ter (GCTT-CGCT) positions, respectively, to generate new level 0 parts (Fig. 1B). In addition, full expression cassettes were made for Suc selection (sacB) and antibiotic resistance (AbRSpec, AbRKan, and AbREry) in level 0 that ligate into positions signal peptide (AATG-AGGT) and coding sequence 2 with stop codon (CDS2 [stop]; AGGT-GCTT), respectively. Marked level 1 modules can be assembled using UP FLANK, DOWN FLANK, sacB, and the required AbR level 0 part. For generating the corresponding unmarked level 1 module, a short 59-bp linker (UNMARK LINKER) can be ligated into the CDS1ns (no stop codon; AATG-GCTT) position for assembly with an UP FLANK and DOWN FLANK (Fig. 1D). Unmarked and marked level 1 modules can then be assembled into level T integrative vectors, with the potential capacity to include multiple knock-out modules in a single level T vector.
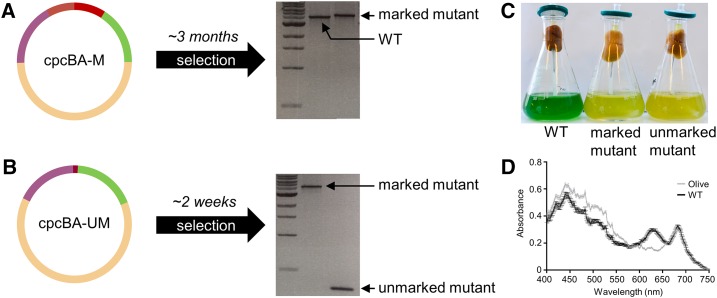
To validate our approach, we constructed the level 0 flanking vectors pC0.024 and pC0.025 and assembled two level T integrative vectors using pUC19-T (cpcBA-M and cpcBA-UM, with and without the sacB and AbR cassettes, respectively) to remove the cpcBA promoter and operon in Synechocystis and generate an “Olive” mutant unable to produce the phycobiliprotein C-phycocyanin (Fig. 3; Supplemental Table S1; Kirst et al., 2014; Lea-Smith et al., 2014). Following transformation with cpcBA-M, we successfully generated a marked ΔcpcBA mutant carrying the sacB and the AbRKan cassettes after selective segregation (∼3 months; Fig. 3A). The unmarked ΔcpcBA mutant was then isolated following transformation of the marked ΔcpcBA mutant with cpcBA-UM and selection on Suc (∼2 weeks; Fig. 3B). Absence of C-phycocyanin in the Olive mutant resulted in a characteristic drop in A625 (Fig. 3D) and a significant reduction in chlorophyll content compared to that in wild-type cells (28.4 ± 0.2 and 48.3 ± 0.2 amol chl cell−1, respectively; Kirst et al., 2014; Lea-Smith et al., 2014).
Figure 3.
Generating knock-out mutants in cyanobacteria. A, Assembled level T vector cpcBA-M (see Fig. 1C) targeting the cpcBA promoter and operon (3,563 bp) to generate a marked ΔcpcBA “Olive” mutant in Synechocystis sp. PCC 6803. Following transformation and segregation on kanamycin (∼3 months), a segregated marked mutant was isolated (wild-type [WT] band is 3,925 bp, marked mutant band is 5,503 bp, 1-kb DNA ladder [New England Biolabs] is shown). B, Assembled level T vector cpcBA-UM (see Fig. 1D) for generating an unmarked ΔcpcBA mutant. Following transformation and segregation on Suc (∼2 weeks), an unmarked mutant was isolated (unmarked band is 425 bp). C, Liquid cultures of wild type and marked and unmarked Olive mutants. D, Spectrum showing the absorbance of the unmarked Olive mutant and wild-type cultures after 72 h of growth. Values are the average of four biological replicates ± se and are standardized to 750 nm.
Generating Knock-In Mutants
Flexibility in designing level 1 insertion cassettes is needed when making knock-in mutants. Thus, for knock-in mutants the upstream and downstream sequences flanking the insertion site, and any required expression or marker cassettes, are first assembled into separate level 1 modules from UP FLANK and DOWN FLANK level 0 parts (Fig. 1, E and F). Seven level 1 modules can be assembled directly into level T (Fig. 2). Therefore, with a single pair of flanking sequences, up to five level 1 expression cassettes could be included in a level T vector.
Linker parts (20 bp) UP FLANK LINKER and DOWN FLANK LINKER were generated to allow assembly of level 0 UP FLANK and DOWN FLANK parts into separate level 1 acceptor vectors. Similarly, level 0 linker parts were generated for sacB and AbR (Fig. 1, H and I). Level 1 vectors at different positions can then be assembled in level T (or M) containing one or more expression cassettes, an AbR of choice, or both sacB and AbR (Fig. 2).
Using this approach, CyanoGate can facilitate the generation of knock-in mutants using a variety of strategies. For example, if retention of a resistance marker is not an experimental requirement (e.g. Liberton et al., 2017), only a single antibiotic resistance cassette needs to be included in level T. Alternatively, a two-step marked-unmarked strategy could be followed, as for generating knock-out mutants.
Whereas knock-out strategies can target particular loci, knock-in approaches often rely on recombination at designated NS within the genome of interest that can be disrupted with no or minimal impact on the growth phenotype (Ng et al., 2015; Pinto et al., 2015). Based on loci reported in the literature, we have assembled a suite of flanking regions to target NS in Synechocystis (designated 6803 NS1–NS4; Pinto et al., 2015), Synechococcus sp. PCC 7002 (PCC 7002 hereafter; designated 7002 NS1 and NS2; Ruffing et al., 2016; Vogel et al., 2017), and NS common to UTEX 2973, PCC 7942 and S. elongatus PCC 6301 (designated 7942 NS1-3; Supplemental Table S1; Bustos and Golden, 1992; Kulkarni and Golden, 1997; Andersson et al., 2000; Niederholtmeyer et al., 2010). Pinto et al. (2015) have qualitatively compared the impact of the four Synechocystis NS assembled here under several different growth conditions, and observed that insertions at 6803 NS3 and NS4 had no significant effect on growth compared to that of wild-type cultures, whereas insertions at NS2 and NS1 had small but significant effects depending on the growth conditions. Several studies have used 6803 NS3, for example, to engineer a Synechocystis strain for the bioremediation of microcystins (Dexter et al., 2018) and the development of T7 polymerase-based synthetic promoter systems (Ferreira et al., 2018). For the two PCC 7002 NS, growth rates with insertions at 7002 NS1 were slightly reduced (Vogel et al., 2017), but not significantly affected with insertions at 7002 NS2 (Ruffing et al., 2016). Insertions at the three 7942 NS reportedly have no phenotypic effect on morphology or growth rate (Clerico et al., 2007; Niederholtmeyer et al., 2010) and have been used to study mRNA stability and translation (Kulkarni and Golden, 1997), circadian rhythms (Andersson et al., 2000), chromosome duplication (Watanabe et al., 2018) and to engineer PCC 7942 for synthesizing heterologous products (Niederholtmeyer et al., 2010; Gao et al., 2016). When using the NS supplied with CyanoGate (or others), we would still recommend a thorough growth analysis under the specific culturing conditions being tested to identify any potential impact of the inserted DNA on growth phenotype.
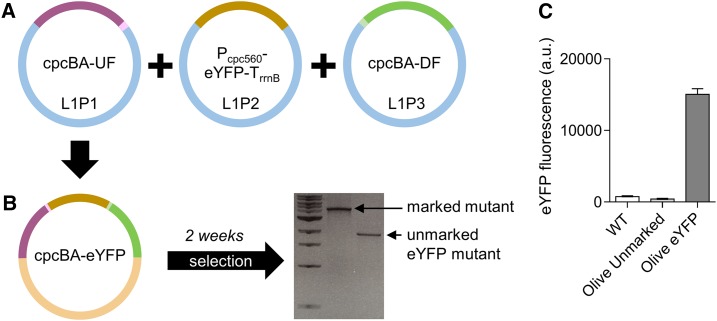
To validate our system, we generated a level T vector carrying the flanking regions for the cpcBA operon and an enhanced yellow fluorescent protein (eYFP) expression cassette (cpcBA-eYFP; Fig. 4, A and B; Supplemental Table S1). We successfully transformed this vector into our marked “Olive” Synechocystis mutant, and generated a stable olive mutant with constitutive expression of eYFP (Olive-eYFP; Fig. 4C).
Figure 4.
Generating knock-in mutants in cyanobacteria. A, Assembly of level 1 modules cpcBA-UF (see Fig. 1E) in the level 1, position 1 acceptor (L1P1), Pcpc560-eYFP-TrrnB (see Fig. 1G) in L1P2, and cpcBA-DF (see Fig. 1F) in L1P3. B, Transfer of level 1 assemblies to level T vector cpcBA-eYFP for generating an unmarked ΔcpcBA mutant carrying an eYFP expression cassette. Following transformation and segregation on Suc (∼3 weeks), an unmarked eYFP mutant was isolated (1,771 bp). C, Fluorescence values are the means ± se of four biological replicates, where each replicate represents the median measurements of 10,000 cells.
Expression Comparison for Promoter Parts in Synechocystis and UTEX 2973
We constructed level 0 parts for a wide selection of synthetic promoters and promoters native to Synechocystis. Promoters were assembled as expression cassettes driving eYFP in replicative level T vector pPMQAK1-T to test for differences in expression when conjugated into Synechocystis or UTEX 2973. We first compared the growth rates of Synechocystis, UTEX, and PCC 7942 (a close relative of UTEX 2973 [Yu et al., 2015]) under a variety of different culturing conditions (Supplemental Fig. S1). We found that growth rates were comparable between Synechocystis and PCC 7942 at temperatures below 40°C regardless of light levels and supplementation with CO2. In contrast, UTEX 2973 grew poorly under those conditions. UTEX 2973 only showed an enhanced growth rate at 45°C under the highest light tested (500 µmol photons m−2 s−1) with CO2, whereas all three strains failed to grow at 50°C. These results confirm that the enhanced growth phenotype reported for UTEX 2973 requires specific conditions as reported by Ungerer et al. (2018a, 2018b). Furthermore, they are consistent with recent reports that this phenotype is linked to an increased stress tolerance, which has been attributed to a small number of nucleotide polymorphisms (Lou et al., 2018; Ungerer et al., 2018b). We proceeded with CyanoGate part characterizations and comparisons under the best conditions achievable for Synechocystis and UTEX 2973 (see “Materials and Methods”; Supplemental Fig. S2A).
Promoters Native to Synechocystis
We assembled several previously reported promoters from Synechocystis in the CyanoGate kit. These include six inducible/repressible promoters (PnrsB, PcoaT, PnirA, PpetE, PisiAB, and ParsB), which were placed in front of the strong, synthetic Synechocystis ribosomal binding site (RBS; Heidorn et al., 2011) as used in Englund et al. (2016; Supplemental Table S1; Supplemental Information S4). PnrsB and PcoaT drive the expression of nickel and cobalt ion efflux pumps and are induced by Ni2+, and Co2+ or Zn2+, respectively (Peca et al., 2008; Blasi et al., 2012; Guerrero et al., 2012; Englund et al., 2016). PnirA, from the nitrate assimilation operon, is induced by the presence of NO3− and/or NO2− (Kikuchi et al., 1996; Qi et al., 2005). PpetE drives the expression of plastocyanin and is induced by Cu2+, which has previously been used for the expression of heterologous genes (Guerrero et al., 2012; Camsund et al., 2014). The promoter of the isiAB operon (PisiAB) is repressed by Fe3+ and activated when the cell is under iron stress (Kunert et al., 2003). ParsB drives the expression of a putative arsenite and antimonite carrier and is activated by AsO2− (Blasi et al., 2012).
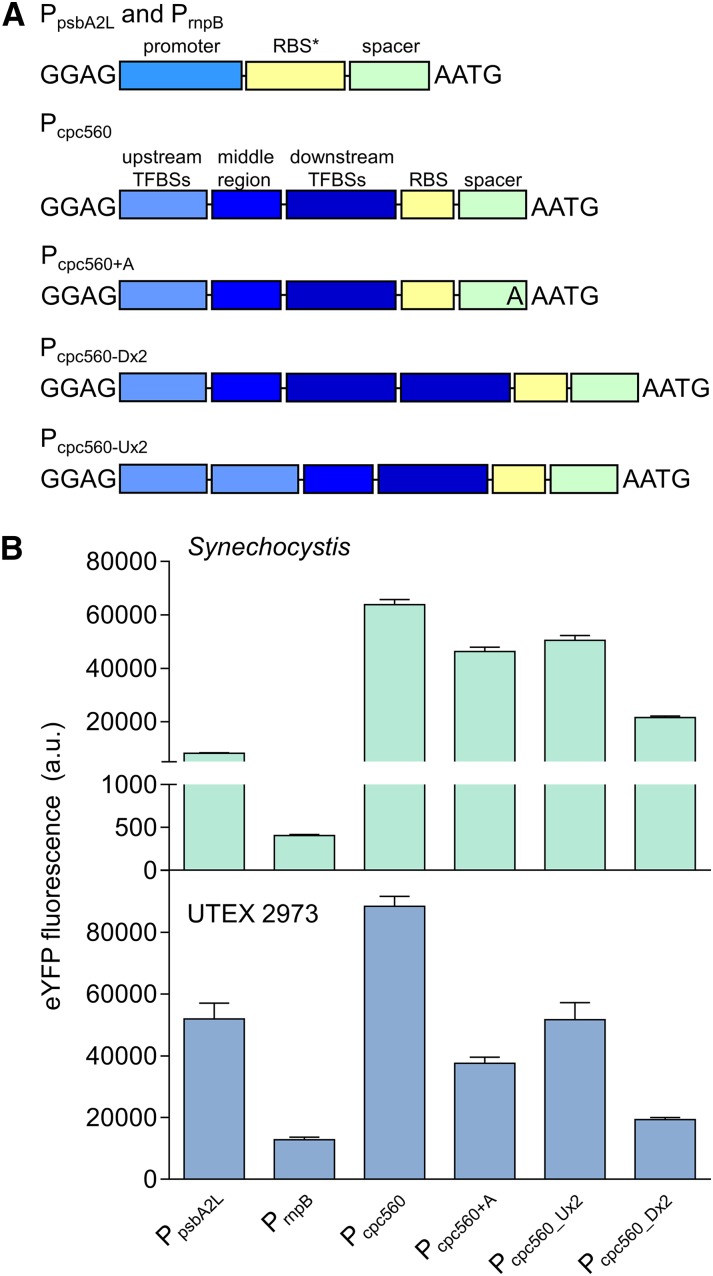
We also cloned the rnpB promoter, PrnpB, from the RNase P gene (Huang et al., 2010), a long version of the psbA2 promoter, PpsbA2L, from the Photosystem II protein D1 gene (Lindberg et al., 2010; Englund et al., 2016) and the promoter of the C-phycocyanin operon, Pcpc560 (also known as PcpcB and PcpcBA; Zhou et al., 2014). PrnpB and PpsbA2L were placed in front of RBS (Fig. 5A; Heidorn et al., 2011). To build on a previous functional characterization of Pcpc560 (Zhou et al., 2014), we assembled four variants of this strong promoter. First, Pcpc560+A consisted of the promoter and the 4-bp MoClo overhang AATG. Second, Pcpc560 was truncated by 1 bp (Fig. 5A, A), so that the start codon was aligned with the native Pcpc560 RBS spacer region length. Zhou et al. (2014) identified 14 predicted transcription factor binding sites (TFBSs) in the upstream region of Pcpc560 (−556 to −381 bp) and removal of this region resulted in a significant loss of promoter activity. However, alignment of the reported TFBSs showed that their locations are in the downstream region of the promoter (−180 to −5 bp). We identified 11 additional predicted TFBSs using Virtual Footprint (Münch et al., 2005) in the upstream region and hypothesized that the promoter activity may be modified by duplicating either of these regions. So, third, we generated Pcpc560_Dx2 containing a duplicated downstream TFBS region. For Pcpc560_Dx2, only the region between −31 to −180 bp was duplicated to avoid repeating the Shine-Dalgarno sequence. Fourth, we duplicated the upstream region to generate Pcpc560_Ux2. We then assembled PrnpB, PpsbA2L, and the four Pcpc560 variants with eYFP and the rrnB terminator (TrrnB) into a level 1 expression cassette, and subsequently into a level T replicative vector (pPMQAK1-T) for expression analysis (Supplemental Table S2).
Figure 5.
Expression levels of cyanobacterial promoters in Synechocystis and UTEX 2973. A, Structure of the cyanobacterial promoters adapted for the CyanoGate kit. Regions of Pcpc560 shown are the TFBSs (−556 to −381 bp), middle region (−380 to −181 bp), and the downstream TFBSs, RBS, and spacer (−180 to −5 bp). B, Expression levels of eYFP driven by promoters in Synechocystis and UTEX 2973 calculated from measurements taken from 10,000 individual cells. Values are the means ± se from at least four biological replicates after 48 h of growth (average OD750 values for Synechocystis and UTEX 2973 cultures were 3.5 ± 0.2 and 3.6 ± 0.2, respectively). See Supplemental Figure S2 for more info.
In Synechocystis the highest-expressing promoter was Pcpc560 (Fig. 5B), which indicated that maintaining the native RBS spacer region for Pcpc560 is important for maximizing expression. Neither Pcpc560_Dx2 nor Pcpc560_Ux2 resulted in higher expression levels compared to that of Pcpc560. Pcpc560_Dx2–driven expression was strongly decreased compared to expression driven by Pcpc560, suggesting that promoter function is sensitive to modification of the downstream region and this region could be a useful target for modulating Pcpc560 efficacy. Previous work in Synechocystis has suggested that modification of the middle region of Pcpc560 (−380 to −181 bp) may also affect function (Lea-Smith et al., 2014). PpsbA2L produced lower expression levels than any variant of Pcpc560 in Synechocystis, whereas PrnpB produced the lowest expression levels. The observed differences in expression levels are consistent with those in other studies with Synechocystis (Camsund et al., 2014; Englund et al., 2016; Liu and Pakrasi, 2018).
In UTEX 2973, the trend in expression patterns was similar to that in Synechocystis (Fig. 5B). However, the overall expression levels of eYFP measured in UTEX 2973 were significantly higher than in Synechocystis. Pcpc560 was increased by 30%, whereas PrnpB showed a 20-fold increase in expression relative to Synechocystis. The relative expression strength of PpsbA2L was also higher than in Synechocystis, and second only to Pcpc560 in UTEX 2973. As promoters derived from PpsbA are responsive to increasing light levels (Englund et al., 2016), the increased levels of expression for PpsbA2L may be associated with the higher light intensities used for growing UTEX 2973 compared to that used for growing Synechocystis. Background fluorescence levels were similar between UTEX 2973 and Synechocystis conjugated with an empty pPMQAK1-T vector (i.e. lacking an eYFP expression cassette), which suggested that the higher fluorescence values in UTEX 2973 were a direct result of increased levels of eYFP protein.
Heterologous and Synthetic Promoters
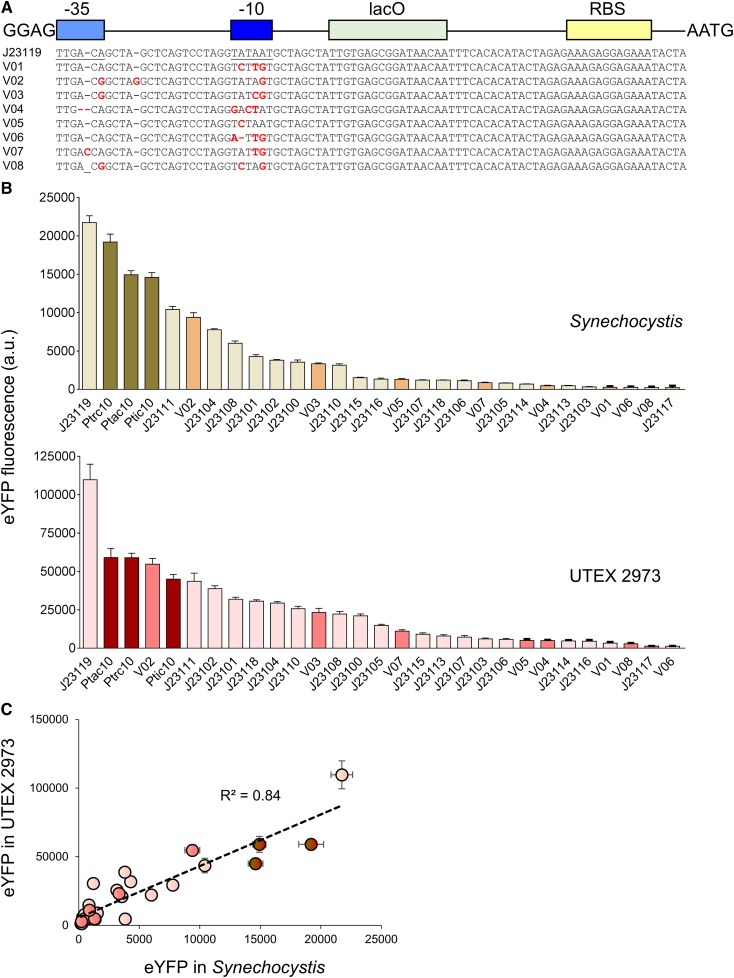
A suite of 20 constitutive synthetic promoters was assembled in level 0 based on the modified BioBricks BBa_J23119 library of promoters (Markley et al., 2015), and the synthetic Ptrc10, Ptic10, and Ptac10 promoters (Supplemental Table S1; Supplemental Information S4; Huang et al., 2010; Albers et al., 2015). We retained the broad-range BBa_B0034 RBS (AAAGAGGAGAAA) and lac operator (lacO) from Huang et al. (2010) for future lacI-based repression experiments (lacI and the PlacIQ promoter are included in the CyanoGate kit; Bahl et al., 1977). We cloned eight new variants (J23119MH_V01-8) with mutations in the canonical BBa_J23119 promoter sequence (Fig. 6A). Additionally, we included the l-arabinose-inducible promoter from E. coli (PBAD; Abe et al., 2014).
Figure 6.
Expression levels of heterologous and synthetic promoters in Synechocystis and UTEX 2973. A, Structure and alignment of eight new synthetic promoters derived from the BioBricks BBa_J23119 library and Ptrc10 promoter design (Huang et al., 2010). B, Expression levels of eYFP driven by promoters in Synechocystis and UTEX 2973 calculated from measurements taken from 10,000 individual cells. C, Correlation analysis of expression levels of synthetic promoters tested in Synechocystis and UTEX 2973. The coefficient of determination (R2) is shown for the J23119 library (red), new synthetic promoters (pink), and trc variants (dark red). Values are the means ± se from at least four biological replicates after 48 h of growth (average OD750 values for Synechocystis and UTEX 2973 cultures were 3.5 ± 0.2 and 3.6 ± 0.2, respectively). See Supplemental Figure S2 for more info.
We encountered an unexpected challenge with random internal deletions in the −35 and −10 regions of some promoters of the BBa_J23119 library and trc promoter variants when cloning them into level 0 acceptors. Similar issues were reported previously for the E. coli EcoFlex kit (Moore et al., 2016) that may relate to the functionality of the promoters and the host vector copy number in E. coli, which consequently resulted in cell toxicity and selection for mutated promoter variants. To resolve this issue, we generated a low copy level 0 promoter acceptor vector compatible with CyanoGate (pSB4K5 acceptor) for cloning recalcitrant promoters (Supplemental Table S1; Supplemental Information S4). Subsequent assemblies in levels 1 and T showed no indication of further mutation.
We then tested the expression levels of eYFP driven by the synthetic promoters in Synechocystis and UTEX 2973 following assembly in pPMQAK1-T (Fig. 6B; Supplemental Table S2). The synthetic promoters showed a 120-fold dynamic range in both cyanobacterial strains. Furthermore, a similar trend in promoter expression strength was observed (R2 = 0.84; Fig. 6C). However, eYFP expression levels were on average 8-fold higher in UTEX 2973 compared to that in Synechocystis. In Synechocystis, the highest expression levels were observed for J23119 and Ptrc10, but these were still ∼50% lower than values for the native Pcpc560 promoters (Fig. 5B). The expression trends for the BBa_J23119 library were consistent with the subset reported by Camsund et al. (2014) in Synechocystis, whereas the observed differences between Ptrc10 and Pcpc560 were similar to those reported by Liu and Pakrasi (2018).
In contrast, the expression levels in UTEX 2973 for J23119 were ∼50% higher than that for Pcpc560. Several synthetic promoters showed expression levels in a similar range to those for the native Pcpc560 promoter variants, including Ptrc10, J23111, and the J23119 variant V02. V02 is identical to J23111 except for an additional “G” between the −35 and −10 motifs, suggesting that small changes in the length of this spacer region may not be critical for promoter strength (similar expression levels were also observed for these two promoters in Synechocystis). In contrast, a single bp difference between J23111 and J23106 in the −35 motif resulted in an 8- and 10-fold reduction in expression in Synechocystis and UTEX 2973, respectively. The results for UTEX 2973 were unexpected, and to our knowledge no studies to date have directly compared these promoters in this strain. Recent work has examined the expression of β-galactosidase using promoters such as Pcpc560 and Ptrc in UTEX 2973 (Li et al., 2018). Li et al. (2018) highlighted that different growth environments (e.g. light levels) can have significant effects on protein expression. Changes in culture density can also affect promoter activity, such that protein expression levels can change during the exponential and stationary growth stages depending on the promoter and expression vector used (Ng et al., 2015; Madsen et al., 2018). Here we tracked eYFP expression levels over time for 3 d during the early and late exponential growth phases for Synechocystis and UTEX 2973. Although expression levels for each promoter fluctuated over time, with peak expression levels at 24 h and 48 h in UTEX 2973 and Synechocystis, respectively, the overall expression trends were generally consistent for the two strains (Supplemental Fig. S2B).
Protein Expression Levels in Synechocystis and UTEX 2973
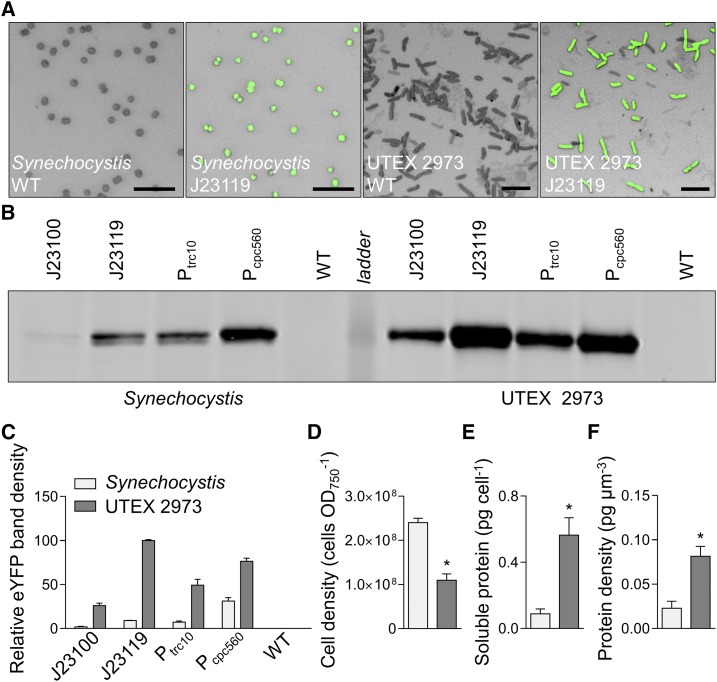
To investigate further the increased levels of eYFP expression observed in UTEX 2973 compared to that in Synechocystis, we examined cell morphology, protein content, and eYFP protein abundances in expression lines for each strain. Confocal image analysis confirmed the coccoid and rod shapes of Synechocystis and UTEX 2973, respectively, and the differences in cell size (Fig. 7A; van de Meene et al., 2006; Yu et al., 2015). Immunoblot analyses of eYFP from protein extracts of four eYFP-expressing strains correlated well with previous flow cytometry measurements (Figs. 5 and 6). eYFP driven by the J23119 promoter in UTEX 2973 produced the highest levels of eYFP protein (Fig. 7, B and C). Although the density of cells in culture was 2-fold higher in Synechocystis compared to that in UTEX 2973 (Fig. 7D), the protein content per cell was 6-fold lower (Fig. 7E). We then estimated the average cell volumes for Synechocystis and UTEX 2973 at 3.91 ± 0.106 µm3 and 6.83 ± 0.166 µm3 (n = 50 each), respectively, based on measurements from confocal microscopy images (Supplemental Fig. S3). Based on those estimates, we calculated that the density of soluble protein per cell was 4-fold higher in UTEX 2973 compared to that in Synechocystis (Fig. 7F). Thus, we hypothesized that the enhanced levels of eYFP observed in UTEX 2973 were a result of the expression system harnessing a larger available amino acid pool. Mueller et al. (2017) have reported that UTEX 2973 has an increased investment in amino acid content compared to that in PCC 7942, which may be linked to higher rates of translation in UTEX 2973. Therefore, UTEX 2973 continues to show promise as a bioplatform for generating heterologous protein products, although future work should study production rates under conditions optimal for faster growth (Lou et al., 2018; Ungerer et al., 2018a). Recent characterization of the UTEX 2973 transcriptome will also assist with native promoter characterizations (Tan et al., 2018).
Figure 7.
Protein expression levels in Synechocystis and UTEX 2973 cells. A, Confocal images of wild-type (WT) strains and mutants expressing eYFP (eYFP fluorescence shown in green on bright field images) driven by the J23119 promoter (bars = 10 µm). B, Representative immunoblot of protein extracts (3 µg protein) from mutants with different promoter expression cassettes (as in Fig. 6) probed with an antibody against eYFP. The protein ladder band corresponds to 30 kD. C, Relative eYFP protein abundance relative to that in UTEX 2973 mutants carrying the J23119 expression cassette. D to F, Cell density (D), protein content per cell (E), and protein density per estimated cell volume (F) for Synechocystis and UTEX 2973. Asterisks (*) indicate significant difference (P < 0.05) as determined by Student’s t test. Values are the means ± se of four biological replicates.
The RK2 Origin of Replication Is Functional in Synechocystis
Synthetic biology tools (e.g. gene expression circuits, CRISPR/Cas-based systems) are often distributed between multiple plasmid vectors at different copy numbers in order to synthesize each component at the required concentration (Bradley et al., 2016). The large RSF1010 vector is able to replicate in a broad range of microbes including gram-negative bacteria such as E. coli and several cyanobacterial species. However, for 25 years it has remained the only nonnative vector reported to be able to self-replicate in cyanobacteria (Mermet-Bouvier et al., 1993). Recently, two small plasmids native to Synechocystis, pCA2.4 and pCB2.4, have been engineered for gene expression (Armshaw et al., 2015; Ng et al., 2015; Liu and Pakrasi, 2018). The pANS plasmid (native to PCC 7942) has also been adapted as a replicative vector, but so far it has been only shown to function in PCC 7942 and Anabaena PCC 7120 (Chen et al., 2016). Similarly, the high copy number plasmid pAQ1 (native to PCC 7002) has been engineered for heterologous expression, but up to now it has only been used in PCC 7002 (Xu et al., 2011). To expand the replication origins available for cyanobacterial research further we tested the capacity for vectors from the Standard European Vector Architecture (SEVA) library to replicate in Synechocystis (Silva-Rocha et al., 2013).
We acquired three vectors driven by three different replication origins (pSEVA421 [RK2], pSEVA431 [pBBR1], and pSEVA442 [pRO1600/ColE1]) and carrying a spectinomycin antibiotic resistance marker. These vectors were domesticated and modified as level T acceptor vectors, assembled, and then transformed into Synechocystis by electroporation or conjugation. Only Synechocystis strains conjugated with vectors carrying RK2 (pSEVA421-T) grew on spectinomycin-containing plates (Supplemental Table S1; Supplemental Information S4). To confirm that RSF1010 and RK2 replication origins can replicate autonomously in Synechocystis, we recovered the pPMQAK1-T or pSEVA421-T vector from lysates of axenic Synechocystis strains previously conjugated with each vector by transformation into E. coli. The identity and integrity of pPMQAK1-T and pSEVA421-T extracted from transformed E. coli colonies were confirmed by restriction digest and Sanger sequencing.
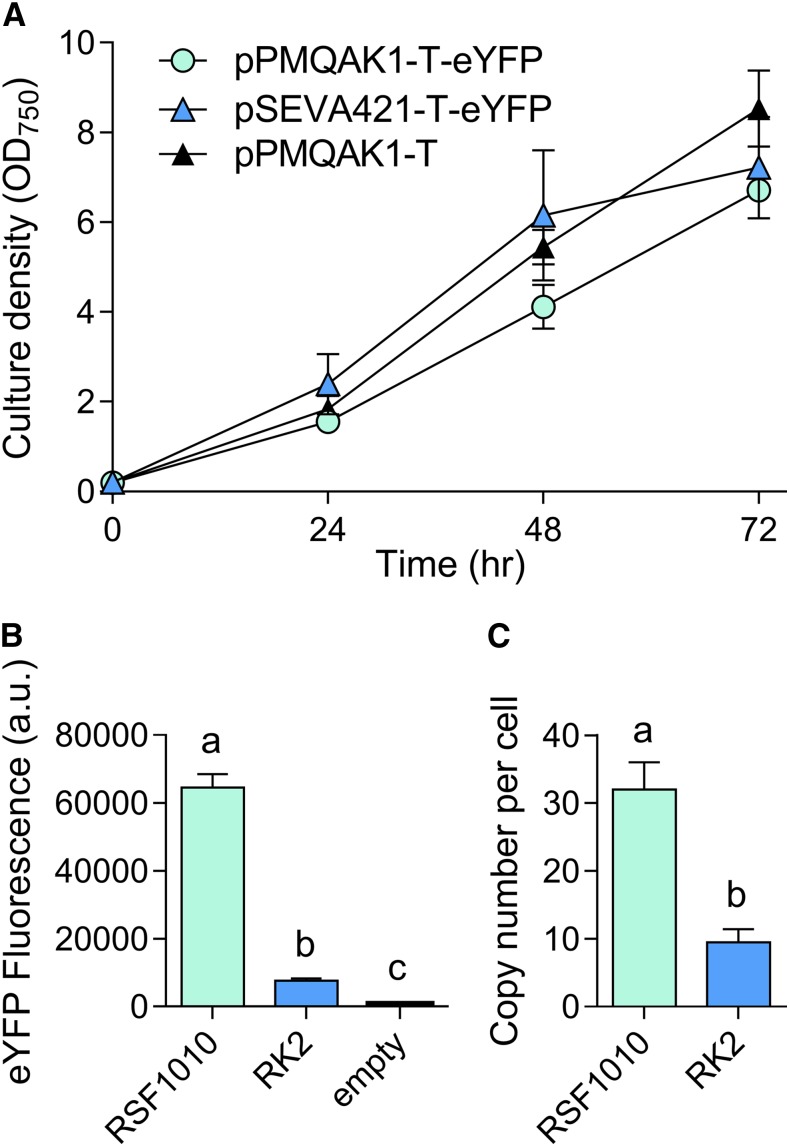
We then assembled two level T vectors with an eYFP expression cassette (Pcpc560-eYFP-TrrnB) to produce pPMQAK1-T-eYFP and pSEVA421-T-eYFP, which were conjugated into Synechocystis (Fig. 8; Supplemental Table S2). Both pPMQAK1-T-eYFP and pSEVA421-T-eYFP transconjugates grew at similar rates in 50 µg mL−1 kanamycin and 5 µg mL−1 spectinomycin, respectively (Fig. 8A). However, eYFP levels were 8-fold lower in pSEVA421-T-eYFP, suggesting that RK2 has a reduced copy number relative to RSF1010 in Synechocystis (Fig. 8B). We measured the heterologous plasmid vector copy number in strains expressing pSEVA421-T or pPMQAK1-T and estimated an average copy number per cell of 9 ± 2 and 31 ± 5, respectively (Fig. 8C). The copy number for pPMQAK1-T was similar to values reported previously for RSF1010-derived vectors in Synechocystis (∼30; Ng et al., 2000). Our results are also consistent with the lower copy numbers in E. coli for vectors with RK2 (4 to 7 copies) compared to those with RSF1010 (10 to 12 copies) replication origins (Frey et al., 1992; Blasina et al., 1996). Furthermore, we compared the genome copies per cell between transformants and wild-type strains and found no significant differences—the average value was 11 ± 2, which is consistent with the typical range of genome copy numbers observed in Synechocystis cells (Zerulla et al., 2016).
Figure 8.
Cell growth and expression levels of eYFP with the RK2 replicative origin in Synechocystis. A, Growth of strains carrying RK2 (vector pSEVA421-T-eYFP), RSF1010 (pPMQAK1-T-eYFP), or empty pPMQAK1-T, with cultures containing appropriate antibiotic selection. Growth was measured as OD750 under a constant illumination of 100 µmol photons m−2 s−1 at 30°C. B, Expression levels of eYFP after 48 h of growth calculated from measurements taken from 10,000 individual cells. C, Plasmid copy numbers per cell after 48 h of growth. Lowercase letters indicating significant difference (P < 0.05) are shown, as determined by ANOVA followed by Tukey’s honestly significant difference tests. Values are the means ± se of four biological replicates.
Gene Repression Systems
CRISPR interference (CRISPRi) is a relatively new but well characterized tool for modulating gene expression at the transcription stage in a sequence-specific manner (Qi et al., 2013; Behler et al., 2018). CRISPRi typically uses a nuclease-deficient Cas9 from Streptococcus pyogenes (dCas9) and has been demonstrated to work in several cyanobacterial species, including Synechocystis (Yao et al., 2016), PCC 7002 (Gordon et al., 2016), PCC 7942 (Huang et al., 2016), and Anabaena sp. PCC 7120 (Higo et al., 2018). A second approach for gene repression uses rationally designed small regulatory RNAs (srRNAs) to regulate gene expression at the translation stage (Na et al., 2013; Higo et al., 2016). The synthetic srRNA is attached to a scaffold to recruit the Hfq protein, an RNA chaperone that is conserved in a wide range of bacteria and cyanobacteria, which facilitates the hybridization of srRNA and target mRNA, and directs mRNA for degradation. The role of cyanobacterial Hfq in interacting with synthetic srRNAs is still unclear (Zess et al., 2016). However, regulatory ability can be improved by introducing Hfq from E. coli into Synechocystis (Sakai et al., 2015). Both CRISPRi- and srRNA-based systems have potential advantages as they can be used to repress multiple genes simultaneously.
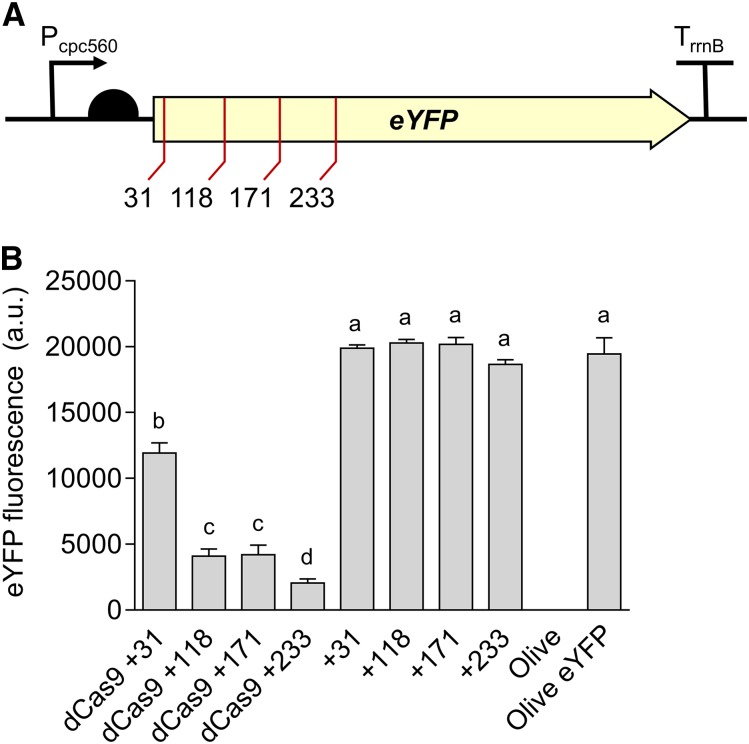
To validate the CRISPRi system, we assembled an expression cassette for dCas9 (Pcpc560-dCas9-TrrnB) on the level 1 position 1 vector pICH47732, and four different sgRNA (single guide RNA) expression cassettes (Ptrc10_TSS-sgRNA-sgRNA scaffold) targeting eYFP on the level 1 position 2 vector pICH47742 (Supplemental Table S2; Engler et al., 2014). For assembly of CRISPRi sgRNA expression cassettes in level 1, we targeted four 18- to 22-bp regions of the eYFP nontemplate strand with an adjacent 3′ protospacer adjacent motif of 5′-NGG-3′, as required by S. pyogenes dCas9 (Fig. 9A). The sgRNA sequences contained no off-target sites in the Synechocystis genome (confirmed by CasOT; Xiao et al., 2014). The sgRNAs were made by PCR using two complementary primers carrying the required overhangs and BsaI sites, and were assembled with the Ptrc10_TSS promoter (pC0.220) and the sgRNA scaffold (pC0.122). Level T vectors were assembled carrying dCas9 and a single sgRNA, or just the sgRNA alone. We subsequently conjugated the Olive-eYFP mutant and tracked eYFP expression.
Figure 9.
Gene regulation system using CRISPRi in Synechocystis. A, Four target regions were chosen as sgRNA protospacers to repress eYFP expression in Olive-eYFP (Fig. 4): “CCAGGATGGGCACCACCC” (+31), “ACTTCAGGGTCAGCTTGCCGT” (+118), “AGGTGGTCACGAGGGTGGGCCA” (+171), and “AGAAGTCGTGCTGCTTCATG” (+233). B, eYFP fluorescence of Olive-eYFP expressing constructs carrying sgRNAs with and without dCas9 (representative of 10,000 individual cells). Untransformed Olive-eYFP and the Olive mutant were used as controls. Lowercase letters indicating significant difference (P < 0.05) are shown, as determined by ANOVA followed by Tukey’s honestly significant difference tests. Values are the means ± se of four biological replicates.
Transconjugates carrying only the sgRNA showed no reduction in eYFP level compared to that in nontransconjugated Olive-eYFP (Fig. 9B). However, all strains carrying dCas9 and a sgRNA showed a decrease in eYFP that ranged from 40% to 90% depending on the sgRNA used. These reductions are similar to those observed previously in PCC 7002 and in Synechocystis (Gordon et al., 2016; Yao et al., 2016) and demonstrated that CRISPRi system is functional in the CyanoGate kit.
CONCLUSION
The CyanoGate kit was designed to increase the availability of well characterized libraries and standardized modular parts in cyanobacteria (Sun et al., 2018). We aimed to simplify and accelerate modular cloning methods in cyanobacterial research and allow integration with the growing number of labs that rely on the established common plant and algal syntax for multipart DNA assembly (Patron et al., 2015; Crozet et al., 2018). Here, we have demonstrated the functionality of CyanoGate in sufficient detail to show that it is straightforward to adopt and functionally robust across two different cyanobacterial species. CyanoGate includes parts for usage in other cyanobacterial species and could likely be utilized also in noncyanobacterial microbes amenable to transformation (e.g. Rhodopseudomonas spp.) and adapted for use in subcellular eukaryotic compartments of prokaryote origin (e.g. chloroplasts; Economou et al., 2014; Doud et al., 2017; Leonard et al., 2018). In addition to the parts discussed, we have also assembled a suite of 21 terminators (Supplemental Table S1). To increase the accessibility and usability of the CyanoGate, we have included the vector maps for all parts and new acceptors (Supplemental Information S4), implemented support for Cyanogate assemblies in the online DNA “Design and Build” portal of the Edinburgh Genome Foundry (dab.genomefoundry.org; Supplemental Information S5), and submitted all vectors as a toolkit for order from Addgene (Addgene Kit #1000000146; www.addgene.org/kits/mccormick-cyanogate).
Standardization will help to accelerate the development of reliable synthetic biology tools for biotechnological applications and promote sharing and evaluation of genetic parts in different species and under different culturing conditions (Patron et al., 2015). Going forward, it will be important to test the performance of different parts with different components (e.g. gene expression cassettes) and in different assembly combinations. Several groups using plant MoClo assembly have reported differences in cassette expression and functionality depending on position and orientations (e.g. Ordon et al., 2017), which highlights a key synthetic biology crux—the performance of a system is not simply the sum of its components (Mutalik et al., 2013; Heyduk and Heyduk, 2018).
The increasing availability of genome-scale metabolic models for different cyanobacterial species and their utilization for guiding engineering strategies for producing heterologous high-value biochemicals has helped to reinvigorate interest in the industrial potential of cyanobacteria (Knoop et al., 2013; Hendry et al., 2016; Mohammadi et al., 2016; Shirai et al., 2016). Future efforts should focus on combining genome-scale metabolic models with synthetic biology approaches, which may help to overcome the production yield limitations observed for cyanobacterial cell factories (Nielsen et al., 2016), and will accelerate the development of more complex and precise gene control circuit systems that can better integrate with host metabolism and generate more robust strains (Bradley and Wang, 2015; Jusiak et al., 2016; Luan and Lu, 2018). The future development of truly “programmable” photosynthetic cells could provide significant advancements in addressing fundamental biological questions and tackling global challenges, including health and food security (Dobrin et al., 2016; Medford and Prasad, 2016; Smanski et al., 2016).
MATERIALS AND METHODS
Cyanobacterial Culture Conditions
Cyanobacterial strains of Synechocystis, UTEX 2973, and Synechococcus elongatus PCC 7942 (PCC 7942 hereafter) were maintained on 1.5% (w/v) agar plates containing BG11 (Blue-Green) medium. Liquid cultures were grown in Erlenmeyer flasks (100 mL) containing BG11 medium (Rippka et al., 1979) supplemented with 10 mm NaHCO3, shaken at 100 rpm and aerated with filter-sterilized water-saturated atmospheric air. Synechocystis and PCC 7942 strains were grown at 30°C with continuous light (100 µmol photons m−2 s−1) and UTEX 2793 strains were grown at 40°C with 300 µmol photons m−2 s−1 in an Infors Multitron-Pro supplied with warm white LED lighting (Infors HT).
Growth Analysis
Growth of Synechocystis, UTEX 2973, and PCC 7942 was measured in a Photon Systems Instrument Multicultivator MC 1000-OD (MC). Starter cultures were grown in a Photon Systems Instrument AlgaeTron AG 230 at 30°C under continuous warm-white light (100 µmol photons m−2 s−1) with air bubbling and shaken at 160 rpm unless otherwise indicated. These were grown to an optical density at 750 nm (OD750) of ∼1.0 and used to seed 80-mL cultures for growth in the MC at a starting OD720 of ∼0.2 (the MC measures culture growth at OD720). Cultures were then grown under continuous warm-white light at 30°C (300 µmol photons m−2 s−1) with air bubbling or 30°C (300 or 500 µmol photons m−2 s−1) or 41°C, 45°C, and 50°C (500 µmol photons m−2 s−1) with 5% CO2 bubbling until the fastest grown strain was at OD720 = ∼0.9, the maximum accurate OD that can be measured with this device. A total of five to six replicate experiments were performed over two separate runs (16 in total), each inoculated from a different starter culture.
Vector Construction
Level 0 Vectors
Native cyanobacterial genetic parts were amplified from genomic DNA using New England Biolabs Q5 High-Fidelity DNA Polymerase (Fig. 1; Supplemental Table S1). Where necessary, native genetic parts were domesticated (i.e. BsaI and BpiI sites were removed) using specific primers. Alternatively, parts were synthesized as Gblocks DNA fragments (Integrated DNA Technology) and cloned directly into an appropriate level 0 acceptor (see Supplemental Information S4 for vector maps; Engler et al., 2014).
Golden Gate assembly reactions were performed with restriction enzymes BsaI (New England Biolabs) or BpiI (Thermo Fisher Scientific), and T4 DNA ligase (Thermo Fisher Scientific; see Supplemental Information S2, S3, and S6 for detailed protocols). Vectors were transformed into One Shot TOP10 chemically competent Escherichia coli (Thermo Fisher Scientific) as per the manufacturer’s instructions. Transformed cultures were grown at 37°C on 1.5% (w/v) LB agar plates or in liquid LB medium shaking at 260 rpm, with appropriate antibiotic selection for levels 0, 1, M and P vectors as outlined in Engler et al. (2014).
Level T Acceptor Vectors and New Level 0 Acceptors
A new level T vector system was designed that provides MoClo-compatible replicative vectors or integrative vectors for genomic modifications in cyanobacteria (Fig. 2; Supplemental Table S1; Supplemental Information S4; Heidorn et al., 2011). For replicative vectors, we modified the pPMQAK1 carrying an RSF1010 replicative origin (Huang et al., 2010) to make pPMQAK1-T, and vector pSEVA421 from the SEVA 2.0 database (seva.cnb.csic.es) carrying the RK2 replicative origin to make pSEVA421-T (Silva-Rocha et al., 2013). Replicative vector backbones were domesticated to remove native BsaI and BpiI sites where appropriate. The region between the BioBrick’s prefix and suffix was then replaced by a lacZ expression cassette flanked by two BpiI sites that produce overhangs TGCC and GGGA, which are compatible with the plant Golden Gate MoClo assembly syntax for level 2 acceptors (e.g. pAGM4673; Engler et al., 2014). For integrative vectors, we domesticated a pUC19 vector backbone and introduced two BpiI sites compatible with a level 2 acceptor (as above) to make pUC19A-T and pUC19S-T. In addition, we made a new low copy level 0 acceptor (pSC101 origin of replication) for promoter parts based on the BioBrick standard vector pSB4K5 (Liu et al., 2018). DNA was amplified using New England Biolabs Q5 High-Fidelity DNA Polymerase (New England Biolabs). All vectors were sequenced following assembly to confirm domestication and the integrity of the MoClo cloning site.
Level 0 Parts for CRISPRi and srRNA
A nuclease deficient Cas9 gene sequence sourced from Addgene (www.addgene.org/44249/) was domesticated and assembled as a level 0 CDS part (Supplemental Tables S1 and S2; Qi et al., 2013). Five promoters of different strengths were truncated to the transcriptional start site (TSS) and cloned into a new level 0 acceptor vector with the unique overhangs GGAG and TAGC (Fig. 1). Two new level 0 parts with the unique overhangs GTTT and CGCT were generated for the sgRNA scaffold and srRNA host factor-I protein handle (based on MicC; Na et al., 2013), respectively. Assembly of level 1 expression cassettes proceeded by combining appropriate level 0 parts with a PCR product for either a srRNA or sgRNA (Fig. 1).
Cyanobacterial Transformation and Conjugation
Transformation with integrative level T vectors was performed as in Lea-Smith et al. (2016). For transformation by electroporation, cultures were harvested during the exponential growth phase (OD750 of ∼0.6) by centrifugation at 4,000 g for 10 min. The cell pellet was washed 3 times with 2 mL of sterile 1 mm HEPES buffer (pH 7.5), resuspended in water with 3–5 µg of level T vector DNA and transferred into a 0.1-cm electroporation cuvette (Scientific Laboratory Suppliers). Resuspended cells were electroporated using an Eppendorf 2510 electroporator (Eppendorf) set to 1200 V. Sterile BG11 (1 mL) was immediately added to the electroporated cells. Following a 1-h incubation at RT, the cells were plated on 1.5% (w/v) agar plates containing BG11 with antibiotics at standard working concentrations to select for transformed colonies. The plates were sealed with parafilm and placed under 15 µmol photons m−2 s−1 light at 30°C for 1 d. The plates were then moved to 30 µmol photons m−2 s−1 light until colonies appeared. After 15–20 d, putative transformants were recovered and streaked onto new plates with appropriate antibiotics for further study.
Genetic modification by conjugation in Synechocystis was facilitated by an E. coli strain (HB101) carrying both mobilizer and helper vectors pRK2013 (ATCC 37159) and pRL528 (www.addgene.org/58495/), respectively (Tsinoremas et al., 1994). For UTEX 2973, conjugation was facilitated by a MC1061 strain carrying mobilizer and helper vectors pRK24 (www.addgene.org/51950/) and pRL528, respectively. Cultures of HB101 and OneShot TOP10 E. coli strains carrying level T cargo vectors were grown for approximately 15 h with appropriate antibiotics. Cyanobacterial strains were grown to an OD750 of ∼1. All bacterial cultures were washed three times with either fresh LB medium for E. coli or BG11 for cyanobacteria prior to use. Synechocystis cultures (100 μl, OD750 of 0.5–0.8) were conjugated by combining appropriate HB101 and the cargo strains (100 μL each) and plating onto HATF 0.45-μm transfer membranes (Merck Millipore) placed on LB:BG11 (1:19) agar plates. For UTEX 2973 conjugations, appropriate MC1061 and the cargo strains (100 μL each) were initially combined and incubated at 30°C for 30 min, then mixed with UTEX 2973 cultures (100 μL, OD750 of 0.5–0.8) and incubated at 30°C for 2 h, and then plated onto transfer membranes as above. Synechocystis and UTEX 2973 transconjugates were grown under culturing conditions outlined above. Following growth on nonselective media for 24 h, the membranes were transferred to BG11 agar plates supplemented with appropriate antibiotics. Colonies were observed within a week for both strains. Chlorophyll content of wild-type and mutant strains was calculated as in Lea-Smith et al. (2013).
Fluorescence Assays
Transgenic strains maintained on agar plates containing appropriate antibiotics were used to inoculate 10-mL seed cultures that were grown to an OD750 of ∼1.0, as measured with a WPA Biowave II spectrometer (Biochrom). Seed cultures were diluted to an OD750 of 0.2, and 2-mL starting cultures were transferred to 24-well plates (Costar Corning Incorporated) for experiments. Synechocystis and UTEX 2973 strains were grown in an Infors Multitron-Pro in the same culturing conditions described above. OD750 was measured using a FLUOstar OMEGA microplate reader (BMG Labtech). Fluorescence of eYFP for individual cells (10,000 cells per culture) was measured by flow cytometry using an Attune NxT Flow Cytometer (Thermo Fisher Scientific). Cells were gated using forward and side scatter, and median eYFP fluorescence was calculated from excitation/emission wavelengths 488 nm/515–545 nm (Kelly et al., 2018) and reported at 48 h unless otherwise stated.
Cell Counts, Soluble Protein, and eYFP Quantification
Synechocystis and UTEX 2973 strains were cultured for 48 h as described above, counted using a hemocytometer, and then harvested for soluble protein extraction. Cells were pelleted by centrifugation at 4,000 g for 15 min, resuspended in lysis buffer (0.1 m potassium phosphate buffer [pH 7.0], 2 mm dithiothreitol, and one Roche cOmplete EDTA-free protease inhibitor tablet per 10 mL [Roche Diagnostics]) and lysed with 0.5 mm glass beads (Thistle Scientific) in a TissueLyser II (Qiagen). The cell lysate was centrifuged at 18,000 g for 30 min and the supernatant assayed for soluble protein content using Pierce 660nm Protein Assay Reagent against BSA standards (Thermo Fisher Scientific). Extracts were subjected to SDS-PAGE in a 4% to 12% (w/v) polyacrylamide gel (Bolt Bis-Tris Plus Gel; Thermo Fisher Scientific) alongside a SeeBlue Plus2 prestained protein ladder (Thermo Fisher Scientific), transferred to a polyvinylidene fluoride membrane, then probed with monoclonal anti-green fluorescent protein serum (AbCAM) at 1:1,000 dilution, followed by LI-COR IRDye 800CW goat anti-rabbit IgG (LI-COR Inc.) at 1:10,000 dilution, then viewed on an LI-COR Odyssey CLx Imager. eYFP protein content was estimated by immunoblotting using densitometry using LI-COR Lite Studio software v5.2 . Relative eYFP protein abundance was estimated by densitometry using LI-COR Lite Studio software v5.2.
Plasmid Vector and Genome Copy Number Determination
The genome copy number and copy number of heterologous self-replicating plasmid vectors in Synechocystis was estimated using a quantitative real-time PCR approach adapted from Zerulla et al. (2016). Cytoplasmic extracts containing total cellular DNA were harvested from Synechocystis cultures after 48 h growth (OD750 = ∼5) according to Zerulla et al. (2016). Cells in 10 mL of culture were pelleted by centrifugation at 4,000 g for 15 min, disrupted by shaking at 30 Hz for 10 min in a TissueLyser II with a mixture of 0.2-mm and 0.5-mm acid washed glass beads (0.35 g each), and then resuspended in dH2O. The culture cell count was determined prior to harvest using a hemocytometer and checked again after cell disruption to calculate the efficiency of cell disruption. A standard curve based on a dilution series of vector DNA was generated and used for a quantitative real-time PCR analysis in parallel with extracts carrying the same vector. Two DNA fragments (∼1 kb) targeting two separate loci (petB and secA) were amplified from isolated genomic DNA from Synechocystis using standard PCR (Pinto et al., 2012). DNA mass concentrations were determined photometrically and the concentrations of DNA molecules were calculated from the known molecular mass. As above, a standard curve based on a dilution series of the two fragments was generated to estimate genome copy number in the extracts (Zerulla et al., 2016). The cycle threshold of the extracts were then plotted against the linear portion of the standard curves to estimate plasmid vector copy number and genome copy number per cell. Oligonucleotides used are summarized in Supplemental Table S3.
Confocal Laser Scanning Microscopy
Cultures were imaged using Leica TCS SP8 confocal microscopy (Leica Microsystems) with a water immersion objective lens (HCX APO L 20x/0.50 W). Excitation/emission wavelengths were 514 nm/527–546 nm for eYFP and 514 nm/605–710 nm for chlorophyll autofluorescence.
SUPPLEMENTAL DATA
The following supplemental materials are available:
Supplemental Figure S1. Comparison of growth for Synechocystis, PCC 7942 and UTEX 2973 under different culturing conditions.
Supplemental Figure S2. Growth and expression levels of heterologous and synthetic promoters in Synechocystis and UTEX 2973.
Supplemental Figure S3. Cell volume calculations for Synechocystis and UTEX 2973 from confocal microscopy images.
Supplemental Table S1. Table of all parts from CyanoGate kit generated in this work.
Supplemental Table S2. List of level T vectors used in this study.
Supplemental Table S3. Sequences of synthetic oligonucleotides used to determine copy number.
Supplemental Information S1. Comparison of Gibson Assembly and Golden Gate Assembly.
Supplemental Information S2. Detailed assembly strategies using the CyanoGate kit.
Supplemental Information S3. Integrative engineering strategies using the CyanoGate kit.
Supplemental Information S4. Sequence maps (.gb files) of the components of the CyanoGate kit.
Supplemental Information S5. Protocol and online interface for building CyanoGate vector assemblies.
Supplemental Information S6. Protocols for MoClo assembly in level −1 through to level T.
Acknowledgments
We thank Conrad Mullineaux (Queen Mary University), Julie Zedler and Poul Erik Jensen (University of Copenhagen) for providing components for conjugation, and Eva Steel (University of Edinburgh) for assistance with assembling the CyanoGate kit.
Footnotes
This work was supported by the PHYCONET Biotechnology and Biological Sciences Research Council (BBSRC) Network in Industrial Biotechnology and Bioenergy (NIBB) and the Industrial Biotechnology Innovation Centre (IBioIC) (R.V., A.J.M., B.W., C.J.H.), a BBSRC EASTBIO CASE PhD studentship (BB/M010996/1 to G.A.R.G.), a United Kingdom BBSRC grant (BB/N007212/1 to B.W.), a Leverhulme Trust research grant (RPG-2015-445 to B.W.), a Consejo Nacional de Ciencia y Tecnología (CONACYT) PhD studentship (to A.A.S.), and a CSIRO Synthetic Biology Future Science Fellowship (to K.V.).
Articles can be viewed without a subscription.
References
- Abe K, Sakai Y, Nakashima S, Araki M, Yoshida W, Sode K, Ikebukuro K (2014) Design of riboregulators for control of cyanobacterial (Synechocystis) protein expression. Biotechnol Lett 36: 287–294 [DOI] [PubMed] [Google Scholar]
- Albers SC, Gallegos VA, Peebles CAM (2015) Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J Biotechnol 216: 36–46 [DOI] [PubMed] [Google Scholar]
- Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS (2000) Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol 305: 527–542 [DOI] [PubMed] [Google Scholar]
- Andreou AI, Nakayama N (2018) Mobius Assembly: A versatile Golden-Gate framework towards universal DNA assembly. PLoS One 13: e0189892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armshaw P, Carey D, Sheahan C, Pembroke JT (2015) Utilising the native plasmid, pCA2.4, from the cyanobacterium Synechocystis sp. strain PCC6803 as a cloning site for enhanced product production. Biotechnol Biofuels 8: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl CP, Wu R, Stawinsky J, Narang SA (1977) Minimal length of the lactose operator sequence for the specific recognition by the lactose repressor. Proc Natl Acad Sci USA 74: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behler J, Vijay D, Hess WR, Akhtar MK (2018) CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol. 36: 996–1010. [DOI] [PubMed] [Google Scholar]
- Blasi B, Peca L, Vass I, Kós PB (2012) Characterization of stress responses of heavy metal and metalloid inducible promoters in synechocystis PCC6803. J Microbiol Biotechnol 22: 166–169 [DOI] [PubMed] [Google Scholar]
- Blasina A, Kittell BL, Toukdarian AE, Helinski DR (1996) Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc Natl Acad Sci USA 93: 3559–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RW, Wang B (2015) Designer cell signal processing circuits for biotechnology. N Biotechnol 32: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RW, Buck M, Wang B (2016) Tools and principles for microbial gene circuit engineering. J Mol Biol 428(5 Pt B): 862–888 [DOI] [PubMed] [Google Scholar]
- Bustos SA, Golden SS (1992) Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC 7942: Evidence for the role of duplicated psbD genes in cyanobacteria. Mol Gen Genet 232: 221–230 [DOI] [PubMed] [Google Scholar]
- Camsund D, Heidorn T, Lindblad P (2014) Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J Biol Eng 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castenholz RW. (2001) Phylum BX. Cyanobacteria. Oxygenic photosynthetic bacteria. In Garrity G, Boone DR, Castenholz RW, eds, Bergey’s Manual of Systematic Bacteriology. Springer, New York, pp 473–599 [Google Scholar]
- Chambers S, Kitney R, Freemont P (2016) The Foundry: The DNA synthesis and construction Foundry at Imperial College. Biochem Soc Trans 44: 687–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Taton A, Go M, London RE, Pieper LM, Golden SS, Golden JW (2016) Self-replicating shuttle vectors based on pANS, a small endogenous plasmid of the unicellular cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 162: 2029–2041 [DOI] [PubMed] [Google Scholar]
- Clerico EM, Ditty JL, Golden SS (2007) Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol 362: 155–171 [DOI] [PubMed] [Google Scholar]
- Crozet P, Navarro FJ, Willmund F, Mehrshahi P, Bakowski K, Lauersen KJ, Pérez-Pérez ME, Auroy P, Gorchs Rovira A, Sauret-Gueto S, et al. (2018) Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth Biol 7: 2074–2086 [DOI] [PubMed] [Google Scholar]
- Dexter J, Dziga D, Lv J, Zhu J, Strzalka W, Maksylewicz A, Maroszek M, Marek S, Fu P (2018) Heterologous expression of mlrA in a photoautotrophic host—Engineering cyanobacteria to degrade microcystins. Environ Pollut 237: 926–935 [DOI] [PubMed] [Google Scholar]
- Dobrin A, Saxena P, Fussenegger M (2016) Synthetic biology: Applying biological circuits beyond novel therapies. Integr Biol 8: 409–430 [DOI] [PubMed] [Google Scholar]
- Doud DFR, Holmes EC, Richter H, Molitor B, Jander G, Angenent LT (2017) Metabolic engineering of Rhodopseudomonas palustris for the obligate reduction of n-butyrate to n-butanol. Biotechnol Biofuels 10: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat DC, Way JC, Silver PA (2011) Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29: 95–103 [DOI] [PubMed] [Google Scholar]
- Economou C, Wannathong T, Szaub J, Purton S (2014) A simple, low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii. Methods Mol Biol 1132: 401–411 [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Englund E, Liang F, Lindberg P (2016) Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci Rep 6: 36640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira EA, Pacheco CC, Pinot F, Pereira J, Lamosa P, Oliveira P, Kirov B, Jaramillo A, Tamagnini P (2018) Expanding the toolbox for Synechocystis sp. PCC 6803: Validation of replicative vectors and characterization of a novel set of promoters. Synth Biol 3: ysy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WKW, Lomas MW, Veneziano D, et al. (2013) Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA 110: 9824–9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J, Bagdasarian MM, Bagdasarian M (1992) Replication and copy number control of the broad-host-range plasmid RSF1010. Gene 113: 101–106 [DOI] [PubMed] [Google Scholar]
- Gao X, Gao F, Liu D, Zhang H, Nie X, Yang C (2016) Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ Sci 9: 1400–1411 [Google Scholar]
- Gordon GC, Korosh TC, Cameron JC, Markley AL, Begemann MB, Pfleger BF (2016) CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab Eng 38: 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero F, Carbonell V, Cossu M, Correddu D, Jones PR (2012) Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS One 7: e50470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn T, Camsund D, Huang HH, Lindberg P, Oliveira P, Stensjö K, Lindblad P (2011) Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol 497: 539–579 [DOI] [PubMed] [Google Scholar]
- Hendry JI, Prasannan CB, Joshi A, Dasgupta S, Wangikar PP (2016) Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour Technol 213: 190–197 [DOI] [PubMed] [Google Scholar]
- Heyduk E, Heyduk T (2018) DNA template sequence control of bacterial RNA polymerase escape from the promoter. Nucleic Acids Res 46: 4469–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo A, Isu A, Fukaya Y, Hisabori T (2016) Efficient gene induction and endogenous gene repression systems for the filamentous cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol 57: 387–396 [DOI] [PubMed] [Google Scholar]
- Higo A, Isu A, Fukaya Y, Ehira S, Hisabori T (2018) Application of CRISPR interference for metabolic engineering of the heterocyst-forming multicellular cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol 59: 119–127 [DOI] [PubMed] [Google Scholar]
- Huang HH, Lindblad P (2013) Wide-dynamic-range promoters engineered for cyanobacteria. J Biol Eng 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Shen CR, Li H, Sung LY, Wu MY, Hu YC (2016) CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb Cell Fact 15: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HH, Camsund D, Lindblad P, Heidorn T (2010) Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res 38: 2577–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immethun CM, DeLorenzo DM, Focht CM, Gupta D, Johnson CB, Moon TS (2017) Physical, chemical, and metabolic state sensors expand the synthetic biology toolbox for Synechocystis sp. PCC 6803. Biotechnol Bioeng 114: 1561–1569 [DOI] [PubMed] [Google Scholar]
- Jusiak B, Cleto S, Perez-Piñera P, Lu TK (2016) Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol 34: 535–547 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. (2004) Diversity and evolutionary history of plastids and their hosts. Am J Bot 91: 1481–1493 [DOI] [PubMed] [Google Scholar]
- Kelly CL, Taylor GM, Hitchcock A, Torres-Méndez A, Heap JT (2018) A rhamnose-inducible system for precise and temporal control of gene expression in cyanobacteria. ACS Synth Biol 7: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kikuchi H, Aichi M, Suzuki I, Omato T (1996) Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol 178: 5822–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Lee SM, Um Y, Sim SJ, Woo HM (2017) Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Front Plant Sci 8: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H, Formighieri C, Melis A (2014) Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size. Biochim Biophys Acta 1837: 1653–1664 [DOI] [PubMed] [Google Scholar]
- Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R (2013) Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLOS Comput Biol 9: e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RD, Golden SS (1997) mRNA stability is regulated by a coding-region element and the unique 5′ untranslated leader sequences of the three Synechococcus psbA transcripts. Mol Microbiol 24: 1131–1142 [DOI] [PubMed] [Google Scholar]
- Kunert A, Vinnemeier J, Erdmann N, Hagemann M (2003) Repression by Fur is not the main mechanism controlling the iron-inducible isiAB operon in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol Lett 227: 255–262 [DOI] [PubMed] [Google Scholar]
- Lea-Smith DJ, Ross N, Zori M, Bendall DS, Dennis JS, Scott SA, Smith AG, Howe CJ (2013) Thylakoid terminal oxidases are essential for the cyanobacterium Synechocystis sp. PCC 6803 to survive rapidly changing light intensities. Plant Physiol 162: 484–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea-Smith DJ, Bombelli P, Dennis JS, Scott SA, Smith AG, Howe CJ (2014) Phycobilisome-deficient strains of Synechocystis sp. PCC 6803 have reduced size and require carbon-limiting conditions to exhibit enhanced productivity. Plant Physiol 165: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea-Smith DJ, Vasudevan R, Howe CJ (2016) Generation of marked and markerless mutants in model cyanobacterial species. J Vis Exp 111: e54001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SP, Perutka J, Powell JE, Geng P, Richhart DD, Byrom M, Kar S, Davies BW, Ellington AD, Moran NA, et al. (2018) Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth Biol 7: 1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sun T, Xu C, Chen L, Zhang W (2018) Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab Eng 48: 163–174 [DOI] [PubMed] [Google Scholar]
- Liberton M, Chrisler WB, Nicora CD, Moore RJ, Smith RD, Koppenaal DW, Pakrasi HB, Jacobs JM (2017) Phycobilisome truncation causes widespread proteome changes in Synechocystis sp. PCC 6803. PLoS One 12: e0173251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg P, Park S, Melis A (2010) Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng 12: 70–79 [DOI] [PubMed] [Google Scholar]
- Liu D, Pakrasi HB (2018) Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microb Cell Fact 17: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Schumacher J, Wan X, Lou C, Wang B (2018) Orthogonality and burdens of heterologous AND Gate gene circuits in E. coli. ACS Synth Biol 7: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou W, Tan X, Song K, Zhang S, Luan G, Li C, Lu X (2018) A specific single nucleotide polymorphism in the ATP synthase gene significantly improves environmental stress tolerance of Synechococcus elongatus PCC 7942. Appl Environ Microbiol 84: e01222-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan G, Lu X (2018) Tailoring cyanobacterial cell factory for improved industrial properties. Biotechnol Adv 36: 430–442 [DOI] [PubMed] [Google Scholar]
- Madsen MA, Semerdzhiev S, Amtmann A, Tonon T (2018) Engineering mannitol biosynthesis in Escherichia coli and Synechococcus sp. PCC 7002 using a green algal fusion protein. ACS Synth Biol 7: 2833–2840 [DOI] [PubMed] [Google Scholar]
- Markley AL, Begemann MB, Clarke RE, Gordon GC, Pfleger BF (2015) Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth Biol 4: 595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AJ, Bombelli P, Bradley RW, Thorne R, Wenzele T, Howe CJ (2015) Biophotovoltaics: oxygenic photosynthetic organisms in the world of bioelectrochemical systems. Energy Environ Sci 8: 1092–1109 [Google Scholar]
- Medford JI, Prasad A (2016) Towards programmable plant genetic circuits. Plant J 87: 139–148 [DOI] [PubMed] [Google Scholar]
- Mermet-Bouvier P, Cassier-Chauvat C, Marraccini P, Chauvat F (1993) Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the general Synechocystis and Synechococcus. Curr Microbiol 27: 323–327 [Google Scholar]
- Mohammadi R, Fallah-Mehrabadi J, Bidkhori G, Zahiri J, Javad Niroomand M, Masoudi-Nejad A (2016) A systems biology approach to reconcile metabolic network models with application to Synechocystis sp. PCC 6803 for biofuel production. Mol Biosyst 12: 2552–2561 [DOI] [PubMed] [Google Scholar]
- Moore SJ, Lai H-E, Kelwick RJR, Chee SM, Bell DJ, Polizzi KM, Freemont PS (2016) EcoFlex: a multifunctional MoClo kit for E. coli synthetic biology. ACS Synth Biol 5: 1059–1069 [DOI] [PubMed] [Google Scholar]
- Mueller TJ, Ungerer JL, Pakrasi HB, Maranas CD (2017) Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci Rep 7: 41569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D (2005) Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21: 4187–4189 [DOI] [PubMed] [Google Scholar]
- Mutalik VK, Guimaraes JC, Cambray G, Mai QA, Christoffersen MJ, Martin L, Yu A, Lam C, Rodriguez C, Bennett G, et al. (2013) Quantitative estimation of activity and quality for collections of functional genetic elements. Nat Methods 10: 347–353 [DOI] [PubMed] [Google Scholar]
- Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY (2013) Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 31: 170–174 [DOI] [PubMed] [Google Scholar]
- Ng AH, Berla BM, Pakrasi HB (2015) Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 81: 6857–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WO, Zentella R, Wang Y, Taylor JS, Pakrasi HB (2000) PhrA, the major photoreactivating factor in the cyanobacterium Synechocystis sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch Microbiol 173: 412–417 [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H, Wolfstädter BT, Savage DF, Silver PA, Way JC (2010) Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol 76: 3462–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Keasling JD (2016) Engineering cellular metabolism. Cell 164: 1185–1197 [DOI] [PubMed] [Google Scholar]
- Nielsen AZ, Mellor SB, Vavitsas K, Wlodarczyk AJ, Gnanasekaran T, Perestrello Ramos H de Jesus M, King BC, Bakowski K, Jensen PE (2016) Extending the biosynthetic repertoires of cyanobacteria and chloroplasts. Plant J 87: 87–102 [DOI] [PubMed] [Google Scholar]
- Ordon J, Gantner J, Kemna J, Schwalgun L, Reschke M, Streubel J, Boch J, Stuttmann J (2017) Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J 89: 155–168 [DOI] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. (2015) Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 208: 13–19 [DOI] [PubMed] [Google Scholar]
- Peca L, Kós PB, Máté Z, Farsang A, Vass I (2008) Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS Microbiol Lett 289: 258–264 [DOI] [PubMed] [Google Scholar]
- Pinto F, Pacheco CC, Ferreira D, Moradas-Ferreira P, Tamagnini P (2012) Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS One 7: e34983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto F, Pacheco CC, Oliveira P, Montagud A, Landels A, Couto N, Wright PC, Urchueguía JF, Tamagnini P (2015) Improving a Synechocystis-based photoautotrophic chassis through systematic genome mapping and validation of neutral sites. DNA Res 22: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG (2017) Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA 114: 5601–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Hao M, Ng WO, Slater SC, Baszis SR, Weiss JD, Valentin HE (2005) Application of the Synechococcus nirA promoter to establish an inducible expression system for engineering the Synechocystis tocopherol pathway. Appl Environ Microbiol 71: 5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae BD, Long BM, Förster B, Nguyen ND, Velanis CN, Atkinson N, Hee WY, Mukherjee B, Price GD, McCormick AJ (2017) Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants. J Exp Bot 68: 3717–3737 [DOI] [PubMed] [Google Scholar]
- Ramey CJ, Barón-Sola Á, Aucoin HR, Boyle NR (2015) Genome Engineering in Cyanobacteria: Where We Are and Where We Need To Go. ACS Synth Biol 4: 1186–1196 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61 [Google Scholar]
- Ruffing AM, Jensen TJ, Strickland LM (2016) Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis. Microb Cell Fact 15: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar KL, Bombelli P, Lea-Smith DJ, Call T, Aro E-M, Müller T, Howe CJ, Knowles TPJ (2018) Enhancing power density of biophotovoltaics by decoupling storage and power delivery. Nat Energy 3: 75–81 [Google Scholar]
- Sakai Y, Abe K, Nakashima S, Ellinger JJ, Ferri S, Sode K, Ikebukuro K (2015) Scaffold-fused riboregulators for enhanced gene activation in Synechocystis sp. PCC 6803. MicrobiologyOpen 4: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D (2013) GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162: 1618–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T, Osanai T, Kondo A (2016) Designing intracellular metabolism for production of target compounds by introducing a heterologous metabolic reaction based on a Synechosystis sp. 6803 genome-scale model. Microb Cell Fact 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de Las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, et al. (2013) The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41: D666–D675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA (2016) Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol 14: 135–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensjö K, Vavitsas K, Tyystjärvi T (2018) Harnessing transcription for bioproduction in cyanobacteria. Physiol Plant 162: 148–155 [DOI] [PubMed] [Google Scholar]
- Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF (2012) Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis. Curr Microbiol 65: 552–560 [DOI] [PubMed] [Google Scholar]
- Sun T, Li S, Song X, Diao J, Chen L, Zhang W (2018) Toolboxes for cyanobacteria: Recent advances and future direction. Biotechnol Adv 36: 1293–1307 [DOI] [PubMed] [Google Scholar]
- Tan X, Yao L, Gao Q, Wang W, Qi F, Lu X (2011) Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng 13: 169–176 [DOI] [PubMed] [Google Scholar]
- Tan X, Hou S, Song K, Georg J, Klähn S, Lu X, Hess WR (2018) The primary transcriptome of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Biotechnol Biofuels 11: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A, Unglaub F, Wright NE, Zeng WY, Paz-Yepes J, Brahamsha B, Palenik B, Peterson TC, Haerizadeh F, Golden SS, et al. (2014) Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res 42: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A, Ma AT, Ota M, Golden SS, Golden JW (2017) NOT gate genetic circuits to control gene expression in cyanobacteria. ACS Synth Biol 6: 2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Kutach AK, Strayer CA, Golden SS (1994) Efficient gene transfer in Synechococcus sp. strains PCC 7942 and PCC 6301 by interspecies conjugation and chromosomal recombination. J Bacteriol 176: 6764–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J, Pakrasi HB (2016) Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep 6: 39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J, Lin P-C, Chen H-Y, Pakrasi HB (2018a) Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio 9: e02327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J, Wendt KE, Hendry JI, Marans CD, Pakrasi HB (2018b) Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc Natl Acad Sci USA 115: E11761–E11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Meene AM, Hohmann-Marriott MF, Vermaas WF, Roberson RW (2006) The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol 184: 259–270 [DOI] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Orzaez D, Patron N (2018) DNA assembly standards: Setting the low-level programming code for plant biotechnology. Plant Sci 273: 33–41 [DOI] [PubMed] [Google Scholar]
- Vioque A. (2007) Transformation of Cyanobacteria. In León R, Galván A, Fernández E, eds, Transgenic Microalgae as Green Cell Factories. Advances in Experimental Medicine and Biology. Vol 616 Springer, New York, pp 12–22 [Google Scholar]
- Vogel AIM, Lale R, Hohmann-Marriott MF (2017) Streamlining recombination-mediated genetic engineering by validating three neutral integration sites in Synechococcus sp. PCC 7002. J Biol Eng 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Eckert C, Maness P-C, Yu J (2018) A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth Biol 7: 276–286 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Noda A, Ohbayashi R, Uchioke K, Kurihara A, Nakatake S, Morioka S, Kanesaki Y, Chibazakura T, Yoshikawa H (2018) ParA-like protein influences the distribution of multi-copy chromosomes in cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 164: 45–56 [DOI] [PubMed] [Google Scholar]
- Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB (2016) CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Fact 15: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S (2012) Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3: 38–43 [DOI] [PubMed] [Google Scholar]
- Wlodarczyk A, Gnanasekaran T, Nielsen AZ, Zulu NN, Mellor SB, Luckner M, Thøfner JFB, Olsen CE, Mottawie MS, Burow M, et al. (2016) Metabolic engineering of light-driven cytochrome P450 dependent pathways into Synechocystis sp. PCC 6803. Metab Eng 33: 1–11 [DOI] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30: 1180–1182 [DOI] [PubMed] [Google Scholar]
- Xu Y, Alvey RM, Byrne PO, Graham JE, Shen G, Bryant DA (2011) Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol Biol 684: 273–293 [DOI] [PubMed] [Google Scholar]
- Yao L, Cengic I, Anfelt J, Hudson EP (2016) Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth Biol 5: 207–212 [DOI] [PubMed] [Google Scholar]
- Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB (2015) Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep 5: 8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerulla K, Ludt K, Soppa J (2016) The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology 162: 730–739 [DOI] [PubMed] [Google Scholar]
- Zess EK, Begemann MB, Pfleger BF (2016) Construction of new synthetic biology tools for the control of gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. Biotechnol Bioeng 113: 424–432 [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang H, Meng H, Zhu Y, Bao G, Zhang Y, Li Y, Ma Y (2014) Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci Rep 4: 4500. [DOI] [PMC free article] [PubMed] [Google Scholar]