Excessive nitrogen supply leads to reactive oxygen species accumulation and requires the function of major transcriptional regulators to maintain physiological balance.
Abstract
Reactive oxygen species (ROS) can accumulate in cells at excessive levels, leading to unbalanced redox states and to potential oxidative stress, which can have damaging effects on the molecular components of plant cells. Several environmental conditions have been described as causing an elevation of ROS production in plants. Consequently, activation of detoxification responses is necessary to maintain ROS homeostasis at physiological levels. Misregulation of detoxification systems during oxidative stress can ultimately cause growth retardation and developmental defects. Here, we demonstrate that Arabidopsis (Arabidopsis thaliana) plants grown in a high nitrogen (N) environment express a set of genes involved in detoxification of ROS that maintain ROS at physiological levels. We show that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) is an important mediator of this response and is required for the expression of detoxification genes. Mutation in HNI9 leads to elevated ROS levels and ROS-dependent phenotypic defects under high but not low N provision. In addition, we identify ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. Our results demonstrate the requirement of a balance between N metabolism and ROS production, and our work establishes major regulators required to control ROS homeostasis under conditions of excess N.
Reactive oxygen species (ROS), generated as byproducts of a large range of enzymatic reactions, are integral to plant metabolism. The dynamics of ROS in plant cells correspond to two distinct scenarios. First, ROS are involved in numerous signaling pathways, and thus their levels affect many developmental and physiological processes such as cell differentiation and response to biotic and abiotic stresses (Noctor et al., 2018). In such cases, variations in ROS are generally dynamic and transient, and these occur at moderate concentrations. On the other hand, ROS can also accumulate in cells at excessive and sustained levels, leading to unbalanced redox states and to potential oxidative stress (Schieber and Chandel, 2014). At the cellular level, excessive accumulation of ROS can trigger oxidation and damage to many essential molecules (Møller et al., 2007). For instance, oxidation of enzymatic proteins often leads to loss of activity; oxidation of DNA can lead to degradation or mutations; and oxidation of lipids leads to disorganization of cellular membranes. Maintenance of ROS homeostasis is therefore essential to preserve cell integrity. ROS levels are also associated with a range of physiological and developmental phenotypes. For instance, functioning of the shoot apical meristem is largely influenced by redox status (Schippers et al., 2016), and many developmental processes involve interactions between ROS and phytohormones (Considine and Foyer, 2014). Several reports have also demonstrated a role for ROS in root growth and development (Tsukagoshi, 2016). As a consequence, mutations leading to excessive ROS production or exogenous application of ROS at high concentrations lead to retardation of root growth in Arabidopsis (Arabidopsis thaliana; Dunand et al., 2007; Tsukagoshi et al., 2010; Zhao et al., 2016). Altogether, these observations demonstrate that controlling ROS levels is crucial for plant growth, development, and physiology.
Given the importance of ROS homeostasis, plants possess a large range of mechanisms to remove or detoxify ROS. A main component of this antioxidant response corresponds to enzymes, such as peroxidases, that use reducing power in oxidation/reduction reactions to decrease the cell redox status (Møller et al., 2007). These enzymes work in complex systems that also involve essential antioxidant molecules, like ascorbic acid (ASC), glutathione, or thiamin, that are known to protect against oxidative stress (Tunc-Ozdemir et al., 2009; Ramírez et al., 2013). At the molecular level, transcriptional induction of ROS-responsive genes in plants constitutes a major part of the response to ROS overproduction (Willems et al., 2016). However, in agreement with the high number of genes encoding components of the antioxidant response in Arabidopsis, a specificity of response occurs according to the signals at the origin of ROS production. Indeed, plant signaling pathways and genes involved in ROS homeostasis and the response to oxidative stress largely vary depending on the type of stress encountered (Apel and Hirt, 2004). Furthermore, the mechanisms that underlie transcriptional regulation and specificity remain to be identified.
ROS production has been observed in plants in response to many environmental factors, with both biotic interactions and abiotic stress contributing to ROS signaling (Baxter et al., 2014). Among abiotic factors, drought, salinity, and heat stress are known to induce the production of ROS (Choudhury et al., 2017). However, these responses correspond to cases where ROS contribute to signaling pathways, but do not accumulate and generate oxidative stress. In contrast, several examples of abiotic stress have been directly linked to elevation of ROS and eventual perturbation of plant redox balance. Such is the case for nutrient deprivation, where potassium, nitrogen (N), or phosphorus starvation rapidly induce the accumulation of H2O2 in roots (Shin and Schachtman, 2004; Shin et al., 2005). This result suggests that the redox status of plants is highly sensitive to changes in the nutritional environment.
Previously, the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) was shown to be involved in the response to high N provision in Arabidopsis. Mutations in HNI9 led to an increase in the transcript level of the nitrate transporter NRT2.1 under high N, a condition under which this gene is normally strongly downregulated (Widiez et al., 2011). However, a direct relationship between HNI9 and NRT2.1 has not been demonstrated. Several reports in plants, animals, and yeast have shown that HNI9 is a positive regulator of gene expression (Yoh et al., 2008; Li et al., 2010; Chen et al., 2012; Wang et al., 2013, 2014), which is inconsistent with the hypothesis of a direct role for HNI9 in NRT2.1 down-regulation. Here, we examine the function of HNI9 in the response of plants to high N provision and demonstrate an unexpected role for HNI9 in the regulation of ROS homeostasis through the induction of a subset of genes involved in detoxification. We also identify the transcription factor ELONGATED HYPOCOTYL5 (HY5) as an important component of this response, and highlight the interaction between plant nutrition and ROS homeostasis.
RESULTS
HNI9 Is Required to Up-regulate the Expression of Genes Involved in Redox Processes in Response to High N Provision
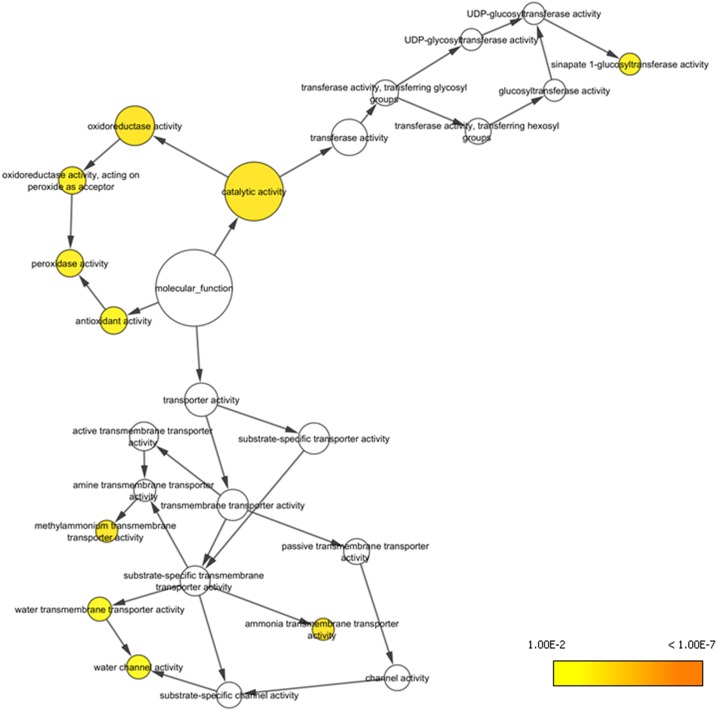
In order to examine the role of HNI9 in the response to high N provision, we compared the transcriptomic data of the wild type and hni9-1 mutant plants under low nitrate (0.3 mm KNO3) and high N (10 mm NH4NO3) provision (Widiez et al., 2011). These distinct conditions were selected for their large differences in N availability, leading to the identification of the hni9 phenotype (Widiez et al., 2011). As HNI9 is a positive regulator of gene expression, we selected genes that were induced by high N in the wild type, and we looked for those that were not induced in the hni9-1 mutant line. Using these criteria, we obtained a list of 108 genes induced under high N in an HNI9-dependent manner (Supplemental Table S1). In order to determine which biological functions are affected by the HNI9 mutation under high N provision, we performed a gene ontology analysis using the list of 108 genes. Among the biological functions overrepresented, we found that several categories related to redox processes, including oxidoreductase activity, peroxidase activity, or antioxidant activity, were the most significantly represented (Fig. 1; Table 1). Indeed, this list includes genes whose functions in general detoxification or antioxidant processes are well described, such as peroxidases, catalases, and NAD(P)-linked oxidoreductases. In addition, upon further manual curation, we observed that many genes present in other Gene Ontology (GO) categories might also encode enzymes or proteins associated with ROS detoxification or antioxidant biosynthetic pathways (Supplemental Table S1). Therefore, we hypothesized that high N provision could disrupt ROS homeostasis in plants, and that HNI9 may be required to induce a set of genes involved in cellular detoxification.
Figure 1.
Genes induced by high N provision in HNI9-dependent manner are associated with redox processes. Functional network realized from the list of 108 genes induced specifically under high N condition (10 mm NH4NO3) in an HNI9-dependent manner. Orange to yellow circles correspond to P values < 0.01 (Table 1). A major hub corresponding to function associated with redox and antioxidant activities emerges at the center of the network
Table 1. GO enrichment identified from HNI9-dependent genes induced by high N (10 mm NH4NO3).
The GO term enrichment analysis was performed in the Cytoscape environment by BINGO software using the Molecular Function ontology file, with P values < 0.01. Details of the analysis are provided in Supplemental File S1.
| GO-ID | P value | No. of genes in the GO category | Description |
|---|---|---|---|
| 3824 | 6.5259E-4 | 50 | catalytic activity |
| 16491 | 6.5259E-4 | 17 | oxidoreductase activity |
| 51739 | 6.5259E-4 | 2 | ammonia transmembrane transporter activity |
| 15200 | 6.5259E-4 | 2 | methylammonium transmembrane transporter activity |
| 16684 | 1.5486E-3 | 5 | oxidoreductase activity, acting on peroxide as acceptor |
| 4601 | 1.5486E-3 | 5 | peroxidase activity |
| 50284 | 2.2260E-3 | 2 | sinapate 1-glucosyltransferase activity |
| 16209 | 3.4940E-3 | 5 | antioxidant activity |
| 15250 | 5.4994E-3 | 3 | water channel activity |
| 5372 | 5.4994E-3 | 3 | water transmembrane transporter activity |
HNI9 Mutation Leads to Higher ROS Levels and to ROS-dependent Phenotypes Under High N Provision
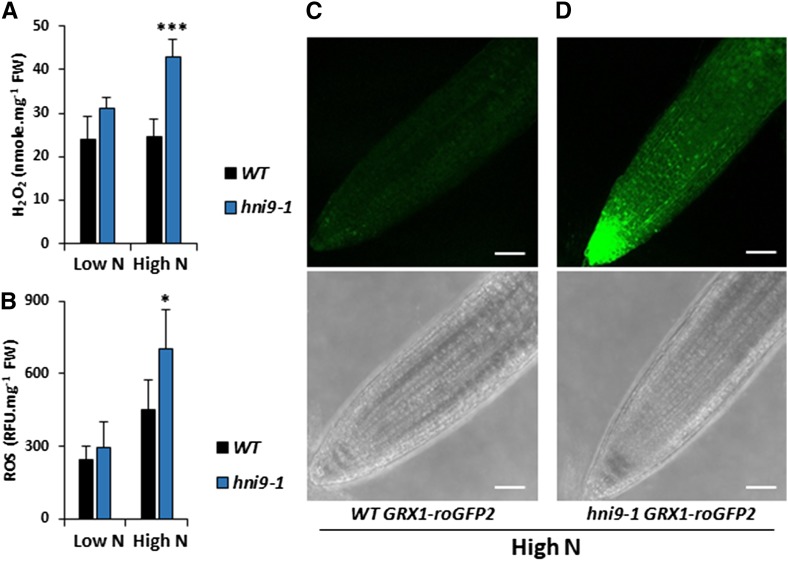
In order to assess the role of HNI9 in ROS homeostasis under high mixed N provision, we measured H2O2 and total ROS levels in the wild type and hni9-1 mutant lines under low nitrate and high N provision. In the wild type, H2O2 levels remained constant, independent of the N level. But total ROS levels slightly increased under high N provision, although not significantly, demonstrating that ROS homeostasis is maintained from low to high N provision (Fig. 2, A and B). In contrast, ROS accumulation was significantly altered in the hni9-1 mutant line. The levels of H2O2 and total ROS in the hni9-1 mutant were similar to the wild type under low N, but they increased significantly under high N provision (Fig. 2, A and B). To confirm these results, we used an Arabidopsis line expressing the glutaredoxin 1–reduction-oxidation sensitive green fluorescent protein 2 (GRX1-roGFP2) construct (hereafter referred to as roGFP2), in order to visualize ROS levels in planta. RoGFP2 is a redox-sensitive probe that allowed us to monitor plant redox status and, in particular, to assess H2O2 levels in vivo (Meyer et al., 2007; Marty et al., 2009). In the wild-type plants grown under high N provision, the fluorescence signal of roGFP2 was very low, in agreement with our measurements of H2O2 and ROS levels (Fig. 2C). In contrast, the fluorescence signal of roGFP2 in hni9-1 plants was strong, corresponding to the elevation of H2O2 and ROS levels measured in this line (Fig. 2D). Quantification of roGFP2 signals in each line validated a strongly significant increase in ROS levels in the hni9-1 mutant, as compared with the wild type (Supplemental Figure S1). Altogether, these results led us to conclude that HNI9 contributes to prevent ROS overaccumulation under high N supply, likely through the induction of a set of genes required to detoxify ROS produced under such conditions.
Figure 2.
ROS levels are higher in hni9-1 under high N provision. A and B, Measurement of H2O2 (A) and ROS (B) in roots of the wild type (WT) and hni9-1 lines under low N (0.3 mm NO3−) and high N (10 mm NH4NO3). Data represent mean ±sd of three biological replicates from independent experiments. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05; ***, P < 0.001). C and D, Visualization of H2O2 levels in vivo using GRX1-roGFP2 probe in the wild type (C) or hni9-1 (D) lines under high N condition. White bars = 50 µm.
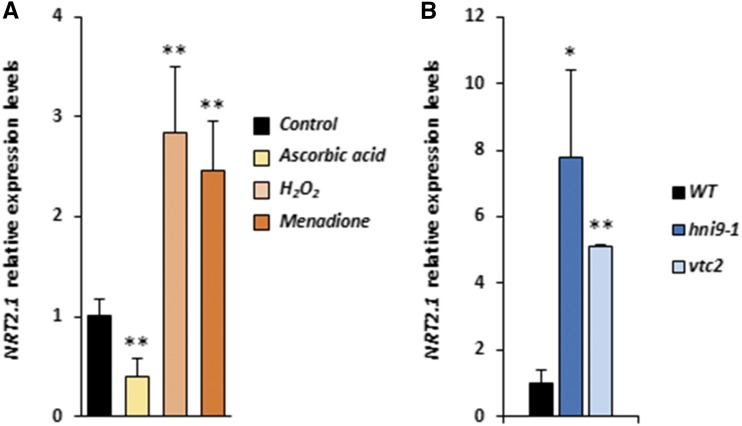
We next sought to determine whether up-regulation of nitrate transporter 2.1 (NRT2.1) expression under high N supply (which is a main phenotype described for the hni9-1 line) was linked with ROS overaccumulation in this mutant. To this end, we investigated the expression of NRT2.1 under conditions associated with altered ROS levels. First, we measured NRT2.1 transcript levels in the wild-type plants treated with ROS-generating or ROS-scavenging chemicals. We used ASC as a ROS scavenger. We also used menadione, a redox-active quinone that causes an elevation of ROS in plant roots (Lehmann et al., 2009), and H2O2 as ROS-generating molecules. Our results showed that ASC treatment caused a decrease in NRT2.1 expression while menadione and H2O2 treatment led to an increase in NRT2.1 transcript levels (Fig. 3A). Next, we measured NRT2.1 transcript levels in an ascorbate-deficient mutant, vtc2, which has known defects in ROS detoxification processes and accumulates significantly higher H2O2 levels than the wild-type plants (Kotchoni et al., 2009). In comparison with the wild type, NRT2.1 transcript levels were significantly higher in the vtc2 mutant line, similar to what we observed for the hni9-1 mutant (Fig. 3B). Altogether, these observations suggest that NRT2.1 expression is responsive to plant redox status, which lends support to the hypothesis that the previously reported hni9-1 phenotype under high N conditions may be due to elevated ROS levels in this mutant.
Figure 3.
NRT2.1 expression is sensitive to ROS homeostasis. A, Relative expression of NRT2.1 in the presence of ASC (400 µM), H2O2 (10 mM), and menadione (100 µM) in roots of the wild-type (WT) plants. Plants were grown on MS/2 medium containing 1 mm NO3− and then transferred on the same medium with or without ASC, H2O2, or menadione for 4 h. B, Relative expression of NRT2.1 in roots of the wild type, hni9-1, and vtc2 mutants. Plants were grown on MS/2 medium containing high N (10 mm NH4NO3). Data represent mean ±sd of three biological replicates from independent experiments. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05; **, P < 0.01).
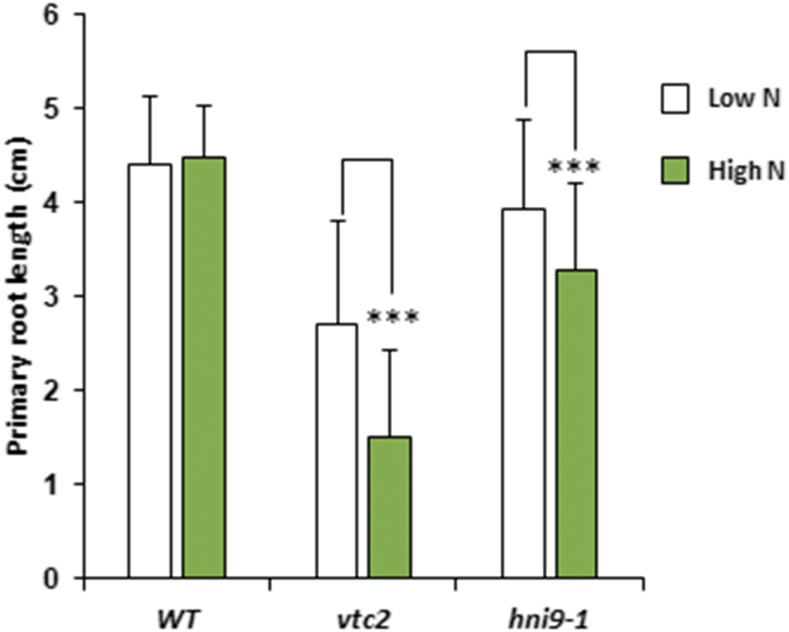
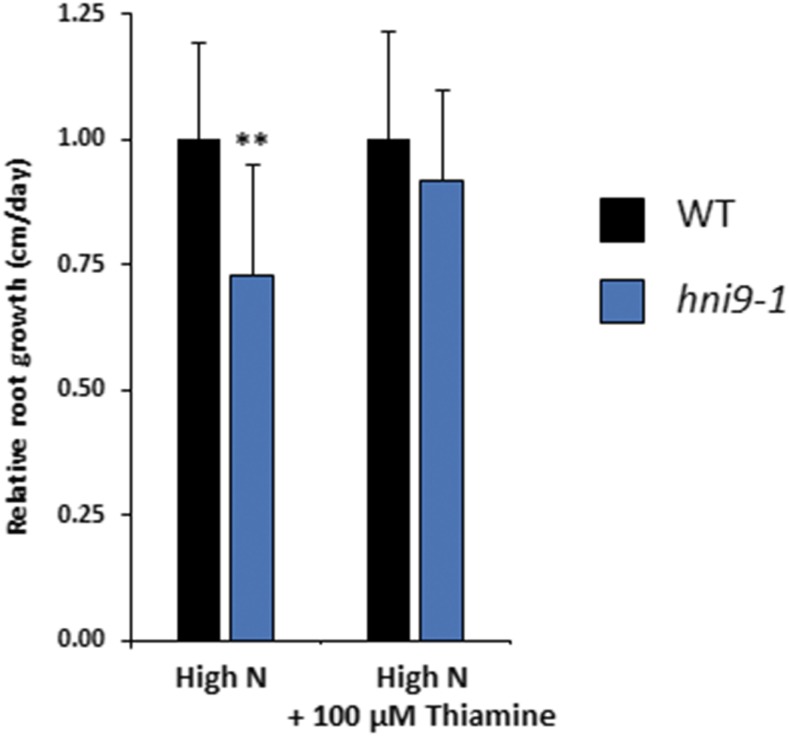
Conversely, we investigated whether well-known phenotypes associated with ROS overaccumulation were also observed in hni9-1 plants under high N supply. In particular, elevated ROS levels have been shown to alter root growth, such that plants grown under conditions of elevated ROS display reduced primary root length (Dunand et al., 2007; Tsukagoshi et al., 2010). Therefore, we asked whether elevated ROS levels in the hni9-1 mutant line under high N provision were correlated with alterations in root growth. We used the vtc2 mutant line, affected in ROS detoxification processes, as a positive control to confirm that elevated ROS levels inhibit root growth under our experimental conditions. Indeed, the vtc2 mutant line showed a decrease in primary root length, as compared with the wild type, regardless of the level of N. However, this decrease in root growth was significantly higher under high N than under low N (Fig. 4). The hni9-1 mutant line also exhibited reduced root growth in comparison to the wild type (Fig. 4), which is consistent with the general role of HNI9 in plant growth and development that had been previously described (Li et al., 2010). Moreover, the decrease in root growth was also significantly higher under high N than under low N (Fig. 4). This also suggests that phenotypic changes due to HNI9 mutation under high N supply may be explained by ROS overaccumulation. Finally, we tested whether external application of an antioxidant molecule could rescue the hni9-1 root growth phenotype. We therefore measured root growth of the wild type and hni9-1 lines under high N conditions, in the presence or absence of thiamin, an antioxidant known to alleviate growth defects caused by oxidative stress (Tunc-Ozdemir et al., 2009). Indeed, we found that the primary root growth defect of hni9-1 was fully abolished in the presence of thiamin (Fig. 5). This demonstrates that the presence of antioxidants can rescue the hni9-1 root growth phenotype, further indicating that HNI9 affects root development under high N supply specifically through altered ROS homeostasis.
Figure 4.
Root growth retardation is more pronounced under high N provision in hni9-1 and vtc2 mutants. Primary root length measurements of 7-d-old plants grown under low (0.3 mm NO3−) and high (10 mm NH4NO3) N provision in the wild type (WT), hni9-1, and vtc2 lines. The extent of root growth reduction is enhanced under high N and correlated with the presence of ROS in the mutant lines. Data represent mean ±sd of at least 10 independent plants. Statistical significance was computed using a two-tailed Student’s t test (***, P < 0.001).
Figure 5.
External antioxidant application rescues the hni9-1 root growth phenotype. Plants were grown under high N (10 mm NH4NO3) for 4 d and transferred to the same medium with or without 100 µM thiamin. Primary root growth of the wild type (WT) and hni9-1 lines was measured after 2 d of growth. Data represent mean ±sd of at least 10 independent plants. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05).
HNI9 Might Be Required to Achieve Full Levels of H3K4me3 Modification at the Loci of Detoxification Genes
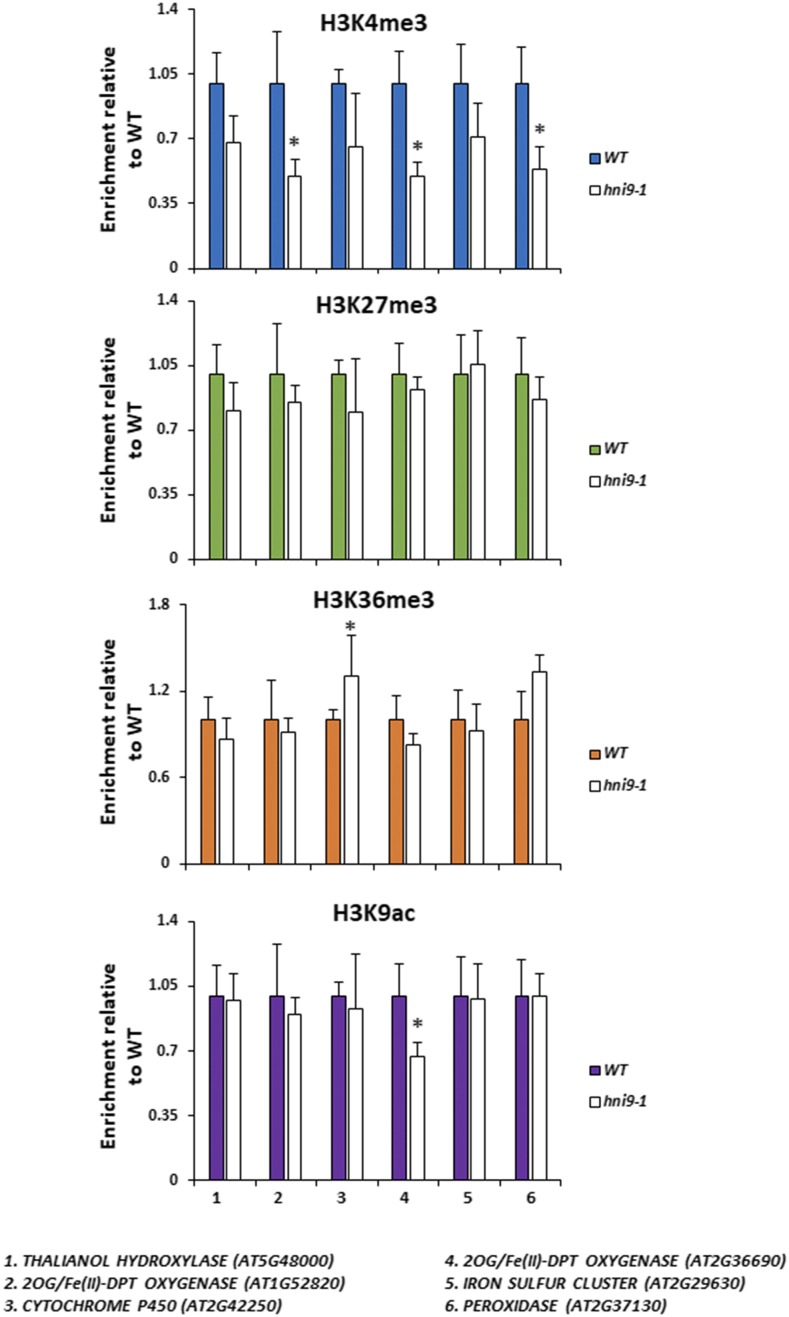
Along with elongation and chromatin remodeling factors, HNI9 is part of a large protein complex that influences the histone modification state of genes (Yoh et al., 2008; Li et al., 2010). Indeed, several reports in plants and animals demonstrated that HNI9 operates at the switch between transcriptional repression and activation, in cooperation with H3K27me3 demethylases and H3K36 methyl-transferases (Li et al., 2013; Wang et al., 2014). Therefore, we tested the hypothesis that HNI9 controls the expression of ROS detoxification genes by modulating their chromatin state in response to N provision. To do so, we assayed the level of three active chromatin marks (H3K4me3, H3K9ac, and H3K36me3) and one repressive chromatin mark (H3K27me3) at the loci of several HNI9-dependent genes implicated in ROS detoxification in response to high N. Representative genes for this analysis were selected from the subset of genes identified by the GO analysis as belonging to the category “oxidoreductase activity” (Supplemental File S1) and displaying a clear induction of their transcript levels by high N provision in an HNI9-dependent manner (Supplemental Fig. S2). Here, the results showed that the level of H3K27me3 and H3K36me3 were not changed in the hni9-1 mutant at the loci of selected HNI9-dependent detoxification genes (Fig. 6). In addition, the levels of H3K9ac (associated with active transcription) were also generally similar between the wild type and hni9-1. However, we observed that H3K4me3 levels were globally lower in the hni9-1 mutant at the loci of several HNI9-dependent ROS detoxification genes (Fig. 6). We therefore conclude that HNI9 may induce the expression of ROS detoxification genes by regulating the profile of H3K4me3.
Figure 6.
Mutation in HNI9 is associated with reduction of the H3K4me3 modification at the loci of detoxification genes. Shown is the ChIP analysis of H3K4me3, H3K27me3, H3K36me3, and H3K9ac in the wild type (WT) and hni9-1 roots of 7-d-old plants grown under high N provision (10 mm NH4NO3). Quantification by qPCR is shown as the percentage of H3K4me3, H3K36me3, H3K27me3, or H3K9ac over H3 at target loci, normalized by the percentage of H3K4me3, H3K36me3, H3K27me3, or H3K9ac over H3 at positive control loci (that is, ACT2 for H3K4me3, H3K36me3, and H3K9ac, and LEC2 for H3K27me3). Data are presented relative to the wild type’s level. Error bars represent ses of the mean based on at least three biological replicates. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05).
Analysis of HNI9-Dependent Genes Involved in ROS Detoxification under High N Conditions Reveals the Role of HY5 in the Control of ROS Homeostasis
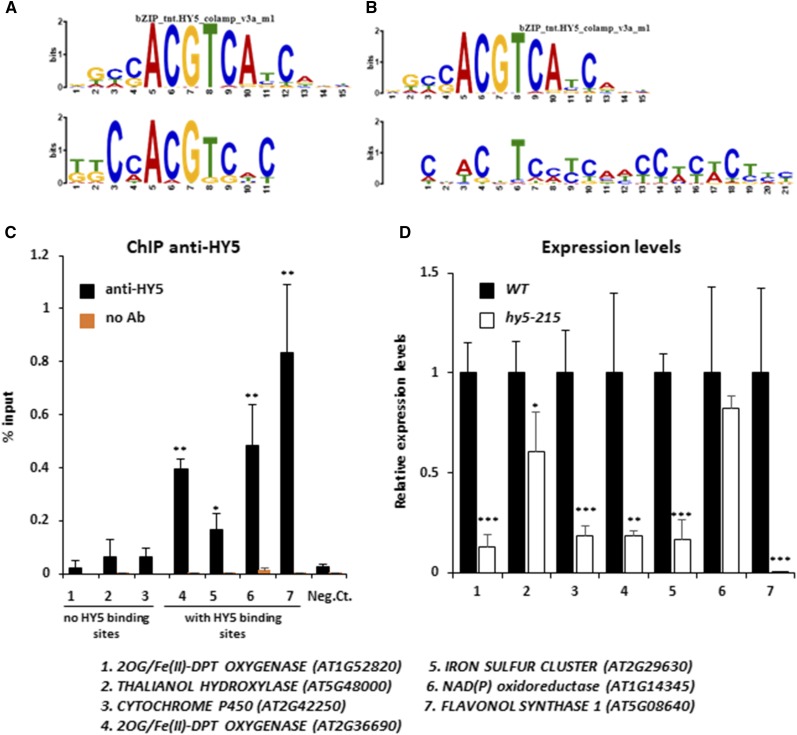
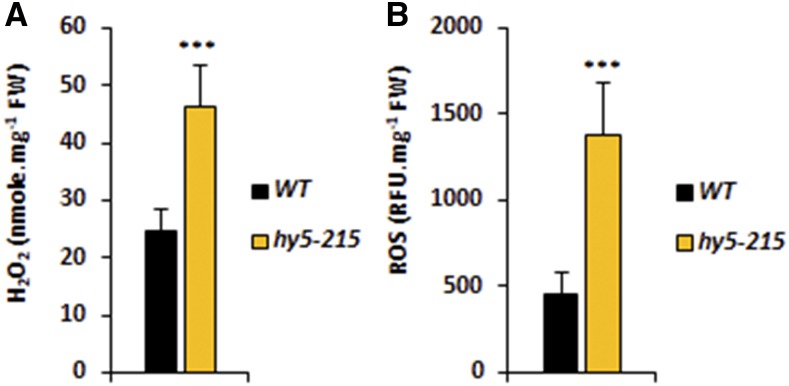
HNI9 is a general regulator of gene expression, as it does not provide by itself any sequence specificity to its target loci. Specificity of response thus requires the action of transcription factors to drive chromatin remodeling complexes to target loci (Li et al., 2010). Therefore, we tested whether a promoter sequence analysis of genes involved in ROS detoxification under high N could help identify such transcription factors. We used the promoter sequences from the 108 genes induced by high N and dependent on HNI9 in order to find putative conserved cis-regulatory elements using MEME software. This analysis revealed substantial enrichment of two cis-regulatory elements showing strong sequence similarity with the binding site consensus of the transcription factor HY5 defined by DNA affinity purification sequencing (Fig. 7, A and B; O’Malley et al., 2016). In total, 45 of the 108 aforementioned genes contained a putative HY5 binding site (Supplemental Tables S2 and S3). Interestingly, it was shown previously that HY5 is involved in the control of ROS production in response to light or temperature treatments (Catalá et al., 2011; Chen et al., 2013; Chai et al., 2015). We first analyzed HY5 transcript levels in response to N provision, and we observed that HY5 expression was slightly higher under a high N condition, but independent of HNI9 function (Supplemental Fig. S3). To explore the possible role of HY5 in the regulation of genes involved in ROS detoxification under a high N condition, we investigated the binding of HY5 to the putative consensus sequences identified in some of the ROS detoxification genes. To this end, we performed chromatin immunoprecipitation followed by quantitative (PCR) on a subset of genes with or without putative HY5 binding sites. The results showed that HY5 indeed binds to the promoter of detoxification genes containing a putative HY5 binding site; no significant binding was observed for genes lacking a putative HY5 binding site, although some are also involved in detoxification processes under high N provision (Fig. 7C). In order to test the effect of HY5 mutation, we next measured the expression of the identified genes (with or without HY5 binding sites) in the wild type and the hy5-215 mutant line. All genes, with one exception, were significantly down-regulated in the hy5-215 mutant line under high N provision (Fig. 7D), strongly supporting a major role of HY5 in the activation of the detoxification program under high N provision. Finally, to test the effect of misregulation of the detoxification program under high N following HY5 mutation, we measured the levels of ROS and H2O2 in the wild type and the hy5-215 mutant. The results clearly showed that ROS and H2O2 levels were higher in hy5-215 than in the wild type, demonstrating the functional importance of HY5 in the detoxification process under high N provision (Fig. 8). Altogether, our results demonstrate the activation of a transcriptional program required for ROS detoxification under high N, which is controlled by the chromatin remodeler HNI9 and the transcription factor HY5.
Figure 7.
HY5 binds to and regulates the expression of genes involved in detoxification under high N provision. A and B, Comparison of first (A) and second (B) conserved motifs discovered by MEME analysis in the promoters of HNI9-dependent genes induced under high N (10 mm NH4NO3), with the HY5 consensus binding site identified by DNA affinity purification sequencing. C, ChIP analysis following HY5 enrichment in the wild-type (WT) roots at the loci of HNI9-dependent genes induced under high N. Quantification by qPCR is shown as the percentage of input. Error bars represent ses of the mean based on at least three biological replicates. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05; **, P < 0.01), in comparison to a negative control (AT4G03900, which showed no relation to N or HY5 signaling). D, Relative expression of genes involved in detoxification that were induced by high N in roots of the wild type and hy5-215. Error bars represent ses of the mean based on three biological replicates from independent experiments. Statistical significance was computed using a two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Figure 8.
ROS levels are higher in hy5-215 under high N provision. Measurement of H2O2 (A) and ROS (B) in roots of the wild type (WT) and hy5-215 under high N (10 mm NH4NO3). Error bars represent ses of the mean based on three biological replicates from independent experiments. Statistical significance was computed using a two-tailed Student’s t test ***, P < 0.001).
DISCUSSION
Plants, like every living organism, must cope with environmental constraints. ROS accumulation has been shown to be involved in numerous signaling pathways or to cause oxidative stress in response to different environmental challenges (Baxter et al., 2014; Choudhury et al., 2017; Noctor et al., 2018). The main finding of our work is that high N provision leads to the generation of ROS in plant roots, which is managed by the transcriptional induction of a specific detoxification program. Interestingly, increasing N availability is often viewed positively and as a favorable condition for plant growth, but as we demonstrate, excessive N provision can, in fact, be detrimental to plants. Indeed, even if efficient detoxification systems exist to maintain ROS homeostasis, their induction and functioning undoubtedly incur costs for plants, and their efficiencies can be limited under certain circumstances. In spite of the demonstration that high N availability leads to ROS accumulation, the physiological cause of ROS production under high N conditions remains to be determined. Nitrate assimilation and, to a lesser extent, ammonium assimilation pathways can be candidates, as they consume a lot of reducing power (Hachiya and Sakakibara, 2017) and affect the cellular redox balance of plants. However, ROS accumulation under high N may also reflect a more general link between nutrition and redox balance. Indeed, several reports have shown that excessive nutrition in animals leads to ROS production and perturbation of cellular redox states (Sies et al., 2005; Samoylenko et al., 2013; Görlach et al., 2015). In the case of plants, nutrient starvation has also been identified as a condition that generates oxidative stress through the production of ROS (Shin and Schachtman, 2004; Shin et al., 2005). In addition, recent reports demonstrated that ROS accumulate early during the N starvation response, and most likely ROS contribute to the regulation of nitrate-responsive genes such as NRT2.1 (Jung et al., 2018; Safi et al., 2018). Therefore, these studies and our present work suggest the existence of thresholds of N provision inside which the cellular redox balance is optimal. This situation is reminiscent of what has been described for iron (Fe) homeostasis. Indeed, excessive Fe provision can also lead to an accumulation of ROS, and specific mechanisms have been described to limit this negative effect (Briat et al., 2010). Nutrient deficiency or excess (at least for N and Fe) would correspond to conditions in which ROS production is above a physiological limit and alters plant redox status. Again, it is interesting to note that the same findings hold true for animals (Görlach et al., 2015).
In our work, we highlight the role of HNI9 in the transcriptional induction of this detoxification program. In Arabidopsis, HNI9 has been mainly associated with brassinosteroid signaling (Li et al., 2010; Wang et al., 2014). In response to brassinosteroids, HNI9 associates with the chromatin remodeling factors EARLY FLOWERING 6, RELATIVE OF EARLY FLOWERING 6, and SET DOMAIN GROUP 8, and also with the transcription factor bri1-EMS-suppressor 1 (Li et al., 2010; Wang et al., 2014). EARLY FLOWERING 6 and RELATIVE OF EARLY FLOWERING 6 are responsible for removing of the repressive chromatin mark H3K27me3, while SET DOMAIN GROUP 8 is thought to catalyze the trimethylation of H3K36, which promotes elongation of transcription (Lu et al., 2011; Crevillén et al., 2014; Li et al., 2015). This suggests a role for HNI9 in complexes associated with transcriptional switches (for example from repression to activation), as demonstrated in animals (Chen et al., 2012; Wang et al., 2013). In our work, HNI9 is mainly linked with differences in H3K4me3 levels. This suggests that HNI9 could be associated with other chromatin complexes or could influence other steps of transcriptional regulation. Nevertheless, it identifies another transcriptional pathway in which HNI9 is essential for the induction of expression of a large set of genes.
In addition, our work identifies HY5 as a major component involved in the induction of the gene network underlying ROS detoxification in response to high N. HY5 is a master regulator of gene expression in Arabidopsis, at the center of several transcriptional networks. It is notably involved in the response to photosynthesis, light, temperature, and hormones (Gangappa and Botto, 2016). Interestingly, HY5 has been also implicated in the control of ROS homeostasis in response to light or cold treatments (Catalá et al., 2011; Chen et al., 2013; Chai et al., 2015). In each case, HY5 is required to suppress ROS accumulation during stressful conditions. However, the set of genes induced by HY5 to balance ROS levels seems to differ from one stimulus to another. This result suggests that other transcription factors, in addition to HY5, could specify the response according to the environmental signal. Interestingly, we found that the expression of every gene of the ROS detoxification network in response to high N that we tested was dependent on HY5, independent of the presence of a HY5 binding site in their promoter. This finding suggests that HY5 could be a direct (and general) regulator of many ROS detoxification genes, but that it may also function as an indirect regulator for a subset of them, for instance by regulating another transcription factor that, in turn, regulates these genes. This would imply the existence of several layers in this gene network, and that HY5 is a master regulator at the top of the network.
In conclusion, we demonstrated how a detoxification program is induced in order to maintain plant redox status under physiological conditions, even under high nutritional provision. However, these detoxification processes are certainly not without cost for plants. Overall, this work demonstrates that excess N availability is not necessarily advantageous for plant physiology and development, and lends support to the existence of an optimum balance between nutrition and ROS homeostasis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) accession used in this study was Col-0. Mutant alleles and transgenic plants used in this study were hni9-1 (Widiez et al., 2011), vct2 (Collin et al., 2008), hy5-215 (Oyama et al., 1997), and GRX1-roGFP2 (Meyer et al., 2007). Experiments were performed using roots from 7-d-old seedlings grown under a long-day photoperiod (16 h light and 8 h dark) on vertical half-strength Murashige and Skoog medium (MS/2) plates without N (PlantMedia), supplemented with 0.8% (w/v) agar, 0.1% (w/v) Suc, 0.5 g/L MES, and the specific concentration of N (0.3 mm KNO3 or 10 mm NH4NO3), as described in the main text and figure legends.
Analysis of Gene Expression by Quantitative PCR
Root samples were frozen and ground in liquid N, and total RNA was extracted using TRI REAGENT (MRC) and DNase treated (RQ1 Promega). Reverse transcription was performed with M-MLV reverse transcriptase (RNase H minus, Point Mutant, Promega) using an anchored oligo(dT)20 primer. Transcript levels were measured by reverse transcription quantitative PCR (LightCycler 480, Roche Diagnostics) using the TB Green Premix Ex Taq (Tli Rnase H plus; TaKaRa). Gene expression was normalized using the ACT2 gene as an internal standard. Sequences of primers used for gene expression analysis are listed in Supplemental File S2.
Chromatin Immunoprecipitation Quantitative PCR
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described in Bellegarde et al. (2018). Chromatin was precipitated with 2.5 μg of antibodies against H3 (Abcam 1791), H3K27me3 (Millipore 07-449), H3K4me3 (Diagenode C15410030), H3K36me3 (Abcam 9050), H3K9ac (Agrisera AS163198), or HY5 (Agrisera AS12 1867). Immunoprecipitated DNA was purified by phenol-chloroform extraction and ethanol precipitation, and the resulting DNA was analyzed by qPCR. For chromatin marker analyses, ChIP experiments were quantified using H3 level as an internal standard, and normalized using ACT7 (H3K4me3, H3K9ac), ACT2 (H3K36me3), or LEC2 (H3K27me3) enrichment. For HY5 binding site analyses, ChIP experiments were normalized to the input levels. Sequences of primers used in qPCR for ChIP experiments are listed in Supplemental File S2.
ROS and H2O2 Assays
Root samples were frozen and ground in liquid N, and ROS and H2O2 were extracted using approximately 30 mg of plant material in 200 µL of phosphate buffer (20 mm K2HPO4, pH 6.5). For ROS measurements, 50 µL of supernatant were mixed with 50 µL of 20 µM DFFDA (Molecular Probes). Reactions were incubated for 30 min at ambient temperature, and fluorescence was detected using 492/527 nm as excitation/emission parameters. H2O2 was measured using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen), as described previously in Brumbarova et al. (2016).
Microscopy
Confocal imaging of Arabidopsis root cells expressing GRX1-roGFP2 was performed using a Leica SP8 Confocal Microscope (Leica). GFP quantifications were done with an Axiovert 200 m microscope (Zeiss) and images were analyzed using ImageJ software. The data represent the fluorescence quantification values measured in the root tip.
Root Growth Analyses
Vertical agar plates containing plants were scanned after 7 d of growth, and root length was analyzed using ImageJ software. For experiments with thiamin, plants were grown on vertical MS/2 plates without N (PlantMedia), supplemented with 0.8% (w/v) agar, 0.1% (w/v) Suc, 0.5 g/L MES, and 10 mm NH4NO3 for 4 d and then transferred to the same medium with or without 100 µM thiamin. The position of the root tip was marked on the back of the fresh plates, and root length was analyzed 2 d after transfer by measuring growth from the marked position.
Determination of Conserved Cis-Regulatory Sequences
Putative cis-regulatory sequences were identified using the MEME suite (Bailey et al., 2009). The primary input was 500 bp of promoter sequences, with the following parameters: 0-order background model, classic discovery mode, 0 or one occurrence per sequence, and motif width between 6 and 50 nucleotides.
Analysis of GO
GO analysis was performed using BINGO in the Cytoscape environment, using Biological Process file, and a significance level of 0.05.
Statistical Analysis
Mean ±se is shown for all numerical values. Statistical significance was computed using a two-tailed Student’s t test. Significance cutoff: *P < 0.05, ** P < 0.01, *** P < 0.001.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HNI9 AT1G32130; HY5 AT5G11260; NRT2.1 AT1G08090; 2OG/Fe(II)-dpt oxygenase AT1G52820; iron sulfur cluster AT2G29630; THALIANOL HYDROXYLASE AT5G48000; NAD(P) AT1G14345; CYTOCHROME P450 AT2G42250; FLAVONOL SYNTHASE 1 AT5G08640; 2OG/Fe(II)-dpt oxygenase AT2G36690; and PEROXIDASE AT2G37130.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of GFP signal from GRX1-0roGFP2 probe in the wild type and hni9-1 lines under high N (10 mm NH4NO3) provision.
Supplemental Figure S2. Expression of genes identified as induced under high N condition in an HNI9-dependent manner.
Supplemental Figure S3. HY5 expression is slightly higher under high N provision, independently of HNI9 function.
Supplemental Table S1. List of 108 genes induced under high N conditions (10 mm NH4NO3) in an HNI9-dependent manner, and the corresponding description of their functions in TAIR
Supplemental Table S2. Genes from the list of 108 genes induced under high N conditions (10 mm NH4NO3) in an HNI9-dependent manner showing the presence of a putative HY5 binding site (motif #1) in their promoter
Supplemental Table S3. Genes from the list of 108 genes induced under high N conditions (10 mm NH4NO3) in an HNI9-dependent manner showing the presence of a putative HY5 binding site (motif #2) in their promoter
Supplemental File S1. Details of GO analysis using BINGO software.
Supplemental File S2. List of primers used in this study
Acknowledgments
We thank members of the lab for discussion. We thank Andreas Meyer and Alexandre Martinière for the GRX1-roGFP2 line, Fredy Barneche for the hy5-215 line, and Jean-François Briat for the vtc2 line. We thank the Montpellier Rio Imaging platform for microscopy observations.
Footnotes
This work was supported by a grant from the National Agency for Research (grant no. ANR14-CE19-0008 IMANA to A.G., An.M., and G.K.). F.B. was the recipient of a PhD fellowship from the Institut National de la Recherche Agronomique Département Biologie et Amélioration des Plantes.
References
- Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Bellegarde F, Herbert L, Séré D, Caillieux E, Boucherez J, Fizames C, Roudier F, Gojon A, Martin A (2018) Polycomb Repressive Complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1. Sci Rep 8: 7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, Cellier F, Gaymard F (2010) New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann Bot 105: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Le C, Bauer P (2016) Hydrogen peroxide measurement in Arabidopsis root tissue using Amplex Red. Bio Protoc 6: 1–11 [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai T, Zhou J, Liu J, Xing D (2015) LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis. Front Plant Sci 6: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, et al. (2012) The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90: 856–867 [DOI] [PubMed] [Google Scholar]
- Collin VC, Eymery F, Genty B, Rey P, Havaux M (2008) Vitamin E is essential for the tolerance of Arabidopsis thaliana to metal-induced oxidative stress. Plant Cell Environ 31: 244–257 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Foyer CH (2014) Redox regulation of plant development. Antioxid Redox Signal 21: 1305–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Görlach A, Dimova EY, Petry A, Martínez-Ruiz A, Hernansanz-Agustín P, Rolo AP, Palmeira CM, Kietzmann T (2015) Reactive oxygen species, nutrition, hypoxia and diseases: Problems solved? Redox Biol 6: 372–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Sakakibara H (2017) Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J Exp Bot 68: 2501–2512 [DOI] [PubMed] [Google Scholar]
- Jung JY, Ahn JH, Schachtman DP (2018) CC-type glutaredoxins mediate plant response and signaling under nitrate starvation in Arabidopsis. BMC Plant Biol 18: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotchoni SO, Larrimore KE, Mukherjee M, Kempinski CF, Barth C (2009) Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiol 149: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Møller BL, Fernie AR, et al. (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406 [DOI] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y (2010) Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen X, Zhong X, Zhao Y, Liu X, Zhou S, Cheng S, Zhou DX (2013) Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25: 4725–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mukherjee I, Thum KE, Tanurdzic M, Katari MS, Obertello M, Edwards MB, McCombie WR, Martienssen RA, Coruzzi GM (2015) The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol 16: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R (2009) The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Noctor G, Reichheld JP, Foyer CH (2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez L, Bartoli CG, Lamattina L (2013) Glutathione and ascorbic acid protect Arabidopsis plants against detrimental effects of iron deficiency. J Exp Bot 64: 3169–3178 [DOI] [PubMed] [Google Scholar]
- Safi A, Medici A, Szponarski W, Marshall-Colón A, Ruffel S, Gaymard F, Coruzzi G, Lacombe B, Krouk G (2018) HRS1/HHOs GARP transcription factors and reactive oxygen species are regulators of Arabidopsis nitrogen starvation response. bioRxiv. https://www.biorxiv.org/content/10.1101/164277v1(date)
- Samoylenko A, Hossain JA, Mennerich D, Kellokumpu S, Hiltunen JK, Kietzmann T (2013) Nutritional countermeasures targeting reactive oxygen species in cancer: From mechanisms to biomarkers and clinical evidence. Antioxid Redox Signal 19: 2157–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS (2014) ROS function in redox signaling and oxidative stress. Curr Biol 24: R453–R462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers JH, Foyer CH, van Dongen JT (2016) Redox regulation in shoot growth, SAM maintenance and flowering. Curr Opin Plant Biol 29: 121–128 [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Sies H, Stahl W, Sevanian A (2005) Nutritional, dietary and postprandial oxidative stress. J Nutr 135: 969–972 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H. (2016) Control of root growth and development by reactive oxygen species. Curr Opin Plant Biol 29: 57–63 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Miller G, Song L, Kim J, Sodek A, Koussevitzky S, Misra AN, Mittler R, Shintani D (2009) Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol 151: 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V (2013) The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J 32: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen J, Xie Z, Liu S, Nolan T, Ye H, Zhang M, Guo H, Schnable PS, Li Z, et al. (2014) Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol Plant 7: 1303–1315 [DOI] [PubMed] [Google Scholar]
- Widiez T, El Kafafi S, Girin T, Berr A, Ruffel S, Krouk G, Vayssières A, Shen W-H, Coruzzi GM, Gojon A, et al. (2011) High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3− uptake is associated with changes in histone methylation. Proc Natl Acad Sci USA 108: 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Mhamdi A, Stael S, Storme V, Kerchev P, Noctor G, Gevaert K, Van Breusegem F (2016) The ROS Wheel: Refining ROS transcriptional footprints. Plant Physiol 171: 1720–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA (2008) The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Sokolov LN, Ye J, Tang CY, Shi J, Zhen Y, Lan W, Hong Z, Qi J, Lu GH, et al. (2016) The LIKE SEX FOUR2 regulates root development by modulating reactive oxygen species homeostasis in Arabidopsis. Sci Rep 6: 28683. [DOI] [PMC free article] [PubMed] [Google Scholar]