A robust pipeline identifies and experimentally validates cis-natural antisense transcripts controlling cognate sense mRNA translation.
Abstract
Cis-Natural Antisense Transcripts (cis-NATs), which overlap protein coding genes and are transcribed from the opposite DNA strand, constitute an important group of noncoding RNAs. Whereas several examples of cis-NATs regulating the expression of their cognate sense gene are known, most cis-NATs function by altering the steady-state level or structure of mRNA via changes in transcription, mRNA stability, or splicing, and very few cases involve the regulation of sense mRNA translation. This study was designed to systematically search for cis-NATs influencing cognate sense mRNA translation in Arabidopsis (Arabidopsis thaliana). Establishment of a pipeline relying on sequencing of total polyA+ and polysomal RNA from Arabidopsis grown under various conditions (i.e. nutrient deprivation and phytohormone treatments) allowed the identification of 14 cis-NATs whose expression correlated either positively or negatively with cognate sense mRNA translation. With use of a combination of cis-NAT stable over-expression in transgenic plants and transient expression in protoplasts, the impact of cis-NAT expression on mRNA translation was confirmed for 4 out of 5 tested cis-NAT:sense mRNA pairs. These results expand the number of cis-NATs known to regulate cognate sense mRNA translation and provide a foundation for future studies of their mode of action. Moreover, this study highlights the role of this class of noncoding RNAs in translation regulation.
A large proportion of the genome of eukaryotes is transcribed into RNA that is not coding for proteins or house-keeping RNAs (e.g. tRNAs, ribosomal RNAs; Djebali et al., 2012). Whereas first being considered as transcriptional noise, noncoding RNAs have emerged as major regulators of gene expression (Rinn and Chang, 2012; Bonasio and Shiekhattar, 2014; Chekanova, 2015; Ransohoff et al., 2018). Besides the well-characterized small RNAs that include short interfering RNAs (siRNAs) and micro RNAs (miRNAs), abundant long noncoding RNAs (lncRNAs) have been identified across a wide spectrum of organisms. LncRNAs are typically defined as capped and polyadenylated transcripts longer than 200 bases that do not contain conserved open reading frame capable of encoding proteins (Rinn and Chang, 2012; Bonasio and Shiekhattar, 2014; Chekanova, 2015; Ransohoff et al., 2018). However, recent studies have indicated that some lncRNAs could associate with ribosomes and, in some cases, generate small peptides (Ji et al., 2015; Hsu et al., 2016; Bazin et al., 2017). Whereas most of the lncRNAs identified by genome-wide studies have yet unknown functions, an increasing number has been shown to be involved in critical biological processes such as X chromosome inactivation in mammals (Brockdorff et al., 1992) or flowering in plants (Liu et al., 2010; Heo and Sung, 2011).
LncRNAs located in intergenic regions relative to coding genes are defined as long intergenic noncoding RNA (lincRNAs), whereas lncRNAs overlapping coding genes and transcribed from the opposite DNA strand are categorized as cis-Natural Antisense Transcripts (cis-NATs; Rinn and Chang, 2012). In addition, lincRNAs able to bind target mRNAs by partial base-pair complementarity are defined as trans-Natural Antisense Transcripts (trans-NAT; Lapidot and Pilpel, 2006). Cis-NATs are widespread in eukaryotes, with 20–70% of coding genes having an associated cis-NAT in Saccharomyces cerevisiae, Drosophila melanogaster, mice, human, and Arabidopsis (Arabidopsis thaliana; Faghihi and Wahlestedt, 2009; Liu et al., 2015). Cis-NATs can overlap completely with their cognate mRNAs or only at the 5′ (head-to-head orientation) or the 3′ end (tail-to-tail orientation).
Whereas various modes of action have been reported for lncRNAs impacting the regulation of target gene expression, the majority involves changes in the steady-state level or structure of mRNA via changes in transcription, mRNA stability, or splicing (Rinn and Chang, 2012; Bonasio and Shiekhattar, 2014; Chekanova, 2015; Ransohoff et al., 2018). This is true for either lincRNAs or cis-NATs and applies to both animal and plant models. Well-characterized mechanisms by which lncRNAs affect gene transcription include recruitment of chromatin or transcription regulators and displacement of transcriptional repressors. Examples of such mechanisms in animals includes inhibition of transcription via histone methylation by HOTAIR (Gupta et al., 2010) or DNA methylation by promoter-associated RNA (Schmitz et al., 2010), stimulation of transcription by the recruitment of the activator Pygopus Homolog 2 by the lincRNA Prostate Cancer Gene Expression Marker 1 (Yang et al., 2013), and the displacement of the repressive glucocorticoid response element by the lincRNA Growth Arrest-Specific 5 (Kino et al., 2010). Similar mechanisms for lncRNAs in plants have been described, such as histone modification at Flowering Locus C triggered by the cis-NAT COOLAIR (Liu et al., 2010) or the intronic lncRNA COLDAIR (Heo and Sung, 2011), as well as the transcriptional activation of Pathogen-Responsive Gene 1 via the recruitment of a Mediator component by lincRNA ELF18-Induced Long-Noncoding RNA 1 (Seo et al., 2017). LincRNA can also interact with splicing factors to regulate alternative splicing, as described for lincRNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 in animals (Tripathi et al., 2010) and Antisense Competitor in plants (Bardou et al., 2014). Moreover, lincRNA can control mRNA stability via interaction with members of the Staufen double-standed RNA-binding proteins in animals (Gong and Maquat, 2011) or inhibition of microRNA action on mRNA degradation via target mimicry, as described for the Arabidopsis Induced by Phosphate Starvation 1 (IPS1) lincRNA involved in the response of plants to inorganic phosphate (Pi) deficiency (Franco-Zorrilla et al., 2007). Another mechanism for the decrease in steady-state mRNA level associated with cis-NAT expression is the generation of siRNA via processing of dsRNA generated by the overlapping cis-NAT with its cognate sense mRNA (Khorkova et al., 2014). However, considering the large number of potential cis-NAT:sense mRNA pairs in animal and plant genomes, relatively few examples of siRNA-mediated effects for cis-NATs have been described, indicating that this mechanism may be less frequently used than initially thought.
In contrast with the effects of lncRNAs on transcription and mRNA levels, examples of modulation of mRNA translation by lncRNAs are rather rare. Examples involving lincRNAs in human cell lines are the inhibition of translation of targets Catenin Beta 1 and JUNB via recruitment of the translational repressor Rck by the lincRNA-p21 (Yoon et al., 2012) and the inhibition of c-Myc translation by the recruitment of the eukaryotic initiation factor eIF4E by lncRNA Growth Arrest-Specific 5 (Hu et al., 2014). Repression of mRNA translation was also demonstrated for the cis-NAT of the PU.1 gene, encoding a transcription factor in mammals (Ebralidze et al., 2008). Recently, three examples for the enhancement of translation by cis-NATs have been described. In rice (Oryza sativa), expression of the cis-NAT of the Pi exporter gene Phosphate 1;2 was shown to enhance the association of the cognate mRNA to polysomes, leading to the accumulation of Phopshate 1;2 protein despite unchanged steady-state level of the corresponding mRNA (Jabnoune et al., 2013). In mice, Ubiquitin Carboxy-Terminal Hydrolase 1 (Uchl1) mRNA translation is enhanced by a cis-NAT that is exported to the cytoplasm upon inhibition of the Target of Rapamycin pathway (Carrieri et al., 2012). Finally, the human regulator of megakaryocyte differentiation RNA Binding Protein 15 (RBM15) is also translationally enhanced by a cis-NAT (Tran et al., 2016). Little is known about the mechanisms of action of the three translation enhancer cis-NATs reported so far. All three pairs are oriented in a head-to-head manner (5′-5′). For RBM15, the region of the antisense overlapping with the sense mRNA 5′ untranslated region (UTR) alone was found to be sufficient to enhance translation (Tran et al., 2016). In contrast, for Uchl1, two elements in the cis-NAT were found to be essential, namely the region overlapping with the 5′ end of Uchl1 mRNA and a nonoverlapping inverted Short Interspersed Nuclear Element (SINE) B2 element, a class of retrotransposable repeat element (Carrieri et al., 2012). More recently, cis-NATs containing distinct SINE elements have been identified in mammals as potential translation enhancers (Schein et al., 2016), whereas expression of some ribosome-associated cis-NATs in plants were correlated with increased mRNA translation (Bazin et al., 2017).
The low number of cis-NATs experimentally validated to influence translation of the cognate mRNA might reflect the fact that most genome-wide studies of cis-NATs examined the correlation between steady-state level of mRNAs and the expression of cis-NATs, an approach that is not suitable for studying translation. In the current study, we took advantage of the polysome profiling method combined with strand-specific RNA sequencing to identify, in Arabidopsis plants, cis-NATs whose expression level were associated with a change of cognate sense mRNA level, as well as translation across a range of experimental conditions. The impact of cis-NAT expression on cognate mRNA translation was further validated by expression of several cis-NATs in transgenic Arabidopsis and/or by transient expression in protoplasts.
RESULTS
Experimental Setup to Identify cis-NATs Associated with Changes in mRNA Level and mRNA Translation
To identify cis-NATs impacting their cognate sense mRNA transcript level as well as mRNA translation, an experimental procedure was set up allowing the quantification of steady-state levels of coding and noncoding RNAs along with the determination of mRNA translation efficiency genome-wide in Arabidopsis seedlings grown under various conditions. Whole seedlings grown in liquid cultures in the presence of a high (1 mM) or low (100 µM) concentration of Pi were analyzed, as well as roots and shoots from seedlings grown on agar-solidified medium supplemented with different phytohormones, namely auxin (indole acetic acid), abscisic acid (ABA), methyl-jasmonate (MeJA), or 1-aminocyclopropane-1-carboxylic acid (ACC), a precursor of ethylene. For each sample, steady-state levels of cis-NATs and mRNAs were determined by strand-specific sequencing of total polyA+ RNA, whereas translation efficiency was assessed for the same sample by sequencing polysome-associated RNA purified by centrifugation through Suc density gradients. Sequencing of each total or polysomal RNA sample yielded between 30 and 60 million paired-end reads. Three independent biological replicates were analyzed for each treatment, and a total of at least 120 million paired-end reads were obtained per condition.
The genes up- or down-regulated in response to the different treatments were identified by pairwise comparisons between hormone-treated or low Pi samples and their corresponding controls. In response to low Pi, 2,991 protein-coding genes (according to The Arabidopsis Information Resource [TAIR]10 annotation) were significantly up-regulated with a fold change >2 and adjusted p-value (adj.pval) <0.1, and 2,149 were significantly down-regulated (Fig. 1A; Supplemental Table S1 and S2). Fewer genes were differentially expressed in response to the different hormone treatments (Supplemental Fig. S1; Supplemental Table S1 and S2). For example, upon auxin treatment, 377 and 120 protein-coding genes were up-regulated in roots and shoots, respectively (Supplemental Table S2). Untreated root and shoot tissues were also compared, and their transcriptomes were dramatically different, as expected for two different organs, with 3,906 and 4,742 protein-coding genes significantly up- or down-regulated, respectively, in roots relative to shoots (Supplemental Table S2).
Figure 1.
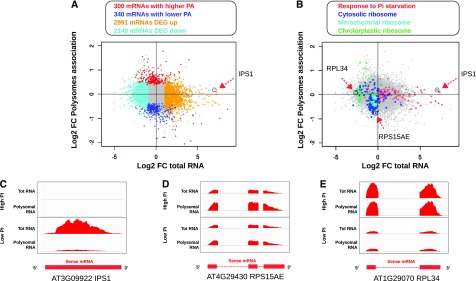
Steady-state mRNA expression level and association with polysomes in response to growth of Arabidopsis under low-Pi conditions. A, Relation between log2-fold change of mRNA steady-state level (x axis) is plotted against the log2-fold change in polysome association (y axis). Coding genes significantly up- or down-regulated at the mRNA steady-state level are colored in yellow and cyan, respectively, whereas mRNA significantly more or less associated with polysomes are colored in red and blue, respectively. The genes not showing any statistical difference are colored in gray. B, Same plot as (A) where genes associated with GO terms “Response to Pi starvation,” “Cytosolic ribosome,” “Mitochondrial ribosome,” and “Chloroplastic ribosome” are colored in pink, dark blue, light blue, and green, respectively. C–E, Normalized RNA-seq coverage plots for the IPS1, RPS15AE, and Ribosomal Protein L34 (RPL34) genes. The two top panels show the coverage plots for total mRNA and polysomal RNA from high Pi samples, and the two bottom panels correspond to low phosphate samples. The schematic exonic organization of each gene is represented by red boxes and lines below the plots.
Quality assessment of the transcriptomic data was first performed by Gene Ontology (GO) term enrichment analyses for each set of up-regulated genes. This analysis confirmed the strong induction of marker genes associated with the different treatments. The genes up-regulated in response to low Pi were significantly enriched for the GO term “cellular response to Pi starvation” (GO:0016036 adj.pval = 5.3 × 10−11; Supplemental Fig. S2A). Among these, IPS1, a known highly induced marker of Pi deficiency, was strongly over-expressed (fold change = 127.9; Fig. 1, A and C). Similarly, the up-regulated genes were significantly enriched in GO terms “response to ABA” (GO:0009737, adj.pval = 1.3 ×10−10), “response to auxin” (GO:0009733, adj.pval = 6.5 × 10−11), “jasmonic acid metabolic process” (GO:0009694, adj.pval = 5.6 × 10−11,), and “ethylene-activated signaling pathway” (GO:0009873, adj.pval = 6.4 × 10−8) in root samples treated with ABA, auxin, MeJA, and ACC, respectively, compared with untreated roots (Supplemental Fig. S2, B–E).
The genes differentially expressed in response to low Pi and ABA treatments were further compared with previously published datasets (Supplemental Fig. S3). Approximately 71% of the genes up-regulated in whole seedlings upon ABA treatment in the study of Song et al. (2016) were also up-regulated in ABA-treated roots and/or shoots in our dataset (941 genes out of 1,327). Similarly, 79% of genes up-regulated in the study of Bazin et al. (2017; 154 / 194) and 30.5% of genes up-regulated either in roots or shoot in Yuan et al. (2016) were common with the genes up-regulated under our low-Pi condition. The lower proportion of common differentially expressed genes with that reported by Yuan et al. (2016) might be explained by the differences in terms of tissue analyzed (root versus whole seedlings) and growth (e.g. liquid versus solid medium and different Pi concentrations) conditions.
Analysis of Differential mRNA Translation
Translation efficiency of coding genes can be estimated by measuring the proportion of mRNA molecules associated with polysomes relative to the amount of total RNA, as previously described (Mustroph et al., 2009; Juntawong et al., 2014). With the use of sequencing data from polysome-associated RNA, the ratio of polysome association (PA) was calculated for each gene by dividing the normalized read count in the polysomal RNA fraction by the normalized read count measured for total RNA steady-state level. The treated samples were compared with that in the corresponding control conditions and loci with a 30% increase or decrease in PA ratio and an adj.pval < 0.1 were considered differentially associated with polysomes, and thus potentially regulated translationally (Supplemental Table S1 and S3). In response to growth under low-Pi conditions, 300 and 340 protein-coding genes were significantly more and less associated with polysomes, respectively, compared with that under high-Pi conditions (Fig. 1A). GO enrichment analyses revealed that the coding genes with a lower association with polysomes in response to low Pi were strongly enriched for ribosomal proteins (GO:0022626cellular component “cytosolic ribosome”, adj.pval = 2.26 × 10−11; Fig. 1B; Supplemental Fig. S4A), such as the cytosolic ribosomal protein RPS15AE (Fig. 1D). This finding was consistent with that of previous reports, where a similar down-regulation of the translation of the ribosomal proteins was observed by ribosome footprints in response to both Pi deficiency (Bazin et al., 2017) and hypoxia (Juntawong et al., 2014), validating both techniques for the analysis of mRNA translation. Of note, most of the genes constituting chloroplastic ribosomal proteins, such as Ribosomal Protein L34 (Fig. 1, B and E) showed a strong decrease in mRNA steady-state level without significant change in PA. The genes encoding mitochondrial ribosomal proteins, on the other hand, were globally less associated with polysomes, similarly to those encoding cytosolic ribosomal proteins (Fig. 1B).
Many genes were also found differentially associated with polysomes when comparing root and shoot tissues (Supplemental Fig. S5; Supplemental Table S3). For example, 946 protein-coding genes were significantly more associated with polysomes in roots and 1,033 in shoots, in untreated samples. Interestingly, the strongest enrichment within the set of genes with higher PA in shoots corresponds to GO:0008380 “mRNA splicing” (adj.pval = 6 × 10−11; Supplemental Fig. S4B). SERRATE and Serine/Arginine-Rich 45 (SR45), for example, were strongly associated with polysomes in shoots and very poorly in roots (fold change PA = 5.3 and 4.2 for SERRATE and SR45, respectively), despite similar steady-state levels of mRNA in both tissues (Supplemental Fig. S5, B–D). Both SERRATE and SR45 have been experimentally validated to participate in splicing, with an additional role for SERRATE in microRNA processing (Laubinger et al., 2008; Zhang et al., 2017).
De Novo Identification of cis-NATs
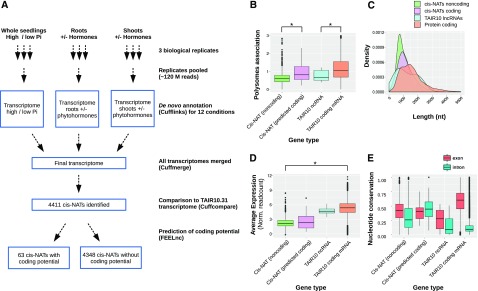
Cis-NATs expressed in response to the different treatments were identified using the pipeline described in “Materials and Methods” (Fig. 2A). In this pipeline, pairs of protein-coding genes having mRNAs that may overlap in a sense-antisense fashion are not included as bona fide cis-NATs. De novo transcriptome annotations corresponding to each of the 12 experimental conditions analyzed were merged, and, after comparison with the TAIR10.31 annotation (Berardini et al., 2015), a novel set of 4,411 cis-NATs were identified. Approximately 9% (374) of these cis-NATs were recently annotated in the Araport11 database (Cheng et al., 2017). We then used the FEELnc tool (Wucher et al., 2017; see “Materials and Methods” and Supplemental Materials and Methods for details) to determine in silico the coding potential of all the newly identified cis-NATs. The large majority of these cis-NATs (98.5%) were lacking coding potential, and only 63 were predicted to be potentially coding. This prediction of coding potential was well supported by our experimental polysome profiling data because the cis-NATs predicted to be coding were significantly more associated with polysomes than seen for the noncoding cis-NATs (Fig. 2B). A similar difference was observed when comparing protein-coding and noncoding genes annotated in the TAIR10 database.
Figure 2.
Identification and characterization of cis-NATs. A, Schematic diagram of the pipeline used for de novo cis-NAT identification from the 12 different experimental conditions. B, Boxplot comparing polysome association of cis-NATs predicted to be noncoding (green) or coding (pink), non-coding RNA (ncRNA;cyan), and protein-coding genes (salmon) annotated in TAIR10 database. C and D, Plots comparing transcript length (C) and RNA steady-state level (average expression reported as normalised readcounts; D) of cis-NATs predicted to be noncoding (green) or coding (pink), ncRNA (cyan), and protein-coding genes (salmon) annotated in TAIR10 database. E, Boxplots comparing the nucleotide conservation across 20 angiosperm genomes within exonic and intronic regions of the four categories of transcripts listed above.
Exploring the conservation across plant genomes of the peptides encoded by the 63 cis-NATs with high coding potential, we identified a group of 10 peptides that were well conserved among plant genomes, and a second group of nine peptides conserved among Brassicaceae species only (Supplemental Fig. S6). The remaining 44 predicted coding cis-NATs were poorly or not conserved. Seven of the cis-NATs encoding conserved peptides (Group I or II) were recently annotated as (putative) protein-coding genes in the Arabidopsis Information Portal 11 database but not in TAIR10. The transcripts encoding these evolutionary conserved peptides should thus likely be considered as protein-coding genes.
Expression of the identified cis-NATs was well supported by published epigenetic profiling data from Jégu et al. (2017). The predicted transcription start sites of the cis-NATs were strongly enriched for the activating histone mark H3K9Ac as well as micrococcal S7 nuclease (Mnase) footprints to the same extent as that in TAIR10 protein-coding loci, confirming the transcriptionally active state of the promoter regions of the cis-NATs (Supplemental Fig. S7). Moreover, 60% of the cis-NATs detected in our dataset overlapped with cis-NATs previously identified in at least one of the three datasets used for comparison: the PlncDB database (Jin et al., 2013; Wang et al., 2014) as well as the work of Yuan et al. (2016) and ; Supplemental Fig. S8).
Cis-NATs were on average shorter, expressed at a lower level, and had a weaker genomic sequence conservation (PHASTcons score) compared with that of TAIR10 annotated noncoding RNA and protein-coding mRNAs (Fig. 2, C–E), consistent with previous reports of cis-NATs in plants and other eukaryotes (Wang et al., 2005; Khorkova et al., 2014; Yuan et al., 2015). Furthermore, the polysome association value of cis-NATs was significantly lower (0.64) compared with that of mRNAs (1.19), but similar to that of the noncoding transcripts annotated in the TAIR10 database (0.54; Fig. 2B).
To validate our pipeline of identification of differentially expressed cis-NATs and protein-coding genes, we analyzed by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) the expression level of six protein-coding genes and six cis-NATs predicted to be up- or down-regulated in response to phosphate starvation. For the 12 genes analyzed, the RT-qPCR results showed a significant increase or decrease in RNA steady-state level in agreement with the RNA sequence data (Supplemental Fig. S9).
Cis-NATs Associated with Changes in Steady-State Level of their Cognate mRNA
To identify cis-NATs potentially regulating the expression of their cognate mRNA at the transcriptional or translational level, we looked for correlation between expression levels of all available cis-NATs (e.g. cis-NATs identified in this study and those included in TAIR10.31, total of 4,846 cis-NATs) and cognate sense mRNA steady-state levels or polysome association across the different experimental conditions analyzed, comparing hormone-treated samples with untreated controls for root and shoot tissues as well as seedlings grown under low- or high-Pi conditions. Untreated root and shoot tissues were also compared to identify cis-NATs potentially regulating tissue-specific gene expression. All cis-NAT:sense mRNA pairs were put into four categories considering their region of overlap, namely overlap in the 5′ end, 3′ end, completely included within the sense region, or cis-NATs that extend beyond the 5′ and 3′ region of the coding sense (overhanging; Supplemental Fig. S10).
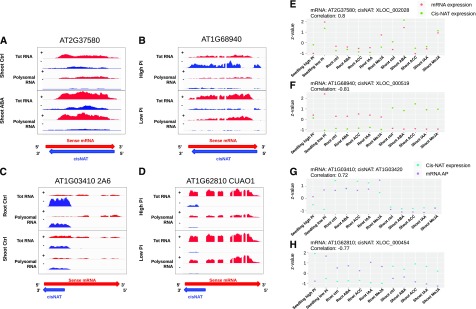
Analysis for potential effects of cis-NAT expression on steady-state sense mRNA level was performed. For each pairwise comparison, the cis-NAT:mRNA pairs were considered correlated if both the cis-NAT and the cognate mRNA were differentially expressed, with a fold change of at least 2 and a false discovery rate < 0.1. A total of 1,310 cis-NATs, including 67 annotated in TAIR10, were differentially expressed in at least one condition (Supplemental Table S1). For 107 of these loci, steady-state level of the cognate mRNA was positively correlated to cis-NAT expression (Table 1; Supplemental Table S4). For example, both the mRNA and the cis-NAT of the locus AT2G37580 were significantly up-regulated upon ABA treatment in shoots (fold-change [FC] = 2.13, adj.pval = 0.029 for the mRNA and FC = 2.06, adj.pval = 1.3 × 10−2 for the cis-NAT; Fig. 3A). We also found 41 pairs with a negative correlation, such as AT1G68940, whose mRNA was up-regulated in response to low-Pi conditions (FC = 2.16, adj.pval = 7.9 × 10−3), whereas the cis-NAT was down-regulated (FC = 0.39, adj.pval = 7.2 × 10−2; Fig. 3B). Pearson correlation coefficient between cis-NAT and mRNA expression was calculated across the 12 experimental conditions analyzed, taking advantage of the whole experimental dataset to identify cis-NATs with stronger positive and negative expression correlation with their cognate sense mRNA. This analysis revealed that 86 cis-NAT:sense mRNA pairs out of 107 had a positive correlation coefficient higher than 0.4, whereas 27 cis-NAT:sense mRNA pairs out of 41 had a negative correlation coefficient lower than −0.4, across all 12 experimental conditions (Fig. 3, E and F; Table 1).
Table 1. Number of cis-NATs correlated with cognate gene steady-state mRNA expression or association with polysomes.
Number of mRNA / cis-NAT pairs with either a positive or negative correlation between cis-NAT and cognate gene steady-state mRNA expression (second and third columns), and number of pairs with positive or negative correlation between cis-NAT expression and cognate gene mRNA PA (fourth and fifth columns). The experimental conditions compared are indicated in the first column where root and shoot tissues are indicated with the prefix R and S, respectively, and untreated control conditions indicated with the suffix ctrl. The figures in brackets show the number of those pairs with a Pearson correlation coefficient > 0.4 or < −0.4 across the 12 experimental correlations.
| Treatment | Positive Correlation cis-NAT / mRNA Expression | Negative Correlation cis-NAT / mRNA Expression | Positive Correlation cis-NAT Expression / mRNA PA | Negative Correlation cis-NAT Expression / mRNA PA |
|---|---|---|---|---|
| Low / high Pi | 40 (33) | 10 (3) | 4 (1) | 1 (1) |
| RIAA / Rctrl | 2 (2) | 0 | 0 | 0 |
| RABA / Rctrl | 17 (13) | 0 | 1 (0) | 1 (0) |
| RMeJA / Rctrl | 6 (4) | 0 | 0 | 0 |
| SABA / Sctrl | 10 (7) | 1 (0) | 0 | 0 |
| SMeJA / Sctrl | 3 (3) | 1 (0) | 0 | 0 |
| Rctrl / Sctrl | 47 (41) | 29 (24) | 3 (2) | 4 (2) |
| Total (unique) | 107 (86) | 41 (27) | 8 (3) | 6 (3) |
Figure 3.
Correlations between expression of cis-NATs and changes in steady-state level or poylsome association of the cognate sense mRNA. A–D, Coverage plots showing the density of RNA-sequence (RNA-seq) reads per position at AT2G37580, AT1G68940, AT1G03410, and CuAO1 loci. The red and blue areas represent the density of reads mapping to the sense mRNA and cis-NATs, respectively. For each experimental condition, the top part corresponds to total RNA-seq reads and the bottom part to polysomal RNA-seq reads. The red and blue arrows at bottom indicate the cis-NAT-mRNA pair orientation. E–H, Correlation plots showing the steady-state level of the coding mRNA (red dots) and cis-NAT (green dots) for AT2G37580 and AT1G68940 loci (E and F, respectively) or the steady-state level of the cis-NAT (cyan dots) and the association with polysomes of the cognate sense mRNA (purple dots) for AT1G03410 and CuAO1 loci (G and H, respectively). The Z-score of normalized read counts calculated from the 12 experimental conditions is represented on the y axis. Pearson correlation coefficients between the two variables shown in each plot are indicated on top of the plots.
Identification of Putative Translation Regulator cis-NATs
To identify cis-NATs influencing the translation of their cognate sense mRNA, we looked for cis-NAT:sense mRNA pairs where the cis-NAT was differentially expressed (fold change > 2 and adj.pval < 0.1) and the sense mRNA that was differentially associated with polysomes (at least 30% increase or decrease, adj.pval < 0.1) in response to treatment. A finer filtering step was also performed using additional criteria such as the size of the overlapping region or the relative level of expression between cis-NAT and mRNA (see “Materials and Methods” for further details). A total of eight cis-NAT:sense mRNA pairs were identified for which cis-NAT differential expression was positively correlated to mRNA differential association with polysomes in at least one pairwise comparison (Supplemental Table S4). For example, AT1G03410 mRNA was more associated with polysomes (FC = 1.79, adj.pval = 0.01) when the cis-NAT was more expressed (FC = 2.71, ajd.pval = 5.6 × 10−18) in untreated roots samples compared with untreated shoots (Fig. 3C). A total of six pairs showed a negative correlation between cis-NAT expression and cognate mRNA association with polysomes (Supplemental Table S4), including the Copper Amine Oxidase 1 (CuAO1) locus whose sense mRNA was more associated with polysomes (FC = 1.68, adj.pval = 0.05) when the cis-NAT was less abundant (FC = 0.29, adj.pval = 1.3 × 10−5) in Pi-deficient seedlings (Fig. 3D). The expression of three out of the eight cis-NAT:sense mRNA pairs with a positive correlation and three out of the six pairs with negative correlation had a Pearson correlation coefficient > 0.4 and < −0.4, respectively, with polysome association of their cognate mRNAs across the 12 experimental conditions (Fig. 3, G and H; Table 1; Supplemental Table S4).
The cis-NATs identified that positively or negatively correlated with sense mRNA steady-state transcript level or polysome association were further analyzed for relation with miRNAs, i.e. presence of miRNA precursor sequence, miRNA target sequence, and potential as a microRNA target mimic (see “Materials and Methods”). Out of the 4,846 cis-NATs analyzed, 14% (682) were predicted to contain at least one miRNA binding site (Supplemental Table S1), including 7 and 14 cis-NATs negatively and positively correlated to cognate mRNA steady-state level, respectively (Supplemental Table S1). Two cis-NATs positively correlated with mRNA polysome association were predicted to contain miRNA binding sites, but none of the cis-NATs with a negative correlation. Only seven cis-NATs were predicted as miRNA precursors, and 69 contained potential miRNA target mimic sites, including two cis-NATs positively correlated with cognate mRNA expression and one cis-NAT positively correlated with mRNA polysome association (Supplemental Table S1). No cis-NAT negatively correlated to mRNA expression or polysome association contained a putative miRNA target mimic site.
We also took advantage of 40 publicly available small RNA datasets to analyze the cis-NATs in relation to siRNAs. We identified 24,119,910 small reads between 18 and 28 nucleotides long mapping to TAIR10 reference genome. Of those, 666,181 mapped to cis-NAT loci and were considered as cis-NAT-siRNAs. Most of them were 21 and 24 nucleotides long (Supplemental Fig. S11A) and the overlapping region of cis-NATs showed a significantly higher density in small RNAs compared with that of nonoverlapping regions (Supplemental Fig. S11B), in agreement with previous reports (Zhou et al., 2009; Yuan et al., 2015). We identified 1336 potential siRNA precursor cis-NATs, with at least five small reads mapping to the overlapping region and a read density at least 2-fold higher in the overlapping region than that in the nonoverlapping region. From this set of 1,336 cis-NATs, 25 belonged to the group of putative transcription enhancers (representing 23% of the 107 candidates), 10 to the group of putative transcription inhibitors (representing 24% of the 41 candidates), one to the group of putative translation enhancers (representing 12.5% of the 8 candidates), and none to the group of putative translation repressors (Supplemental Table S1).
We also looked for the presence of transposable elements or inverted repeats within the cis-NATs identified (see “Materials and Methods”; Supplemental Table S1). Approximately 10.5% of the cis-NATs (i.e. 508) contained at least part of transposable element sequences, including one positively correlated and one negatively correlated to mRNA translation. Transposable element sequences were found in 15 out of the 107 cis-NATs correlated with cognate mRNA steady-state level. Similarly, we found that 121 cis-NATs contained inverted repeats, including one in a cis-NAT positively correlated with cognate mRNA translation and one in a cis-NATs negatively correlated with cognate mRNA steady-state level.
Experimental Validation of cis-NAT Regulation of Cognate Sense mRNA Translation
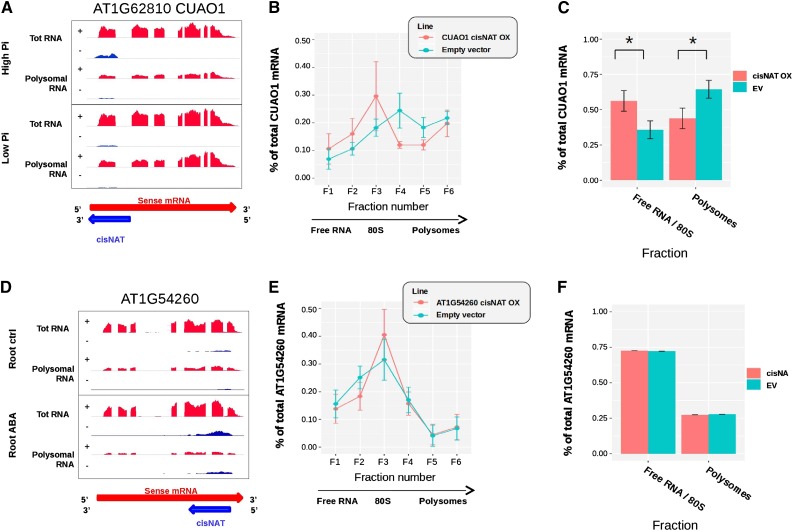
A positive or negative correlation between cis-NAT expression and cognate mRNA association with polysomes could indicate translation enhancement or repression of the mRNA by the cis-NAT. To experimentally validate such a potential translation regulation activity, two cis-NATs with potential translation enhancer activity and two with putative translation repressor activity were cloned after the CaMV35S promoter and used to transform Arabidopsis to produce transgenic lines over-expressing the cis-NATs in trans. Two independent transgenic lines were selected for each cis-NAT construct with a robust (>10-fold) over-expression of the transgenic cis-NAT compared with the steady-state level of the endogenous cis-NAT in wild-type lines but without a significant change of steady-state level of the endogenous cognate mRNA (Supplemental Fig. S12). Polysome association of each cognate sense mRNA was analyzed by Suc density gradient in the lines over-expressing the cis-NATs compared with that in a control line transformed with an empty vector. The distribution of mRNAs along the Suc density gradient was determined by RT-qPCR (Figs. 4, B and E, and 5, B and E; Supplemental Fig. S13). To quantify the changes in terms of association with polysomes in a more robust manner, the proportion of mRNA present either in fractions containing free mRNA or monosomes (fractions 1 to 3) versus fractions containing polysomal mRNAs (fractions 4 to 6) was calculated for each of the eight independent biological replicates (Figs. 4, C and F, and 5, C and F). This analysis revealed that over-expression of the cis-NAT associated with the CuAO1 locus was associated with a decrease in translation of the cognate mRNA (Fig. 4, B and C), in agreement with the negative correlation between cis-NAT steady-state level and mRNA polysome association (Fig. 4A). In contrast, polysome association of the mRNA of locus At1g54260, encoding a potential transcription factor, was not significantly changed by the over-expression of its cis-NAT in trans (Fig. 4, E and F), despite the positive correlation (Fig. 4D).
Figure 4.
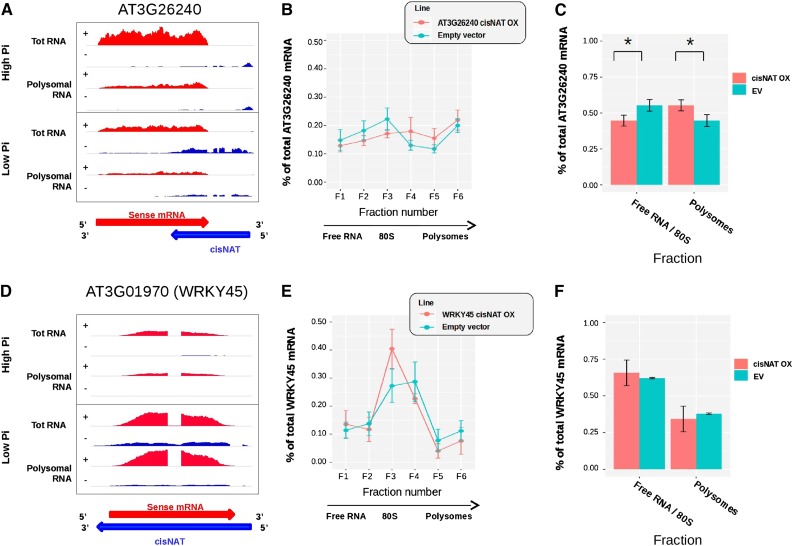
Expression of putative translation repressor cis-NATs in transgenic Arabidopsis. A–D, Coverage plots showing the density of RNA-sequence (RNA-seq) reads per position for the CuAO1 (A) and AT1G54260 (D) loci, with the red and blue areas representing the sense mRNA and cis-NATs, respectively. Tot RNA, total RNA; Root ctrl, root control. B and E, Polysome profiles showing the proportion of endogenous mRNA in each of the six fractions of the Suc gradient for transgenic lines over-expressing (OX) the cis-NAT (red) versus that in control lines transformed with an empty vector (EV; turquoise) for the CuAO1 (B) and AT1G54260 (E) sense mRNA-cis-NAT pair. C and F, Proportion of mRNA present in the first three fractions (free RNA and monosomes) and in the last three fractions (polysomes) of the gradient. Determinations were in transgenic lines OX cis-NAT (red) and in control lines transformed with an EV (turquoise) for the CuAO1 (C) and AT1G54260 (F) sense mRNA-cis-NAT pair. Data in B, C, E, and F represent the average of 8 independent biological replicates obtained with 2 independent transgenic lines. The error bars represent the confidence intervals with alpha = 0.05. Asterisks indicate significant differences (Student’s t test with p-value < 0.05).
Figure 5.
Expression of putative translation activator cis-NATs in transgenic Arabidopsis. A and D, Coverage plots showing the density of RNA-sequence (RNA-seq) reads per position for the AT3G26240 (A) and WRKY45 (D) loci, with the red and blue areas representing the sense mRNA and cis-NATs, respectively. Tot RNA, total RNA; Root ctrl, root control. B and E, Polysome profiles showing the proportion of endogenous mRNA in each of the six fractions of the Suc gradient for transgenic lines over-expressing (OX) the cis-NAT (red) versus that in control lines transformed with an empty vector (EV; turquoise) for the AT3G26240 (B) and WRKY45 (E) sense mRNA-cis-NAT pair. C and F, Proportion of mRNA present in the first three fractions (free RNA and monosomes) and in the last three fractions (polysomes) of the gradient, in transgenic lines OX cis-NAT (red) and in control lines transformed with an EV (turquoise) for the AT3G26240 (C) and WRKY45 (F) sense mRNA-cis-NAT pair. Data in B, C, E, and F represent the average of 8 independent biological replicates obtained with 2 independent transgenic lines. The error bars represent the confidence intervals with alpha = 0.05. Asterisks indicate significant differences (Student’s t test with p-value < 0.05).
Similar analysis performed for two cis-NATs that displayed positive correlation with their cognate mRNA translation (Fig. 5, A and D) showed that lines over-expressing the cis-NAT to locus At3g26240 showed a reproducible shift of its cognate mRNA toward the heavy polysome fractions in eight independent biological replicates, indicating a stimulatory effect of cis-NAT expression on translation (Fig. 5, B and C). In contrast, over-expression of the cis-NAT to locus WRKY45 (AT3G01970) did not change significantly the polysome profile of its cognate sense mRNAs (Fig. 5, E and F).
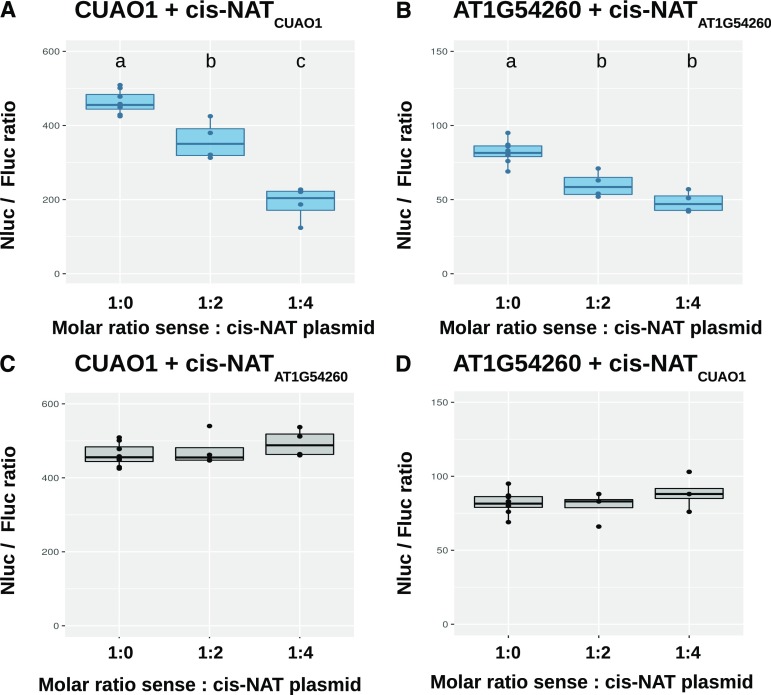
A protoplast cotransformation system was developed to independently validate the effects of cis-NAT expression on the translation of their cognate sense mRNA. Protoplasts were transformed with a plasmid containing a sense-coding gene fused to NanoLuc luciferase (Nluc), in the presence or absence of a plasmid expressing the cognate cis-NAT. The sense-Nluc vectors contained also an independent expression cassette for the firefly luciferase (Fluc), used as an internal transformation and loading control (see “Materials and Methods” for further details). The ratio Nluc:Fluc activity was used to assess the effect of each selected cis-NAT on its sense-encoded protein accumulation. Increasing the molar amounts of cis-NATCuAO1 resulted in a corresponding decrease in the expression of the CuAO1-Nluc fusion protein as detected by a decrease in the Nluc:Fluc ratio (Fig. 6A). Importantly, this inhibitory effect was not observed at the transcript level (Supplemental Fig. S14). Similarly, increasing amounts of cis-NATAT1G54260 resulted in reduction in expression of the AT1G54260-Nluc fusion protein (Fig. 6B) without an effect on transcript level (Supplemental Figure S14), although the effect was less pronounced than the one observed for the cis-NATCuAO1-sense construct pair. These later results are in contrast with the lack of significant effect observed in stable transgenic lines overexpressing cis-NATAT1G54260 and analyzed by quantification of polysomal mRNA in Suc gradient fractions (Fig. 4, E and F). Such discrepancy may reflect the higher variability of the transgenic-polysomal approach or the higher sensitivity of the protoplast transformation method. To further validate the specificity of the effects of cis-NAT on translation of the corresponding sense mRNA, cis-NATCuAO1 was cotransformed with the AT1G54260-Nluc construct, and cis-NATAT1G54260 was cotransformed with the CuAO1-Nluc construct. In this cis-NAT swap experiment, no effect on Nluc:Fluc ratio were observed (Fig. 6, C and D), revealing that the inhibitory effect of cis-NATAT1G54260 and cis-NATCuAO1 on translation was specific to their cognate sense genes.
Figure 6.
Transient expression of putative translation repressor cis-NATs in protoplasts. Arabidopsis leaf protoplasts were cotransformed with a plasmid combining a sense mRNA-NanoLuc luciferase (Nluc) fusion and a firefly luciferase (Fluc) along with various molar ratios of an independent plasmid for expression of a cis-NAT. The ratio of Nluc over Fluc activity is plotted for each combination of sense and cis-NAT plasmids. A, Coexpression of CuAO1-Nluc fusion with its cognate cis-NAT. B, Coexpression of AT1G54260-Nluc with its cognate cis-NAT. C, Coexpression of CuAO1-Nluc with the cis-NAT to AT1G54260. D, Coexpression of AT1G54260-Nluc with the cis-NAT to CuAO1. Statistically significant differences (Student’s t test, p-value < 0.05; four biological replicates) between treatments are indicated by distinct letters above the boxes.
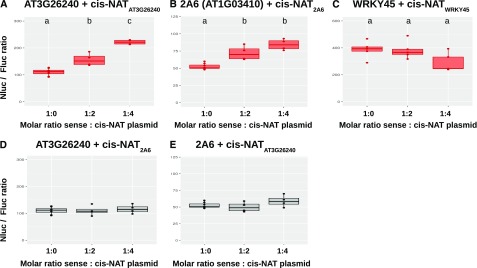
The protoplast system was also used to test the two cis-NATs acting as potential translational enhancers that were previously analyzed in transgenic plants, namely the cis-NATs to AT3G26240 and WRKY45, as well as an additional third candidate, 2A6 (AT1G03410; Fig. 7). There was a significant increase in Nluc:Fluc ratio upon addition of increasing amount of cis-NATAT3G26240 to its cognate sense construct, without a corresponding increase in transcript levels (Fig. 7A; Supplemental Fig. S14), confirming the translational enhancement of this cis-NAT. Similar to the results obtained with stable transgenic plants (Fig. 5, E and F), there was no significant effect of addition of cis-NATWRKY45 on the expression of the WRKY45-Nluc construct (Fig. 7C). Although we did not generate transgenic lines to test the effect of cis-NAT2A6 on the translation of its cognate sense mRNA, protoplast analysis revealed an increasing Nluc:Fluc ratio associated with the addition of cisNAT2A6 to its corresponding sense 2A6-Nluc construct, without changes in mRNA levels (Fig. 7B; Supplemental Fig. S14), revealing a similar translational enhancement effect as those observed with cis-NATAT3G26240. A swap experiment performed between the cis-NATAT3G26240 and cis-NAT2A6 showed no enhancement effect on unrelated sense-Nluc fusion (Fig. 7, D and E), confirming that the stimulatory effect of cis-NATAT3G26240 and cis-NAT2A6 on translation was specific to their cognate sense genes.
Figure 7.
Transient expression of putative translation activator cis-NATs in protoplasts. Arabidopsis leaf protoplasts were cotransformed with a plasmid combining a sense mRNA-NanoLuc luciferase (Nluc) fusion and a firefly luciferase (Fluc) along with various molar ratios of an independent plasmid for expression of a cis-NAT. The ratio of Nluc over Fluc activity is plotted for each combination of sense and cis-NAT plasmids. A, Coexpression of AT3G26240-Nluc fusion with its cognate cis-NAT. B, Coexpression of 2A6 (AT1G03410)-Nluc with its cognate cis-NAT. C, Coexpression of WRKY45 (AT3G01970)-Nluc with its cognate cis-NAT. D, Coexpression of AT3G26240-Nluc with the cis-NAT to 2A6. E, Coexpression of 2A6-Nluc with the cis-NAT to AT3G26240. Statistically significant differences (Student’s t test, p-value < 0.05; four biological replicates) between treatments are indicated by distinct letters above the boxes.
Over-expression of the putative translation inhibitor cis-NATCuAO1 in transgenic lines probably resulted in lower levels of endogenous CuAO1 protein, although we were not able to detect and quantify reliably CuAO1 protein by targeted mass spectrometry using N15-labeled plants (Hart-Smith et al., 2017). Because Arabidopsis CuAO1 knock-down mutants were shown to be impaired in nitric oxide (NO) production induced by polyamines (Wimalasekera et al., 2011), we undertook to quantify NO production upon spermidine treatment in two independent cis-NATCuAO1 over-expressing transgenic lines along with a CuAO1 T-DNA knock-down mutant. NO production was strongly impaired in CuAO1 knock-down mutant, in agreement with the previous results of Wimalasekera et al. (2011), but also in the cis-NATCuAO1 over-expressing line #1 (Supplemental Fig. S15). The second cis-NAT over-expressing line also showed a reproducible reduction in NO production compared with that in the Col-0 control, although the associated p-value was above 0.05 (0.13).
DISCUSSION
Out of a total set of 4,846 cis-NATs identified in this study or annotated in TAIR10, 157 (3.24%) were found to have a potential to regulate the expression of their cognate sense mRNA based on positive or negative correlations with either the steady-state mRNA level or polysome association. The great majority of those potential regulatory cis-NATs (147 out of 157; Table 1) were associated with changes in the transcript level of the cognate sense mRNA, with a stronger bias toward concordant expression (107 out of 147). This bias toward concordance is somewhat surprising because negative effects of cis-NAT expression on steady-state mRNA level is more commonly described in the literature than positive effects (Khorkova et al., 2014). It is possible that phenotypes associated with the disruption of cis-NATs with discordant expression pattern may be more apparent than for concordant expression pattern. Furthermore, coexpression of cis-NAT and cognate sense mRNA could, in many cases, be simply a consequence of local changes in chromatin state encompassing a whole locus that would equally affect the access of the transcription machinery to both the sense and antisense promoters, and thus would not be associated with a regulatory mechanism for controlling sense mRNA expression. However, numerous examples exist in the literature showing that increased expression of a cis-NAT may negatively affect sense mRNA steady-state level via changes in histone marks localized primarily at the promoters of the sense genes (Khorkova et al., 2014). Whereas fewer examples of similar local effect only on the promoter activity of the sense genes have been described for cis-NATs having concordant expression pattern (Mondal et al., 2010), more examples may be found through a more systematic analysis of this group of cis-NATs in Arabidopsis.
Noncoding RNAs, and particularly lincRNAs, can regulate the expression of coding mRNA by either masking a miRNA binding site via base pairing or by acting as a miRNA mimic (Wang et al., 2013; Cho and Paszkowski, 2017). At least one example has been described in animals for a cis-NAT masking a miRNA binding site present in the cognate sense gene (Faghihi et al., 2010). In the current study, 600 sense genes associated with a cis-NAT were found to be a potential target for a microRNA and 69 cis-NATs were found to contain a sequence that could act as a miRNA mimic (Supplemental Table S1). Of those, only two cis-NATs were found in the group associated with changes in steady-state mRNA level (both concordant) and only one associated with changes in mRNA polysome association, namely cis-NATWRKY45. However, the effect of cis-NATWRKY45 in its cognate sense RNA could not be experimentally validated either in transgenic plants nor in protoplasts (Figs. 5D and 7C). Thus, whereas it is possible that some plant cis-NATs may function in miRNA masking or as a miRNA mimic, it would not appear to be common.
Overlap between cis-NATs and their cognate sense mRNAs can potentially generate siRNAs leading to gene silencing. There are several examples of cis-NATs down-regulating the cognate mRNA level via a siRNA-mediated silencing pathway, including in Arabidopsis (Held et al., 2008; Ron et al., 2010). It is thus possible that some of the cis-NATs identified in this study as potentially down-regulating sense transcript levels and associated with cis-NAT-siRNAs may act through a siRNA pathway. There is also an example in Arabidopsis where cis-NAT expression leads to an increase in cognate sense mRNA transcript via the generation of cis-NAT-siRNAs that inhibit the action of a microRNA targeting the same cognate sense mRNA, thus leading to an increase in sense mRNA level (Gao et al., 2015). Such a mechanism could potentially apply to three genes having cis-NATs identified as potential transcription enhancers and associated with cis-NAT-siRNAs, namely At1g23090, At2g44430, and At2g45850, which could be targeted by miR826a, miR838, and miR837, respectively. In contrast, the only gene with a cis-NAT generating cis-NAT-siRNAs that belongs to the group of potential translation regulators does not harbor miRNA targets.
In silico analysis of the set of cis-NATs identified a small group of 63 cis-NATs that had a higher coding potential and that were more associated with polysomes than were other noncoding RNAs. Further analysis revealed that 19 of those cis-NATs could encode polypetides that were conserved either mainly in Brassicaceae (Group II, 9 cis-NATs) or more broadly in plants (Group I, 10 cis-NATs; Supplemental Fig. S6). Only one out of these 63 cis-NATs was positively correlated with change in sense mRNA steady-state level (cis-NATAT1G69260), but none were correlated with changes in mRNA translation (Supplemental Table S1). Association of mRNA with polysomes does not directly show if the RNA is being actively translated into a polypetide. However, analysis of RNA translation by ribosome footprint in both plants and animals have revealed that a remarkably broad spectrum of RNAs previously thought to be noncoding are actively being translated by ribosomes (Aspden et al., 2014; Ji et al., 2015; Hsu et al., 2016; Bazin et al., 2017). Recent analysis of noncoding RNAs in Arabidopsis roots revealed that 568 out of 1,676 cis-NATs had ribosome footprints consistent with translation (Bazin et al., 2017). Interestingly, translation of a small ORF present in a trans-acting siRNA was shown to enhance trans-acting siRNA production (Bazin et al., 2017), suggesting that whereas many lincRNAs and cis-NATs may indeed be translated into peptides, the act of translation itself rather than the specific sequence of the polypeptide may, in some cases, be the predominant mechanism of regulation. In that context, four cis-NATs found by Bazin et al. (2017) to have ribosome footprints are included in the group of 19 cis-NATs with coding potential that are well conserved in plants (Supplemental Fig. S6). These four cis-NATs may be good candidates for transcripts coding for biologically active polypeptides.
In contrast with numerous reports of the effects of cis-NAT expression on sense gene transcription and/or transcript stability, very few examples of cis-NATs affecting the translation of their cognate sense mRNA have been described. Repression of mRNA translation by a cis-NAT has only been described for the PU.1 gene in mouse (Ebralidze et al., 2008), whereas cis-NATs enhancing cognate mRNA translation have been reported for the rice PHO1.2, the mouse Uchl1, and the human RBM15 genes (Carrieri et al., 2012; Jabnoune et al., 2013; Tran et al., 2016). A major goal of this work has thus been to systematically explore the role of cis-NAT expression on translation of their cognate mRNA in Arabidopsis. A total of 14 candidate cis-NATs with putative repressive or stimulatory effects on cognate mRNA translation were found, which is 10-fold less compared to the number of cis-NATs with effects on mRNA steady-state level. Analysis of the configuration of the cis-NATs relative to the sense mRNA showed that cis-NATs associated with either translational stimulation or repression had a higher proportion of head-to-head configuration compared with that of all other cis-NATs (Supplemental Fig. S10), although the low number of cis-NATs associated with translation makes this distinction not statistically significant. Furthermore, some of the Arabidopsis cis-NATs that were experimentally confirmed to affect translation have other configurations, such as tail-to-tail (AT1G54260, AT3G26240).
The effects of cis-NAT expression on cognate mRNA translation were experimentally tested by either stable transformation in plants and/or transient expression in leaf protoplasts for 5 of the 14 candidates. Four of those were validated, namely two cis-NATs mediating translational repression (CuAO1 and AT1G54260) and two cis-NATs mediating translational stimulation (AT3G26240 and AT1G03410). These results highlight the robustness of the experimental pipeline used to identify the candidates.
CuOA1 encodes a copper amine oxidase involved in the catabolism of polyamines (Wimalasekera et al., 2011). CuAO1 has been shown to be involved in the generation of nitric oxide, a key signaling molecule involved in a wide range of functions in plants, including seed germination, root development, and ABA-induced stomatal closure (Besson-Bard et al., 2008). The Arabidopsis cuao1-1 T-DNA knock-down mutant shows reduced production of NO after treatment with spermidine (Wimalasekera et al., 2011). Several stress conditions are known to induce NO synthesis, including phosphate deficiency (Sun et al., 2016). Although proteomic experiments could not reliably quantify the amount of CuAO1 protein in transgenic lines overexpressing cis-NATCuAO1, the same lines did show reduced NO production to levels similar to that of the cuao1-1 mutant, supporting an inhibitory effect of cis-NATCuAO1 expression on CuAO1 production (Supplemental Figure S15).
AT1g54260 harbors a highly conserved central globular domain (GH1) present in the linker histone H1, proteins that perform important functions on chromatin structure and influencing accessibility of transacting factors to DNA (Hergeth and Schneider, 2015; Kotliński et al., 2017). The GH1 domain is known to bind DNA, and the AT1G54260 protein belongs to the winged helix family of DNA binding proteins. Beside histones H1, proteins containing GH1 domain have been shown to binds to DNA, including at the telomeres, and potentially act at various level in the regulation of chromatin structure (Zhou et al., 2016). The cis-NAT to AT1G54260 is up-regulated by both ABA and low Pi (Supplemental Table S1). Modulation of AT1G54260 protein synthesis via expression of its cis-NAT could thus have broad impact on chromatin structure and gene regulation under various stress.
AT3G26240 encodes a protein of unknown function. AT1g03410 (2A6) encodes a protein containing a domain associated with oxoglutarate and iron-dependent dioxygenase. In plants, enzymes containing this domain catalyze the formation of plant hormones, such as ethylene, gibberellins, anthocyanidins, and pigments such as flavones. The cis-NATAT1G03410 is of particular interest because it corresponds to a retroelement of the Sadhu family (Rangwala and Richards, 2010). The stimulatory activity of the cis-NAT of the mouse Uchl1 on translation was shown to be dependent on the SINEB2 retroelement (Carrieri et al., 2012). Sadhu retroelements resemble SINEs in their structure, except that they do not contain similarity to known noncoding RNAs, such as 5SrRNA or tRNAs (the SINEB2 element is derived from a tRNA; Weiner, 2002). Whereas the cis-NATs of both Uchl1 and AT1g03410 are in the head-to-head configuration, the SINEB2 element of cis-NATUchl1 is located at the nonoverlapping 3′ end of the cis-NAT and cis-NATAT1G03410 is almost completely overlapping with the 5′UTR region of the sense mRNA except for the last 56 nucleotides (Supplemental Fig. S16; Carrieri et al., 2012). Whether SINEB2 elements and Sadhu retrotransposon stimulates mRNA translation by a similar mechanism remains to be determined.
The success of the validation methods relying on stable expression of cis-NATs in transgenic plants or transient expression in protoplast reveals that the effect of cis-NAT expression on sense mRNA translation can occur in trans. This implies that the cis-NAT produced from a distinct locus must be sufficiently stable to locate and anneal to its target mRNA and recruit or sequester factors that affect translation. This may, however, not always be the case, because the effects of some cis-NATs on mRNA transcript level have been found to occur only in cis and not in trans (Fedak et al., 2016; Rosa et al., 2016). Thus experimental validation of some cis-NATs for regulation of sense mRNA translation may, in some cases, require other methods working in cis, such as precise mutation of the cis-NAT locus by CRISPR/Cas9.
In conclusion, the experimental pipeline described in this work identified and validated a number of novel cis-NATs in Arabidopsis that influence cognate sense mRNA translation. Although the proportion of cis-NATs associated with changes in mRNA translation was relatively low compared with the total number of cis-NATs expressed in the genome, it is likely that more candidates will be found when plants are grown under different experimental conditions that lead to greater spectrum of cis-NAT expression. Considering that a broad range of mechanisms have been identified for the effect of lincRNAs and cis-NATs on transcriptional regulation, it is likely that the mechanisms through which cis-NATs enhance or repress translation will also be quite diverse.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) seeds (Col-0) were germinated in half-strength Murashige and Skoog (MS) liquid medium containing 1 mm (high) or 100 uM (low) Pi. On d 5 and d 6 after germination, the medium was replaced to maintain a constant level of Pi. On d 7, whole seedlings were harvested and used for total RNA extraction and polysome profiling. Arabidopsis seeds were also germinated on agar-solidified half-strength MS medium for 10 d, after which the seedlings were flooded with a solution of half-strength MS containing 5 µM indole acetic acid, 10 µM ABA, 10 µM MeJA, 10 µM ACC, or no hormone for the untreated control. After 3 h of incubation, roots and shoots were split and harvested separately. For each of the 12 experimental conditions, 3 independent biological replicates were carried out at different times.
Total and Polysomal RNA Extraction
Plant samples (whole seedlings, roots, or shoots) were flash frozen and ground in a mortar and pestle, and the polysomes were extracted essentially as described in Mustroph et al. (2009) with minor modifications (see Supplemental Materials and Methods).
Library Preparation and RNA Sequencing
From each total and polysomal RNA sample, strand specific libraries were prepared using the TruSeq Stranded Total RNA kit (Illumina) and polyA+ RNAs were selected according to manufacturer's instructions. The libraries were sequenced on a HiSeq 2500 Illumina sequencer and about 30 million of paired-end reads per sample were obtained. In total, about 120 million reads were obtained for each of the 12 experimental conditions.
Identification of cis-NATs and Analysis of their Coding Potential
To identify cis-NATs, the paired-end reads from the 3 replicates were pooled together and uniquely mapped to the TAIR10 genome using Hisat2 (Kim et al., 2015). For each of the 12 conditions, the transcriptome was determined de novo with Cufflinks (Trapnell et al., 2010), using the TAIR10.31 annotation as guide. The 12 annotation files obtained were merged using the Cuffmerge tool (Trapnell et al., 2010). This transcriptome was then compared with TAIR10.31 using Cuffcompare (Trapnell et al., 2010), and transcripts antisense to TAIR10.31 coding genes (class_code_x) were considered as putative cis-NATs. The read count for each TAIR10.31 protein coding gene and each identified cis-NATs was determined using HTSeq-count (mode Union) and the identified cis-NATs with a ratio read count cis-NAT / coding gene < 0.01 were discarded as false positives likely because of imperfect strand specificity of the library preparation protocol (99.9%).
The “FEELnc codpot” module from FEELnc (version 0.01; Wucher et al., 2017) was used to identify cis-NATs that could potentially be coding for polypeptides (see Supplemental Material and Methods).
Characterization of cis-NATs
Basic features such as length or guanine-cytosine content of transcripts, average steady-state levels, or polysome association were determined for each cis-NAT using custom functions written in Python. To analyze the nucleotide conservation, PHASTcons scores where extracted from the 20 angiosperm genome alignment as described by Hupalo and Kern (2013). For each transcript, the average PHASTcons score was calculated for exonic and intronic sequences. The presence of inverted repeats was determined using the einverted program (European Molecular Biology Open Software Suite [EMBOSS]; http://emboss.bioinformatics.nl/cgi-bin/emboss/einverted) using default parameters. The presence of miRNA binding sites within cis-NATs and coding transcripts was determined using psRNATarget server (http://plantgrn.noble.org/psRNATarget/) with an expectation ≤ 3 and unpaired energy ≤ 25. Potential miRNA precursors were identified by comparing the cDNA sequences of cis-NATs against a database of miRNA hairpins downloaded from miRBase (http://www.mirbase.org).
The presence of potential miRNA target mimic sites was determined using custom python functions following the rules edicted in Wu et al. (2013), namely (1) perfect nucleotide pairing was required at the second to eighth positions of miRNA sequence, (2) bulges were only permitted at the 5′ end ninth to 12th positions of miRNA sequence, and (3) it should be composed of only three nucleotides. No more than 3 mismatches or G/U pairs were allowed in pairing regions (not considering the bulge).
Analysis of siRNAs that could be generated by cis-NATs was essentially performed according to the method described by Yuan et al. (2015) using the Arabidopsis small RNA dataset available on the Gene Expression Omnibus. Briefly, the small reads 18–28 nucleotides long were mapped to the TAIR10 reference genome using bowtie. For each cis-NAT locus, the length and density in small RNAs was calculated for overlapping and nonoverlapping regions by dividing the number of mapped small reads by the length of the region using custom scripts and the python library pysam.
The presence of transposable elements within cis-NAT transcripts was determined by comparing the cis-NATs sequences against a database containing all transposable elements annotated in TAIR10 using Blastn with a cutoff of evalue = 1e-12 and percent identify >50.
Quantification of TAIR10 and Identified Loci and Identification of Differentially Expressed Genes
For each experimental condition and biological replicate, the read count of TAIR10 as well as identified loci was determined with HTSeq-count (mode Union; Anders et al., 2015), and normalized with DESeq2 (Love et al., 2014). A gene was considered differentially expressed comparing two conditions if the adj.pval was < 0.1 and the fold change > 2 or < 0.5.
Validation for Differentially Expressed Genes by RT-qPCR
Arabidopsis seedlings were grown in liquid cultures in the presence of a high or low concentration of inorganic phosphate as described above in the “Plant materials” section. Total RNA was extracted from whole seedlings with Trizol following manufacturer’s instructions. One microgram of RNA was then used for reverse-transcription using the M-MLV Reverse Transcriptase (Promega) and oligo d(T)15 as primer using manufacturer's instructions. RT-qPCR analysis to measure mRNA steady-state level was completed using SYBR select Master Mix (Applied Biosystems) with a primer set specific of the gene of interest as well as a primers specific of ACT2 gene used as reference. Log2 fold changes were calculated by the ΔΔCt method.
Determination of PA Ratio
To estimate the translation efficiency for each gene, the PA ratio was determined using Xtail package (Xiao et al., 2016), which calculates the ratio between read count from polysomal RNA sample and total RNA sample. Genes with a Xtail adj.pval < 0.1 and at least a 30% increase or decrease of the AP ratio were considered differentially associated with polysomes.
Identification of cis-NATs Influencing Steady-State Level or PA of Cognate Sense mRNA
The candidate regulatory cis-NATs were identified by pairwise comparisons between whole seedlings grown under high- or low-Pi conditions, roots, or shoots treated with phytohormones and appropriate untreated controls, as well as between untreated root and shoot tissues, using a series of criteria. Only the pairs coding gene/cis-NAT overlapping by at least 50 nucleotides and with a normalized read count for both coding gene and cis-NAT > 20 were considered. A cis-NAT was considered positively correlated to its cognate coding mRNA expression if both cis-NAT and coding mRNA were either up-regulated or down-regulated (fold change > 2 and adj.pval < 0.1) between the two conditions compared. It was considered negatively correlated if one partner was up-regulated, whereas the other was down-regulated (fold change > 2 and adj.pval < 0.1) between the two conditions compared. To identify the putative translation regulatory cis-NATs, only the pairs for which the coding gene was differentially translated with fold change > 1.3 and adj.pval < 0.1 between the two conditions compared, and with fold-change of mRNA steady-state level < 3 were kept. From these pairs, the cis-NATs had to be differentially expressed, with fold change > 2 and adj.pval < 0.1 and the ratio read count cis-NAT / read count coding gene had to be above 0.2, in at least one condition. The cis-NATs up-regulated when their cognate mRNA was more associated with polysomes were considered as putative translation enhancers, whereas cis-NATs up-regulated when their cognate mRNA was less associated with polysomes were considered as putative translation repressors.
Pearson correlation coefficient between mRNA and cis-NAT steady-state level was also calculated across the 12 experimental conditions analyzed for each candidate pair with a positive or negative correlation between cis-NAT and mRNA expression. Similarly, the correlation between PA ratio, and cis-NAT steady-state level was also calculated across the 12 experimental conditions for each translation regulator cis-NAT candidate. The candidate pairs with a correlation factor > 0.4 or < −0.4 were considered as the most robust candidates.
Creation and Analysis of Transgenic Lines Over-Expressing Putative Translation Regulatory cis-NATs
To create transgenic plants over-expressing the candidate translation regulator cis-NATs, the genomic sequence encompassing the transcribed region was cloned into the vector pFAST-R02 (Shimada et al., 2010), in the correct orientation, to allow synthesis of the cis-NAT transcript under the control of the cauliflower mosaic virus 35S promoter. The constructs were introduced into Arabidopsis by Agrobacterium tumefaciens-mediated transformation using floral dipping (Clough and Bent, 1998). Transgenic lines over-expressing the different cis-NAT constructs or transformed with empty vector were grown for 10 d on agar-solidified half-strength MS medium containing Basta as a selection marker. Whole seedlings were crushed in liquid nitrogen and total RNA was extracted using standard procedure.
To purify polysomes, 10-d-old seedlings were ground into powder in liquid nitrogen and 2 volumes of polysome extraction buffer were added. The mixture was incubated for 15 min on ice, centrifuged at 16,000 g to pellet debris and 200 µL of supernatant were loaded on top of 5-mL Suc gradients. After 75 min of centrifugation at 55,000 rpm in a SW55 rotor (Beckman) at 4°C, the gradients were collected and split into 6 fractions. For each fraction, 500 µL was transferred into a new eppendorf tube and RNA was extracted with 1 mL of Trizol according to manufacturer's instructions. An additional step of acetate ammonium precipitation with ethanol and washing was added to remove remaining salt and phenol traces. For each sample, 300 ng of RNA was then used for reverse transcription using the M-MLV Reverse Transcriptase (Promega) and oligo d(T)15 as primer using manufacturer's instructions. To analyze WRKY45 mRNA, due to the full overlap between cis-NAT and mRNA, the reverse transcription was performed in the same conditions but using a mix of reverse primers specific to WRKY45 and ACT2 mRNA instead of oligo d(T)15. RT-qPCR analysis to quantify the relative amount of endogenous mRNA in each fraction was performed with a primer set specific for the gene of interest as well as a primer specific for the ACT2 gene used as reference. The results are presented as relative proportion of endogenous mRNA in each fraction of the gradient, as described in Faye et al. (2014). The average of 8 independent biological replicates obtained with 2 independent transgenic lines is reported. To be able to quantify in a more robust manner the changes in terms of polysome association, the sum of the proportions of mRNA in fractions 1–3 and fractions 4–6 were calculated to compare the proportion of mRNA not or poorly translated, e.g. free mRNA (fraction 1 and 2) or associated with monosomes (fraction 3), versus the proportion of mRNA efficiently translated, e.g. associated with low (fraction 4) or high polysomes (fractions 5–6).
Transient Translation Assays in Arabidopsis Protoplasts
Plasmids used for protoplast transformation were assembled using BsaI-based Golden Gate cloning, and the final constructs contained a recombination site for Gateway cloning. A Gateway destination vector, for cloning and expression of sense-coding genes, included a C-terminal in-frame fusion with a foot-and-mouth disease virus 2A peptide, followed by fusion with NanoLuc luciferase (Nluc; plasmid nLucFlucGW, GenBank MH552885). Additionally, an independent expression cassette driving firefly luciferase (Fluc) was also included in this vector. Another Gateway destination vector, for cloning and expression of antisense noncoding genes, was produced without any fusion or additional expression cassette (plasmid RHIP1pGW, GenBank MH552886). Both Gateway destination vectors expressed the cloned gene, sense or antisense, under control of the same promoter (1.1 kb genomic sequence upstream of AT4G26410) and terminator (250 bp downstream of AT5G59720). Genomic sequences for sense-coding genes (from 5′UTR to last codon, without STOP) and antisense-noncoding genes were cloned via Gateway cloning into their corresponding vector.
Protoplasts were produced and transformed essentially as described by Yoo et al. (2007) with minor modifications (see Supplemental Material and Methods). Protoplasts were harvested by centrifugation at 6,000 g for 1 min, and resuspended in 1X Passive Lysis buffer (Promega, E1941). The lysate was cleared by centrifugation and used for luminescence quantification using Nano-Glo Dual-Luciferase Reporter Assay System (Promega, N1610), according to the manufacturer’s instructions. Luminescence values for Nluc fused to sense-coding gene were normalized against Fluc to control for loading and transfection efficiency. Statistical significant differences (Student’s t test, p-value < 0.05) in ratio Nluc:Fluc were used to assess the effect of antisense noncoding gene coexpression.
Quantification of NO Production
NO production was quantified in 10-d-old seedlings treated with spermidine following the procedure described in Wimalasekera et al. (2011). Briefly, 5 to 6 seedlings were equilibrated in 3 mL of MES buffer (30 mm MES, 0.1 mm CaCl2, 1 mm KCl) for 2 h. Then 4,5-diaminofluorescein diacetate and spermidine or dimethyl sulfoxide was added to the medium. After 30 min incubation at 24°C under light with shaking, 100 µL of medium was transferred to 96-well plate and fluorescence was quantified. Eight independent biological replicates were analyzed, and the florescence was normalized by milligram of fresh weight of spermidine-treated seedlings or untreated control.
Accession Numbers
The data reported in this paper have been deposited in the Gene Expression Omnibus database, https//www.ncbi.nlm.nih.gov/geo (accession no. GSE116553). The processed data tables (Supplemental Table S1 and S4) are included as additional files for this article. The sequence of created plasmids used in this study has been submitted to GenBank, MH552885 and MH552886.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Steady state mRNA expression level and association with polysomes in response to treatment with phytohormones.
Supplemental Figure S2. GO terms enriched in the set of genes up-regulated in plants grown in low Pi conditions or treated with various phytohormones.
Supplemental Figure S3. Analysis of the degree of overlap between independent studies analyzing gene expression in response of low Pi or ABA treatment.
Supplemental Figure S4. GO terms enriched in the set of genes with changes in polysome association.
Supplemental Figure S5. Steady state mRNA expression level and association with polysomes in untreated roots compared to shoots.
Supplemental Figure S6. Evolutionary conservation of cis-NAT encoded peptides.
Supplemental Figure S7. Analysis of histone acetylation and nucleosome occupancy near the transcription start site of cis-NATs.
Supplemental Figure S8. Analysis of the degree of overlap in cis-NATs identified in distinct studies.
Supplemental Figure S9. Validation of differentially expressed genes by RT-qPCR.
Supplemental Figure S10. Proportion of the different types of orientation for the cis-NAT – sense mRNA pairs.
Supplemental Figure S11. Analysis of cis-NAT-siRNAs.
Supplemental Figure S12. Quantification of the endogenous cognate mRNA in cis-NAT overexpressing lines.
Supplemental Figure S13. Polysome profile.
Supplemental Figure S14. Levels of sense mRNA-NanoLuc luciferase (Nluc) fusion transcripts in transiently transformed protoplasts.
Supplemental Figure S15. Quantification in NO production upon spermidine treatment.
Supplemental Figure S16. Organization of the cis-NAT:mRNA pair at AT1G03410 locus.
Supplemental Table S1. Summary of features associated with each transcript.
Supplemental Table S2. Genes differentially expressed in various conditions.
Supplemental Table S3. Number of mRNAs differentially associated with polysomes.
Supplemental Table S4. RNAseq and polysome profiling data relative to putative transcription or translation regulatory cis-NATs.
Supplementary Material and Methods. Supplementary information on various methods.
Acknowledgments
We thank the University of Lausanne Genomic Technology Facility for high-throughput sequencing support and the members of the Sinergia consortium, Jacques Rougemont (EPFL), and Bulak Arpat (SIB) for useful discussions, as well as Aidan Tay for help with statistical analysis. No conflict of interest declared.
Footnotes
This work was supported by a Swiss National Science Foundation (Schweizerische Nationalfonds) Sinergia grant (CRSII3_154471 to I.L.H. and Y.P.). The Swiss Institute of Bioinformatic (SIB) and Vital-IT Center for high-performance computing of the SIB are supported by University of Lausanne and the Swiss Federal Government.
Articles can be viewed without a subscription.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MAS, Brocard M, Couso JP (2014) Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3: e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JWS, Crespi M (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30: 166–176 [DOI] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J (2017) Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA 114: E10018–E10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R (2014) Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48: 433–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457 [DOI] [PubMed] [Google Scholar]
- Chekanova JA. (2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Cho J, Paszkowski J (2017) Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 6: e30038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. (2012) Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, Dayaram T, Klupp J, et al. (2008) PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 22: 2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G III, Wahlestedt C (2010) Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 11: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye MD, Graber TE, Holcik M (2014) Assessment of selective mRNA translation in mammalian cells by polysome profiling. J Vis Exp 28: e52295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedak H, Palusinska M, Krzyczmonik K, Brzezniak L, Yatusevich R, Pietras Z, Kaczanowski S, Swiezewski S (2016) Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc Natl Acad Sci USA 113: E7846–E7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gao W, Liu W, Zhao M, Li WX (2015) NERF encodes a RING E3 ligase important for drought resistance and enhances the expression of its antisense gene NFYA5 in Arabidopsis. Nucleic Acids Res 43: 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart-Smith G, Reis RS, Waterhouse PM, Wilkins MR (2017) Improved quantitative plant proteomics via the combination of targeted and untargeted data acquisition. Front Plant Sci 8: 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Penning B, Brandt AS, Kessans SA, Yong W, Scofield SR, Carpita NC (2008) Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci USA 105: 20534–20539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hergeth SP, Schneider R (2015) The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 16: 1439–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li F-W, Rothfels CJ, Ohler U, Benfey PN (2016) Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci USA 113: E7126–E7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Lou Z, Gupta M (2014) The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One 9: e107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalo D, Kern AD (2013) Conservation and functional element discovery in 20 angiosperm plant genomes. Mol Biol Evol 30: 1729–1744 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégu T, Veluchamy A, Ramirez-Prado JS, Rizzi-Paillet C, Perez M, Lhomme A, Latrasse D, Coleno E, Vicaire S, Legras S, et al. (2017) The Arabidopsis SWI/SNF protein BAF60 mediates seedling growth control by modulating DNA accessibility. Genome Biol 18: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K (2015) Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4: e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Liu J, Wang H, Wong L, Chua N-H (2013) PLncDB: Plant long non-coding RNA database. Bioinformatics 29: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111: E203–E212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorkova O, Myers AJ, Hsiao J, Wahlestedt C (2014) Natural antisense transcripts. Hum Mol Genet 23(R1): R54–R63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliński M, Knizewski L, Muszewska A, Rutowicz K, Lirski M, Schmidt A, Baroux C, Ginalski K, Jerzmanowski A (2017) Phylogeny-based systematization of Arabidopsis proteins with histone H1 globular domain. Plant Physiol 174: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y (2006) Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep 7: 1216–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua N-H (2015) Long noncoding RNA transcriptome of plants. Plant Biotechnol J 13: 319–328 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C (2010) Characterization of the RNA content of chromatin. Genome Res 20: 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, Richards EJ (2010) The structure, organization and radiation of Sadhu non-long terminal repeat retroelements in Arabidopsis species. Mob DNA 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19: 143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Duncan S, Dean C (2016) Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun 7: 13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein A, Zucchelli S, Kauppinen S, Gustincich S, Carninci P (2016) Identification of antisense long noncoding RNAs that function as SINEUPs in human cells. Sci Rep 6: 33605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Sun HX, Park BS, Huang CH, Yeh SD, Jung C, Chua NH (2017) ELF18-INDUCED LONG-NONCODING RNA associates with Mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29: 1024–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Bi Y, Tao J, Huang S, Hou M, Xue R, Liang Z, Gu P, Yoneyama K, Xie X, et al. (2016) Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ 39: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Tran NT, Su H, Khodadadi-Jamayran A, Lin S, Zhang L, Zhou D, Pawlik KM, Townes TM, Chen Y, Mulloy JC, Zhao X (2016) The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep 17: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang I-C, Kean MJ, Xu J, Chua N-H (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H (2013) Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25: 69–80 [DOI] [PubMed] [Google Scholar]
- Weiner AM. (2002) SINEs and LINEs: The art of biting the hand that feeds you. Curr Opin Cell Biol 14: 343–350 [DOI] [PubMed] [Google Scholar]
- Wimalasekera R, Villar C, Begum T, Scherer GFE (2011) COPPER AMINE OXIDASE1 (CuAO1) of Arabidopsis thaliana contributes to abscisic acid- and polyamine-induced nitric oxide biosynthesis and abscisic acid signal transduction. Mol Plant 4: 663–678 [DOI] [PubMed] [Google Scholar]
- Wu H-J, Wang Z-M, Wang M, Wang X-J (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. (2017) FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res 45: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Zou Q, Liu Y, Yang X (2016) Genome-wide assessment of differential translations with ribosome profiling data. Nat Commun 7: 11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. (2013) LncRNA-dependent mechanisms of androgen receptor-regulated gene activation programs. Nature 500: 598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M (2012) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Wang J, Harrison AP, Meng X, Chen D, Chen M (2015) Genome-wide view of natural antisense transcripts in Arabidopsis thaliana. DNA Res 22: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang Y, Dong J, Sun Y, Lim BL, Liu D, Lu ZJ (2016) Systematic characterization of novel lncRNAs responding to phosphate starvation in Arabidopsis thaliana. BMC Genomics 17: 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-N, Shi Y, Powers JJ, Gowda NB, Zhang C, Ibrahim HMM, Ball HB, Chen SL, Lu H, Mount SM (2017) Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana. BMC Genomics 18: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W (2009) Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 19: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hartwig B, James GV, Schneeberger K, Turck F (2016) Complementary activities of TELOMERE REPEAT BINDING proteins and polycomb group complexes in transcriptional regulation of target genes. Plant Cell 28: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]