A cytochrome P450 enzyme contributes to the production of 2-phenylethanol and nitrogen-containing volatiles in Plumeria.
Abstract
Plumeria (Plumeria rubra), well known for its brightly colored and fragrant flowers, emits a number of floral volatile organic compounds (VOCs). Plumeria flowers emit a total of 43 VOCs including nine phenylpropanoids/benzenoids, such as 2-phenylethanol (2PE), benzaldehyde, 2-phenylacetaldehyde (PAld), (E/Z)-phenylacetaldoxime (PAOx), benzyl nitrile (BN), and 2-phenylnitroethane (PN). To identify genes and pathways involved in the production of the major compound 2PE, we analyzed the plumeria floral transcriptome and found a highly expressed, flower-specific gene encoding a cytochrome P450 family 79D protein (PrCYP79D73), which catalyzed the formation of (E/Z)-PAOx. Feeding experiments with deuterated phenylalanine or deuterated (E/Z)-PAOx showed that (E/Z)-PAOx is an intermediate in the biosynthesis of 2PE, as are two nitrogen-containing volatiles, BN and PN, in plumeria flowers. Crude enzyme extracts from plumeria flowers converted l-phenylalanine to (E/Z)-PAOx, PAld, 2PE, BN, and PN. The biosynthesis of these compounds increased with addition of PrCYP79D73-enriched microsomes but was blocked by pretreatment with 4-phenylimidazole, an inhibitor of cytochrome P450 enzymes. Moreover, overexpression of PrCYP79D73 in Nicotiana benthamiana resulted in the emission of (E/Z)-PAOx as well as PAld, 2PE, BN, and PN, all of which were also found among plumeria floral VOCs. Taken together, our results demonstrate that PrCYP79D73 is a crucial player in the biosynthesis of the major floral VOC 2PE and other nitrogen-containing volatiles. These volatiles may be required for plant defense as well as to attract pollinators for the successful reproduction of plumeria.
The different scents emitted by flowers are associated with different unique combinations of low-molecular-weight volatile organic compounds (VOCs). Floral VOCs can be broadly classified into terpenoids, phenylpropanoids/benzenoids, and volatile esters (Croteau and Karp, 1991). In some cases, floral VOCs serve as a signal to trigger plant defense strategy and repel herbivores (Fürstenberg-Hägg et al., 2013), whereas in other cases, they act as attractants for pollinators to ensure plant reproduction (Fürstenberg-Hägg et al., 2013).
2-Phenylethanol (2PE) is one of the floral VOCs emitted by numerous flowers, including rose (Rosa spp.; Sakai et al., 2007), petunia (Petunia spp.; Verdonk et al., 2003), and champak (Magnolia champaca; Dhandapani et al., 2017), and fruits such as cantaloupe melon (Cucumis melo) and tomato (Solanum lycopersicum; Aubert et al., 2005). Plants use 2PE as a chemical signal to communicate with insects, moths, beetles, and flies (Imai et al., 1998; Zhu et al., 2005; Bengtsson et al., 2006). One of the most common pollinators of roses is the beetle Hoplia communis. Among all the volatile compounds emitted by rose flowers, this insect is most attracted to 2PE (Imai et al., 1998). On the other hand, female Chrysoperla carnea insects use 2PE as a signal for oviposition sites (Zhu et al., 2005). Besides pollinator attraction, 2PE aids plant defense indirectly by attracting natural enemies of pests (Lawrence et al., 2008). Moreover, 2PE itself possesses antimicrobial properties (Zhu et al., 2011; Liu et al., 2014). Thus, flowers may emit 2PE for dual purposes: (1) to attract pollinators and (2) to protect flowers from attack by herbivores and microbes, thus ensuring successful plant reproduction.
In plants, 2PE is synthesized in two major steps: (1) conversion of l-Phe into 2-phenylacetaldehyde (PAld) followed by (2) subsequent conversion of PAld into 2PE (Qi et al., 2015). Although few genes and pathways have been identified for the first step, only a single enzyme, phenylacetaldehyde reductase (PAR), is known to convert PAld to 2PE (Tieman et al., 2007; Chen et al., 2011). For the first step, pyridoxal-5′-phosphate-dependent aromatic amino acid decarboxylase (AADC) converts l-Phe to the intermediate compound PAld in roses (Sakai et al., 2007). On the other hand, petunia contains phenylacetaldehyde synthase (PAAS) for the conversion of l-Phe into PAld (Kaminaga et al., 2006). A third enzyme named aromatic amino acid aminotransferase (AAAT) has been identified in rose (Hirata et al., 2012), which catalyzes the formation of phenylpyruvic acid from l-Phe; phenylpyruvic acid can then be converted into PAld by phenylpyruvate decarboxylase.
In addition to the above-mentioned enzymes for the first step, Sakai et al. (2007) proposed an alternate pathway involving oxidative decarboxylation of l-Phe by the cytochrome P450 (CYP450) enzymes of the CYP79 family to produce the intermediate compound phenylacetaldoxime (PAOx). However, attempts to detect even trace amounts of PAOx in the petals of rose flowers were unsuccessful (Sakai et al., 2007). Even though multiple studies reported the conversion of l-Phe to PAOx by CYP79 family enzymes (Wittstock and Halkier, 2000; Irmisch et al., 2013; Yamaguchi et al., 2014), the aldoxime formed was either used as a defense compound against herbivores or served as a precursor for production of other defense compounds, such as nitrogen (N)-containing volatiles. For instance, aldoximes were produced upon gypsy moth (Lymantria dispar) damage and herbivory treatment, suggesting their roles as defense compounds in poplar (Populus trichocarpa) and maize (Zea mays), respectively (Irmisch et al., 2013, 2015). PAOx can also serve as a precursor for the biosynthesis of defense compounds, such as benzylglucosinolates in Arabidopsis (Arabidopsis thaliana; Wittstock and Halkier, 2000), and other N-containing volatiles, such as 2-methylbutyronitrile, 3-methylbutyronitrile, benzyl nitrile (BN), and 2-phenylnitroethane (PN), in poplar (Irmisch et al., 2013). Therefore, the results so far suggest that CYP79 enzyme-mediated formation of PAOx is closely related to direct defense against insects, moths, caterpillars, and fungi (Wittstock and Halkier, 2000; Irmisch et al., 2013, 2015).
Plumeria (Plumeria rubra), commonly known as frangipani or plumeria, belongs to the family Apocynaceae and is widely cultivated as a garden plant around the world. This ornamental tree is well known for its brilliantly colored, five-petaled aromatic flowers. Plumeria flower extracts are used in traditional medicine for the treatment of diabetes mellitus (Shinde et al., 2014) and bacterial infection (Egwaikhide et al., 2009). In addition, essential oils from these flowers are widely used in perfumery, aromatherapy, and cosmetics (Verma, 2016). Plumeria flowers grown in different geographical regions show remarkable qualitative and quantitative differences in floral VOCs (Omata et al., 1991, 1992; Tohar et al., 2006). Compared with other popular ornamental flowers like rose (Koning-Boucoiran et al., 2015), orchid (Phalaenopsis equestris; Cai et al., 2015), and lily (Lilium spp.; Li et al., 2016), the lack of genome and transcriptome sequence information for plumeria has prevented the identification of the key genes and pathways related to floral scent. Therefore, it is difficult to examine the ecological roles of VOCs of plumeria flowers and to improve the quality and quantity of its essential oil by genetic engineering.
High-throughput genomics, transcriptomics, and metabolomics have been used to identify genes involved in the biosynthesis of floral VOCs from diverse aromatic flowers, such as ylang ylang (Cananga odorata), syringa (Syringa oblata), and champak (Jin et al., 2015; Zheng et al., 2015; Dhandapani et al., 2017). Here, we sequenced the plumeria flower transcriptome and obtained more than 41,000 de novo assembled unigenes. A large set of unigenes allowed the identification of genes related to floral VOC pathways in plumeria. Most importantly, we identified a flower-specific CYP450 enzyme, PrCYP79D73, that catalyzes the formation of PAOx. The latter further converted not only into PAld and 2PE but also into N-containing BN and PN in plumeria flowers. Our results show that, unlike rose and petunia, plumeria uses PrCYP79D73 for the production of 2PE. These results uncover a pathway involving the production of 2PE by CYP450.
RESULTS
Chemical Composition Profile of Plumeria Flowers
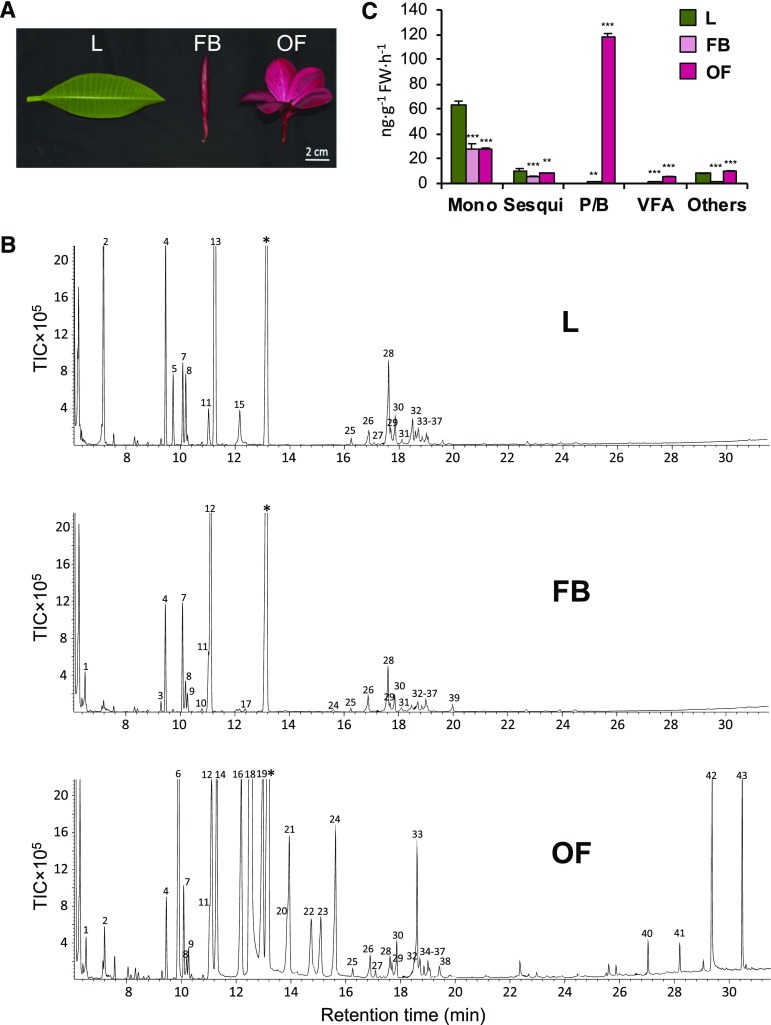
To investigate the VOC profile of plumeria grown in Singapore (1.29°N, 103.77°E), we collected headspace volatiles emitted from leaves, floral buds, and open flowers (Fig. 1A) and analyzed them by gas chromatography-mass spectrometry (GC-MS). We found a total of 43 emitted compounds with qualitative and quantitative differences among the three tissues (Fig. 1B; Table 1). Open flowers emitted the highest amount of VOCs at the rate of 207.6 ng g−1 fresh weight h−1 with 35 compounds, among which phenylpropanoids/benzenoids (70.1%) overshadowed terpenoids (20.8%) and volatile esters (3.2%; Fig. 1C). Within the class of phenylpropanoids/benzenoids, 2PE (35.67%), benzaldehyde (11.77%), PAld (5.82%), BN (7.01%), PN (3.56%), (E)-PAOx (1.62%), and (Z)-PAOx (1.84%) accounted for ∼65% of total flower VOCs (Table 1). On the other hand, floral buds emitted 24 different compounds at the rate of 33.1 ng g−1 fresh weight h−1. Unlike open flowers, VOCs of floral buds were composed mainly of terpenoids, such as eucalyptol (51.68%), α-pinene (9.07%), sabinene (8.98%), limonene (6.69%), and β-caryophyllene (5.38%), accounting for more than 80% of total bud VOC composition (Fig. 1; Table 1). Leaves emitted a total of 21 compounds at the rate of 81.8 ng g−1 fresh weight h−1 with monoterpenoids (77.12%), such as β-ocimene (55.53%), α-pinene (8.46%), and sabinene (3.24%), as major constituents (Fig. 1C).
Figure 1.
VOCs composition of plumeria tissues. A, Photograph of plumeria leaf (L), floral bud (FB), and open flower (OF). Bar = 2 cm. B, Gas chromatograms showing qualitative and quantitative differential VOC emissions by plumeria tissues. The peak numbers correspond to the compounds mentioned in Table 1. Asterisks denote the internal standard camphor. TIC, Total ion chromatogram. C, Classification of VOCs emitted by plumeria tissues. Mono, Monoterpenoids; Sesqui, sesquiterpenoids; P/B, phenylpropanoids/benzenoids; VFA, volatile fatty acids; FW, fresh weight. Means and se are shown (n = 3). Student’s t test was used to calculate the significant differences of floral buds and open flowers as compared with the leaves: **, P < 0.01 and ***, P < 0.001.
Table 1. VOCs composition emitted from plumeria tissues.
| No.a | Compound | RTb | RIc | Formula | Concentrationd | RAe | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | Bud | Flower | Leaf | Bud | Flower | |||||
| 1 | Ethyl propanoate | 6.504 | 706 | C5H10O2 | – | 0.92 ± 0.03 | 0.86 ± 0.10 | – | 2.77 | 0.41 |
| 2 | 8-Aminocaprylic acid | 7.175 | 773 | C5H11NO | 8.01 ± 0.05 | – | 1.00 ± 0.02 | 9.79 | – | 0.48 |
| 3 | β-Thujene | 9.289 | 925 | C10H16 | – | 0.29 ± 0.07 | – | – | 0.89 | – |
| 4 | α-Pinene | 9.455 | 935 | C10H16 | 6.92 ± 0.19 | 3.03 ± 1.59 | 2.04 ± 0.13 | 8.46 | 9.07 | 0.98 |
| 5 | Camphene | 9.727 | 952 | C10H16 | 2.39 ± 0.40 | – | – | 2.92 | – | – |
| 6 | Benzaldehyde | 9.896 | 962 | C7H6O | – | – | 24.26 ± 0.09 | – | – | 11.77 |
| 7 | Sabinene | 10.084 | 973 | C10H16 | 2.65 ± 0.21 | 2.99 ± 0.13 | 2.44 ± 0.19 | 3.24 | 8.98 | 1.17 |
| 8 | β-Pinene | 10.193 | 980 | C10H16 | 2.25 ± 0.01 | 0.69 ± 0.04 | 0.40 ± 0.01 | 2.75 | 2.08 | 0.19 |
| 9 | β-Myrcene | 10.259 | 984 | C10H16 | – | 0.42 ± 0.04 | 0.63 ± 0.02 | – | 1.26 | 0.30 |
| 10 | 6-Methyl-5-octen-2-one | 10.783 | 1,015 | C9H16O | – | 0.14 ± 0.02 | – | – | 0.43 | – |
| 11 | Limonene | 11.034 | 1,030 | C10H16 | 1.36 ± 0.04 | 2.23 ± 0.08 | 2.24 ± 0.03 | 1.66 | 6.69 | 1.08 |
| 12 | Eucalyptol | 11.107 | 1,034 | C10H18O | – | 17.24 ± 1.71 | 10.36 ± 0.10 | – | 51.68 | 4.98 |
| 13 | β-Ocimene | 11.264 | 1,043 | C10H16 | 45.43 ± 2.48 | – | – | 55.53 | – | – |
| 14 | 2-Phenylacetaldehyde | 11.299 | 1,045 | C8H8O | – | – | 12.09 ± 0.12 | – | – | 5.82 |
| 15 | β-Linalool | 12.166 | 1,096 | C10H18O | 2.10 ± 0.00 | – | – | 2.56 | – | – |
| 16 | Geranyl benzoate | 12.194 | 1,098 | C17H22O2 | – | – | 12.51 ± 0.12 | – | – | 6.02 |
| 17 | 2H-Pyran-3(4H)-one, 6-ethenyldihydro-2,2,6-trimethyl- | 12.382 | 1,109 | C10H16O2 | – | 0.14 ± 0.02 | – | – | 0.43 | – |
| 18 | 2-Phenylethyl alcohol | 12.569 | 1,120 | C8H10O | – | – | 74.16 ± 1.06 | – | – | 35.67 |
| 19 | Benzyl nitrile | 12.979 | 1,144 | C8H7N | – | – | 14.57 ± 0.26 | – | – | 7.01 |
| 20 | α-Terpineol | 13.853 | 1,196 | C10H18O | – | – | 2.91 ± 0.11 | – | – | 1.40 |
| 21 | Methyl salicylate | 13.945 | 1,201 | C8H8O3 | – | – | 5.84 ± 0.11 | – | – | 2.81 |
| 22 | (E)-Phenylacetaldoxime | 14.748 | 1,251 | C8H9NO | – | – | 3.36 ± 0.25 | – | – | 1.62 |
| 23 | (Z)-Phenylacetaldoxime | 15.094 | 1,272 | C8H9NO | – | – | 3.81 ± 0.23 | – | – | 1.84 |
| 24 | 2-Phenylnitroethane | 15.632 | 1,305 | C8H9NO2 | – | 0.11 ± 0.05 | 7.41 ± 0.29 | – | 0.34 | 3.56 |
| 25 | Elemene isomer | 16.254 | 1,346 | C15H24 | 0.24 ± 0.05 | 0.11 ± 0.01 | 0.25 ± 0.02 | 0.29 | 0.32 | 0.12 |
| 26 | α-Copaene | 16.898 | 1,388 | C15H24 | 0.99 ± 0.28 | 0.85 ± 0.02 | 0.95 ± 0.14 | 1.21 | 2.55 | 0.46 |
| 27 | β-Elemene | 17.086 | 1,400 | C15H24 | 0.11 ± 0.03 | – | 0.21 ± 0.02 | 0.14 | – | 0.10 |
| 28 | β-Caryophyllene | 17.623 | 1,437 | C15H24 | 4.41 ± 0.29 | 1.79 ± 0.03 | 0.72 ± 0.04 | 5.39 | 5.38 | 0.35 |
| 29 | (E)-α-Bergamotene | 17.700 | 1,442 | C15H24 | 0.31 ± 0.02 | 0.14 ± 0.05 | 0.30 ± 0.01 | 0.37 | 0.42 | 0.15 |
| 30 | (E)-β-Farnesene | 17.862 | 1,453 | C15H24 | 1.31 ± 0.06 | 0.60 ± 0.13 | 1.23 ± 0.00 | 1.60 | 1.81 | 0.59 |
| 31 | α-Caryophyllene | 18.117 | 1,471 | C15H24 | 0.27 ± 0.06 | 0.17 ± 0.04 | – | 0.33 | 0.51 | – |
| 32 | Germacrene D | 18.497 | 1,497 | C15H24 | 1.44 ± 0.05 | 0.29 ± 0.00 | 0.59 ± 0.01 | 1.77 | 0.88 | 0.28 |
| 33 | α-Farnesene | 18.603 | 1,504 | C15H24 | 0.27 ± 0.06 | – | 3.68 ± 0.02 | 0.33 | 0.15 | 1.77 |
| 34 | Bicyclogermacrene | 18.712 | 1,512 | C15H24 | 0.68 ± 0.11 | 0.34 ± 0.04 | 0.51 ± 0.01 | 0.83 | 1.01 | 0.25 |
| 35 | (Z)-γ-Bisabolene | 18.860 | 1,523 | C15H24 | 0.26 ± 0.11 | 0.14 ± 0.00 | 0.28 ± 0.04 | 0.31 | 0.42 | 0.13 |
| 36 | δ-Cadinene | 19.008 | 1,533 | C15H24 | 0.25 ± 0.09 | 0.30 ± 0.08 | 0.36 ± 0.09 | 0.31 | 0.90 | 0.17 |
| 37 | (E)-γ-Bisabolene | 19.069 | 1,538 | C15H24 | 0.17 ± 0.13 | – | 0.19 ± 0.02 | 0.20 | 0.28 | 0.09 |
| 38 | (E)-Nerolidol | 19.423 | 1,563 | C15H26O | – | – | 0.38 ± 0.04 | – | – | 0.18 |
| 39 | β-Caryophyllene epoxide | 19.992 | 1,604 | C15H24O | – | 0.22 ± 0.03 | – | – | 0.67 | – |
| 40 | Stearyl acetate | 27.041 | 2,196 | C20H40O2 | – | – | 0.86 ± 0.09 | – | – | 0.42 |
| 41 | 1-[(Trimethylsilyl)oxy]-2-methylanthraquinone | 28.195 | 2,309 | C18H18O3Si | – | – | 0.87 ± 0.02 | – | – | 0.42 |
| 42 | Advastab 405 | 29.369 | 2,430 | C23H32O2 | – | – | 10.38 ± 0.34 | – | – | 4.99 |
| 43 | Ethylhexyl phthalate | 30.481 | 2,550 | C24H38O4 | – | – | 4.98 ± 0.10 | – | – | 2.40 |
Compounds are listed in the same order as their elution from the HP-5MS column.
Retention time in minutes.
Retention indices calculated using C7-C30 n-alkanes on the HP-5MS column.
Data (ng g−1 fresh weight h−1) are means ± se of three replicates.
Relative abundance in percentage.
The composition of VOCs varied immensely among leaves, floral buds, and open flowers. We found that open plumeria flowers emitted nearly 6 and 2.5 times more total VOCs than floral buds and leaves, respectively. Moreover, leaves and buds produced mainly monoterpenes with relatively few sesquiterpenes and no benzenoid/phenylpropanoids, whereas open flowers produced and emitted many phenylpropanoids (Fig. 1; Table 1).
De Novo Assembly and Functional Annotation of the Plumeria Flower Transcriptome
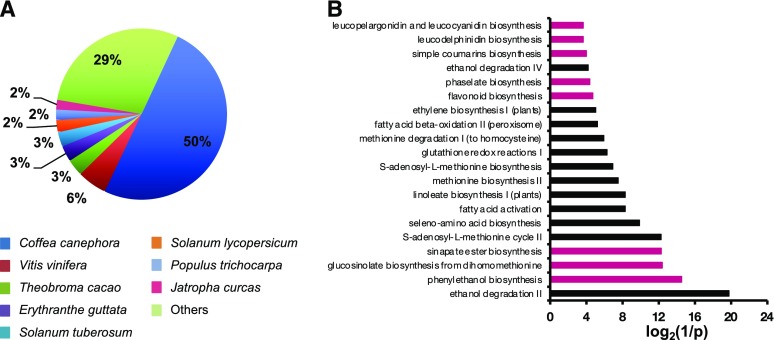
Our VOC analysis suggested that a large volume of phenylpropanoids emitted from open plumeria flowers likely contributes to its sweet floral fragrances. This speculation prompted us to search for genes involved in the biosynthesis of major floral VOCs of plumeria flowers. Therefore, we performed RNA sequencing (RNA-seq) of a complementary DNA (cDNA) library constructed from RNAs of open plumeria flowers and obtained more than 142 million independent reads. After removal of adaptor sequences, FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) analysis showed that the quality score of 30 (q30) for the remaining reads was 82.1%. Due to the lack of a plumeria reference genome sequence, the reads were de novo assembled using Trinity software (Grabherr et al., 2011). The assembly resulted in 41,088 unigenes with nucleotide sequences totaling 303,632,763 bp. The sequence length of the shortest contig at 50% of the total assembly sizewas 2,356 bp (Table 2). Over 61% of unigenes were more than 500 bp in length. Proteins encoded by unigene sequences were first searched against different protein databases using BLASTX. Overall, 19,600 unigenes (47.7%) were annotated using a BLASTX (with the E-value cutoff of 10−5) search against the following protein databases: nonredundant from the National Center for Biotechnology Information (NCBI), Arabidopsis, rice (Oryza sativa), and grape (Vitis vinifera). The species distribution analysis showed that the top hits for 50% of the unigenes were from coffee (Coffea canephora) followed by grape (5.6%) and cacao (Theobroma cacao; 3.2%; Fig. 2A).
Table 2. Summary of the de novo assembly of the plumeria flower transcriptome.
| No. of Reads | No. of Unigenes | N50a | No. Annotated | Percentage Annotated |
|---|---|---|---|---|
| 142,069,174 | 41,088 | 2,356 bp | 19,600 | 47.7 |
N50 is the sequence length of the shortest contig at 50% of the total assembly size.
Figure 2.
Analysis of the de novo assembled transcriptome of plumeria. A, Species distribution of the top BLASTX hits. B, Top 20 plant metabolic pathways highly active in plumeria open flowers. Pathways that originate from the general phenylpropanoid pathway are shown in pink.
The Phenylpropanoid Pathway Is Highly Active in Plumeria Flowers
To identify relevant metabolic pathways, the genes expressed in plumeria flowers were mapped to different pathways using a comprehensive plant metabolic pathway database, the PlantCyc protein database. Figure 2B shows that eight of the top 20 pathways identified by PlantCyc (phenylethanol biosynthesis, glucosinolate biosynthesis from dihomomethionine, sinapate ester biosynthesis, flavonoid biosynthesis, phaselate biosynthesis, simple coumarin biosynthesis, leucodelphinidin biosynthesis, and leucopelargonidin and leucocyanidin biosynthesis) originated from the general phenylpropanoid pathway.
Analysis of Transcript Levels of Methylerythritol 4-Phosphate, Mevalonate, and Shikimate Pathway Genes in Plumeria
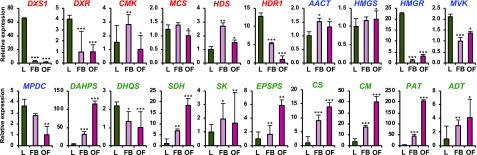
Because our GC-MS analysis of plumeria tissues showed distinct VOC compositions, we investigated the expression levels of genes encoding enzymes for the initial reactions of methylerythritol 4-phosphate (MEP), mevalonate (MVA), and shikimate pathways, which are the main precursor pathways for the production of monoterpenoids/sesquiterpenoids and phenylpropanoids/benzenoids (McGarvey and Croteau, 1995; Fraser and Chapple, 2011). Most of the genes involved in these pathways could be identified from the RNA-seq of plumeria flowers and included full-length open reading frames (ORFs) with high homology to their orthologs from other plants. The genes encoding the first, second, and last enzymes of the MEP pathway, 1-deoxy-d-xylulose 5-phosphate synthase, 1-deoxy-d-xylulose 5-phosphate reductoisomerase, and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, were clearly expressed at higher levels in leaves when compared with floral buds and open flowers (Fig. 3). Among the genes involved in the MVA pathway, only hydroxymethylglutaryl-CoA reductase showed a nearly 20-fold higher expression in leaves than in floral buds and open flowers. 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHPS), shikimate dehydrogenase (SDH), chorismate mutase (CM), and prephenate aminotransferase (PAT), which are required for phenylpropanoid/benzenoid production, were expressed at the highest levels in open flowers, followed by floral buds and then leaves. Overall, the expression levels of the genes encoding enzymes of the MEP, MVA, and shikimate pathways associated well with the VOC profile of plumeria tissues.
Figure 3.
Reverse transcription quantitative PCR (RT-qPCR) analysis showing the expression levels of MEP (red), MVA (blue), and shikimate (green) pathway genes in plumeria leaves (L), floral buds (FB), and open flowers (OF). Actin was used as a reference gene. DXS1, 1-Deoxy-d-xylulose 5-phosphate synthase1; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; CMK, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; MCS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; HDR1, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase1; AACT, acetyl-coenzyme A acetyltransferase; HMGS, hydroxymethylglutaryl-coenzyme A synthase; HMGR, hydroxymethylglutaryl-coenzyme A reductase; MVK, mevalonate kinase; MPDC, mevalonate diphosphate decarboxylase; DAHPS, 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase; DHQS, 3-dehydroquinate synthase; SDH, shikimate dehydrogenase; SK, shikimate kinase; EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase; CS, chorismate synthase; CM, chorismate mutase; PAT, prephenate aminotransferase; ADT, arogenate dehydratase. Results represent means ± se of three technical repetitions and three biological replicates. Student’s t test was used to calculate the significant differences of floral buds and open flowers as compared with the leaves: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Identification and Sequence Analysis of PrCYP79D73
2PE is the most abundant VOC in plumeria open flowers, accounting for 35.67% of total VOCs (Fig. 1; Table 1). AADC and PAAS catalyze the conversion of l-Phe to PAld in rose and petunia, respectively (Kaminaga et al., 2006; Sakai et al., 2007). Additionally, rose possesses an alternative pathway for 2PE biosynthesis involving AAAT (Hirata et al., 2012).
To unravel the biosynthetic route of 2PE in plumeria flowers, we examined our transcriptome data and searched for homologs of PAAS, AADC, and AAAT or genes that show the highest level of amino acid similarity. Only a single unigene (unigene71187) from the plumeria flower transcriptome had 57% to 59% amino acid similarity with AADC, AAAT, and PAAS from rose and petunia PAAS (Supplemental Fig. S1A; Supplemental Table S1). However, unigene71187 was originally annotated as Tyr decarboxylase in our BLASTX analysis against protein databases, which revealed 85% identity with Tyr decarboxylase1 (XP_019225727) from Nicotiana attenuata. Moreover, its expression level was not high in plumeria flowers, with less than 1,000 reads mapping to this unigene and 2.33 fragments per kilobase million (Supplemental Fig. S1B; Supplemental Data File S1). These results suggested that unigene71187 might not be involved in 2PE biosynthesis.
Therefore, we searched for other genes that could be related to the production of 2PE in plumeria flowers. An important clue came from studies showing the conversion of l-Phe to PAOx by CYP450 family 79 genes in Arabidopsis, poplar, and maize (Wittstock and Halkier, 2000; Sakai et al., 2007; Irmisch et al., 2013) and the hypothetical pathway proposed by Sakai et al. (2007) for 2PE biosynthesis. Using the CYP79 genes from Arabidopsis, maize, and poplar for BLASTX search against our plumeria transcriptome, one unigene (unigene72960) emerged as the top hit against all of these genes (Supplemental Table S2). This gene, which was annotated as Phe N-monooxygenase, was the third most highly expressed unigene in plumeria flowers, with more than 1.7 million reads mapping and 12.78 fragments per kilobase million. Thus, unigene72960 containing the complete ORF was designated as CYP79D73 by the CYP450 nomenclature committee (Nelson, 2009).
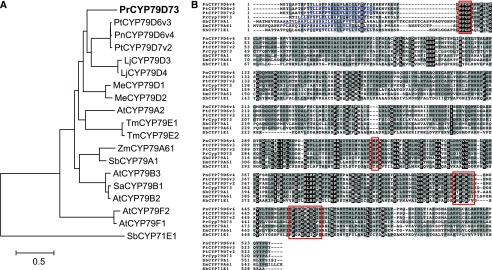
The plumeria CYP79D73 (PrCYP79D73) encodes a protein of 525 amino acids that is closely related to poplar PtCYP79D6v3, PtCYP79D7v2, and PnCYP79D6v4, with about 60% to 61% amino acid sequence identities (Fig. 4A). PrCYP79D73 contained the three motifs that are conserved in most P450 enzymes: (1) an N-terminal Pro-rich motif, (2) a heme-binding motif with the conserved Cys residue, and (3) a PERF motif (Fig. 4B). Although the heme-binding motif of PrCYP79D73 (SFSTGRRGCPG) showed two amino acid substitutions compared with the consensus sequence PFGxGRRxCxG of the A-type P450s (Durst and Nelson, 1995), these substitutions were highly conserved in other CYP79 family enzymes (Irmisch et al., 2013, 2015). In addition, the replacement of PERF and TT/S motifs with PERH and NP, respectively, was found in PrCYP79D73, both of which are unique features of CYP79 family enzymes (Durst and Nelson, 1995). This amino acid sequence analysis suggested that PrCYP79D73 is likely one of the typical CYP79 family enzymes.
Figure 4.
Amino acid sequence analysis of PrCYP79D73. A, Phylogenetic analysis of PrCYP79D73 and CYP79 enzymes from other plant species. The maximum likelihood tree was obtained using MEGA 6.0. CYP71E1 from sorghum (Sorghum bicolor) was chosen as an outgroup. The accession numbers of all the proteins shown in this tree are given in Supplemental Table S3. B, Alignment of amino acid sequences of PrCYP79D73 and CYP79 enzymes from other plant species using ClustalW. Amino acid sequences identical in all sequences are shown in white with black background. Residues with similar side chains are shown in black with dark gray background. Sequence motifs typical for CYP79 proteins are boxed in red. The predicted N-terminal signal sequences for endoplasmic reticulum (ER) transmembrane anchoring are underlined in blue.
Expression and Subcellular Localization of PrCYP79D73
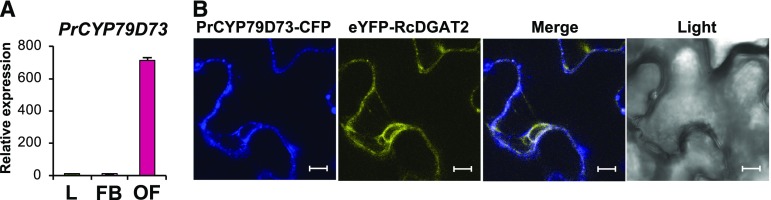
Since PrCYP79D73 was the third most abundant transcript in the plumeria flower transcriptome, we examined its expression levels in other tissues. Figure 5A shows that PrCYP79D73 expression was very low in leaves and floral buds but extremely high in open flowers. These results suggested that PrCYP79D73 may play an important role in open flowers rather than in leaves or floral buds.
Figure 5.
Expression levels and subcellular localization of PrCYP79D73. A, RT-qPCR analysis of PrCYP79D73 in plumeria leaves (L), floral buds (FB), and open flowers (OF). Results represent means ± se of three technical repetitions and three biological replicates. B, Subcellular localization of PrCYP79D73 observed in the leaves of 4-week-old N. benthamiana plants after infiltration with A. tumefaciens strains harboring PrCYP79D73-CFP, eYFP-RcDGAT2, and p19. The leaves were observed with a Carl Zeiss Exciter 5 confocal microscope. Bars = 10 μm.
Most P450s are anchored to the membrane of the ER by an N-terminal 25- to 30-amino acid signal sequence. PrCYP79D73 was predicted to be targeted to ER membranes (Fig. 4B). To confirm the ER localization of PrCYP79D73, we transiently expressed cyan fluorescent protein-fused PrCYP79D73 (PrCYP79D73-CFP) along with the ER membrane marker, RcDGAT2, tagged with the enhanced yellow fluorescent protein (eYFP-RcDGAT2) in leaves of 4-week-old Nicotiana benthamiana plants via Agrobacterium tumefaciens-mediated infiltration. Figure 5B shows that PrCYP79D73 was exclusively associated with the ER, as the subcellular localization of PrCYP79D73-CFP and eYFP-RcDGAT2 matched in N. benthamiana cells.
PrCYP79D73 Produces (E)- and (Z)-Isomers of PAOx
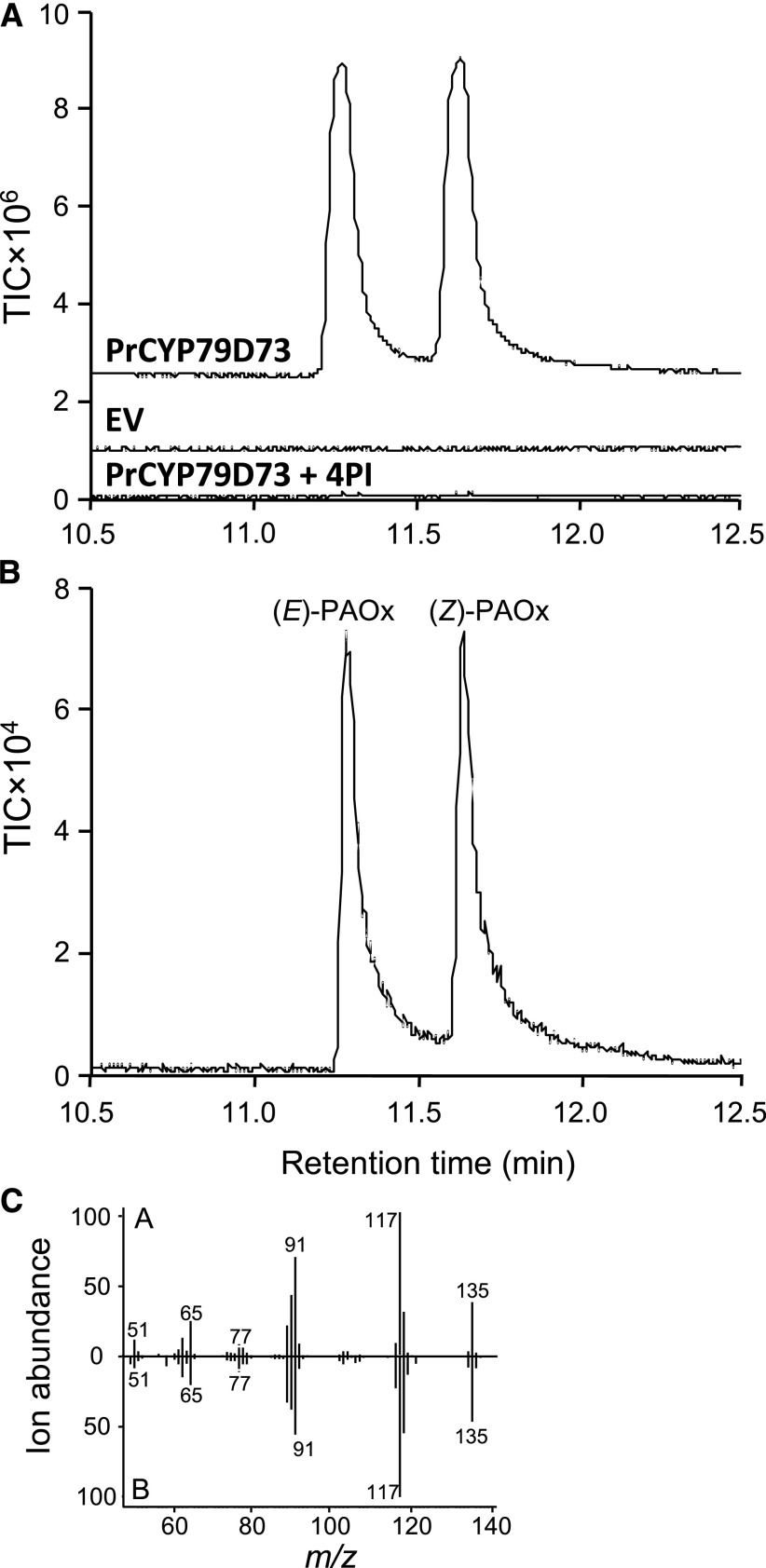
Yeast (Saccharomyces cerevisiae WAT11 strain) that expresses Arabidopsis thaliana NADPH-cytochrome P450 reductase1 has been routinely used for testing the activity of plant CYP450s (Pompon et al., 1996; Irmisch et al., 2015). Thus, we transformed WAT11 cells with the pESC-Leu-2d vector carrying the full-length ORF of PrCYP79D73 under the control of the yeast GAL10 promoter, which is subjected to both Glc repression and Gal induction (Lohr et al., 1995). Expression of PrCYP79D73 in the transformed yeast cells upon Gal treatment was confirmed by RT-PCR (Supplemental Fig. S2). For metabolite assays, microsomes prepared from WAT11 cells expressing PrCYP79D73 were incubated with l-Phe and the electron-donating cosubstrate NADPH. Product analysis using GC/quadrupole time-of-flight mass spectrometry (Q-TOF) showed that PrCYP79D73 converted l-Phe to (E)- and (Z)-isomers of PAOx (Fig. 6A, PrCYP79D73). These peaks were identified by comparison of the retention time and mass spectra of chemically synthesized (E/Z)-PAOx (Fig. 6, B and C). Moreover, pretreatment of microsomes with 4PI, a widely used heme inhibitor of CYP450 enzymes (Podust et al., 2001; Correia and Ortiz de Montellano, 2005), completely blocked the formation of (E/Z)-PAOx (Fig. 6A, PrCYP79D73+4PI). These results demonstrate that PrCYP79D73 catalyzes the formation of (E/Z)-PAOx. A control assay using microsomes prepared from WAT11 cells expressing the empty pESC-Leu-2d vector (referred to as empty vector) did not produce (E/Z)-PAOx (Fig. 6A, EV).
Figure 6.
In vitro characterization of PrCYP79D73. A, GC/Q-TOF analysis of reaction products obtained by incubation of l-Phe and NADPH with yeast microsomes prepared from PrCYP79D73 (PrCYP79D73)- or empty vector (EV)-expressing WAT11 cells. PrCYP79D73+4PI, PrCYP79D73-enriched microsomes were pretreated with 4-phenylimidazole (4PI) prior to in vitro assay. The products were confirmed by comparison of retention times and mass spectra with those of standards. TIC, Total ion chromatogram. B, TIC of authentic (E/Z)-PAOx standard. C, Head-to-tail comparison of mass spectra of the peaks from A with the authentic standard in B. m/z, Mass-to-charge ratio.
PrCYP79D73-Mediated 2PE Biosynthesis
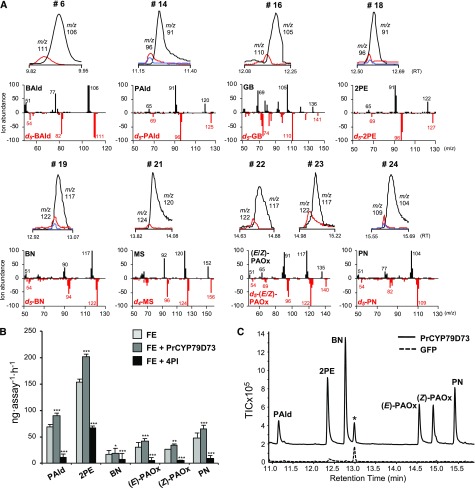
Sakai et al. (2007) proposed the hypothetic pathway involving CYP79 for 2PE biosynthesis. Here, we found that PrCYP79D73 was specifically expressed in plumeria flowers in which 2PE was highly accumulated. To investigate if (E/Z)-PAOx produced by PrCYP79D73 could be used for 2PE biosynthesis in plumeria flowers, volatiles emitted by fresh intact flowers incubated with deuterated (d5)-Phe or d5-(E/Z)-PAOx in glass vials were analyzed by headspace-GC/Q-TOF. Figure 7A shows that nine phenylpropanoids/benzenoids, benzaldehyde, PAld, geranyl benzoate, 2PE, BN, (E)-PAOx, (Z)-PAOx, methyl salicylate, and PN, were produced by d5-Phe supplemented into plumeria flowers, all of which were among the VOC profile of plumeria flowers. Among them, only d5-isomers of PAld, 2PE, BN, and PN were synthesized in the flowers incubated with d5-(E/Z)-PAOx (Fig. 7A). These results indicate that PrCYP79D73 mediates the biosynthesis of 2PE and two N-containing volatiles, BN and PN, in plumeria flowers.
Figure 7.
In vivo characterization of PrCYP79D73. A, Peak analysis of VOCs emanated from plumeria flowers incubated with d5-l-Phe or d5-(E/Z)-PAOx overnight to assess d-labeled isomer content. The peak numbers in the gas chromatograms correspond to the compounds mentioned in Table 1. Black lines, Natural compound; red lines, d-labeled isomer produced upon incubation with d5-l-Phe; blue lines, d-labeled isomer produced upon incubation with d5-(E/Z)-PAOx. A head-to-tail comparison of mass spectra of the natural compounds (top) and their d-labeled isomers (bottom) is shown below each peak. BAld, Benzaldehyde; GB, geranyl benzoate; MS, methyl salicylate; m/z, mass-to-charge ratio; RT, retention time (min). B, GC/Q-TOF analysis of reaction products obtained by incubation of crude enzyme extracts from plumeria flowers with l-Phe (FE), crude enzyme extracts with PrCYP79D73-enriched microsomes and l-Phe (FE+PrCYP79D73), and crude enzyme extracts pretreated with 10 μm 4PI and l-Phe (FE+4PI). Results represent means ± se of two technical repetitions and three biological replicates. Student’s t test was used to calculate the significant differences of FE+PrCYP79D73 and FE+4PI as compared with FE: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. C, Volatile emission of N. benthamiana plants transiently overexpressing PrCYP79D73. Plants were infiltrated with A. tumefaciens containing 35S:GFP (control) or 35S:PrCYP79D73. Volatiles were collected using the push-pull headspace method and analyzed by GC-MS. The asterisk denotes the internal standard camphor. TIC, Total ion chromatogram.
In addition, we performed CYP450 enzyme assays using crude protein extracts from plumeria flowers. Figure 7B shows that total protein extract from flowers synthesized PAld, 2PE, BN, (E/Z)-PAOx, and PN in the presence of l-Phe and NADPH. Addition of microsomes enriched in PrCYP79D73 to the above reaction mixture increased the quantity of these compounds, whereas pretreatment of crude enzymes with 4PI inhibited the production of these compounds (Fig. 7B), confirming again the function of PrCYP79D73 in the biosynthesis of 2PE in plumeria flower.
To further demonstrate the ability of PrCYP79D73 to produce 2PE in vivo, N. benthamiana plants were infiltrated with an A. tumefaciens strain carrying PrCYP79D73 under the control of a cauliflower mosaic virus 35S (CaMV 35S) promoter. A strain carrying the gene encoding GFP under the control of the CaMV 35S promoter was used as a negative control. As the plants infiltrated with GFP showed bright fluorescence 3 d postinfiltration (dpi), volatiles were collected from control plants and PrCYP79D73-overexpressing plants by the push-pull headspace method and analyzed by GC-MS. Figure 7C shows that N. benthamiana plants expressing PrCYP79D73 released (E/Z)-PAOx, which was consistent with the results obtained via in vitro assay. Moreover, PrCYP79D73-overexpressing N. benthamiana plants also produced PAld, 2PE, BN, and PN, demonstrating that PrCYP79D73 leads to the production of 2PE in planta. None of these compounds was emitted by the control plants expressing GFP (Fig. 7C).
DISCUSSION
Plumeria Open Flowers Emit Mainly Phenylpropanoids
In general, floral scent production increases as flowers mature and reaches a peak in fully opened flowers (Guterman et al., 2002; Jin et al., 2015). Our analysis of plumeria VOCs also showed that fully opened flowers emit the highest quantity of volatiles, over 60% of which is composed of phenylpropanoids/benzenoids such as 2PE (35.67%), benzaldehyde (11.77%), and PAld (5.82%; Fig. 1; Table 1). Two volatiles, 2PE and PAld, are among the major components of essential oils from different varieties of plumeria grown in Brazil, Hawaii, and Nigeria (Omata et al., 1991, 1992; Lawal et al., 2015). Similar to Rosa hybrida ‘Hoh-Jun’ (Sakai et al., 2007), petunia (Verdonk et al., 2003), and skypilot (Polemonium viscosum; Galen et al., 2011), 2PE is likely of major importance to the typical scent and fragrance of plumeria flowers.
Our species distribution analysis of plumeria RNA-seq showed that more than 50% of unigenes have the highest homology to those of coffee (Fig. 2A), reflecting the closer evolutionary relationship between the two species. Therefore, it is not surprising that coffee flowers also emit significant amounts of PAld, 2PE, BN, PAOx, and PN (Emura et al., 1997). Our results suggest that the biosynthetic pathways for the production of these phenylpropanoids might be identical in coffee and plumeria.
Among the shikimate pathway genes, DAHPS, SDH, CM, and PAT were highly expressed in open flowers, followed by floral buds and the lowest expression levels in leaves (Fig. 3). The high expression levels of shikimate pathway genes may explain the high accumulation levels of l-Phe in open flowers. Similarly, these genes are predominantly expressed in petunia petals that emit largely volatile phenylpropanoids and benzenoids (Maeda et al., 2011).
PrCYP79D73 Mediates 2PE Biosynthesis in Plumeria
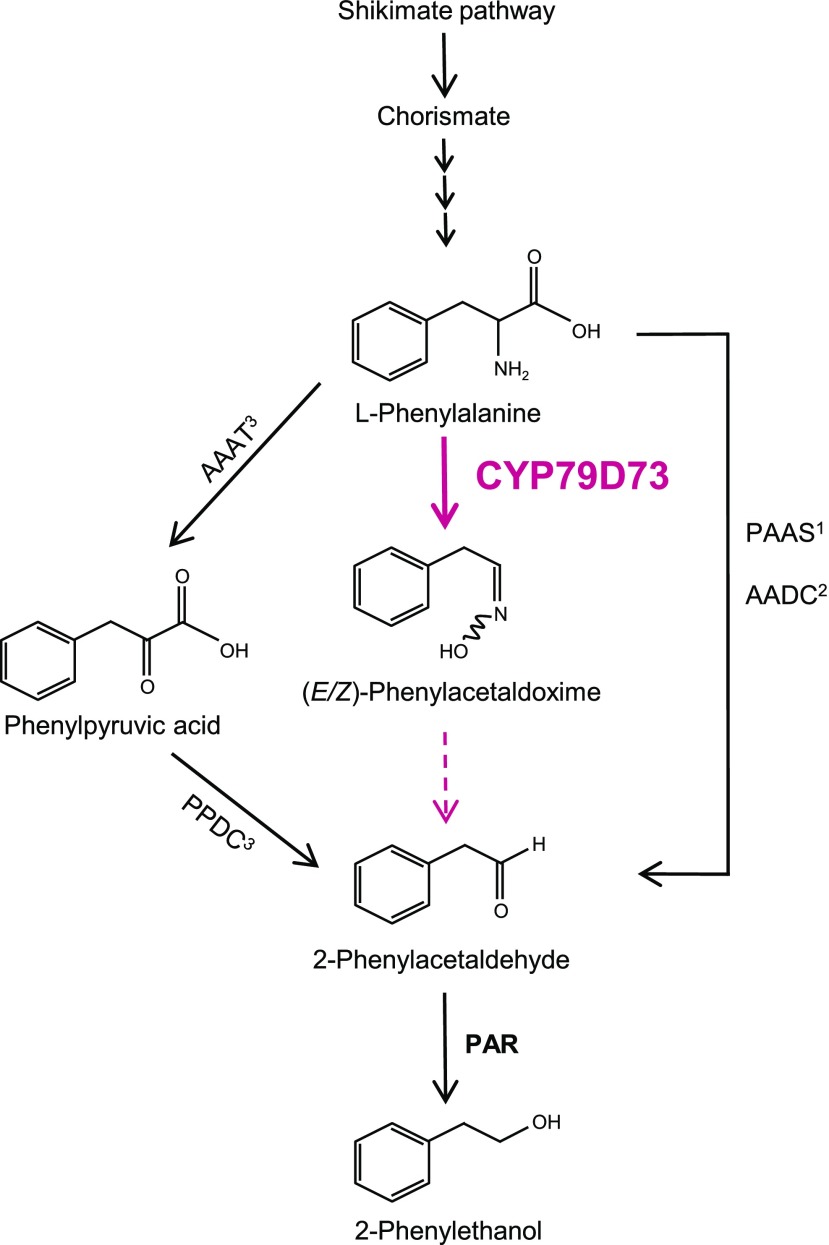
Our claim that PrCYP79D73 is a critical player for 2PE biosynthesis in plumeria flowers (Fig. 8) is supported by the following lines of evidence. (1) The homologs of PAAS/AADC and AAAT that are involved in the production of 2PE in rose and petunia were not found in the plumeria flower transcriptome. (2) Expression of PrCYP79D73 was significantly higher in open flowers in comparison with the neighboring floral buds and leaves (Fig. 3). (3) PrCYP79D73 produced (E/Z)-PAOx in vitro (Fig. 6; Supplemental Fig. S3). (4) Intact plumeria flowers fed with d5-(E/Z)-PAOx emitted d5-isomers of PAld and 2PE (Fig. 7A). (5) By adding microsomes from PrCYP79D73-overexpressed WAT11 strain to the crude protein extract from plumeria flower, the production of (E/Z)-PAOx, PAld, and 2PE from l-Phe increased (Fig. 7B). Moreover, the formation of (E/Z)-PAOx, PAld, and 2PE from l-Phe was blocked by 4PI, an inhibitor of CYP450 enzymes (Fig. 7B). This implies that, unlike rose and petunia, PAAS/AADC/AAAT might not be crucial for 2PE biosynthesis in plumeria. (6) PAld and 2PE were detected by overexpression of PrCYP79D73 in N. benthamiana (Fig. 7C), suggesting the conversion of (E/Z)-PAOx to yield PAld (Sakai et al., 2007) and further conversion to 2PE by a reductase of N. benthamiana. (7) Lack of emission of (E/Z)-PAOx, PAld, and 2PE from plumeria floral buds and leaves can be explained by the very low levels of PrCYP79D73 transcripts (Fig. 5A). This indicates a strong temporal and spatial regulation of 2PE biosynthesis in plumeria. Correlation between 2PE production and expression levels of key genes such as PAAS and AAAT has been observed in petunia and rose (Kaminaga et al., 2006; Hirata et al., 2016). (8) A plumeria unigene (PrPAR), which was preferentially expressed in open flowers and homologous to the rose PAR, catalyzed the conversion of PAld to 2PE in the presence of NADPH (Supplemental Fig. S4; Supplemental Table S1).
Figure 8.
2PE biosynthesis in flowers. The 2PE biosynthetic pathways are shown for petunia (1Kaminaga et al., 2006), rose (2Sakai et al., 2007; 3Hirata et al., 2012), and plumeria (this study). The 2PE biosynthetic pathway in plumeria flowers is highlighted in pink. Solid arrows indicate reactions previously reported, stacked arrows indicate the presence of multiple steps, and the dotted arrow suggests a nonenzymatic reaction. PPDC, Phenylpyruvic acid decarboxylase.
PrCYP79D73 Is Also Involved in the Production of N-Containing Volatile Compounds
Besides 2PE biosynthesis, PrCYP79D73 also aids the production of other floral compounds such as BN and PN in plumeria flowers (Fig. 1; Table 1). PrCYP79D73-mediated production of these N-containing volatiles was confirmed by transiently overexpressing PrCYP79D73 in N. benthamiana plants (Fig. 7). Similar results were obtained from the overexpression of ZmCYP79A61, PtCYP79D6v3, and PtCYP79D7v2 in N. benthamiana plants (Irmisch et al., 2013, 2015). Formation of N-containing compounds like BN and PN from an aldoxime precursor has been previously described (Irmisch et al., 2014). Thus, a certain level of (E/Z)-PAOx formed by the activity of PrCYP79D73 in plumeria flowers could be used as a precursor for the biosynthesis of BN and PN, along with the most-abundant plumeria floral volatile, 2PE.
PrCYP79D73 May Have Important Roles in Both Pollinator Attraction and Defense
Even though the overexpression of PrCYP79D73, ZmCYP79A61, PtCYP79D6v3, and PtCYP79D7v2 in N. benthamiana yielded similar results, PrCYP79D73 differs significantly from other CYP79 enzymes characterized thus far in several ways. First, PrCYP79D73 is among the most highly expressed genes in plumeria open flowers, whereas the expression levels of ZmCYP79A61, PtCYP79D6v3, PtCYP79D7v2, and AtCYP79A2 were very low or undetected in wild-type plants (Wittstock and Halkier, 2000; Irmisch et al., 2013, 2015). Second, in agreement with the expression levels, large amounts of chemical compounds synthesized by PrCYP79D73 activity were detected in plumeria open flowers, whereas the products of other CYP79 enzymes were not detected under normal conditions in maize, poplar, and Arabidopsis (Wittstock and Halkier, 2000; Irmisch et al., 2013, 2015). Finally, even when the levels of ZmCYP79A61, PtCYP79D6v3, and PtCYP79D7v2 transcripts increased upon mechanical damage/herbivory attack, it resulted mainly in the production of defense compounds, such as aldoximes, benzylglucosinolates, and other N-containing volatiles, and not much of 2PE and PAld (Wittstock and Halkier, 2000; Irmisch et al., 2013, 2015), compounds that are known to be popular pollinator attractants (Dobson, 2006).
The following observations suggest that the main role of PrCYP79D73 in plumeria is likely to produce 2PE for pollination. (1) Large amounts of 2PE are emitted by plumeria flowers (Fig. 1; Table 1). (2) PrCYP79D73 is predominantly expressed in plumeria open flowers rather than in floral buds or leaves (Fig. 5A). (3) Plumeria flowers are pollinated by moths (Haber, 1984). (4) 2PE is well known for attracting moths (Bengtsson et al., 2006).
On the other hand, PrCYP79D73 might also be involved in flower defense to a certain extent through the production of N-containing volatiles. BN and PN are well-known defense molecules against larvae infestation (Irmisch et al., 2014). In addition, these nitrogenous volatiles might act as precursors for the biosynthesis of other defense compounds, including glucosinolates and cyanogenic glycosides (Sibbesen et al., 1995).
CONCLUSION
Using transcriptome and metabolome data from plumeria open flowers, we identified a P450 enzyme of the CYP79 family, PrCYP79D73, that catalyzes the formation of (E/Z)-PAOx both in vitro and in vivo. Furthermore, we showed that PrCYP79D73 contributes to the PAOx-dependent formation of PAld, 2PE, BN, and PN in plumeria open flowers, all of which are known to have ecological functions. Whereas PAOx, BN, and PN might play a role in defense against herbivores and pathogens, PAld and 2PE might aid in attracting moths and other pollinators for successful reproduction of plumeria. Therefore, plumeria may have evolved to express PrCYP79D73 in flowers as a consequence of adaptation to its environment.
MATERIALS AND METHODS
Plant Materials
Young leaves of about 10 to 15 cm in length, floral buds with heights similar to those of the open flowers, and mature flowers with open petals were collected from a single plumeria (Plumeria rubra) tree grown on the National University of Singapore campus several times a year for VOC and RT-qPCR analyses. For RNA-seq, open flowers were collected during the month of November. All sample collection was conducted between 8 to 9 am. Samples unattacked by herbivores and/or aphids and appearing fresh were used for the studies. For VOC collection, samples were immediately placed in a push-pull headspace collection system. For total RNA extraction, samples were frozen in liquid N and stored at −80°C until use. Nicotiana benthamiana plants grown for 4 weeks in a greenhouse under long-day conditions (16 h of light/8 h of dark) were used for subcellular localization and in vivo assays.
Collection and Analysis of VOCs
VOCs were collected from freshly collected plumeria leaves, floral buds, and open flowers (10 g each) using a push-pull headspace collection system and analyzed by GC-MS as described by Dhandapani et al. (2017). Briefly, samples were placed in a glass jar, and charcoal-filtered air was pushed into the jars with the help of a compressed air pump. VOCs from the headspace of the samples were then collected in a Hayesep Q trap (80/100 mesh size; Restek) for 6 h. After the addition of camphor as an internal control, VOCs were extracted twice from the sorbent trap with 100 μL of hexane. VOCs were then identified and quantitated by GC-MS.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from homogenized leaf, floral bud, and open flower samples using Spectrum Plant Total RNA kits (Sigma-Aldrich). RNase-free DNase I (Roche) treatment was carried out for 15 min on a column to avoid DNA contamination. A Nanodrop spectrophotometer (ND-1000; Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer with RNA 6000 Nano Labchip Kit (Agilent Technologies) were used to determine the quantity and quality of RNA, respectively. An open flower RNA sample with RNA integrity number greater than 7 was sent to the Rockefeller University Genomics Resource Center for next-generation sequencing using an Illumina HiSeq2000 sequencing system. Single-stranded cDNAs were generated from the total RNA of leaves, floral buds, and flowers using Moloney murine leukemia virus reverse transcriptase (Promega). cDNAs were stored at −20°C until use.
De Novo Assembly and Analysis of RNA-Seq Data
The quality of the raw reads from the RNA-seq experiment was evaluated by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). After trimming adaptor sequences and removing low-quality reads, the Trinity method (Grabherr et al., 2011) was used to assemble the remaining high-quality reads de novo. Functional annotations for the unigenes were carried out by sequence similarity comparisons against the nonredundant protein database of NCBI and protein databases of rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), and grape (Vitis vinifera) with BLASTX (E value cutoff ≤ 1e−5). Biosynthetic pathways highly active in plumeria flowers were identified by mapping the unigenes to the plant metabolic pathway database (PlantCyc; http://www.plantcyc.org).
Expression Analysis by RT-qPCR
Expression levels of various genes in plumeria leaves, floral buds, and open flowers were analyzed using RT-qPCR. Primers were designed using the Primer3 program (http://bioinfo.ut.ee/primer3-0.4.0/; Supplemental Table S4). The transcript level of Actin was used for normalization. Primer-dimer formation and genomic DNA contamination were ruled out by including non-reverse transcriptase-treated templates and nontemplate control. RT-qPCR products of a few random genes were cloned and sequenced for verification. The relative expression levels of the unigenes were calculated using the 2−∆∆CT method. All RT-qPCR experiments were carried out in triplicate. SDS 2.4 software (Applied Biosystems) was used for analysis of the results.
Phylogenetic Tree Construction
The deduced amino acid sequence of PrCYP79D73 was aligned with CYP450 enzymes from other plants using ClustalW (gap open, 3; gap extension, 1.8; Gonnet; penalties, on; gap separation, 4; cutoff, 30%) in MEGA 6.0 software. The tree was constructed using a maximum likelihood algorithm (LG+G model) and was evaluated by a bootstrap analysis with 1,000 replicates.
Vector Construction and Agrobacterium tumefaciens Transformation
For expression of PrCYP79D73 in N. benthamiana, the ORF of PrCYP79D73 with or without the stop codon was amplified from plumeria flower cDNA by PCR. Purified products were cloned into pENTR vector using TOPO clonase (Invitrogen), and product identity was verified by sequencing. To generate a CFP fusion construct for subcellular localization, pENTR containing PrCYP79D73 without the stop codon construct was integrated into the pBA-DC-CFP expression vector under the control of the CaMV 35S promoter using LR clonase (Invitrogen). For functional analysis in vivo, PrCYP79D73 with the stop codon was integrated into the pBA-DC expression vector. Plasmids harboring PrCYP79D73-YFP and PrCYP79D73 were transformed into A. tumefaciens GV3101 strain by electroporation (Bio-Rad) and grown on Luria-Bertani (LB) agar plates containing spectinomycin (100 μg mL−1) and gentamycin (20 μg mL−1) at 28°C for 2 d.
For expression of PrPAR in Escherichia coli, the full-length ORF of PrPAR was amplified from plumeria flower cDNA by PCR. Purified products were cloned into the pENTR vector using TOPO clonase (Invitrogen). Product identity was verified by sequencing. pENTR containing PrPAR was then integrated into the destination vector pDEST17 using LR clonase (Invitrogen) to generate the 6×His fusion construct.
Heterologous Expression of PrCYP79D73 in Saccharomyces cerevisiae
The full-length ORF of PrCYP79D73 was amplified from open flower cDNA with gene-specific primer sets having additional NotI/BglII restriction sites. The purified PCR product was then cloned into the pESC-Leu-2d vector, and the sequence alignment was verified by sequencing. Similarly, the full-length ORF of PtCYP79D6v3 was also cloned into the pESC-Leu-2d vector. The resulting constructs and empty pESC-Leu-2d (empty vector) were transformed into the S. cerevisiae strain WAT11 by the lithium acetate method (Pompon et al., 1996), spread on solid selection medium (26.7 g L−1 minimal synthetic-defined base medium [Clontech], 0.69 g L−1 Leu dropout supplement [Clontech], and 20 g L−1 bacto agar), and grown at 30°C for 2 d. A single transformed yeast colony for each construct was inoculated into 15 mL of liquid selection medium and grown overnight at 30°C and 180 rpm. One OD of this culture was used to inoculate 50 mL of yeast extract, peptone, dextrose, and adenine full medium (Clontech), which was grown until the OD reached ∼5. At this stage, the cultures were induced by the addition of Gal to a final concentration of 2% (w/v) and cultured for another 15 to 18 h.
CYP450 Activity Assay
Microsomes for CYP450 activity assays were prepared from yeast cells as previously described (Pompon et al., 1996; Irmisch et al., 2013). Three hundred microliters of the reaction mixture containing 20 μL of microsomes, 1 mm NADPH, and 1 mm l-Phe (Sigma-Aldrich) in 75 mm sodium phosphate buffer (pH 7) was incubated for 30 min at 25°C and 300 rpm in a glass vial. Microsomes prepared from yeast cells transformed with the empty vector served as negative controls. Assays were stopped by placing on ice, and the reaction products were extracted using 200 μL of hexane for 30 min. For CYP450 inhibition assays, microsomes were pretreated with different concentrations of 4PI at room temperature for 5 min before addition to the reaction mixtures. The hexane extracts were concentrated to 100 µL using an N gas evaporator, and 1 µL of the extract was analyzed by an Agilent 7890B GC/Q-TOF system equipped with an HP-5MS column (Agilent Technologies). (E/Z)-PAOx was synthesized by condensation of PAld (Sigma-Aldrich) with hydroxylamine as described by Irmisch et al. (2013).
Subcellular Localization
For subcellular localization of PrCYP79D73, a single colony of A. tumefaciens transformed with PrCYP79D73-CFP construct was inoculated into 10 mL of LB medium (100 μg mL−1 spectinomycin and 20 μg mL−1 gentamycin) and cultured overnight (220 rpm, 28°C). The ER membrane marker, eYFP-RcDGAT2, was generated by fusing type 2 diacylglycerol acyltransferase from Ricinus communis with eYFP (Tian et al., 2014). A single colony of A. tumefaciens harboring the eYFP-RcDGAT2 construct was inoculated into 10 mL of LB medium (100 μg mL−1 kanamycin and 20 μg mL−1 gentamycin) and cultured overnight (220 rpm, 28°C). The cells were harvested by centrifugation (4,000g, 10 min) and resuspended in infiltration buffer containing 10 mm MgCl2, 10 mm MES, and 0.2 mm acetosyringone to reach a final OD600 of 0.4. After shaking for at least 2 h at room temperature, the PrCYP79D73 culture was mixed with eYFP-RcDGAT2 and RNA silencing suppressor p19 cultures in a ratio of 2:2:1 and infiltrated into the leaves of 4-week-old N. benthamiana plants using a needleless syringe. At 3 dpi, leaves were observed with a confocal laser-scanning upright microscope (Carl Zeiss, LSM 5 Exciter) using a standard filter set and analyzed by LSM image browser (Carl Zeiss).
In Vivo Assays
For feeding experiments, multiple glass vials with 300 μL of the reaction mixture containing 20 μL of microsomes enriched in PrCYP79D73, 1 mm NADPH, and 1 mm d5-l-Phe (Sigma-Aldrich) in 75 mm sodium phosphate buffer (pH 7) each were incubated for 30 min at 25°C at 300 rpm. Assays were stopped by placing on ice, and the reaction products were extracted using 200 μL of hexane for 30 min. The hexane extracts were pooled, concentrated using an N gas evaporator, and resuspended in water as 1 mg mL−1 solution and stored at 4°C until use. Intact open flowers were incubated overnight at 25°C individually in glass vials containing 1 mm d5-Phe or d5-(E/Z)-PAOx. A glass vial containing water served as the negative control. Thereafter, volatiles present in the vials were analyzed using headspace-GC/Q-TOF. As the natural compounds and their d-labeled isomers coeluted, d-labeled isomers in each peak were identified using the selected ion monitoring method. The most abundant ion of each compound was used as a monitor.
Crude enzymes from plumeria flowers and leaves were extracted by homogenizing the tissues in liquid N and resuspended in a buffer containing 50 mm Tris-HCl (pH 8), 150 mm sodium chloride, 10% (v/v) glycerol, 1% (v/v) Nonidet P-40, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Roche). CYP450 assay using crude protein extracts was carried out as described above in “CYP450 Activity Assay.”
For the transient overexpression assay, a single colony of A. tumefaciens transformed with PrCYP79D73 was cultured and processed as described above. After shaking for at least 2 h at room temperature, the PrCYP79D73 and p19 cultures were mixed in a ratio of 4:1 and infiltrated into leaves of N. benthamiana plants. Leaves infiltrated with GFP served as the negative control. At 3 dpi, VOCs were collected for 6 h using the push-pull headspace method and analyzed by GC-MS.
PrPAR Functional Assay
For in vitro functional analysis, plasmids containing the final His-PrPAR construct were transformed into E. coli C41 competent cells. E. coli extracted after isopropyl β-d-1-thiogalactopyranoside induction was incubated with Ni-NTA Sepharose resin (Qiagen) to purify recombinant protein. For the PrPAR functional assay, 300 μL of the reaction mixture containing 100 mm potassium phosphate buffer (pH 7), 20 μg of purified His-PrPAR recombinant protein, 1 mm NADPH, and 2.5 mm PAld was incubated for 30 min at 30°C at 300 rpm in a glass vial. His-SUMO and heat-inactivated His-PrPAR proteins were used as negative controls. Assays were stopped by placing on ice, and the reaction products were extracted using 200 μL of hexane for 30 min. One microliter of the extract was analyzed by the GC/Q-TOF system equipped with an HP-5MS column (Agilent Technologies). The peak was identified by comparing retention times and mass spectra of an authentic 2PE standard (Sigma-Aldrich).
Accession Numbers
The raw RNA-seq data from this work are available in the DDBJ: DNA Bank of Japan under accession number DRA005989 (http://trace.ddbj.nig.ac.jp/DRASearch/submission?acc=DRA005989). The nucleotide sequences of PrCYP79D73 and PrPAR are available in the NCBI (https://www.ncbi.nlm.nih.gov) under accession numbers MG459015 and MK523384, respectively. A voucher specimen of this species has been deposited in the Singapore herbarium under barcode SING0230615.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Deduced amino acid sequence alignment and expression analysis of Tyr decarboxylase.
Supplemental Figure S2. Expression of PrCYP79D73 in Gal-induced WAT11 cells.
Supplemental Figure S3. Biosynthesis of d5-labeled (E/Z)-PAOx using microsomes prepared from PrCYP79D73-expressing WAT11 cells.
Supplemental Figure S4. Characterization of plumeria phenylacetaldehyde reductase.
Supplemental Table S1. Accession numbers of proteins used in the amino acid sequence alignments.
Supplemental Table S2. Results of homologous gene search from plumeria flower transcriptome.
Supplemental Table S3. Accession numbers of proteins used in the PrCYP79D73 phylogenetic analysis.
Supplemental Table S4. List of primers used in this study.
Supplemental Data File S1. Functional annotation of plumeria unigenes using BLASTX against the multiple protein databases.
Acknowledgments
We thank Dr. Tobias G. Köllner (Max Planck Institute for Chemical Ecology) for the pESC-Leu vector and S. cerevisiae WAT11 strain and Dr. Yin Zhongchao (Temasek Life Sciences Laboratory) for the A. tumefaciens GV3101 strain harboring eYFP-RcDGAT2.
Footnotes
This research was supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Synthetic Biology Research and Development Programme (Award No: SBP-P3).
References
- Aubert C, Baumann S, Arguel H (2005) Optimization of the analysis of flavor volatile compounds by liquid-liquid microextraction (LLME): Application to the aroma analysis of melons, peaches, grapes, strawberries, and tomatoes. J Agric Food Chem 53: 8881–8895 [DOI] [PubMed] [Google Scholar]
- Bengtsson M, Jaastad G, Knudsen G, Kobro S, Bäckman AC, Pettersson E, Witzgall P (2006) Plant volatiles mediate attraction to host and non-host plant in apple fruit moth, Argyresthia conjugella. Entomol Exp Appl 118: 77–85 [Google Scholar]
- Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, Chen LJ, He Y, Xu Q, Bian C, et al. (2015) The genome sequence of the orchid Phalaenopsis equestris. Nat Genet 47: 65–72 [DOI] [PubMed] [Google Scholar]
- Chen XM, Kobayashi H, Sakai M, Hirata H, Asai T, Ohnishi T, Baldermann S, Watanabe N (2011) Functional characterization of rose phenylacetaldehyde reductase (PAR), an enzyme involved in the biosynthesis of the scent compound 2-phenylethanol. J Plant Physiol 168: 88–95 [DOI] [PubMed] [Google Scholar]
- Correia MA, Ortiz de Montellano PR (2005) Inhibition of cytochrome P450 enzymes. In Ortiz de Montellano PR, ed, Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer, Boston, pp 247–322 [Google Scholar]
- Croteau R, Karp F (1991) Origin of natural odorants. In Muller P, Lanmparsky D, eds, Perfume: Art, Science and Technology. Elsevier Applied Sciences, New York, pp 101–126 [Google Scholar]
- Dhandapani S, Jin J, Sridhar V, Sarojam R, Chua NH, Jang IC (2017) Integrated metabolome and transcriptome analysis of Magnolia champaca identifies biosynthetic pathways for floral volatile organic compounds. BMC Genomics 18: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson HE. (2006) Relationship between floral fragrance composition and type of pollinator. In Dudareva N, Pichersky E, eds, Biology of Floral Scent. CRC Press, Boca Raton, FL, pp 147–198 [Google Scholar]
- Durst F, Nelson DR (1995) Diversity and evolution of plant P450 and P450-reductases. Drug Metabol Drug Interact 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Egwaikhide PA, Okeniyi SO, Gimba CE (2009) Screening for anti-microbial activity and phytochemical constituents of some Nigerian medicinal plants. J Med Plants Res 3: 1088–1091 [Google Scholar]
- Emura M, Nohara I, Toyoda T, Kanisawa T (1997) The volatile constituents of the coffee flower (Coffea arabica L.). Flavour Fragrance J 12: 9–13 [Google Scholar]
- Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J, Zagrobelny M, Bak S (2013) Plant defense against insect herbivores. Int J Mol Sci 14: 10242–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen C, Kaczorowski R, Todd SL, Geib J, Raguso RA (2011) Dosage-dependent impacts of a floral volatile compound on pollinators, larcenists, and the potential for floral evolution in the alpine skypilot Polemonium viscosum. Am Nat 177: 258–272 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman I, Shalit M, Menda N, Piestun D, Dafny-Yelin M, Shalev G, Bar E, Davydov O, Ovadis M, Emanuel M, et al. (2002) Rose scent: Genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14: 2325–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber WA. (1984) Pollination by deceit in a mass-flowering tropical tree Plumeria rubra L. (Apocynaceae). Biotropica 16: 269–275 [Google Scholar]
- Hirata H, Ohnishi T, Ishida H, Tomida K, Sakai M, Hara M, Watanabe N (2012) Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J Plant Physiol 169: 444–451 [DOI] [PubMed] [Google Scholar]
- Hirata H, Ohnishi T, Tomida K, Ishida H, Kanda M, Sakai M, Yoshimura J, Suzuki H, Ishikawa T, Dohra H, et al. (2016) Seasonal induction of alternative principal pathway for rose flower scent. Sci Rep 6: 20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Maekawa M, Tsuchiya S, Fujimori T (1998) Field attraction of Hoplia communis to 2-phenylethanol, a major volatile component from host flowers, Rosa spp. J Chem Ecol 24: 1491–1497 [Google Scholar]
- Irmisch S, McCormick AC, Boeckler GA, Schmidt A, Reichelt M, Schneider B, Block K, Schnitzler JP, Gershenzon J, Unsicker SB, et al. (2013) Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense. Plant Cell 25: 4737–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, McCormick AC, Günther J, Schmidt A, Boeckler GA, Gershenzon J, Unsicker SB, Köllner TG (2014) Herbivore-induced poplar cytochrome P450 enzymes of the CYP71 family convert aldoximes to nitriles which repel a generalist caterpillar. Plant J 80: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Zeltner P, Handrick V, Gershenzon J, Köllner TG (2015) The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol 15: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Kim MJ, Dhandapani S, Tjhang JG, Yin JL, Wong L, Sarojam R, Chua NH, Jang IC (2015) The floral transcriptome of ylang ylang (Cananga odorata var. fruticosa) uncovers biosynthetic pathways for volatile organic compounds and a multifunctional and novel sesquiterpene synthase. J Exp Bot 66: 3959–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaga Y, Schnepp J, Peel G, Kish CM, Ben-Nissan G, Weiss D, Orlova I, Lavie O, Rhodes D, Wood K, et al. (2006) Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J Biol Chem 281: 23357–23366 [DOI] [PubMed] [Google Scholar]
- Koning-Boucoiran CF, Esselink GD, Vukosavljev M, van’t Westende WP, Gitonga VW, Krens FA, Voorrips RE, van de Weg WE, Schulz D, Debener T, et al. (2015) Using RNA-Seq to assemble a rose transcriptome with more than 13,000 full-length expressed genes and to develop the WagRhSNP 68k Axiom SNP array for rose (Rosa L.). Front Plant Sci 6: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal OA, Ogunwande IA, Opuku AR (2015) Chemical composition of essential oils of Plumeria rubra L. grown in Nigeria. Eur J Med Plants 6: 55–61 [Google Scholar]
- Lawrence SD, Novak NG, Ju CJ, Cooke JE (2008) Potato, Solanum tuberosum, defense against Colorado potato beetle, Leptinotarsa decemlineata (Say): Microarray gene expression profiling of potato by Colorado potato beetle regurgitant treatment of wounded leaves. J Chem Ecol 34: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Li W, Liu X, Lu Y (2016) Transcriptome comparison reveals key candidate genes in response to vernalization of Oriental lily. BMC Genomics 17: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng Y, Yang M, Liu Y, Chen K, Long CA, Deng X (2014) Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol 14: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J (1995) Transcriptional regulation in the yeast GAL gene family: A complex genetic network. FASEB J 9: 777–787 [DOI] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N (2011) Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat Chem Biol 7: 19–21 [DOI] [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7: 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR. (2009) The cytochrome p450 homepage. Hum Genomics 4: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata A, Yomogida K, Nakamura S, Hashimoto S, Arai T, Furukawa K (1991) Volatile components of Plumeria flowers. Part 1. Plumeria rubra forma acutifolia (Poir.) Woodson cv. ‘Common Yellow.’ Flavour Fragrance J 6: 277–279 [Google Scholar]
- Omata A, Shoji N, Seiji H, Kiyoshi F (1992) Volatile components of Plumeria flowers. Part 2. Plumeria rubra L. cv. ‘Irma Bryan.’ Flavour Fragrance J 7: 33–35 [Google Scholar]
- Podust LM, Poulos TL, Waterman MR (2001) Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc Natl Acad Sci USA 98: 3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Qi G, Wang D, Yu L, Tang X, Chai G, He G, Ma W, Li S, Kong Y, Fu C, et al. (2015) Metabolic engineering of 2-phenylethanol pathway producing fragrance chemical and reducing lignin in Arabidopsis. Plant Cell Rep 34: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Sakai M, Hirata H, Sayama H, Sekiguchi K, Itano H, Asai T, Dohra H, Hara M, Watanabe N (2007) Production of 2-phenylethanol in roses as the dominant floral scent compound from L-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci Biotechnol Biochem 71: 2408–2419 [DOI] [PubMed] [Google Scholar]
- Shinde PR, Patil P, Bairagi VA (2014) Phytopharmacological review of Plumeria species. Sch Acad J Pharm 3: 217–227 [Google Scholar]
- Sibbesen O, Koch B, Halkier BA, Møller BL (1995) Cytochrome P-450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J Biol Chem 270: 3506–3511 [DOI] [PubMed] [Google Scholar]
- Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, et al. (2014) The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26: 497–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Loucas HM, Kim JY, Clark DG, Klee HJ (2007) Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 68: 2660–2669 [DOI] [PubMed] [Google Scholar]
- Tohar N, Mohd MA, Jantan I, Awang K (2006) A comparative study of the essential oils of the genus Plumeria Linn. from Malaysia. Flavour Fragrance J 21: 859–863 [Google Scholar]
- Verdonk JC, Ric de Vos CH, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC (2003) Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62: 997–1008 [DOI] [PubMed] [Google Scholar]
- Verma S. (2016) Multipurpose ornamental plant Plumeria rubra Linn (Apocynaceae). Int J Sci Res Sci Eng Technol 2: 646–649 [Google Scholar]
- Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J Biol Chem 275: 14659–14666 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Yamamoto K, Asano Y (2014) Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in L-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume Sieb. et Zucc. Plant Mol Biol 86: 215–223 [DOI] [PubMed] [Google Scholar]
- Zheng J, Hu Z, Guan X, Dou D, Bai G, Wang Y, Guo Y, Li W, Leng P (2015) Transcriptome analysis of Syringa oblata Lindl. inflorescence identifies genes associated with pigment biosynthesis and scent metabolism. PLoS ONE 10: e0142542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Obrycki JJ, Ochieng SA, Baker TC, Pickett JA, Smiley D (2005) Attraction of two lacewing species to volatiles produced by host plants and aphid prey. Naturwissenschaften 92: 277–281 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zhou H, Hu Y, Tang J, Su M, Guo Y, Chen Q, Liu B (2011) Antityrosinase and antimicrobial activities of 2-phenylethanol, 2-phenylacetaldehyde and 2-phenylacetic acid. Food Chem 124: 298–302 [Google Scholar]