The gene SlHAK10 encodes a mycorrhiza-specific potassium transporter of the KT/KUP/HAK family that mediates K+ uptake at the intraradical symbiotic interface in tomato.
Abstract
Most terrestrial plants form a root symbiosis with arbuscular mycorrhizal (AM) fungi, which receive fixed carbon from the plant and enhance the plant’s uptake of mineral nutrients. AM symbiosis improves the phosphorous and nitrogen nutrition of host plants; however, little is known about the role of AM symbiosis in potassium (K+) nutrition. Here, we report that inoculation with the AM fungus Rhizophagus irregularis improved tomato (Solanum lycopersicum) plant growth and K+ acquisition and that K+ deficiency has a negative effect on root growth and AM colonization. Based on its homology to a Lotus japonicus AM-induced K+ transporter, we identified a mycorrhiza-specific tomato K+ transporter, SlHAK10 (Solanum lycopersicum High-affinity Potassium Transporter10), that was exclusively expressed in arbuscule-containing cells. SlHAK10 could restore a yeast K+ uptake-defective mutant in the low-affinity concentration range. Loss of function of SlHAK10 led to a significant decrease in mycorrhizal K+ uptake and AM colonization rate under low-K+ conditions but did not affect arbuscule development. Overexpressing SlHAK10 from the constitutive cauliflower mosaic virus 35S promoter or the AM-specific Solanum melongena Phosphate Transporter4 not only improved plant growth and K+ uptake but also increased AM colonization efficiency and soluble sugar content in roots supplied with low K+. Our results indicate that tomato plants have a SlHAK10-mediated mycorrhizal K+ uptake pathway and that improved plant K+ nutrition could increase carbohydrate accumulation in roots, which facilitates AM fungal colonization.
Potassium (K+) is one of the three essential macronutrients, constituting 2% to 10% of plant dry matter (Leigh and Wyn Jones, 1984; Amrutha et al., 2007; Yang et al., 2009). As the most abundant cation in living plant cells, K+ plays crucial roles in many fundamental processes, including photosynthesis, osmoregulation, membrane transport, enzyme activation, and anion neutralization, and consequently affects overall plant growth and development (Amrutha et al., 2007; Yang et al., 2009). As a result of leaching losses, chemical fixation, and the relatively low diffusion rate in soil, the availability of K+ drastically fluctuates in large areas of agricultural land worldwide and is often a limiting factor for crop quality and yields (Rengel and Damon, 2008).
Plant cells need to maintain their cytoplasmic K+ concentrations at approximately 100 mm (Leigh and Wyn Jones, 1984; Walker et al., 1996), but the K+ concentration in the rhizosphere varies drastically and rarely exceeds 0.01 to 1 mm (Maathuis, 2009; White, 2013). This indicates the requirement for specialized transport systems for the uptake of K+ and for its internal redistribution within plants. Transport of K+ through plant membranes can be mediated either by K+ channels, which use the membrane potential and electrochemical gradient to facilitate K+ transport, or by secondary transporters with different affinities located in the plasma or organelle membranes (Gierth et al., 2005; Voelker et al., 2006; Li et al., 2018). Genes encoding plant K+ transporters are classified into four major families: KT/HAK/KUP (K+ Transporter/High-affinity K+ transporter/K Uptake Permease), HKT (High-affinity K+/Na+ Transporter), KEA (K+ Exchange Antiporters), and CHX (Cation/H+ Exchanger; Uozumi et al., 2000; Cellier et al., 2004; Kunz et al., 2014; Aranda-Sicilia et al., 2016; Wang and Wu, 2017). The KT/KUP/HAK family comprises most of the plant K+ transporters identified so far, and members of this family are further divided into four major clusters (I–IV; Rubio et al., 2000; Grabov, 2007; Gupta et al., 2008; Wang and Wu, 2013; Nieves-Cordones et al., 2016). Multiple transporters in cluster I, such as AtHAK5, HvHAK1, OsHAK1, and OsHAK5, have high-affinity K+ uptake capacity and rapid up-regulation in response to K+ deficiency (Schachtman and Schroeder, 1994; Santa-María et al., 1997; Gierth et al., 2005; Yang et al., 2014; Chen et al., 2015). The cluster II members of the KT/KUP/HAK family showed great divergence in sequence and function (Elumalai et al., 2002; Yang et al., 2009; Osakabe et al., 2013). Transporters in this cluster have been characterized to mediate both high- and low-affinity K+ uptake. AtKUP1 from Arabidopsis (Arabidopsis thaliana) is a typical cluster II member of the KT/KUP/HAK family that functions as a dual-affinity K+ transporter (Fu and Luan, 1998). Much less has been reported regarding the potential functions or physiological roles of the cluster III and IV transporters (Han et al., 2016).
Arbuscular mycorrhizal (AM) symbiosis, formed between AM fungi of Glomeromycotina and roots of more than 80% of land plants, represents one of the most important symbiotic interactions in nature (Smith and Smith, 2011; Kobae et al., 2016). Formation of an AM symbiosis improves growth status for the plant partner, especially under nutrient-poor growing conditions (Govindarajulu et al., 2005; Javot et al., 2007; Jin et al., 2012; Casieri et al., 2013; Courty et al., 2016; Garcia et al., 2016). In the nonmycorrhizal (NM) plants, mineral nutrients can only be directly taken up from the rhizosphere via root epidermis and root hairs. Once colonized by AM fungi, the mycorrhizal plants thus possess another indirect uptake pathway, called the mycorrhizal pathway, via the AM fungal hyphae into root cortical cells (Kiers et al., 2011; Smith and Smith, 2012; Sieh et al., 2013; Giovannetti et al., 2014).
The role of phosphate (Pi) nutrition in AM symbiosis has always been a major focus. It has been repeatedly demonstrated that the mycorrhizal pathway is key for Pi uptake (Harrison et al., 2002; Paszkowski et al., 2002; Smith et al., 2003; Nagy et al., 2006; Chen et al., 2007). Multiple plant transporters responsible for transporting Pi across the intraradical symbiotic interfaces have been identified (Paszkowski et al., 2002; Nagy et al., 2005; Yang et al., 2012; Xie et al., 2013; Chen et al., 2014). Although the contribution of AM fungi to the transport of other nutrients is less clear, increasing evidence has shown that plants can acquire substantial amounts of nitrogen (N) and sulfur (S) through the symbiotic uptake pathway (Mäder et al., 2000; Allen and Shachar-Hill, 2009; Koegel et al., 2013; Giovannetti et al., 2014).
Compared with the relatively large number of reports on symbiotic Pi and N uptake, much less is known about the effects of AM symbiosis on plant K+ acquisition. Nonetheless, enrichment of K+ in different tissues of several mycorrhizal plants, including maize (Zea mays), lettuce (Lactuca sativa), and wheat (Triticum aestivum), could be traced in a small number of studies (Garcia and Zimmermann, 2014), leading to speculation that plants may also have a symbiotic K+ uptake pathway. This hypothesis gained some support from the transcriptome analysis of both AM partners. On the fungal side, four sequences from an EST library of Rhizophagus irregularis were identified as K+ transporters (Casieri et al., 2013). On the plant side, a KT/KUP/HAK transporter (Lj4g3v3116360.1; hereafter referred to as LjHAK) was shown to be up-regulated in AM roots of Lotus japonicus compared with the NM roots (Guether et al., 2009). A recent study using whole-genome RNA sequencing of Medicago truncatula mycorrhizal roots under K+ deficiency revealed the up-regulation of several genes encoding putative transporters, including a putative K+/H+ exchanger, in mycorrhizal plants under K+ deprivation (Garcia et al., 2017). However, in several transcriptome studies, no K+ transporter was observed to be up-regulated in M. truncatula mycorrhizal roots (Gomez et al., 2009; Gaude et al., 2012). Thus, it is tempting to investigate whether the gene regulation involved in the transfer of K+ from the AM fungus to the plant is conserved across different plant species.
In this study, we performed extensive searches of putative K+ transporters in the recently released transcriptomes for several mycorrhizal plants (Güimil et al., 2005; Fiorilli et al., 2009; Gomez et al., 2009; Gaude et al., 2012; Garcia et al., 2017; Sugimura and Saito, 2017), which resulted in the identification of a member of the KT/KUP/HAK family from tomato (Solanum lycopersicum) named SlHAK10 (Solyc03g097860.1.1) as the putative orthologue of the AM-induced LjHAK. SlHAK10 was shown to be induced 500-fold in AM roots compared with the NM roots (Sugimura and Saito, 2017). Except for LjHAK and SlHAK10, none of the other K+ transporters from the KT/KUP/HAK family or other families showed observable up-regulation in the transcriptomes of tomato and L. japonicus mycorrhizal roots. Genome-wide hunting and expression analysis of all the tomato KT/KUP/HAK genes confirmed that SlHAK10 was the sole one having AM-inducible expression in this family. Tissue-specific and functional analyses revealed that SlHAK10 was expressed exclusively in the cells containing arbuscules and could mediate K+ uptake through the mycorrhizal pathway. These findings thus indicate that there might be a mycorrhizal K+ uptake pathway in mycorrhizal plants, at least in tomato, through activating the AM-induced K+ transporter(s), such as SlHAK10 and its potential orthologues.
RESULTS
AM Symbiosis Improves the K+ Accumulation in Tomato Plants
In this study, to investigate the potential roles of AM formation in plant K+ acquisition, tomato plants were grown in a sand-based pot culture in the presence or absence of AM fungus (R. irregularis) supplied with 2 mm K+ as a high-K+ condition and with 0.2 mm K+ as a low-K+ condition. The plant samples were harvested at 5 weeks postinoculation for determining the shoot and root biomass, K+ accumulation, and fungal colonization rates.
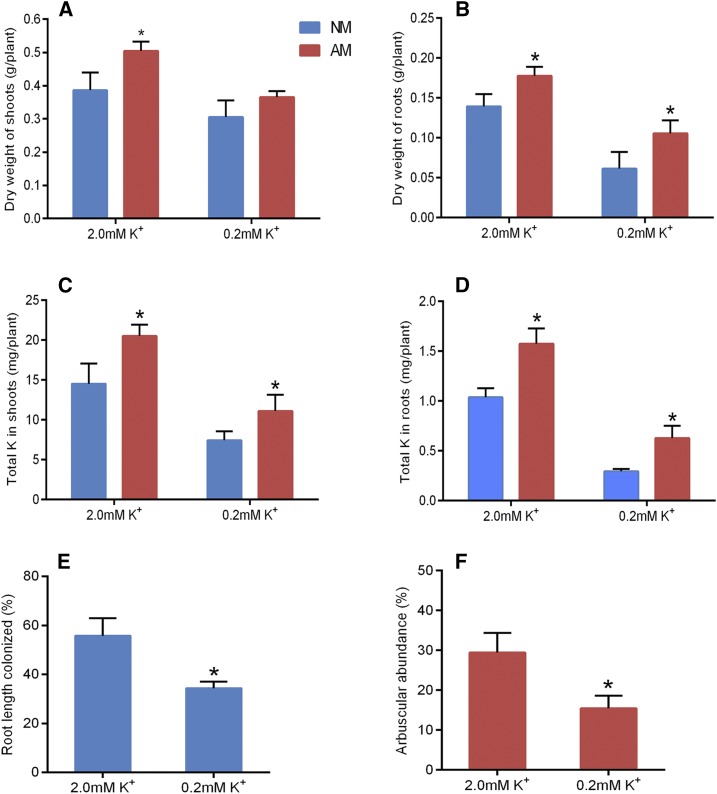
The plants treated with low-K+ solution showed a significant growth suppression in both shoots and roots over than the plants supplied with high K+ (Fig. 1, A and B). Under low K+, root growth was more inhibited compared with shoot growth, leading to a typical decrease in root-shoot ratio (Supplemental Fig. S1A). Compared with the NM plants, the mycorrhizal plants (AM plants) showed increased shoot and root biomass under the high-K+ condition (Fig. 1, A and B). The shoot K+ content in AM plants grown under high K+ did not differ significantly with that in the NM plants (Supplemental Fig. S1B), but the root K+ content (Supplemental Fig. S1C) and the total K+ accumulation in both shoots and roots (Fig. 1, C and D) increased by more than 20% in the AM plants. The shoot biomass of AM plants supplied with low K+ had no significant difference compared with that of the NM control plants (Fig. 1A). By contrast, a significant increase in root biomass (Fig. 1B) and root-shoot ratio (Supplemental Fig. S1), as well as K+ accumulation in both shoots and roots (Fig. 1, C and D), could be observed under the low-K+ condition.
Figure 1.
Impact of different K+ applications and AM fungal colonization on tomato plant growth and K+ acquisition. A and B, Shoot (A) and root (B) biomass of AM and NM plants. C and D, K+ accumulation in shoots (C) and roots (D) of AM and NM plants. E and F, Total colonization rate (E) and arbuscule abundance (F) under high-/low-K+ supply conditions. AM, AM fungus-colonized plants; NM, nonmycorrhizal plants. Error bars indicate se (n = 5). Asterisks indicate significant differences (P < 0.05).
The plants supplied with low K+ had a significantly lower AM colonization rate and arbuscule incidence compared with the plants grown under high K+ (Fig. 1, E and F). An assessment of mycorrhization efficiency by different concentrations of K+ application was performed, which led to the suggestion that the K+-limiting growth condition might have a negative effect on AM colonization in tomato (Supplemental Fig. S2).
Identification and Characterization of a Tomato AM-Induced Potassium Transporter
A previous study based on microarray analysis revealed a putative AM-induced K+ transporter gene (Lj4g3v3116360.1) of the KT/KUP/HAK family in L. japonicus (named LjHAK in this study; Guether et al., 2009). To identify the potential homologues responsive to AM symbiosis in tomato, the coding sequence of LjHAK was employed for BLAST search against the tomato genomic database (http://solgenomics.net/), which led to the identification of 21 nonallelic sequences with substantial identities and similar structures to the tomato K+ transporter genes (Supplemental Fig. S3).
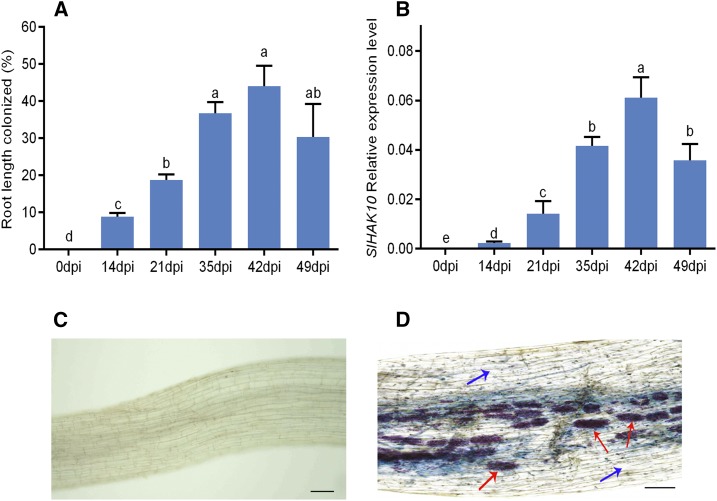
A phylogenetic tree constructed with these putative tomato K+ transporters (named SlHAK1 to SlHAK20 in this study) and homologues from several other plant species revealed a cluster of eight SlHAK transporters with the AM-induced LjHAK in group II of the KT/KUP/HAK family. Reverse transcription quantitative PCR (RT-qPCR) analysis revealed that, with the exception of SlHAK10, the closest orthologue of LjHAK, which was highly induced in the tomato mycorrhizal roots, none of the other SlHAK genes showed significantly up-regulated responses to AM symbiosis (Supplemental Fig. S4). The mycorrhiza-inducible expression property of SlHAK10 was further confirmed by a recent study on comparative transcriptome analysis between L. japonicus and tomato, in which SlHAK10 was shown to be induced 500-fold in AM roots (Sugimura and Saito, 2017). A time-course expression analysis further proved the high correlation between the transcript abundance of SlHAK10 and colonization intensity (Fig. 2, A and B). The transcripts of SlHAK10 were barely detectable in other tissues, including leaf, flower, and fruits, and showed no conspicuous induction to other environmental factors, such as K+ and Pi deficiency, as well as high-Na+ stress (Supplemental Fig. S5).
Figure 2.
Expression analysis of SlHAK10 in response to AM symbiosis. A, Quantification of AM fungal colonization at different sampling time points. dpi, Days postinoculation. B, Time-course expression of SlHAK10 in mycorrhizal roots of tomato plants. Values are means of three biological replicates with se. Different letters indicate significant differences (P < 0.05). C and D, Histochemical GUS staining of tomato roots expressing pSlHAK10::GUS in the absence (C) and presence (D) of R. irregularis inoculation. Red arrows indicate arbuscules. Blue arrows denote noncolonized cells in mycorrhizal roots. Bars = 50 μm.
To investigate the SlHAK10 expression pattern in detail, a 2,281-bp promoter fragment of SlHAK10 was fused to the GUS reporter gene and introduced into tomato plants. The resulting transgenic plants were then inoculated with R. irregularis. Five weeks after inoculation, the GUS activity could be detected only in the mycorrhizal roots. Colocalization of GUS expression and AM fungal structure by overlay of Magenta-GUS with Trypan Blue staining revealed that GUS activity driven by the SlHAK10 promoter was confined to distinct cells containing arbuscules (Fig. 2, C and D). No GUS staining could be observed in the neighboring noncolonized cells, consistent with the results reported for some other AM-specific nutrient transporters, such as Pi and NH4+ (Kobae et al., 2010; Chen et al., 2011).
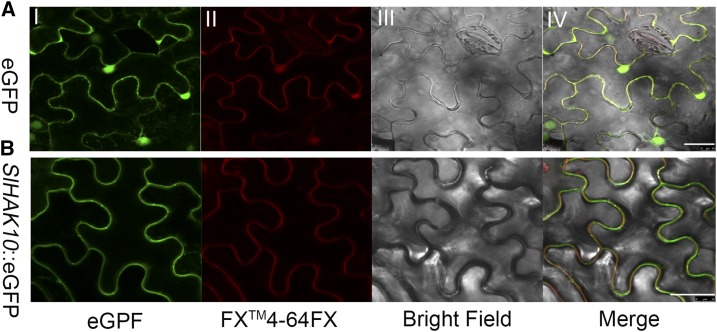
To determine whether SlHAK10 functions as a membrane-localized protein, its subcellular localization was investigated. The coding sequence of SlHAK10 was fused in frame with the 5′ end of the eGFP reporter gene under the control of the 2× cauliflower mosaic virus 35S (CaMV35S) promoter (2×35S::SlHAK10::eGFP). The construct 35S::eGFP was used as a control. Both constructs were transiently expressed in the tobacco (Nicotiana benthamiana) leaf epidermal cells via Agrobacterium tumefaciens-mediated infiltration. Samples were stained using FM4-64FX, a plasma membrane-specific dye. Confocal microscopy showed an overlap signal (yellow) of the GFP fluorescence (green) and the FM4-64FX fluorescence (red) in the cells expressing the 2×35S::SlHAK10::eGFP chimeric gene (Fig. 3B), indicating that AM-specific SlHAK10 might encode a plasma membrane-localized K+ transporter.
Figure 3.
Subcellular localization analysis of SlHAK10. Confocal laser scanning microscopy images show tobacco leaf epidermal cells transiently expressing 35S::eGFP (A) or 35S::SlHAK10-GFP (B). I, Confocal images under GFP channel only. II, FM4-64FX dye-induced red fluorescence showing the position of the plasma membrane. III, Bright field. IV, Merged images of GFP fluorescence (green) and FM4-64FX fluorescence (red). Bars = 50 μm.
Functional Characterization of SlHAK10 in Yeast
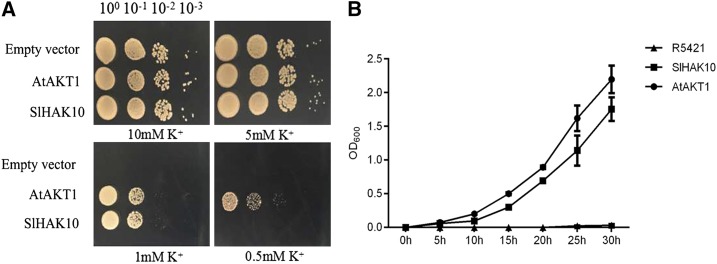
The K+ transport capacity of SlHAK10 was initially evaluated by determining its ability to complement a yeast mutant. Full-length SlHAK10 was introduced into the auxotrophic yeast mutant strain R5421 (MATα ura3-52 leu2 trk1Δ his3Δ200 his4-15 trk2Δ1::pCK64). R5421 is defective in K+ uptake and could not grow under low-K+ culture conditions. AtAKT1, a K+ channel from Arabidopsis, was also transformed into R5421 as a positive control (Hirsch et al., 1998; Wang and Wu, 2013). The yeast growth assays were carried out on arginine phosphate (AP) medium containing different concentrations of K+. It was shown that, under high-K+ conditions (5 and 10 mm), all the tested yeast strains could grow well, and no visible difference was observed between the yeasts transformed with empty vector (pYES2) versus the vector expressing SlHAK10. Along with the decline of K+ concentration from 5 to 1 mm, the growth of R5421 harboring empty vector was almost entirely suppressed. Although the R5421 cells transformed with SlHAK10 were also not able to grow at 0.5 mm K+, the yeast mutant expressing SlHAK10 could indeed rescue its growth defect at 1 mm K+ (Fig. 4A). Growth kinetic analysis showed that the yeast R5421 cells expressing AtAKT1 and SlHAK10 grew much faster than the yeast cells harboring empty vector in liquid AP medium containing 1 mm KCl (Fig. 4B), suggesting that SlHAK10 has K+ transport activity.
Figure 4.
Functional complementation of SlHAK10 for K+ acquisition in the K+ uptake-defective yeast strain R5421. A, Growth status of R5421 cells expressing AtAKT1, empty vector (pYES2), and SlHAK10 on AP medium supplemented with 10, 5, 1, and 0.5 mm KCl. Each transformant was precultured in liquid AP medium containing 100 mm KCl. The 1:10 serial dilutions of yeast cells were spotted on the AP medium and then incubated at 30°C for 6 d. B, Growth kinetic analysis of the yeast R5421 cells expressing AtAKT1, empty vector, and SlHAK10 cultured in liquid AP medium containing 1 mm KCl. Errors bars represent sd of five biological replicates.
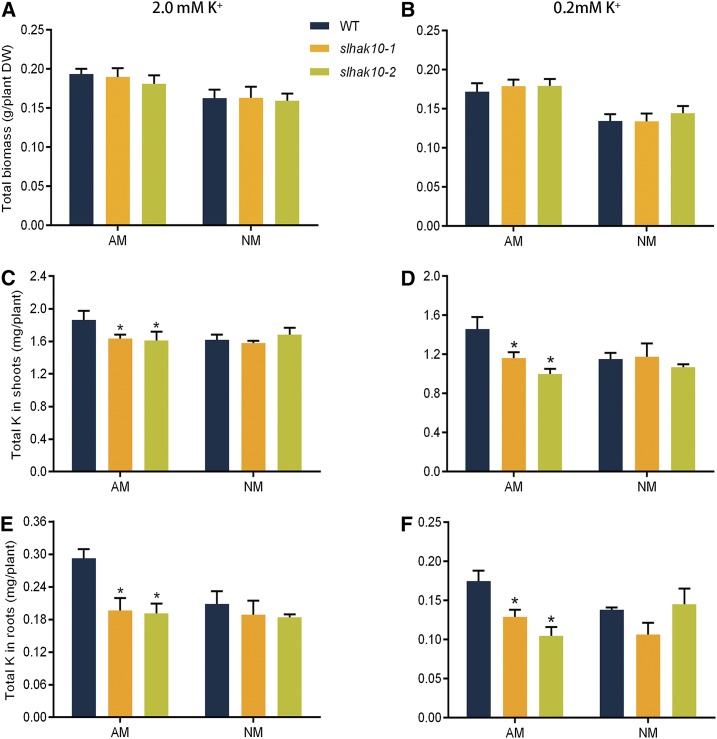
Loss-of-Function Mutation of SlHAK10 Decreases Mycorrhizal K+ Uptake But Does Not Affect Arbuscule Morphology
As SlHAK10 is an AM-responsive K+ transporter in tomato, it is reasonable to speculate that SlHAK10 might have a contribution to the symbiotic K+ uptake and/or AM formation. To test this hypothesis, we first generated mutant lines of SlHAK10 by the CRISPR-Cas9 system. Two independent homozygous mutant lines with one nucleotide deletion and insertion, respectively, in the coding sequence of SlHAK10 were identified (Supplemental Fig. S6). The two mutant lines and wild-type plants were cultivated in a compartmented culture system containing a middle root/fungal compartment (RFC) and two hyphal compartments (HCs; Supplemental Fig. S7A). The RFC and HC were separated by 30-μm nylon mesh. All three compartments were filled with a 4:1 mixture of sterilized sand and low-K+ soil (the soil contains 30.2 mg kg−1 available K and 18.5 mg kg−1 available Pi). Two plants (mutant or wild type) were grown in the RFC in the presence or absence of R. irregularis for 5 weeks. All the plants were watered weekly with nutrient solution containing either 2 mm K+ as a high-K+ condition or 0.2 mm K+ as a low-K+ condition. To monitor whether fungal hyphae could reach and take up nutrients from HCs, additional 15N-labled Ca(NO3)2 was introduced into the two HCs by watering. Compared with the inoculated wild-type and mutant plants, 15N accumulation in all the noninoculated plants was extremely low (Supplemental Fig. S7B), suggesting that fungal hyphae can absorb nutrients from HCs and almost no ion diffusion across the nylon meshes occurred. In all the treatments, the mutant plants showed a comparable total biomass with the wild-type plants (Fig. 5, A and B). No significant difference in the K+ accumulation could be detected between the NM wild-type and mutant plants grown under both the K+ supply conditions (Fig. 5, C–F), while the K+ contents decreased by nearly 20% in both shoots and roots of the mycorrhizal mutant plants compared with those in the mycorrhizal wild-type plants (Fig. 5, D and F). As it is well established that AM fungal colonization has a dominant contribution to plant Pi nutrition, the Pi contents in these plants were also determined. As expected, all the inoculated wild-type and mutant plants contained significantly higher Pi in both shoots and roots than those in the NM plants (Supplemental Fig. S8, A–D). The Pi contents in mycorrhizal mutant plants grown under low K+ were significantly decreased compared with those in the mycorrhizal wild-type plants (Supplemental Fig. S8, B and D). The mycorrhizal mutant plants also contained a significantly lower Pi in their roots than the mycorrhizal wild-type plants grown under high K+ (Supplemental Fig. S8C).
Figure 5.
Effects of mutation of SlHAK10 on the total biomass and K+ accumulation in tomato grown under a compartmented culture condition. All the compartments were filled with a 4:1 mixture of sand and low-K+ soil (the soil contains 30.2 mg kg−1 available K and 18.5 mg kg−1 available Pi). Wild-type (WT) and mutant plants were grown in the RFC for AM fungal inoculation. Plants were watered weekly with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. The total biomass (A and B) and K+ contents in shoots (C and E) and roots (D and F) of the wild-type and mutant plants were determined at 5 weeks postinoculation. AM, Mycorrhizal plants; DW, dry weight; NM, nonmycorrhizal plants. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
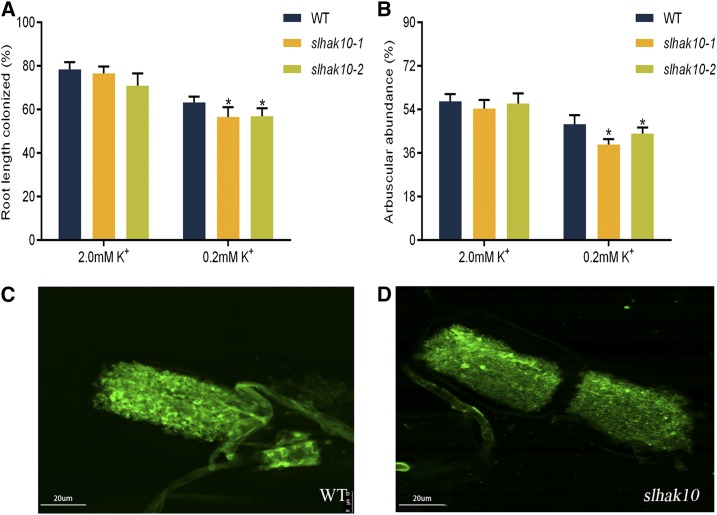
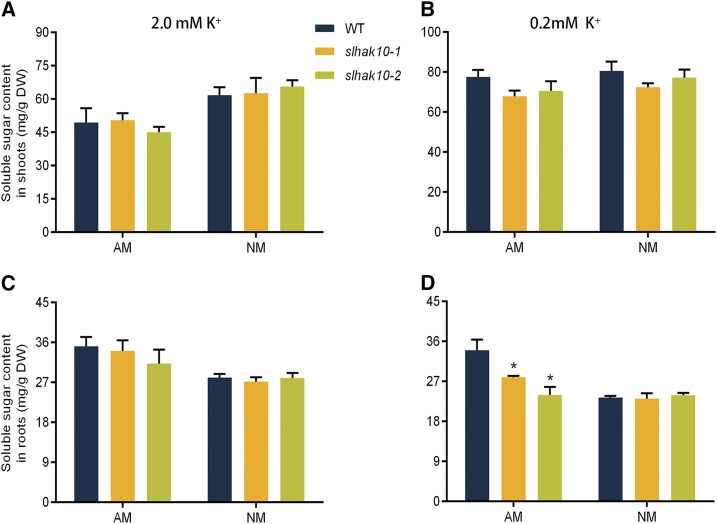
To determine whether SlHAK10 might be involved in the regulation of AM symbiosis, the degree of AM colonization in the mycorrhizal roots of the wild type and mutant lines was assessed. Compared with wild-type plants, both the total colonization rate and arbuscule abundance were significantly reduced in the mutant lines grown under the low-K+ supply condition (Fig. 6, A and B), while the arbuscule morphology in the mutant lines was not significantly impaired (Fig. 6, C and D). These findings suggest that SlHAK10 is involved in mycorrhizal K+ uptake but is not essential for arbuscule development. Several recent studies have pointed to an important role of the host-derived fatty acids in maintaining AM symbiosis (Bravo et al., 2017; Jiang et al., 2017, 2018; Keymer et al., 2017; Luginbuehl et al., 2017). The assay of fatty acid contents in the mycorrhizal roots of wild-type and slhak10 mutant plants showed no significant difference under both the K+ supply conditions (Supplemental Fig. S9). A further investigation of the soluble sugar content revealed that in the absence of AM fungal colonization, both the wild type and slhak10 mutants supplied with low K+ contained significantly higher soluble sugar in their shoots, but a lower amount in their roots, compared with plants grown under high K+ (Fig. 7). The soluble sugar content in the mycorrhizal roots of mutant plants was significantly lower than that in the wild-type mycorrhizal roots under the low-K+ supply condition (Fig. 7, C and D).
Figure 6.
Effects of mutation of SlHAK10 on the AM colonization and arbuscule morphology. Wild-type (WT) and mutant plants were grown in a compartmented culture system. Plants were watered weekly with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. The total root length colonization (A) and arbuscule abundance (B) in the mycorrhizal roots of wild-type and mutant plants were assessed at 5 weeks postinoculation. Arbuscules were stained with WGA-AF-488 in the mycorrhizal roots of wild-type (C) and mutant (D) plants. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
Figure 7.
The contents of soluble sugar in the wild-type (WT) and slhak10 mutant plants. Wild-type and mutant plants were grown in a compartmented culture system. Plants were watered weekly with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. The soluble sugar concentrations in the shoots (A and B) and roots (C and D) of wild-type and mutant plants were determined at 5 weeks postinoculation. AM, Mycorrhizal plants; DW, dry weight; NM, nonmycorrhizal plants. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
Overexpressing SlHAK10 Improves K+ Uptake and Mycorrhizal Colonization under K+ Deficiency Conditions
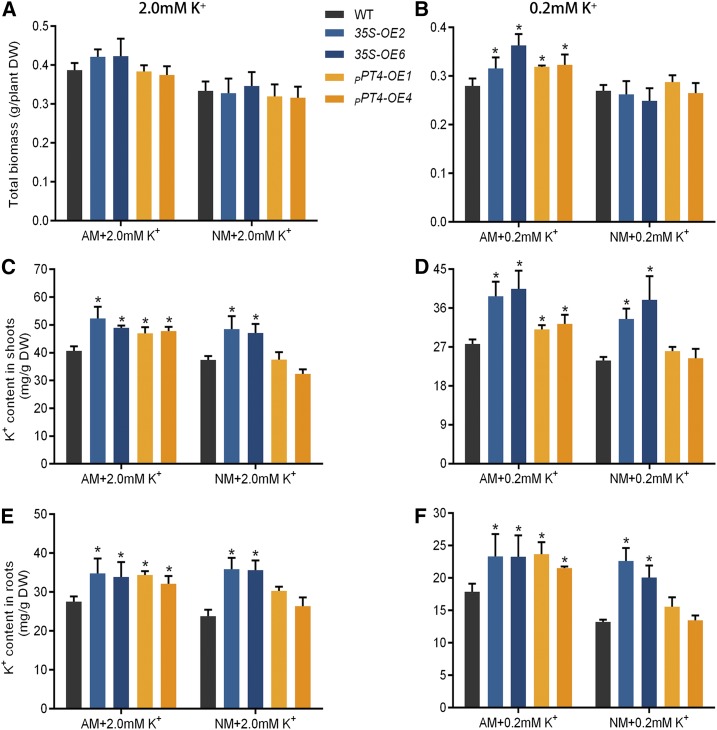
To assess whether overexpression of SlHAK10 can facilitate K+ uptake, we further generated transgenic tomato plants overexpressing SlHAK10 under the control of either the CaMV35S promoter or the AM-specific promoter pSmPT4. We have previously proven that pSmPT4, the promoter of an eggplant (Solanum melongena) Pi transporter, could drive GUS reporter expression exclusively in AM fungus-colonized cells (Chen et al., 2007, 2011). RT-qPCR analysis showed that the expression of SlHAK10 was greatly increased in the mycorrhizal roots of the overexpression (OE) lines (Supplemental Fig. S10). Four independent transgenic lines (35S-OE2 and 35S-OE6, overexpression driven by the CaMV35S promoter; and pSmPT4-OE1 and pSmPT4-OE4, overexpression driven by the pSmPT4 promoter) were thus selected for analysis of K+ absorption in sand-based pot culture inoculated with R. irregularis for 5 weeks. All the plants were irrigated with nutrient solution containing either 2 mm K+ as a high-K+ condition or 0.2 mm K+ as a low-K+ condition. In the presence of AM colonization, overexpressing SlHAK10 could significantly increase the biomass of plants grown under the low-K+ condition (Fig. 8, A and B). Compared with wild-type plants, all the 35S-OE lines showed a significantly higher K+ accumulation in both shoots and roots under both K+ supply conditions, irrespective of the presence or absence of AM inoculation (Fig. 8, C–F). It is worth noting that the NM 35S-OE plants showed a comparable K content in both shoots and roots to the mycorrhizal 35S-OE plants, suggesting that constitutive overexpression of SlHAK10 may mask the contribution of the mycorrhizal K uptake pathway mediated by the native SlHAK10. This speculation seems to be reasonable, as the transcripts of SlHAK10 were increased nearly 100-fold in the mycorrhizal roots of 35S-OE lines (Supplemental Fig. S10). With respect to the pSmPT4-OE lines, increased K+ uptake, as indicated by significantly higher shoot and root K+ contents compared with the wild-type plants, could be detected only in the mycorrhizal plants. As expected, no significant difference in either plant biomass or K+ content could be detected between the NM wild-type and pSmPT4-OE plants grown under both the K+ conditions (Fig. 8).
Figure 8.
Effects of the overexpression of SlHAK10 on tomato growth and K+ acquisition under a sand-based pot culture condition. For 35S-OE, SlHAK10 was overexpressed under the control of the constitutive CaMV35S promoter; for pPT4-OE, SlHAK10 was overexpressed under the control of the mycorrhiza-specific Pi transporter promoter pSmPT4. Two-week-old seedlings were grown in sand-based pot culture for AM fungal inoculation for 5 weeks. The plants were treated with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. AM, Mycorrhizal plants; DW, dry weight; NM, nonmycorrhizal plants. A and B, Total biomass. C and D, K+ concentration in shoots. E and F, K+ concentration in roots. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
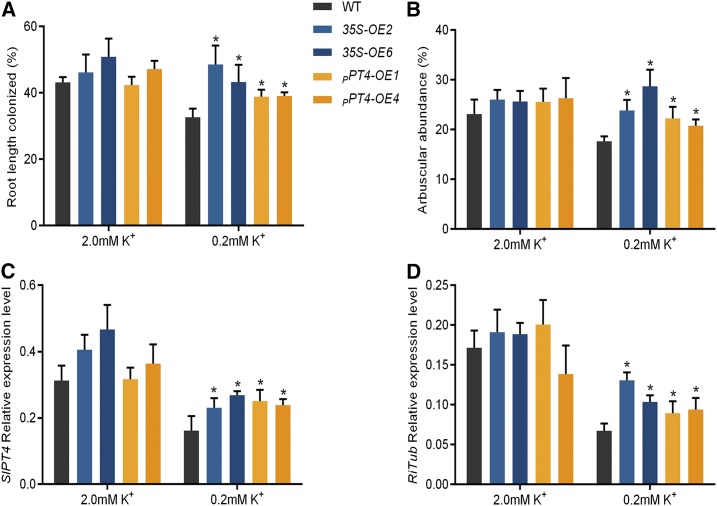
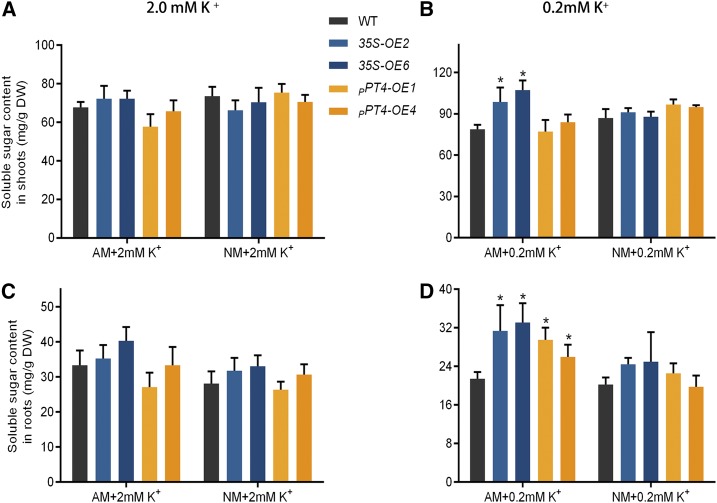
The wild type plants grown under low-K+ conditions showed a significant decrease in AM colonization compared with the plants grown under the high-K+ conditions (Fig. 9). Overexpressing SlHAK10 from either promoter significantly improved the total fungal colonization and arbuscule abundance under the low-K+ condition. Consistent with this, the Pi contents in the mycorrhizal overexpressing lines grown under low K+ were also significantly increased compared with the wild-type plants (Supplemental Fig. S11). The increased mycorrhizal colonization in the roots of SlHAK10-OE plants could also be seen in the increased expression of SlPT4 and RiTub (Fig. 9, C and D), the two marker genes used for evaluating AM colonization (Floss et al., 2013; Liao et al., 2015). A further assay of the soluble sugar contents showed that the SlHAK10-OE plants grown under low K+ contained significantly higher soluble sugar contents in their mycorrhizal roots compared with the wild-type plants (Fig. 10).
Figure 9.
Effects of overexpression of SlHAK10 on AM colonization efficiency under a sand-based pot culture condition. Two-week-old seedlings were grown in sand-based pot culture for AM fungal inoculation. The plants were treated with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. A and B, The total root length colonization (A) and arbuscule abundance (B) in the mycorrhizal roots of wild-type (WT) and SlHAK10-overexpressing plants were assessed at 5 weeks postinoculation. C and D, The transcript levels of two AM-related marker genes, SlPT4 (LePT4) and RiTub from tomato and R. irregularis, respectively. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
Figure 10.
The contents of soluble sugar in the wild-type (WT) and SlHAK10-overexpressing plants. Two-week-old seedlings were grown in sand-based pot culture for AM fungal inoculation. The plants were treated with either high-K+ (2 mm K+) or low-K+ (0.2 mm K+) solution. The soluble sugar concentrations in the shoots (A and B) and roots (C and D) of wild-type and SlHAK10-overexpressing plants were determined at 5 weeks postinoculation. DW, Dry weight. Error bars indicate se (n = 5). The asterisks indicate significant differences (P < 0.05).
DISCUSSION
Contributing Effects of AM Fungal Colonization on Tomato Plant Growth and K+ Acquisition
Improving the availability of mineral nutrients for plants is one of the major contributions of AM symbiosis. Although the role of AM symbiosis in plant K+ nutrition received much less attention than Pi and N (Mäder et al., 2000; Nagy et al., 2006; Chen et al., 2007; Javot et al., 2007), an increased K accumulation in distinct tissues upon AM symbiosis has been reported related to diverse plant species (Perner et al., 2007; Baslam et al., 2013; Oliveira et al., 2016). Physiological analysis of the AM-associated responses of M. truncatula plants revealed a significant increase of root biomass and K+ content in AM plants under the K+ deprivation condition (Garcia et al., 2017). In this study, we also observed that the AM tomato plants had a higher root biomass by more than 20% than the NM plants under the low-K+ condition (Fig. 1B). However, in the high-K+ treatment, the AM plants showed more growth advantages, with shoot and root biomass increased by nearly 40% compared with the NM control plants (Fig. 1, A and B). Moreover, inoculation with AM fungus resulted in a much higher K+ accumulation in AM plants than the NM plants, irrespective of K+ regimen (Fig. 1, C and D). These findings highlighted the positive roles of AM fungal colonization in tomato plant growth and K+ acquisition. However, it could not be ruled out that the increased K+ accumulation in mycorrhizal tomato plants might be a partial consequence of an improved Pi uptake, as earlier studies have proposed K+ to be one of the major counter-ions of polyphosphates, which forms a phosphate reserve for the host plant in AM symbiosis (Bücking and Heyser, 1999). A recent study of the K nutrition of ectomycorrhizal Pinus pinaster showed that altered K+ transfer toward the host plant could affect fungal Pi translocation to the shoots, suggesting that interactions would also take place during the uptake of K+ and Pi by ectomycorrhizal plants (Garcia et al., 2014).
SlHAK10 Is Involved in the Mycorrhizal K+ Uptake at the Intraradical Symbiotic Interface
The increased K+ accumulation in mycorrhizal tomato plants is a strong hint of the presence of an AM symbiotic pathway for K+ uptake in tomato. SlHAK10, the AM-induced K+ transporter, was proposed to be the suitable candidate for K+ uptake at the symbiotic interface, as its expression was specifically confined in the cells containing arbuscules (Fig. 2). Its capacity in K+ transport was demonstrated in both the heterologous yeast system and transgenic plants. Overexpression of SlHAK10 with the constitutive CaMV35S promoter led to a significant increase of K+ uptake under both the high-K+ (2 mm) and low-K+ (0.2 mm) conditions, regardless of the presence or absence of AM fungal colonization (Fig. 8).
The evidence for the action of SlHAK10 in mycorrhizal K+ uptake was obtained through the analysis of slhak10 mutants in the compartmented culture in the inoculation condition. Knockout of SlHAK10 significantly reduced the uptake of K+ from the HCs, resulting in lower K+ contents in both shoots and roots of the slhak10 mutants relative to the wild-type plants (Fig. 5). More indirect evidence was obtained through the overexpression of SlHAK10 in tomato under the control of a strong, AM-specific promoter, pSmPT4. Overexpressing SlHAK10 exclusively in mycorrhizal roots resulted in a significant increase of K+ content in both shoots and roots of the mycorrhizal transgenic plants compared with the wild-type plants (Fig. 8). Interestingly, the 35S-OE mycorrhizal plants seemed to have a higher K+ content in shoots than the pSmPT4-OE plants grown under low K+ (Fig. 8D). Such a discrepancy in K+ accumulation might be caused by different expression efficiency and tissue localization of SlHAK10 controlled by the two kinds of promoters. A previous study demonstrated that blocking mycorrhizal Pi transport by silencing the AM-induced Pi transporter MtPT4 in M. truncatula could severely impair arbuscule formation (Javot et al., 2007). The absence of defective arbuscular morphology in the slhak10 mutants observed in this study (Fig. 5) suggests that transport of K+ across the symbiotic interface might not be a requirement for arbuscule development. Even so, the identification of an orthologue of SlHAK10 in L. japonicus that also showed an inducible response to AM symbiosis (Guether et al., 2009) is evidence that the regulatory mechanism regarding the AM-mediated K+ uptake pathway might be conserved across different plant families.
Improving Plant K+ Nutrition May Facilitate AM Colonization under the K+ Deficiency Condition
The availability of mineral nutrients, in particular Pi, has a strong impact on AM colonization (Nagy et al., 2009; Breuillin et al., 2010; Nouri et al., 2014; Johnson et al., 2015). Pi and N starvation was also characterized as being able to induce a stress-responsive transcriptional reprogramming that is beneficial for AM colonization (Bonneau et al., 2013). However, the effect of plant K+ nutrition on AM colonization has not been well characterized thus far. A recent study regarding the mycorrhizal M. truncatula plants showed that K+ availability did not affect the establishment of AM symbiosis (Garcia et al., 2017). However, the conclusions derived from different symbiotic systems were not always consistent, as in some other studies a positive effect on AM colonization by K+ application has been proposed (Zhang et al., 2017).
In this study, we found that mycorrhizal tomato plants supplied with high K+ had a significantly higher AM colonization rate and arbuscule incidence than the K+-deficient plants (Fig. 1F; Supplemental Fig. S1). Previous studies have shown that in K+ starvation plants, the export of carbohydrates from source leaves and the allocation to roots were heavily interrupted, leading to a severe repression in root growth and a typical decrease in root-shoot ratio (Cakmak et al., 1994a, 1994b; Deeken et al., 2002; Hermans et al., 2006; Kanai et al., 2007; Pettigrew, 2008; Cai et al., 2012). In tomato, it has been revealed that K+ deficiency caused a decreased sugar accumulation in roots (Kanai et al., 2007). In this study, we also observed a significant increase of soluble sugar contents in shoots but a decrease in roots of both the wild type and mutants grown under K+ deficiency (Fig. 7). Additionally, overexpressing SlHAK10 by either the constitutive CaMV35S promoter or the AM-specific pSmPT4 promoter under K+ deficiency led to not only an improvement in K+ absorption but also a significant increase in AM colonization and soluble sugar accumulation in roots (Figs. 8–10). Considering that maintenance of AM symbiosis requires the transfer of substantial amounts of plant-fixed carbohydrates to AM fungi, it is reasonable to speculate that improving K+ uptake under K+ deficiency may promote carbohydrate allocation to roots, which facilitates root growth and AM colonization. Such a hypothesis could gain indirect support from an earlier study reporting that silencing a fungal monosaccharide transporter, MST2, with a broad substrate spectrum, impaired mycorrhiza formation in M. truncatula mycorrhizal roots (Helber et al., 2011).
In conclusion, in this study, we characterized the potential interactions between K+ nutrition and AM symbiosis in tomato and assessed the function of a K+ transporter, SlHAK10, in both the heterologous yeast system and transgenic plants. We demonstrated that there is a SlHAK10-mediated mycorrhizal K+ uptake pathway in tomato and that improving plant K+ nutrition under the low-K+ condition may increase carbohydrate accumulation in roots and facilitate AM fungal colonization. As a mutually stimulating mechanism has been revealed to exist during the simultaneous exchange of C and Pi/N between the AM partners, it would be interesting to investigate in the future whether the free-market model is also applicable to the exchange of K and C during AM symbiosis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum ‘MicroTom’) plants were used in this study. Seeds of wild-type and transgenic plants were surface sterilized with 10% (v/v) sodium hypochlorite for 10 min and then germinated in a growth chamber programmed for 16 h of light at 28°C and 8 h of dark at 20°C. The seedlings were then transplanted to sterilized quartz sand for a 2-week culture irrigated with full-strength nutrient solution containing the following: 1 mm NH4+, 4 mm NO3−, 2 mm K+, 1 mm Pi, 0.75 mm Ca2+, 0.5 mm Mg2+, 0.25 mm Cl−, 0.5 mm SO42−, 20 μm Fe2+, 9 μm Mn2+, 46 μm BO33−, 8 μm Zn2+, 3 μm Cu2+, and 0.03 μm MoO42−. The plantlets were then transferred to either sand-based pot culture or a compartmented culture system for inoculation with AM fungi.
For sand-based pot culture, three plantlets were transplanted to a 3-dm3 pot filled with sterilized sand. A sand-based mycorrhizal inoculum containing Rhizophagus irregularis (previously named Glomus intraradices) was used for colonization. Each plantlet was inoculated with 1.2 g of inoculum containing approximately 200 spores around the roots. The NM control plants were obtained by inoculation with autoclaved inoculum. The plants were supplied with nutrient solution including either 2 mm K+ as a high-K+ treatment or 0.2 mm K+ as a low-K+ treatment. To guarantee high mycorrhizal colonization, the plants in pot culture were watered with a relatively low concentration (50 μm) of Pi.
A compartmented culture system was employed to investigate the contribution of SlHAK10 to mycorrhizal K uptake by using two loss-of-function slhak10 mutant lines. The culture system contains an RFC and two HCs (Supplemental Fig. S7A). All three compartments were filled with a 4:1 mixture of sand and soil (the soil contains 30.2 mg kg−1 available K and 18.5 mg kg−1 available Pi). Two mutant lines or wild-type plants were grown in the RFC in the presence or absence, respectively, of R. irregularis inoculation for 5 weeks. All the plants were watered weekly with nutrient solution containing either 2 mm K+ as a high-K+ condition or 0.2 mm K+ as a low-K+ condition. To prevent ion diffusion between the RFC and HCs, the RFC was separated from the HCs by two iron gauzes with a 0.5-cm air gap. A 30-μm nylon mesh was placed between the two iron gauzes to prevent roots from entering the HCs. To monitor whether fungal hyphae could reach and take up nutrients from HCs, additional 15N-labeled Ca(NO3)2 was introduced into the two HCs by watering, and 15N accumulation in the mycorrhizal plants and NM plants was determined.
RT-qPCR
For performing RT-qPCR analysis, approximately 2 μg of total RNA from each sample was used to synthesize cDNA using a reverse transcription kit (Takara), and the synthesized cDNAs were used as templates for the following RT-qPCR analysis. The PCR was conducted on an Applied Biosystems Plus Real-Time PCR System using the SYBER premix ExTaq kit (Takara). The specific primer pairs for each of the HAK genes are listed in Supplemental Table S1. The relative transcript abundance of each target gene was standardized to the transcript level of a tomato constitutive Actin gene (Chen et al., 2014).
Identification of HAK Genes in Tomato
Members of the KT/KUP/HAK gene family in the tomato genome were identified using the BLASTN and TBLASTN algorithms. To identify the potential HAK genes in the tomato genome, the coding sequence of the AM-induced Lotus japonicus K+ transporter (Lj4g3v3116360.1; Guether et al., 2009) was employed for the BLAST search against the tomato genomic sequence database (www.sgn.cornell.edu). Sequences with a query over 50% and E value less than −10 were taken as the candidates. All obtained sequences were submitted to the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) for further confirmative analysis.
Subcellular Localization Analysis
For subcellular localization analysis of SlHAK10 by agroinfiltration of tobacco (Nicotiana benthamiana) leaves, the full-length coding sequence of SlHAK10 without the stop codon was first ligated to an intermediate vector, pSAT6AEGFP-N1. The SlHAK10-GFP fusion construct as well as GFP alone were then subcloned into the expression vector pRCS2-ocs-nptII with the rare restriction enzyme PI-PspI. The plasmids were then transformed into the Agrobacterium tumefaciens strain EHA105. The bacterial cells were harvested by centrifugation and resuspended in a solution (pH 5.7) containing 10 mm MES, 10 mm MgCl2, and 200 µm acetosyringone. Cell suspensions at an optical density (600 nm) of 0.1 were infiltrated into the leaves of tobacco using a needle-free syringe. GFP fluorescence in the transformed leaves was imaged using confocal microscopy (Leica Confocal TCS-SP8) after 48 to 72 h.
Construction of a Binary Vector and Plant Transformation
To construct the SlHAK10 promoter vector, a 2,281-bp-long promoter fragment of SlHAK10 immediately upstream of the translation start ATG was amplified by PCR to introduce HindIII and BamHI restriction sites at the end of the 5′ and 3′ regions, respectively. After digestion with the two restriction enzymes, the amplified fragments were cloned into binary vector pBI121 to replace the CaMV35S promoter in front of the GUS reporter gene. The generated vector was used for tomato transformation by the EHA105 strain.
To construct the SlHAK10 constitutive overexpression vector, the coding sequence of SlHAK10 was amplified and cloned into the binary vector pCAMBIA 1300 using the ClonExpress II One Step Cloning Kit (Vazyme Biotech). The resulting construct (named 35S-SlHAK10), in which SlHAK10 was controlled by the CaMV35S promoter, was introduced into A. tumefaciens EHA105 strain for transformation of tomato and rice (Oryza sativa) plants. To construct the SlHAK10 AM-specific overexpression vector, an 865-bp-long promoter fragment of the AM-specific Pi transporter gene SmPT4 (Chen et al., 2011) was amplified and cloned into the 35S-SlHAK10 construct to replace the CaMV35S promoter. The recombinant construct (named pSmPT4-SlHAK10) was also introduced into the EHA105 strain for transformation of tomato plants.
The CRISPR/Cas9 gene knockout constructs were generated using the commercial vector VK005-03 (Viewsolid Biotech). Two spacers were designed to target two different sites in exons of SlHAK10. For site 1, we used the oligonucleotide pair 5′-TGGATCCTTGCCATATCAAATC-3′ and 5′-AACGATTTGATATGGCAAGGAT-3′, and for site 2, we used the oligonucleotides 5′-TGGACAGACCTAAGTCCCGGACT-3′ and 5′-AACAGTCCGGGACTTAGGTCTGT-3′. The oligonucleotide pairs were first annealed to produce a double-stranded fragment with three-nucleotide 5′ overhangs at both ends and then ligated into the linearized vector VK005-03.
A. tumefaciens-mediated transformation of tomato plants was performed according to the protocol described by Sun et al. (2006). The transformation of rice plants was carried out as described previously (Upadhyaya et al., 2000).
Functional Characterization of SlHAK10 in Yeast
The full-length coding sequences of SlHAK10 and AtAKT1 were inserted into yeast expression vector p-YESII under the control of the Met promoter (Bernstein et al., 2013). The resulting vectors were transformed into the yeast mutant strain R5421 (MATα ura3-52 leu2 trk1Δ his3Δ200 his4-15 trk2Δ1::pCK64), in which two endogenous K+ transporter genes (TRK1 and TRK2) were deleted (Li et al., 2014). The yeast cell transformation was performed using the LiAC/single-stranded DNA/polyethylene glycol method, and the growth assays on AP medium were performed as described by Horie et al. (2011).
Determination of the Concentrations of K, P, and Soluble Sugars
The dried plant material was digested with 98% H2SO4 and 30% hydrogen peroxide as described previously (Chen et al., 2007). The K+ concentration in the solution was measured by an ICP emission spectrometer (Optima 2100DV; Perkin-Elmer) as described by Cai et al. (2012). The total Pi concentration was measured using the Molybdate Blue method as described previously (Chen et al., 2007). The measurement of soluble sugar concentrations was conducted based on the method of Hansen and Møller (1975), as described in detail by Chen et al. (2015).
Analysis of Fatty Acid Contents
Extraction of fatty acid methyl esters from tomato roots was performed using the modified method of Browse et al. (1986) and Jiang et al. (2017). Briefly, 50 mg of root samples was finely ground in liquid N and then transferred into 2-mL centrifugation tubes containing 1 mL of 5% (v/v) H2SO4 in methanol. The samples were then heated to 80°C for 1 h. After cooling, extraction solution containing 30 μL of methyl nonadecanoate, 0.3 mL of hexane, and 1 mL of 0.9% NaCl was added to the samples and then shaken vigorously for 5 min. After centrifugation, the hexane phase containing the fatty acid methyl esters was collected and used for determination by gas chromatography-mass spectrometry (GC-FID, 7890A Plus GC; Agilent). The parameters for the gas chromatography-mass spectrometry assay were used as described by Jiang et al. (2017).
Detection of Mycorrhizal Colonization and Histochemical GUS Staining
The examination of mycorrhizal fungal colonization was performed using the magnified line intersection method (McGonigle et al., 1990). Root samples were cut randomly into 1-cm pieces and cleared for 3 h at 70°C in 10% (w/v) KOH solution, and then the root segments were stained in 0.3% Trypan Blue at 90°C for 2 h. Ten 1-cm root segments were mounted on microscope slides, and each of these segments was investigated at 10 randomly chosen spots, which were scored for the presence of hyphae, arbuscule, vesicle, or none (absence of any fungal structures). For each root system, three to four microscope slides were employed. The total colonization level (root length colonization percentage) was calculated by the formula (number of intersections cutting through any fungal structures)/(total number of intersections examined); and the arbuscular colonization level (arbuscule abundance percentage) was calculated by the formula (number of intersections cutting through at least one arbuscule)/(total number of intersections examined). At least three different root systems were evaluated for each genotype. Histochemical staining of the GUS activity in transgenic plants was performed as described previously (Karandashov et al., 2004).
Statistical Analysis
The data were analyzed by ANOVA (SPSS 16.0; SPSS), followed by Tukey’s test (P < 0.05) to test differences between different plant genotypes and treatments. The data represent means ± se of five independent biological replicates.
Accession Numbers
Sequence data from this article can be found in the aramemnon data libraries (http://aramemnon.botanik.uni-koeln.de/) under the following accession numbers: SlHAK1 (Solyc08g015680), SlHAK2 (Solyc06g050170), SlHAK3 (Solyc12g096580), SlHAK4 (Solyc12g017910), SlHAK5 (Solyc12g005670), SlHAK6 (Solyc02g087000), SlHAK7 (Solyc04g025880), SlHAK8 (Solyc05g005740), SlHAK9 (Solyc10g047270), SlHAK10 (Solyc03g097860), SlHAK11 (Solyc01g105150), SlHAK12 (Solyc05g005720), SlHAK13 (Solyc09g074800), SlHAK14 (Solyc04g008450), SlHAK15 (Solyc09g074790), SlHAK16 (Solyc12g005680), SlHAK17 (Solyc06g051830), SlHAK18 (Solyc02g031840), SlHAK19 (Solyc02g068590), SlHAK20 (Solyc04g025990), and SlHAK21 (Solyc09g074820).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of different K+ applications and AM fungal colonization on root-shoot ratio and K+ uptake.
Supplemental Figure S2. Effects of different K+ applications on AM colonization.
Supplemental Figure S3. Phylogenetic analysis and exon/intron organization of tomato HAK transporters.
Supplemental Figure S4. Expression analysis of tomato HAK genes in response to AM symbiosis.
Supplemental Figure S5. Tissue-specific expression analysis of SlHAK10.
Supplemental Figure S6. Screening the homozygous mutant lines of SlHAK10 by PCR sequencing.
Supplemental Figure S7. Effects of mutation of SlHAK10 on tomato growth under a compartmented culture condition.
Supplemental Figure S8. Effects of mutation of SlHAK10 on mycorrhizal Pi uptake in tomato plants grown under a compartmented culture condition.
Supplemental Figure S9. Fatty acid contents in the mycorrhizal roots of wild-type and slhak10 mutant plants.
Supplemental Figure S10. SlHAK10 is overexpressed in mycorrhizal roots of transgenic tomato plants.
Supplemental Figure S11. Effects of overexpression of SlHAK10 on tomato Pi acquisition under a sand-based pot culture condition.
Supplemental Table S1. List of the primers used for RT-qPCR analysis in this study.
Acknowledgments
We thank Dr. Rick Gaber from Northwestern University for kindly providing the yeast mutant R5421.
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (31572188, 31372121, and 31272225) and the Fundamental Research Funds for the Central Universities (KYZ201868), by the Graduate Student Scientific Research and Innovation Projects in Jiangsu Province (KYZZ16_0379), the Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource Utilization, and the Innovative Research Team Development Plan of the Ministry of Education of China (IRT1256).
Articles can be viewed without a subscription.
References
- Allen JW, Shachar-Hill Y (2009) Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol 149: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrutha RN, Sekhar PN, Varshney RK, Kishor PB (2007) Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci 172: 708–721 [Google Scholar]
- Aranda-Sicilia MN, Aboukila A, Armbruster U, Cagnac O, Schumann T, Kunz HH, Jahns P, Rodríguez-Rosales MP, Sze H, Venema K (2016) Envelope K+/H+ antiporters AtKEA1 and AtKEA2 function in plastid development. Plant Physiol 172: 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslam M, Garmendia I, Goicoechea N (2013) The arbuscular mycorrhizal symbiosis can overcome reductions in yield and nutritional quality in greenhouse-lettuces cultivated at inappropriate growing seasons. Sci Hortic 164: 145–154 [Google Scholar]
- Bernstein JD, Okamoto Y, Kim M, Shikano S (2013) Potential use of potassium efflux-deficient yeast for studying trafficking signals and potassium channel functions. FEBS Open Bio 3: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau L, Huguet S, Wipf D, Pauly N, Truong HN (2013) Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol 199: 188–202 [DOI] [PubMed] [Google Scholar]
- Bravo A, Brands M, Wewer V, Dörmann P, Harrison MJ (2017) Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol 214: 1631–1645 [DOI] [PubMed] [Google Scholar]
- Breuillin F, Schramm J, Hajirezaei M, Ahkami A, Favre P, Druege U, Hause B, Bucher M, Kretzschmar T, Bossolini E, et al. (2010) Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J 64: 1002–1017 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Bücking H, Heyser W (1999) Elemental composition and function of polyphosphates in ectomycorrhizal fungi: An x-ray microanalytical study. Mycol Res 103: 31–39 [Google Scholar]
- Cai J, Chen L, Qu H, Lian J, Liu W, Hu Y, Xu G (2012) Alteration of nutrient allocation and transporter genes expression in rice under N, P, K, and Mg deficiencies. Acta Physiol Plant 34: 939–946 [Google Scholar]
- Cakmak I, Hengeler C, Marschner H (1994a) Partitioning of shoot and root dry matter and carbohydrates in bean plants suffering from phosphorus, potassium and magnesium deficiency. J Exp Bot 45: 1245–1250 [Google Scholar]
- Cakmak I, Hengeler C, Marschner H (1994b) Changes in phloem export of sucrose in leaves in response to phosphorus, potassium and magnesium deficiency in bean plants. J Exp Bot 45: 1251–1257 [Google Scholar]
- Casieri L, Ait Lahmidi N, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, Courty PE, Garcia K, Charbonnier M, Delteil A, et al. (2013) Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza 23: 597–625 [DOI] [PubMed] [Google Scholar]
- Cellier F, Conéjéro G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F (2004) Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J 39: 834–846 [DOI] [PubMed] [Google Scholar]
- Chen A, Hu J, Sun S, Xu G (2007) Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831 [DOI] [PubMed] [Google Scholar]
- Chen A, Gu M, Sun S, Zhu L, Hong S, Xu G (2011) Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol 189: 1157–1169 [DOI] [PubMed] [Google Scholar]
- Chen A, Chen X, Wang H, Liao D, Gu M, Qu H, Sun S, Xu G (2014) Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol 14: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hu Q, Luo L, Yang T, Zhang S, Hu Y, Yu L, Xu G (2015) Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ 38: 2747–2765 [DOI] [PubMed] [Google Scholar]
- Courty PE, Doidy J, Garcia K, Wip D, Zimmermann SD (2016) The transportome of mycorrhizal systems. In Martin F, ed, Molecular Mycorrhizal Symbiosis. Wiley Blackwell Publishing, Hoboken, NJ [Google Scholar]
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May ST, Hedrich R (2002) Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334–344 [DOI] [PubMed] [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorilli V, Catoni M, Miozzi L, Novero M, Accotto GP, Lanfranco L (2009) Global and cell-type gene expression profiles in tomato plants colonized by an arbuscular mycorrhizal fungus. New Phytol 184: 975–987 [DOI] [PubMed] [Google Scholar]
- Floss DS, Levy JG, Lévesque-Tremblay V, Pumplin N, Harrison MJ (2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 110: E5025–E5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HH, Luan S (1998) AtKuP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 10: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K, Zimmermann SD (2014) The role of mycorrhizal associations in plant potassium nutrition. Front Plant Sci 5: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K, Delteil A, Conéjéro G, Becquer A, Plassard C, Sentenac H, Zimmermann S (2014) Potassium nutrition of ectomycorrhizal Pinus pinaster: Overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of both K+ and phosphorus in the host plant. New Phytol 201: 951–960 [DOI] [PubMed] [Google Scholar]
- Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty PE (2016) Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci 21: 937–950 [DOI] [PubMed] [Google Scholar]
- Garcia K, Chasman D, Roy S, Ané JM (2017) Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol 173: 1811–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F (2012) Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J 69: 510–528 [DOI] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M, Tolosano M, Volpe V, Kopriva S, Bonfante P (2014) Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol 204: 609–619 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar-Hill Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823 [DOI] [PubMed] [Google Scholar]
- Grabov A. (2007) Plant KT/KUP/HAK potassium transporters: Single family-multiple functions. Ann Bot 99: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Güimil S, Chang HS, Zhu T, Sesma A, Osbourn A, Roux C, Ioannidis V, Oakeley EJ, Docquier M, Descombes P, et al. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA 102: 8066–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q (2008) KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genomics 280: 437–452 [DOI] [PubMed] [Google Scholar]
- Han M, Wu W, Wu WH, Wang Y (2016) Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol Plant 9: 437–446 [DOI] [PubMed] [Google Scholar]
- Hansen J, Møller I (1975) Percolation of starch and soluble carbohydrates from plant tissue for quantitative determination with anthrone. Anal Biochem 68: 87–94 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N (2011) A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23: 3812–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI (2011) K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol 156: 1493–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D, et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie Q, Wang W, Yang J, Zhang X, Yu N, Zhou Y, Wang E (2018) Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol Plant 11: 1344–1359 [DOI] [PubMed] [Google Scholar]
- Jin H, Liu J, Liu J, Huang X (2012) Forms of nitrogen uptake, translocation, and transfer via arbuscular mycorrhizal fungi: A review. Sci China Life Sci 55: 474–482 [DOI] [PubMed] [Google Scholar]
- Johnson NC, Wilson GW, Wilson JA, Miller RM, Bowker MA (2015) Mycorrhizal phenotypes and the law of the minimum. New Phytol 205: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Kanai S, Ohkura K, Adu-Gyamfi JJ, Mohapatra PK, Nguyen NT, Saneoka H, Fujita K (2007) Depression of sink activity precedes the inhibition of biomass production in tomato plants subjected to potassium deficiency stress. J Exp Bot 58: 2917–2928 [DOI] [PubMed] [Google Scholar]
- Karandashov V, Nagy R, Wegmüller S, Amrhein N, Bucher M (2004) Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 101: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer A, Pimprikar P, Wewer V, Huber C, Brands M, Bucerius SL, Delaux PM, Klingl V, Röpenack-Lahaye EV, Wang TL, et al. (2017) Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6: 29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, et al. (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Tamura Y, Takai S, Banba M, Hata S (2010) Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol 51: 1411–1415 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Ohmori Y, Saito C, Yano K, Ohtomo R, Fujiwara T (2016) Phosphate treatment strongly inhibits new arbuscule development but not the maintenance of arbuscule in mycorrhizal rice roots. Plant Physiol 171: 566–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegel S, Ait Lahmidi N, Arnould C, Chatagnier O, Walder F, Ineichen K, Boller T, Wipf D, Wiemken A, Courty PE (2013) The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol 198: 853–865 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Gierth M, Herdean A, Satoh-Cruz M, Kramer DM, Spetea C, Schroeder JI (2014) Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc Natl Acad Sci USA 111: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26: 3387–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu G, Alli A, Yu L (2018) Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin Cell Dev Biol 74: 133–141 [DOI] [PubMed] [Google Scholar]
- Liao D, Chen X, Chen A, Wang H, Liu J, Liu J, Gu M, Sun S, Xu G (2015) The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol 56: 674–687 [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ (2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Mäder P, Vierheilig H, Streitwolf-Engel R, Boller T, Frey B, Christiand P, Wiemken A (2000) Transport of 15N from a soil compartment separated by a polytetrafluoroethylene membrane to plant roots via the hyphae of arbuscular mycorrhizal fungi. New Phytol 146: 155–161 [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Nagy R, Karandashov V, Chague V, Kalinkevich K, Tamasloukht M, Xu G, Jakobsen I, Levy AA, Amrhein N, Bucher M (2005) The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J 42: 236–250 [DOI] [PubMed] [Google Scholar]
- Nagy R, Vasconcelos MJ, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol (Stuttg) 8: 186–197 [DOI] [PubMed] [Google Scholar]
- Nagy R, Drissner D, Amrhein N, Jakobsen I, Bucher M (2009) Mycorrhizal phosphate uptake pathway in tomato is phosphorus-repressible and transcriptionally regulated. New Phytol 181: 950–959 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Ródenas R, Chavanieu A, Rivero RM, Martinez V, Gaillard I, Rubio F (2016) Uneven HAK/KUP/KT protein diversity among angiosperms: Species distribution and perspectives. Front Plant Sci 7: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri E, Breuillin-Sessoms F, Feller U, Reinhardt D (2014) Phosphorus and nitrogen regulate arbuscular mycorrhizal symbiosis in Petunia hybrida. PLoS ONE 9: e90841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RS, Ma Y, Rocha I, Carvalho MF, Vosátka M, Freitas H (2016) Arbuscular mycorrhizal fungi are an alternative to the application of chemical fertilizer in the production of the medicinal and aromatic plant Coriandrum sativum L. J Toxicol Environ Health A 79: 320–328 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Arinaga N, Umezawa T, Katsura S, Nagamachi K, Tanaka H, Ohiraki H, Yamada K, Seo SU, Abo M, et al. (2013) Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner H, Schwarz D, Bruns C, Mäder P, George E (2007) Effect of arbuscular mycorrhizal colonization and two levels of compost supply on nutrient uptake and flowering of pelargonium plants. Mycorrhiza 17: 469–474 [DOI] [PubMed] [Google Scholar]
- Pettigrew WT. (2008) Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol Plant 133: 670–681 [DOI] [PubMed] [Google Scholar]
- Rengel Z, Damon PM (2008) Crops and genotypes differ in efficiency of potassium uptake and use. Physiol Plant 133: 624–636 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-María GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9: 2281–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658 [DOI] [PubMed] [Google Scholar]
- Sieh D, Watanabe M, Devers EA, Brueckner F, Hoefgen R, Krajinski F (2013) The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol 197: 606–616 [DOI] [PubMed] [Google Scholar]
- Smith SE, Smith FA (2011) Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu Rev Plant Biol 62: 227–250 [DOI] [PubMed] [Google Scholar]
- Smith SE, Smith FA (2012) Fresh perspectives on the roles of arbuscular mycorrhizal fungi in plant nutrition and growth. Mycologia 104: 1–13 [DOI] [PubMed] [Google Scholar]
- Smith SE, Smith FA, Jakobsen I (2003) Functional diversity in arbuscular mycorrhizal (AM) symbioses: The contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol 162: 511–524 [Google Scholar]
- Sugimura Y, Saito K (2017) Comparative transcriptome analysis between Solanum lycopersicum L. and Lotus japonicus L. during arbuscular mycorrhizal development. Soil Sci Plant Nutr 63: 127–136 [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schunmann PHD, Taylor W, Waterhouse PM, Wang MB (2000) Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust J Plant Physiol 27: 201–210 [Google Scholar]
- Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K (2006) Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J 48: 296–306 [DOI] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA 93: 10510–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 4.1–4.26 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu WH (2017) Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol 39: 123–128 [DOI] [PubMed] [Google Scholar]
- White PJ. (2013) Improving potassium acquisition and utilization by crop plants. J Plant Nutr Soil Sci 176: 305–316 [Google Scholar]
- Xie X, Huang W, Liu F, Tang N, Liu Y, Lin H, Zhao B (2013) Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol 198: 836–852 [DOI] [PubMed] [Google Scholar]
- Yang SY, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N, et al. (2012) Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24: 4236–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, et al. (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Gao Q, Sun C, Li W, Gu S, Xu C (2009) Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). J Genet Genomics 36: 161–172 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wei S, Hu W, Xiao L, Tang M (2017) Arbuscular mycorrhizal fungus Rhizophagus irregularis increased potassium content and expression of genes encoding potassium channels in Lycium barbarum. Front Plant Sci 8: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]