Abstract
Neutralization of HIV-1 primary isolates has been a tremendous challenge for AIDS vaccine development. Here, we identify a single amino acid change (T198P) in gp120 that alters the neutralization sensitivity of the primary isolate DH012 to antibodies against multiple neutralization epitopes that include the V3, CD4-induced, and CD4 binding sites in gp120. This mutation is located in the V1/V2 stem region that forms the third β strand (β3) of the bridging sheet of gp120. The conformation of variable loops, especially V1/V2 and V3, was proposed to regulate the accessibility of these neutralization epitopes. The results of this study indicate a direct association between the V1/V2 and V3 loops of DH012 gp120. The single amino acid mutation T198P in the β3 severely compromises the interaction between the V1/V2 and V3 loops. These results suggest that interaction of V1/V2 and V3 can mask the neutralization epitopes and that the β3 plays a critical role in determining the neutralization sensitivity by modulating the interaction. This study provides an insight into why primary isolates are relatively resistant to antibody neutralization and might facilitate the development of anti-HIV strategies against HIV-1 infection.
Human immunodeficiency virus type 1 (HIV-1) infection causes AIDS. HIV-1 infection generally provokes a vigorous antibody response to the envelope glycoproteins gp120 and gp41. In natural infection, antisera that can neutralize a broad spectrum of HIV-1 primary isolates are uncommon and of low titer. Likewise, vaccine sera are generally ineffective against primary isolates (1–5). The relative resistance of HIV-1 primary isolates seems to be because of a decreased accessibility of the relevant neutralization epitopes on the envelope glycoproteins (6–9). The observations of differential binding of mAbs to primary isolates versus T cell line-adapted virus support the notion that the relative constraints of the primary isolate envelope might contribute to neutralization resistance (10–13).
The ability of HIV-1 to evade immune responses is likely the major reason that allows the virus to establish persistent infection. The structure of the envelope glycoproteins is responsible for the ability of HIV-1 to evade the humoral immune response. The variable regions of gp120 might play an important role in neutralization resistance of HIV-1 primary isolates because there is a remarkable similarity between the gp120 cores from primary and laboratory-adapted HIV-1 strains (14). The V1/V2 loop was proposed to functionally interact with the V3 loop, as mutations in V1/V2 could modulate the conformation of the V3 loop (15) and, in some cases, alter antibody binding or neutralization sensitivity of HIV-1 to anti-V3 antibodies (16, 17). The V1/V2 loop was suggested to mask neutralization epitopes in the CD4 binding site and those of CD4-induced sites in gp120 of primary isolates (13, 18). In addition, V1/V2 was proposed to cooperate with V3 to determine the neutralization-resistant phenotype and to interact with chemokine receptors (19–21). These studies suggest that V1/V2 can functionally interact, or is located in close proximity, with the V3 loop. However, whether there is a direct physical interaction between the V1/V2 and V3 loops is unknown, as the crystal structure of a gp120 core was determined devoid of these V regions (22). The crystal structure of the gp120 core displays a unique structure called the bridging sheet that is situated in proximity to both the V1/V2 and V3 loops (22). The bridging sheet contains four antiparallel β strands (β2, β3, β20, and β21) that come from discontinuous regions of gp120. It is proposed that the bridging sheet is exposed after CD4 binding and is involved in subsequent envelope/chemokine receptor interaction.
Previously, we have found that the V1/V2, V3, and bridging sheet regions determine the neutralization sensitivity of DH012 viruses (6, 23). To further understand the role of the interplays among the V1/V2, V3, and bridging sheet regions in neutralization sensitivity, we have used the primary isolate DH012 as a model system to demonstrate how the primary isolate envelope evades neutralization by various antibodies and soluble CD4.
Materials and Methods
Cell Lines and Viruses.
The human T lymphoblastoid cell lines, CEM and MT4, were maintained in RPMI 1640 medium containing 10% FBS and 100 units/ml penicillin and streptomycin. The construction and characterization of the chimeric virus NLDH120 has been described (24). The selection and characterization of the neutralization-resistant DH012mu virus was also reported recently (6).
Cloning and Sequencing of the Neutralization-Resistant DH012 Envelope.
The human chromosomal DNA containing the neutralization-resistant virus genome was prepared by extracting chromosomal DNA from the virus-infected MT4 cells. The neutralization-resistant HIV-1 envelope sequence was amplified by using PCR. The 5′ primer used in the PCR was located in the junction of the gp120 signal peptide and the N terminus of the gp120 sequence (5′-GATGATCTGTAGTGCTGCAG-3′). The 3′ primer was located in the cytoplasmic domain of gp41 (5′-CGTCCCAGAAGTTCCAC-3′). The 2.2-kb PCR product was cloned into a TA vector pCR3.1 (Invitrogen) for sequence analysis.
Site-Directed Mutagenesis.
A quick-exchange mutagenesis kit (Stratagene) was used to introduce mutations into the NLDH120 envelope sequence by following the protocol provided by the manufacturer. The NLDH120 variant M1-NLDH was obtained by introducing a mutation into NLDH120 with the two mutagenesis primers 5′-CATTGGGAAATCGATGAGAGAAGAAATAAAAAACTG-3′ and 5′-CAG TTTTTTATTTCTTCTCTCATCGATTTCCCAATG-3. Likewise, M2-NLDH was created with the primers 5′-GCTATAGGTTGATCAGCTGTAACCCCTCAACCCTTA C-3′ and 5′-GTAAGGGTTGAGGGGTTACAGCTGATCAACCTATAGC-3′. The M3 mutation of M3-NLDH was introduced by the two primers 5′-GTATTTTATACAACCGGTAAAATAGTAGGAG-3′ and 5′-CTCCTACTATTTTACCGGTTGTATAAAATAC-3′.
Construction of Recombinant Proteins.
The procedure for construction of recombinant proteins containing various domains of gp120 was similar to that which we have described for the gp41 recombinant proteins (25). The gp120 DNA fragments used in this study were cloned into the EcoRI/HindIII site of the expression vector pMal-p2. The gp120 sequence of the recombinant protein MBri was obtained by using PCR with the upstream primer 5′-TCAGTTGAATTCAAACAAATTATA-AACATGTGGCAGGAAGTAGGAAAAGCAATGTAT-GCCCCTGGGAAGCCATGTGTAAAATTAAC-3′ and the downstream primer 5′-TAGACCAAGCTTTTATGGACAGGCCTGTGTAAGGGTTGAGG-3′. The upstream and downstream primers used in a PCR to create M-V1/V2 were 5′-TCAGTTGAATTCCCACTCTGTGTTACTTTACATTGC-3′ and 5′-TAGACCAAGCTTTTAACTTATCAACCTATAGCTAG-3′. A quick-exchange mutagenesis kit (Stratagene) was used to introduce mutations into M2–MBri by using MBri as templates. The same primers that were used to generate M2–NLDH were used to introduce the M2 mutation.
Results
Development and Mutations of Neutralization-Resistant DH012.
Like many primary isolates, DH012 is relatively resistant to neutralization by HIV-1-positive sera. Nevertheless, we have previously reported that this virus is very sensitive to serum obtained from a DH012-infected chimpanzee (6, 23). The virus (DH012mu) that escapes the neutralizing serum C1206 replicated well in the presence of the chimpanzee-neutralizing serum at a 100-fold dilution (Fig. 1A). On the other hand, the chimpanzee serum can totally block DH012 replication at a 1,000-fold dilution (Fig. 1B). The neutralization escape mutant was at least 100-fold less sensitive to the neutralizing DH012 serum (6). There are only three amino acid differences between the entire envelope sequences of wild-type DH012 and the neutralization escape mutant as indicated in Fig. 1C. The differences are a G152E (M1) mutation in the V1/V2 loop, a T198P (M2) change in the β3 of the bridging sheet, and an E320K (M3) mutation in the C terminus of the V3 loop. To determine the effect of each mutation on neutralization sensitivity, each of the mutations was reintroduced into a chimeric virus NLDH120 that contained the wild-type DH012 envelope in the genetic background of NL4–3. The envelope subunit gp120 alone is sufficient to confer the neutralization sensitivity to the neutralizing chimpanzee serum C1206 (23). Introduction of all three mutations into NLDH120 resulted in a marked reduction (133-fold) in sensitivity to neutralization by this serum (Fig. 2A). Among the three mutations, the T198P mutation had the most impact on neutralization by the chimpanzee serum. This mutation resulted in a 9.3-fold reduction in the neutralization sensitivity to the chimpanzee serum. The mutation in the V3 loop alone, E320K, did not have a significant effect on neutralization by the chimpanzee serum. However, this V3 mutation did contribute to neutralization resistance, as a virus that contains only the M1 and M2 mutations is 2-fold more sensitive than M123-NLDH to the chimpanzee serum C1206 (data not shown). These results are consistent with our previous report that multiple envelope regions are involved in the neutralization of NLDH120 by the sera from infected chimpanzees.
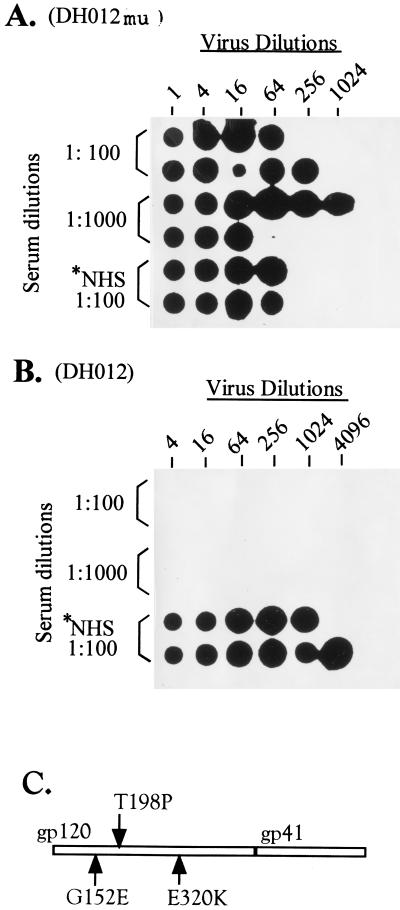
Figure 1.
Mutations in the envelope of the neutralization-resistant DH012. DH012mu is the neutralization-resistant mutant derived from culturing DH012 in the presence of escalating concentrations of the chimpanzee serum C1206 (6). The viruses were initially grown in MT4 cells in the presence of C1206 at 1,000-fold dilution. The concentration of C1206 was subsequently elevated to 1/500, 1/200, and 1/100. DH012mu is the virus that grows in the presence of C1206 at 100-fold dilution. The detailed protocol for the neutralization has been described (24, 25). Normal human serum (NHS) was included in the assay as a negative control. (A) Dot blot analysis of HIV-1 reverse transcription activity in the assay samples. The chimpanzee serum C1206 at 100 and 1,000-fold dilutions was used in this neutralization assay. Each positive dot indicates successful virus replication. (B) DH012 virus neutralization assays were carried out as described above. DH012 is very sensitive to the chimpanzee serum C1206 because virus replication was not detected in the presence of C1206 at 1,000-fold dilution. (C) The envelope sequence of DH012mu including gp120 and gp41 was determined. The arrows indicate three mutations in gp120 of the neutralization-resistant mutant DH012mu. *NHS, normal human serum.
Figure 2.
Effect of each mutation of DH012mu on neutralization sensitivity. The neutralization sensitivity was determined by using a Magi assay previously described (6). (A) Neutralization sensitivity of each NLDH120 mutant to the chimpanzee serum C1206. M1-NLDH is the NLDH120 mutant that contains the G152E mutation, M2-NLDH contains the T198P mutation, M3-NLDH contains the E320K mutation, and M123-NLDH contains all three mutations. (B) The neutralization sensitivity of these mutants was tested against the guinea pig serum NC75 that contains neutralizing anti-V3 antibodies (6). The serum dilution that inhibits 50% of the blue cells, for example a reduction of 400 to 200 blue cells, is defined as the neutralization titer of the serum. The neutralization titer derived from each virus was used to determine the relative sensitivity of the viruses. The neutralization titers of C1206 against NLDH120 and M123-NLDH are 28,000 and 210, respectively. Therefore, M123-NLDH is 133-fold more resistant to C1206 than NLDH120.
A Single Amino Acid Change Alters Neutralization Sensitivity to Multiple Neutralizing Antibodies.
In contrast to the C1206 neutralization, these mutants did not exhibit resistance to sera derived from guinea pigs immunized with baculovirus-expressed DH012 gp120. Indeed, the mutants that are resistant to C1206 became very sensitive to the guinea pig sera (Fig. 2B). For example, M123–NLDH is 20-fold more sensitive to the guinea pig serum NC75. The increased sensitivity to the guinea pig sera was because of the presence of neutralizing anti-V3 antibodies in the guinea pig sera and an increased accessibility of the V3 loop in the mutants that escape neutralizing antibodies in the chimpanzee serum (6). The chimpanzee serum does not contain detectable anti-V3 antibodies (6). Unlike resistance to C1206, the increased neutralization sensitivity of the DH012 mutants to the guinea pig serum NC75 is mainly because of the T198P mutation in β3 (Fig. 2B). The M1 and M3 mutations did not significantly change neutralization sensitivity of the viruses to the guinea pig serum (Fig. 2B). Because the T198P mutation is not located in the V3 loop, it is possible that the T198P mutation alters the conformation of the primary isolate DH012 envelope so that neutralization epitopes such as the V3, CD4 binding site, and CD4-induced epitopes (CD4i) become accessible to neutralizing antibodies.
To test this possibility, the sensitivity of the DH012 mutants to reagents that target the CD4 binding site and CD4i was determined. The neutralization-resistant mutant M123-NLDH is 24-, 6-, and 15-fold more sensitive to soluble CD4, IgGb12, and 17b, respectively (Fig. 3 A–C). The mAbs IgGb12 and 17b target the CD4 binding site and CD4i, respectively. Similar to the anti-V3 activity in the guinea pig serum, M2 alone could account for the increased neutralization sensitivity. This conformational effect seems to be specific to the cryptic sites in gp120 because the T198P mutation did not change neutralization sensitivity of the DH012 mutants to 2F5 and 2G12 (Fig. 3D). The mAb 2F5 targets gp41, and 2G12 is believed to recognize the surface sugar moiety in gp120. Both of the mAbs can neutralize a broad spectrum of HIV-1 primary isolates.
Figure 3.
Sensitivity of NLDH120 mutants to soluble CD4 and mAbs. The neutralization sensitivity of the NLDH120 and mutants was determined with the same Magi assay that has been described in the Fig. 2 legend. The viruses were used to infect Magi cells in the presence of (A) soluble CD4, (B) IgGb12, (C) 17b, and (D) 2G12 and 2F5. IgGb12, 2G12, and 2F5 are mAbs that neutralize a broad spectrum of HIV-1 primary isolates.
Direct Association of V1/V2 with V3 and the Role of the Bridging Sheet in This Association.
These results indicate that a single amino acid change, T198P, in the bridging sheet region has a significant impact on the accessibility of the V3, CD4 binding site, and CD4-induced epitopes of the DH012 envelope. The bridging sheet is situated in close proximity with both the V1/V2 and V3 loops based on the model predicted by the crystal structure of the gp120 core (22). Therefore, we hypothesize that V1/V2 and V3 of DH012 can physically associate with each other, and the bridging sheet modulates this interaction. If this indeed is the case, the association of the V1/V2 and V3 loops could explain the observations that the V3, CD4 binding site, and CD4-induced epitopes are less accessible in wild-type DH012. The latter two regions are predicted to be located spatially between the V1/V2 and V3 loops. An association between V1/V2 and V3 is likely to affect the accessibility of these sites. To test this hypothesis, we used recombinant gp120 fragments to determine the interactions between the V1/V2 and V3 regions, as well as the role of the β3 mutation in the interaction. We have previously used a similar approach to show the interaction of two heptad repeats in the ectodomain of HIV-1 gp41 and the critical role of this interaction in HIV-1 entry (25). The association of the two heptad repeats of gp41 was later demonstrated by using x-ray crystallography (26, 27).
The recombinant proteins MBri and M2–MBri contain the bridging sheet and V1/V2 regions (Fig. 4A). The arrangement of the β strands and V1/V2 of the recombinant proteins is not based on the order in which they appear in the primary sequence of gp120. Instead, they were constructed in a sequence that might best mimic the tertiary structure of gp120 predicted by x-ray crystallography (22). Therefore, β20–β21 is placed N terminal to β2–β3. The fact that MBri and M2–MBri interact well with the conformational mAbs IgGb12 (Fig. 4B) suggests that these recombinant proteins at least retain certain fractions of the authentic gp120 structure. Addition of free MBri or M2–MBri effectively competed against the binding of IgGb12 to the recombinant proteins on ELISA plates. The epitope of IgGb12 is located in the CD4 binding site (28). The V1/V2 stem and β21 of the bridging sheet were shown to be involved in CD4 binding. It is interesting that the binding of IgGb12 to M2–Mbri that contains the T198P mutation in β3 of the bridging sheet is reduced (Fig. 4B). These two recombinant proteins also interacted well with the conformational antibody 17b (C.B.Z. and C.H.C., unpublished results). The mAb 17b makes substantial contact to the amino acids of all four β-strands of the bridging sheet (22). Because both of the recombinant proteins contain the 17b and IgGb12 epitopes, it will be of interest to test whether these reagents can induce antibodies directed to the CD4 binding site and CD4-induced epitope.
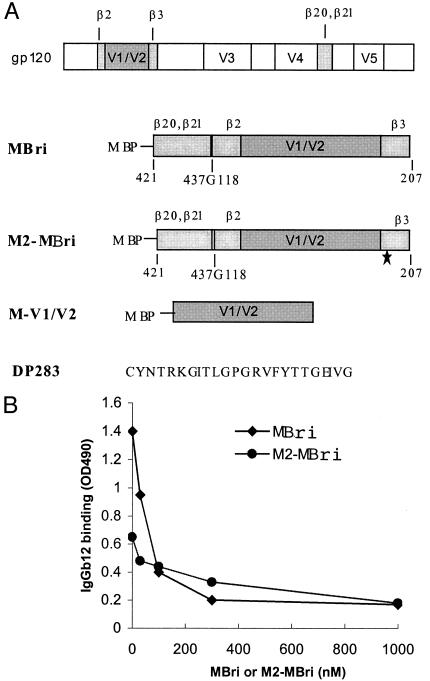
Figure 4.
(A) Construction of recombinant gp120 fragments. The relative position of the four β strands, β2, β3, β20, and β21, is indicated in the schematic presentation of gp120. A glycine residue was added to MBri between β21 and β2 as a linker to join two gp120 fragments that span from amino acids 421–437 and 118–207 as indicated (437G118). The amino acid numbering system used here is the same as that described by Kwong et al. (22). M2–MBri is identical to MBri, except for an amino acid change that corresponds to the T198P mutation (* under the β3 region indicates this mutation). DP283 is a DH012 V3 peptide. (B). Reactivity of IgGb12 to recombinant gp120 fragments. The binding of IgGb12 to the recombinant proteins was determined by using an ELISA assay as described (25). IgGb12 (10 μg/ml) was added to ELISA plates coated with the recombinant proteins MBri or M2–MBri in the presence of various concentrations of the free MBri or M2–MBri, respectively, as indicated.
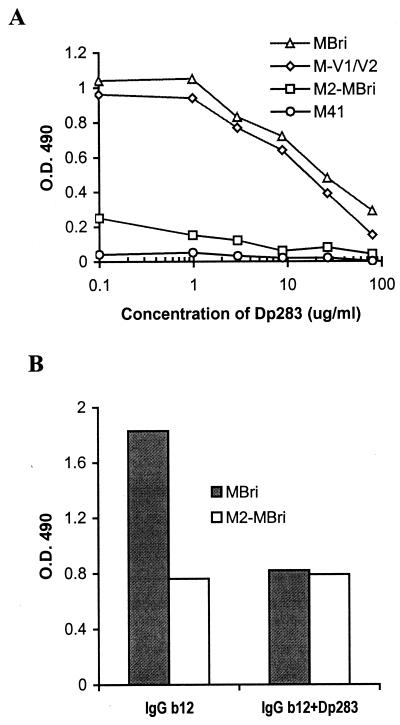
The ability of direct binding between V1/V2 and V3 of DH012 is demonstrated in Fig. 5A. The recombinant protein M-V1/V2 binds very well to the V3 peptide DP283 on ELISA plates. Excess amounts of the V3 peptides in solution effectively compete for the binding. MBri, which is the construct that contains both the V1/V2 loop and the bridging sheet (Fig. 4), behaves similarly to M-V1/V2 in its ability to interact with the V3 peptide DP283 (Fig. 5A). On the contrary, the T198P mutation dramatically decreased the ability of MBri to interact with the V3 peptide DP283 (Fig. 5A). The recombinant protein M41, which contains the ectodomain of HIV-1 gp41 (25), did not exhibit any reactivity with the V3 peptide. These results support our hypothesis that V1/V2 can physically interact with V3, and this interaction can be modulated by the β3 motif of the bridging sheet.
Figure 5.
Association of recombinant gp120 fragments to V3 peptide and the effect of this binding on antibody reactivity. (A) To determine binding between the recombinant proteins and the V3 peptide, DP283 was coated on an ELISA plate. The recombinant proteins were then added to interact with the V3 peptide. The binding of recombinant proteins to the V3 peptide was monitored by antibodies to the maltose binding protein. We have used the same approach to determine the binding between a gp41 peptide DP178 and gp41 fragments (25). M41 is a recombinant protein with the ectodomain of gp41 cloned downstream of the maltose binding protein. (B) The recombinant proteins MBri and M2–MBri were coated on ELISA plates. The ELISA assay was carried out as described in Fig. 4. IgGb12 at 10 μg/ml was used to interact with the recombinant proteins in the presence or absence of the V3 peptide DP283 at 20 μg/ml.
Interaction of V1/V2 and V3 Masks Neutralization Epitopes.
We next tested whether the IgGb12 epitope on MBri can be masked by the presence of the V3 peptide DP283. The binding of IgGb12 to MBri on ELISA plates was significantly reduced in the presence of excess amounts of the V3 peptide (Fig. 5B). On the contrary, the V3 peptide does not significantly affect the binding of IgGb12 to M2–MBri. These results are consistent with the notion that the interaction between V1/V2 and V3 can mask the CD4 binding site of DH012 gp120.
Discussion
The results of this study reveal a direct interaction between the V1/V2 and V3 regions of DH012 gp120. This interaction may explain why the wild-type DH012 primary isolate is relatively resistant to neutralization by antibodies such as IgGb12, 17b, and anti-V3 antibodies. The model in Fig. 6 illustrates that the V3, CD4i, and CD4 binding sites are masked by the interaction between V1/V2 and V3. The mutation in the β3 strand of the DH012 bridging sheet disrupts the interaction, which results in the exposure of epitopes in the V3, CD4i, and CD4 binding sites of DH012 gp120. The β3 motif, or perhaps the entire bridging sheet, might function as a molecular switch that opens or closes the door for neutralizing antibodies. Although the CD4i and CD4 binding sites are relatively conserved among HIV-1 isolates, whether the model demonstrated in Fig. 6 applies to other HIV-1 primary isolates remains to be determined. Although the model depicts intramolecular interaction within a monomeric gp120, intermolecular interaction between the V1/V2 and V3 regions might occur in the native trimeric structure of gp120. The close proximity between the V2 and V3 loops has been described to correlate with the acquisition of neutralization resistance in a simian HIV (13). Our results support the notion that the V2 and V3 loops are in close proximity and further demonstrate that a direct V1/V2 and V3 association might be responsible for the proximity.
Figure 6.
A model of DH012 V1/V2 and V3 interaction. The relative location of each domain, the CD4 binding site (orange), CD4-induced epitopes (green), bridging sheet (yellow), V1/V2 (blue), and V3 (pink), was shown in the models for NLDH120 and M2-NLDH. * in B indicates a single amino acid mutation in the bridging sheet, either T198P or N197K.
The critical role of β3 of the bridging sheet in modulating the interaction between V1/V2 and V3 is further evidenced by another DH012 mutant that has a N197K mutation (data not shown). This N197K DH012 mutant behaves similarly to the T198P mutant in neutralization sensitivity, antibody binding, and its effects on the interaction between V1/V2 and V3. The asparagine 197 of gp120 is a glycosylation site that has been implicated in modulating the ability of V1/V2 to occlude the CCR5 binding region of gp120 in a recent report by Kolchinski et al. (29). Mutation of the asparagine 197 was suggested to be associated with a movement of the V1/V2 loop, which controls the exposure of the CCR5 binding region of gp120 (29).
The ability of the single amino acid mutations in β3 to alter neutralization sensitivity and the interaction of V1/V2 and V3 suggest that β3, or the bridging sheet, might determine the relative global location of the V1/V2 and V3 loops on the surface of gp120. The dramatic conformational effect of the T198P mutation might arise from the ability of the proline to bend the β3 strand or to disrupt the interaction of β3 with other β strands in the bridging sheet. In addition to the conformational effects, the bridging sheet might modulate the interaction in an interplay involving V1/V2, V3, and the bridging sheet itself. This speculation is supported by a report that mutations in β21 of the bridging sheet enhance mAb binding to V3 (30).
The interaction of V1/V2 and V3 may enable HIV-1 to mask its neutralization epitopes. However, exposure of these epitopes is essential for the viral entry. Therefore, this interaction is likely to be well regulated to maintain optimal viral infectivity. One of the factors that regulate the interaction between V1/V2 and V3 might be the binding of CD4 to gp120. It is conceivable that a conformational change involving the bridging sheet after CD4 binding can alter the interaction between V1/V2 and V3. Understanding the structure and functional relationship of gp120 is likely to facilitate the development of anti-HIV-1 entry strategies including drugs and vaccines against the virus infection.
Acknowledgments
We thank Dr. Malcolm Martin for providing the chimpanzee neutralizing sera and Dr. Christopher Aiken for critical review and discussion of this manuscript. This study was supported by National Institutes of Health Grants AI40856 and AI44281.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Belshe R B, Gorse G J, Mulligan M J, Evans T G, Keefer M C, Excler J L, Duliege A M, Tartaglia J, Cox W I, McNamara J, et al. AIDS. 1998;12:2407–2415. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Belshe R B, Graham B S, Keefer M C, Gorse G J, Wright P, Dolin R, Matthews T J, Weinhold K J, Bolognesi D P, Sposto R. J Am Med Assoc. 1994;272:475–480. doi: 10.1001/jama.272.6.475. [DOI] [PubMed] [Google Scholar]
- 3.Mascola J R, Snyder S W, Weislow O S, Belay S M, Belshe R B, Schwartz D H, Clements M L, Doloin R, Graham B S, Gorse G J, et al. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 4.Bures R, Gaitan A, Zhu T, Graziosi C, McGrath K M, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, et al. AIDS Res Hum Retroviruses. 2000;16:2019–2035. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 5.Graham B S, McElrath M J, Connor R I, Schwartz D H, Gorse G J, Keefer M C, Mulligan M J, Matthews T J, Wolinsky S M, Montefiori D C, et al. J Infect Dis. 1998;177:310–319. doi: 10.1086/514209. [DOI] [PubMed] [Google Scholar]
- 6.Chen C H, Jin L, Zhu C B, Holz-Smith S, Matthews T J. J Virol. 2001;75:6700–6704. doi: 10.1128/JVI.75.14.6700-6704.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouts T R, Binley L M, Trkola A, Robinson J E, Moore J P. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parren P W, Moore J P, Burton D R, Sattentau Q J. AIDS. 1999;13A,Suppl.:S137–S162. [PubMed] [Google Scholar]
- 9.Sullivan N, Sun Y, Binley J, Lee J, Barbas C F, Parren P W, Burton D R, Sodroski J. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bou-Habib D C, Roderiquez G, Oravecz T, Berman P W, Lusso P, Norcross M A. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamatatos L, Cheng-Mayer C. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Si Z H, Moore J P, Sodroski J. J Virol. 2000;74:11955–11962. doi: 10.1128/jvi.74.24.11955-11962.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong P D, Wyatt R, Majeed S, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Struct Fold Des. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 15.Koito A, Stamatatos L, Cheng-Mayer C. Virology. 1995;206:878–884. doi: 10.1006/viro.1995.1010. [DOI] [PubMed] [Google Scholar]
- 16.Lee M K, Martin M A, Cho M W. AIDS Res Hum Retroviruses. 2000;16:765–775. doi: 10.1089/088922200308765. [DOI] [PubMed] [Google Scholar]
- 17.Liao H X, Etemad-Moghadam B, Montefiori D C, Sun Y, Sodroski J, Scearce R M, Doms R W, Thomasch J R, Robinson S, Letvin N L, Haynes B F. J Virol. 2000;74:254–263. doi: 10.1128/jvi.74.1.254-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etemad-Moghadam B, Sun Y, Nicholson E K, Karlsson G B, Schenten D, Sodroski J. J Virol. 1999;73:8873–8879. doi: 10.1128/jvi.73.10.8873-8879.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labrosse B, Treboute C, Brelot A, Alizon M. J Virol. 2001;75:5457–5464. doi: 10.1128/JVI.75.12.5457-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho M W, Lee M K, Chen C H, Matthews T J, Martin M A. J Virol. 2000;74:9749–9754. doi: 10.1128/jvi.74.20.9749-9754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holz-Smith S L, Sun I C, Jin L, Matthews T J, Lee K H, Chen C H. Antimicrob Agents Chemother. 2001;45:60–66. doi: 10.1128/AAC.45.1.60-66.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 27.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 28.Roben P, Moore J P, Thali M, Sodroski J, Barbas C F, Burton D R. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolchinski P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. J Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore J P, Thali M, Jameson B A, Vignaux F, Lewis G K, Poon S W, Charles M, Fung M S, Sun B, Durda P J. J Virol. 1993;67:4785–4796. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]