Abstract
A review of the recent progress in plant genetic engineering for disease resistance highlights future challenges and opportunities in the field.
BACKGROUND
Modern agriculture must provide sufficient nutrients to feed the world’s growing population, which is projected to increase from 7.3 billion in 2015 to at least 9.8 billion by 2050. This goal is made even more challenging because of crop loss to diseases. Bacterial and fungal pathogens reduce crop yields by about 15% and viruses reduce yields by 3% (Oerke and Dehne, 2004). For some crops, such as potatoes, the loss caused by microbial infection is estimated to be as high as 30% (Oerke and Dehne, 2004). As an alternative to the application of chemical agents, researchers are altering the genetic composition of plants to enhance resistance to microbial infections.
Conventional breeding plays an essential role in crop improvement but usually entails growing and examining large populations of crops over multiple generations, a lengthy and labor-intensive process. Genetic engineering, which refers to the direct alteration of an organism’s genetic material using biotechnology (Christou, 2013), possesses several advantages compared with conventional breeding. First, it enables the introduction, removal, modification, or fine-tuning of specific genes of interest with minimal undesired changes to the rest of the crop genome. As a result, crops exhibiting desired agronomic traits can be obtained in fewer generations compared with conventional breeding. Second, genetic engineering allows for interchange of genetic material across species. Thus, the raw genetic materials that can be exploited for this process is not restricted to the genes available within the species. Third, plant transformation during genetic engineering allows the introduction of new genes into vegetatively propagated crops such as banana (Musa sp.), cassava (Manihot esculenta), and potato (Solanum tuberosum). These features make genetic engineering a powerful tool for enhancing resistance against plant pathogens.
Most cases of plant genetic engineering rely on conventional transgenic approaches or the more recent genome-editing technologies. In conventional transgenic methods, genes that encode desired agronomic traits are inserted into the genome at random locations through plant transformation (Lorence and Verpoorte, 2004). These methods typically result in varieties containing foreign DNA. In contrast, genome editing allows changes to the endogenous plant DNA, such as deletions, insertions, and replacements of DNA of various lengths at designated targets (Barrangou and Doudna, 2016). Depending on the type of edits introduced, the product may or may not contain foreign DNA. In some areas of the world, including the United States (USDA, 2018), Argentina (Whelan and Lema, 2015), and Brazil (CTNBio, Brazil, 2018), genome-edited plants that do not contain foreign DNA are not subject to the additional regulatory measures applied to transgenic plants (Orozco, 2018). Regardless of differences in regulatory policies, both conventional transgenic techniques and genome editing continue to be powerful tools for crop improvement.
Plants have evolved multilayered defense mechanisms against microbial pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). For example, preformed physical and physiological barriers and their reinforcements prevent potential pathogens from gaining access inside the cell (Collinge, 2009; Uma et al., 2011). Plasma membrane-bound and intracellular immune receptors initiate defense responses upon the perception of pathogens either directly through physically interacting with pathogen-derived immunogens or indirectly by monitoring modifications of host targets incurred by pathogens (Jones and Dangl, 2006; Bentham et al., 2017; Zhang et al., 2017; Kourelis and van der Hoorn, 2018). Plant-derived antimicrobial peptides and other compounds can suppress pathogenicity by direct detoxification or through inhibition of the activity of virulence factors (Kitajima and Sato, 1999; Thomma et al., 2002; Ahuja et al., 2012). Plants also employ RNA interference (RNAi) to detect invading viral pathogens and target the viral RNA for cleavage (Rosa et al., 2018).
On the other side, successful pathogens have evolved strategies to overcome the defense responses of their plant hosts. For instance, many bacterial and fungal pathogens produce and release cell-wall-degrading enzymes (Kubicek et al., 2014). Pathogens can also deliver effectors into the host cytoplasm (Xin and He, 2013; Franceschetti et al., 2017); some of these effectors suppress host defenses and promote susceptibility. To counteract plant RNAi-based defenses, almost all plant viruses produce viral suppressors of RNAi (Zamore, 2004). Some viruses also hijack the host RNAi system to silence host genes to promote viral pathogenicity (Wang et al., 2012).
Some aspects of host-microbe interactions provide opportunities for genetic engineering for disease resistance (Dangl et al., 2013). For example, genes that encode proteins capable of breaking down mycotoxins (Karlovsky, 2011) or inhibiting the activity of cell-wall-degrading enzymes (Juge, 2006) can be introduced into plants. Plants can be engineered to synthesize and secrete antimicrobial peptides or compounds that directly inhibit colonization (Osusky et al., 2000). The RNAi machinery can be exploited to confer robust viral immunity by targeting viral RNA for degradation (Rosa et al., 2018). Natural or engineered immune receptors that recognize diverse strains of a pathogen can be introduced individually or in combination for robust, broad-spectrum disease resistance (Fuchs, 2017). Essential defense hub regulatory genes can be reprogrammed to fine-tune defense responses (Pieterse and Van Loon, 2004). Susceptible host targets can be modified to evade recognition and manipulation by pathogens (Li et al., 2012). Similarly, host decoy proteins, which serve to trap pathogens, can be modified through genetic engineering for altered specificity in pathogen recognition (Kim et al., 2016). For a more comprehensive overview of various aspects of engineered disease resistance in plants, the readers are referred to previously published review articles (Collinge et al., 2010; Cook et al., 2015).
Increased knowledge regarding the molecular mechanism underlying plant-pathogen interactions and advancements in biotechnology have provided new opportunities for engineering resistance to microbial pathogens in plants. In this review, we describe recent breakthroughs in genetic engineering in plants for enhanced disease resistance and highlight several strategies that have proven successful in field trials.
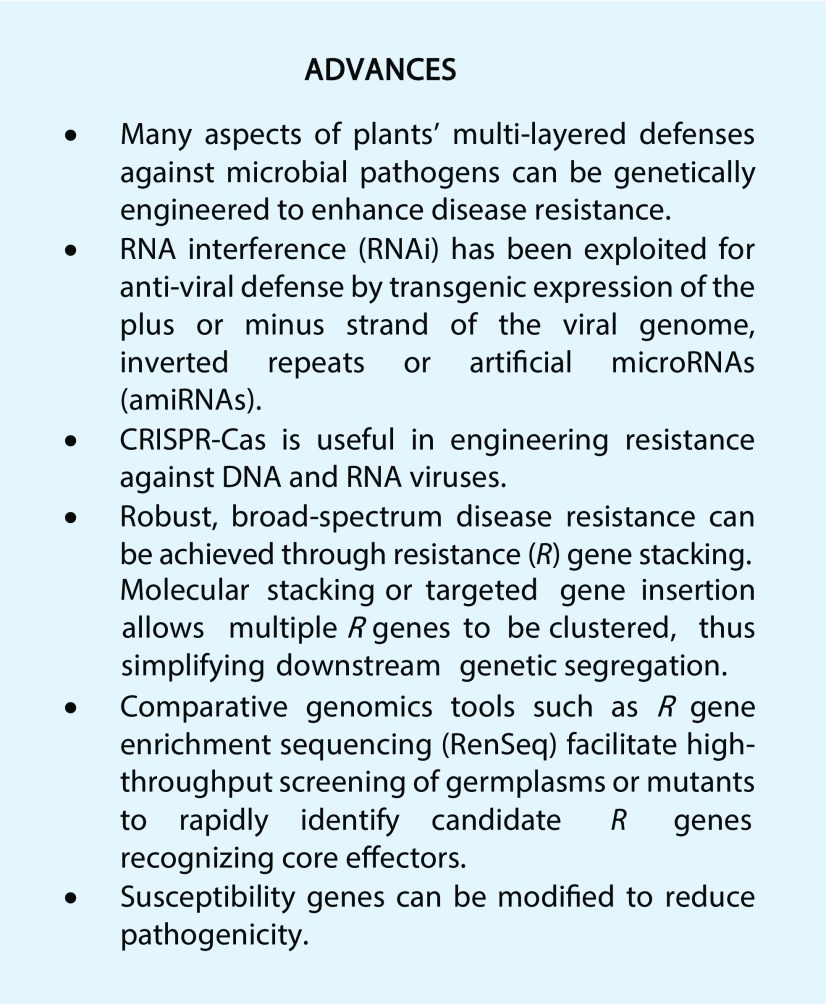
HIGHLIGHTS OF RECENT BREAKTHROUGHS IN GENETIC ENGINEERING FOR DISEASE RESISTANCE IN PLANTS
Pathogen-Derived Resistance and RNAi
Researchers have long observed that transgenic plants expressing genes derived from viral pathogens often display immunity to the pathogen and its related strains (Lomonossoff, 1995). These results led to the hypothesis that ectopic expression of genes encoding wild-type or mutant viral proteins could interfere with the viral life cycle (Sanford and Johnston, 1985). More recent studies demonstrated that this immunity is mediated by RNAi, which plays a major role in antiviral defense in plants.
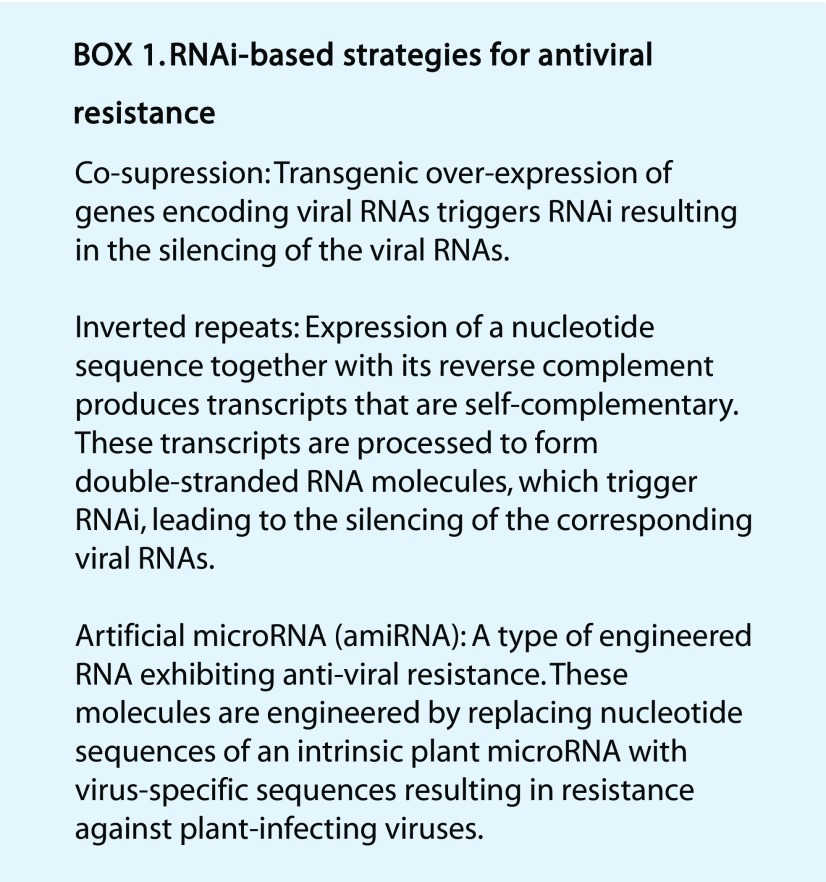
The detailed molecular mechanism of RNAi in antiviral defense has been described in several excellent reviews (Wang et al., 2012; Katoch and Thakur, 2013; Galvez et al., 2014). Activation of RNAi has proven to be an effective approach for engineering resistance to viruses (Lindbo and Dougherty, 2005; Lindbo and Falk, 2017) as they rely on the host cellular machinery to complete their life cycle. Most plant viruses contain single-stranded RNA as their genetic information. Double-stranded RNA (dsRNA) replicative intermediates often form during the replication of the viral genome mediated by RNA-dependent RNA polymerase, triggering RNAi in the host.
Transgenic overexpression of viral RNA often leads to the formation of dsRNA, which also triggers RNAi (Galvez et al., 2014). This process is often known as cosuppression (Box 1). By far, most existing cases of genetically engineered crops with resistance to viral pathogens that have been approved for cultivation and consumption were generated using this strategy (Table 1). Notably, transgenic squash (Cucurbita sp.) and papaya (Carica papaya) varieties created using this approach have been grown commercially in the United States for more than 20 years. Field data indicate that disease resistance achieved through RNAi-mediated strategies is highly durable (Fuchs and Gonsalves, 2007). The RNAi strategy has also been effective in generating squash varieties that are resistant to multiple viral species (Table 1). These varieties were produced by gene stacking (i.e. transgenic expression of two distinct RNAi constructs targeted to different viruses; Tricoli et al., 1995).
Table 1. Events of genetically engineered food crops with enhanced disease resistance to microbial pathogens that have been approved for commercial production.
Data collected from The International Service for the Acquisition of Agri-biotech Applications (ISAAA) and the U.S. Department of Agriculture-Animal and Plant Health Inspection Service (USDA-APHIS). MOA, Ministry of Agriculture, China; ARS, Agriculture Research Service; EMBRAPA, the Brazilian Agricultural Research Corporation; CTNBio, the Brazilian National Technical Commission on Biosafety.
| Crop Species | Developer | Initial Approval | Target Pathogen | Gene(s) Expresseda | References |
|---|---|---|---|---|---|
| Squash | Seminis and Monsanto | 1994 USDA | Watermelon mosaic virus 2 and zucchini yellow mosaic virus | Coat proteins | Tricoli et al., 1995 |
| Squash | Seminis and Monsanto | 1996 USDA | Cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus | Coat proteins | Tricoli et al., 1995 |
| Papaya | Cornell University and the University of Hawaii | 1996 USDA | Papaya ringspot virus | Coat protein | Gonsalves, 1998, 2006 |
| Potato | Monsanto | 1998 USDA | Potato leafroll virus | Replicase and helicase | Thomas et al., 1997; Kaniewski and Thomas, 2004 |
| Sweet Pepper | Beijing University | 1998 MOA | Cucumber mosaic virus | Coat protein | Zhu et al., 1996 |
| Potato | Monsanto | 1999 USDA | Potato virus Y | Coat protein | Newell et al., 1991; Kaniewski and Thomas, 2004 |
| Tomato | Beijing University | 1999 MOA | Cucumber mosaic virus | Coat protein | Yang et al., 1995 |
| Papaya | South China Agricultural University | 2006 MOA | Papaya ringspot virus | Replicase | Ye and Li, 2010 |
| Plum | USDA/ARS | 2007 USDA | Plum pox virus | Coat protein | Scorza et al., 1994; Ilardi and Tavazza, 2015 |
| Papaya | University of Florida | 2009 USDA | Papaya ringspot virus | Coat protein | Davis and Ying, 2004 |
| Bean | EMBRAPA | 2011 CTNBio | Bean golden mosaic virus | +, − RNA of viral replication protein | Bonfim et al., 2007; Tollefson, 2011 |
| Potato | J.R. Simplot | 2015 USDA | Phytophthora infestans (late blight) | Resistance protein | Foster et al., 2009 |
Only genes relevant to pathogen resistance are listed.
The knowledge that dsRNA effectively induces RNAi (Lindbo and Dougherty, 2005) inspired the design of transgenes encoding inverted repeat sequences, the transcripts of which form dsRNA (Box 1; Waterhouse et al., 1998). This strategy was used to develop a transgenic common bean variety exhibiting resistance to the DNA virus Bean golden mosaic virus (BGMV; Bonfim et al., 2007). BGMV contains single-stranded DNA as its genetic material, which is converted to dsDNA in the host, transcribed, and translated to produce the essential proteins required for its replication (Hanley-Bowdoin et al., 2013). To generate small interfering RNA targeting the viral transcripts, sense and antisense sequences of part of the BGMV replication gene AC1 were directionally cloned into an intron-spliced hairpin RNA expression cassette (Bonfim et al., 2007). The resulting genetically engineered bean exhibited strong and robust resistance in greenhouse conditions (Bonfim et al., 2007) as well as field conditions (Aragão and Faria, 2009). The BGMV-resistant bean was deregulated in 2011 in Brazil (Table 1), which allows farmers to grow the crop. It is to date the only example of a deregulated genetically engineered crop showing resistance to a DNA virus. Compared with the cosuppression strategy, transgenic expression of an artificially designed inverted repeat allows simultaneous generation of heterogenous small interfering RNAs targeting multiple transcripts in a relatively simple manner.
The discovery of microRNAs (miRNAs), a class of endogenous noncoding regulatory RNAs (Ambros, 2001; Reinhart et al., 2002) led to further refinements of genetic engineering for viral resistance. The miRNA machinery (Xie et al., 2015) has been exploited in engineering resistance to RNA viruses by replacing specific nucleotides in the miRNA-encoding genes to alter targeting specificity (Box 1). Such artificial miRNAs have been used for engineering resistance to a wide range of plant viral pathogens including Turnip mosaic virus (Niu et al., 2006), Cucumber mosaic virus (Qu et al., 2007; Duan et al., 2008; Zhang et al., 2011), Potato virus X, and Potato virus Y (Ai et al., 2011). These results suggest that artificial-miRNA-based antiviral strategies are highly promising. Disease resistance engineered by this strategy awaits future field tests.
Harnessing CRISPR-Cas to Target Viral Pathogens Directly
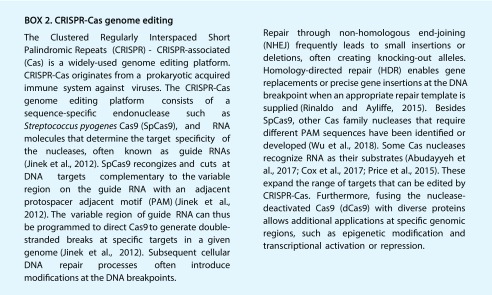
In recent years, the bacterially derived clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) technology has proven to be a promising approach for engineering resistance against plant viruses (Box 2; Wright et al., 2016). In many bacterial species, CRISPR-Cas acts as antiviral defense machinery. In this process, an RNA-guided nuclease (often a Cas protein) cleaves at specific target sites on the substrate viral DNA or RNA, leading to their degradation. The specificity of the cleavage is governed by base complementarity between the CRISPR RNA and the target DNA or RNA molecules. A number of Cas proteins with sequence-specific nuclease activity have been identified (Wu et al., 2018). For example, the RNA-guided endonuclease Cas9 from Streptococcus pyogenes (SpCas9) induces double-stranded breaks in DNA in vivo (Jinek et al., 2012) and the RNA-guided RNases Cas13a from Leptotrichia shahii (LshCas13a) or Leptotrichia wadei (LwaCas13a) target RNA in vivo (Abudayyeh et al., 2017; Cox et al., 2017). Cas9 from Francisella novicida (FnCas9) cleaves both DNA and RNA in vivo (Price et al., 2015).
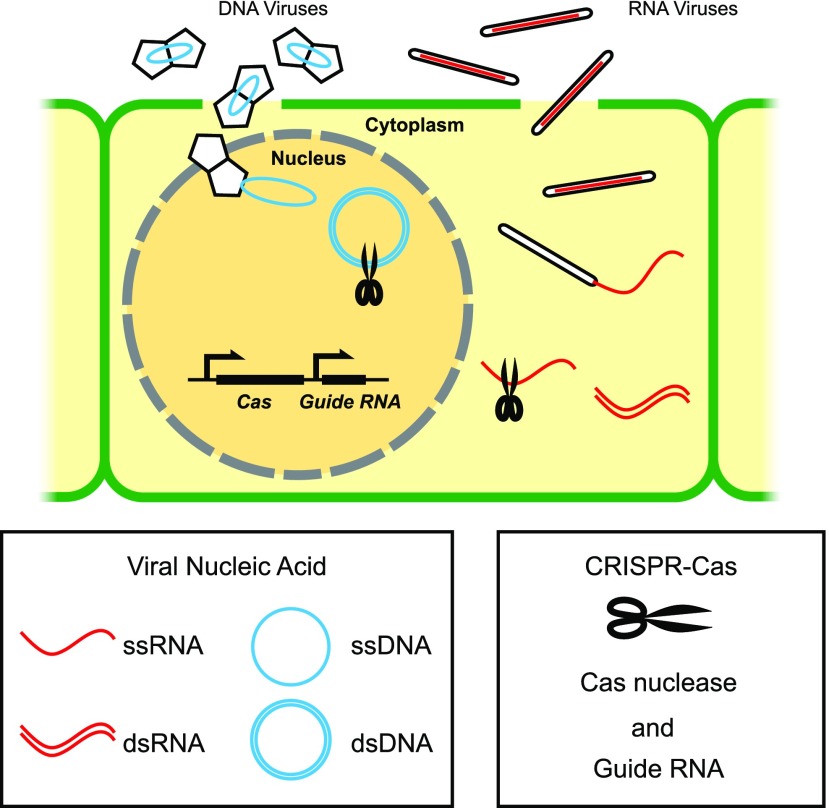
The ability of CRISPR-Cas to act like molecular scissors, creating breaks at sequence-specific targets in the substrate DNA or RNA molecules makes it an excellent candidate tool for engineering for antiviral defense in plants. CRISPR-Cas platforms based on Cas9 or Cas13a have been successfully harnessed to engineer resistance to DNA viruses or RNA viruses in planta (Fig. 1).
Figure 1.
CRISPR-Cas platforms for engineering viral resistance in plants. Plant viruses complete their life cycles in the host cells. Viral particles with geminate capsids, and rod-shaped capsids are used as examples to illustrate DNA viruses and RNA viruses, respectively. One of the Cas nuclease genes (SpCas9, FnCas9, or LshCas13a) and the matching guide RNA(s) are expressed transgenically to generate sequence-specific ribonucleoproteins that target viral DNA or RNA substrates for degradation. SpCas9 targets nuclear dsDNA, while FnCas9 and LshCas13a have been used to target single-stranded RNA (ssRNA) substrates in the cytoplasm. For simplicity, only one generic Cas nuclease is drawn. ssDNA, single-stranded DNA.
The Tomato yellow leaf curl virus (TYLCV) belongs to Geminiviridae, a major family of plant viruses with single-stranded circular DNA genomes (Dry et al., 1993). The viral genome forms double-stranded intermediates during replication inside the host cell nuclei. Overexpression of SpCas9 and artificially designed guide RNAs targeting various regions of TYLCV conferred resistance to the virus in Nicotiana benthamiana and tomato (Solanum lycopersicum; Ali et al., 2015; Mahfouz et al., 2017). One potential caveat of this approach is that alterations to the viral DNA sequence may occur near the cleavage target due to DNA repair within the host cell. These mutations could shield the viral DNA from recognition by the guide RNA. Among all the guide RNAs used against TYLCV, the ones targeting the stem-loop sequence within the replication of origin were the most effective, possibly because of the reduced occurrence of viable escapee variants of the virus with mutations in this region (Ali et al., 2015, 2016). A separate lab study demonstrated that overexpression of SpCas9 and guide RNAs in N. benthamiana and Arabidopsis (Arabidopsis thaliana) conferred resistance to the geminivirus family member Beet severe curly top virus (Ji et al., 2015).
CRISPR-Cas has also been used to engineer resistance to RNA viruses, which comprise most known plant viral pathogens (Mahas and Mahfouz, 2018). For example, stable expression of the RNA-targeting nuclease Cas13a and the corresponding guide RNA in N. benthamiana conferred resistance to the RNA virus Turnip mosaic virus (Aman et al., 2018). Using Cas13a to target viral RNA substrates does not induce DNA breakage and thus would not introduce undesired off-target mutations to the host genome (Abudayyeh et al., 2017). Similarly, FnCas9 has been used to engineer resistance against RNA viruses Cucumber mosaic virus and Tobacco mosaic virus in N. benthamiana and Arabidopsis (Zhang et al., 2018).
Although a field test of CRISPR-Cas-based antiviral resistance on crop species has not been reported, the laboratory studies have demonstrated its potential as an antiviral tool. To ensure durable resistance, it is important to consider potential viral evasions from the surveillance by the specific guide RNA used. Choosing genomic targets essential for the replication or movement of the viral pathogen minimizes viral evasions (Ali et al., 2016). Multiplexing the guide RNAs can also improve the robustness. In addition, it has been hypothesized that the use of Cas12a (also known as Cpf1) may reduce the occurrence of escapee viral variants because mutations caused by CRISPR-Cas12a are less likely to abloish the recognition of the target by the orginal guide RNA (Ali et al., 2016). Future efforts on improving CRISPR-Cas for antivirus resistance may also be focused on establishing an in planta adaptive immunity by exploiting the spacer acquisition machinery in the CRISPR-Cas adaptive immune system in prokaryotes (McGinn and Marraffini, 2019).
Deploying Resistance Genes for Broad-Spectrum and Durable Resistance
Pioneering genetic studies in plant-pathogen interaction using flax and flax rust fungus by Flor in the early 1940s (Flor, 1956) established the classic “gene-for-gene” theory, which states that the outcome of any given plant-pathogen interaction is largely determined by a resistance (R) locus from the host and the matching avirulence factor (avr) from the pathogen (Flor, 1971). When both the R gene in the plant host and the cognate avr in the pathogen are present, the plant-pathogen interaction is incompatible and the host exhibits full resistance to the pathogen (Flor, 1971). The effectiveness of R gene-mediated resistance was first demonstrated by British scientist Rowland Biffen in wheat (Triticum sp.) breeding in the early twentieth century (Biffen, 1905).
Since that time, numerous R genes have been cloned and introduced into varieties of the same species (De Wit et al., 1985), across species boundaries (Song et al., 1995) and across genera (Tai et al., 1999). For example, the introduction of the maize (Zea mays) R gene Rxo1 into rice (Oryza sativa) conferred resistance to the bacterial streak pathogen Xanthomonas oryzae pv. oryzicola under lab conditions (Zhao et al., 2005). In tomato, multiyear fields trials under commercial type growth conditions have demonstrated that tomatoes expressing the pepper Bs2 R gene confer robust resistance to Xanthomonas sp. causing the bacterial spot disease (Horvath et al., 2015; Kunwar et al., 2018). Wheat transgenically expressing various alleles of the wheat resistance locus Pm3 exhibited race-specific resistance to powdery mildew in the field (Brunner et al., 2012). Potatoes transgenically expressing wild potato R gene RB or Rpi-vnt1.1 display strong field resistance to Phytophthora infestans, the causal agent of potato late blight (Halterman et al., 2008; Bradeen et al., 2009; Foster et al., 2009; Jones et al., 2014). Notably, transgenic potato expressing Rpi-vnt1.1 developed by J.R. Simplot is to date the only case of a genetically engineered crop with enhanced resistance to a nonviral pathogen that has been approved for commercial use (Table 1).
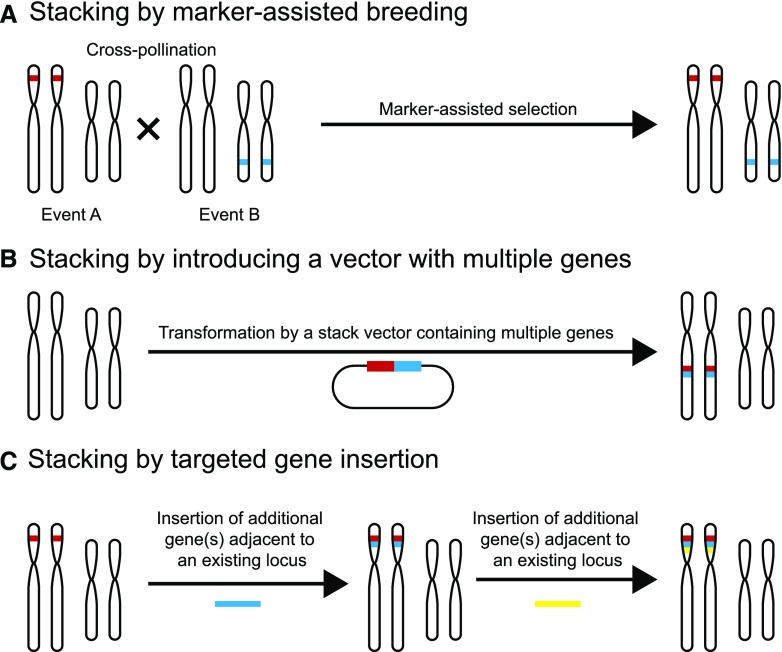
Successful pathogens often evade detection by host R genes (Jones and Dangl, 2006). Thus, disease resistance conferred by a single R gene often lacks durability in the field because pathogens can evolve to evade recognition by mutating the corresponding avr gene. For improved durability and to broaden the resistance spectrum, multiple R genes are often introduced simultaneously, which is commonly known as stacking (Li et al., 2001; Fuchs, 2017; Mundt, 2018). Resistance conferred by stacked R genes is predicted to be long lasting, as the evolution of a pathogen strain that could overcome resistance conferred by multiple R genes simultaneously is a low occurrence event. One approach to stack R genes is by cross breeding preexisting R loci (Fig. 2). Breeders can then use marker-assisted selection to identify the progeny with the desired R gene composition (Das and Rao, 2015). For example, bacterial blight in rice, caused by the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), is a serious disease in much of Asia and parts of Africa (Niño-Liu et al., 2006). Through cross breeding and marker-assisted selection, three R genes that confer resistance to bacterial blight in rice, Xa21, Xa5, and Xa13 were introduced into a deep-water rice cultivar called Jalmagna (Pradhan et al., 2015). The resulting line with the stacked R genes exhibited a higher level of field resistance to eight Xoo isolates tested (Pradhan et al., 2015). Although marker-assisted selection has largely improved the efficiency of the selection process, combining multiple loci through this approach can still be highly time consuming.
Figure 2.
Methods of R gene stacking. A, Stacking by marker-assisted breeding is performed by cross pollinating individuals with existing trait loci followed by marker-assisted selection for progeny with combined trait loci. B, Stacking can be completed by combining multiple genes into a single stack vector and introducing them together as a single transgenic event. C, Stacking by targeted insertion aims at placing new gene(s) adjacent to an existing locus. This process can be performed iteratively to stack large numbers of genes. In B and C, the stacked genes are genetically linked and thus can be easily introduced into a different genetic background as a single locus through breeding.
As an alternative to gene stacking through marker-assisted selection, scientists can assemble multiple R gene cassettes onto one plasmid and then introduce this R gene cluster en bloc at a single genetic locus through plant transformation (Fig. 2; Dafny-Yelin and Tzfira, 2007; Que et al., 2010). This approach, called molecular stacking, simplifies the selection process, as all the R genes introduced this way are inherited as a single genetic locus. As an example of molecular stacking, Zhu et al. (2012) introduced molecular stacking of three broad-spectrum potato late blight R genes Rpi-sto1, Rpi-vnt1.1, and Rpi-blb3 through Agrobacterium-based transformation of a susceptible potato cultivar. The resulting triple gene transformants displayed broad-spectrum resistance equivalent to the sum of the strain-specific resistance conferred by all three individual Rpi genes under greenhouse conditions (Zhu et al., 2012). In a related study, a single DNA fragment harboring Rpi-vnt1.1 and Rpi-sto1 was introduced into three different potato varieties through Agrobacterium-mediated transformation (Jo et al., 2014). The R gene-stacked lines showed broad-spectrum late blight resistance because of the introduction of both R genes (Jo et al., 2014). Notably, no foreign DNA such as a selectable marker gene was included in the inserted DNA fragment in the double gene study (Jo et al., 2014), which may reduce the number of regulatory approvals needed for the resulting products.
Resistance to the late blight pathogen in both the double gene-stacked and the triple gene-stacked potatoes mentioned above was confirmed under field conditions (Haverkort et al., 2016). It is estimated that proper spatial and temporal deployment of late blight R gene stacks in potatoes can reduce the amount of fungicide used on this crop by over 80% (Haverkort et al., 2016). Similarly, in a field study of two African highland potato varieties in Uganda, Ghislain et al. (2018) reported that significant field resistance to the late blight pathogen could be achieved by the molecular stacking of three R genes (RB, Rpi-blb2, and Rpi-vnt1.1). The yields of these R gene stacked potato varieties were three times higher than the national average. These results demonstrate that resistance achieved by this strategy does not negatively affect yield (Ghislain et al., 2018). The above studies show the simplicity and effectiveness of molecular stacking for engineering broad-spectrum disease resistance, especially in vegetatively propagated crop species, where breeding stacks are not practical.
Despite the advantages of molecular stacking, the number of genes that can be introduced through molecular stacking is often restrained by the limit in the length of the DNA insert that can be put into a vector (Que et al., 2010). This limitation can be overcome if DNA fragments can be sequentially inserted at the same genomic target (Fig. 2). Recent breakthroughs in genome-editing technologies in plants enable such targeted insertion of DNA fragments in diverse crop species (Puchta and Fauser, 2013; Rinaldo and Ayliffe, 2015; Kumar et al., 2016; Voytas, 2017). This technology allows multiple R gene cassettes to be inserted at a single locus in multiple rounds (Ainley et al., 2013). As genome-editing platforms in plants continue to be improved, the efficiency of targeted insertion, the size limit of the DNA inserts, and the number of applicable plant species are increasing. Future advancements in targeted gene insertion would offer new opportunities for stacking of large numbers of R genes and engineered viral resistance at a single locus for broad-spectrum, durable disease resistance and convenience in breeding.
Enriching the Known Repertoire of Immune Receptors
Plants possess immune receptors that perceive pathogens and trigger cellular defense responses. Discovery of novel immune receptors recognizing major virulence factors will enrich the repertoire of known immune receptor genes that may be deployed in the field. The nucleotide-binding leucine-rich repeats (NLR) family proteins comprise a major category of intracellular immune receptors conserved across the plant and animal kingdoms (Maekawa et al., 2011; Li et al., 2015; Bentham et al., 2017; Zhang et al., 2017). A distinct hallmark for NLR-mediated defense is the onset of localized programmed cell death known as the hypersensitive response, which plays a crucial role in restricting the movement of pathogens (del Pozo and Lam, 1998; Pontier et al., 1998). The hypersensitive response is often used as a marker to screen diverse plant germplasm for novel functional NLRs recognizing known effectors (Vleeshouwers and Oliver, 2014). During the screening, core virulence factors are delivered into plant leaves either as transiently expressed genes through Agro-infiltration (Du et al., 2014) or as genes expressed in laboratory bacterial strains with a functional secretion system (Zhang and Coaker, 2017).
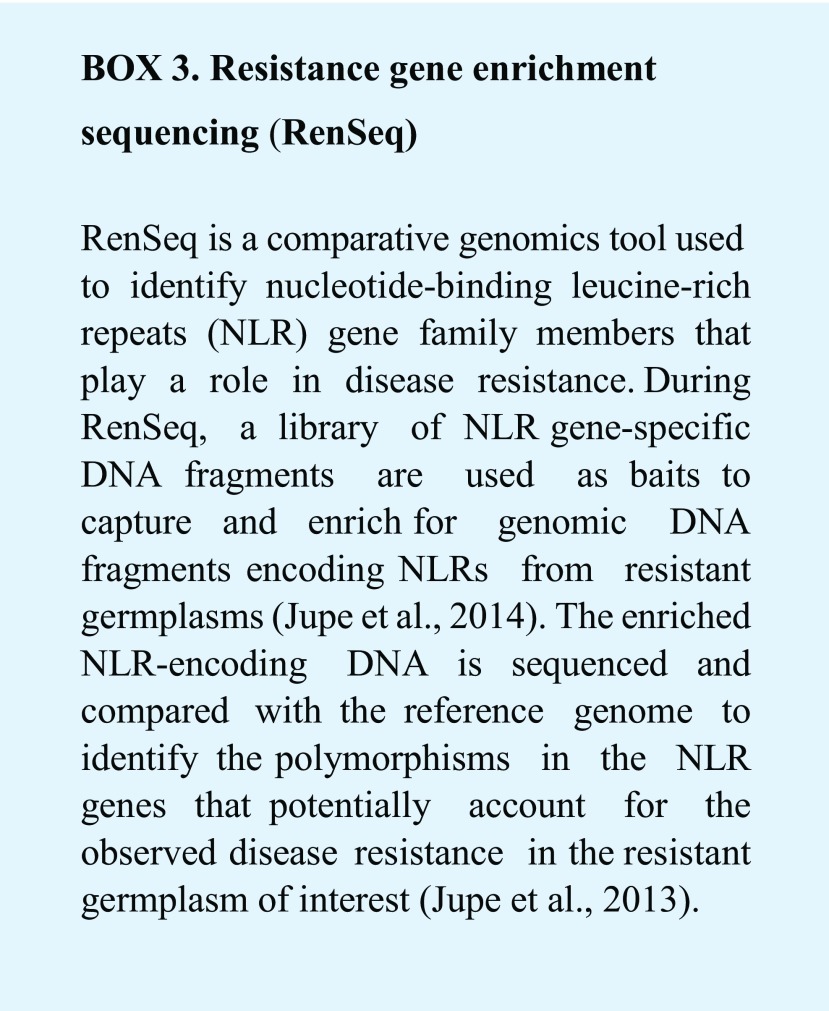
Once a collection of germplasm exhibiting various degrees of resistance to a particular pathogen strain is identified, comparative genomics tools such as resistance gene enrichment sequencing (RenSeq; Jupe et al., 2013, 2014) can be applied to identify genomic variants in NLR genes that are linked to disease phenotypes (Box 3). This leads to the cloning of new NLR genes and their potential deployment in crop protection through genetic engineering. For example, RenSeq was successfully applied in the accelerated identification and cloning of an anti-P. infestans NLR gene Rpi-amr3i from Solanum americanum, a wild potato relative (Witek et al., 2016). Transgenic expression of Rpi-amr3i in potato conferred full resistance to P. infestans in greenhouse conditions (Witek et al., 2016). In a more recent study, Chen et al. described the rapid mapping of a newly identified anti-potato-late-blight NLR gene Rpi-ver1 by RenSeq in the wild potato species Solanum verrucosum (Chen et al., 2018).
In addition to its application in potato research, RenSeq has also been employed in the cloning of wheat NLR resistance genes against the stem rust fungus Puccinia graminis f. sp. tritici. As an example, Arora et al. (2019) used RenSeq to survey accessions of a wild progenitor species of bread wheat, Aegilops tauschii ssp. strangulata for trait-linked. This led to the rapid cloning of four stem rust (Sr) resistance genes (Arora et al., 2019). In a related study, Steuernagel et al. (2016) applied RenSeq to a mutagenized hexaploid bread wheat population to identify mutations disrupting resistance to the stem rust fungus. The study revealed the identity of two stem rust NLR genes, Sr22 and Sr45, which confer resistance to commercially important races of the stem rust pathogen (Steuernagel et al., 2016). Although future field experiments are required to evaluate the potential of the deployment of these newly identified NLR genes, the above lab studies demonstrate that RenSeq is a powerful tool to rapidly identify novel NLR genes.
In addition to the identification of useful immune receptors from diverse plant germplasm, researchers have attempted to engineer known immune receptors for new ligand specificity. For example, the fusion of the ectodomain of the Arabidopsis pattern recognition receptor EF-Tu receptor (EFR) to the intracellular domain of the phylogenetically related rice receptor Xa21 yielded a functional chimeric receptor in both rice and Arabidopsis (Holton et al., 2015; Schwessinger et al., 2015). This receptor triggers defense markers when transgenic tissues are treated with elf18, the ligand of EFR, although whole-plant resistance to the microbe was weak or undetectable (Holton et al., 2015; Schwessinger et al., 2015). Conversely, expression of a fusion receptor consisting of the ectodomain of Xa21 and the cytoplasmic domain of EFR in rice conferred robust resistance to Xoo expressing the cognate ligand of Xa21 (Thomas et al., 2018). Two related studies reported that chimeric receptors generated by fusing the rice chitin-binding protein Chitin Elicitor-Binding Protein and the intracellular protein kinase domain of the rice receptor-like kinase Xa21 or Pi-d2 confers disease resistance to the rice blast fungus (Kishimoto et al., 2010; Kouzai et al., 2013). Although these studies have not been advanced to field trials, they demonstrate that domain swapping among immune receptors may be an attractive approach in engineering broadened recognition specificity.
NLRs often perceive pathogens indirectly by monitoring the modification of host target proteins by pathogen-derived virulence factors (Jones and Dangl, 2006). Some of these host target proteins have evolved to serve as decoys that are targeted by virulence factors (van der Hoorn and Kamoun, 2008). For example, the NLR protein RESISTANCE TO PSEUDOMONAS SYRINGAE 5 (RPS5), activates defense responses in Arabidopsis (Simonich and Innes, 1995). Defense is triggered when the plant decoy protein PBS1, a kinase, is cleaved by the protease AvrPphB secreted by the bacterial pathogen Pseudomonas syringae into the plant cell (Ade et al., 2007). Kim et al. (2016) altered the protease cleavage site in PBS1 to expand the resistance spectrum of RPS5 so that it could recognize other pathogens. For example, they found that they could remove the cleavage site of PBS1 that is recognized by the AvrPphB protease and replace it with cleavage site variants recognized by other pathogen proteases such as the AvrRpt2 protease from Pseudomonas syringae, or the Nla protease from Turnip mosaic virus (Kim et al., 2016). The engineered forms of PBS1 are cleaved in vivo by these proteases, which activates RPS5 defense in response to the corresponding pathogen strains (Kim et al., 2016). These successful examples make decoy modification an attractive approach to engineer resistance to new pathogens (Kourelis et al., 2016).
Altered specificity and activity of immune receptors can also be achieved by directed molecular evolution (Lassner and Bedbrook, 2001). During this process, immune receptor genes are subjected to mutagenesis to generate multiple variants, which are screened for their immune functions. The process can be performed iteratively for continuous improvement in the activity of the gene product. Two pioneering studies demonstrated the potential value of directed molecular evolution in expanding the pool of available immune receptor genes. In a search for gain-of-function mutants of the potato late blight NLR gene R3a, Segretin et al. (2014) screened a library of R3a variants generated through PCR-based mutagenesis and identified R3a mutants recognizing additional Avr3a isoforms from P. infestans and the related blight pathogen Phytophthora capsici. In a follow-up study, the newly identified mutation in R3a was transferred into its tomato ortholog I2 and expanded the recognition spectrum of the NLR encoded when transiently expressed in N. benthaminana (Giannakopoulou et al., 2015).
Recently, a CRISPR-based mutagenesis platform known as EvolvR was developed for the directed evolution of a precisely defined window of genomic DNA in vivo (Halperin et al., 2018; Sadanand, 2018). EvolvR employs the mutagenesis activity of a fusion of the Cas9 nickase and an error-prone DNA polymerase to introduce nucleotide diversification at specific genomic regions defined by guide RNAs (Halperin et al., 2018). For example, EvolvR may potentially be used to develop novel alleles of the rice immune receptor Xa21 with the ability to recognize a broader array of Xoo strains, which secrete ligand variants (Pruitt et al., 2015; Luu et al., 2019). Variants of Xa21 receptors carrying amino acid substitutions predicted to alter ligand specificity can be screened for and introduced into susceptible rice cultivars. Future development of high-throughput functional screening methods to quickly identify gene variants conferring resistance phenotypes would broaden the application of directed molecular evolution for enhanced disease resistance in plants.
Modifying Susceptibility Genes to Attenuate Pathogenicity
Many plant pathogens hijack host genes to facilitate the infection process. These targeted host genes are often referred to as susceptibility genes (van Schie and Takken, 2014). The modification or removal of host susceptibility genes may, therefore, be an effective strategy to achieve resistance by preventing their manipulation by the pathogens. RNAi and genome-editing platforms (Box 2) enable efficient modification of endogenous susceptibility genes in a relatively simple manner (Rinaldo and Ayliffe, 2015).
The highly conserved eukaryotic translation initiation factor 4E (eIF4E) in many plant species is manipulated by a number of plant viruses to facilitate the viral infection process (Robaglia and Caranta, 2006). Naturally occurring recessive alleles of eIF4E confer viral resistance due to abolished protein-protein interaction with a specialized viral virulence protein (Cavatorta et al., 2008). Based on this knowledge, Cavatorta et al. (2011) demonstrated that transgenic overexpression of a site-directed-mutagenized viral-resistant allele of the eIF4E conferred resistance to Potato virus Y in potato. In a more recent study, Chandrasekaran et al. (2016) knocked out the eIF4E gene in cucumber using CRISPR-Cas and obtained homozygous mutant plants that are resistant to a variety of viral pathogens under greenhouse conditions.
The SWEET family genes encode cross-membrane sugar transporters, many of which are exploited by plant pathogens for virulence (Chen et al., 2010; Streubel et al., 2013). Often, when SWEET genes are activated, more sugar is transported outside of the cell and made available to the bacteria (Chen et al., 2010, 2012). For example, the promoters of rice SWEET11 and SWEET14 are targets of various transcriptional activator-like (TAL) effectors from Xoo (Yang et al., 2006; Antony et al., 2010). Li et al. (2012) used a TAL effector-like nuclease to mutate predicted TAL effector-binding sites within the SWEET14 promoter (Li et al., 2012). The resulting rice exhibits enhanced resistance to at least two strains of Xoo due to the lack of induction of SWEET14 by the pathogens. In a follow-up study, Zhou et al. (2014) successfully generated rice lines carrying mutations in the promoter of SWEET11 using CRISPR-Cas. Mutations within the SWEET14 promoter in rice protoplasts by CRISPR-Cas have also been demonstrated (Jiang et al., 2013). In addition to the genome-editing approach, RNAi has been employed to knock down SWEET11 in rice for enhanced resistance to an Xoo strain carrying the cognate TAL effector (Yang et al., 2006).
The disease susceptibility gene for citrus bacterial canker disease, Citrus sinensis Lateral Organ Boundary1 (CsLOB1) encodes a transcription factor that regulates plant growth and development (Hu et al., 2014). Deletion of the effector binding element within the promoter of the susceptibility gene CsLOB1 by CRISPR-Cas conferred resistance to the bacterial canker pathogen Xanthomonas citri spp. citri (Xcc) in orange (Citrus sinensis; Peng et al., 2017). Similarly, disrupting the coding region of CsLOB1 by CRISPR-Cas in grapefruit (Citrus paradisi) resulted in enhanced resistance to Xcc (Jia et al., 2017). In both studies, the plants were raised in controlled environmental conditions. The effectiveness of resistance gained through mutating CsLOB1 in citrus plants awaits further evaluation by field experiments.
Susceptibility genes are often conserved among plant species (Huibers et al., 2013). Therefore, knowledge of susceptibility genes in one plant species can facilitate the discovery of new susceptibility genes in another plant species. The Arabidopsis susceptibility gene DOWNY MILDEW RESISTANT6 (DMR6) encodes an oxygenase, and its expression is up-regulated during pathogen infection (Van Damme et al., 2005; van Damme et al., 2008; Zeilmaker et al., 2015). Through phylogeny and gene expression analysis, Thomazella et al. (2016) identified a single DMR6 ortholog in tomato (SlDMR6-1). Knocking out SlDMR6-1 in tomato using CRISPR-Cas led to increased resistance to the bacterial pathogen P. syringae under greenhouse conditions (Thomazella et al., 2016). In another study, Sun et al. (2016) selected 11 Arabidopsis genes that had been previously shown to correlate with susceptibility to one or more of six common plant pathogens. In a search for gene products with similar amino acid sequences within the potato genome, they identified 11 candidate susceptibility genes in potato (Sun et al., 2016). Silencing of six of these candidate genes individually in potato using RNAi conferred enhanced or complete resistance to the late blight pathogen in controlled environmental conditions (Sun et al., 2016). These studies show that advancements in Arabidopsis biology and identification of an increasing number of susceptibility genes have enabled the discovery of additional susceptibility genes in a large variety of crop species.
Modifying susceptibility genes to thwart virulence strategies of pathogens holds great potential in crop protection. Because a given allele that confers susceptibility to one pathogen may confer resistance to another or have other essential biological functions (Lorang et al., 2012; McGrann et al., 2014), it is important to evaluate the potential side-effects of modifying susceptibility genes. It remains to be determined whether the observed resistance in the instances discussed in the laboratory studies is robust or durable under field conditions or if the plants perform well in the field.
CONCLUSION
Since the first report of a genetically engineered crop conferring resistance to disease in 1986, a virus-resistant tobacco expressing a viral coat protein gene (Abel et al., 1986), there have been tremendous breakthroughs both in labs and in the field. In terms of successful field applications, antiviral resistance has had a clear head start with most of the commercialized genetically engineered crops for disease resistance thus far falling into this category. However, with new technologies that enable rapid identification of novel immune receptor genes, such as RenSeq and directed molecular evolution, the pool of deployable genes for enhanced resistance to other microbes has expanded substantially. In parallel, advancements in molecular stacking and targeted gene insertion through genome editing are expected to play a major role in generating broad-spectrum resistance against both viral and nonviral pathogens in the near future. Furthermore, the increasingly versatile, accurate, and efficient genome-editing tools such as CRISPR-Cas enable accurate modification of endogenous genes for disease resistance, including, but not limited to, susceptibility and the decoy genes.
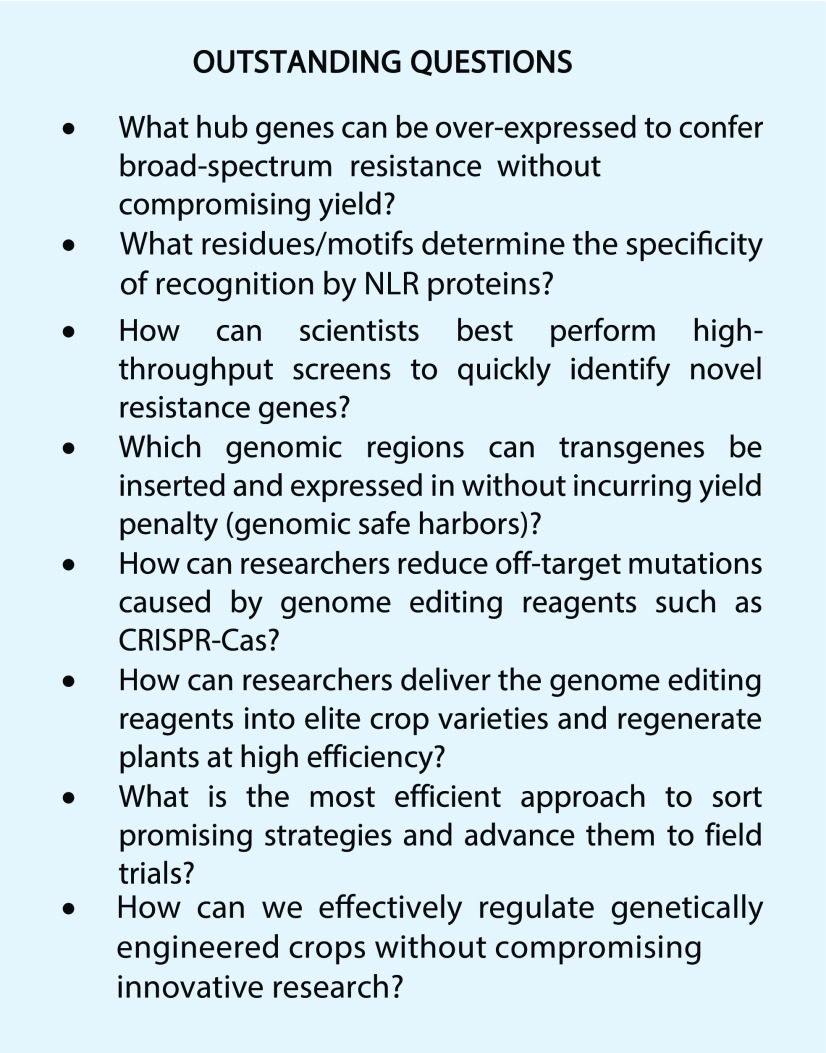
In the Outstanding Questions Box, we present some examples of challenges faced in genetic engineering for resistance. Answers to these questions will fill important knowledge gaps and can be used to develop new strategies for engineering plant disease resistance.
Acknowledgments
We thank Dr. Mawsheng Chern and Dr. Michael Steinwand for valuable suggestions on the manuscript.
Footnotes
This work was supported by research grants from the Innovative Genomics Institute (49411), the National Institutes of Health (R01GM122968) and the United States Department of Agriculture (NIFA 2017-67013-26590) to P.C.R.
Articles can be viewed without a subscription.
References
- Abel PP, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT, Beachy RN (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232: 738–743 [DOI] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017) RNA targeting with CRISPR-Cas13. Nature 550: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Ai T, Zhang L, Gao Z, Zhu CX, Guo X (2011) Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol (Stuttg) 13: 304–316 [DOI] [PubMed] [Google Scholar]
- Ainley WM, Sastry-Dent L, Welter ME, Murray MG, Zeitler B, Amora R, Corbin DR, Miles RR, Arnold NL, Strange TL, et al. (2013) Trait stacking via targeted genome editing. Plant Biotechnol J 11: 1126–1134 [DOI] [PubMed] [Google Scholar]
- Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Ali S, Tashkandi M, Zaidi SS, Mahfouz MM (2016) CRISPR/Cas9-mediated immunity to geminiviruses: Differential interference and evasion. Sci Rep 6: 26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. (2001) microRNAs: tiny regulators with great potential. Cell 107: 823–826 [DOI] [PubMed] [Google Scholar]
- Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22: 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragão FJL, Faria JC (2009) First transgenic geminivirus-resistant plant in the field. Nat Biotechnol 27: 1086–1088, author reply 1088–1089 [DOI] [PubMed] [Google Scholar]
- Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, Johnson R, Enk J, Periyannan S, Singh N, et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat Biotechnol 37: 139–143 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34: 933–941 [DOI] [PubMed] [Google Scholar]
- Bentham A, Burdett H, Anderson PA, Williams SJ, Kobe B (2017) Animal NLRs provide structural insights into plant NLR function. Ann Bot 119: 827–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffen RH. (1905) Mendel’s laws of inheritance and wheat breeding. J Agric Sci 1: 4–48 [Google Scholar]
- Bonfim K, Faria JC, Nogueira EOPL, Mendes EA, Aragão FJL (2007) RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant Microbe Interact 20: 717–726 [DOI] [PubMed] [Google Scholar]
- Bradeen JM, Iorizzo M, Mollov DS, Raasch J, Kramer LC, Millett BP, Austin-Phillips S, Jiang J, Carputo D (2009) Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol Plant Microbe Interact 22: 437–446 [DOI] [PubMed] [Google Scholar]
- Brunner S, Stirnweis D, Diaz Quijano C, Buesing G, Herren G, Parlange F, Barret P, Tassy C, Sautter C, Winzeler M, et al. (2012) Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Cavatorta JR, Savage AE, Yeam I, Gray SM, Jahn MM (2008) Positive Darwinian selection at single amino acid sites conferring plant virus resistance. J Mol Evol 67: 551–559 [DOI] [PubMed] [Google Scholar]
- Cavatorta J, Perez KW, Gray SM, Van Eck J, Yeam I, Jahn M (2011) Engineering virus resistance using a modified potato gene. Plant Biotechnol J 9: 1014–1021 [DOI] [PubMed] [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17: 1140–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chen X, Lewandowska D, Armstrong MR, Baker K, Lim TY, Bayer M, Harrower B, McLean K, Jupe F, Witek K, et al. (2018) Identification and rapid mapping of a gene conferring broad-spectrum late blight resistance in the diploid potato species Solanum verrucosum through DNA capture technologies. Theor Appl Genet 131: 1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Christou P. (2013) Plant genetic engineering and agricultural biotechnology 1983-2013. Trends Biotechnol 31: 125–127 [DOI] [PubMed] [Google Scholar]
- Collinge DB. (2009) Cell wall appositions: The first line of defence. J Exp Bot 60: 351–352 [DOI] [PubMed] [Google Scholar]
- Collinge DB, Jørgensen HJ, Lund OS, Lyngkjaer MF (2010) Engineering pathogen resistance in crop plants: current trends and future prospects. Annu Rev Phytopathol 48: 269–291 [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BPHJ (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CTNBio, Brazil (2018) National Biosafety Technical Commission Normative Resolution No. 16. https://agrobiobrasil.org.br/wp-content/uploads/2018/05/Normative-Resolution-16-of-January-15-2018.pdf (March 12, 2019)
- Dafny-Yelin M, Tzfira T (2007) Delivery of multiple transgenes to plant cells. Plant Physiol 145: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Rao GJN (2015) Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front Plant Sci 6: 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MJ, Ying Z (2004) Development of papaya breeding lines with transgenic resistance to Papaya ringspot virus. Plant Dis 88: 352–358 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Lam E (1998) Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol 8: R896. [DOI] [PubMed] [Google Scholar]
- De Wit PJ, Hofman AE, Velthuis GC, Kuć JA (1985) Isolation and characterization of an elicitor of necrosis isolated from intercellular fluids of compatible interactions of Cladosporium fulvum (syn. Fulvia fulva) and tomato. Plant Physiol 77: 642–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry IB, Rigden JE, Krake LR, Mullineaux PM, Rezaian MA (1993) Nucleotide sequence and genome organization of tomato leaf curl geminivirus. J Gen Virol 74: 147–151 [DOI] [PubMed] [Google Scholar]
- Du J, Rietman H, Vleeshouwers VG (2014) Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J Vis Exp 3: e50971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Wang CH, Fang RX, Guo HS (2008) Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J Virol 82: 11084–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. (1956) The complementary genic systems in flax and flax rust. Adv Genet 8: 29–54 [Google Scholar]
- Flor HH. (1971) Current status of gene-for-gene concept. Annu Rev Phytopathol 9: 275–296 [Google Scholar]
- Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen E, Jones JDG (2009) Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact 22: 589–600 [DOI] [PubMed] [Google Scholar]
- Franceschetti M, Maqbool A, Jimenez-Dalmaroni MJ, Pennington HG, Kamoun S, Banfield MJ (2017) Effectors of filamentous plant pathogens: Commonalities amid diversity. Microbiol Mol Biol Rev 81: e0006616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M. (2017) Pyramiding resistance-conferring gene sequences in crops. Curr Opin Virol 26: 36–42 [DOI] [PubMed] [Google Scholar]
- Fuchs M, Gonsalves D (2007) Safety of virus-resistant transgenic plants two decades after their introduction: lessons from realistic field risk assessment studies. Annu Rev Phytopathol 45: 173–202 [DOI] [PubMed] [Google Scholar]
- Galvez LC, Banerjee J, Pinar H, Mitra A (2014) Engineered plant virus resistance. Plant Sci 228: 11–25 [DOI] [PubMed] [Google Scholar]
- Ghislain M, Byarugaba AA, Magembe E, Njoroge A, Rivera C, Román ML, Tovar JC, Gamboa S, Forbes GA, Kreuze JF, et al. (2018) Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol J, doi:10.1111/pbi.13042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulou A, Steele JF, Segretin ME, Bozkurt TO, Zhou J, Robatzek S, Banfield MJ, Pais M, Kamoun S (2015) Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol Plant Microbe Interact 28: 1316–1329 [DOI] [PubMed] [Google Scholar]
- Gonsalves D. (1998) Control of papaya ringspot virus in papaya: A case study. Annu Rev Phytopathol 36: 415–437 [DOI] [PubMed] [Google Scholar]
- Gonsalves D. (2006) Transgenic papaya: Development, release, impact and challenges. Adv Virus Res 67: 317–354 [DOI] [PubMed] [Google Scholar]
- Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV, Dueber JE (2018) CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560: 248–252 [DOI] [PubMed] [Google Scholar]
- Halterman DA, Kramer LC, Wielgus S, Jiang J (2008) Performance of transgenic potato containing the late blight resistance gene RB. Plant Dis 92: 339–343 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, Vossen JH, Visser RGF (2016) Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res 59: 35–66 [Google Scholar]
- Holton N, Nekrasov V, Ronald PC, Zipfel C (2015) The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog 11: e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DM, Pauly MH, Hutton SF, Vallad GE, Scott JW, Jones JB, Stall RE, Dahlbeck D, Staskawicz BJ, Tricoli D, et al. (2015) The pepper Bs2 gene confers effective field resistance to bacterial leaf spot and yield enhancement in Florida tomatoes. Acta Hortic 1069: 47–51 [Google Scholar]
- Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer WB, Yang B, White FF, Wang N, Jones JB (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci USA 111: E521–E529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibers RP, Loonen AE, Gao D, Van den Ackerveken G, Visser RG, Bai Y (2013) Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One 8: e67467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardi V, Tavazza M (2015) Biotechnological strategies and tools for Plum pox virus resistance: Trans-, intra-, cis-genesis, and beyond. Front Plant Sci 6: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1: 15144. [DOI] [PubMed] [Google Scholar]
- Jia H, Zhang Y, Orbović V, Xu J, White FF, Jones JB, Wang N (2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo KR, Kim CJ, Kim SJ, Kim TY, Bergervoet M, Jongsma MA, Visser RGF, Jacobsen E, Vossen JH (2014) Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol 14: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JD, Witek K, Verweij W, Jupe F, Cooke D, Dorling S, Tomlinson L, Smoker M, Perkins S, Foster S (2014) Elevating crop disease resistance with cloned genes. Philos Trans R Soc Lond B Biol Sci 369: 20130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N. (2006) Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci 11: 359–367 [DOI] [PubMed] [Google Scholar]
- Jupe F, Witek K, Verweij W, Sliwka J, Pritchard L, Etherington GJ, Maclean D, Cock PJ, Leggett RM, Bryan GJ, et al. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J 76: 530–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe F, Chen X, Verweij W, Witek K, Jones JD, Hein I (2014) Genomic DNA library preparation for resistance gene enrichment and sequencing (RenSeq) in plants. Methods Mol Biol 1127: 291–303 [DOI] [PubMed] [Google Scholar]
- Kaniewski WK, Thomas PE (2004) The potato story. AgBioForum 7: 41–46 [Google Scholar]
- Karlovsky P. (2011) Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl Microbiol Biotechnol 91: 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoch R, Thakur N (2013) Advances in RNA interference technology and its impact on nutritional improvement, disease and insect control in plants. Appl Biochem Biotechnol 169: 1579–1605 [DOI] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, Innes RW (2016) Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351: 684–687 [DOI] [PubMed] [Google Scholar]
- Kishimoto K, Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y (2010) Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J 64: 343–354 [DOI] [PubMed] [Google Scholar]
- Kitajima S, Sato F (1999) Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J Biochem 125: 1–8 [DOI] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL, Sueldo DJ (2016) Decoy engineering: The next step in resistance breeding. Trends Plant Sci 21: 371–373 [DOI] [PubMed] [Google Scholar]
- Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y (2013) Expression of the chimeric receptor between the chitin elicitor receptor CEBiP and the receptor-like protein kinase Pi-d2 leads to enhanced responses to the chitin elicitor and disease resistance against Magnaporthe oryzae in rice. Plant Mol Biol 81: 287–295 [DOI] [PubMed] [Google Scholar]
- Kubicek CP, Starr TL, Glass NL (2014) Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52: 427–451 [DOI] [PubMed] [Google Scholar]
- Kumar S, Barone P, Smith M (2016) Gene targeting and transgene stacking using intra genomic homologous recombination in plants. Plant Methods 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar S, Iriarte F, Fan Q, Evaristo da Silva E, Ritchie L, Nguyen NS, Freeman JH, Stall RE, Jones JB, Minsavage GV, et al. (2018) Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 108: 1402–1411 [DOI] [PubMed] [Google Scholar]
- Lassner M, Bedbrook J (2001) Directed molecular evolution in plant improvement. Curr Opin Plant Biol 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Li X, Kapos P, Zhang Y (2015) NLRs in plants. Curr Opin Immunol 32: 114–121 [DOI] [PubMed] [Google Scholar]
- Li ZK, Sanchez A, Angeles E, Singh S, Domingo J, Huang N, Khush GS (2001) Are the dominant and recessive plant disease resistance genes similar? A case study of rice R genes and Xanthomonas oryzae pv. oryzae races. Genetics 159: 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo JA, Dougherty WG (2005) Plant pathology and RNAi: a brief history. Annu Rev Phytopathol 43: 191–204 [DOI] [PubMed] [Google Scholar]
- Lindbo JA, Falk BW (2017) The impact of “coat protein-mediated virus resistance” in applied plant pathology and basic research. Phytopathology 107: 624–634 [DOI] [PubMed] [Google Scholar]
- Lomonossoff GP. (1995) Pathogen-derived resistance to plant viruses. Annu Rev Phytopathol 33: 323–343 [DOI] [PubMed] [Google Scholar]
- Lorang J, Kidarsa T, Bradford CS, Gilbert B, Curtis M, Tzeng SC, Maier CS, Wolpert TJ (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science 338: 659–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence A, Verpoorte R (2004) Gene transfer and expression in plants. Methods Mol Biol 267: 329–350 [DOI] [PubMed] [Google Scholar]
- Luu DD, Joe A, Chen Y, Parys K, Bahar O, Pruitt R, Chan LJG, Petzold CJ, Long K, Adamchak C, et al. (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc Natl Acad Sci USA 201818275. 10.1073/pnas.1818275116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Mahas A, Mahfouz M (2018) Engineering virus resistance via CRISPR-Cas systems. Curr Opin Virol 32: 1–8 [DOI] [PubMed] [Google Scholar]
- Mahfouz M, Tashkandi M, Ali Z, Aljedaani F, Shami A (2017) Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. bioRxiv 237735, doi:10.1101/237735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA (2019) Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol 17: 7–12 [DOI] [PubMed] [Google Scholar]
- McGrann GRD, Stavrinides A, Russell J, Corbitt MM, Booth A, Chartrain L, Thomas WTB, Brown JKM (2014) A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. J Exp Bot 65: 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt CC. (2018) Pyramiding for resistance durability: Theory and practice. Phytopathology 108: 792–802 [DOI] [PubMed] [Google Scholar]
- Newell CA, Rozman R, Hinchee MA, Lawson EC, Haley L, Sanders P, Kaniewski W, Tumer NE, Horsch RB, Fraley RT (1991) Agrobacterium-mediated transformation of Solanum tuberosum L. cv. ‘Russet Burbank’. Plant Cell Rep 10: 30–34 [DOI] [PubMed] [Google Scholar]
- Niño-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7: 303–324 [DOI] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Oerke EC, Dehne HW (2004) Safeguarding production: Losses in major crops and the role of crop protection. Crop Prot 23: 275–285 [Google Scholar]
- Orozco P. (2018) Argentina and Brazil Merge Law and Science to Regulate New Breeding Techniques. https://allianceforscience.cornell.edu/blog/2018/01/argentina-and-brazil-merge-law-and-science-to-regulate-new-breeding-techniques/ (March 12, 2019)
- Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S (2000) Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat Biotechnol 18: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J 15: 1509–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon LC (2004) NPR1: The spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7: 456–464 [DOI] [PubMed] [Google Scholar]
- Pontier D, Balagué C, Roby D (1998) The hypersensitive response. A programmed cell death associated with plant resistance. C R Acad Sci III 321: 721–734 [DOI] [PubMed] [Google Scholar]
- Pradhan SK, Nayak DK, Mohanty S, Behera L, Barik SR, Pandit E, Lenka S, Anandan A (2015) Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice (N Y) 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS (2015) Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA 112: 6164–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, Thomas N, Liu F, Albert M, Robinson MR, Chan LJ, Luu DD, Chen H, et al. (2015) The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv 1: e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57: 629–637 [DOI] [PubMed] [Google Scholar]
- Qu J, Ye J, Fang R (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81: 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Chilton MD, de Fontes CM, He C, Nuccio M, Zhu T, Wu Y, Chen JS, Shi L (2010) Trait stacking in transgenic crops: Challenges and opportunities. GM Crops 1: 220–229 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo AR, Ayliffe M (2015) Gene targeting and editing in crop plants: A new era of precision opportunities. Mol Breed 35: 40 [Google Scholar]
- Robaglia C, Caranta C (2006) Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Rosa C, Kuo YW, Wuriyanghan H, Falk BW (2018) RNA interference mechanisms and applications in plant pathology. Annu Rev Phytopathol 56: 581–610 [DOI] [PubMed] [Google Scholar]
- Sadanand S. (2018) EvolvR-ing to targeted mutagenesis. Nat Biotechnol 36: 819. [DOI] [PubMed] [Google Scholar]
- Sanford JC, Johnston SA (1985) The concept of parasite-derived resistance—deriving resistance genes from the parasite’s own genome. J Theor Biol 113: 395–405 [Google Scholar]
- Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR, et al. (2015) Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog 11: e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorza R, Ravelonandro M, Callahan AM, Cordts JM, Fuchs M, Dunez J, Gonsalves D (1994) Transgenic plums (Prunus domestica L.) express the plum pox virus coat protein gene. Plant Cell Rep 14: 18–22 [DOI] [PubMed] [Google Scholar]
- Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JIB, Banfield MJ, Kamoun S (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol Plant Microbe Interact 27: 624–637 [DOI] [PubMed] [Google Scholar]
- Simonich MT, Innes RW (1995) A disease resistance gene in Arabidopsis with specificity for the avrPph3 gene of Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 8: 637–640 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JDG, et al. (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34: 652–655 [DOI] [PubMed] [Google Scholar]
- Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013) Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200: 808–819 [DOI] [PubMed] [Google Scholar]
- Sun K, Wolters AM, Vossen JH, Rouwet ME, Loonen AE, Jacobsen E, Visser RG, Bai Y (2016) Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res 25: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NC, Oksenberg N, Liu F, Caddell D, Nalyvayko A, Nguyen Y, Schwessinger B, Ronald PC (2018) The rice XA21 ectodomain fused to the Arabidopsis EFR cytoplasmic domain confers resistance to Xanthomonas oryzae pv. oryzae. PeerJ 6: e4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Kaniewski WK, Lawson EC (1997) Reduced field spread of potato leafroll virus in potatoes transformed with the potato leafroll virus coat protein gene. Plant Dis 81: 1447–1453 [DOI] [PubMed] [Google Scholar]
- Thomazella D, Brail Q, Dahlbeck D, Staskawicz B (2016) CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv, doi: 10.1101/064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Tollefson J. (2011) Brazil cooks up transgenic bean. Nature 478: 168. [DOI] [PubMed] [Google Scholar]
- Tricoli DM, Carney KJ, Russell PF, Mcmaster JR, Groff DW, Hadden KC, Himmel PT, Hubbard JP, Boeshore ML, Quemada HD (1995) Field-evaluation of transgenic squash containing single or multiple virus coat protein gene constructs for resistance to cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus. Biotechnology (N Y) 13: 1458–1465 [Google Scholar]
- Uma B, Rani TS, Podile AR (2011) Warriors at the gate that never sleep: Non-host resistance in plants. J Plant Physiol 168: 2141–2152 [DOI] [PubMed] [Google Scholar]
- USDA (2018) Secretary Perdue Issues USDA Statement on Plant Breeding Innovation. USDA press release no. 0070.18. https://www.usda.gov/media/press-releases/2018/03/28/secretary-perdue-issues-usda-statement-plant-breeding-innovation (March 12, 2019)
- Van Damme M, Andel A, Huibers RP, Panstruga R, Weisbeek PJ, Van den Ackerveken G (2005) Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol Plant Microbe Interact 18: 583–592 [DOI] [PubMed] [Google Scholar]
- van Damme M, Huibers RP, Elberse J, Van den Ackerveken G (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J 54: 785–793 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Kamoun S (2008) From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CCN, Takken FLW (2014) Susceptibility genes 101: How to be a good host. Annu Rev Phytopathol 52: 551–581 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, Oliver RP (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27: 196–206 [DOI] [PubMed] [Google Scholar]
- Voytas D. (2017) Optimizing gene targeting in plants. In Vitro Cell Dev Biol Anim 53 (Suppl 1): S23. [Google Scholar]
- Wang MB, Masuta C, Smith NA, Shimura H (2012) RNA silencing and plant viral diseases. Mol Plant Microbe Interact 25: 1275–1285 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95: 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan A, Lema M (2015) Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food 6: 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JDG (2016) Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol 34: 656–660 [DOI] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA (2016) Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 164: 29–44 [DOI] [PubMed] [Google Scholar]
- Wu WY, Lebbink JHG, Kanaar R, Geijsen N, van der Oost J (2018) Genome editing by natural and engineered CRISPR-associated nucleases. Nat Chem Biol 14: 642–651 [DOI] [PubMed] [Google Scholar]
- Xie M, Zhang S, Yu B (2015) microRNA biogenesis, degradation and activity in plants. Cell Mol Life Sci 72: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, He SY (2013) Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103: 10503–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RC, Xu HL, Yu WG, Lu CG, Long MS, Liu CQ, Pan NS, Chen ZL (1995) Transgenic tomato plants expressing cucumber mosaic virus coat protein (CMV-cp) and their resistance to cucumber mosaic virus (CMV). Acta Agric Jiangsu 11: 42–46 [Google Scholar]
- Ye C, Li H (2010) 20 Years of transgenic research in China for resistance to Papaya ringspot virus. Transgenic Plant J 4: 58–63 [Google Scholar]
- Zamore PD. (2004) Plant RNAi: How a viral silencing suppressor inactivates siRNA. Curr Biol 14: R198–R200 [DOI] [PubMed] [Google Scholar]
- Zeilmaker T, Ludwig NR, Elberse J, Seidl MF, Berke L, Van Doorn A, Schuurink RC, Snel B, Van den Ackerveken G (2015) DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J 81: 210–222 [DOI] [PubMed] [Google Scholar]
- Zhang M, Coaker G (2017) Harnessing effector-triggered immunity for durable disease resistance. Phytopathology 107: 912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zheng Q, Yi X, An H, Zhao Y, Ma S, Zhou G (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J 16: 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li H, Zhang J, Zhang C, Gong P, Ziaf K, Xiao F, Ye Z (2011) Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res 20: 569–581 [DOI] [PubMed] [Google Scholar]
- Zhang X, Dodds PN, Bernoux M (2017) What do we know about NOD-like receptors in plant immunity? Annu Rev Phytopathol 55: 205–229 [DOI] [PubMed] [Google Scholar]
- Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102: 15383–15388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42: 10903–10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li Y, Vossen JH, Visser RGF, Jacobsen E (2012) Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res 21: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Ou-Yang WJ, Zhang YF, Chen ZL (1996) Transgenic sweet pepper plants from Agrobacterium mediated transformation. Plant Cell Rep 16: 71–75 [DOI] [PubMed] [Google Scholar]