ABI5 interacts with MYB49 and represses its function by preventing its binding to the downstream genes bHLH38, bHLH101, HIPP22, and HIPP44, resulting in inactivation of IRT1 and reduced Cd uptake.
Abstract
Abscisic acid (ABA) reduces accumulation of potentially toxic cadmium (Cd) in plants. How the ABA signal is transmitted to modulate Cd uptake remains largely unclear. Here, we report that the basic region/Leu zipper transcription factor ABSCISIC ACID-INSENSITIVE5 (ABI5), a central ABA signaling molecule, is involved in ABA-repressed Cd accumulation in plants by physically interacting with a previously uncharacterized R2R3-type MYB transcription factor, MYB49. Overexpression of the Cd-induced MYB49 gene in Arabidopsis (Arabidopsis thaliana) resulted in a significant increase in Cd accumulation, whereas myb49 knockout plants and plants expressing chimeric repressors of MYB49:ERF-associated amphiphilic repression motif repression domain (SRDX49) exhibited reduced accumulation of Cd. Further investigations revealed that MYB49 positively regulates the expression of the basic helix-loop-helix transcription factors bHLH38 and bHLH101 by directly binding to their promoters, leading to activation of IRON-REGULATED TRANSPORTER1, which encodes a metal transporter involved in Cd uptake. MYB49 also binds to the promoter regions of the heavy metal-associated isoprenylated plant proteins (HIPP22) and HIPP44, resulting in up-regulation of their expression and subsequent Cd accumulation. On the other hand, as a feedback mechanism to control Cd uptake and accumulation in plant cells, Cd-induced ABA up-regulates the expression of ABI5, whose protein product interacts with MYB49 and prevents its binding to the promoters of downstream genes, thereby reducing Cd accumulation. Our results provide new insights into the molecular feedback mechanisms underlying ABA signaling-controlled Cd uptake and accumulation in plants.
Cadmium (Cd) is a nonnutrient metal that is potentially toxic even at low concentrations in plants (Clemens et al., 2002; Yoshihara et al., 2014; Clemens and Ma, 2016; Sasaki et al., 2016). Exposure of plants to Cd increases endogenous abscisic acid (ABA) levels in plant cells (Sharma and Kumar, 2002). Several studies have demonstrated that the application of ABA is a promising strategy for reducing Cd accumulation in crops (Hsu and Kao, 2003; Uraguchi et al., 2009; Fan et al., 2014). For instance, Fan et al. (2014) reported that the decrease in Cd accumulation by ABA treatment correlates with the down-regulation of ABA-inhibited IRON-REGULATED TRANSPORTER1 (IRT1) expression in roots. The same authors also showed that Cd uptake in an irt1 mutant did not respond to ABA application, suggesting an important role of ABA in controlling Cd uptake through IRT1. However, how ABA signaling affects Cd uptake by modulating IRT1 expression remains largely unclear.
Uptake of Cd from soil into roots requires several different heavy metal transporter families, including the cation diffusion facilitator family, heavy metal ATPase family, natural resistance-associated macrophage protein family, yellow stripe1-like (YSL) family, ATP-binding cassette family, multidrug and toxic compound extrusion family, cation/H+ exchanger family, zinc (Zn)-regulated transporters, and iron (Fe)-regulated transporter-like protein (ZIP) family (Guerinot, 2000; Eren and Argüello, 2004; Hirschi, 2004; Wu et al., 2016). IRT1 is a ZIP transporter that functions as a divalent cation transporter, transporting Fe2+, Zn2+, and Cd2+ in plants (Vert et al., 2002; Kobayashi and Nishizawa, 2012). Several studies have reported the important role of IRT1 in Cd accumulation and tolerance. Loss of function of the irt1 mutant markedly reduced Cd accumulation (Vert et al., 2002; Fan et al., 2014). These studies suggested that in addition to its role in Fe transportation, IRT1 is a critical transporter that is responsible for Cd uptake and accumulation in plants.
Plants have evolved fine-tuned mechanisms to protect cells from Cd toxicity by partitioning Cd into vacuoles or trapping free Cd2+ in the cytosol (Tehseen et al., 2010; Clemens and Ma, 2016). In eukaryotes, Cd2+ that enters the cells is chelated by small-molecule ligands and protein molecular chaperones, such as dehydrins (Xu et al., 2008) and heavy metal-associated isoprenylated plant proteins (HIPPs). Studies from several independent laboratories have demonstrated the roles of HIPPs in heavy metal transport, accumulation, and detoxification (Chu et al., 2005; Tehseen et al., 2010; Zhao et al., 2013). The Arabidopsis (Arabidopsis thaliana) hipp20hipp21hipp22 triple mutant accumulated less Cd and was more sensitive to Cd toxicity than wild-type plants (Tehseen et al., 2010).
Modulation of Cd transport and responsive gene expression is an important mechanism for Cd accumulation in plants. Metal-responsive elements (MREs) were originally shown to modulate metallothionein gene expression in animals. Sun et al. (2015) found that a bean (Phaseolus vulgaris) short PvSR2 transcript (S-PvSR2) functions as a plant MRE-binding transcription factor (TF) to regulate Cd tolerance by directly regulating the feedback-insensitive anthranilate synthase α-subunit gene ASA2 expression in tobacco (Nicotiana tabacum). However, no homologous gene of S-PvSR2 has been found in other plant species. In the past 10 years, several Fe-, copper (Cu)-, and Zn-deficiency response element-binding TFs have been identified (Kropat et al., 2005; Assunção et al., 2010; Sommer et al., 2010). Arabidopsis bZIP19 and bZIP23 regulate Zn deficiency adaptation by directly binding to Zn-deficiency response elements of target genes (Assunção et al., 2010). The Cu response regulator CRR1 regulates Cu deficiency tolerance by directly binding to Cu-responsive elements in Chlamydomonas reinhardtii (Sommer et al., 2010). Arabidopsis SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 also regulates Cu deficiency tolerance by directly binding to Cu-responsive elements within the promoter regions of FERRIC REDUCTION OXIDASE4 (FRO4), FRO5 (Bernal et al., 2012), and COPPER TRANSPORTER6 (Jung et al., 2012). Rice (Oryza sativa) OsIDEF1 regulates Fe homeostasis by directly binding to the Fe-deficiency response element of target genes (Kobayashi et al., 2007, 2012). Given that most of these target genes are also responsible for Cd uptake and tolerance, these TFs may potentially be involved in Cd homeostasis.
In addition to direct binding with MREs, some TFs are also involved in modulating metal accumulation and detoxification in plants. For example, Arabidopsis MYB10 and MYB72 regulate Fe deficiency tolerance by regulating the expression of the nicotianamine synthases NAS2 and NAS4 in roots (Palmer et al., 2013). The biosynthesis of phytosiderophore nicotianamine is also regulated by basic helix-loop-helix (bHLH) TFs and the Met salvage pathway (Aprile et al., 2018). Several studies have demonstrated that the bHLH TF FER-like iron deficiency-induced TF (FIT) forms heterodimers with four Ib subgroups of the bHLH TFs bHLH38, bHLH39, bHLH100, and bHLH101 to activate FRO2 and IRT1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005, 2008; Wang et al., 2013). The IVc subgroup of the bHLH TFs bHLH34, bHLH104, and bHLH105 functions as homodimers or heterodimers to nonredundantly regulate the Fe-deficiency response by directly targeting the bHLH38, bHLH39, bHLH100, bHLH101, and POPEYE (PYE) genes (Zhang et al., 2015). Expression of the bHLH38, bHLH39, bHLH100, bHLH101, bHLH104, and PYE genes could be regulated by Cd, and coexpression of FIT with bHLH38 and bHLH39 or overexpression of bHLH104 increased Cd sequestration in the roots (Wu et al., 2012; Yao et al., 2018), indicating the role of this module in Cd accumulation and tolerance.
In this study, we found that the basic region/Leu zipper TF ABSCISIC ACID-INSENSITIVE5 (ABI5) is involved in Cd accumulation. Overexpression of ABI5 reduces, whereas loss of function in the abi5 mutant increases, Cd accumulation in Arabidopsis, indicating the involvement of ABI5 in modulating Cd uptake. To identify transcriptional regulators involved in ABI5-mediated Cd accumulation, we searched for ABI5-interacting proteins and identified the Cd-induced R2R3-MYB TF MYB49. Here, we examined the function and the roles of an uncharacterized R2R3-MYB member, MYB49, in Arabidopsis. We found that Arabidopsis MYB49 regulates Cd accumulation by directly regulating bHLH38, bHLH101, HIPP22, and HIPP44 expression.
RESULTS
ABI5 Affects Cd Accumulation
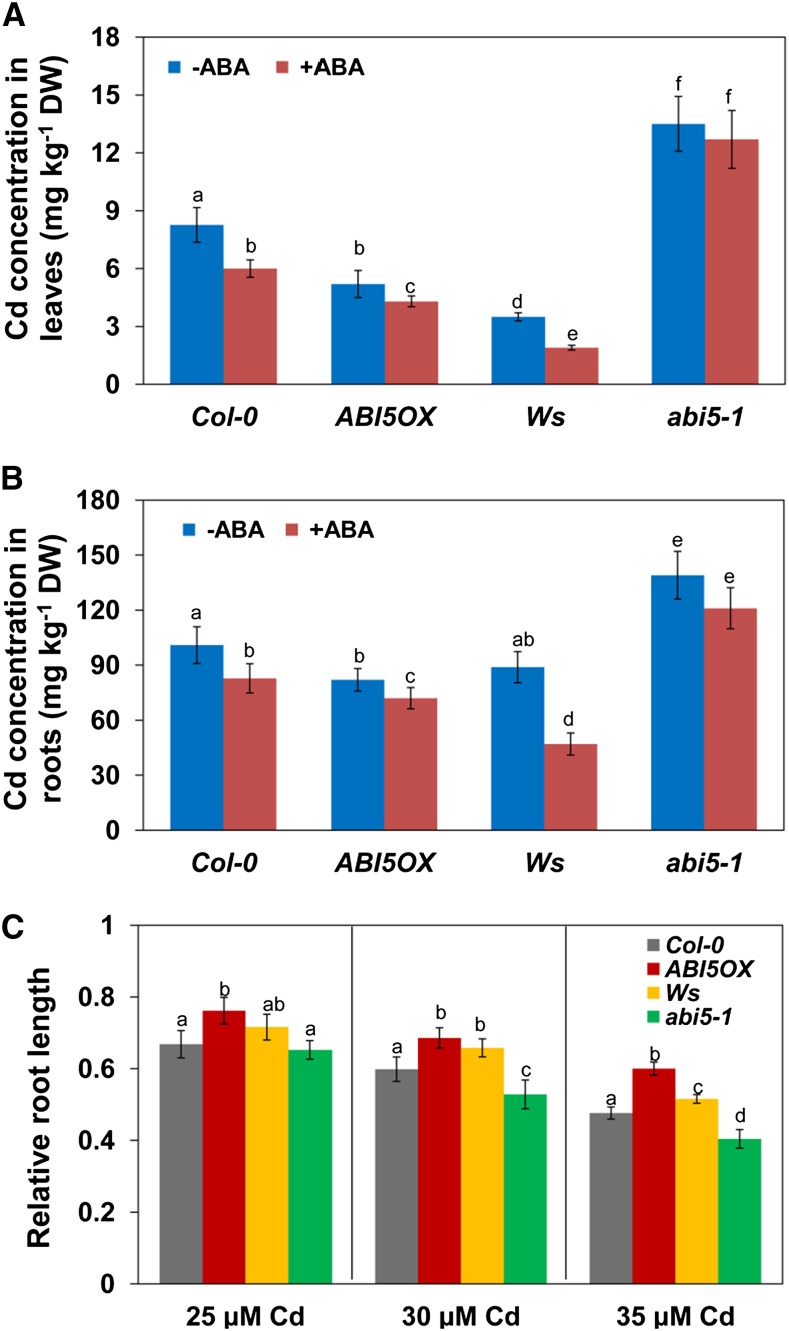
To determine whether ABI5 is involved in Cd accumulation, we first analyzed the Cd content in overexpression and mutant plants. Overexpression of ABI5 (ABI5OX; Columbia-0 [Col-0] background) reduced Cd accumulation (by 37% and 18.7% in leaves and roots, respectively), whereas the abi5 mutant (Wassilewskija [Ws] background) showed increased Cd accumulation (by 285.8% and 56.4% in leaves and roots, respectively) when compared with wild-type plants (Fig. 1, A and B). ABA treatment markedly reduced Cd accumulation in Col-0, ABI5OX, and Ws plants; in contrast, ABA-repressed Cd accumulation is not affected in the abi5 mutant (Fig. 1, A and B).
Figure 1.
ABI5 is involved in ABA-mediated Cd accumulation. A and B, Four-week-old hydroponics-grown Col-0, ABI5OX, Ws, and abi5-1 seedlings were transferred to fresh medium containing 10 µm CdCl2 with or without 1 µm ABA for 2 d. The leaves (A) and roots (B) were harvested and subjected to Cd content analysis by inductively coupled plasma atomic emission spectroscopy (ICP-AES). Error bars represent the sd (n = 6). DW, Dry weight. C, Five-day-old Arabidopsis Col-0, ABI5OX, Ws, and abi5-1 seedlings germinated on one-half-strength Murashige and Skoog (1/2 MS) medium were transferred to fresh medium supplemented with or without 25 to 35 μm CdCl2 for continued growth for 4 d, and the relative PR growth was measured. Three independent experiments were performed using at least 20 plants per experiment. Error bars represent the sd (n = 3). Different letters indicate that values were significantly different at P < 0.01 according to Tukey’s test.
We then investigated primary root (PR) growth in a Cd-containing medium. PR growth was inhibited by 33.2%, 40.1%, and 52.4% in 25, 30, and 35 µm CdCl2-treated Col-0 seedlings, respectively, whereas it was inhibited by 23.7%, 31%, and 39.9% in 25, 30, and 35 µm CdCl2-treated ABI5OX seedlings, respectively. PR growth was inhibited by 28.4%, 34.2%, and 48.4% in 25, 30, and 35 µm CdCl2-treated Ws seedlings, respectively, whereas it was inhibited by 34.7%, 47.2%, and 59.6% in 25, 30, and 35 µm CdCl2-treated abi5 mutant seedlings, respectively (Fig. 1C). These data indicated that overexpression of ABI5 resulted in less reduction, whereas the loss of function in the abi5 mutant resulted in greater reduction in PR growth in response to Cd toxicity when compared with wild-type Col-0 and Ws plants. Taken together, these data indicated that ABI5 is involved in ABA-mediated Cd accumulation.
The ABI5 Protein Interacts with the R2R3-MYB TF MYB49
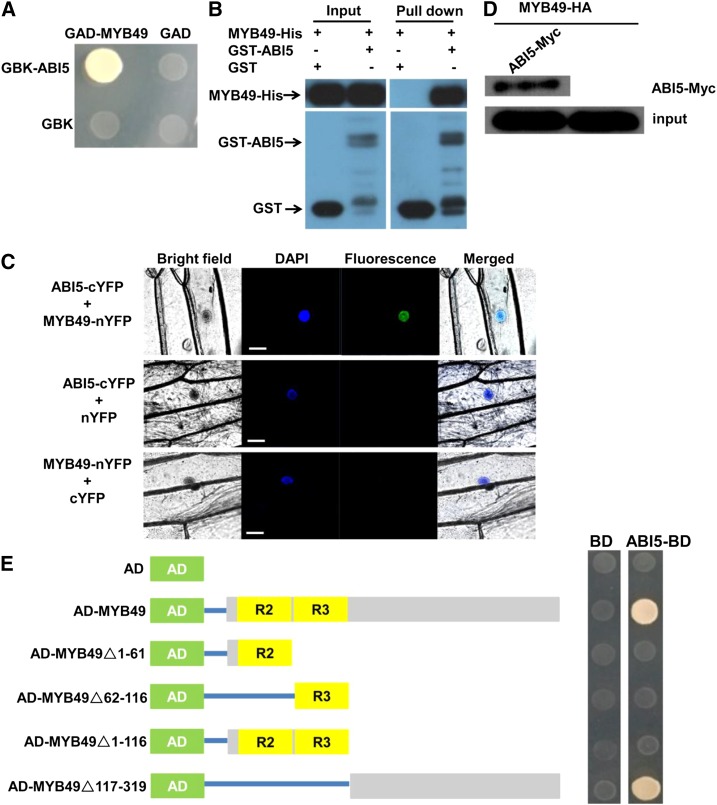
The above results indicated that ABI5 negatively regulates Cd accumulation. To identify transcriptional regulators potentially involved in ABI5-mediated Cd accumulation, we searched for ABI5 interaction proteins from the BioGRID interaction database (https://thebiogrid.org/) and identified the R2R3-MYB TF MYB49. Reverse transcription-quantitative PCR (RT-qPCR) analysis indicated that the gene expression of MYB49 was markedly induced by Cd treatment (Supplemental Fig. S1). We then confirmed the interaction by yeast two-hybrid (Y2H) assays. The MYB49 protein was fused to the pGADT7 (amino acids 768–881 of the Galectin 4 [GAL4] activation domain) vector (AD-MYB49), and the ABI5 protein was introduced into the pGBKT7 (amino acids 1-147 of the GAL4 DNA binding domain) vector (binding domain [BD]-ABI5). As shown in Figure 2A, the MYB49 protein interacted with the ABI5 protein in yeast cells. We also confirmed the interaction between MYB49 and ABI5 by an in vitro pull-down assay. The pull-down results showed that the glutathione S-transferase (GST)-fused ABI5 could retain MYB49-His, whereas GST alone could not (Fig. 2B). Next, we used bimolecular fluorescence complementation (BiFC) assays to further determine the interaction between MYB49 and ABI5 in plant cells (Fig. 2C). The ABI5 protein was fused to the C-terminal region of yellow fluorescent protein (ABI5-cYFP), and the MYB49 protein was fused to the N-terminal region of yellow fluorescent protein (MYB49-nYFP). A strong YFP fluorescence was observed in the nuclei when ABI5-cYFP was cobombarded with MYB49-nYFP in onion (Allium cepa) epidermal cells. The interaction between MYB49 and ABI5 was also corroborated by a coimmunoprecipitation (Co-IP) assay (Fig. 2D). Taken together, these data indicated that MYB49 interacts with ABI5 in vitro and in vivo.
Figure 2.
ABI5 interacts with MYB49 in vitro and in vivo. A, Y2H assay shows the interaction between ABI5 and MYB49. Transformed yeast cells were grown on synthetic dextrose (SD)-Trp/-His/-Leu/-Ade medium. Y2H assay was performed three times with the same result. B, An in vitro pull-down assay shows the interaction between ABI5 and MYB49. MYB49-His protein was incubated with immobilized GST or GST-ABI5 protein, and immunoprecipitated fractions were detected by anti-His antibody. The assay was performed three times with the same result. C, BiFC analysis of the interaction between ABI5 and MYB49. DAPI, 4′,6-Diamidino-2-phenylindole. Fluorescence was observed in the nuclei of onion epidermal cells that resulted from complementation of the N-terminal region of YFP fused with MYB49 (MYB49-nYFP) and the C-terminal region of YFP fused with ABI5 (ABI5-cYFP). No signal was observed from the negative controls. BiFC was repeated three times independently with the same result. Blue and green fluorescence represent DAPI and GFP channels, respectively. Bars = 50 µm. D, Co-IP assay showing the interaction between ABI5 and MYB49. MYB49-HA was immunoprecipitated using an anti-HA antibody, and coimmunoprecipitated Myc-ABI5 was detected using an anti-Myc antibody. Protein input for MYB49-HA in the immunoprecipitated complex is shown. The assay was performed three times with the same result. E, The C-terminal part of MYB49 contributes to the interaction of MYB49 with ABI5. The diagram at left shows full-length and truncated MYB49 constructs with specific deletions. At right, the interactions are shown by the ability of transformed yeast cells to grow on SD-Trp/-His/-Leu/-Ade medium, and empty pGADT7 was used as a negative control. Y2H assay was performed three times with the same result. AD (green boxes), pGADT7 prey vector; R2 and R3 (yellow boxes), R2 and R3 domains of MYB49 protein; gray boxes, MYB49 protein fragments; BD, pGBKT7; ABI5-BD, pGBKT7-MYB49. Blue lines indicate that the boxes (fragments) were linked to AD vector.
We also determined which domain of the MYB49 protein is responsible for interacting with the ABI5 protein. For this purpose, the MYB49 protein was divided into four parts: N-terminal part I (amino acids 1–63) containing the MYB R2 motif; part II (amino acids 64–116) containing the MYB R3 motif; part III (amino acids 1–116) containing both the R2 and R3 motifs; and C-terminal part IV (amino acids 117–319). Y2H analysis revealed that deletion of the C-terminal part of MYB49 eliminated the physical interaction with ABI5, whereas deleting the N-terminal parts (R2R3 motif) did not affect the interaction (Fig. 2E). These data indicated that the C-terminal region of MYB49 contributes to the interaction between MYB49 and ABI5.
Arabidopsis MYB49 Expression and Transactivational Activity Analysis
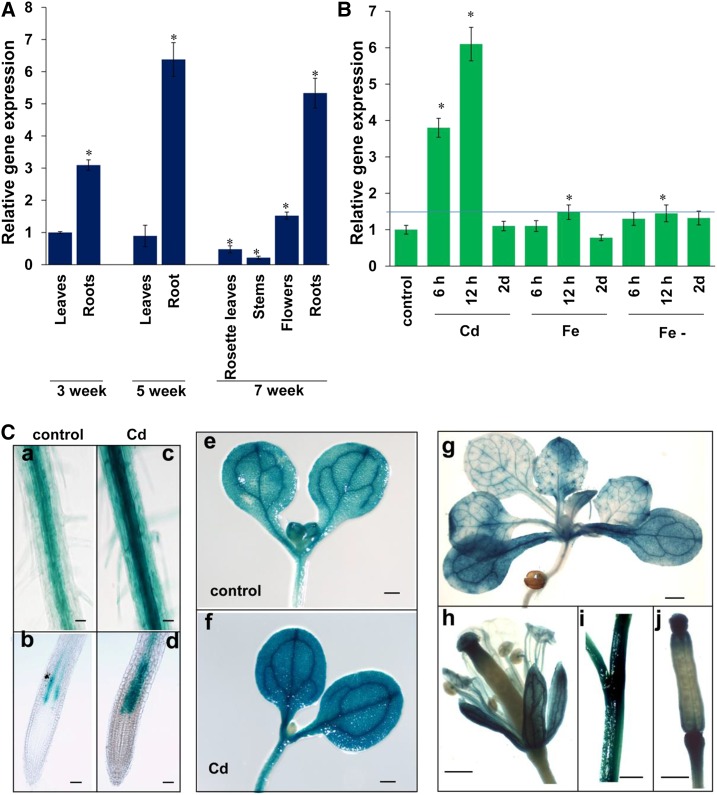
We analyzed the expression of MYB49 in different tissues at different growth stages by RT-qPCR analysis. As shown in Figure 3A, MYB49 is mainly expressed in roots. We also investigated whether the expression of MYB49 is affected by heavy metal toxicity and found that MYB49 expression was strongly induced by Cd toxicity; however, the expression of MYB49 was only slightly induced by excess Fe and Fe deficiency (Fig. 3B; Supplemental Fig. S1).
Figure 3.
Expression pattern of MYB49. A, Relative expression of MYB49 in various tissues at different growth stages. The expression level of MYB49 in the leaves of 3-week-old seedlings was set to 1. Error bars represent the sd (n = 3). Asterisks indicate significant differences from the expression level of the leaves in 3-week-old seedlings (Student’s t test, P < 0.01). B, Responses of MYB49 expression to heavy metal stresses. Seven-day-old Arabidopsis Col-0 seedlings germinated on 1/2 MS medium were transferred to fresh medium supplemented with 30 μm Cd, 100 μm Fe, or Fe-deficient medium for 6 h, 12 h, or 2 d, and the expression level of MYB49 was determined by RT-qPCR. The expression level of MYB49 in untreated seedlings was set to 1. Error bars represent the sd (n = 3). The blue line represents a value of relative gene expression of 1.5. Asterisks indicate significant differences from the control (Student’s t test, P < 0.01). C, MYB49 expression patterns detected in the MYB49 promoter:GUS transgenic line. Images show the following: GUS staining observed in roots without (a and b) or with 30 μm Cd (c and d); cotyledons and shoots without (e) or with 30 μm Cd (f); 2-week-old seedling (g); flower (h); stem (i); and siliques (j). Three independent experiments were performed using at least 20 plants per experiment with similar results. Bars = 100 µm (a–d) and 0.5 mm (e–j).
To further evaluate the tissue-specific and developmental expression profiles of MYB49 in detail, we used the MYB49promoter:GUS transgenic line that fused a 1.5-kb promoter sequence upstream of the ATG start codon to the GUS reporter gene. GUS activity could be detected in the entire root, especially in vascular tissue. Consistent with the RT-qPCR results (Fig. 3B), remarkably enhanced GUS activity was detected in the cortex and epidermal cells in the roots upon Cd stress (Fig. 3C). However, almost no activity was detected in the root tips. In addition, GUS activity was detected in cotyledons, shoots, and young leaves. In mature plants, GUS activity could be detected in flowers, siliques, and stems (Fig. 3C).
We analyzed the subcellular localization of MYB49 using the MYB49 promoter:MYB49-GFP (green fluorescent protein) construct and confirmed the nuclear localization of MYB49 in both transiently expressed tobacco leaves (Supplemental Fig. S2A) and roots of MYB49 promoter:MYB49-GFP transgenic Arabidopsis seedlings (Supplemental Fig. S2B).
The above results showed that MYB49 is localized to the nucleus and were consistent with the predicted function of MYB49 as a TF. We thus examined the transcriptional activation ability of MYB49 using a yeast assay system. MYB49 was fused to the GAL4 DNA-binding domain in the pGBKT7 vector. pAD and pGBKT7 were used as the positive and negative controls, respectively (Supplemental Fig. S3A). These plasmids were then transformed into the yeast strain AH109. pMYB49 could grow in SD/-Trp-His-Ade medium (Supplemental Fig. S3B), indicating that MYB49 may function as a transcriptional activator.
MYB49 Increases Cd Accumulation in Plants
To investigate the physiological roles of MYB49 in plants, we first generated transgenic Arabidopsis plants constitutively expressing MYB49 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. These overexpression lines (OX49) showed obviously elevated expression of MYB49 (Supplemental Fig. S4A).
Because there are no available myb49 knockout or knockdown mutants in Arabidopsis seed stocks, we generated knockout alleles of MYB49 by using CRISPR/Cas9 technology to target the MYB49 genomic sequence. We obtained two independent alleles, one that contains a 1-bp insertion (added a C base at 106-107 bp in the coding sequence [CDS]) and the other that loses 8 bp (lost TCCCGGCA at 98-99 bp in the CDS), causing a premature stop, and we named them myb49.1 and myb49.2, respectively (Supplemental Fig. S5).
Given the expected redundancy presented by other R2R3-MYB transcriptional factor families, we also constructed lines with chimeric repressors of MYB49:ERF-associated amphiphilic repression motif repression domain (SRDX49) using chimeric repressor silencing technology (Supplemental Fig. S4B) to compare their phenotype with that of OX49-overexpressing lines.
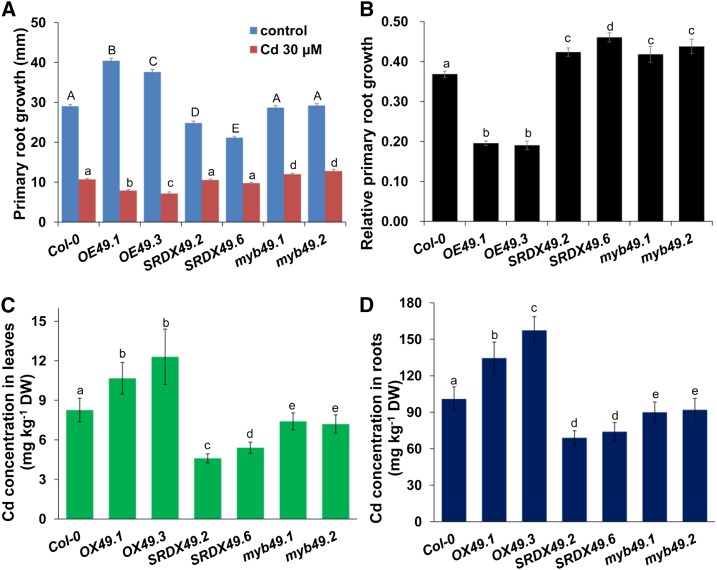
We first investigated root system growth in response to Cd toxicity. Five-day-old Arabidopsis seedlings germinated on one-quarter-strength MS medium were transferred to fresh medium supplemented with or without 30 μm Cd for continued growth for 4 d, and the growth of PR was measured. The PR in the OX49 lines is longer than in Col-0 plants under normal conditions, whereas it is shorter in SRDX49 lines (Fig. 4A). Overexpression of MYB49 resulted in stronger inhibition, whereas in the SRDX49 lines, myb49.1, and myb49.2 exhibited less inhibition of PR growth upon Cd stresses (Fig. 4, A and B), suggesting that MYB49 increases the sensitivity to Cd toxicity.
Figure 4.
Phenotypic analysis of OX49 and SRDX49 lines. A and B, Growth of 5-d-old Arabidopsis Col-0, OX49, and SRDX49 seedlings germinated on 1/2 MS medium and then transferred to fresh medium supplemented with 30 μm Cd for 5 d. A, PR length. B, Relative PR growth of seedlings treated with 30 μm Cd compared with untreated seedlings. Three independent experiments were performed using at least 20 plants per experiment. Error bars represent the sd (n = 3). C and D, Four-week-old hydroponics-grown Col-0, OX49, myb49, and SRDX49 seedlings were transferred to fresh medium containing 10 µm CdCl2 for 2 d, and the leaves (C) and roots (D) were harvested and subjected to Cd content analysis by ICP-AES. Error bars represent the sd (n = 6). Different letters indicate that values were significantly different at P < 0.01 according to Tukey’s test. DW, Dry weight.
Next, we analyzed Cd contents in hydroponically grown OX49, SRDX49, and myb49 lines. The Cd concentrations in both leaves and roots were substantially increased in OX49 plants, whereas they were decreased in the SRDX49 and myb49 lines (Fig. 4, C and D).
Mineral analysis of seedlings under normal conditions showed that the concentrations of Zn, Fe, magnesium (Mg), and sulfur (S) were higher in the OX49 plants, whereas the P concentration was slightly but insignificantly higher in SRDX49 plants, and the concentrations of calcium (Ca), manganese (Mn), and copper (Cu) did not significantly differ among the OX49, SRDX49, and myb49 plants (Supplemental Fig. S6). These data indicated that MYB49 activation affects mineral accumulation in plants.
MYB49 Regulates the Genes Involved in Metal Accumulation
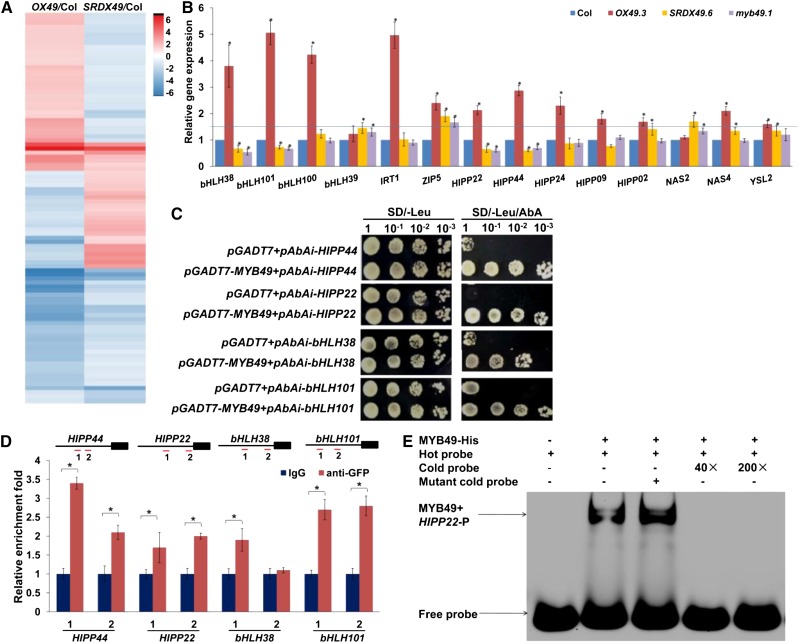
The above results indicated that OX49 and SRDX49 plants show opposite effects on Cd accumulation. To further elucidate the molecular mechanisms underlying MYB49-mediated Cd accumulation, we analyzed the RNA sequencing data from 7-d-old seedlings of the Col-0, OX49 (OX49.3), and SRDX49 (SRDX49.6) lines. Nine samples were sequenced using the BGISEQ-500 platform, which on average generated approximately 23.6 Mb of data per sample. All sequencing data were archived at the Short Read Archive of the National Center for Biotechnology Information under accession number SRP154279. The average mapping ratio with the reference genome is 98.3%, and the average mapping ratio with the gene is 96.9%. A total of 25,160 genes were detected. Differentially expressed genes (DEGs) were identified by comparison with the Col-0 control (log2 fold change ≥ 1 and adjusted P ≤ 0.05), and we obtained 714 and 770 DEGs in the OX49 and SRDX49 plants, respectively (Fig. 5A; Supplemental Tables S1 and S2).
Figure 5.
MYB49 regulates genes involved in metal accumulation. A, Hierarchical clustering analysis diagram showing the DEGs in OX49 and SRDX49 seedlings. B, RT-qPCR analysis of the genes involved in heavy metal accumulation and tolerance in 7-d-old Arabidopsis Col-0, OX49, SRDX49, and myb49 seedlings germinated on 1/2 MS medium and then transferred to fresh medium containing 30 μm Cd for 12 h. The expression levels of the indicated genes in wild-type Col-0 were set to 1. Error bars represent the sd (n = 3). Asterisks indicate significant differences from the control (Student’s t test, P < 0.01). The blue line represents a value of relative gene expression of 1.5. C, A Y1H assay showed that MYB49 bound to the promoters of bHLH38, bHLH101, HIPP22, and HIPP44. Transformed yeast cells were grown on SD-Leu/aureobasidin A (AbA) medium. The assay was performed three times with the same result. D, ChIP-qPCR assays of MYB49-DNA complexes. The scheme of the primer design for each gene is shown at top. The black boxes represent the gene-coding regions, and the black lines represent the promoter regions. Red lines mark the locations of amplicons amplified in the ChIP-qPCR. The promoter fragment enrichment following ChIP-qPCR was performed in the absence (IgG) or presence (anti-GFP) of anti-GFP antibody. Error bars represent the sd (n = 3). Asterisks indicate significant differences from the control (Student’s t test, P < 0.01). E, EMSA binding of MYB49 to the HIPP22 promoter region harboring MYB49-binding sites. Two biological experiments were performed with similar results.
We investigated the genes involved in heavy metal accumulation and tolerance and found that MYB49 activation affected the expression of several metal transporters, heavy metal transport/detoxification superfamily proteins, and Fe deficiency-responsive bHLH TFs (Supplemental Table S3). Using RT-qPCR, we confirmed that four genes, bHLH38, bHLH101, HIPP22, and HIPP44, showed elevated expression in the OX49 line, whereas the expression of these genes was decreased in SRDX49 and myb49 plants compared with Col-0 plants under Cd stress (Fig. 5B). Four genes, bHLH100, IRT1, HIPP24, and HIPP09, showed increased expression in the OX49 lines (the values of relative gene expression > 1.5), whereas the expression of these genes was largely unaffected in SRDX49 and myb49 plants compared with Col-0 plants under Cd stress. The data suggested that these genes are likely to be involved in MYB49-mediated Cd accumulation and tolerance in Arabidopsis by either direct or indirect pathways.
To investigate whether MYB49 directly regulates these Cd accumulation- and tolerance-associated genes by binding to their promoters, we first examined the binding of MYB49 to the promoters in a separate yeast one-hybrid (Y1H) assay. MYB49 bound to the promoters of bHLH38, bHLH101, HIPP22, and HIPP44 (Fig. 5C).
We then performed chromatin immunoprecipitation (ChIP)-qPCR against bHLH38, bHLH101, HIPP22, and HIPP44 using the MYB49promoter:MYB49-GFP line. The transgenic lines showed a Cd-sensitive phenotype in PR growth, indicating that the MYB49-GFP fusion protein retained its biological function. By using the AC elements of MYB49 in the promoters according to Kelemen et al. (2015), we designed primers for the ChIP-PCR assays. We observed enrichment of MYB49-GFP at the promoters of bHLH38, bHLH101, HIPP22, and HIPP44 (Fig. 5D).
To further confirm the results, we chose the HIPP22 gene to perform an electrophoretic mobility shift assay (EMSA) to test for the physical interaction of MYB49 with the promoter sequence of HIPP22. Analysis of the promoter of MYB49 identified a series of connected MYB-binding sites containing two AC-element ACCWHH and two MYB-core TNGTT[G/A] sequences in the −227- to −188-bp upstream promoter regions (Kelemen et al., 2015). As shown in Figure 5E, MYB49 interacted with the promoter sequence of the HIPP22 gene, and the interaction was significantly reduced when unlabeled promoter fragments (cold probe) were added in excess, indicating the specificity of the interaction.
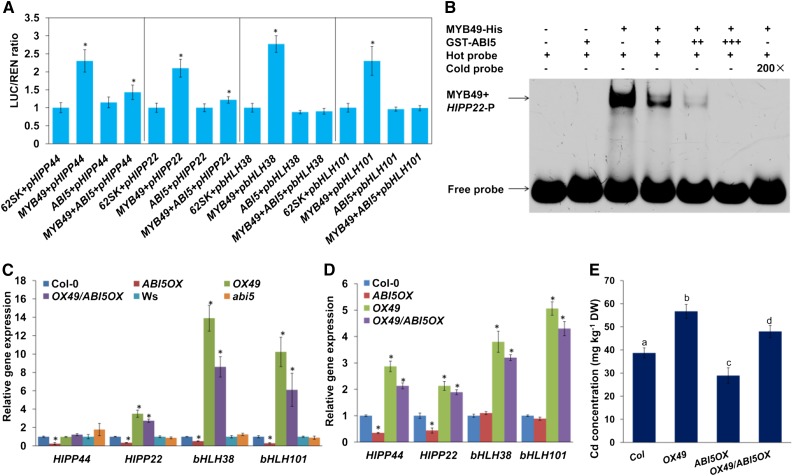
We next examined the function of MYB49 in promoting the transcription of bHLH38, bHLH101, HIPP22, and HIPP44 by analyzing MYB49-mediated transcriptional activity using a protoplast cotransfection luciferase reporter assay. As shown in Figure 6A, MYB49 induced transcription from the promoters of bHLH38, bHLH101, HIPP22, and HIPP44. Taken together, these data indicated that MYB49 directly regulates the expression of bHLH38, bHLH101, HIPP22, and HIPP44.
Figure 6.
ABI5 negatively regulates MYB49-mediated Cd accumulation. A, Transient dual-luciferase reporter assays show that the activation of bHLH38, bHLH101, HIPP22, and HIPP44 expression by MYB49 is repressed by ABI5. Protoplast transient expression assays used the 1.5-kb promoter fragments of the bHLH38, bHLH101, HIPP22, and HIPP44 genes. The luciferase luminescence intensities were quantitated following transfection with different vectors: 62SK represents empty pGreenII 62-SK vector; MYB49 represents the pGreenII 62-SK-MYB49 vector; ABI5 represents the pGreenII 62-SK-AtABI5 vector; pbHLH38, pbHLH101, pHIPP22, and pHIPP44 represent pGreenII 0800-LUC-pbHLH38, pGreenII 0800-LUC-pbHLH101, pGreenII 0800-LUC-pHIPP22, and pGreenII 0800-LUC-pHIPP44 vectors, respectively. Renilla luciferase (REN) was used for normalization. Error bars represent the sd (n = 3). Asterisks indicate significant differences from the control (Student’s t test, P < 0.01). B, The binding of MYB49 to the HIPP22 promoter region harboring MYB49-binding sites was analyzed using EMSA in the presence of ABI5. Two biological experiments were performed with similar results. C and D, RT-qPCR analysis of the expression of bHLH38, bHLH101, HIPP22, and HIPP44 genes in 7-d-old Arabidopsis Col-0, ABI5OX, Ws, abi5-1, OX49, and OX49/ABI5OX seedlings germinated on 1/2 MS medium and then transferred to fresh medium without (C) or with (D) 30 μm Cd for 1 d. The expression levels of the indicated genes in wild-type Col-0 or Ws were set to 1. Error bars represent the sd (n = 3). Asterisks indicate significant differences from the control (Student’s t test, P < 0.01). E, Four-week-old hydroponics-grown Col-0, OX49, ABI5OX, and OX49/ABI5OX seedlings were transferred to fresh medium containing 10 µm CdCl2 for 2 d, and Cd contents were then determined by ICP-AES. Error bars represent the sd (n = 6). Different letters indicate that values were significantly different at P < 0.01 according to Tukey’s test. DW, Dry weight.
Because bHLH38, bHLH101, and their downstream IRT1 genes were Fe deficiency-responsive genes, we wondered whether MYB49 is also involved in the Fe-deficiency response. To address this question, we first investigated Fe deficiency tolerance in the OX49, SRDX49, and myb49 lines. We did not observe obvious differences among the OX49, SRDX49, and myb49 lines (Supplemental Fig. S7A). We thus analyzed the gene expression in Fe-deficient seedlings. OX49 plants exhibited higher expression of the bHLH38, bHLH101, and IRT1 genes than Col-0 plants under normal conditions (Supplemental Fig. S7B). Consistent with this result, OX49 plants exhibited higher Fe concentrations than Col-0 plants under normal growth conditions (Supplemental Fig. S6). Fe deficiency markedly induced the expression of these genes, while both OX49 and SRDX49 plants showed similar levels compared with Col-0 plants under Fe-deficient conditions (Supplemental Fig. S7B).
ABI5 Negatively Regulates MYB49-Mediated Cd Accumulation
To investigate the role of the interaction between ABI5 and MYB49, we cotransfected 35S:ABI5 with 35S:MYB49 together with bHLH38-LUC, bHLH101-LUC, HIPP22-LUC, or HIPP44-LUC reporter plasmids. Cotransfection of ABI5 and MYB49 repressed bHLH38, bHLH101, HIPP22, and HIPP44 expression compared with MYB49 transfection alone (Fig. 6A), suggesting that the interaction between ABI5 and MYB49 inhibits MYB49’s ability to transcriptionally regulate its target genes.
To further confirm the results, we analyzed the effect of ABI5 on the physical interaction of MYB49 with the promoter sequence of its target gene using EMSA. As shown in Figure 6B, the interaction of MYB49 with the promoter sequence of HIPP22 was significantly reduced when ABI5 was added in excess. These data demonstrated that the ABI5-MYB49 interaction decreases MYB49 DNA-binding activity.
Consistent with the cotransfection assay and EMSA results, RT-qPCR analysis indicated that the expression levels of bHLH38, bHLH101, HIPP22, and HIPP44 were markedly lower in 35S:ABI5 plants than in wild-type plants (Fig. 6, C and D).
To further elucidate the physiological role of the interaction between ABI5 and MYB49, we generated OX49/ABI5OX plants by crossing OX49 with ABI5OX and then analyzed the Cd contents in these plants. The concentration of Cd was lower in OX49/ABI5OX plants than in OX49 plants (Fig. 6E). Furthermore, the gene expression of bHLH38, bHLH101, HIPP22, and HIPP44 was lower in OX49/ABI5OX than in OX49 plants (Fig. 6, C and D). Taken together, these data indicated that ABI5 negatively regulates MYB49-mediated Cd accumulation.
HIPP44 Is Involved in Cd Accumulation in Plants
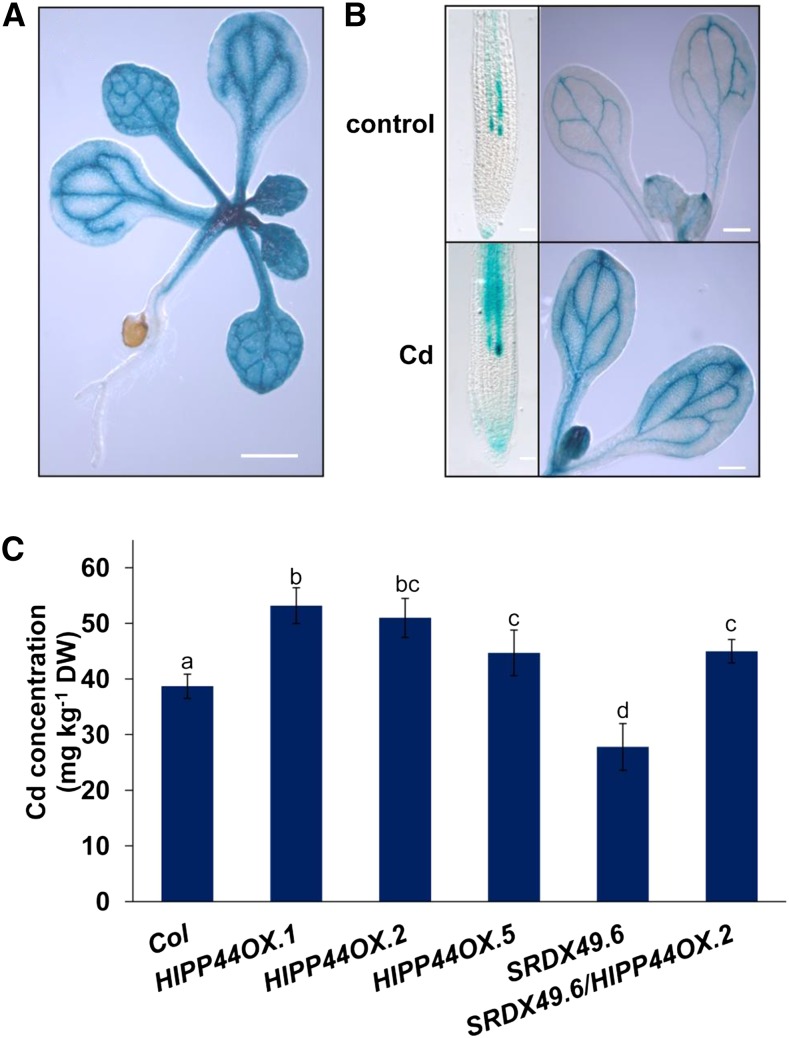
HIPPs play important roles in modulating heavy metal transport and detoxification (Chu et al., 2005; Tehseen et al., 2010; Zhao et al., 2013). The above results indicated that MYB49 directly regulated HIPP22 and HIPP44 expression. Previous studies have demonstrated that HIPP22 increases Cd accumulation (Tehseen et al., 2010). We thus wondered whether HIPP44 is also involved in this process. To address this question, we first investigated the expression patterns of HIPP44 using the HIPP44promoter:GUS transgenic line (Fig. 7A). Cd treatment enhanced GUS activity both in roots and leaves (Fig. 7B). Next, we investigated the role of HIPP44 in Cd accumulation in plants. Because there is no available hipp44 mutant in Arabidopsis seed stocks, we generated HIPP44-overexpressing transgenic Arabidopsis plants under the control of the CaMV 35S promoter (Supplemental Fig. S8). The Cd concentration was higher in HIPP44OX plants than in Col-0 plants (Fig. 7C).
Figure 7.
HIPP44 regulates Cd accumulation in Arabidopsis. A, HIPP44 expression patterns detected in the 2-week-old HIPP44promoter:GUS transgenic line. Bar = 1 mm. B, GUS staining of the root tips, cotyledons, and shoots of HIPP44promoter:GUS transgenic seedlings without or with 30 μm Cd for 12 h. Bars = 0.1 mm (roots) and 0.5 mm (shoots). Three independent experiments were performed using at least 20 plants per experiment with similar results. C, Four-week-old hydroponics-grown Col-0, HIPP44OX, SRDX49, and SRDX49/HIPP44OX seedlings were transferred to fresh medium containing 10 µm CdCl2 for 2 d, and Cd contents were then determined by ICP-AES. Error bars represent the sd (n = 6). Different letters indicate that values were significantly different at P < 0.01 according to Tukey’s test. DW, Dry weight.
Phenotypic and molecular analyses demonstrated that MYB49 positively regulates Cd accumulation through direct activation of HIPP44 expression. To further confirm this conclusion, we explored the genetic relationship between MYB49 and HIPP44. The SRDX49 plant was crossed with the HIPP44OX plant, and the Cd contents were examined. The Cd concentration in SRDX49/HIPP44OX plants was higher than that in SRDX49 plants (Fig. 7C). Taken together, these results indicated that MYB49 acts upstream of HIPP44 to regulate Cd accumulation.
DISCUSSION
ABA reduces Cd accumulation in plants (Hsu and Kao, 2003; Uraguchi et al., 2009; Fan et al., 2014). Previous studies have demonstrated that IRT1 plays an important role in modulating Cd accumulation in response to ABA (Fan et al., 2014). However, how the ABA signal reduces Cd uptake and whether ABA inhibits IRT1 expression by negatively regulating the bHLH38/bHLH39/bHLH100/bHLH101-IRT1 pathway remain unknown. Here, we found that ABI5 is involved in ABA-reduced Cd accumulation in Arabidopsis by interacting with MYB49.
Previous studies have shown that two R2R3-MYB TFs, MYB10 and MYB72, could be induced by Cd, excess Zn, and Fe deficiency in Arabidopsis and modulate Fe accumulation and distribution by directly binding to the NAS2 and NAS4 promoters (Palmer et al., 2013). Here, we demonstrate that the R2R3-MYB TF MYB49 positively regulates Cd uptake and accumulation. MYB49 is markedly induced in the roots within 6 h of exposure to Cd toxicity (Fig. 3, B and C). Overexpression of MYB49 increased Cd accumulation in plants (Fig. 4, C and D). The increased Cd content in OX49 roots may contribute to the increased Cd sensitivity of PR growth to Cd toxicity compared with that of wild-type plants (Fig. 4, A and B). We also found that overexpression of MYB49 increases the concentrations of Zn, Fe, Mg, and S in normal-grown seedlings (Supplemental Fig. S6), suggesting that overexpression of MYB49 increases nutrient uptake in plants. Supporting these results, we found that OX49 lines showed longer PR than Col-0 seedlings under normal conditions (Fig. 4A). These findings indicated that MYB49 improves plant growth by positively regulating mineral uptake and accumulation in plants. Another possibility is that the overexpression of MYB49 in Arabidopsis enhances root growth and thus improves mineral uptake and accumulation. However, whether and how MYB49 directly regulates root system development need further elucidation.
Our results indicate that MYB49 directly regulates Cd accumulation in plants. Several lines of evidence support these conclusions. First, MYB49 affects the expression of genes involved in heavy metal uptake, transport, and tolerance (Fig. 5B; Supplemental Table S3). Second, using Y1H and ChIP-qPCR, we confirmed that MYB49 directly regulates the expression of bHLH38, bHLH101, HIPP22, and HIPP44 by binding to their promoters (Fig. 5, C and D). Previous studies have demonstrated that in addition to regulating the Fe-deficiency response, the IVc subgroup of bHLH TFs, which includes bHLH38, bHLH39, bHLH100, and bHLH101, also increases Cd accumulation by regulating IRT1 expression (Wu et al., 2012; Wang et al., 2013; Yao et al., 2018). Indeed, we observed a remarkable induction of IRT1 expression in OX49 plants (Fig. 5B). These results indicated that MYB49 increases Cd uptake by modulating the bHLH38/bHLH39/bHLH100/bHLH101-IRT1 pathway. Third, overexpression of HIPP44 increased Cd accumulation in plants, and genetic analysis demonstrated that SRDX49/HIPP44OX could not alter the Cd accumulation phenotype of HIPP44OX, indicating that MYB49 increases Cd accumulation in a HIPP44-dependent manner.
The involvement of HIPPs in modulating Cd accumulation has been shown previously (Tehseen et al., 2010). HIPPs are metallochaperones that transport metal ions inside the cell and are involved in heavy metal homeostasis and detoxification (Chu et al., 2005; Tehseen et al., 2010; Zhao et al., 2013; Zschiesche, 2013). Arabidopsis HIPP20, HIPP22, HIPP26, and HIPP27 rescued the Cd-sensitive ycf1 yeast mutant, and the hipp20hipp21hipp22 triple mutant was more sensitive to Cd and accumulated less Cd than Col-0 plants (Tehseen et al., 2010). In addition to HIPP44, six HIPP genes, HIPP02, HIPP09, HIPP13, HIPP22, HIPP24, and HIPP34, showed increased expression in OX49 plants (Fig. 5B; Supplemental Table S3). Among these HIPP genes, HIPP22 and HIPP44 showed markedly elevated expression in OX49 plants and reduced expression in myb49 and SRDX49 plants under Cd treatment (Fig. 5B). Further analysis confirmed that MYB49 binds directly to the promoters of HIPP22 and HIPP44. Taken together, these data indicated that MYB49 positively regulates Cd accumulation by modulating HIPP gene expression.
Overexpression of MYB49 markedly induced the expression of several Fe-deficiency response genes (Fig. 5B; Supplemental Table S3). Consistent with this result, OX49 plants exhibited higher Fe concentrations than Col-0 plants under normal growth conditions (Fig. 4E). Fe deficiency strongly induced the expression of bHLH38, bHLH101, and IRT1; however, in contrast to the results in normal conditions or Cd treatment that OX49 plants exhibited higher expression of these genes than Col-0 and SRDX49 plants, both OX49 and SRDX49 plants showed similar expression levels of these genes under Fe-deficient conditions (Supplemental Fig. S7B). Consistent with the gene expression results, all the overexpression, knockout, and SRDX lines exhibited similar phenotypes under Fe-deficient conditions (Supplemental Fig. S7A). Gene expression analysis of MYB49 showed that MYB49 is poorly responsive to Fe deficiency (Fig. 3B). Taken together, these data indicated that MYB49 is not involved in the Fe-deficiency response.
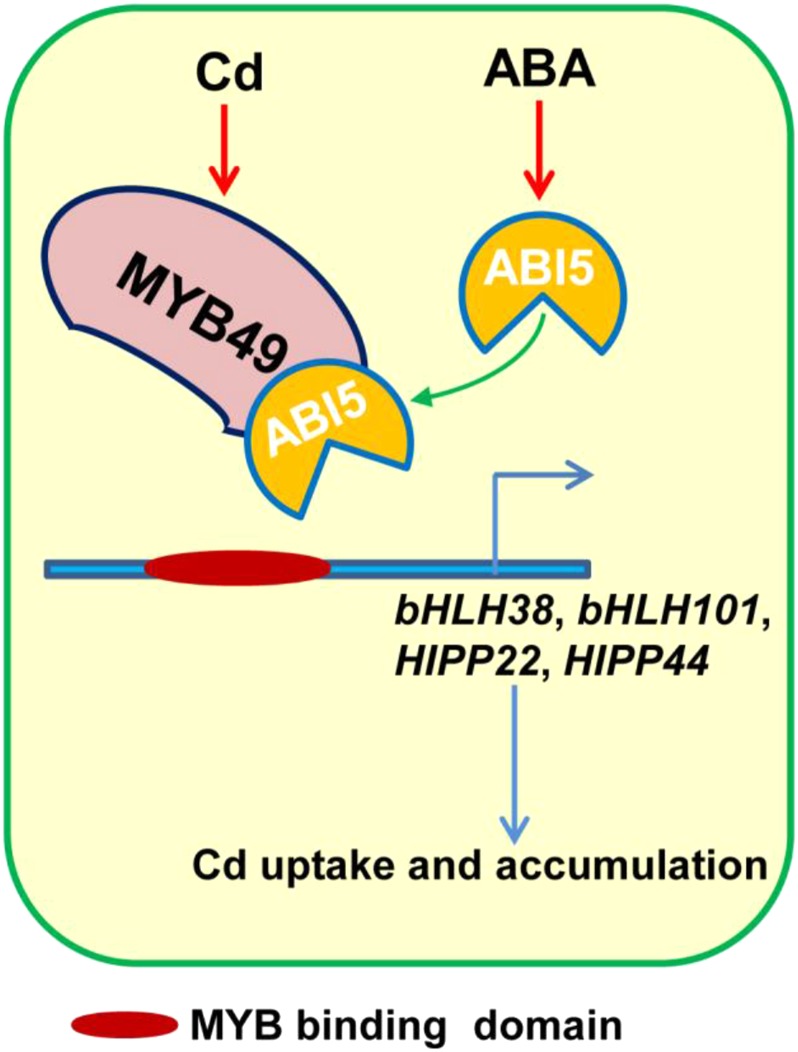
For the last 20 years, ABI5 has been widely known for its functions as a central ABA signaling molecule in modulating seed germination and early seedling growth in response to abiotic stresses (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001; Nakamura et al., 2001). However, accumulating evidence has indicated that ABI5 also plays a critical role in the vegetative stage of development (Brocard et al., 2002; Kong et al., 2013). The expression of ABI5 could be observed in roots, leaves, and flowers in mature plants (Brocard et al., 2002; Kong et al., 2013; Skubacz et al., 2016). ABI5 is involved in ABA-mediated lateral root growth inhibition (Signora et al., 2001; De Smet et al., 2003; Shkolnik-Inbar and Bar-Zvi, 2010; Skubacz et al., 2016). ABI5 also represses photosynthesis by activating chlorophyll degradation, thereby mediating ABA-induced leaf senescence (Sakuraba et al., 2014; Su et al., 2016). In this study, we found that ABI5 is involved in ABA-repressed Cd accumulation. Overexpression of ABI5 reduced Cd accumulation, while the abi5 mutant showed increased Cd accumulation. Using Y2H, Co-IP, pull-down, and BiFC assays, we confirmed the physical interaction between ABI5 and MYB49 (Fig. 2). ABI5 likely interferes with the transcriptional function of MYB49. First, transient expression assays demonstrated that ABI5 suppresses the transcriptional regulatory activation of MYB49 on bHLH38, bHLH101, HIPP22, and HIPP44 expression (Fig. 6A). Second, EMSA further confirmed that the interaction of ABI5 and MYB49 inhibits the binding of MYB49 to the promoter sequence of the HIPP22 gene (Fig. 6B). Third, the RT-qPCR analysis showed that the expression levels of MYB49 target genes were decreased in ABI5OX plants (Fig. 6C). To further understand how the ABI5-MYB49 interaction regulates Cd accumulation, we investigated its effects on Cd contents. We found that the expression of the four MYB49 targets, bHLH38, bHLH101, HIPP22, and HIPP44, was lower in OX49/ABI5OX than in OX49 plants (Fig. 6, C and D). Consistent with the gene expression results, the Cd content was lower in OX49/ABI5OX plants than in OX49 plants (Fig. 6E). Taken together, our results indicated that ABA-induced ABI5 represses the transcriptional activity of Cd-induced MYB49 by preventing its binding to the promoters of downstream genes, thereby reducing Cd accumulation in plants (Fig. 8).
Figure 8.
A proposed model of ABA signaling-controlled Cd uptake in Arabidopsis through ABI5-MYB49 interaction. The interaction of ABA-induced ABI5 with Cd-induced MYB49 prevents the binding of MYB49 to the promoter regions of its downstream genes, resulting in their down-regulation, thereby reducing Cd uptake and accumulation.
In summary, our findings showed that MYB49 positively regulates Cd accumulation in plants by directly regulating bHLH38, bHLH101, HIPP22, and HIPP44 expression in Arabidopsis. Considering that Cd toxicity induces endogenous ABA accumulation in plants, we suggest that a feedback mechanism based on physical interactions between MYB49 and ABI5 controls Cd uptake and accumulation in plant cells. The results of this study have enabled us to understand the mechanisms underlying the plant response to Cd stress through the actions of MYB49 and ABI5. Additionally, our findings also provide ideas for generating transgenic plants with reduced or enhanced Cd uptake for growing in Cd-contaminated areas or removing accumulated Cd from Cd-contaminated soils.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized with 50% (v/v) bleach, rinsed five times with sterile water, and then plated on 1/2 MS medium containing 1% (w/v) Suc and 0.65% (w/v) agar, which was adjusted to pH 5.75. The seeds were stratified for 2 d at 4°C and then transferred to a growth chamber at 22°C with a 16-h-light/8-h-dark cycle. For agar plate culture, 5-d-old Arabidopsis seedlings germinated on 1/2 MS medium were transferred to fresh medium supplemented with or without 25 to 35 μm CdCl2 for continued growth for 4 d, and the PR growth was measured. The relative PR lengths of the seedlings treated with CdCl2 are presented relative to the untreated control values. For hydroponic culture, 4-week-old one-quarter-strength MS liquid medium (without Suc and vitamin)-grown seedlings were transferred to fresh medium containing 10 µm CdCl2 with or without 1 µm ABA for 2 d.
Constructs
The full-length MYB49 coding region was amplified from Arabidopsis cDNA using MYB49-F/MYB49-R and MYB49-F/MYB49-SRDX and then transferred into the pCAMBIA1300 vector with a CaMV 35S promoter to generate constructs for MYB49 overexpression (OX49) and chimeric repressor (SRDX49) lines, respectively. To generate the MYB49promoter:GUS construct, we amplified a 1.5-kb promoter sequence upstream of the ATG start codon of the MYB49 gene from Arabidopsis genomic DNA using pMYB49-F and pMYB49-R and then transferred the construct into the pCAMBIA1381 vector. To generate the MYB49promoter:MYB49-GFP construct, we amplified the 1.5-kb promoter sequence along with the MYB49 coding region from Arabidopsis genomic DNA using gMYB49-F and gMYB49-R and then transferred the construct into the pCAMBIA1302 vector. The generated constructs were used to generate transgenic Arabidopsis plants (Col-0 background) using Agrobacterium tumefaciens-mediated floral dip transformation.
CRISPR/Cas9-Mediated Knockout of MYB49
A 20-bp small guide RNA (sgRNA) sequence (AAACGCACGGUCCCGGCAAA) in the first exon of MYB49 was chosen as a protospacer sequence for gene editing. The sgRNA containing the MYB49-protospacer was inserted in pCAMBIA1300-Cas9, yielding pCAMBIA1300-Cas9-MYB49, which was then transformed into the Col-0 background. Hygromycin-resistant plants from the T1 generation were tested for the insertion of the 35S-hyB-sgRNA-gRNA scaffold-pUbi-Cas9 by PCR with M49-G1F and M49-G1R. Targeted gene mutations were detected by sequencing of the PCR products using M49-G1SeqF (Supplemental Table S4). Plants with an identified MYB49 mutation were allowed to self-fertilize, and homozygous plants in the T3 generation were detected by sequencing using the same technique (Supplemental Fig. S5).
Cd and Nutrient Content Analysis
Treated seedlings were immersed for 2 h with 1 mm EDTA solution and then thoroughly rinsed eight times with distilled water as described previously (Liu et al., 2016b). The leaves and roots were harvested and oven dried at 75°C for 3 to 4 d. The dried samples were digested in nitric acid for 3 to 5 d at room temperature and then boiled for 1 to 2 h until completely digested. After adding 4 mL of Millipore-filtered deionized water and briefly centrifuging the solution, the Cd, Mg, Mn, Zn, Cu, Fe, K, P, S, and Ca contents were determined using ICP-AES. Each experiment was repeated six times.
RT-qPCR
Total RNA was isolated using an RNAiso Plus kit (TaKaRa). RT was performed using the PrimeScript RT Reagent Kit with genomic DNA Eraser (TaKaRa). RT-qPCR was performed using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). We used ACTIN2 (At3g18780) and Elongation Factor 1a (At5g60390) as internal controls for RT-qPCR normalization according to GeNorm (Czechowski et al., 2005; Liu et al., 2016a). The specific primers are listed in Supplemental Table S4. The reactions were performed on three biological replicates with three technical repetitions.
Y2H Assays
The full-length CDS of ABI5 was amplified and cloned into the bait plasmid pGBKT7 (BD) vector, and the CDS of MYB49 was cloned into the prey vector pGADT7 (AD) and transformed into yeast strain Y2HGold cells according to the Matchmaker user’s manual protocol (Clontech). The Y2H assay was performed as described previously (Jiang et al., 2019). Yeast transformants were selected by growth on synthetic dropout nutrient medium SD/-Leu/-His/-Ade/-Trp (Clontech) supplemented with 40 µg mL–1 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside at 30°C for 3 to 5 d to determine possible interactions between proteins. The primers used for Y2H are summarized in Supplemental Table S4. Each assay was performed three times with the same result.
Co-IP Assay
The full-length CDSs of MYB49 and ABI5 were amplified and cloned into tagging vectors with HA or Myc tags under the control of the CaMV 35S promoter, respectively. Co-IP assays were performed by transient expression in tobacco (Nicotiana tabacum) by agroinfiltration as described by Jiang et al. (2014). Briefly, total protein was extracted from tobacco leaves using an extraction buffer containing 0.1 m Tris-HCl (pH 7.5), 0.2 m NaCl, 20% (v/v) glycerol, 5 mm EDTA, 0.01 m β-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride and immunoprecipitated using anti-HA antibody. The coimmunoprecipitated ABI5-Myc was then detected using an anti-Myc antibody. The primers used for the clones are listed in Supplemental Table S4.
In Vitro Pull-Down Assay
The full-length CDSs of MYB49 and ABI5 were amplified and cloned into the pET30a and pGEX4T-1 vectors, respectively, and then introduced into Rosetta (DE3) chemically competent cells for expression of MYB49-His and GST-ABI5 proteins by using 0.1 mm isopropyl-β-thiogalactopyranoside induction. Soluble GST or GST-ABI5 fusion proteins were extracted and immobilized using a glutathione HiCap matrix (Qiagen). MYB49-His protein was incubated with immobilized GST or GST-ABI5 protein, and the interaction was detected by western-blot analysis using an anti-His antibody (Sigma-Aldrich) as described by Zhao and Wang (2004). The primers used for clones are listed in Supplemental Table S4.
BiFC Assay
The full-length CDSs of MYB49 and ABI5 were amplified and cloned into pUC-SPYNE and pUC-SPYCE vectors, respectively, to generate N-terminal in-frame fusions with nYFP and C-terminal in-frame fusions with cYFP constructs. BiFC assays were performed by biolistic bombardment into onion (Allium cepa) epidermal cells as described by Zhu et al. (2011). After overnight expression, YFP fluorescence was observed by a confocal laser-scanning microscope (Zeiss). BiFC was repeated three times independently. The primers used for clones are listed in Supplemental Table S4.
Y1H Assay
The Y1H experiment was performed following the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech). The promoter fragments were amplified from Arabidopsis genomic DNA and fused into the pAbAi vector. Then, the pBait-AbAi constructs were integrated into the genome of the yeast strain Y1HGold and streaked onto SD/-Ura agar medium to select positive colonies (Konishi and Yanagisawa, 2013). The CDS of AtMYB49 was cloned into the prey vector pGADT7 (AD) and transformed into the yeast strain Y1HGold (pBait-AbAi). Yeast transformants were tested on SD/-Leu/aureobasidin A medium. Primers used in this assay are listed in Supplemental Table S4.
ChIP-qPCR
The seedlings of 35S:MYB49-GFP (∼2.5 g) were washed three times with distilled water and immediately ground in liquid nitrogen. The chromatin DNA was isolated and then sonicated. Chromatin solution (300 µL) was diluted to 3 mL using ChIP dilution buffer (containing 10% Triton, 1 µm EDTA, 16.7 µm Tris-Cl, pH 8, and 167 µm NaCl) and divided into two other tubes equally. Then, 40 µL of protein G-agarose beads (Upstate Biotechnology) was added to each tube, and the mixture was incubated for 1 h at 4°C. The solutions were transferred into two preprepared fresh tubes; one tube had 10 µL of anti-GFP antibody (ab290; Abcam) with a 1:150 dilution, and the other tube was a negative control with no additions. Fifty microliters of protein G-agarose beads was added into the two tubes for immunoprecipitation, and the samples were incubated and rotated gently at 4°C overnight. The immunoprecipitated complex was eluted using TE buffer (10 µm Tris-Cl, pH 8, and 1 µm EDTA), and proteinase K (10 mg mL−1) was added. The elution was then performed with 5 m NaCl to reverse the cross-linking. DNA was extracted with phenol:chloroform (1:1, v/v), and sterile distilled water (20 µL) was added to dissolve the sample. The purified DNA was used for PCR. The primer sequences used here are listed in Supplemental Table S4.
EMSA
EMSA was carried out according to the LightShift Chemiluminescent EMSA Kit (GS009; Beyotime). The MYB49-His and GST-ABI5 proteins were induced by isopropyl-β-thiogalactopyranoside and purified using Ni-NTA agarose (Qiagen) and a glutathione HiCap matrix (Qiagen), respectively. A specific fragment in the HIPP22 promoters containing predicted MYB-binding sites was used as a DNA probe for EMSA. Labeled probes were incubated with purified protein in binding buffer (10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm EDTA, 5 mm dithiothreitol, and 5% [v/v] glycerol) at room temperature for 30 min. The reaction products were electrophoresed on an 8% PAGE gel in 0.5× Tris-borate-EDTA buffer for approximately 1 h. The sequences of wild-type and mutant probes used here are listed in Supplemental Table S4. Two biological experiments were performed with similar results.
Dual-Luciferase Reporter System
Vectors pGreen II 0800-Luciferase (LUC) and pGreen II 62-SK were used in this study. The dual-luciferase reporter assay was performed following the manufacturer’s instructions (Promega) and as described by Hellens et al. (2005). Briefly, Arabidopsis protoplasts were prepared for cotransfection with the constructs (Hellens et al., 2005; Yoo et al., 2007). After the protoplasts were cultured for 16 h under low-light conditions, the LUC-Renilla luciferase (REN) activity ratio was determined for the dual-luciferase reporter system (Promega). The LUC-REN ratio indicates transcriptional activity. The primers used here are listed in Supplemental Table S4. Three biological repeats were performed.
Statistical Analyses
The experimental data are presented as means ± sd. For variance analysis, Statistical Package for the Social Sciences (SPSS) was used to assess statistically significant differences (P < 0.01) based on Student’s t tests or Tukey’s test. Values at P < 0.01 were statistically significant.
Accession Numbers
Sequence data for the genes described in this article can be found in The Arabidopsis Information Resource database (https://www.arabidopsis.org) under the following accession numbers: MYB49 for At5g54230, ABI5 for At2g36270, HIPP02 for At5g26690, HIPP09 for At5g24580, HIPP22 for At1g22990, HIPP24 for At4g08570, HIPP44 for At4g10465, bHLH38 for At3g56970, bHLH39 for At3g56980, bHLH100 for At2g41240, bHLH101 for At5g04150, IRT1 for At4g19690, ZIP5 for At1g05300, NAS2 for At5g56080, NAS4 for At1g56430, and YSL2 for At5g24380.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Responses of MYB49 expression to Cd treatment.
Supplemental Figure S2. Subcellular localization of MYB49.
Supplemental Figure S3. Transactivational activity assay.
Supplemental Figure S4. Generation of OX49 and SRDX49 transgenic plants.
Supplemental Figure S5. Generation of myb49.1 and myb49.2 knockout lines by CRISPR/Cas9.
Supplemental Figure S6. Mineral concentrations in Col-0, OX49.3, and SRDX49.6 seedlings.
Supplemental Figure S7. MYB49 did not affect the Fe-deficiency response.
Supplemental Figure S8. HIPP44 expression levels in HIPP44OX lines.
Supplemental Table S1. DEGs in OX49/Col-0.
Supplemental Table S2. DEGs in SRDX49/Col-0.
Supplemental Table S3. Expression of metal accumulation-related genes in OX49 and SRDX49.
Supplemental Table S4. List of primers.
Acknowledgments
We thank the Public Technology Service Center of the Xishuangbanna Tropical Botanical Garden of the Chinese Academy of Sciences for providing research facilities.
Footnotes
This work was supported by the China National Natural Sciences Foundation (31772383), the National Key Research and Development Program of China (2016YFC0501901), the Basic Research Program of Qinghai Province (2019-ZJ-7033), and the Yunnan Province Foundation for Academic Leader (2014HB043).
References
- Aprile A, Sabella E, Vergine M, Genga A, Siciliano M, Nutricati E, Rampino P, De Pascali M, Luvisi A, Miceli A, et al. (2018) Activation of a gene network in durum wheat roots exposed to cadmium. BMC Plant Biol 18: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assunção AG, Herrero E, Lin YF, Huettel B, Talukdar S, Smaczniak C, Immink RG, van Eldik M, Fiers M, Schat H, et al. (2010) Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc Natl Acad Sci USA 107: 10296–10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL (2005) A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol 139: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Ma JF (2016) Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu Rev Plant Biol 67: 489–512 [DOI] [PubMed] [Google Scholar]
- Clemens S, Palmgren MG, Krämer U (2002) A long way ahead: Understanding and engineering plant metal accumulation. Trends Plant Sci 7: 309–315 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H (2003) An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J 33: 543–555 [DOI] [PubMed] [Google Scholar]
- Eren E, Argüello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P(IB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136: 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan SK, Fang XZ, Guan MY, Ye YQ, Lin XY, Du ST, Jin CW (2014) Exogenous abscisic acid application decreases cadmium accumulation in Arabidopsis plants, which is associated with the inhibition of IRT1-mediated cadmium uptake. Front Plant Sci 5: 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol 122: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML. (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD. (2004) The calcium conundrum: Both versatile nutrient and specific signal. Plant Physiol 136: 2438–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, Kao CH (2003) Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ 26: 867–874 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Jiang J, Xi H, Dai Z, Lecourieux F, Yuan L, Liu X, Patra B, Wei Y, Li S, Wang L (2019) VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J Exp Bot 70: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HI, Gayomba SR, Rutzke MA, Craft E, Kochian LV, Vatamaniuk OK (2012) COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J Biol Chem 287: 33252–33267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen Z, Sebastian A, Xu W, Grain D, Salsac F, Avon A, Berger N, Tran J, Dubreucq B, Lurin C, et al. (2015) Analysis of the DNA-binding activities of the Arabidopsis R2R3-MYB transcription factor family by one-hybrid experiments in yeast. PLoS ONE 10: e0141044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, Nishizawa NK (2007) The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc Natl Acad Sci USA 104: 19150–19155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Itai RN, Aung MS, Senoura T, Nakanishi H, Nishizawa NK (2012) The rice transcription factor IDEF1 directly binds to iron and other divalent metals for sensing cellular iron status. Plant J 69: 81–91 [DOI] [PubMed] [Google Scholar]
- Kong Y, Chen S, Yang Y, An C (2013) ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett 587: 3076–3082 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang R, Zhang P, Chen Q, Luo Q, Zhu Y, Xu J (2016a) The nitrification inhibitor methyl 3-(4-hydroxyphenyl)propionate modulates root development by interfering with auxin signaling via the NO/ROS pathway. Plant Physiol 171: 1686–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Wang RL, Zhang P, Sun LL, Xu J (2016b) Involvement of reactive oxygen species in lanthanum-induced inhibition of primary root growth. J Exp Bot 67: 6149–6159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Kim D, Kim YS, Hörtensteiner S, Paek NC (2014) Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett 588: 3830–3837 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF (2016) Transporters involved in mineral nutrient uptake in rice. J Exp Bot 67: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Sharma SS, Kumar V (2002) Responses of wild type and abscisic acid mutants of Arabidopsis thaliana to cadmium. J Plant Physiol 159: 1323–1327 [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Skubacz A, Daszkowska-Golec A, Szarejko I (2016) The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 7: 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Huang G, Zhang Q, Wang X, Li C, Tao Y, Zhang S, Lai J, Yang C, Wang Y (2016) The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci 247: 93–103 [DOI] [PubMed] [Google Scholar]
- Sun N, Liu M, Zhang W, Yang W, Bei X, Ma H, Qiao F, Qi X (2015) Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco. Plant Physiol 167: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehseen M, Cairns N, Sherson S, Cobbett CS (2010) Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2: 556–564 [DOI] [PubMed] [Google Scholar]
- Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S (2009) Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot 60: 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Wu D, Yamaji N, Yamane M, Kashino-Fujii M, Sato K, Ma JF (2016) The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol 172: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Chen C, Du J, Liu H, Cui Y, Zhang Y, He Y, Wang Y, Chu C, Feng Z, et al. (2012) Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol 158: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang YX, Wei W, Han L, Guan ZQ, Wang Z, Chai TY (2008) BjDHNs confer heavy-metal tolerance in plants. Mol Biotechnol 38: 91–98 [DOI] [PubMed] [Google Scholar]
- Yao X, Cai Y, Yu D, Liang G (2018) bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J Integr Plant Biol 60: 691–702 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Suzui N, Ishii S, Kitazaki M, Yamazaki H, Kitazaki K, Kawachi N, Yin YG, Ito-Tanabata S, Hashida SN, et al. (2014) A kinetic analysis of cadmium accumulation in a Cd hyper-accumulator fern, Athyrium yokoscense and tobacco plants. Plant Cell Environ 37: 1086–1096 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015) The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang X (2004) Arabidopsis phospholipase Dalpha1 interacts with the heterotrimeric G-protein α-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279: 1794–1800 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhou H, Li Y (2013) UBIQUITIN-SPECIFIC PROTEASE16 interacts with a HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN27 and modulates cadmium tolerance. Plant Signal Behav 8: e25680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiesche W. (2013) Das Metallbindeprotein HIPP3 als regulatorische Komponente pflanzlicher Stressantworten und Entwicklung. PhD thesis.Universitäts- und Landesbibliothek Sachsen-Anhalt, Halle (Saale), Germany [Google Scholar]