Arabidopsis protein phosphatase PP7-like (PP7L) is a predominantly nuclear-localized protein that promotes chloroplast translation and development, thus contributing to abiotic stress tolerance.
Abstract
Chloroplast biogenesis is indispensable for proper plant development and environmental acclimation. In a screen for mutants affected in photosynthesis, we identified the protein phosphatase7-like (pp7l) mutant, which displayed delayed chloroplast development in cotyledons and young leaves. PP7L, PP7, and PP7-long constitute a subfamily of phosphoprotein phosphatases. PP7 is thought to transduce a blue-light signal perceived by crys and phy a that induces expression of SIGMA FACTOR5 (SIG5). We observed that, like PP7, PP7L was predominantly localized to the nucleus in Arabidopsis (Arabidopsis thaliana), and the pp7l phenotype was similar to that of the sig6 mutant. However, SIG6 expression was unaltered in pp7l mutants. Instead, loss of PP7L compromised translation and ribosomal RNA (rRNA) maturation in chloroplasts, pointing to a distinct mechanism influencing chloroplast development. Promoters of genes deregulated in pp7l-1 were enriched in PHYTOCHROME-INTERACTING FACTOR (PIF)-binding motifs and the transcriptome of pp7l-1 resembled those of pif and CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) signalosome complex (csn) mutants. However, pif and csn mutants, as well as cop1, cryptochromes (cry)1 cry2, and phytochromes (phy)A phyB mutants, do not share the pp7l photosynthesis phenotype. PhyB protein levels were elevated in pp7l mutants, but phyB overexpression plants did not resemble pp7l. These results indicate that PP7L operates through a different pathway and that the control of greening and photosystem biogenesis can be separated. The lack of PP7L increased susceptibility to salt and high-light stress, whereas PP7L overexpression conferred resistance to high-light stress. Strikingly, PP7L was specifically recruited to Brassicales for the regulation of chloroplast development. This study adds another player involved in chloroplast biogenesis.
Chloroplasts are the site of photosynthesis and are essential for plant growth and development. They are the prototypes of a diverse family of organelles, the plastids, which are found in plants and various algae (Keeling, 2010). As descendants of cyanobacteria-like progenitors, plastids of higher plants are semiautonomous organelles that have retained a reduced genome of ∼100 genes (Kleine et al., 2009). However, they also contain ∼2,000–3,000 nuclear-encoded proteins (Abdallah et al., 2000; Yu et al., 2008). Consequently, chloroplast multiprotein complexes, such as the photosynthetic apparatus and the chloroplast gene expression machinery, comprise a mixture of proteins encoded by chloroplast and nuclear genes, and chloroplast function therefore requires stringent coordination of the expression of the two genomes (Woodson and Chory, 2008). This is achieved by bidirectional exchange of information between the organelles. During retrograde signaling, chloroplasts send out signals that inform the nucleus about their developmental and physiological status so that nuclear gene expression can be adjusted accordingly (Bobik and Burch-Smith, 2015; Chan et al., 2016; Kleine and Leister, 2016). Because most chloroplast proteins are encoded in the nucleus, the nucleus exercises anterograde control over the plastids by regulating various functions, in particular chloroplast gene expression (Tiller and Bock, 2014; Schmitz-Linneweber et al., 2015; Shikanai, 2015).
In true leaves, chloroplasts develop from meristematic proplastids when the leaf primordia emerge; in cotyledons, chloroplasts develop from proplastids/eoplasts or etioplasts present in mesophyll tissue within the embryo (Waters and Langdale, 2009; Liebers et al., 2017). These precursors are rapidly converted into chloroplasts upon exposure to light. Thus, the development of photosynthetically competent chloroplasts in angiosperm seedlings is strictly dependent on exposure to light (Pogson and Albrecht, 2011) and involves the so-called chloroplast biogenic signaling pathway, which triggers massive changes in the nuclear transcriptome, with at least 20% of the genome showing light-regulated differential expression during seedling development (Jiao et al., 2007; Jarvis and López-Juez, 2013). Light is perceived mainly by two families of photoreceptors—the phytochromes (phys) and the cryptochromes (crys)—which are sensitive to red/far-red and UVA/blue light, respectively, and undergo conformational changes upon light perception that enable them to interact with downstream signaling partners (Jiao et al., 2007). The latter include PHYTOCHROME-INTERACTING FACTOR (PIF)-like transcription factors of the basic helix–loop–helix (bHLH) family and basic Leu zipper (bZIP) transcription factors, such as ELONGATED HYPOCOTYL5 (HY5) and HY5-like HYH (Jiao et al., 2007; Leivar and Monte, 2014). PIFs mainly repress photomorphogenesis genes in the dark, whereas HY5 and HYH activate those same genes upon illumination. Hence, PIFs and HY5/HYH have antagonistic functions in the regulation of e.g. protochlorophyllide production and reactive oxygene species (ROS)-responsive genes during the dark-to-light transition (Chen et al., 2013). In the dark, HY5 is targeted for degradation by the ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) in the nucleus (Osterlund et al., 2000). Therefore, COP1 suppresses photomorphogenic development when translocated to the nucleus (Yi and Deng, 2005) and its nuclear localization is regulated by a component of the COP9 signalosome complex (CSN; Wang et al., 2009). Conversely, upon light exposure, phys trigger PIF degradation and COP1 inactivation (Gommers and Monte, 2018). Strikingly, the HEMERA (HMR)/transcriptionally active chromosome protein 12 (pTAC12) not only contributes to phy signaling in the nucleus, but also forms part of the chloroplast’s pTAC and is involved in plastid gene expression (Chen et al., 2010).
In higher plants, transcription of plastid DNA is carried out by a nuclear-encoded phage-type RNA polymerase and a plastid-encoded prokaryotic-like RNA polymerase (PEP; Pfannschmidt et al., 2015). For its function, the multisubunit enzyme PEP requires a set of polymerase-associated proteins and sigma factors (SIGs), all of which are encoded in the nucleus (Chi et al., 2015; Pfannschmidt et al., 2015). In Arabidopsis (Arabidopsis thaliana), there are six different SIGs, designated as SIG1–SIG6. Depending on their promoter selectivity, SIGs may have overlapping functions as well as specialized roles (Chi et al., 2015). SIG5, for example, effectively mediates blue-light signaling by activating transcription from the “blue-light–responsive” psbD promoter (Nagashima et al., 2004), as its own expression is strictly dependent on blue light. The response to blue-light signals in the nucleus is assumed to involve the perception of blue light by crys and phyA, and transduction of the signal by the nucleus-localized protein phosphatase PP7, which in turn leads to the induction of SIG5 expression (Møller et al., 2003; Chi et al., 2015). PP7 is a member of the family of Ser/Thr-specific phosphoprotein phosphatases (PPPs; Farkas et al., 2007). The Arabidopsis PPP family comprises 26 members, which can be assigned to seven subfamilies. PPPs have roles in abscisic acid, auxin, and brassinosteroid signaling, phototropism, regulating the target of rapamycin pathway, cell stress responses. and flowering time (Uhrig et al., 2013; Lillo et al., 2014). For example, phy-associated Ser/Thr protein phosphatase, which belongs to the type-6 subfamily, dephosphorylates phyA in vitro and delays flowering (Kim et al., 2002). Members of the PPP family are present in all eukaryotes. However, the type-7 subfamily is unique to plants. In Arabidopsis, this subfamily contains three members—PP7, “long PP7,” and “inactive PP7” (Farkas et al., 2007; Uhrig et al., 2013). Only PP7 has been characterized in detail, and shown to regulate blue-light (Møller et al., 2003) as well as red/far-red light signaling (Genoud et al., 2008). Long PP7 is also designated “MAINTENANCE OF MERISTEMS-LIKE3” and encodes a protein bearing a putative aminotransferase domain in addition to the PP7 domain. The phenotype of mail3-2, in which a transfer-DNA (T-DNA) is inserted in the PP7 domain, is indistinguishable from that of wild-type plants (Ühlken et al., 2014).
Here, we characterize PP7-like (pp7l) mutants, the first of which was identified in a screen for mutants defective in photosynthesis. The PP7L gene encodes the PP7 homolog PP7L (previously designated as “inactive PP7”; Farkas et al., 2007). PP7L is localized to the nucleus, and is a positive regulator of protein synthesis in the developing chloroplast. However, it does not act by modulating SIG factor gene expression like PP7. Instead, the pp7l mutant is shown here to be defective in chloroplast ribosomal RNA (rRNA) maturation, and consequently in mRNA translation. Promoter analysis of genes deregulated in the pp7l-1 mutant and database analysis of conditions or mutations associated with gene expression changes similar to those seen in pp7l-1 suggested a tentative association of PP7L with PIFs and other light signaling components, but neither pif, csn, cop1, cry1 cry2, or phyA phyB mutants display a photosynthesis phenotype. Moreover, although phyB levels are enhanced in pp7l mutants, overexpression of phyB does not induce a photosynthesis phenotype. Seed germination of pp7l mutants was reduced by exposure to salt and high light, whereas overexpression of PP7L rendered 4-week–old plants more tolerant to high light.
RESULTS
Identification and Phenotypic Analysis of Mutants for the PP7L Locus
Screening of an Arabidopsis mutant collection carrying insertions of the maize transposable element En-1 (Wisman et al., 1998) for lines that show alterations in the effective quantum yield of PSII, designated ΦII, resulted in the recovery of a set of mutants with defects in photosynthesis (Varotto et al., 2000). In one of these (ZIGIA line V2-880), the effective (ΦII) and maximum (Fv/Fm) quantum yields of PSII were significantly reduced in emerging leaves of 3-week–old plants compared to the wild type (Col-0), implying a defect in energy transfer within PSII. Isolation of the genomic sequence flanking the En-1 transposon enabled identification of the insertion site in the second exon of the gene AT5G10900 (Supplemental Fig. S1A). AT5G10900 is listed in the UniProtKB database (http://www.uniprot.org/uniprot/Q9LEV0) as “Ser/Thr-protein phosphatase 7 (PP7) inactive homolog.” Thus, AT5G10900 was designated “PP7L” and the mutant was named “pp7l:En-1.” Because the pp7l:En-1 mutant germinated poorly, further mutant lines were identified with the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress). In these lines, named “pp7l-1” (SALK_018295), “pp7l-2” (SALK_033071), and “pp7l-3” (SALK_022053), the pROK2 T-DNA was inserted in exon 4, intron 5, and exon 7 of PP7L at positions 651, 1951, and 2251 relative to the start codon, respectively (Supplemental Fig. S1, A and B). In all identified pp7l mutants, both overall growth rates and Fv/Fm values were reduced in emerging leaves of 3-week–old soil-grown plants compared to the wild type (Supplemental Fig. S1A).
According to The Arabidopsis Information Resource genome annotation 10, AT5G10900 is a single-copy gene with three predicted transcript splice forms AT5G10900.1, AT5G10900.2, and AT5G10900.3, which differ only at their 3′ ends (Supplemental Fig. S1B). To confirm that the altered expression of PP7L was responsible for the mutant phenotype, reverse transcription quantitative PCR (RT-qPCR) was conducted. We found that the PP7L transcript was barely detectable in the pp7l-2 mutant and undetectable in pp7l-3, but its level was elevated in pp7l-1 (Supplemental Fig. S1C). The overexpression of the 3′ segment of PP7L in the pp7l-1 allele can be explained by the orientation of the T-DNA integration in the pROK2 vector in the 5′RB (right border)–T-DNA–left border 3′ direction, because this vector contains the 35S promoter on the left border site (Baulcombe et al., 1986) and can potentially activate flanking genomic sequences (Ulker et al., 2008). However, because of these differences in PP7L transcript accumulation, all pp7l mutant lines were transformed with a genomic DNA fragment comprising the coding sequence from the start to the stop codon of AT5G10900 fused upstream of the enhanced green fluorescence protein (eGFP) reporter gene, which was placed under the control of the Cauliflower Mosaic Virus 35S promoter. Independent transgenic lines for all three mutants were obtained (overexpression of PP7L [oe-PP7L] ppl7l-1 to -3) and their phenotypic and chlorophyll (Chl) fluorescence analyses showed that the wild-type function had been restored in all oe-PP7L lines (Supplemental Fig. S2A). This shows that the pp7l phenotype is indeed caused by the insertions within the AT5G10900 gene. The striking dark-green coloration of pp7l leaves (Fig. 1A) was found not to be caused by a higher overall Chl content. In fact both (Chl a+b) and the Chl a/b ratio were slightly, but not significantly, reduced compared to the wild type (Supplemental Fig. S2B). Instead, the darker color of pp7l leaves, which is most prominent on the abaxial side, can be attributed to the approximately eightfold-higher content of anthocyanins (compared to wild-type levels) found in these mutants (Fig. 1B).
Figure 1.
Phenotypic characterization of pp7l T-DNA insertion mutants. A, Phenotypes of 3-week–old wild-type (Col-0) and the different pp7l mutant plants grown in LD (16-h light/8-h dark) conditions. The maximum quantum yield of PSII (Fv/Fm) was measured with an imaging Chl fluorometer (Imaging PAM). Scale bar = 1 cm. B, The abaxial surface of leaves of plants grown in LD conditions reveals enhanced anthocyanin accumulation in pp7l plants. Anthocyanins were extracted and quantified photometrically, and amounts are reported in arbitrary units (a.u.). Data are shown as mean values ± sd from three different plant pools. Each pool contained >20 plants. Significant differences between the data pairs were identified by Tukey’s test, and significant differences (P < 0.05) with respect to Col-0 are indicated by asterisks.
PP7L Is Localized to the Nucleus and the Cytosol
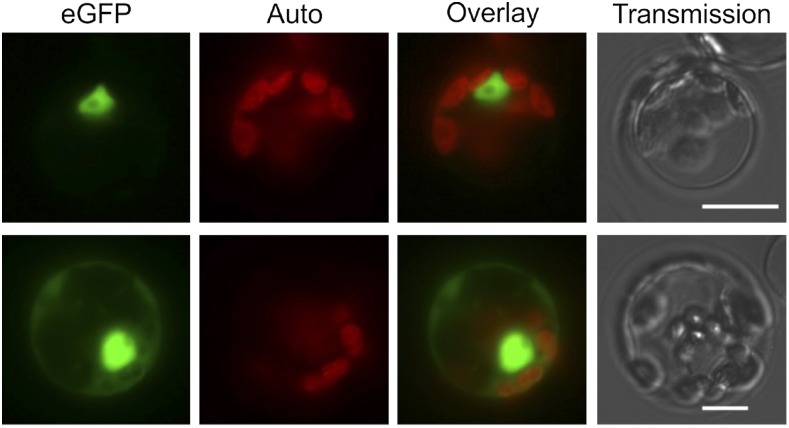
The pp7l mutants show a photosynthesis phenotype, which at first sight suggests chloroplast localization of the PP7L protein. However, computational prediction of the subcellular localization of PP7L by the algorithms implemented in SUBA4 (Hooper et al., 2017; http://suba.plantenergy.uwa.edu.au) did not result in a chloroplast localization, although various predictors assigned PP7L either to the nucleus, cytosol, or mitochondria, and the SUBAcon algorithm predicted a nuclear localization for PP7L. A putative nuclear localization signal (NLS) was indeed found by the cNLS mapper (Kosugi et al., 2009; http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi), which predicts a bipartite NLS (ISKRVLDSKLEACKFAFLKLSAVKTTRMK) spanning the region from amino acids 6 to 34. To experimentally examine the subcellular localization of PP7L, protoplasts were transformed with the 35S:PP7L-eGFP construct that had been used for complementation of the pp7l alleles. Visualizing eGFP fluorescence in the protoplasts confirmed that PP7L-eGFP was targeted to the nucleus (Fig. 2). Moreover, several protoplasts displayed eGFP fluorescence in the cytosol, indicating a dual (nuclear and cytosolic) localization of PP7L.
Figure 2.
PP7L is localized to the nucleus and the cytosol. Fluorescence microscopy of Arabidopsis protoplasts transiently expressing PP7L fused to eGFP. The eGFP fluorescence (green) and chloroplast autofluorescence (red) are shown together in the overlay picture. Scale bars = 10 µm.
PP7L Is a Positive Regulator of Photosynthesis at Young Developmental Stages
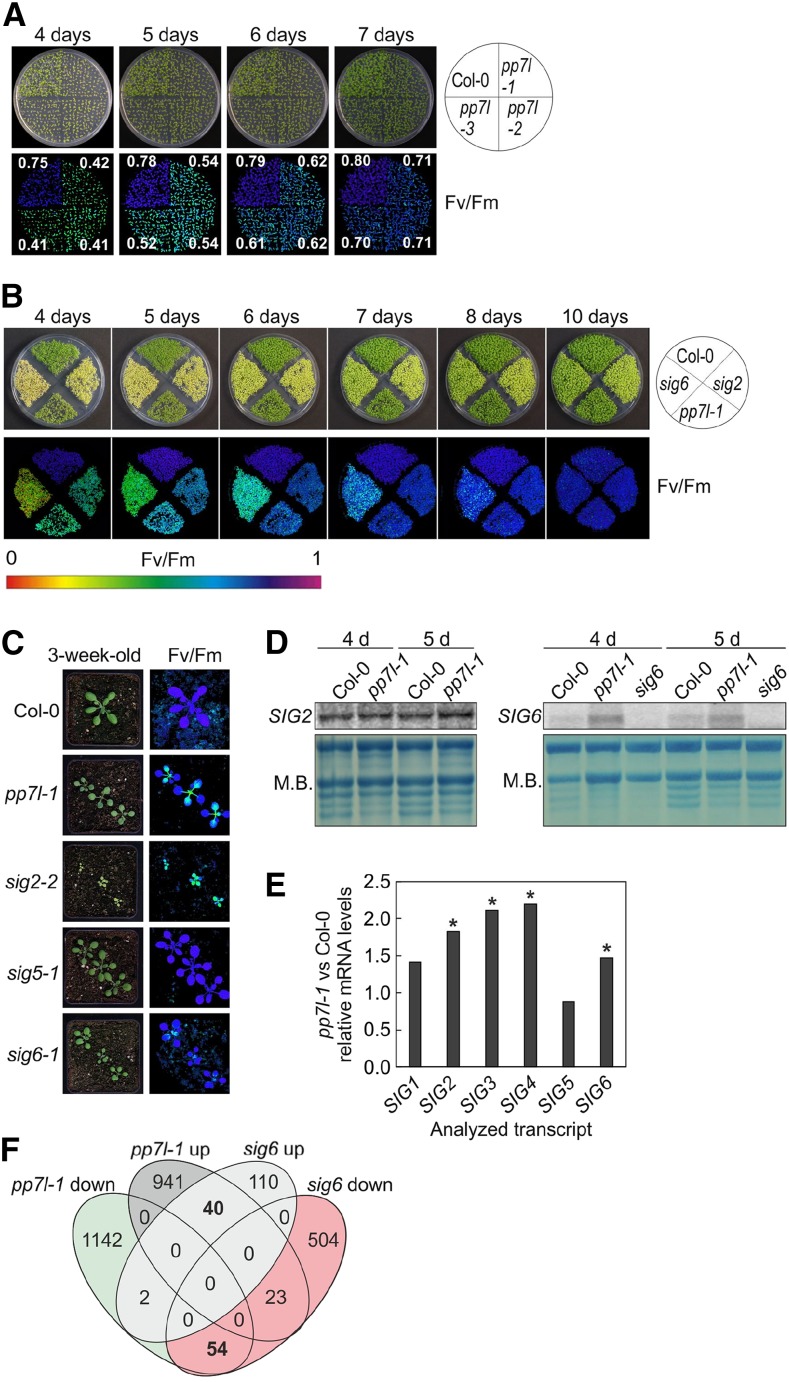
In addition to the drop in Fv/Fm seen in emerging leaves of pp7l mutants, reduced photosynthetic activity was observed during seedling development. In 4-, 5-, 6-, and 7-d–old seedlings, Fv/Fm was significantly depressed in all pp7l mutants to ∼0.41, 0.54, 0.62, and 0.71, respectively, compared to the respective wild-type values of 0.75, 0.78, 0.79, and 0.80 (Fig. 3A), and this depressed Fv/Fm recovered completely after 10 d (Fig. 3B). Interestingly, this phenotype is reminiscent of those of sig2 and sig6 seedlings (Fig. 3B) and of young leaves of the sig6 mutant (Fig. 3C). It was previously shown that sig2 and sig6 mutant seedlings are pale green and accumulate fewer plastid- and nuclear-encoded transcripts for photosynthesis proteins, whereas sig1, sig3, sig4, and sig5 seedlings appear more like wild type (Woodson et al., 2013); for the sig5 mutant, see Figure 3C. Because PP7 has been implicated in the induction of SIG5 expression (reviewed in Chi et al., 2015), we asked whether PP7L might similarly act through SIG-mediated gene expression changes. Because sig2 and sig6 seedlings show a transient photosynthesis phenotype that resembles the pp7l phenotype, SIG2 and SIG6 mRNA levels were studied in 4- and 5-d–old wild-type and pp7l-1 seedlings by RNA gel-blot analysis. In fact, we found that levels of SIG2 and SIG6 transcripts were increased and not decreased in pp7l-1 (Fig. 3D).
Figure 3.
Comparative analysis of pp7l-1 and sig (sig2 and sig6) mutants. A, Phenotypes of 4- to 7-d–old Col-0 and the different pp7l mutant seedlings together with Imaging PAM pictures showing maximum quantum yields of PSII (Fv/Fm) sds are provided in Supplemental Table S1. B, Phenotypes and Imaging PAM images representing the Fv/Fm activity of 4- to 10-d–old Col-0, pp7l-1 and sig2 and sig6 mutant seedlings. C, Phenotypes of 3-week–old Col-0, pp7l-1, and sig mutant plants and Imaging PAM pictures showing maximum quantum yields of PSII (Fv/Fm). Scale bars = 1 cm. D, Northern-blot analysis of SIG2 and SIG6 transcripts in 4- and 5-d–old Col-0 and pp7l-1 plants. For the detection of SIG6 mRNA levels, the sig6 mutant was included as a control, because several unspecific bands were detected with the SIG6 probe. Total RNAs were fractionated in a formaldehyde-containing denaturing gel, transferred onto a nylon membrane, and probed with (α-32P)dCTP-labeled cDNA fragments specific for the different SIG transcripts. The rRNA was visualized by staining the membrane with methylene blue (M.B.) and served as a loading control. E, Graph representing the changes (relative to Col-0 values) in SIG mRNA levels in pp7l-1, as determined by lncRNA-Seq analysis. Significant differences (DESeq2 analysis running with the fit type set to “parametric”; adjusted P < 0.05) with respect to Col-0 are denoted by asterisks. The sd was not calculated, because differentially expressed genes were identified with “parametric” and not “mean” fit settings in DESeq2. F, Venn diagrams depicting the degree of overlap between the sets of genes whose expression levels were altered by at least twofold (up or down) in the pp7l‐1 mutant compared with the sig6 mutant.
To obtain an overall insight into the RNA expression pattern of nuclear- and organelle-encoded genes when PP7L function is compromised, RNAs isolated from 4-d–old wild-type and pp7l-1 mutant seedlings grown in long-day (LD; 16-h light/8-h dark) conditions were subjected to long non-coding (lnc)RNA sequencing. With this method, the elevated levels of SIG2 (1.8-fold) and SIG6 (1.7-fold) mRNA were confirmed. Also, SIG1, SIG3, and SIG4 transcripts were slightly increased, whereas SIG5 mRNA levels were almost unchanged (0.9-fold) in pp7l-1 (Fig. 3E; Supplemental Table S2). Moreover, comparison of whole genome transcriptome changes in pp7l-1 with previously published mRNA-sequencing (Seq) data for sig6 showed that pp7l-1 and sig6 mutants do not provoke similar gene expression changes (Fig. 3F).
In summary, PP7L does not positively regulate SIG gene expression. Therefore, PP7L likely acts through a mechanism that is different from that of PP7.
PP7L Is Involved in the Regulation of Chloroplast Gene Expression
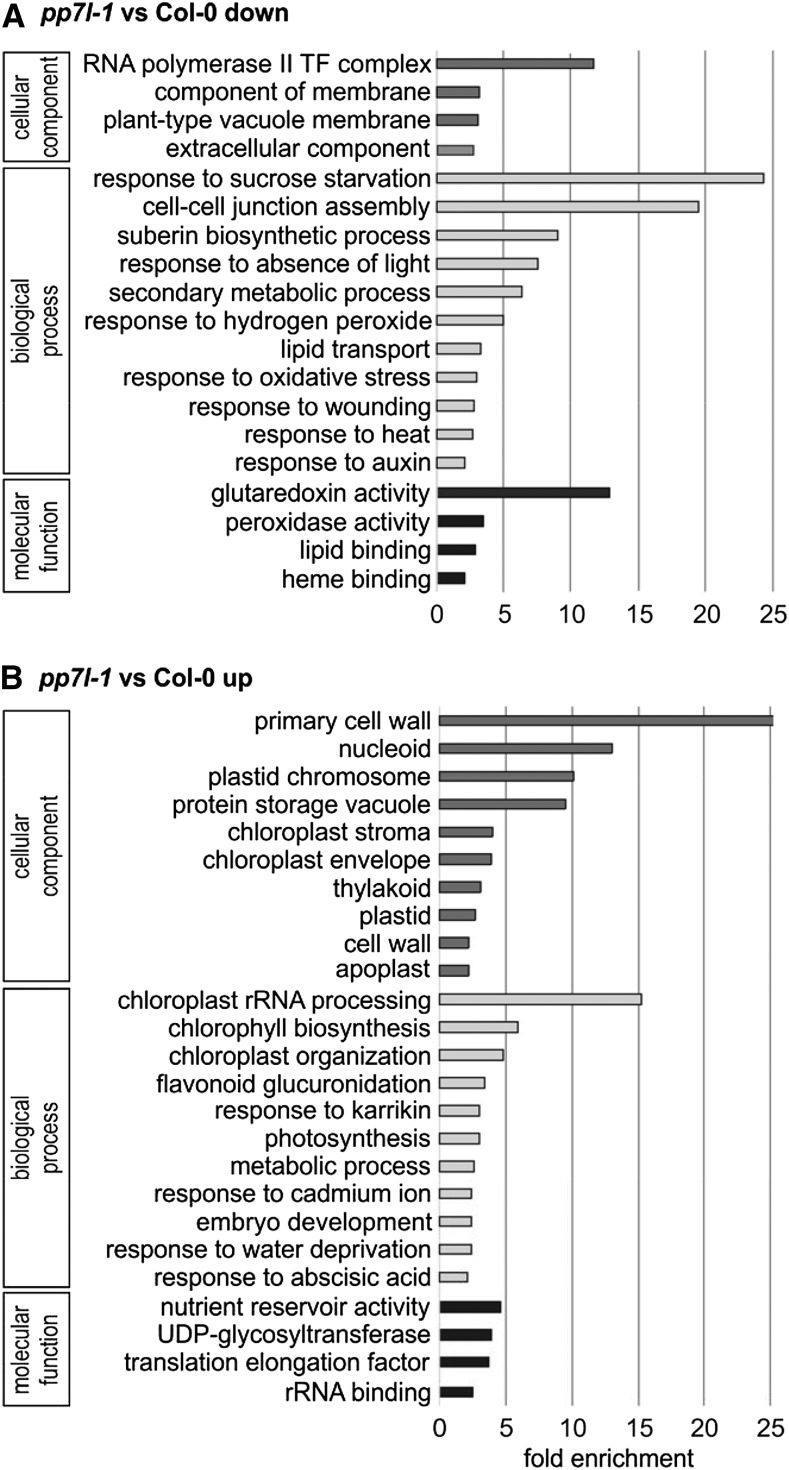
The lncRNA-Seq analysis revealed substantial changes in gene expression in 4-d–old pp7l-1 seedlings, specifically 1,198 and 1,004 genes whose mRNA levels were significantly (more than twofold) reduced or elevated, respectively (Supplemental Table S2). Genes encoding enzymes of the phenylpropanoid pathway leading to anthocyanin like DIHYDROFLAVONOL4-REDUCTASE (TRANSPARENT TESTA3), ANTHOCYANIDIN SYNTHASE, and the key enzyme CHALCONE SYNTHASE were upregulated 14-, seven-, and fourfold, respectively, a finding that is consistent with higher anthocyanin accumulation in pp7l mutants (see Fig. 1B). Although photosynthetic activity is reduced in pp7l mutants (see Figs. 1A and 3A), no enrichment for genes related to photosynthesis or the chloroplast could be identified among those downregulated in pp7l-1 mutant seedlings with the gene ontology (GO) analysis tool DAVID (Huang et al., 2009; Fig. 4A). Instead, under the categories “cellular component,” “biological process,” and “molecular function,” DAVID detected an enrichment of genes encoding proteins for the “RNA polymerase II transcription factor complex,” “response to sucrose starvation,” and “glutaredoxin activity,” respectively (Fig. 4A). These groups encompass genes encoding bHLH transcription factors, or extracellular proteins like protease inhibitor/seed storage/lipid transfer family proteins, xyloglucan endotransglucosylase/hydrolase proteins, and peroxidase superfamily proteins.
Figure 4.
GO analysis of genes whose expression is reduced (A) or elevated (B) in pp7l-1 seedlings compared to Col-0 grown for 4 d in LD conditions. GO annotations for the cellular component, biological process, and molecular function categories were extracted from DAVID (Huang et al., 2009). GO terms with a >2-fold change and a Benjamini corrected value of <0.05 are shown. TF, transcription factor.
However, among the elevated transcripts within the “cellular component” category, most component categories were associated with the chloroplast, in particular “nucleoid,” “plastid chromosome,” “chloroplast stroma,” “envelope,” and “thylakoid” (Fig. 4B). In the category “biological process,” the GOs “chloroplast rRNA processing” and “chlorophyll biosynthesis” were the most highly enriched (Fig. 4B). In particular, the rise in transcripts for proteins involved in chloroplast RNA metabolism has already been noted in mutants and conditions that impair protein synthesis in the chloroplast (Leister and Kleine, 2016).
Investigation of plastid-encoded transcripts in the lncRNA-Seq data showed that transcripts of only five genes were reduced by more than twofold. These encode subunit 8 of the cytochrome b6 f complex, several PSII proteins (D1, PsbK, PsbI), and the PSI protein PsaJ (Supplemental Table S2). Conversely, expression levels of 44 plastid genes rose by more than twofold. Among these are genes encoding tRNAs, ribosomal proteins, subunits of RNA polymerase and ATP synthase, and the PSII proteins CP47, D2, and PsbT.
Taken together, these transcriptome changes point to a role for PP7L in posttranscriptional processes in the chloroplast.
Lack of PP7L Impairs Chloroplast Translation and Development
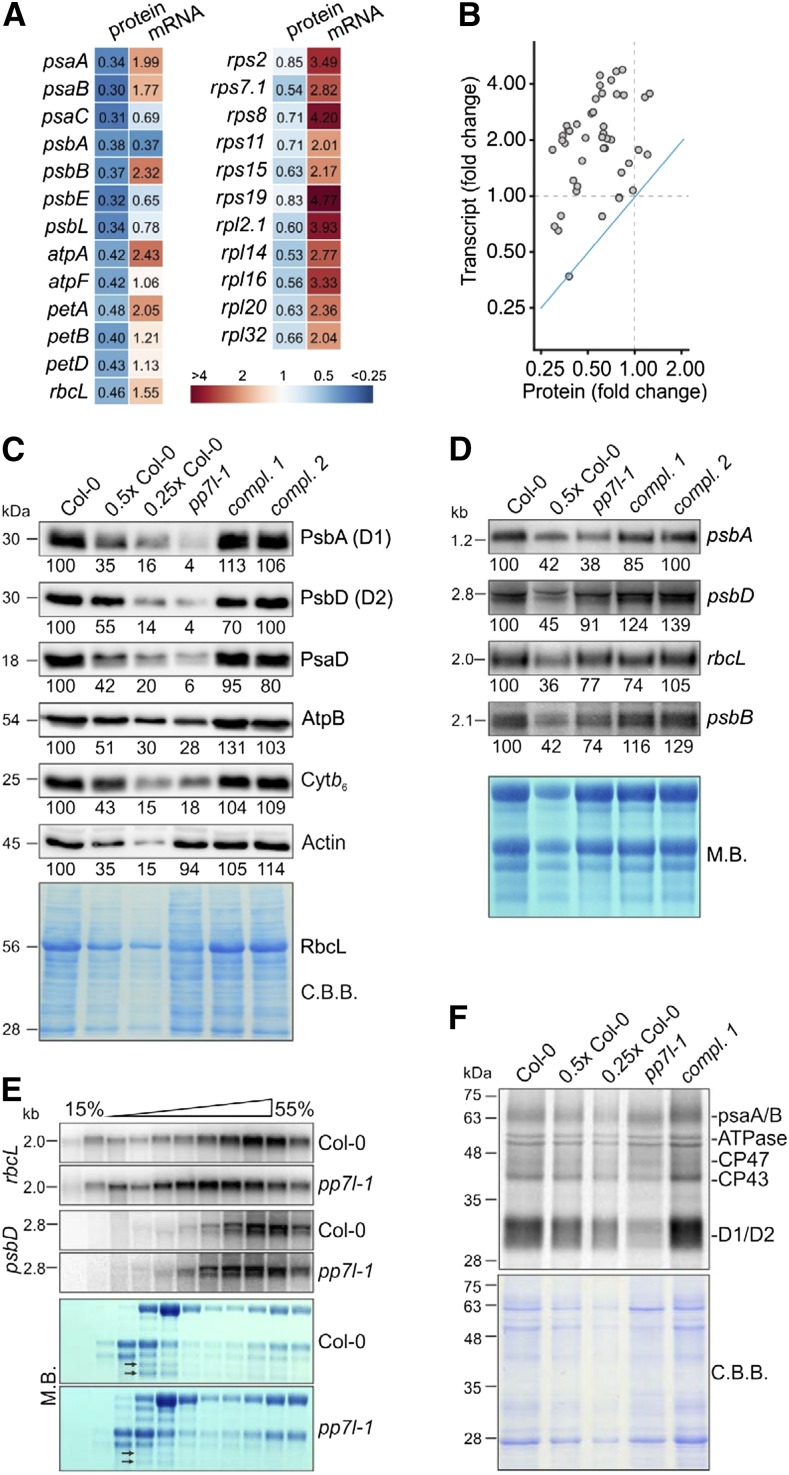
To investigate whether the defect in photosynthetic activity found in the pp7l mutants was a consequence of reduced accumulation of photosynthetic proteins, shot-gun proteomics was performed on total protein extracts from 4-d–old wild-type and pp7l-1 seedlings grown under LD conditions. In the pp7l-1 mutant, chloroplast-encoded subunits of PSI and PSII, the chloroplast ATP synthase, the cytochrome b6 f complex, the large subunit of Rubisco, and also ribosomal proteins accumulated to lower levels than that in wild type (Fig. 5, A and B; Supplemental Table S3). Western-blot analyses confirmed this for representative subunits of the different photosynthesis complexes (Fig. 5C), and also confirmed that protein levels were restored in oe-PP7L pp7l-1 and -2 lines. In contrast, lncRNA-Seq indicated that most of the corresponding transcripts—except psbA—accumulated to higher levels than those seen for the protein levels in the pp7l-1 mutant (Fig. 5, A and B; Supplemental Table S2), which was confirmed by RNA gel-blot analysis for representative instances (Fig. 5D). A scatter plot demonstrates that the relative changes in protein amounts are negatively correlated with those in the levels of the corresponding mRNAs (Fig. 5B). For example, psbD mRNA was nearly at wild-type levels (protein: ∼10% of wild type) and rbcL transcripts were only slightly reduced if at all (RNA-Seq data: 100% wild type; RNA gel blot: 75% wild type) but amounts of the corresponding proteins were drastically reduced (∼15% of wild type; Fig. 5, C and D). Thus, the reduced amounts of chloroplast-encoded proteins in the pp7l-1 mutant cannot be explained by a relative dearth of their transcripts. Closer inspection of Figs. 3D and 5D revealed, however, that amounts of chloroplast rRNAs were abnormally low in pp7l-1 seedlings, as indicated by methylene-blue staining of membranes. In the chloroplasts of land plants, the rRNAs are encoded in the rRNA operon, which contains the genes for tRNAs for Ile, Ala, and Arg, and for 16S, 23S, 4.5S, and 5S rRNAs (Supplemental Fig. S3A). This gene cluster is transcribed as a single molecule and processed via endonucleolytic cleavage and exonucleolytic trimming events (Strittmatter and Kössel, 1984). Indeed, RNA gel-blot analyses showed that in all pp7l alleles, levels of mature 23S rRNA were lower than in the wild type, whereas unprocessed precursors overaccumulated and 16S rRNA was unaffected (Supplemental Fig. S3B). This, together with the fact that amounts of chloroplast ribosomal proteins were reduced in pp7l-1 (Fig. 5A), suggests that pp7l mutants might experience a general reduction in chloroplast translation capacity—an inference that is compatible with the overall analysis of lncRNA-Seq data. To assess this possibility, the effects of the pp7l-1 mutation on the translational efficiency of chloroplast transcripts were further examined by analyzing the association of psbD and rbcL mRNAs with polysomes. Seedling extracts were fractionated in Suc gradients under conditions that preserve polysome integrity, and mRNAs were identified by hybridization with specific probes. In general, efficiently translated RNAs migrate deep into the gradient because almost all are associated with ribosomes. Methylene blue staining of rRNAs in wild type and pp7l-1 polysome gradients showed similar, though not identical distributions, indicating that there was no general difference in ribosome distribution between mutant and wild-type plants. However, psbD and rbcL mRNA were shifted toward nonpolysomal fractions in pp7l-1 as compared to the wild type (Fig. 5E), indicating that these mRNAs were less likely to be associated with polysomes in the pp7l-1 mutant and that at least some of the mRNAs in pp7l-1 chloroplasts are less engaged in translation.
Figure 5.
Accumulation of chloroplast-encoded proteins is perturbed at steps downstream of chloroplast transcription in pp7l-1. A, Heat-map showing relative protein (determined by shot-gun proteomics) and transcript levels (determined by lncRNA-Seq) in 4-d–old pp7l-1 seedlings compared to Col-0 grown in LD conditions. Only a selection of all data are shown. B, Scatter plot showing the anticorrelation between changes in the levels of mRNAs and their protein products. The blue line indicates the theoretical 1:1 correlation. C, Immunoblot analysis of representative thylakoid proteins. Total protein extracts from 4-d–old wild-type (Col-0), pp7l-1, and oe-PP7L pp7l complemented (compl.) seedlings were fractionated by SDS-PAGE, and blots were probed with antibodies raised against individual subunits of photosynthetic complexes. Decreasing levels of wild-type proteins were loaded in the lanes marked Col-0, 0.5× Col-0, and 0.25× Col-0 (1× Col = 5 μg). Actin and the Coomassie Brilliant Blue (C.B.B.)-stained membrane served as loading controls. Please note that RbcL protein levels are decreased in pp7l-1 seedlings. Thus they are also reduced on the C.B.B.-stained membrane. D, Steady-state levels of transcripts of photosynthetic genes. Total RNA was isolated from 4-d–old Col-0, pp7l-1, and oe-PP7L pp7l (compl.) seedlings, and aliquots (7 and 3.5 μg from Col-0; 7 µg from all other lines) were fractionated in a formaldehyde-containing denaturing gel, transferred onto a nylon membrane, and probed with (α-32P)dCTP-labeled cDNA fragments specific for transcripts encoding individual subunits of PSII (psbA, psbB, psbD) and rbcL. rRNA was visualized by staining the membrane with methylene blue (M.B.) and served as a loading control. E, Chloroplast mRNAs accumulate in nonpolysomal fractions in pp7l-1 plants. Whole-cell extracts from Col-0 and pp7l-1 plants were fractionated in linear 0.44–1.6 m (15% to 55%) Suc gradients by ultracentrifugation. Gradients were divided into 11 fractions, and RNA was isolated from equal volumes. RNA blots were hybridized with [α-32P]dCTP-labeled cDNA fragments specific for psbD and rbcL. Ribosomal RNAs were stained with M.B. Arrows indicate the difference in 23S rRNA accumulation. F, In vivo pulse-labeling of thylakoid membrane proteins with [35S]Met in the presence of cycloheximide indicates that translation occurs at reduced rates in pp7l-1 chloroplasts. Proteins were resolved by SDS-PAGE after pulse-labeling for 30 min, and visualized by autoradiography. The C.B.B.-stained membrane served as a loading control.
The effects of the pp7l-1 mutation on the translation of chloroplast transcripts were further investigated by in vivo labeling, which allows one to quantify de novo protein synthesis. To this end, synthesis of plastid-encoded thylakoid membrane proteins was monitored by pulse labeling of wild-type, pp7l-1 mutant, and oe-PP7L pp7l-1 seedlings in the presence of cycloheximide, which inhibits the translation of nuclear-encoded proteins. After pulse labeling for 30 min, de novo synthesis of the D1 and D2 proteins was found to be strongly reduced in pp7l-1 relative to the wild type (Fig. 5F)—although psbD (encoding the D2 protein) transcript levels were notably high (Fig. 5D; Supplemental Table S2).
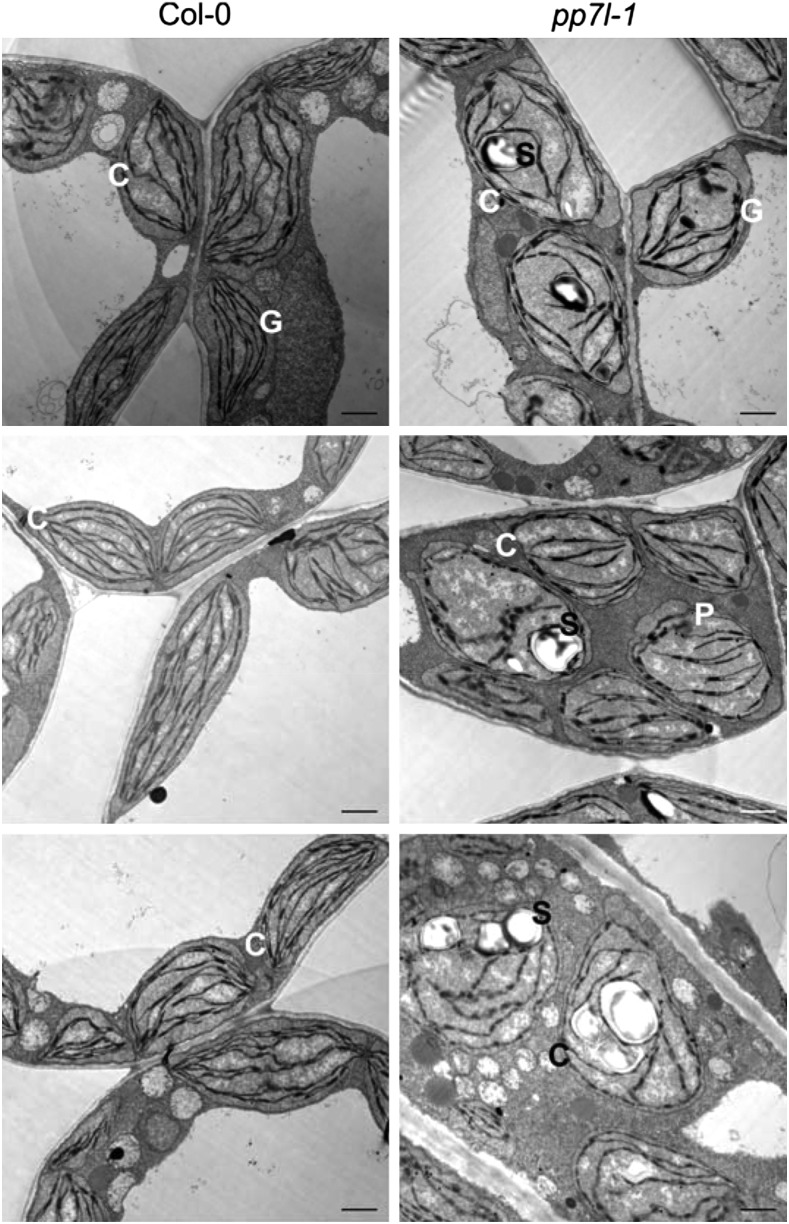
To further investigate the role of PP7L in chloroplast development, the ultrastructure of pp7l chloroplasts was investigated. Cotyledons from 4-d–old wild-type and pp7l-1 seedlings were fixed, embedded, sectioned, and examined for the morphology of chloroplasts by transmission electron microscopy (TEM). Thin sections of wild-type cotyledons revealed normally developed chloroplasts with their internal structures intact. Because the plant material was directly fixed after 8 h in the dark, wild-type chloroplasts did not show any starch granules and, moreover, the stroma and grana thylakoids were well structured (Fig. 6). The chloroplasts of the pp7l-1 mutant contained large starch granules and an elevated number of plastoglobuli. Moreover, the thylakoids in these chloroplasts were less well ordered, indicating a delay in chloroplast development in the pp7l-1 mutant.
Figure 6.
Chloroplast development is disturbed in the pp7l-1 mutant. TEM images of ultrathin sections of 4-d–old wild-type and pp7l-1 cotyledons. The wild type shows chloroplasts with a well-developed thylakoid system and grana stacks. Chloroplasts in the pp7l-1 mutant contain large starch grains, although the seedlings were stored in darkness before the cotyledons were thin-sectioned. Moreover, pp7l-1 chloroplasts contain more plastoglobuli and the thylakoid system is less developed. Bar = 1 μm. C, chloroplast; G, grana; P, plastoglobule; S, starch grain.
Together, these findings suggest that chloroplast transcription is not primarily affected in pp7l, but hint at a disturbance of subsequent processes in chloroplast gene expression that would account for the reduction in synthesis of chloroplast proteins and the delay in chloroplast development in the pp7l mutants.
Photosynthesis Is Not Affected in Photomorphogenesis and Light Signaling Mutants
In addition to driving photosynthesis, light is a major factor in controlling the expression of photosynthesis genes (Berry et al., 2013). The most prominent light-sensing proteins that mediate light-responsive gene expression are phys and cry1 and cry2. Crys mediate blue-light induction of nuclear genes that encode chloroplast photosynthesis proteins or components of the chloroplast transcriptional apparatus (reviewed in Yu et al., 2010). Diverse developmental and physiological processes are controlled by phyA and phyB, including Chl synthesis (McCormac and Terry, 2002). In fact, 4-d–old pp7l seedlings contain less Chl than do wild-type seedlings (Fig. 7A). Moreover, photoactivated crys (Yu et al., 2010) and phys (Li et al., 2011) repress the COP1-dependent degradation of transcription factors that control the expression of light-responsive genes, and Chl content is reduced in cop1-4 seedlings just as in pp7l-1 seedlings (Fig. 7A). Fv/Fm measurements are a simple read-out for PSII function. However, to our knowledge, these measurements have not been extensively performed on cry, phy, and cop1 mutants grown under normal light conditions, and Fv/Fm values can only be extracted from the controls used in stress studies of adult plants (Kleine et al., 2007; Rusaczonek et al., 2015). Thus, we determined Fv/Fm values in 4-week–old wild-type, cop1-4, cry1-304 cry2-1, phyA-211 phyB-9, and—as a control—pp7l-1 mutant plants grown under LD conditions. As expected, Fv/Fm was reduced in the younger leaves of pp7l-1 plants, but none of the other mutants showed any decrease in the maximum quantum yield of PSII (Supplemental Fig. S4).
Figure 7.
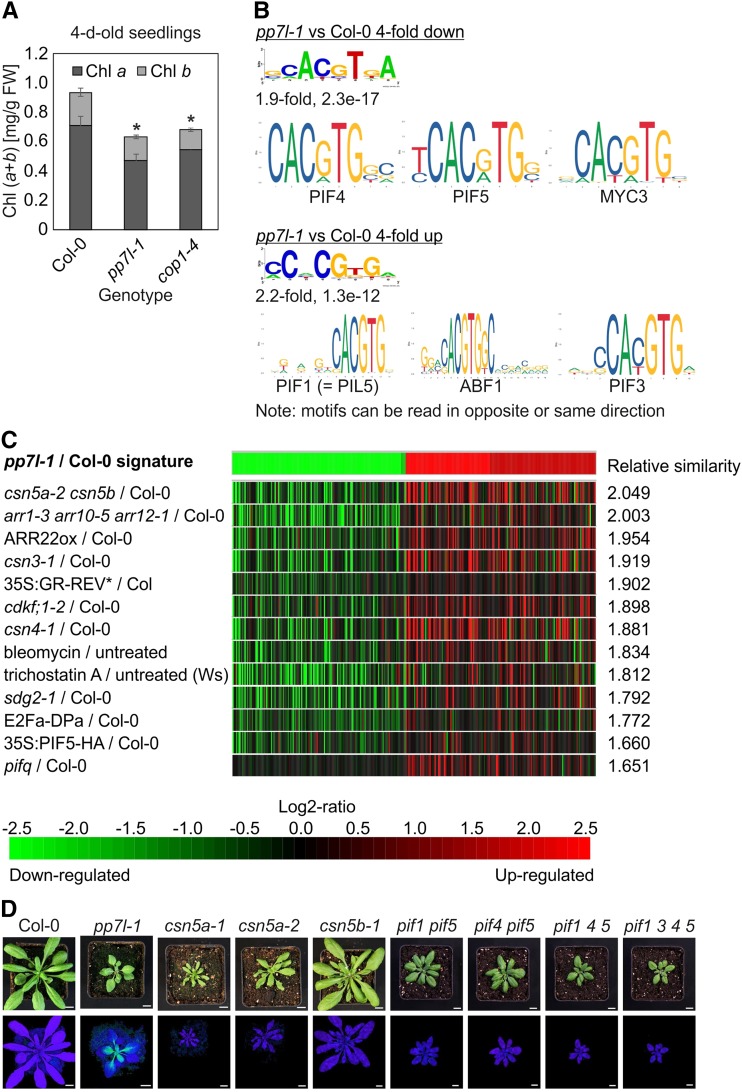
Chl content of 4-d–old pp7l-1 and cop1-4 seedlings, and identification of cis-elements in pp7l-1 deregulated genes and conditions that have a similar effect on the transcriptome to those seen in the pp7l-1 mutant. A, Determination of total Chl (Chl a + b) content. Note that Chl is significantly reduced in 4-d–old pp7l-1 seedlings, in contrast to its essentially wild-type content in 4-week–old pp7l-1 plants (see Supplemental Fig. S2B). Pigments were acetone-extracted, measured spectrophotometrically, and concentrations were determined as described in Jeffrey and Humphrey (1975). Data are shown as mean values ± sd from three different plant pools. Each pool contained >20 plants. The significant difference (Tukey’s test; P < 0.05) between the mutants and Col-0 was identified by Tukey’s test, and is denoted by an asterisk. B, Sequence logos of the most significantly identified cis-elements of genes whose expression was reduced or elevated in the pp7l-1 mutant compared to Col-0. The names of the putative transcription factors binding to the identified cis-elements, together with their respective logos, are also shown. C, Identification of perturbations that induce similar changes in gene expression to those identified in the pp7l-1 mutant. The gene expression changes of the 200 most strongly up- or downregulated genes in 4-d–old pp7l-1 seedlings were submitted to the Genevestigator Signature Tool (Hruz et al., 2008). Mutants or conditions that triggered the most similar gene expression profiles were identified by Pearson distance clustering. Note that the higher the relative similarity parameter, the higher the similarity to the pp7l-1 signature. D, Phenotypes of 4-week–old Col-0, pp7l-1, and the various csn and pif mutant plants. In the bottom row, Imaging PAM pictures depict the maximum quantum yields of PSII (Fv/Fm). Scale bars = 1 cm.
Promoters of Genes Deregulated in pp7l-1 Contain PIF-Related Cis-Acting Elements
To study the transcriptional networks activated by lack of PP7L, the lncRNA-Seq data were investigated in more detail. Cis-acting elements that are overrepresented in promoters of differentially regulated genes in the pp7l-1 mutant were identified by scanning 1,000-basepair (bp) promoter sequences for 8-bp cis-elements with the expectation-maximization–based program Amadeus (Linhart et al., 2008). Because a very large set of genes is differentially regulated in the pp7l-1 mutant, we focused on genes with more than 4-fold up- or downregulation (363 down; 188 up). The putative cis-elements thus identified were used to search the JASPAR plant database (http://jaspar.genereg.net/; Khan et al., 2018) for Arabidopsis transcription factors known to bind to similar sequences. The highest similarity between queried motifs is defined by the lowest E-value. The motifs with the lowest E-value found by JASPAR for both the promoters of up- and downregulated gene sets contained the G-box motif CACGTG (Fig. 7B). The G-box motif is highly conserved and is bound by bHLH and bZIP transcription factors in organisms ranging from yeasts to humans. In plants, these two transcription factor families have expanded massively; e.g. in Arabidopsis, >100 bHLH transcription factors have been described (Carretero-Paulet et al., 2010). Sequences flanking the G-boxes define the specificity of protein binding (Williams et al., 1992) and enable one to predict the in vitro binding of bZIP homodimers (Ezer et al., 2017). In addition, a number of studies have experimentally determined the binding motifs of several transcription factors (Becker et al., 2017). In fact, among the downregulated gene set, binding sites for the phytochrome-interacting factors PIF4 and PIF5, as well as the bHLH transcription factor MYC3, were identified by JASPAR, whereas interaction sites for PIF1, PIF3, and the abscisic-acid response element binding factor 1 were found in the upregulated gene set (Fig. 7B).
To obtain more information on the putative function of PP7L, the list of genes that are differentially expressed in the pp7l-1 mutant was submitted to the Genevestigator database (Hruz et al., 2008; https://www.genevestigator.com/gv/) to identify mutants and conditions associated with similar transcriptome profiles. The most similar transcriptomic responses were found in the csn5a-2 csn5b mutant (Dohmann et al., 2008), in which the function of the CSN multiprotein complex is compromised, and the pif1 pif3 pif4 pif5 quadruple mutant (Leivar et al., 2008; Fig. 7C). These results prompted us to test whether the Fv/Fm was reduced in the emerging leaves of 4-week–old csn5a-1, csn5a-2, csn5b-1, pif1 pif5, pif4 pif5, pif1 pif4 pif5, and pif1 pif3 pif4 pif5 quadruple mutants, as it is in the pp7l mutants. The csn5a csn5b double mutant was not included, because it ceases growing at earlier stages of development (Dohmann et al., 2005). However, although all mutants—with the exception of the csn5b-1 mutant—displayed a growth phenotype similar to that of pp7l-1, none of them exhibited any change (relative to that in the wild type) in the Fv/Fm value (Fig. 7D).
phyB mRNA and Protein Levels Are Elevated in pp7l-1
Inspection of the lncRNA-Seq data revealed that transcripts coding for several phy/PIF signaling-associated proteins like pTAC12 (Chen et al., 2010), GOLDEN2-LIKE1 (Martín et al., 2016), and NUCLEOSIDE DIPHOSPHATE KINASE2 (Shen et al., 2005; Supplemental Table S2) accumulate to higher levels in pp7l-1 mutants. Moreover, PHYB mRNA levels were twofold-increased (Supplemental Table S2), a finding that was confirmed for all three pp7l alleles by RT-qPCR (Supplemental Fig. S5A). To test whether the elevated PHYB mRNA levels were reflected in corresponding changes in phyB protein levels, immunoblot analysis using antiphyB antibodies was performed on total protein extracts isolated from 4-d–old seedlings grown under LD conditions. Indeed, in three independent experiments, the phyB protein level was found to be ∼2.5-fold elevated in the pp7l-1 mutant, but was similar to that in wild type in oe-PP7L pp7l-1 (Supplemental Fig. S5B). Therefore, we asked whether the pp7l Fv/Fm phenotype might be a consequence of the overexpression of phyB. For this purpose, a line was used in which phyB was overexpressed under the control of the 35S promoter in the No-0 background (designated “ABO”; Wagner et al., 1991). However, the maximum quantum yield of PSII was not reduced in 3- and 4-d–old ABO seedlings (Supplemental Fig. S5C).
We therefore conclude that the reduced photosynthetic capacity of the pp7l mutants is not caused (solely) by their abnormally high levels of phyB.
PP7L Is Involved in the Tolerance to Salt and High-Light Stress
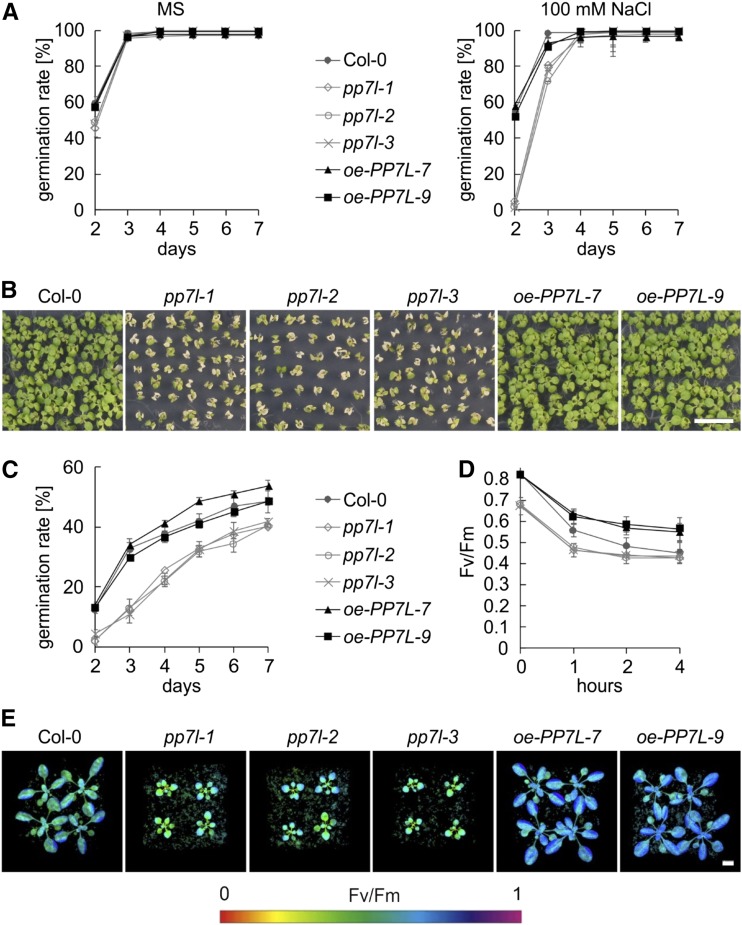
Perturbations of organellar gene expression frequently result in the alteration of acclimation and tolerance responses to adverse growth conditions (Leister et al., 2017), which include the tolerance to salt (Lee et al., 2014) and high-light stress (Coll et al., 2009). To investigate whether PP7L might play a role in the response to the aforementioned stresses, seeds of wild type, pp7l mutants, and PP7L overexpression lines (oe-PP7L; Supplemental Fig. S6) were germinated on Murashige and Skoog (MS) medium supplemented with 100 mm NaCl and on MS medium without supplement as control (Fig. 8A). Additionally, seeds were germinated under high light (1,000 μmol photons m−2 s−1; Fig. 8C), and germination rates were followed from d 2 to d 7. After 2 d on control MS medium under growth light, 60% of wild-type and oe-PP7L line seeds germinated (Fig. 8A). Germination rates of pp7l mutants (48%) were slightly delayed, but after 3 d all lines displayed an ∼100% germination rate. The delay in germination of pp7l mutants was pronounced when seeds were challenged with 100 mm NaCl: Only 2% to 5% of pp7l mutants germinated after 2 d, whereas oe-PP7L lines and wild type displayed ∼55% germination (Fig. 8A). Although after 4 d all lines displayed nearly 100% germination, the enhanced sensitivity of the pp7l mutants to NaCl was manifested after 2 weeks; the growth of pp7l seedlings was substantially delayed and cotyledons appeared bleached (Fig. 8B).
Figure 8.
PP7L affects salt and high-light tolerance. A, Seed germination of wild type (Col-0), pp7l mutants and oe-PP7L lines on MS medium without NaCl (“MS”), and on MS supplemented with 100-mm NaCl. The data represent mean values ± sd of three independent experiments, each performed with 100 seeds per treatment and genotype. B, Phenotypes of wild type (Col-0), pp7l mutants, and oe-PP7L lines grown for 2 weeks on MS supplemented with 100 mm NaCl. Scale bar = 1 cm. C, Germination of wild-type (Col-0), pp7l mutants, and oe-PP7L line seeds grown under high light (1,000 µmol m−2 s−1). The data represent mean values ± sd of three independent experiments, each performed with 100 seeds per treatment and genotype. D, Graph displaying Fv/Fm values of 4-week–old (Col-0), pp7l mutants, and oe-PP7L lines treated for the indicated time with high light (1,000 µmol m−2 s−1). The data represent mean values ± sd of three independent experiments. E, Imaging PAM pictures showing the maximum quantum yields of PSII (Fv/Fm) of 4-week–old plants subjected to 1 h of high light (1,000 µmol m−2 s−1). Scale bar = 1 cm.
When seeds were germinated under high light, the germination rates of pp7l mutants were lower than those of the wild-type and the oe-PP7L lines during the whole time course (Fig. 8C). The oe-PP7L-7 line germinated even slightly faster than the wild type. To investigate the high-light phenotype in adult plants, 4-week–old plants were shifted to high light and the maximum quantum yield of PSII was measured after 1, 2, and 4 h (Fig. 8, D and E). In the wild type, high light caused a drop of the Fv/Fm value from 0.81 to 0.56, 0.48, and 0.45 after 1, 2, and 4 h of treatment, respectively. The pp7l mutant displayed photoinhibition that was slightly enhanced compared to that in the wild type. However, at 0.67, the Fv/Fm value of the pp7l mutants was already significantly lower in normal growth conditions (Fig. 8, D and E). Importantly, overexpression of PP7L rendered plants more high-light–tolerant with higher Fv/Fm values under high-light treatment compared to that of the wild type.
These results show that PP7L plays a positive role in seed germination especially under adverse growth conditions and that overexpression of PP7L renders plants more high-light–tolerant.
PP7L Evolved 43–92 Million Years Ago in the Lineage Leading to Brassicales
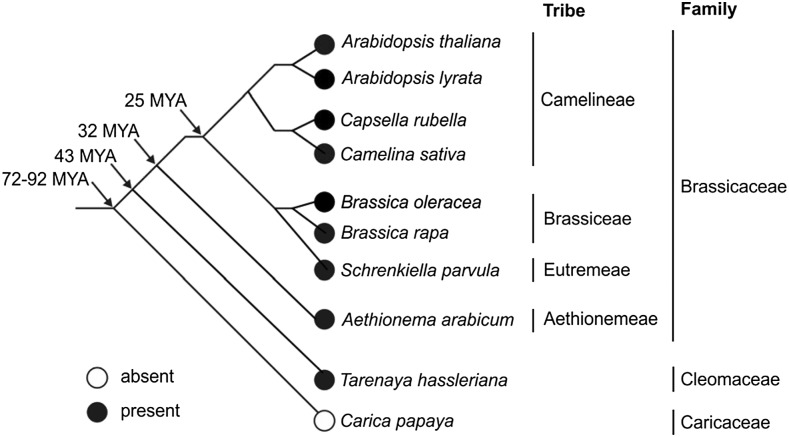
The type-7 subfamily of PPPs is specific to plants (Farkas et al., 2007; Uhrig et al., 2013). The increasing number of plant genomes sequenced in recent years (see for example: http://plants.ensembl.org) allowed us to refine the analysis of the evolution of the type-7 subfamily. To this end, type-7 members were identified by BLAST searches in the PLAZA database (https://bioinformatics.psb.ugent.be/plaza/; Van Bel et al., 2018) and PP7 was found to be ubiquitously present in the plant families included in the database. PP7L and long PP7, however, were only present in Arabidopsis lyrata, Arabidopsis, Brassica oleracea, Brassica rapa, Capsella rubella, Schrenkiella parvula, and Tarenaya hassleriana (Fig. 9). These plant species are members of the Brassicaceae or Cleomaceae (T. hassleriana) families, which are grouped in the order of Brassicales. To expand this analysis to more Brassicales species and to species beyond the Brassicales, the NCBI Blastp suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) was used to identify further sequences homologous to the PP7L protein sequence. In fact, PP7L was also detected in Camelina sativa and Aethionema arabicum, both of which belong to the Brassicaceae, but not in Carica papaya, which belongs to the Caricaceae family. The Brassicaceae and Caricaceae diverged from a common ancestor ∼72–92 million years ago (MYA; Wikström et al., 2001; Edger et al., 2018). In light of the absence of PP7L in C. papaya, and its presence in T. hassleriana, it can be concluded that PP7L evolved after the divergence of Caricaceae from the other Brassicales families, but before the separation of Cleomaceae from Brassicaceae, i.e. between 43 and 92 MYA. Therefore, regulation of chloroplast and plant development in Brassicales differs, at least with respect to the elements that are dependent on PP7L, from that in species descended from other plant orders.
Figure 9.
Presence of PP7L in Brassicales. The phylogenetic tree for a part of the Brassicales species is based on the information from Wikström et al. (2001) and Edger et al. (2018). Arrows indicate estimated dates of divergence (in MYA) at different nodes; open circle, PP7L absent; black circle, PP7L present.
DISCUSSION
Lack of PP7L leads to pleiotropic phenotypic effects, which include a small rosette size, increased anthocyanin levels (Fig. 1), susceptibility to salt and high-light stress (Fig. 8) and perturbed chloroplast development (Fig. 6), accompanied by a general reduction in the expression of proteins synthesized in the chloroplast (Fig. 5). Using the maximum quantum yield of PSII (Fv/Fm) as a proxy for chloroplast development, we have shown that chloroplast development is disturbed in cotyledons (Fig. 3), as well as in emerging true leaves (Fig. 1) in pp7l mutants, a finding that was further corroborated by ultrastructural analysis of cotyledons of 4-d–old seedlings (Fig. 6).
PP7L Acts as a Positive Regulator of Chloroplast Development in Seedlings and in Emerging True Leaves
The different cytological origins of chloroplasts in cotyledons and leaves (Waters and Langdale, 2009; Jarvis and López-Juez, 2013; Van Dingenen et al., 2016), together with differences between the programs of gene expression triggered by light in the cotyledons and the shoot apex (López-Juez et al., 2008), suggest that chloroplasts in cotyledons develop through mechanisms that are at least partly distinct from those in leaves. Moreover, various mutants have been isolated in which the greening of cotyledons and leaves are differentially affected (Waters and Langdale, 2009). Thus, chloroplast development mutants have been described that exhibit variegated phenotypes in true leaves, but develop normal green cotyledons. Among these, the best-characterized chloroplast biogenesis mutants are those designated as immutans mutants and variegated (var; Putarjunan et al., 2013). The genes IMMUTANS and VAR code for the plastid terminal oxidase and the AtFtsH2 subunit of the thylakoid FtsH metalloprotease complex, respectively. The FtsH complex is mainly involved in turnover of the PSII reaction-center D1 protein. Conversely, the three snowy cotyledon (sco) mutants, namely sco1 (Albrecht et al., 2006), sco2/cyo1 (Shimada et al., 2007; Albrecht et al., 2008), and sco3 (Albrecht et al., 2010), have white or pale green cotyledons but green leaves. The SCO class appears not to have a common specific function that causes the cotyledon-specific mutant phenotypes, because its members include proteins involved in different processes, such as plastid protein translation (SCO1; Albrecht et al., 2006), folding of Cys-rich thylakoid proteins (SCO2/CYO1; Shimada et al., 2007), or association with microtubules and the peroxisome (SCO3; Albrecht et al., 2010). Seeds of many oilseed plants like Arabidopsis are green due to the presence of photosynthetically active chloroplasts during embryogenesis, which dedifferentiate into nonphotosynthetic eoplasts during the desiccation phase (Liebers et al., 2017). Immature green sco embryos dissected from siliques are green (Ruppel and Hangarter, 2007; Albrecht et al., 2008, 2010), and precocious germination of sco2 and sco3 mutants rescues their sco cotyledon phenotype (Albrecht et al., 2008, 2010). Thus, it was suggested that the defects are specific to the development of chloroplasts from proplastids/eoplasts in cotyledons, and SCO proteins were assumed to be cotyledon-specific factors (Waters and Langdale, 2009). However, under short-day conditions, smaller and pale-green true leaves can be observed in sco2 mutants, and disruption of SCO2 in Lotus japonicus results not only in paler cotyledons but also in variegated true leaves (Zagari et al., 2017).
Two of the six nuclear-encoded SIGs that interact with PEP have also been proposed to have a specific function in the development of cotyledon chloroplasts (Privat et al., 2003; Ishizaki et al., 2005). Although the cotyledons of SIG2 antisense plants are Chl-deficient, the true leaves are green (Privat et al., 2003); similarly, sig6-1 null mutants exhibit delayed greening of cotyledons, but the true leaves were described as normal (Ishizaki et al., 2005). Under our growth conditions, the true leaves of sig6-1 also appeared normal in color, but a more detailed analysis of photosynthesis by Imaging pulse amplitude modulation (PAM) revealed that chloroplast development was perturbed (Fig. 3C). In consequence, some proteins that were previously classified as being specific for chloroplast development in cotyledons or true leaves are less specific than was once thought. More sensitive phenotyping methods like Imaging PAM analysis (this study) or the application of diverse growth conditions (as in the case of sco2; Zagari et al., 2017) will be instrumental in the future analysis of mutants that appear to be specifically affected at certain developmental stages.
The Role of PP7L in Chloroplast Development Is Not Mediated by the Principal Photomorphogenesis Pathways
It has been noted before that light and chloroplast signaling pathways converge at some point (Ruckle et al., 2007; Leister et al., 2014; Martín et al., 2016). In agreement with this, we have identified PIF-related cis-elements in the promoters of pp7l-deregulated genes (Fig. 7B), and the pp7l gene expression signature resembles the changes in gene expression induced by csn and pif mutations (Fig. 7C). Moreover, in pp7l, transcripts coding for several phy/PIF signaling-associated proteins (Supplemental Table S2) accumulate to a greater extent than that in wild type, and phyB expression is itself elevated at the transcript (Supplemental Fig. S5A; Supplemental Table S2) and protein level (Supplemental Fig. S5B). In seedlings, chloroplasts develop from etioplasts that develop from proplastids/eoplasts in darkness (Pribil et al., 2014). Upon exposure to light, photoactivated phys inhibit PIFs and the proteasome, thus inducing transcriptional reprogramming, which in turn promotes photomorphogenesis, including greening (Xu et al., 2015). Consequently, Chl content is reduced in phyB and phyA phyB seedlings (Neff and Chory, 1998), and Chl content is also reduced in cop1-4 seedlings and pp7l-1 seedlings (Fig. 7A). Therefore, we tested the possibility that PP7L acts through the phy/PIF or proteasome pathway. However, none of the tested mutants lacking the main PIFs, photoreceptors, or the COP and CSN proteins associated with photomorphogenesis show any change in Fv/Fm (Supplemental Fig. S4). In addition, overexpression of phyB does not result in a reduction of photosynthesis (Supplemental Fig. S5). This indicates that (1) PP7L operates through a different pathway to regulate chloroplast development, and (2) that greening and photosystem biogenesis are controlled by separate systems.
Photosynthetic activity in pp7l mutants during seedling development is most strongly affected in young seedlings (Fig. 3A; Supplemental Table S1), recovers gradually during subsequent seedling development, and is essentially normal after 10 d (Fig. 3B). Recently, the linear developmental program of thylakoid biogenesis in cotyledons was documented by TEM, RNA-Seq, and photosynthesis protein level analysis (Liang et al., 2018). When seeds are germinated under continuous light, thylakoid biogenesis begins between 24 and 36 h after imbibition (HAI), and PSII complexes are first inserted into the thylakoid membrane at 36 HAI, when polysomes attach to the flattening thylakoids (Liang et al., 2018). Cotranslational insertion of PSII by thylakoid-membrane-bound polysomes further enriches PSII-light-harvesting complex II at 60 HAI (2.5 d; Liang et al., 2018). Protein subunits of the PSI complex are detected only after 84 HAI, suggesting that PSI assembly is initiated at ∼84 HAI, corresponding to 3.5 d and thus matching our results in which full photosynthetic activity is reached between days 3 and 4. Importantly, the results of Liang et al. (2018) suggest that thylakoid-bound polysomes mediate critical stages of chloroplast biogenesis. Indeed, this is supported by the fact that a disturbance in chloroplast ribosome assembly in the rh3-4 mutant, in which the DEAD-box RNA helicase RH3 is defective (Asakura et al., 2012), causes a delay in thylakoid development (Liang et al., 2018). In pp7l, fewer mature chloroplast 23S rRNA molecules (Figs. 3D and 5D; Supplemental Fig. S3) and smaller amounts of ribosomal proteins accumulate (Fig. 5A; Supplemental Table S3). Thus, we hypothesize that the delay in thylakoid development in the pp7l mutant (Fig. 6) is caused by a perturbation of rRNA maturation and accumulation of chloroplast ribosomal proteins, which results in a reduction in the numbers of functional ribosomes and a decrease in protein synthesis in the chloroplast (Fig. 5, E and F).
PP7L Contributes to the Tolerance to Abiotic Stresses
Plants as sessile organisms are constantly exposed to environmental changes and stresses, and they consequently evolved mechanisms to cope with different stresses. Because (1) pp7l mutants are more susceptible to salt and high-light treatment and (2) overexpression of PP7L enhances the capacity of plants to tolerate high light intensities (see Fig. 8), PP7L is clearly involved in salt and high-light tolerance. The reasons for the multiple stress susceptibility of pp7l mutants is not plainly evident, but one plausible explanation is that it can be ascribed at least in part to the perturbations of chloroplast gene expression observed in pp7l mutants (see Fig. 5). In fact, it was already noted before that changes in chloroplast gene expression can trigger alterations in tolerance responses, presumably via retrograde signaling (Leister et al., 2017). Interestingly, an impairment in chloroplast gene expression results not automatically in stress susceptibility, but can render plants even more resistant to stresses. This is exemplified by the “happy on norflurazon” mutants hon5 and hon23 affected in the ClpR4 subunit of the chloroplast-localized Clp protease complex, and a protein with homology to prokaryotic translation initiation factor2, cpIF2, respectively; they display a constitutive stress response that enhances resistance against a combined low-temperature/high-light stress (Saini et al., 2011). Moreover, lack of the major chloroplast FtsH complex component FtsH2 in the var2 mutant leads to a reduced capacity to acclimate to high-light stress (Miura et al., 2007). When a mutation in FUG1, which encodes a chloroplast protein with homology to cpIF2, is introduced into var2, the high-light sensitivity of var2 is suppressed—pointing to an importance of the balance between chloroplast protein synthesis and degradation in stress acclimation (Miura et al., 2007).
How Does the Nucleocytosolic Compartment Influence Chloroplast Development?
Chloroplast proteins are synthesized both in the nucleocytosolic compartment and the organelle, which necessitates coordination of the expression of nuclear and chloroplast genomes. Additionally, gene expression in the chloroplast itself is a complex process that depends on the supply of a plethora of nuclear-encoded proteins. Primary transcripts are produced by PEP and nuclear-encoded phage-type RNA polymerase (Liere et al., 2011). Subsequently, these RNAs undergo processing, splicing, editing, turnover, and maturation (Stern et al., 2010; Stoppel and Meurer, 2012) before they are translated (Tiller and Bock, 2014). Lack of nuclear-encoded proteins at early stages of chloroplast development impacts on phenotypes in various degrees, ranging from no visible phenotype to pale green cotyledons and rosette leaves to seedling- and embryo-lethality (Barkan and Small, 2014; Kleine and Leister, 2015). In screens for mutants with altered chloroplast biology at this stage, most studies have identified chloroplast-localized proteins involved in chloroplast development and showing a photosynthesis phenotype. Exceptions include PALE GREEN1 (Hsieh et al., 2017) and SCO3 (Albrecht et al., 2010), which are localized to mitochondria and peroxisomes, respectively. Interestingly, the cytoskeleton is altered in sco3-1, and microtubule inhibitors have similar effects on chloroplast biogenesis to sco3-1 (Albrecht et al., 2010), suggesting a role for the cytoskeleton and peroxisomes in chloroplast biogenesis. Thus, it is increasingly clear that, in addition to the already known pathways, the development of chloroplasts requires a number of other processes.
The import of photosynthetic preproteins is mediated by transclocon at the outer envelope membrane of chloroplasts (Toc)33 and Toc159 proteins. Accordingly, the plastid protein import1 (ppi1; toc33) and ppi2 (toc159) mutants are defective in chloroplast biogenesis (Bauer et al., 2000; Kubis et al., 2003). Moreover, not only the process of protein uptake by the chloroplast, but preimport steps such as quality control by the cytosolic ubiquitin–proteasome system (Ling et al., 2012) and the establishment of the appropriate balance between chloroplast and cytosolic translation (Wang et al., 2018), affect chloroplast development.
Notably, alterations in chloroplast protein import in the ppi2 and translocon at the inner chloroplast envelope 56-1 mutants result in decreased accumulation of plastid ribosomal proteins (Köhler et al., 2016) as in the pp7l-1 mutant (Fig. 5A; Supplemental Table S3). However, reduced ribosomal protein accumulation is not a feature common to all albino/pale green (apg) mutants, because ribosomal subunits accumulate to wild-type levels in the apg1, apg2, and apg3 mutants, which are defective in proteins that participate in diverse chloroplast functions (Motohashi et al., 2012). Because Toc159 is an import receptor for photosynthesis-related proteins, a straightforward explanation for the ppi2 photosynthesis phenotype is that it is caused by a defect in the import of the photosynthesis proteins. However, the mechanism must be more complex, because in the ppi2 mutant many nuclear-encoded photosynthesis proteins are downregulated at the transcript level, which points to the activation of retrograde signaling (Bischof et al., 2011). This is not the case—at least for the vast majority of mRNAs for photosynthesis proteins—in the pp7l-1 mutant (Supplemental Table S2) nor in the sco2/cyo1 mutant (Shimada et al., 2007). Why mutants with similar molecular chloroplast phenotypes do not trigger similar gene expression profiles in the nucleus remains elusive.
It is interesting to note that, although the mRNA levels of PHYB, SIG2, SIG6, GLK1, and Chl biosynthesis genes are upregulated in pp7l, greening in pp7l is defective (Fig. 7A), questioning whether the elevated cytosolic mRNA levels reflect functional protein levels. Among the elevated nuclear-encoded transcripts, the GO category “chloroplast rRNA processing” is, in addition to “chlorophyll biosynthesis,” the most highly enriched term (Fig. 4). Processing of 23S rRNA is defective in the pp7l mutant (Supplemental Fig. S3), which might be a secondary effect of reduced chloroplast protein translation. Because chloroplast rRNA maturation requires a plethora of nuclear-encoded proteins (Stoppel and Meurer, 2012), this finding could also support the concept of negative correlation of mRNA and protein levels. Indeed, an increasing number of observations indicate that mRNA transcript levels and protein synthesis are often correlated only to a limited degree (e.g. Oelze et al., 2014). Before a protein destined for the chloroplast reaches the quality-control and import stages, it must first undergo a series of complex processes. The expression level of its mRNA is itself determined by the relative rates of transcription and degradation (Roy and von Arnim, 2013), and the decoding of mRNA by the ribosome involves a plethora of initiation, elongation, and termination factors (Moore et al., 2016). Once the protein is finally made, it may also require posttranslational modification or be degraded by the proteasome (Arsova et al., 2018). This shows that many steps from primary transcript to protein translation and modification can account for a negative correlation between mRNA and protein abundance. The remaining key questions are which nucleo(cytosolic) process is exactly disturbed in the pp7l mutant provoking the delayed chloroplast development, and which transcript changes in pp7l are attributable to retrograde signaling activated by the delay in chloroplast development. Of note is the 8-fold-higher anthocyanin content in the pp7l mutant (Fig. 1B). Anthocyanin accumulation is also induced by abiotic stresses like high light (Kleine et al., 2007), cold (Schulz et al., 2015), or specifically by Suc (Solfanelli et al., 2006). Photosynthesis-derived Suc is exported from chloroplasts to other cellular compartments or to nonphotosynthetic sink tissues, and carbon supply can be sensed through sugar signaling (Wingler, 2018). Despite the obvious linkage of photosynthesis and energy metabolism and signaling, little is known about the connections between cytoplasmic and nucleus-localized key regulators of cellular metabolism like the SNF1-related protein kinase1 and organelles (Wurzinger et al., 2018). Moreover, mRNA levels of many photosynthetic genes are dependent on SNF1-related protein kinase1 activity (Zhang et al., 2009). One possible PP7L function would be that PP7L is at the receiver site of sugar signaling in the nucleus. The perturbed starch mobilization (Fig. 6) and the small growth phenotypes of the pp7l mutant (Fig. 1) would be in accordance with sugar-signaling regulators playing a central role in the regulation of starch metabolism (Sakr et al., 2018; Wurzinger et al., 2018). With its predicted function as an inactive Ser/Thr phosphatase, it is unlikely that it is a transcription factor and future studies will have to clarify the molecular function of PP7L.
The Brassicaceae Arabidopsis is a member of the order Brassicales, which is known for the production of mustard oil (glucosinate) compounds (Cardinal-McTeague et al., 2016). This order contains, in addition to several model plants, many species that are of economical relevance, such as mustard (Brassica juncea), broccoli (Brassica oleracea var. italica), cabbage (Brassica oleracea var. capitata), kale (Brassica oleracea var. sabellica), rapeseed (Brassica napus), and caper (Capparis spinosa). Because PP7L evolved specifically in the order Brassicales (Fig. 9) and because of the prominent phenotype of the pp7l mutant, it appears that PP7L is specific for the regulation of chloroplast and plant development in Brassicales. Other plant orders might (1) have recruited a protein different from PP7L or (2) might differ in their chloroplast and plant development and therefore not require PP7L. Thus, further investigation of PP7L should help to elucidate the aspects of chloroplast and plant development specific to this order, which are of potential significance for the improvement of agronomically valuable crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The pp7l:En-1 mutant (ZIGIA line V2-880) was identified in the mutant collection of Wisman et al. (1998) based on alterations in the effective quantum yield of PSII (ΦII). The mutants pp7l-1 (SALK_018295), pp7l-2 (SALK_033071), and pp7l-3 (SALK_022053) were identified in the SIGnAL database (Alonso et al., 2003). The sig2-2 (SALK_045706) mutant allele has been described previously (Kanamaru et al., 2001), as have the mutants sig5-1 (SALK_049021; (Tsunoyama et al., 2004), sig6-1 (SAIL_893_C09; Woodson et al., 2013), cop1-4 (McNellis et al., 1994), cry1-304 cry2-1 (Mockler et al., 1999), csn5a-1, csn5a-2, csn5b-1 (Dohmann et al., 2005), pif1pif5, pif4pif5, pif1pif4pif5, pif1pif3pif4pif5 (Pfeiffer et al., 2014), phyB-9 and phyA-211 phyB-9 (Su et al., 2015), and ABO (Shinomura et al., 1998). All mutants except of ABO are in the Col-0 background.
Arabidopsis (Arabidopsis thaliana) plants were routinely grown on potting soil (Stender) under controlled greenhouse conditions and on a 16-/8-h light/dark cycle (daylight was supplemented with illumination from HQI Powerstar 400W/NDL, providing a total fluence of ∼180 µmol photons m−2 s−1 on leaf surfaces). Wuxal Super fertilizer (8% N, 8% P2O5, and 6% K2O; MANNA) was used according to the manufacturer’s instructions. Where indicated, seedlings were grown on 0.8% (w/v) agar (Sigma-Aldrich) containing 1% (w/v) Suc at 22°C under 100 μmol photons m−2 s−1 provided by white fluorescent lamps.
Complementation of pp7l Mutants, Expression, and Intracellular Localization of eGFP Fusions
To complement the mutant phenotype, the sequence of PP7L (AT5G10900) without the stop codon was amplified from genomic DNA with the primers indicated in Supplemental Table S4. This PCR product was introduced into the donor vector pDONR207 by the BP clonase reaction (Invitrogen) as described in the Gateway manual. After sequence analysis of the recombined DNA sequence, the attL recombination clonase reaction (Invitrogen) was performed to introduce the fusion construct into the destination vector pB7FWG2 (35S promoter, eGFP), generating 35S:PP7L-GFP. Subsequently, the construct was introduced into Col-0, pp7l-1, pp7l-2, and pp7l-3 backgrounds. Transformed seedlings were selected with the herbicide Basta. For transformations into pp7l mutants, plants showing a wild-type phenotype were selected. Then expression of PP7L mRNA levels in different genotypes was analyzed by RT-PCR using an appropriate primer combination (Supplemental Table S4).
Subcellular localization analysis of fused proteins was conducted in protoplasts isolated from wild type. Protoplasts were imaged with a Fluorescence Axio Imager microscope (Zeiss) after transformation as described in Dovzhenko et al. (2003). Fluorescence was excited with the X-Cite Series 120 fluorescence lamp (EXFO) and images were collected at 500–550 nm (eGFP fluorescence) and 670–750 nm (Chl autofluorescence).
Chl a Fluorescence Measurements
Chl fluorescence parameters were measured using an imaging Chl fluorometer (Imaging PAM, M-Series; Walz). The method employed involved preprogrammed treatments of 30-min dark periods to determine Fv/Fm, for which F0 was measured at a low frequency of pulse-modulated measuring light, whereas Fm was obtained at saturation pulses of ∼2,700 µmol photons m−2 s−1, for a duration of 0.8 s. The calculations and plotting of the parameters were performed using the ImagingWin software.
Chl Concentration Measurements
Chl content was measured according to Jeffrey and Humphrey (1975). Briefly, 100 mg of fresh tissue was ground in 1 mL of cold 80% (v/v) acetone and centrifuged at 12,000 g for 5 min at 4°C and the supernatant was saved. Another 1 mL of 80% (v/v) acetone was added, and the extract was centrifuged again. The procedure was repeated until the green color of the pellet was no longer discernible. Chl concentrations were calculated from spectroscopic absorbance measurements at 663, 646, and 710 nm.
Anthocyanin Measurement
Measurement of anthocyanin content was conducted according to Neff and Chory (1998). Briefly, 100-mg samples of seedlings of each genotype that had been exposed to diverse light treatments were incubated with 300 µL of 1% (v/v) HCl in methanol at 4°C overnight in the dark with shaking. Then 200 µL of distilled water and 500 µL of chloroform were added, vortexed, and briefly centrifuged to separate anthocyanins from Chls. The total anthocyanin content was determined by measuring A530 and A657 of the aqueous phase using a spectrophotometer (Amersham Biosciences). The equation (A530−0.25 × A657) × TV/(FW × 1000) was used to quantify the relative amount of anthocyanin, where TV = total volume of the extract (in milliliters) and FW = fresh weight of seedlings (in grams).
Polysome Analysis
Polysome isolation was performed according to Barkan (1993). Seedlings (200 mg) were ground in liquid N using a mortar and pestle. Subsequently, the microsomal membranes were extracted and solubilized with 0.5% (w/v) sodium deoxycholate. The solubilized material was then layered onto 15/55% (w/v) Suc step-gradients (corresponding to 0.44/1.6 M) and centrifuged at 250,000 g for 65 min at 4°C. The step gradient was fractionated, and the mRNA associated with polysomes was extracted with phenol/chloroform/isoamyl alcohol (25:24:1), precipitated at room temperature with 95% (v/v) ethanol, and subjected to RNA gel-blot analysis.
In Vivo Labeling of Thylakoid Proteins
Four-d–old seedlings were preincubated in 1 mL of labeling solution containing 20 mg/mL cycloheximide, 10 mm ris(hydroxymethyl)aminomethane (TRIS), 5 mm MgCl2, 20 mm KCl (pH 6.8), and 0.1% (v/v) TWEEN®20 (a polyethylene glycol sorbitan monolaurate) under illumination at a fluence rate of 80 μmol photons m−2·s−1 for 30 min to block cytosolic translation. Then labeling was performed in the same solution containing 1 mCi mL−1 [35S]Met under the same lighting conditions for 30 min. After labeling, the thylakoid membranes were isolated and the proteins were fractionated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by fluorography (Variable Mode Imager Typhoon TRIO; Amersham Biosciences).
Protein Isolation and Western Blotting
Seedling samples (100 mg) were ground in liquid N and total proteins were solubilized in protein extraction buffer (0.125 m TRIS, 1% SDS, 10% [v/v] glycerol, 0.05 m sodium metabisulfite) containing 1 mm phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma-Aldrich). Cell debris was removed by centrifugation for 10 min and the supernatant was boiled with SDS loading buffer at 95°C for 10 min. Equal amounts of total proteins were fractionated on 10% SDS-PA gels and transferred topolyvinylidene fluoride membranes (Millipore) using a semidry method. The primary antibodies directed against PsbA (1:4,000), PsbD (1:5,000), RbcL (1:10,000), PsaD (1:1,000), AtpB (1:4,000), and cytb6 (1: 10,000) used in this study were obtained from Agrisera. The B6-B3 monoclonal antibody specific for phyB is described in Hirschfeld et al. (1998). Signals were detected with the PierceECL Western Blotting Kit (Thermo Fisher Scientific) and quantified using the software ImageJ (National Institutes of Health).
Nucleic Acid Extraction
Leaf tissue (100 mg) was homogenized in extraction buffer containing 200 mm TRIS/HCl (pH 7.5), 25 mm NaCl, 25 mm ethylenediaminetetraacetic acid, and 0.5% (w/v) SDS. After centrifugation, DNA was precipitated from the supernatant by adding isopropyl alcohol. After washing with 70% (v/v) ethanol, the DNA was dissolved in distilled water.
For RNA isolation, frozen tissue was ground in liquid N. Total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA samples were treated with DNase I (New England BioLabs) and quantified with a Nanodrop spectrophotometer (Thermo Fisher Scientific). Isolated RNA was stored at −80°C until further use.
RNA Gel-Blot Analysis
For northern blotting, total RNA was extracted using Trizol reagent (Invitrogen). Aliquots (7 μg) of each sample were electrophoresed on a 1.5% (w/v) agarose gel containing formaldehyde, and transferred to a nylon membrane (Hybond-N+; Amersham Biosciences). The membrane was hybridized overnight at 65°C with 32P-labeled cDNA fragments probes. Primers used to amplify the probes are listed in Supplemental Table S4. Signals were quantified with a phosphoimager (Typhoon; Amersham Biosciences) using the program ImageQuant (GE Healthcare).
cDNA Synthesis and RT-qPCR Analysis
Total RNA was extracted with the Direct-zol RNA Kit (Zymo Research) according to the manufacturer’s protocol, and 2 μg of the RNA was employed to synthesize cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR analysis was performed on a Bio-Rad iQ5 real-time PCR instrument with the iQ SYBR Green Supermix (Bio-Rad). The gene-specific primers used for this assay are listed in Supplemental Table S4. Each sample was quantified in triplicate and normalized using AT4G36800, which codes for an RUB1-conjugating enzyme (RCE1) as an internal control.
Sample Preparation for Quantitative Proteomics
Total proteins were extracted from 4-d–old wild-type and pp7l-1 seedlings (four biological replicates for each genotype) by grinding 200 mg of frozen plant material into a fine powder in liquid N. After suspending the powder in 1 mL of extraction buffer (100 mm N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid, pH 7.5, 150 mm NaCl, 10 mm dithiothreitol, 1% [w/v] SDS, 1× Roche cOmplete Protease Inhibitor Cocktail), cell debris was removed by centrifugation at 10,000 g for 30 min. Proteins were then precipitated with chloroform–methanol (Wessel and Flügge, 1984), pellets were solubilized in 6 m guanidine hydrochloride and protein concentration was determined by bicinchoninic assay (Pierce BCA Protein Assay Kit; Thermo Fisher Scientific). Proteome aliquots of 100 μg were reduced with 10 mm dithiothreitol (30 min, 37°C), alkylated with 50 mm iodoacetamide (30 min, room temperature, in the dark), and recovered by chloroform-methanol precipitation before digestion with trypsin (SERVA) at a proteome/enzyme ratio 100:1 (w/w). After overnight incubation at 37°C, stable-isotope dimethyl labeling was performed in solution according to Boersema et al. (2009). Briefly, the samples were labeled separately with 20 mm NaCH3CN and either 40 mm CH2O (wild type, “light formaldehyde”) or 40 mm 13CD2O (pp7l-1, “heavy formaldehyde”) for 4 h at 37°C. Excess reagent was quenched by adding 100 mm of TRIS at pH 6.8 (1 h, 37°C). After acidification with 2% (v/v) formic acid, equal amounts of light and heavy labeled samples were combined to generate four duplex samples. Before liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, all samples were desalted with home-made C18 stage tips (Rappsilber et al., 2007), eluates were vacuum-dried to near-dryness and stored at −80°C before LC-MS/MS analysis.
LC-MS/MS
LC-MS/MS was performed on a nano-LC-system (Ultimate 3000 RSLC; Thermo Fisher Scientific) coupled to an Impact II high resolution quadrupole time of flight (Bruker) using a CaptiveSpray nano electrospray ionization source (Bruker Daltonics). The nano-LC system was equipped with an Acclaim Pepmap nano-trap column (C18, 100 Å, 700 μm × 2 cm; Thermo Fisher Scientific) and an Acclaim Pepmap RSLC analytical column (C18, 100 Å, 75 μm × 50 cm; Thermo Fisher Scientific). The peptide mixture was fractionated by applying a linear gradient of 5% to 45% acetonitrile at a flow rate of 250 nL/min over a period of 150 min. MS1 spectra were acquired at 3 Hz with a mass range from m/z 200–2000, with the Top-18 most intense peaks selected for MS/MS analysis using an intensity-dependent spectra acquisition time between 4 and 16. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD012105.
MS Data Analysis
MS raw files were analyzed with MaxQuant software (version 1.6.0.13; https://maxquant.org/) and peak lists were searched against the Arabidopsis UniProt database (version February 2017) using the Andromeda search engine with default settings. Cys carbamidomethylation was set as a fixed modification, and acetylation of protein N-termini and Met oxidation were set as variable modifications. Lys and N-terminal demethylation (light formaldehyde +28.03 D; heavy formaldehyde +34.06 D) were set as labels, and “requantified” was enabled.
Statistical Analysis
All statistical and bioinformatics analysis were performed using the softwares Perseus (version 1.6.0.2) and Microsoft Office Excel. Two-tailed Student’s t‐test (P < 0.05) was used to define which protein groups are significantly changed and a Box-plot analysis of the ratio of the frequency of occurrence of each peptide in pp7l-1 mutant versus wild-type samples was used to identify significant (P ≤ 0.05) outliers from the mean ratio of all peptides.
lncRNA-Seq and Data Analysis
Total RNA from plants was isolated using Trizol (Invitrogen) and purified using Direct-zol RNA MiniPrep Plus columns (Zymo Research) according to the manufacturer’s instructions. RNA integrity and quality was assessed by a 2100 Bioanalyzer (Agilent). rRNA depletion, generation of RNA-Seq libraries, and 150-bp paired-end sequencing on an HiSeq 2500 system (Illumina) were conducted at Novogene Biotech with standard Illumina protocols. Three independent biological replicates were used per genotype.
RNA-Seq reads were analyzed on the Galaxy platform (https://usegalaxy.org/). After grooming FASTQ files, adaptors were removed with Trimmomatic (Bolger et al., 2014), and sequencing quality was accessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to The Arabidopsis Information Resource genome annotation 10 with the gapped-read mapper TopHat 2.1.1 (Kim et al., 2013) set for Forward Read unstranded libraries and adjusting the maximum intron length to 5,000 bp. Reads were counted with featureCounts (Liao et al., 2014) with the help of the gene annotation in Araport11 (www.araport.org/data/araport11). Differentially expressed genes were obtained with DESeq2 (Love et al., 2014) running with the fit type set to “parametric,” and applying a twofold change cutoff and an adjusted P value < 0.05.
Sequencing data have been deposited to Gene Expression Omnibus (Edgar et al., 2002) under the accession number GSE122495.
TEM
The plant material was directly fixed after 4-d–old seedlings had been kept for 8 h in darkness. Cotyledons were fixed using 75-mm cacodylate buffer containing 2.5% (v/v) glutaraldehyde. Postfixation was carried out using 1% (w/v) osmium tetroxide in water for 1 h followed by en-bloc staining with 1% (w/v) uranyl acetate. After dehydration in a graded acetone series, the sample material was gradually infiltrated with Spurr’s Resin. After polymerization, the embedded samples were ultrathin-sectioned and poststained with lead citrate. TEM was carried out using a model no. EM912 (Zeiss) with an integrated OMEGA-filter in the zero-loss mode and with an acceleration voltage of 80 kV. For image acquisition, we used a Tröndle 2k × 2k slow-scan charge-coupled device camera together with the respective software package (Tröndle Restlichtverstärkersysteme).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers provided in Supplemental Table S4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of pp7l T-DNA insertion mutants.
Supplemental Figure S2. Chl content and complementation of pp7l T-DNA insertion mutants.
Supplemental Figure S3. rRNA maturation in 4-d–old Col-0 and pp7l mutant seedlings.
Supplemental Figure S4. Phenotypes of 4-week–old Col-0, pp7l-1, cop1-4, and the different photoreceptor (cry1-304 cry2-1 and phyA-211 phyB-9) mutant plants.
Supplemental Figure S5. PhyB levels are elevated in pp7l mutants, but overexpression of phyB does not result in a pp7l phenotype.
Supplemental Figure S6. Overexpression of PP7L in Col-0.
Supplemental Table S1. SDS of the Fv/Fm values measured in the different pp7l mutant seedlings presented in Figure 3A.
Supplemental Table S2. Genes whose transcript levels differed by more than twofold relative to Col-0 in 4-d–old LD-grown pp7l-1 seedlings.
Supplemental Table S3. Relative protein accumulation in the pp7l-1 mutant compared to Col-0 grown for 4 d in LD conditions as determined by shotgun proteomics.
Supplemental Table S4. Primers used in this study.
Acknowledgments
We thank Claus Schwechheimer, Ute Höcker, Christian Schmitz-Linneweber, Ferenc Nagy, and Eva Adam for providing the csn, cop1, sig5, and P35S:PHYB-YFP phyB-9 seeds, respectively. We thank Paul Hardy for critical reading of the manuscript and Elisabeth Gerick for technical assistance.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (project C01 no. KL 2362/1-1 to T.K. and project C05 no. TRR175 to D.L.).
Articles can be viewed without a subscription.
References
- Abdallah F, Salamini F, Leister D (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 5: 141–142 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol Biol 60: 507–518 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K (2008) Snowy cotyledon 2: The identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Simková K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ (2010) The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22: 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arsova B, Watt M, Usadel B (2018) Monitoring of plant protein post-translational modifications using targeted proteomics. Front Plant Sci 9: 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ (2012) Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol 159: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Saunders GR, Bevan MW, Mayo MA, Harrison BD (1986) Expression of biologically-active viral satellite RNA from the nuclear genome of transformed plants. Nature 321: 446–449 [Google Scholar]
- Becker MG, Walker PL, Pulgar-Vidal NC, Belmonte MF (2017) SeqEnrich: A tool to predict transcription factor networks from co-expressed Arabidopsis and Brassica napus gene sets. PLoS One 12: e0178256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JO, Yerramsetty P, Zielinski AM, Mure CM (2013) Photosynthetic gene expression in higher plants. Photosynth Res 117: 91–120 [DOI] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, Troesch R, Vidi PA, Roschitzki B, Hirsch-Hoffmann M, Hennig L, Kessler F, Gruissem W, et al. (2011) Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of n-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik K, Burch-Smith TM (2015) Chloroplast signaling within, between and beyond cells. Front Plant Sci 6: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484–494 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal-McTeague WM, Sytsma KJ, Hall JC (2016) Biogeography and diversification of Brassicales: A 103-million-year tale. Mol Phylogenet Evol 99: 204–224 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix–loop–helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Jiang J, Zhang L (2015) Plastid Sigma factors: Their individual functions and regulation in transcription. Biochim Biophys Acta 1847: 770–778 [DOI] [PubMed] [Google Scholar]
- Coll NS, Danon A, Meurer J, Cho WK, Apel K (2009) Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol 50: 707–718 [DOI] [PubMed] [Google Scholar]
- Dohmann EM, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, Isono E, Schmid M, Schwechheimer C (2008) Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol 147: 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovzhenko A, Dal Bosco C, Meurer J, Koop HU (2003) Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma 222: 107–111 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Hall JC, Harkess A, Tang M, Coombs J, Mohammadin S, Schranz ME, Xiong Z, Leebens-Mack J, Meyers BC, et al. (2018) Brassicales phylogeny inferred from 72 plastid genes: A reanalysis of the phylogenetic localization of two paleopolyploid events and origin of novel chemical defenses. Am J Bot 105: 463–469 [DOI] [PubMed] [Google Scholar]
- Ezer D, Shepherd SJK, Brestovitsky A, Dickinson P, Cortijo S, Charoensawan V, Box MS, Biswas S, Jaeger KE, Wigge PA (2017) The G-box transcriptional regulatory code in Arabidopsis. Plant Physiol 175: 628–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Genoud T, Santa Cruz MT, Kulisic T, Sparla F, Fankhauser C, Métraux JP (2008) The protein phosphatase 7 regulates phytochrome signaling in Arabidopsis. PLoS One 3: e2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CMM, Monte E (2018) Seedling establishment: A dimmer switch-regulated process between dark and light signaling. Plant Physiol 176: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45(D1): D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Wang HT, Hung TH, Tseng CC, Chung TY, Hsieh MH (2017) The Arabidopsis thiamin-deficient mutant pale green1 lacks thiamin monophosphate phosphatase of the vitamin B1 biosynthesis pathway. Plant J 91: 145–157 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes Sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jeffrey SW, Humphrey GF (1975) New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz 167: 191–194 [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H (2001) An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol 42: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018) JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46(D1): D260–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kang JG, Yang SS, Chung KS, Song PS, Park CM (2002) A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D (2015) Emerging functions of mammalian and plant mTERFs. Biochim Biophys Acta 1847: 786–797 [DOI] [PubMed] [Google Scholar]
- Kleine T, Leister D (2016) Retrograde signaling: Organelles go networking. Biochim Biophys Acta 1857: 1313–1325 [DOI] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D (2009) DNA transfer from organelles to the nucleus: The idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Köhler D, Helm S, Agne B, Baginsky S (2016) Importance of translocon subunit Tic56 for rRNA processing and chloroplast ribosome assembly. Plant Physiol 172: 2429–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Baldwin A, Patel R, Razzaq A, Dupree P, Lilley K, Kurth J, Leister D, Jarvis P (2003) The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee HJ, Kim DH, Jeon Y, Pai HS, Kang H (2014) A nuclear-encoded chloroplast protein harboring a single CRM domain plays an important role in the Arabidopsis growth and stress response. BMC Plant Biol 14: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Kleine T (2016) Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol Plant 157: 297–309 [DOI] [PubMed] [Google Scholar]
- Leister D, Romani I, Mittermayr L, Paieri F, Fenino E, Kleine T (2014) Identification of target genes and transcription factors implicated in translation-dependent retrograde signaling in Arabidopsis. Mol Plant 7: 1228–1247 [DOI] [PubMed] [Google Scholar]
- Leister D, Wang L, Kleine T (2017) Organellar gene expression and acclimation of plants to environmental stress. Front Plant Sci 8: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Zhu N, Mai KK, Liu Z, Tzeng D, Osteryoung KW, Zhong S, Staehelin LA, Kang BH (2018) Thylakoid-bound polysomes and a dynamin-related protein, FZL, mediate critical stages of the linear chloroplast biogenesis program in greening Arabidopsis cotyledons. Plant Cell 30: 1476–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liebers M, Grübler B, Chevalier F, Lerbs-Mache S, Merendino L, Blanvillain R, Pfannschmidt T (2017) Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during Plant development. Front Plant Sci 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Lillo C, Kataya AR, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z, Jonassen EM (2014) Protein phosphatases PP2A, PP4 and PP6: Mediators and regulators in development and responses to environmental cues. Plant Cell Environ 37: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Linhart C, Halperin Y, Shamir R (2008) Transcription factor and microRNA motif discovery: The Amadeus platform and a compendium of metazoan target sets. Genome Res 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ (2002) Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J 32: 549–559 [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Møller SG, Kim YS, Kunkel T, Chua NH (2003) PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Gossmann N, Dietz KJ (2016) Redox regulation of cytosolic translation in plants. Trends Plant Sci 21: 388–397 [DOI] [PubMed] [Google Scholar]
- Motohashi R, Rödiger A, Agne B, Baerenfaller K, Baginsky S (2012) Common and specific protein accumulation patterns in different albino/pale-green mutants reveals regulon organization at the proteome level. Plant Physiol 160: 2189–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Shikanai T, Fujiwara M, Kanamaru K, Takahashi H, Tanaka K (2004) The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol 45: 357–368 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelze ML, Muthuramalingam M, Vogel MO, Dietz KJ (2014) The link between transcript regulation and de novo protein synthesis in the retrograde high light acclimation response of Arabidopsis thaliana. BMC Genomics 15: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S (2015) Plastid RNA polymerases: Orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot 66: 6957–6973 [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Labs M, Leister D (2014) Structure and dynamics of thylakoids in land plants. J Exp Bot 65: 1955–1972 [DOI] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S (2003) Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol Biol 51: 385–399 [DOI] [PubMed] [Google Scholar]
- Putarjunan A, Liu X, Nolan T, Yu F, Rodermel S (2013) Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth Res 116: 437–453 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Roy B, von Arnim AG (2013) Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11: e0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel NJ, Hangarter RP (2007) Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusaczonek A, Czarnocka W, Kacprzak S, Witoń D, Ślesak I, Szechyńska-Hebda M, Gawroński P, Karpiński S (2015) Role of phytochromes A and B in the regulation of cell death and acclimatory responses to UV stress in Arabidopsis thaliana. J Exp Bot 66: 6679–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LL, Clarke AK, Straus M, Apel K (2011) ‘happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65: 690–702 [DOI] [PubMed] [Google Scholar]
- Sakr S, Wang M, Dédaldéchamp F, Perez-Garcia MD, Ogé L, Hamama L, Atanassova R (2018) The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int J Mol Sci 19: 2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Lampe MK, Sultan LD, Ostersetzer-Biran O (2015) Organellar maturases: A window into the evolution of the spliceosome. Biochim Biophys Acta 1847: 798–808 [DOI] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK (2015) Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ 38: 1658–1672 [DOI] [PubMed] [Google Scholar]
- Shen Y, Kim JI, Song PS (2005) NDPK2 as a signal transducer in the phytochrome-mediated light signaling. J Biol Chem 280: 5740–5749 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2015) RNA editing in plants: Machinery and flexibility of site recognition. Biochim Biophys Acta 1847: 779–785 [DOI] [PubMed] [Google Scholar]
- Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya K (2007) Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Hanzawa H, Schäfer E, Furuya M (1998) Mode of phytochrome B action in the photoregulation of seed germination in Arabidopsis thaliana. Plant J 13: 583–590 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Meurer J (2012) The cutting crew—Ribonucleases are key players in the control of plastid gene expression. J Exp Bot 63: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Strittmatter G, Kössel H (1984) Cotranscription and processing of 23S, 4.5S and 5S rRNA in chloroplasts from Zea mays. Nucleic Acids Res 12: 7633–7647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Hou P, Song M, Zheng X, Guo L, Xiao Y, Yan L, Li W, Yang J (2015) Synergistic and antagonistic action of phytochrome (Phy) A and PhyB during seedling de-etiolation in Arabidopsis thaliana. Int J Mol Sci 16: 12199–12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T (2004) Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA 101: 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ühlken C, Horvath B, Stadler R, Sauer N, Weingartner M (2014) MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J 78: 107–120 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Moorhead GB (2013) Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci 18: 505–513 [DOI] [PubMed] [Google Scholar]
- Ulker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouché N, Edwards R, Sanders D, Knight H, Knight MR (2008) Getting the most out of publicly available T-DNA insertion lines. Plant J 56: 665–677 [DOI] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K (2018) PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46(D1): D1190–D1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dingenen J, Blomme J, Gonzalez N, Inzé D (2016) Plants grow with a little help from their organelle friends. J Exp Bot 67: 6267–6281 [DOI] [PubMed] [Google Scholar]
- Varotto C, Pesaresi P, Maiwald D, Kurth J, Salamini F, Leister D (2000) Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38: 497–504 [Google Scholar]
- Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Tepperman JM, Quail PH (1991) Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhao J, Jia M, Xu N, Liang S, Shao J, Qi Y, Liu X, An L, Yu F (2018) Balance between cytosolic and chloroplast translation affects leaf variegation. Plant Physiol 176: 804–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Piqueras R, Cao K, Deng XW, Wei N (2009) Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J 58: 655–667 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: Calibrating the family tree. Proc Biol Sci 268: 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Foster R, Chua NH (1992) Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 4: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A. (2018) Transitioning to the next phase: The role of sugar signaling throughout the plant life cycle. Plant Physiol 176: 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzinger B, Nukarinen E, Nägele T, Weckwerth W, Teige M (2018) The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol 176: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E (2015) Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci 20: 641–650 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1—From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu QB, Li G, Wang G, Sun JC, Wang PC, Wang C, Mi HL, Ma WM, Cui J, Cui YL, et al. (2008) Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res 18: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagari N, Sandoval-Ibañez O, Sandal N, Su J, Rodriguez-Concepcion M, Stougaard J, Pribil M, Leister D, Pulido P (2017) SNOWY COTYLEDON 2 promotes chloroplast development and has a role in leaf variegation in both Lotus japonicus and Arabidopsis thaliana. Mol Plant 10: 721–734 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]