Genomic and biochemical analyses identify targets of the flowering time regulator FD at the genome-wide scale and shed light on the requirement for interaction with the florigen FLOWERING LOCUS T.
Abstract
The transition to flowering is a crucial step in the plant life cycle that is controlled by multiple endogenous and environmental cues, including hormones, sugars, temperature, and photoperiod. Permissive photoperiod induces the expression of FLOWERING LOCUS T (FT) in the phloem companion cells of leaves. The FT protein then acts as a florigen that is transported to the shoot apical meristem, where it physically interacts with the Basic Leucine Zipper Domain transcription factor FD and 14-3-3 proteins. However, despite the importance of FD in promoting flowering, its direct transcriptional targets are largely unknown. Here, we combined chromatin immunoprecipitation sequencing and RNA sequencing to identify targets of FD at the genome scale and assessed the contribution of FT to DNA binding. We further investigated the ability of FD to form protein complexes with FT and TERMINAL FLOWER1 through interaction with 14-3-3 proteins. Importantly, we observed direct binding of FD to targets involved in several aspects of plant development. These target genes were previously unknown to be directly related to the regulation of flowering time. Our results confirm FD as a central regulator of floral transition at the shoot meristem and provide evidence for crosstalk between the regulation of flowering and other signaling pathways, such as pathways involved in hormone signaling.
The floral transition represents a crucial checkpoint in the plant life cycle at which the shoot apical meristem (SAM) ceases to produce only leaves and begins producing reproductive organs. As the commitment to this developmental phase transition is usually irreversible for a given meristem, plants have evolved several pathways to integrate environmental and endogenous stimuli to ensure flowering is induced at the correct time. A rich literature has identified hormones, sugars, temperature, and day length (photoperiod) as main factors in flowering time regulation (for review, see Srikanth and Schmid, 2011; Romera-Branchat et al., 2014; Song et al., 2015). Photoperiod in particular has been shown to regulate flowering time in many plant species and, depending on the light requirements, short day (SD), long day (LD), and day-neutral plants have been distinguished. In thale cress Arabidopsis (Arabidopsis thaliana), LD promotes flowering but plants will eventually flower even under noninductive SD.
It has long been known that in day-length responsive species, inductive photoperiod is mainly perceived in leaves where it results in the formation of a long-distance signal, or florigen, that moves to the SAM to induce the transition to flowering (An et al., 2004; Corbesier et al., 2007; Mathieu et al., 2007). The molecular nature of florigen has eluded identification for the better part of a century. However, recently FLOWERING LOCUS T (FT) and related genes, which encode phosphatidylethanolamine-binding proteins (PEBPs), have been identified as evolutionarily conserved candidates (Corbesier et al., 2007; Mathieu et al., 2007). Under inductive photoperiod, FT is expressed in leaf phloem companion cells (PCCs) and there is good evidence that the FT protein is loaded into the phloem sieve elements and transported to the SAM (for a review, see Srikanth and Schmid, 2011; Song et al., 2015). At the SAM, FT interacts with FD and 14-3-3 proteins and the resulting florigen-activation complex (FAC) is thought to control the correct expression of flowering time and floral homeotic genes to promote the transition of the vegetative meristem into a reproductive inflorescence meristem (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011).
FD belongs to the Basic Leucine Zipper Domain (bZIP) transcription factor (TF) family (Jakoby et al., 2002) and is mainly expressed at the SAM (Abe et al., 2005; Schmid et al., 2005; Wigge et al., 2005). It has been proposed that, to interact with FT and 14-3-3 proteins, FD must be phosphorylated at Thr-282 (T282; Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011). Recently, two calcium-dependent kinases expressed at the SAM, CALCIUM DEPENDENT PROTEIN KINASE6 (CPK6) and CPK33, have been shown to phosphorylate FD (Kawamoto et al., 2015). FD interacts not only with FT but also with other members of the PEBP protein family. Interestingly, some of the six PEBP proteins encoded in the Arabidopsis genome regulate flowering in opposition (Kim et al., 2013). FT and its paralog TWIN SISTER OF FT (TSF) promote flowering. Mutations in tsf enhance the late flowering phenotype of ft in LD, but TSF also has distinct roles in SD (Yamaguchi et al., 2005). Other members of the PEBP protein family, most prominently TERMINAL FLOWER1 (TFL1), oppose the flower-promoting function of FT and TSF, and repress flowering. The Arabidopsis ortholog of CENTRORADIALIS (ATC) has been shown to act as a SD-induced floral inhibitor that is expressed mostly in the vasculature, but was undetectable at the SAM (Huang et al., 2012). Furthermore, ATC has been suggested to move long distances and can interact with FD to inhibit APETALA1 (AP1) expression. ATC has thus been proposed to antagonize the flower-promoting effect of FT (Huang et al., 2012). Similarly, orthologs of ATC in rice (Oryza sativa) have been recently shown to antagonize the function of FT-like protein (Kaneko-Suzuki et al., 2018). Finally, BROTHER OF FT, which like ATC is strongly expressed in the leaf vasculature, can interact with FD in the nucleus, interfering with FT function under high salinity by inhibiting AP1 expression, thereby delaying flowering (Yoo et al., 2010; Ryu et al., 2014).
TFL1 differs from FT only in 39 nonconserved amino acids but as mentioned above, has an opposite biological function: TFL1 represses flowering while FT is a floral promoter (Ahn et al., 2006). It has been demonstrated that substitutions of a single amino acid (TFL1-H88; FT-Y85) or exchange of the segment B encoded by the fourth exon are sufficient to impose TFL1-like activity onto FT, and vice versa (Hanzawa et al., 2005; Ahn et al., 2006; Ho and Weigel, 2014). Similar to FT, TFL1 also interacts with FD, both in yeast-2-hybrid assays as well as in plant nuclei (Wigge et al., 2005; Hanano and Goto, 2011). Together, these findings suggest that activating FD-FT and repressive FD-TFL1 complexes compete for binding to the same target genes (Ahn et al., 2006). This hypothesis is further supported by the observation that TFL1 apparently acts to repress transcription (Hanano and Goto, 2011), whereas FT seems to function as a transcriptional (co)activator (Wigge et al., 2005). However, evidence that these protein complexes in fact share interactors such as 14-3-3 proteins, or control the same targets, remains sparse.
FD has been reported as a direct and indirect regulator of important flowering time and floral homeotic genes such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SPL3), SPL4, SPL5, LEAFY (LFY), AP1, and FRUITFULL (FUL). Several flowering time pathways contribute to SOC1 regulation. Indeed, it has been proposed that expression of SOC1 can be directly promoted by the FD-FT complex (Lee and Lee, 2010). However, SOC1 expression can also be activated independently from FD-FT probably through the SPL3, SPL4, and SPL5 proteins (Moon et al., 2003; Wang et al., 2009; Lee and Lee, 2010), which have been shown to be directly or indirectly activated by the FD-FT complex (Jung et al., 2012). The activation of floral homeotic genes such as AP1 and FUL in response to FD-FT activity at the SAM can at least in part be explained by the direct activation of the floral meristem identify gene LFY through SOC1 (Moon et al., 2005; Yoo et al., 2005; Jung et al., 2012). In addition, it has also been proposed that the FD-FT complex can promote the expression of AP1 and FUL by directly binding to their promoters (Abe et al., 2005; Teper-Bamnolker and Samach, 2005; Wigge et al., 2005). Taken together, these results support a central role for FD in integrating different pathways to ensure the correct timing of flowering. However, FD targets have not yet been identified at the genome scale, nor has the requirement for protein complex formation for FD function in Arabidopsis been addressed systematically.
Here we identify direct and indirect targets of FD at the genome scale using chromatin immunoprecipitation sequencing (ChIP-seq) and RNA-seq in wild type as well as in ft-10 tsf-1 double mutants. Our results demonstrate that FD can bind to DNA in vivo even in the absence of FT/TSF. However, FD binding to a subset of targets, which includes many important flowering time and floral homeotic genes, was reduced in the ft-10 tsf-1 double mutant, strongly supporting a role for FT/TSF in modulating the binding of FD to DNA and the expression of functionally important target genes. In addition, we report the effects of FD phosphorylation on protein complex formation with FT and TFL1 via 14-3-3 proteins in vitro and show how phosphorylation of FD affects flowering time in planta. Finally, our ChIP-seq experiments identified hundreds of previously unknown FD target genes, both in the PCCs as well as at the SAM. For example, we observed that FD directly binds to and regulates the expression of genes in hormone signaling pathways. These newly identified FD target genes represent a precious resource not only to enhance our knowledge of the photoperiod pathway but also to better understand the integration of different signaling pathways at the transcriptional level. Taken together, our findings support a role for FD as a central integrator of flowering time and provide important data to guide future research on the integration of diverse signaling pathways at the SAM.
RESULTS
FD Binds G-Box Motifs When Expressed in PCCs
FD is normally expressed at the SAM whereas its interaction partner FT is expressed in leaf PCCs. As most 14-3-3 proteins are ubiquitously expressed at moderate to high levels and have also been detected in PCCs (Schmid et al., 2005; Deeken et al., 2008), we reasoned that expression of FD from the PCC-specific SUC2 promoter would maximize FAC complex formation and enable us to investigate the role of FT in modulating FD transcriptional activity.
We performed ChIP-seq on independent biological duplicates in a stable pSUC2::GFP:FD reporter line, which shows no discernible phenotype when compared to Col-0 background. A pSUC2::GFP:NLS line, in which the GFP protein is fused to the nuclear localization signal (NLS), was used as a control. A total of 2,068 and 3,236 genomic regions showing significant enrichment (peaks) were identified in the first and second replicate, respectively (Supplemental Fig. S1A).
In the individual replicates, the majority of the peaks mapped to promoter regions (65.1% and 63.8%, respectively), followed by intergenic regions (16% and 16.8%), transcriptional terminator sites (9.2% and 10.7%), exons (6.4% and 5.6%), introns (2.4% and 2.3%), 5′-untranslated regions (UTRs; 0.5% and 0.3%), and 3′-UTRs (0.4% and 0.5%; Supplemental Fig. S1D).
The relative enrichment of peaks mapping to promoter regions is in agreement with what is expected from a transcriptional regulator. In both replicates, the majority of the peaks are located between 600-bp and 300-bp upstream of the nearest transcription start site (TSS; Supplemental Fig. S1, G and J). Overlapping results from the two biological replicates identified 1,754 high-confidence peaks shared in both experiments (Supplemental Fig. S1A; Supplemental Data Set S1). Similar to the individual experiments, these high-confidence peaks mostly mapped to the promoter regions (66.8%; Fig. 1A). Only this subset of peaks, which includes important flowering time and flower development genes such as AP1, FUL, LFY, SOC1, SEPALLATA1 (SEP1), and SEP2, SEP3, was used for further analysis.
Figure 1.
Identification of FD targets by pSUC2::GFP:FD ChIP-seq in Col-0 and ft-10 tsf-1 and pFD::GFP:FD ChIP-seq in fd-2. A, Annotation of high-confidence peaks found in two biological replicates in Col-0 and ft-10 tsf-1. TTS, transcription terminator site. B, Four-set Venn diagram representing the overlapping peaks among all the biological replicates from Col-0 and ft-10 tsf-1. The majority of the peaks (1,530) are shared between the two genetic backgrounds. C, Nucleotide logo of the predicted FD binding site. D, Correlation heatmap calculated on a binding matrix based on ChIP-seq reads counts for Col-0 and ft-10 tsf-1 samples (affinity scores). The presence/absence of FT and TSF is sufficient to discriminate the two genetic backgrounds. E, DB peaks between Col-0 and ft-10 tsf-1. Red dots indicate differentially bound peaks with a false discovery rate < 0.05. Blue dots represent peaks that were not significantly differentially bound. F, Reads from Col-0, ft-10 tsf-1 and control (ctrl) sample mapped against selected flowering-related genes. G, Annotation of high-confidence peaks identified by ChIP-seq in two biological replicates in pFD::GFP:FD fd-2. H, Nucleotide logo of the predicted FD binding site at the SAM.
De novo motif analysis of the 1,754 high-confidence peaks using MEME-ChIP (Machanick and Bailey, 2011) revealed that peak regions showed a strong enrichment of G-boxes (CACGTG), which is a canonical bZIP binding site (Supplemental Fig. S1M). The subset of 1,754 peak regions was associated with 1,676 unique genes, with 68 genes containing more than one FD binding site. Taken together, these results demonstrate that, when misexpressed in the PCCs, FD is capable of binding to G-box elements in a large number of genes that are involved in diverse aspects of the plant life cycle.
FT and TSF Enhance Binding of FD to DNA
To test whether FT and its paralog TSF are required for FD to bind to DNA, the pSUC2::GFP:FD reporter and pSUC2::GFP:NLS control constructs were transformed into the ft-10 tsf-1 mutant background. Results from two independent biological replicates show that FD is capable of binding to DNA even in the absence of FT and TSF. Most peaks (63% and 62.1% in the first and second biological replicate, respectively) mapped to promoter regions within 600- and 300-bp nucleotides upstream of the nearest TSS (Supplemental Fig. S1, E, H, and K). Overall, these results are very similar to those observed for pSUC2::GFP:FD in Col-0 (Fig. 1A; Supplemental Fig. S1, E, H, K, and N). Comparison between the two biological replicates identified 2,696 common peaks in ft-10 tsf-1 mutant that mapped to 2,504 unique genes (Supplemental Fig. S1B; Supplemental Data Set S2).
Surprisingly, overlapping the sets of genomic regions bound by FD with high confidence in wild-type (1,754) and ft-10 tsf-1 (2,696) backgrounds identified 1,530 shared peaks (Fig. 1B; Supplemental Data Set S3), suggesting that FD is capable of binding to the majority of its targets in the absence of FT and TSF. Motif enrichment analysis of sequence comprising the 1,530 shared peaks revealed that FD maintained a strong preference for binding to G-box motifs (Fig. 1C).
Analysis of differentially bound (DB) regions revealed that, although FT and TSF were not required for FD to bind DNA, their presence increased the enrichment of FD on a subset of target loci and this difference in binding was sufficient to discriminate Col-0 and ft-10 tsf-1 (Fig. 1D). A total of 885 DB regions with a false discovery rate < 0.05 were observed between wild type and ft-10 tsf-1 and almost all of these loci showed higher enrichment in wild type (Fig. 1E, Supplemental Data Set S4). Interestingly, this subset includes important floral homeotic genes such as AP1, SEP1, SEP2, and FUL, as well as two members of the SPL gene family, SPL7 and SPL8. We also found FD bound to the second exon of LFY, a master regulator of flower development (Fig. 1F). In addition, we detected binding to loci encoding genes involved in the regulation of gibberellic acid (GA) biosynthesis and degradation such as GIBBERELLIN 2-OXIDASE 4 (GA2OX4), GA2OX6, and GIBBERELLIN 3-OXIDASE 1 (GA3OX1) as well as to three key components of the circadian clock, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIME FOR COFFEE (TIC; Supplemental Data Set S4).
To test the robustness of our results and for any possible bias due to the use of the different genetic backgrounds Col-0 and ft-10 tsf-1 as controls, peaks were called again using pSUC2::GFP:NLS in Col-0 as single negative control. Analysis identified 917 DB loci (Supplemental Fig. S2), which is comparable to the 885 DB loci from the previous analysis (Fig. 1E). In addition, affinity test analysis clustered by genotype rather than the control used (Supplemental Fig. S2), ruling out strong bias due to the usage of different genetic backgrounds for peak calling.
Importantly, FD is capable of inducing the known FAC target gene AP1 in leaves when expressed under the pSUC2 promoter, suggesting that a functional FAC can be formed in the PCCs when FD is present (Supplemental Fig. S3A). The finding that AP1 expression could only be observed in the Col-0 background but not in pSUC2::GFP:FD ft-10 tsf-1 further supports this idea. However, in contrast to AP1, we failed to detect induction of SOC1 in the PCCs of pSUC2::GFP:FD (Supplemental Fig. S3A), suggesting that other cofactor(s) specifically expressed at the SAM might be required to fully activate FD target gene expression.
FD Phosphorylation Is Required for Complex Formation and to Promote Flowering
To confirm the binding of FD to G-boxes we performed electrophoretic mobility shift assays (EMSAs) using the bZIP domain of the Arabidopsis FD protein (FD-C) and a probe consisting of a 30-bp fragment from the SEP3 promoter containing a G-box that we had identified as FD target region in our ChIP-seq (Fig. 1F). We observed weak binding of FD-C, but failed to detect higher order complexes when 14-3-3, FT, or both were added (Fig. 2A). In contrast, a clear supershift with 14-3-3 and FT was observed when a phosphomimic variant of FD-C, FD-C_T282E, was used (Fig. 2B). Interestingly, TFL1, which is similar to FT in structure (Ahn et al., 2006) but delays flowering, was capable of forming a complex with 14-3-3 and wild-type FD-C (Fig. 2A). Similar results were obtained with the full-length version of FD (Supplemental Fig. S4A). Taken together, these results demonstrate that Arabidopsis FD is capable of binding to DNA without FT, confirming results from our ChIP-seq experiments. Furthermore, this indicates that the unphosphorylated form of FD, in complex with 14-3-3 proteins, can interact with TFL1.
Figure 2.
The C-terminal part of FD binds to a G-box in the SEP3 promoter in vitro. A, EMSA of the wild-type form of FD-C in combination with 14-3-3ν, FT, and TFL1. B, EMSA using the phosphomimic version of FD-C (FD-C_T282E) in combination with 14-3-3ν, FT, and TFL1. First lanes to the left in (A) and (B) show the free probe without any added proteins. Plus signs (+) above the lanes = which proteins were used in a specific EMSA reaction. Asterisk (*) = higher order complexes.
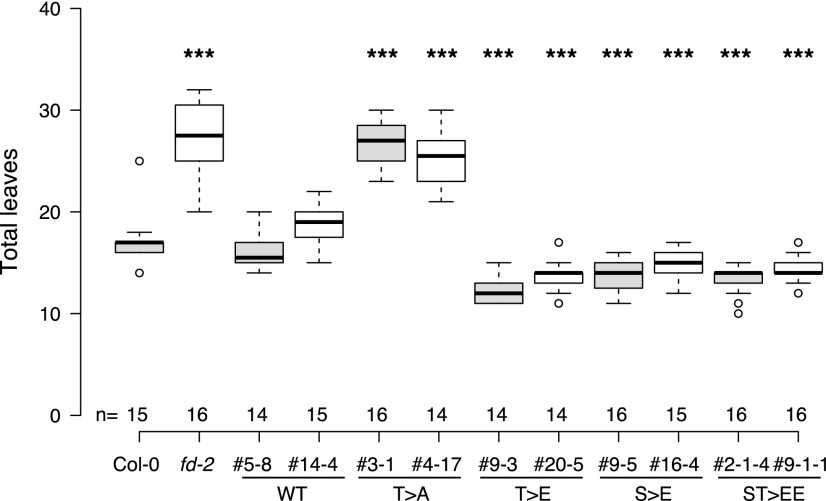
To investigate the importance of FD phosphorylation in vivo we complemented the fd-2 mutant with pFD::FD, pFD::FD-T282E, and pFD::FD-T282A (which cannot be phosphorylated) and determined flowering time of homozygous transgenic plants. Plants transformed with the wild-type version of FD rescued the late flowering phenotype of fd-2, indicating that the rescue construct was fully functional (Fig. 3). In contrast, plants transformed with the T282A version flowered with the same number of leaves as fd-2, demonstrating that FD needs to be phosphorylated to induce flowering. Interestingly, plants transformed with the T282E phosphomimic version of FD flowered even earlier than wild type (Fig. 3), indicating that control of FD phosphorylation is important for its function in vivo. To test whether Ser-281 (S281), which is located next to T282, constitutes a potential FD phosphorylation site, we complemented fd-2 with pFD::FD-S281E and pFD::FD-S281E/T282E constructs. Interestingly, these lines flowered as early as plants transformed with the phosphomimic version T282E (Fig. 3), indicating that S281 may be a FD phosphorylation site but that mimicking double-phosphorylation of S281/T282 does not accelerate flowering any further. These in vivo results are in agreement with our in vitro EMSA results and confirm that phosphorylation of FD is required for its function and must be finely regulated to avoid either premature or delayed flowering. It should be noted, however, that the phosphomimic version of the C-terminal fragment of FD (as used in the EMSA analyses) is insufficient to fully rescue the late flowering of fd-2 (Supplemental Fig. S3B), suggesting that the N-terminal region of FD, even though it does not contain any known functional domains, nevertheless contributes to FD function.
Figure 3.
Phosphorylation of FD at T282 modulates flowering time in Arabidopsis. Box plot reporting the flowering time of control plants (Col-0), the fd-2 mutant, and the fd-2 mutant transformed with either wild-type (WT) FD cDNA under the control of the FD promoter (pFD::FD), nonphosphorable FD (T > A, pFD::FD_T282A), or phosphomimic versions of FD (T > E, pFD::FD_T282E; S > E, pFD::FD-S281E; ST > EE; pFD::FD_ S281E/T282E) as number of leaves formed by the main meristem (total leaves). Results are shown for two independent homozygous lines per construct. Number of plants (n) analyzed per genotype is indicated. Statistical significance was calculated using an unpaired Student’s t test compared to Col-0. ***P < 0.01.
Targets of FD at the SAM
The rationale for carrying out the initial ChIP-seq experiments in PCCs was to maximize the likelihood of FAC formation and to study the contribution of FT/TSF to FD DNA binding. However, because our ChIP-seq and EMSA results indicated that FD-FT interaction is not required for FD to bind to DNA, we decided to determine direct targets of FD in its natural context at the SAM.
To this end we performed ChIP-seq using a fd-2 mutant complemented with a pFD::GFP:FD construct (Supplemental Fig. S3C). We performed ChIP-seq using two independent biological replicates from apices of 16-d-old plants grown in LD conditions. In the two replicates, we could identify 703 and 1,222 FD-bound regions, respectively, of which 595 were shared between the replicates (Supplemental Fig. S1C; Supplemental Data Set S5). Of these, 69.7% mapped to core promoter regions within 300–600-bp upstream of the nearest TSS, 15.8% to intergenic regions, followed by lesser percentages to transcription terminator site (6.2%), exons (5.9%), introns (1.8%), and 5′-UTRs (0.5%; Fig. 1G; Supplemental Fig. S1, F, I, and L). Similar to the situation in our PCC-specific ChIP-seq analyses we found G-box sequences as the most overrepresented TF binding sites under the peak regions (Fig. 1H; Supplemental Fig. S1O). The 595 peak regions shared between the replicates mapped to 572 individual genes, which we consider high-confidence in vivo targets of FD at the SAM and include important flowering-related genes such as AP1, FUL, SOC1, and SEP3.
The precise location of the FD binding site in the AP1 promoter has been discussed controversially (Wigge et al., 2005; Benlloch et al., 2011). Taking into account all six ChIP-seq datasets, we were able to extract a 64-bp sequence covering the peak summits on the AP1 promoter (Fig. 4, A and B). Interestingly, this sequence lies ∼100 bp downstream of the C-box that had previously been implicated in FD binding to AP1 (Wigge et al., 2005), but contains several other palindromic sequences. However, none of these sequences is a bona fide G-box. We selected three potential binding sites within the 64-bp sequence and tested them, along with the upstream C-box, by EMSA for FD binding (Fig. 4C; Supplemental Fig. S4B). Results show that only the phosphomimic version of FD-C (FD-C_T282E) in combination with 14-3-3 can bind to all four DNA sequences tested. Furthermore, we detected a supershift for all palindromic sites tested, including the C-box, when we added TFL1. In contrast, for FT an additional shift resembling the pattern obtained with the G-box in SEP3 promoter was only observed for “site 2” (Figs. 2B and 4C). Closer inspection of the nucleotide sequences of the probes used for the G-box in the SEP3 promoter and the “site 2” in the AP1 promoter revealed that the possible FD binding site in the AP1 promoter (GTCGAC) is also present in the SEP3 promoter, where it overlaps with the G-box (Fig. 4D). Interestingly, in the context of the SEP3 probe, full-length FD and FD-C tolerated mutating the core of the G-box from CG to GC, whereas CG to TA mutations as well as converting the G-box to a C-box (GACGTC) abolished binding (Supplemental Fig. S4D). To further test if “site 2” on the AP1 promoter constitutes a real FD binding site, we mutated its core from CG to TA and checked whether this was sufficient to abolish binding. Results show that indeed the binding of FD to this mutated version of “site 2” was strongly reduced, except in the presence of TFL1 (Supplemental Fig. S4E).
Figure 4.
Mapping of the FD binding site in the AP1 promoter. A, Normalized reads from six ChIP-seq experiments mapped on the AP1 locus. Horizontal bars below the peaks indicate regions of significant enrichment. The position of the summit is indicated for each peak. The C-box previously implicated in FD binding is located upstream (to the right) of peak summits in all six experiments. B, Nucleotide sequence encompassing the six peak summits shows several palindromic regions representing putative binding sites of FD on the AP1 promoter. The distance between the closest potential FD binding site under the ChIP-seq peaks and the C-box is 92 bp. Black triangles indicate the summits of the six separate ChIP-seq experiments. Putative FD binding sites are underlined and numbered from 1 to 4. C, EMSA of the phosphomimic version of FD-C (FD-C T282E) in combination with 14-3-3v, FT, and TFL1 using the four putative binding sites reported in (B). Free probes are not visible because gels were running longer to maximize the distance between shifted probes. Colored squares indicate shifted probes. D, Comparison of the probes used for EMSA: the G-box in SEP3 promoter (Fig. 2) and the binding site 2 in AP1 promoter. The putative FD binding site (site 2) in the AP1 promoter, which is also conserved in SEP3 promoter where it overlaps with the G-box, is marked in red.
Taken together, our findings exclude the C-box as the FD binding site in the AP1 promoter. Furthermore, our results suggest that in vitro FD can bind to other motifs besides the G-box, possibly through interaction with partners other than 14-3-3 and FT/TSF, and we characterized a possible new binding site (GTCGAC) that could constitute the true FD binding site in the AP1 promoter.
Differentially Expressed Genes at the SAM and Direct Targets of FD
To test which of the 595 high-confidence binding sites we had identified by ChIP-seq at the SAM were in fact transcriptionally regulated by FD, we performed RNA-seq on apices from fd-2 mutants and the pFD::GFP:FD fd-2 rescue line. Twenty-one-day–old SD-grown seedlings were shifted to LD to induce synchronous flowering and apices were harvested on the day of the transfer to LD (T0), as well as 1, 2, 3, and 5 d after the shift (T1, T2, T3, T5) from three independent biological replicates.
Differentially expressed (DE) genes were determined for each time point and genes with an adjusted P value < 0.1 were selected as significantly DE. We identified in total 1,759; 583; 2,421; 924; and 153 DE genes in T0, T1, T2, T3, and T5, respectively, corresponding to 4,189 unique genes (Fig. 5A; Supplemental Data Set S6). PCA analysis showed that the first and second principal components, which explain 37% and 21% of the total variance, corresponded to the different time points and genotypes, respectively (Supplemental Fig. S5A). The best separation between the genotypes in the PCA was observed at T3 and T5, indicating that FD contributes to the transcriptional changes at the SAM mainly after exposure to two LDs. This observation is in agreement with the expression profile of FD, which in the pFD::GFP:FD rescue line increased after T2 (Supplemental Fig. S5B). In contrast, FD expression remained low in the fd-2 mutant, indicating the validity of our experimental approach (Supplemental Fig. S5B).
Figure 5.
RNA-seq results at the SAM. A, Scatter plots of DE genes between the fd-2 mutant and pFD::GFP-FD fd-2 (control) at five time points before and during the transition to photoperiod-induced flowering. T0–T5 indicate day of sample collection before (T0) and 1, 2, 3, and 5 d after shifting plants to LD. Red dots indicate DE genes with an adjusted P value < 0.1. B, Venn diagrams showing the overlap between 572 unique FD target genes identified by ChIP-seq and DE genes found by RNA-seq at the SAM at five time points before (T0) and during (T1–T5) the transition to flowering. C, Venn diagram showing the overlap between 572 unique FD target genes identified by ChIP-seq at the SAM (two biological replicates) and genes found to be differentially expressed at the SAM by RNA-seq at least at one time point (T0, T1, T2, T3, and T5; three biological replicates per time point). A total of 135 genes were classified as putative direct targets of FD. Statistical significance was calculated using the Fisher’s exact test. *P = 1.03E-07.
Next, we intersected the list of genes that were bound by FD at the SAM (572) with the list of DE genes (4,189). In total, 135 (23.6%) of the 572 FD-bound genes were significantly DE at the SAM during the transition to flowering at least at one timepoint, indicating that these genes are transcriptionally regulated by FD, which is more than expected by chance (Fig. 5, B and C; Supplemental Data Set S7). Among these 135 directly bound and DE genes, we observed several previously known FD-regulated flowering time and floral homeotic genes including AP1, FUL, and SOC1 (Fig. 1F; Supplemental Fig. S6A). In addition, this set of 135 high-confidence FD targets contained also the MADS box gene SEP3, the promoter of which is bound by FD and which is downregulated in fd-2 mutant (Fig. 1F; Supplemental Fig. S6A). Interestingly, we did not observe binding of FD to any of the other members of SEP gene family in ChIP-seq samples from the SAM, although we did detect FD binding in promoter regions of SEP1 and SEP2, but not SEP4, in ChIP-seq from seedlings in which FD had been misexpressed from the SUC2 promoter. One possible explanation for this is that the ChIP-seq at the SAM apparently worked less efficiently and identified fewer FD targets than in leaves (595 versus 1,754), which might result in a larger number of false negatives. In agreement with this interpretation, SEP1 is downregulated in the fd-2 mutant background (Supplemental Fig. S6), indicating that FD directly or indirectly regulates the expression of SEP1 at the SAM. Interestingly, we also found FD bound to TPR2, a member of the TOPLESS (TPL)-related gene family. TPL and its family members (TPR1, TPR2, TPR3, and TPR4) are strong transcriptional corepressors that interact with other proteins throughout the plant to modulate gene expression (Causier et al., 2012). TPR2 is downregulated in the fd-2 mutant throughout the floral transition from T0 to T5 (Supplemental Fig. S6), indicating FD might regulate development at the SAM through TPR2 in a photoperiod-independent manner.
Gene ontology (GO) analysis of these 135 genes that were bound and differentially expressed by FD revealed significant enrichment in several biological process categories (Supplemental Fig. S7), including “flower development” and “maintenance of inflorescence meristem identity,” as expected for a flowering time regulator such as FD. More surprisingly, however, genes related to the “response to hormone” category were also significantly overrepresented (Supplemental Data Set S8). Among the 27 genes in this category are four genes that are best known for their role in jasmonate signalingMYC2, JASMONATE-ZIM-DOMAIN3 (JAZ3), JAZ6, and JAZ9, three genes directly connected to auxin signaling AUXIN RESPONSE FACTOR18 (ARF18), WES1, and DWARF IN LIGHT1, four genes involved in abscisic acid signaling ALDEHYDE DEHYDROGENASE 3I1, GLYCINE-RICH DOMAIN PROTEIN1, HIGHLY ABA-INDUCED PP2C GENE1, and PROTEIN PHOSPHATASE 2CA, and the flowering-related gene SOC1, which is well-known to be regulated by gibberellins (Supplemental Data Set S8). Closer inspection of the expression profiles of these 27 candidate genes revealed that ARF18 showed a trend similar to SOC1, being strongly induced after T2 in Col-0 but not in fd-2. The four jasmonate-related genes showed a peculiar expression profile in fd-2: an increase from T0 to T1, a decrease in T2, another increase in T3, and decreasing again in T5. Because this peculiar expression profile was observed in three JAZ genes, we checked the remaining genes in this family and found that 11 out of 13 displayed a similar pattern (Supplemental Fig. S6). Furthermore, this profile was also observed in three other genes, DOWN MILDEW RESISTANT6 (DMR6), EPITHIOSPECIFIER PROTEIN (ESP), and TARGET OF EARLY ACTIVATION TAGGED EAT2 (TOE2), which have previously been implicated in pathogen resistance and the jasmonate pathway (Supplemental Fig. S7B). Taken together, these results suggest that FD plays an active role not only in the regulation of flowering time but also functions to connect different hormone signaling pathways.
Validation of FD Targets
We selected a subset of putative FD direct target genes and determined their expression in early flowering FD overexpression lines (p35S::FD) and Col-0. To minimize any bias due to the early flowering of p35S::FD, experiments were carried out in vegetative 7-d-old LD-grown seedlings. For validation, we selected genes known to play a major role in floral transition, genes that according to GO are involved in flowering time and floral development, and other genes that showed a marked differential expression in fd-2 but for which a role in flowering time regulation had not previously been studied in detail. Reverse transcription quantitative-PCR (RT-qPCR) assays confirmed that both SOC1 and AP1 were strongly upregulated in p35S::FD (Fig. 6). Although we had only found SEP3 to be bound by FD in the SAM ChIP-seq analysis, we tested expression of all four SEP genes (SEP1–SEP4) in the p35S::FD line. SEP3 was the only SEP gene that was strongly induced in seedlings in response to FD overexpression, while SEP1 showed only moderate induction. In contrast, expression of SEP2 and SEP4 did not show differences between p35S::FD and Col-0 (Fig. 6). Interestingly, SEP1, SEP2, and SEP3 were also bound by FD in PCC-specific ChIP-seq in seedlings and SEP1 and SEP3 exhibited strong differential expression in RNA-seq (Supplemental Fig. S6). ASYMMETRIC LEAVES, which has been demonstrated to be involved in flowering time by regulation of FT expression in leaves (Song et al., 2012), did not show substantial difference in expression between Col-0 and p35S::FD. We also tested two FRIGIDA-like genes, FRI-like 4a and FRI-like 4b, of which FRI-like 4b showed a decreased expression in p35S::FD. In addition, we also tested two genes, MYC2 and AFR1, which were bound by FD in both the pSUC2 and pFD ChIP-seq experiments, differentially expressed at the SAM, but not differentially bound in ft-10 tsf-1 mutant, that is, not directly influenced by the presence of FT and TSF, for their contribution to flowering time regulation. MYC2 showed no differences in expression in p35S::FD compared to Col-0, whereas AFR1 was upregulated in p35S::FD (Fig. 6). To genetically test the role of these two genes in the regulation of flowering, we isolated T-DNA insertion lines and determined their flowering time under LD at 23°C. Both myc2 and afr1 were significantly early flowering, both as days to flowering and total leaf number, compared to wild type (Fig. 7), confirming their role in regulating the floral transition.
Figure 6.
Validation of FD targets in Col-0 and p35S::FD. RT-qPCR analysis of 12 putative direct targets of FD. RNA was isolated from 15 to 20 7-d-old Col-0 and p35S::FD seedlings. Error bars = ±sd from three biological replicates.
Figure 7.
Flowering time of myc2 and afr1. Flowering time of homozygous myc2 and afr1 T-DNA insertion lines was scored as days to flowering (A) and total leaves (B). Number (n) of plants per genotype is indicated. Statistical significance was calculated using an unpaired Student’s t test compared to Col-0. ***P < 0.01, **P < 0.05. Unfilled circles represent outliers.
DISCUSSION
FD was originally identified as a component of the photoperiod-dependent flowering pathway in Arabidopsis based on the late flowering phenotype of the loss-of-function mutant (Koornneef et al., 1991). FD, which encodes a bZIP TF, is expressed at the SAM before the floral transition but does not seem to induce flowering on its own. Later, it was demonstrated that FD physically interacts with FT, the florigen, and that this interaction is important for FD’s function as a promoter of flowering (Abe et al., 2005; Wigge et al., 2005). In addition, FD was found to also interact with TFL1, which is normally expressed in the SAM and antagonizes the function of FT as floral activator. This and other findings led to the hypothesis that FD is held in an inactive state in the vegetative SAM through interaction with TFL1. When FT is induced in the PCCs and transported to the SAM in response to inductive photoperiod, FT competes with TFL1 for interaction with FD, eventually resulting in the formation of transcriptionally active FD-FT complexes (Ahn et al., 2006). However, the exact molecular mechanisms of FD action and its genome-wide targets remained largely unknown. Here we employed biochemical, genomic, and transcriptomic approaches to clarify the role of FD in the regulation of flowering transition in Arabidopsis.
We found that neither FT nor TSF are required for FD to bind to DNA but that their presence increases the enrichment of FD on a subset of target loci, which encode for known flowering time and floral homeotic genes such as AP1, SEP1, SEP2, and FUL. Our data are compatible with the model described by Ahn et al. (2006), according to which FT acts as a transcriptional coactivator. Without FT, FD would still be capable of binding to DNA but would not activate transcription. This hypothesis is supported by our finding that FD is capable of binding to DNA by itself in vitro. However, we cannot exclude the possibility that the binding of FD to DNA we observed in the pSUC2::GFP:FD ft-10 tsf-1 reporter line could be mediated by the floral repressors BROTHER OF FT and ATC, both of which are expressed in the leaf vasculature and are known to interact with FD (Yoo et al., 2010; Huang et al., 2012; Ryu et al., 2014).
In this context, our EMSA results are of particular interest as they demonstrate that, at least in vitro, TFL1 is capable of interacting with unphosphorylated FD via 14-3-3 proteins, suggesting that the transcriptionally inactive ternary FD/14-3-3/TFL1 complex could be the ground state at the SAM. Only after FD has been phosphorylated can FT, together with 14-3-3 proteins, form an active FAC to induce flowering. This requirement for phosphorylation of T282 of FD adds another safeguard to the system that might help to prevent disastrous premature induction of flowering. Our results clearly suggest that phosphorylation is important for FD function and add to our understanding concerning the role of FD phosphorylation, which had mostly been based on the analyses of a FD/14-3-3/Hd3a complex in rice using a short FD peptide (Taoka et al., 2011; Kaneko-Suzuki et al., 2018).
Which kinases regulate phosphorylation of FD in vivo has been a matter of debate, but recently two calcium-dependent kinases, CPK6 and CPK33, have been shown to phosphorylate FD (Kawamoto et al., 2015). Building on this, we show that expression of a nonphosphorable version of the FD protein (T282A) under the control of the pFD promoter failed to rescue the late flowering of fd-2. In contrast, expression of a phosphomimic version of FD (T282E) resulted in early flowering when expressed in fd-2. Similar results were obtained using a S281E phosphomimic FD. These results indicate that the phosphorylation of FD must be tightly controlled to prevent premature flowering. Interestingly, both CPK6 and CPK33 are more strongly expressed in transition apices than they are in vegetative apices (Schmid et al., 2005), which would be in agreement with an activation of FD by these two kinases during floral induction.
Somewhat surprisingly, we observed that the C-terminal part of the FD protein, which includes the bZIP domain and the phosphorylation site, was sufficient to trigger complex formation with FT, TFL1, and 14-3-3 proteins. This suggests that the N-terminal region of FD, which is predicted to be highly unstructured and contains a stretch of 25 amino acids containing 19 Ser residues, might be dispensable for FD/14-3-3/FT complex formation. However, the N-terminal region of FD is evolutionarily conserved, indicating that it may contribute to FD function. This notion is supported by our observation that expression of the C-terminal part of FD in plants only partially restored the late flowering of fd-2 mutants.
Part of the flowering promoting activity of FD can probably be expressed through its effect on members of the SEP gene family of MADS-domain TFs, which are required for the activity of the A-, B-, C-, and D-class floral homeotic genes (for review, see Theissen et al., 2016). In addition to its function as a floral homeotic gene, SEP3 has also been reported to promote flowering by accumulation in leaves under FT regulation (Teper-Bamnolker and Samach, 2005) and as a downstream target of the miR156-SPL3-FT module in response to ambient temperature (Hwan Lee et al., 2012). However, how SEP3 is regulated at the SAM has remained unclear. Interestingly, we found that FD bound strongly to the SEP3 promoter and SEP3 is downregulated in the fd-2 mutant. As FD also binds to the promoter and activated expression of the A-class gene AP1, FD activity might be sufficient to induce formation of sepals, which form the outmost floral whorl, and which according to the quartet model require the formation of a SEP/AP1 complex (Theissen et al., 2016). However, it should be noted that fd mutants do not display notable homeotic defects, indicating that FD is clearly not the only factor regulating SEP3 and AP1 expression. Furthermore, binding of FD to AP1 is unlikely to be mediated by a C-box as previously suggested (Wigge et al., 2005; Taoka et al., 2011) as the summits of the ChIP-seq peaks do not cover this region of the AP1 promoter. Interestingly, this region contains several palindromic sequences, one or more of which most likely mediate FD binding to the AP1 promoter.
Another interesting outcome of our analyses is the indication that FD might contribute to the regulation of other processes in the plant besides flowering. In particular, we found that FD directly regulated the expression of genes involved in several hormone signaling pathways. For example, we observed FD binding to the promoter of MYC2, a basic helix–loop–helix TF that plays a key role in jasmonate response. It has been shown that MYC2 forms a complex with JAZ proteins and the TPL corepressor, and that this interaction is dependent on NOVEL INTERACTOR OF JAZ proteins (Pauwels et al., 2010). In this context it is noteworthy that FD also bound directly to the promoter of TPR2 and that TPR2 was strongly downregulated in fd-2. This finding indicates that FD not only regulates MYC2 but also at least some of the interacting TPL-like transcriptional corepressors. Finally, we also observed strong binding of FD to (and misexpression of) a number of JAZ genes in either PCCs and/or the SAM in our ChIP-seq and RNA-seq data. Taken together, this indicates that FD may control the expression of three core components of jasmonate signaling: MYC2, TPR2, and several JAZ genes. These results support earlier findings that had reported a link between jasmonate signaling components and flowering time regulation. JAZ proteins have been shown to regulate flowering in leaves through the direct interaction with the floral repressors TOE1 and TOE2, which is also bound by FD and differentially expressed in fd-2, and the regulation of FLC that negatively regulates FT expression (Zhai et al., 2015). Moreover, MYC2 has also been reported to affect flowering time by regulating FT expression in leaves (Zhai et al., 2015; Wang et al., 2017). However, previous publications had reported contradictory results concerning the flowering phenotype of the myc2 mutant, ranging from late flowering (Gangappa and Chattopadhyay, 2010) to early flowering (Wang et al., 2009) or no obvious effect (Major et al., 2017). In our conditions the myc2 mutant exhibited early flowering compared to Col-0, in agreement with the report from Wang et al. (2009); Fig. 7). We also identified ARF18, a member of the auxin response factor family, as a direct target of FD. Notably, the expression of ARF18 is strongly induced after T2 in Col-0 but not in fd-2, and this pattern is the same as for known direct FD targets, for example AP1 and SOC1. Moreover, ARF18 is also induced at the SAM during the floral transition (Schmid et al., 2005), providing further evidence for a possible link between FD and ARF18. In summary, our findings suggest a link between the photoperiodic pathway gene FD and hormone signaling pathways. Although further experiments will be necessary to better understand this connection, we hypothesize that linking hormone signaling to flowering time through FD regulation might allow plants to fine-tune their flowering time response to abiotic and biotic stresses.
Apart from connecting FD with hormone signaling, we characterized another target gene in more detail, AFR1. This gene encodes a putative histone deacetylase subunit and had previously been shown to negatively affect the expression of FT in the leaves. Further, afr1 mutations cause early flowering (Fig. 7; Gu et al., 2013). Our results suggest that FD might modulate flowering through ARF1-mediated regulation of chromatin. However, such regulation would most likely not be mediated by FT, as FT is normally not expressed at the SAM.
Taken together, our results support the role of FD as a key regulator of photoperiod-induced flowering and the expression of A- and E-class floral homeotic genes in Arabidopsis. Furthermore, FD might play an important role in coordinating the crosstalk between the photoperiod pathway and hormone signaling pathways and provide a convergence point for diverse environmental and endogenous signaling pathways.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Thale cress Arabidopsis (Arabidopsis thaliana) accession Col-0 was used as wild-type. Mutants investigated in this study are: fd-2 (SALK_013288), ft-10 (GABI_290E08), tsf-1 (SALK_087522), myc2 (SALK_017005), and arf1 (SALK_026979; Supplemental Table S1). Seeds were stratified for 3 d in 0.1% agar in the dark at 4°C and directly planted on soil. Plants were grown on soil at 23°C and 65% relative humidity under wide spectrum fluorescent lights with a fluence rate of 125–175 μmol m−2 s−1. LDs and SDs are defined as 16-h light/8-h dark and 8-h light/16-h dark, respectively. Plants used for flowering time measurements were grown in a randomized design to reduce location effects in the growth chambers. Statistical significance of differences in flowering time between genotypes was calculated using an unpaired Student’s t test.
DNA Vectors and Plant Transformation
DNA vectors used in this study are listed in Supplemental Table S2. Coding sequences (CDS) were amplified by PCR from cDNA and cloned into either pGREEN-IIS vectors for flowering time studies or pET-M11 vectors for protein expression. Final constructs were transformed by electroporation in Agrobacterium tumefaciens and Arabidopsis plants of accession Col-0 and fd-2 were transformed by the floral dip method. BASTA treatment (0.1% [v/v]) was used for screening for transgenic lines.
ChIP and ChIP-seq
Approximately 1.5 g of seedlings (pSUC2::GFP:FD and pSUC2::GFP:NLS in both Col-0 and ft-10 tsf-1) or 300 mg of manually dissected apices (pFD::GFP:FD in fd-2; Col-0) from 16-d-old plants grown on soil under LD 23°C were harvested and fixed in 1% formaldehyde under vacuum for 1 h. ChIP was performed as described in Kaufmann et al. (2010) with the following minor changes: Sonication was performed using a Covaris E220 system (conditions: intensity 200 W, duty 20, cycles 200, time 120 s), incubation time with antibody was increased to over-night, incubation time with protein-A agarose beads was increased to 4 h, and purification of DNA after decrosslinking was performed with a MinElute Reaction Cleanup Kit (Qiagen).
Anti-GFP (ab290; AbCam) was used for immuno-precipitation. ChIP-seq libraries were prepared using a TruSeq ChIP Library Preparation Kit (Illumina) and the software BluePippin (Sage Science) was used for gel size selection of fragments between 200 and 500 bp. Final concentration and size distribution of the libraries were tested with Qubit and BioAnalyzer (Agilent High Sensitivity DNA Kit). Libraries were sequenced on a HiSeq3000 system (Illumina) using the 50-bp single end kit.
RNA Extraction, RNA-Seq, and Expression Analysis
For RNA-seq, Col-0 and fd-2 plants were grown for 21 d under SDs at 23°C and then shifted to LDs at 23°C. RNA was extracted from manually dissected apices collected the day of the shift (T0) and 1, 2, 3, and 5 d after shifting (T1, T2, T3, and T5, respectively) using the RNeasy Plant Kit (Qiagen) according to the manufacturer’s instructions. RNA integrity and quantification were determined on a BioAnalyzer system (Agilent). One microgram of RNA was used to prepare libraries using the TruSeq RNA Library Prep Kit (Illumina). All libraries were quality controlled and quantified by Qubit and Bioanalyzer and run on a HiSeq3000 (Illumina) with a 50-bp single end kit. Validation of the selected FD targets was performed in 7-d-old seedlings grown on soil under LDs at 23°C.
RNA was extracted using the RNeasy Plant Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthetized using the RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. RT-qPCRs were performed on a CFX96 Touch Real-time PCR Detection System (BioRad) using LightCycler 480 SYBR Green I Master (Roche). Oligonucleotides used as primers for RT-qPCR are listed in Supplemental Table S3.
ChIP-Seq and RNA-Seq Analysis
Raw data from ChIP-seq were trimmed of adapters and aligned to the Arabidopsis genome (TAIR10 release) using the software package BWA (Li and Durbin, 2010). The software MACS2 was used to call peaks using default parameters (Zhang et al., 2008). Mapped reads from samples expressing GFP:NLS under the same promoter of the GFP:FD (e.g. pSUC2) in seedlings experiments or Col-0 without any vector in apices experiments were used for normalization. Differentially bound analyses were carried out using the R package “DiffBind” using default parameters (Stark, 2011; Ross-Innes et al., 2012).
For the analysis of RNA-seq data, sequencing reads mapping to rRNAs were filtered out using Sortmerna (Kopylova et al., 2012) and the remaining reads were trimmed of adapters using Trimmomatic (Bolger et al., 2014). Alignment to the Arabidopsis genome was performed with STAR (Dobin et al., 2013) and read-counted with HTSeqCount (Anders et al., 2015). Differential expression analysis was performed using DESeq2 with default parameters (Love et al., 2014).
EMSAs
CDS of both the wild-type version as well as the phosphomimic variant (T282) of FD and its C-terminal domain (FD-C, amino acids: 203–285), 14-3-3ν (At3g02520; GRF7), FT, and TFL1 were amplified by PCR to generate N-terminal 6×-His-tag CDS, which were cloned into pETM-11 expression vectors by restriction enzyme cloning. All plasmids were transformed into Escherichia coli strain Rosetta plysS and proteins were induced with 1-mm isopropyl β-D-1-thiogalactopyranoside at 37°C over-night. Cell lysis was performed by sonication and proteins were purified using His-60 columns (Clontech) and eluted in 50 mm of sodium P buffer at pH 8.0, 300 mm of NaCl, and 300 mm of Imidazole. EMSA was performed using 5′-Cy3-labeled, double-stranded oligos of 30 bp covering the G-box contained in the SEP3 promoter as a probe (Eurofins). For probe synthesis, single-strand oligos were annealed in annealing buffer (10 mm of Tris at pH 8.0, 50 mm of NaCl, and 1 of mm EDTA at pH 8.0). Binding reactions were carried out in buffer containing 10 mm of Tris at pH 8.0, 50 mm of NaCl, 10 of μm ZnSO4, 50 mm of KCl, 2.5% glycerol, and 0.05% NP-40 in a total volume of 20 μL. The binding reaction was kept in the dark at room temperature for 20 min and then loaded in native 8% polyacrylamide gel and run in 0.5× Tris/Borate/EDTA buffer at 4°C in the dark. Results were visualized using a Typhoon imaging system.
Accession Numbers
RNA-seq and ChIP-seq data have been deposited at the European Nucleotide Archive under accession numbers PRJEB24873 and PRJEB24874, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. ChIP-seq summary statistics for the different biological replicates: pSUC2::GFP:FD in Col-0 and ft-10 tsf-1 mutant background, pFD::GFP:FD in fd-2 mutant background.
Supplemental Figure S2. Verification of comparability of controls used for normalization of FD (pSUC2::GFP:FD) ChIP-seq in Col-O (wild type) and ft-10 tsf-1 seedlings.
Supplemental Figure S3. Effect of misexpression of FD on gene expression and flowering time.
Supplemental Figure S4. EMSAs to test FD binding to the SEP3 and AP1 promoters.
Supplemental Figure S5. Summary of RNA-seq results.
Supplemental Figure S6. Expression profile of selected FD target genes.
Supplemental Figure S7. GO analysis on the subset of 135 direct genes of FD.
Supplemental Table S1. List of mutants and oligonucleotides for genotyping used in the study.
Supplemental Table S2. List of vectors used in the study.
Supplemental Table S3. List of oligonucleotides used for RT-qPCR in the study.
Supplemental Data Set S1. List of 1,754 FD-bound peaks identified in seedlings expressing pSUC2::GFP:FD in Col-0.
Supplemental Data Set S2. List of 2,696 FD-bound peaks identified in seedlings expressing pSUC2::GFP:FD in ft-10 tsf-1.
Supplemental Data Set S3. List of 1,530 FD-bound peaks detected in seedlings expressing pSUC2::GFP:FD in either Col-0 or ft-10 tsf-1.
Supplemental Data Set S4. List of 885 peaks that were differentially bound in seedlings expressing pSUC2::GFP:FD in either Col-0 or ft-10 tsf-1.
Supplemental Data Set S5. List of 595 shared FD-bound peaks in apices of the pFD::GFP:FD fd-2 rescue line.
Supplemental Data Set S6. List of DE genes.
Supplemental Data Set S7. List of 135 potential direct targets of FD.
Supplemental Data Set S8. List of 27 genes related to “response to hormone” category within the subset of the 135 direct targets of FD.
Acknowledgments
We thank Diana Saez for help in isolating homozygous myc2 and afr1 mutants, the Protein Expertise Platform at the Chemical Biological Center at Umeå University for help with protein purification, and Nicolas Delhomme from the Umeå Plant Science Centre Bioinformatics Facility for assistance with submission of sequencing data.
Footnotes
This work was supported by the Sonderforschungsbereich 1101 (Collaborative Research Centre 1101 project grant no. SFB1101/1-B04), VINNOVA, the Knut and Alice Wallenberg Foundation (research project grant no. 2016-0025 to M.S.), the Humboldt Foundation (long-term doctoral fellowship to S.C.), the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (ERC grant no. 679056 to L.Y.), and the UK Biological and Biotechnology Research Council (BBSRC grant no. BB/P013511/1 to the John Innes Centre).
Articles can be viewed without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Kim MC, Sayou C, Thévenon E, Parcy F, Nilsson O (2011) Integrating long-day flowering signals: A LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J 67: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Chattopadhyay S (2010) MYC2, a bHLH transcription factor, modulates the adult phenotype of SPA1. Plant Signal Behav 5: 1650–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, He Y (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol 11: e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K (2011) Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WW, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NC, Jane WN, Chen J, Yu TS (2012) Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J 72: 175–184 [DOI] [PubMed] [Google Scholar]
- Hwan Lee J, Joon Kim J, Ahn JH (2012) Role of SEPALLATA3 (SEP3) as a downstream gene of miR156-SPL3-FT circuitry in ambient temperature-responsive flowering. Plant Signal Behav 7: 1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F; bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM (2012) The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka K (2018) TFL1-Like proteins in rice antagonize rice FT-Like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol 59: 458–468 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kawamoto N, Sasabe M, Endo M, Machida Y, Araki T (2015) Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci Rep 5: 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Park TI, Yoo SJ, Jun AR, Ahn JH (2013) Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation. J Exp Bot 64: 1715–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H (2012) SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217 [DOI] [PubMed] [Google Scholar]
- Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major IT, Yoshida Y, Campos ML, Kapali G, Xin XF, Sugimoto K, de Oliveira Ferreira D, He SY, Howe GA (2017) Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol 215: 1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh S-S, Lee H, Choi K-R, Hong CB, Paek N-C, Kim S-G, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46: 292–299 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M, Andrés F, Coupland G (2014) Flowering responses to seasonal cues: What’s new? Curr Opin Plant Biol 21: 120–127 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Lee HJ, Seo PJ, Jung JH, Ahn JH, Park CM (2014) The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol Plant 7: 377–387 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Song YH, Lee I, Lee SY, Imaizumi T, Hong JC (2012) CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J 69: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: Time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: All roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark RBG. (2011) DiffBind: Differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Melzer R, Rümpler F (2016) MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 143: 3259–3271 [DOI] [PubMed] [Google Scholar]
- Wang H, Li Y, Pan J, Lou D, Hu Y, Yu D (2017) The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol Plant 10: 1461–1464 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH (2010) BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J 63: 241–253 [DOI] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C (2015) Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27: 2814–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]