A triad of Arabidopsis MYB transcription factors controls the production of flavonols and additional phenylpropanoids that are embedded as pollen coat material.
Abstract
The pollen wall is a complex, durable structure essential for plant reproduction. A substantial portion of phenylpropanoids (e.g. flavonols) produced by pollen grain tapetal cells are deposited in the pollen wall. Transcriptional regulation of pollen wall formation has been studied extensively, and a specific regulatory mechanism for Arabidopsis (Arabidopsis thaliana) pollen flavonol biosynthesis has been postulated. Here, metabolome and transcriptome analyses of anthers from mutant and overexpression genotypes revealed that Arabidopsis MYB99, a putative ortholog of the petunia (Petunia hybrida) floral scent regulator ODORANT1 (ODO1), controls the exclusive production of tapetum diglycosylated flavonols and hydroxycinnamic acid amides. We discovered that MYB99 acts in a regulatory triad with MYB21 and MYB24, orthologs of emission of benzenoids I and II, which together with ODO1 coregulate petunia scent biosynthesis genes. Furthermore, promoter-activation assays showed that MYB99 directs precursor supply from the Calvin cycle and oxidative pentose-phosphate pathway in primary metabolism to phenylpropanoid biosynthesis by controlling TRANSKETOLASE2 expression. We provide a model depicting the relationship between the Arabidopsis MYB triad and structural genes from primary and phenylpropanoid metabolism and compare this mechanism with petunia scent control. The discovery of orthologous protein triads producing related secondary metabolites suggests that analogous regulatory modules exist in other plants and act to regulate various branches of the intricate phenylpropanoid pathway.
The plant male gametophyte, or pollen grain, is produced within the anthers of angiosperms. In Arabidopsis (Arabidopsis thaliana), the mature pollen grain contains a large vegetative cell that is destined to form the pollen tube and two identical sperm cells that will eventually be released into the embryo sac when pollen development is complete. The two sperm cells are engulfed within the cytoplasm of the vegetative cell, and the whole tricellular structure is enclosed by a thick, sculptured, multilayered pollen wall (Owen and Makaroff, 1995). The pollen wall protects sperm cells from harsh environmental conditions such as temperature changes, extreme dehydration, and UV light. It also acts as a physical barrier, guarding the male sperm within from microbial attacks and mechanical damage. Besides its protective roles, the pollen wall can facilitate pollination by attracting desired vectors responding to its morphology, chemical components, or other physical properties. Lastly, the pollen wall can trigger fertilization, as it is crucial for pollen hydration that induces pollen tube growth; alternatively, it can prevent fertilization by discriminating between self and nonself pollen (Ariizumi and Toriyama, 2011).
Even though pollen surface morphology displays remarkable diversity between plant species, its fundamental structure is mostly conserved among taxa (Fraser et al., 2012). Pollen walls commonly exhibit two distinct layers: the intine, an inner layer similar to the primary walls of common plant cells; and the exine, an outer layer mostly consisting of a robust material called sporopollenin and a sticky material named tryphine that coats the pollen wall. The outer exine or its sporopollenin component is responsible for the unique morphological patterns displayed by pollen grains and is considered to be the most complex and tough among plant extracellular matrices (Jiang et al., 2013). Due to its durable structure, little is known about sporopollenin composition. Fourier transform infrared and NMR data combined with research on exine mutants suggest that sporopollenin is composed of complex biopolymers derived mainly from long-chain fatty acids and phenolics such as p-coumaric acid (Ahlers et al., 1999; Bubert et al., 2002; Steemans et al., 2010; Shi et al., 2011; Quilichini et al., 2015). The phenolic monomers form ester and ether linkages to in-chain hydroxy lauric acids (Morant et al., 2007), making sporopollenin similar to cutin, suberin, and lignin polymers (Scott, 1994).
An orchestrated cascade of events involving both sporophytic and gametophytic tissues is responsible for exine synthesis and its correct deposition (Sun et al., 2007). Biosynthesis of sporopollenin occurs in the tapetum, which is the innermost somatic layer surrounding the developing pollen within the anther locule. In early stages of anther development, the tapetum undergoes endomitosis as it receives signals from the developing pollen. It becomes extremely dense, with two nuclei, numerous vacuoles, ribosomes, mitochondria, and endoplasmic reticulum indicative of its high metabolic activity. Subsequently, the tapetum provides the developing pollen with proteins, lipids, and other nutrients during pollen development. During the tetrad stage of the developing pollen and after sporopollenin deposition, the tapetum undergoes programmed cell death, and following cellular degradation, all cell remnants are released within the anther locule (Blackmore et al., 2007). The tapetal degenerative debris form a sticky material, the tryphine, composed mostly of short- and long-fatty acids, hydroxycinnamic acid amides (HCAAs), a mixture of flavonoids and carotenoids (Jessen et al., 2011; Fellenberg et al., 2012b), and several types of proteins that take part in tapetal cell death (e.g. Cys proteases), pollen-stigma communication (e.g. caleosins and acetyl cholinesterases), and pollen tube growth (e.g. expansins and glucanases; Rejón et al., 2016). Tryphine is deposited in the cavities of the outer exine to form the pollen coat. Finally, once the exine structure is complete, intine synthesis commences and a fully integrated pollen wall is formed.
Transcriptional regulation of pollen wall development has been studied extensively, particularly through the investigation of male-sterile mutants in Arabidopsis and rice (Oryza sativa; Shi et al., 2015). An array of transcription factors has been identified, including the basic helix-loop-helix (bHLH) proteins DYSFUNCTIONAL TAPETUM1 (Zhang et al., 2006)/UNDEVELOPED TAPETUM1 (Jung et al., 2005) and ABORTED MICROSPORES (AMS; Xu et al., 2010)/TAPETUM DEGENERATION RETARDATION (Li et al., 2006), the PHD-type transcription factor MALE STERILITY1 (MS1; Ito et al., 2007)/PERSISTANT TAPETAL CELL1 (Li et al., 2011), and the R2R3-MYB factors MYB35 (Zhu et al., 2008) and MYB80/MS188 (Phan et al., 2011). Knockout mutants of these genes display aberrations in tapetum and/or pollen wall development. Through RNA in situ hybridizations of mutant plants, a genetic pathway for the regulation of pollen wall development was established in Arabidopsis (Zhu et al., 2011). DYSFUNCTIONAL TAPETUM1 is upstream of MYB35, followed by AMS, MS188, and finally MS1. A similar genetic pathway was partially verified in rice (Shi et al., 2015). Previous studies implicated Arabidopsis MYB99 as being regulated by MS1 (Ito et al., 2007; Yang et al., 2007a). Expression of MYB99 is specific to inflorescences, and RNA in situ hybridization localized MYB99 expression to the tapetum at stages 8 and 9 of anther development, following the tetrad stage and during sporopollenin biosynthesis and tryphine formation. myb99 knockout mutants have smaller siliques and less viable pollen compared with wild-type plants (Alves-Ferreira et al., 2007). Yet, neither the actual activity nor the genes regulated by MYB99 during pollen wall development have been reported to date.
Recent work in tomato (Solanum lycopersicum) demonstrated that accumulation of flavonols and hydroxycinnamates following heterologous expression of the Arabidopsis MYB12 transcription factor (AtMYB12) in fruit is mediated by channeling flux of primary metabolism toward Phe synthesis (Zhang et al., 2015). An interesting finding in that report was the direct promoter binding and activation of ENOLASE (ENO) in the glycolytic pathway and 3-DEOXY-D-ARABINOHEPTULOSONATE 7-PHOSPHATE SYNTHASE in the shikimate pathway by the MYB12 factor. Apart from direct control over core metabolism genes, AtMYB12 also regulates genes in core phenylpropanoid and flavonoid biosynthetic pathways, such as PHENYLALANINE AMMONIA LYASE (PAL), CHALCONE SYNTHASE (CHS), and FLAVANONE 3-HYDROXYLASE (F3H). Regulatory control of the interface between primary and secondary metabolism at the transcriptional level has only been described in intermediate, precursor pathways (e.g. the shikimate and mevalonate pathways; Aharoni and Galili, 2011). One of the early examples suggesting such transcriptional control was ORCA3 activity in Catharanthus roseus, in which a jasmonate-responsive AP2 domain transcription factor appears to coregulate terpenoid indole alkaloid biosynthetic genes as well as upstream genes such as TRYPTOPHAN DECARBOXYLASE required for indole metabolism, GERANIOL 10-HYDROXYLASE, and a CYTOCHROME P450 REDUCTASE required for monoterpene biosynthesis (Vom Endt et al., 2007). In another example, the petunia (Petunia hybrida) ODORANT1 (ODO1), an R2R3-type MYB transcription factor, controls genes of the shikimate pathway, channeling precursors for Phe biosynthesis and consequently the production of benzenoids, phenylpropanoid pathway-derived floral scent components (Verdonk et al., 2005). ODO1 transcription is controlled by two other R2R3-MYB factors related by sequence, EMISSION OF BENZENOIDS I and II (EOBI and EOBII), which together with ODO1 coregulate genes of the benzenoid biosynthetic pathway (Van Moerkercke et al., 2011; Spitzer-Rimon et al., 2012). EOBI and EOBII are phylogenetically related to the Arabidopsis MYB21 and MYB24 transcription factors that promote stamen growth and development (Song et al., 2011). The involvement of MYB21 and MYB24 protein putative orthologs in phenylpropanoid metabolism has been demonstrated to a certain extent in several plant species in addition to petunia. In Arabidopsis, the ectopic expression of either one of these genes triggers up-regulation of PAL transcripts (Shin et al., 2002; Yang et al., 2007b). Their orthologs in both snapdragon (Antirrhinum majus) and pea (Pisum sativum) directly regulate the transcription of PAL as well as other phenylpropanoid pathway genes (Moyano et al., 1996; Uimari and Strommer, 1997). Nevertheless, in all these reports, the class of metabolites in the phenylpropanoid pathway regulated by these factors was not revealed.
The Arabidopsis MYB99 protein, a close ortholog of petunia ODO1, was the main focus of this study. This regulatory protein is present in Arabidopsis tapetum cells, and its expression is regulated by the MS1 transcription factor. The absence of volatile benzenoids in Arabidopsis flowers suggests a role for MYB99 in controlling the production of a different class of secondary metabolites. Detailed metabolic and gene expression experiments in anthers of mutant and overexpression genotypes revealed that, similar to petunia ODO1, MYB99 is involved in the phenylpropanoid pathway. Yet, it controls the biosynthesis of different metabolite classes associated with the biosynthesis of pollen wall components (tryphine and possibly sporopollenin) in the tapetum. We also discovered that MYB99 controls the expression of TRANSKETOLASE2 (TK2), encoding a key upstream enzyme of primary metabolism, as well as genes of the phenylpropanoid pathway. Remarkably, MYB99 acts as part of a regulatory triad with MYB21 and MYB24, orthologs of EOBI and EOBII, respectively, controlling anther phenylpropanoid production in Arabidopsis. The occurrence of orthologous protein triads in two distinct species points to a conserved regulatory mechanism that controls diverse branches of the phenylpropanoid metabolic network.
RESULTS
Arabidopsis MYB99 Is One of Six MYB Family Subclade Members Associated with Branches of the Composite Phenylpropanoid Pathway
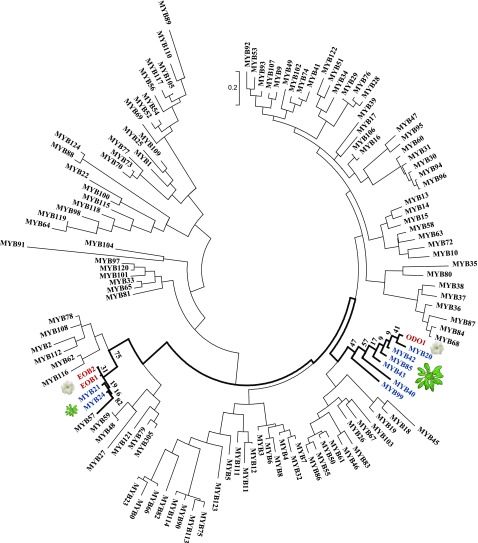
A tapetum-specific regulatory mechanism for flavonol biosynthesis has been suggested in Arabidopsis (Stracke et al., 2010). The myb11/myb12/myb111 triple mutant, which does not accumulate glycosylated flavonols in various organs of the plant, apparently retains its pollen flavonol composition. To learn more about the regulation of pollen flavonol production in Arabidopsis, we examined other members of the R2R3-type MYB protein family. Phylogenetic analysis representing 124 Arabidopsis R2R3-type MYB family proteins revealed a subclade of six proteins associated with branches of the composite phenylpropanoid pathway (Fig. 1). In Arabidopsis, MYB42 is predicted to be a regulator of phenylpropanoid metabolism (Rogers et al., 2005; Alves-Ferreira et al., 2007), while MYB85 overexpression leads to ectopic deposition of lignin in stem epidermal and cortical cells (Zhong et al., 2008). Orthologs of MYB42 and MYB85 in poplar (Populus tomentosa) activate the lignin biosynthetic pathway during cell wall formation (Li et al., 2015a). Japanese cedar tree (Cryptomeria japonica) orthologs of Arabidopsis MYB20 and MYB43 are implicated in the regulation of secondary cell wall biosynthesis in the xylem (Mishima et al., 2014). An ortholog of MYB99 in snapdragon affects monolignol production in transgenic tobacco (Nicotiana tabacum) by activating core structural genes in the phenylpropanoid pathway (Tamagnone et al., 1998). Additionally, sequence alignment indicated that the conserved domain responsible for the interactions with R-like bHLH proteins was absent in the R2R3 domain of this subclade members (Supplemental Fig. S1). Considering the functional evidence from members of the Arabidopsis subclade and their orthologs as well as the association between MYB99 and MS1 (Yang et al., 2007a), we selected MYB99 as a candidate for controlling the production of pollen flavonols in the tapetum.
Figure 1.
Schematic representation of the evolutionary relationship of the 126 Arabidopsis R2R3-MYB family proteins and the petunia ODO1, EOBI, and EOBII proteins. Arabidopsis proteins relevant to this research are marked in blue, while petunia proteins are marked red. Protein sequences were aligned using the MUSCLE algorithm, and then the unconserved regions in N and C termini were removed in an unbiased manner. The tree was inferred using the maximum-likelihood method and 1,000 bootstraps using MEGA 7. The MYB99 subclade, MYB21/MYB24, and EOBI/EOBII are highlighted. Bootstrap values are presented as percentages, and the scale bar indicates 0.2 amino acid substitutions per position.
Pollen Viability and Exine Architecture Are Compromised in myb99 Mutants
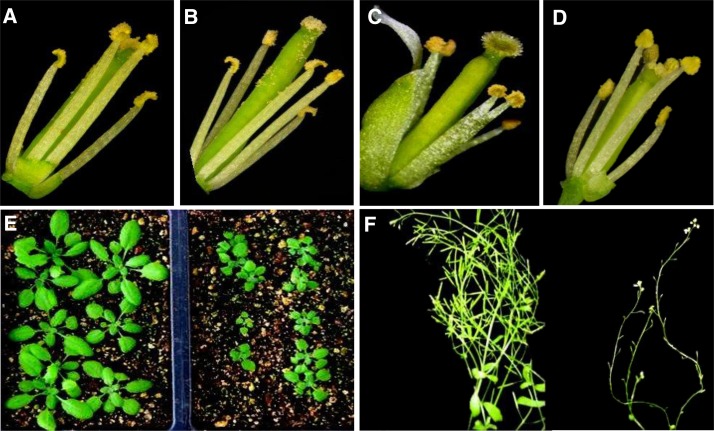
To evaluate the involvement of MYB99 in pollen development, two transfer DNA (T-DNA) insertion lines, myb99-1 (SALK_003193) and myb99-2 (SALK_052877), were obtained and selected for homozygosity (Supplemental Data Set S1). In both mutants, T-DNA insertions were localized to the second exon of MYB99, resulting in loss of function of these genes in homozygous mutant plants. We performed reverse transcription quantitative PCR (RT-qPCR) on isolated anthers obtained from preanthesis flowers of these two mutant lines, employing oligonucleotides amplifying the region upstream of the insertion. Significant down-regulation of MYB99 expression was detected in both mutants (Supplemental Fig. S2). It was previously reported that myb99-1 forms smaller siliques with only a few viable seeds (Alves-Ferreira et al., 2007); however, under our growth conditions, siliques developed normally and no sterility could be observed in the mutant lines. Examination of postanthesis flowers revealed that myb99 stamen filaments were shorter than those in wild-type flowers, pointing to defects in stamen development (Fig. 2, A and B).
Figure 2.
Phenotypic characterization of plants with altered MYB99 expression. A, Open wild-type flower after removal of sepals and petals. B, myb99 knockout mutant with shorter stamen filaments. C, Stamen filaments of a MYB99-overexpressing plant with dwarf phenotype are shorter than the gynoecium, disrupting pollen adhesion to the stigma. D, Normal-looking anthers from MYB99-ox1. E, Plants overexpressing MYB99 (right) exhibit stunted growth when compared with wild-type plants (left). F, Sterile MYB99-overexpressing plant with undeveloped siliques (right) compared with a wild-type plant (left) of the same age.
To further evaluate the effect of MYB99 deficiency on pollen wall formation, pollen grains obtained from anthers of wild-type plants and myb99 mutants were examined for their viability by Alexander staining (Alexander, 1969). While wild-type pollen grains appeared viable (stained in red), about 20% of the pollen grains (n = 114) obtained from myb99-1 and myb99-2 were aborted (stained in turquoise; Fig. 3, A and B). This indicated that pollen viability in myb99 mutants is compromised. We subsequently examined mutant and wild-type pollen grains in their native state (either stuck to the anther locule or trapped by the stigma surface) under wet-mode environmental scanning electron microscopy (SEM). This analysis confirmed the occurrence of pollen displaying either distorted or normal morphology in myb99. While normal-appearing pollen adhered to the stigma in both the wild type and myb99, we observed distorted pollen grains on the anther locule of myb99 but not in wild-type plants. Malformations in pollen wall architecture were evident in myb99 distorted pollen grains, which appeared shriveled and collapsed. In the most severe cases, we observed irregular patterning of the pollen surface (Fig. 3, C and D).
Figure 3.
Malformation of pollen grains derived from myb99 mutants. A, Pollen grains of wild-type plants after Alexander staining. Wild-type pollen was stained red, indicative of viable pollen with an intact exine wall. B, Arrows point to aborted pollen grains isolated from myb99 mutants after Alexander staining. C, SEM of pollen grains adhered to the anther locule derived from wild-type plants displaying normal morphology. D, Pollen grains of myb99-1 mutants that adhered to the anther locule appeared malformed and collapsed. Arrows point to pollen surface malformations.
Flavonol Glycoside and HCAA Levels Are Decreased in myb99 Anthers
To examine the effect of MYB99 deficiency on anther metabolism, we carried out untargeted metabolic analysis on dissected anthers (stages 8 and 9; Sanders et al., 1999) of myb99-1 and wild-type preanthesis flowers (see “Materials and Methods”). Using an ultra-HPLC device coupled to a quadrupole time-of-flight mass spectrometer (positive and negative ionization modes), we detected a total of 221 mass signals that were significantly decreased in the myb99 mutant anthers and only nine mass signal ions that showed an increase compared with the wild type. Analysis of these mass signals resulted in putative identification of 24 metabolites that were significantly decreased in myb99 mutant anthers, 20 of which were annotated by using exact mass calculations and comparing their relative retention times, UV spectra, and mass fragments with previously published data. Confirmation for mass fragmentation spectra of each metabolite was obtained by performing tandem mass spectrometry daughter ion scans on all of the annotated compounds (Supplemental Data Set S2). Remarkably, the 20 annotated metabolites were all products of phenylpropanoid metabolism. These could be categorized into two major chemical classes: (1) flavonol glycosides, such as kaempferol, quercetin, or isorhamnetin conjugated with up to three sugar moieties; and (2) polyamine conjugates, which incorporate a spermidine backbone that is frequently acylated with phenylpropanoids and/or sugars, forming HCAAs. The most prominent changes were detected for naringenin chalcone and the pollen-specific flavonols kaempferol and quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides (Table 1). Remarkably, we observed a 3-fold increase in one of the detected HCAAs, N-caffeoyl-N′-feruloyl spermidine, although this compound likely represents a small part of the total HCAA content in Arabidopsis pollen. A total of six flavonol glycosides and 13 HCAAs were decreased in myb99-1 anthers. Targeted metabolic analysis of myb99-1 anthers performed with an ultra-HPLC device coupled to a triple-quadrupole mass spectrometer and multiple reaction monitoring confirmed a significant reduction in levels of naringenin chalcone. The complete list of fold change values for each compound can be found in Supplemental Table S1.
Table 1. Annotation and selected analytical data for metabolites identified by comparative nontargeted profiling of the wild type versus myb99, myb21, myb24, and myb21/myb24 knockout mutant lines.
The major and specific pollen flavonols are underlined, and metabolites that showed significant changes compared with the wild type are marked with D for decreased and I for increased (P < 0.05, calculated using one-way ANOVA) or with a dash when unchanged. Metabolite quantification was verified using the multiple reaction monitoring mode in an ultra-HPLC device coupled to a triple-quadrupole mass spectrometer.
| Metabolite Name | Elemental Composition | myb99 | myb21 | myb24 | myb21/myb24 |
|---|---|---|---|---|---|
| Naringenin chalcone | C15H12O5 | D | I | D | D |
| Isorhamnetin dideoxyhexose | C28H32O15 | – | D | D | – |
| Isorhamnetin dihexose | C28H32O17 | D | D | D | D |
| Isorhamnetin-hexose-deoxyhexose | C28H32O16 | D | D | D | – |
| Isorhamnetin-pentose-deoxyhexose | C27H30O15 | – | – | D | – |
| Kaempferol deoxyhexose-hexose-deoxyhexose | C33H40O19 | – | – | D | – |
| Kaempferol dideoxyhexose | C27H30O14 | – | – | D | – |
| Kaempferol tri-O-hexoside | C33H40O21 | D | D | D | D |
| Kaempferol 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides | C27H30O16 | D | – | D | – |
| Quercetin deoxyhexose pentose | C26H28O15 | – | – | D | – |
| Quercetin deoxyhexose-hexose-deoxyhexose | C33H40O20 | – | – | D | – |
| Quercetin dideoxyhexose | C27H30O15 | – | – | D | – |
| Quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides | C27H30O17 | D | – | D | – |
| Quercetin-hexose-deoxyhexose | C27H30O16 | – | D | D | – |
| N-Feruloyl spermidine | C17H27N3O3 | – | D | D | D |
| N-Caffeoyl-N′-feruloyl spermidine | C26H33N3O6 | I | D | D | D |
| N-Feruloyl-N′-sinapoyl spermidine | C28H37N3O7 | D | D | D | D |
| N-Feruloyl-N′-(5-hydroxyferuloyl)-spermidine | C27H35N3O7 | D | D | D | D |
| N-Feruloyl-N′-(5-hydroxyferuloyl)-N″-sinapoylspermidine | C38H45N3O12 | D | D | D | D |
| N-(5-Hydroxyferuloyl)-N′,N″-bis-sinapoyl spermidine | C39H47N3O12 | D | D | D | D |
| N-(5-Hydroxyferuloyl)-N′-sinapoyl spermidine | C28H37N3O8 | D | D | D | D |
| N-(p-Coumaroyl)-N′-(5-hydroxyferuloyl)-N″-sinapoyl | C37H43N3O11 | D | D | D | D |
| N,N′-Bis-feruloyl spermidine1 | C27H35N3O6 | – | D | D | D |
| N,N′-Bis-feruloyl spermidine2 | C27H35N3O6 | D | D | D | D |
| N,N′-Bis-feruloyl spermidine-O-hexoside1 | C33H45N3O11 | D | D | D | D |
| N,N′-Bis-feruloyl spermidine-O-hexoside2 | C33H45N3O11 | D | D | D | D |
| N,N′-Bis-feruloyl-N″-(5-hydroxyferuloyl)spermidine | C37H43N3O10 | D | D | D | D |
| N,N'-Bis-coumaroylspermidine-O-hexoside | C31H41N3O9 | – | D | D | D |
| N,N′-Bis-(5-hydroxyferuloyl)-N″-sinapoyl spermidine | C38H45N3O12 | D | D | D | D |
| N,N′N″-Tris-feruloyl spermidine | C37H43N3O9 | D | D | D | D |
| N,N′,N″-Tris-(5-hydroxyferuloyl)-spermidine | C37H43N3O12 | D | D | D | D |
To rescue the metabolic phenotype observed in myb99 mutant anthers, we generated Arabidopsis myb99-1 plants expressing the MYB99 gene fused to the human influenza hemagglutinin tag (HA-tag) at its N terminus (MYB99-HA) under the control of its putative native promoter sequence (myb99-1/MYB99-HA). The presence of the MYB99-HA-tagged protein was confirmed in Arabidopsis floral buds using an anti-HA-tag antibody (Supplemental Fig. S3A). Targeted metabolic analysis of wild-type, myb99-1, and myb99-1/MYB99-HA floral buds confirmed that decreased accumulation of the pollen-specific flavonol glycosides kaempferol and quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides could be restored by reintroducing the MYB99 gene into the myb99-1 mutant background (Supplemental Fig. S3B).
Hydroxycinnamic Acid Esters Accumulate in Leaves of MYB99-Overexpressing Plants
Arabidopsis plants overexpressing MYB99 driven by the Cauliflower mosaic virus 35S promoter were subsequently generated (termed MYB99-ox). First-generation plants had two distinct phenotypes: those developing normally, which could not be distinguished from the wild type, and others showing varying degrees of dwarfism in early growth stages and sterility in the later stages (Fig. 2E). Dwarfism in Arabidopsis is typically linked with reduction of lignin content and/or stress response. Another possible explanation for this phenotype is accumulation of flavonoids such as flavonol glycosides or acylated anthocyanin (Besseau et al., 2007). These flavonoids (not a decrease in lignin content) affected auxin transport, subsequently resulting in dwarfed plants. Furthermore, mature transgenic plants showed reduced fertility and in some cases were completely sterile, possessing smaller siliques that contained either few or no seeds at all (Fig. 2F). To examine the cause of sterility, we performed reciprocal crosses between wild-type and MYB99-ox plants. In all pollination events (n = 5), fertilization was successful, indicating that sterility was not caused by defects in pollen development or in the function of the female gametophyte. Inspection of postanthesis flowers revealed that the stamen filaments of transgenic plants failed to reach the stigma and, as a result, pollen adherence to the stigma did not occur (Fig. 2C).
To examine the metabolic alterations in MYB99-overexpressing plants, we selected transgenic plants (lines MYB99-ox1 and MYB99-ox2) that were most similar in their size and appearance to wild-type plants and displayed MYB99 transcript overexpression (Supplemental Fig. S2). These transgenic plants were fertile and did not show any developmental impairment in the anthers of postanthesis flowers (Fig. 2D). Untargeted metabolic analysis of leaves from wild-type and MYB99-ox plants resulted in the identification of 105 differential mass signals; 102 of these showed a significant increase in the leaves of the transgenic plants compared with the wild type, and only three ions showed a decrease. From this list, we putatively identified and annotated 11 metabolites that accumulated in MYB99-ox plants, 10 of which were products of p-coumaric acid metabolism (Table 2). The CINNAMATE 4-HYDROXYLASE (C4H) enzyme catalyzes the hydroxylation of cinnamic acid to p-coumaric acid, which is further used for the production of the hydroxycinnamate intermediates ferulic and caffeic acids and subsequently sinapic acid. These acids can then be esterified with various sugar molecules, resulting in the biosynthesis of a variety of hydroxycinnamic acid esters (HCAEs). Leaves of MYB99-ox lines accumulated p-coumaric acid and eight HCAEs (Table 2). An increase in a diglycosylated quercetin flavonol was evident in MYB99-ox leaves, demonstrating the impact of MYB99 on the production of flavonol glycosides as well as HCAEs. Interestingly, overexpression of MYB99 also led to an increased level of the Met-derived glucosinolate 8-methylsulfinyloctyl glucosinolate.
Table 2. Annotation and selected analytical data for compounds identified by comparative nontargeted metabolite profiling of leaves derived from the wild type and MYB99 overexpression lines.
The metabolites listed were up-regulated in the MYB99 overexpression lines. Significant (P < 0.05, calculated using one-way ANOVA) fold change values presented are the result of four biological replicates from MYB99-ox1 and the wild type. Similar changes were also detected when MYB99-ox2 was compared with the wild type.
| Metabolite Name | Elemental Composition | MYB99-ox1 |
|---|---|---|
| p-Coumaric acid | C9H8O3 | 2.51 |
| Hydrocinnamic acid hexose | C15H20O8 | 2.41 |
| Hydroxyferuloyl hexose1 | C16H20O10 | 24.72 |
| Hydroxyferuloyl hexose2 | C16H19O10 | 4.89 |
| Sinapoyl hexose1 | C17H22O10 | 4.99 |
| Sinapoyl hexose2 | C17H22O10 | 3.93 |
| Disinapoylglucose1 | C28H32O14 | 8.27 |
| Disinapoylglucose2 | C28H32O14 | 3.64 |
| Disinapoylglucose3 | C28H32O14 | 6.39 |
| Quercetin hexose-deoxyhexose | C27H30O16 | 2.40 |
| 8-Methylsulfinyloctyl glucosinolate | C16H31NO10S3 | 3.14 |
Overexpression of MYB99 Up-Regulates Gene Expression Associated with Both Primary and Secondary Metabolism in Leaves
To examine the transcriptional changes that occurred in MYB99-overexpressing plants, we performed transcriptome analysis of leaves derived from 18-d-old wild-type and MYB99-ox1 plants. We detected a set of 401 genes that were significantly (P < 0.05, calculated with one-way ANOVA) altered in MYB99-ox1; 265 and 136 genes were up-regulated and down-regulated, respectively (Supplemental Data Set S3). Since overexpression of MYB99 in leaves resulted in the accumulation of HCAEs and flavonol glycosides, we examined the expression of genes associated with the phenylpropanoid pathway. We subsequently identified up-regulation of 13 structural genes participating in phenylpropanoid metabolism, including those associated with the core pathway such as PAL, C4H, and 4-COUMARATE:COENZYME A LIGASE (4CL; Table 3). The respective enzymes encoded by these three genes catalyze the initial reactions of the general phenylpropanoid pathway and are therefore essential for the production of both HCAEs and flavonol glycosides. We also detected an increase in expression of CINNAMOYL:COENZYME A REDUCTASE (CCR) family genes, which included the previously characterized CCR1 and two more genes with putative CCR activity, which we termed CCR3 and CCR4. In Arabidopsis, the CCR enzyme participates in lignin biosynthesis by increasing monolignol production via the conversion of p-coumaroyl-CoA to coumaraldehyde (Lauvergeat et al., 2001). However, freehand sectioning of MYB99-overexpressing plant stems followed by phloroglucinol staining did not reveal any evidence of ectopic lignification (Supplemental Fig. S4). The highest changes in expression were detected for CHS, which displayed over a 60-fold increase in expression as compared with the wild type. The CHS protein catalyzes the conversion of p-coumaroyl-CoA to naringenin chalcone, which drives phenylpropanoid metabolism toward flavonol biosynthesis. Subsequent reactions in flavonol biosynthesis are performed by F3H and CAFFEATE O-METHYLTRANSFERASE (OMT; Muzac et al., 2000), both of which were up-regulated in this experiment. Interestingly, the CAFFEOYL-COENZYME A 3-O-METHYLTRANSFERASE (CCoAOMT) protein, which was previously shown to localize to tapetal cells in Arabidopsis, where it contributes to the production of HCAAs (Fellenberg et al., 2012a), was also up-regulated (Table 3).
Table 3. Selected genes that were up-regulated in MYB99-ox1 compared with the wild type in mature leaves as detected by microarray analysis.
| Gene Name | Locus | Description | Metabolic Pathway | MYB99-ox1 > Wild Type |
|---|---|---|---|---|
| ENO1 | AT1G74030 | Phosphoenolpyruvate enolase | Glycolysis | 7.6 |
| G6PD6 | AT5G40760 | Glc-6-P 1-dehydrogenase | Glycolysis | 4.7 |
| PFPα1 | AT1G20950 | Phosphofructokinase family gene | Glycolysis | 2.5 |
| PFPα2 | AT1G76550 | Phosphofructokinase family gene | Glycolysis | 4.4 |
| PFPβ1 | AT1G12000 | Phosphofructokinase family gene | Glycolysis | 3.6 |
| PGK | AT1G79550 | Phosphoglycerate kinase | Glycolysis | 2.3 |
| PGD2 | AT3G02360 | 6-Phosphogluconate dehydrogenase | OPPP | 2.4 |
| TK2 | AT2G45290 | Transketolase | OPPP and Calvin cycle | 16.9 |
| RP3E | AT1G63290 | Ribulose-phosphate 3-epimerase | OPPP | 3.7 |
| 3DQS | AT5G66120 | 3-Dehydroquinate synthase | Shikimate | 4.1 |
| DHQ-SDH | AT3G06350 | Dehydroquinate-shikimate dehydrogenase | Shikimate | 3.6 |
| EPSPS | AT1G48860 | 5-Enolpyruvylshikimate-3-phosphate synthase | Shikimate | 3.6 |
| EMB1144 | AT1G48850 | Chorismate synthase, putative | Shikimate | 3.1 |
| PAL1 | AT2G37040 | Phe ammonia lyase | Phenylpropanoid | 22.5 |
| PAL2 | AT3G53260 | Phe ammonia lyase | Phenylpropanoid | 14.2 |
| PAL4 | AT3G10340 | Phe ammonia lyase | Phenylpropanoid | 36.5 |
| C4H | AT2G30490 | Cinnamic acid 4-hydroxylase | Phenylpropanoid | 3.5 |
| 4CL2 | AT3G21240 | 4-Coumarate:CoA ligase | Phenylpropanoid | 2.8 |
| 4CL3 | AT1G65060 | 4-Coumarate:CoA ligase | Phenylpropanoid | 14.1 |
| CCR1 | AT1G15950 | Cinnamoyl-CoA reductase | Phenylpropanoid | 2.6 |
| CCR3 | AT5G14700 | Putative cinnamoyl-CoA reductase activity | Phenylpropanoid | 9.5 |
| CCR4 | AT2G23910 | Putative cinnamoyl-CoA reductase activity | Phenylpropanoid | 2.4 |
| CHS | AT5G13930 | Chalcone synthase | Phenylpropanoid | 65.8 |
| F3H | AT3G51240 | Flavanone 3-hydroxylase | Phenylpropanoid | 12 |
| OMT1 | AT5G54160 | Caffeate O-methyltransferase | Phenylpropanoid | 3.8 |
| CCoAOMT1 | AT4G34050 | Caffeoyl-CoA 3-O-methyltransferase | Phenylpropanoid | 2.7 |
The up-regulation of genes in phenylpropanoid metabolism was complemented by the up-regulation of genes associated with primary metabolism, such as in glycolysis, the oxidative pentose phosphate pathway (OPPP), and the Calvin cycle (Table 3). Products of these three pathways are used as substrates for enzymes in the shikimate pathway, ultimately leading to the production of Phe. The highest level of up-regulation of genes participating in primary metabolism was detected for TK. In plants, TK catalyzes reversible reactions in the Calvin cycle and OPPP, and by doing so it directly regulates the levels of erythrose-4-phosphate, one of the substrates required for the shikimate pathway. We also detected three shikimate pathway genes that were up-regulated in this experiment, 3-DEHYDROQUINATE SYNTHASE (3DQS), 5-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS), and DEHYDROQUINATE-SHIKIMATE DEHYDROGENASE (DHQ-SDH), as well as a putative chorismate synthase. Next, we examined the list of down-regulated genes in MYB99-ox1 plants; however, we could not detect any decrease in primary metabolism- or phenylpropanoid-related genes. Taken together, these results demonstrated that overexpression of MYB99 resulted in up-regulation (in leaves) of a suite of genes participating in both primary and phenylpropanoid metabolism, resulting in the activation of the entire metabolic network associated with the phenylpropanoid pathway.
MYB99 Coregulates Primary and Phenylpropanoid Metabolism in Anthers
Transcriptome analysis was subsequently performed on manually dissected anthers harvested prior to anthesis from myb99-1, MYB99-ox1, and wild-type plants. A total of 4,247 genes were down-regulated in the myb99-1 mutant [log2 (myb99-1/wild type) < −0.35], while 2,513 genes were up-regulated in MYB99-ox1 [log2 (MYB99-ox1/wild type) > 0.35]. A concise set of 148 transcripts showed altered expression in both data sets that corresponded with the expression of MYB99 (i.e. decreased in myb99-1 and increased in MYB99-ox1; Supplemental Data Set S4). The 148-gene set included core phenylpropanoid genes such as PAL1, PAL2, 4CL2, and CAFFEOYL SHIKIMATE ESTERASE (CSE), which plays a major role in lignin production (Table 4; Vanholme et al., 2013). We also identified two UDP-GLUCURONOSYLTRANSFERASE (UGT) genes (UGT72B1 and UGT73B2) that are potentially involved in the glycosylation of anther flavonols. The ability of these UGT proteins to glycosylate phenylpropanoids was previously demonstrated using in vitro enzyme assays, although never with relation to specific flavonoids or pollen development. While UGT72B1 was shown earlier to catalyze the glycosylation of monolignols (Lin et al., 2016), UGT73B2 was reported to accept various flavonoids as substrates for glycosylation, preferably at the 3-hydroxyl group (Kim et al., 2006). We used the Arabidopsis eFP browser that plots gene expression data onto a diagrammed representation of an Arabidopsis plant (Winter et al., 2007) to examine the expression profiles of UGT72B1 and UGT73B2. According to the eFP browser, the expression of UGT73B2 began prior to flower anthesis and was highest in the stamens, while the expression of UGT72B1 was not specific to any plant tissue and was highest in young leaves (Supplemental Fig. S5). This expression pattern of UGT73B2 corresponded well to the expression profile of MYB99, and thus the UGT73B2 enzyme appeared as a strong candidate for the glycosylation of pollen flavonols. Altered MYB99 expression also affected the transcription of the stamen growth regulator MYB21 (Table 4). Notably, a previous study showed that jasmonate-induced stamen growth is mediated via MYB21 and MYB24 and that myb21, myb24, and myb21/24 all suffer from lower fertility due to male sterility (Song et al., 2011).
Table 4. Selected genes that were both down-regulated in the anthers of myb99-1 and up-regulated in the anthers of MYB99-ox1 when compared with the wild type (detected in transcriptome analysis).
| Gene Name | Locus | Description | Function | Reference |
|---|---|---|---|---|
| UGT73B2 | AT4G34135 | UDP-glucosyltransferase | Kaempferol 3-O-UDP-Glc transferase | Kim et al. (2006) |
| MYB21 | AT3G27810 | R2R3-MYB family transcription factor | Stamen development | Song et al. (2011) |
| 4CL2 | AT3G21240 | 4-Coumarate:CoA ligase | Core phenylpropanoid metabolism | Li et al. (2015b) |
| UGT72B1 | AT4G01070 | UDP-glycosyltransferase | Monolignol glycosylation | Lin et al. (2016) |
| PAL1 | AT2G37040 | Phe ammonia lyase | Core phenylpropanoid metabolism | Rohde et al. (2004); Huang et al. (2010) |
| PAL2 | AT3G53260 | Phe ammonia lyase | Core phenylpropanoid metabolism | Rohde et al. (2004) |
| CSE | AT1G52760 | Caffeoyl shikimate esterase | Lignin production | Vanholme et al. (2013) |
In order to support the gene expression analysis and identify additional potential downstream targets of MYB99, we performed coexpression analysis using the ATTED-II Network Drawer (Obayashi et al., 2009). The gene encoding TK2 (see above) was highly coexpressed with three core phenylpropanoid pathway genes, PAL, HYDROXYCINNAMOYL TRANSFERASE, and 4CL, as well as the lignin biosynthesis gene CSE, indicating its importance in phenylpropanoid biosynthesis. Interestingly, we could not detect any phenylpropanoid pathway genes coexpressed with TK1 (Supplemental Fig. S6). The two LESS ADDHESIVE POLLEN genes, LAP5 and LAP6, reported to take part in pollen fatty acid metabolism in the tapetum for sporopollenin production (Dobritsa et al., 2010) were highly coexpressed with MYB99, suggesting these genes as potential MYB99 downstream targets. An additional UGT, namely UGT79B6, appeared as a possible candidate, as it was previously shown to be responsible for the addition of a second Glc group to pollen flavonols in Arabidopsis tapetum prior to flower anthesis (Yonekura-Sakakibara et al., 2014). Finally, another potential gene that could be downstream of MYB99 was MYB24, which according to previous reports acts together with MYB21 in relation to stamen and petal development (Reeves et al., 2012).
To corroborate the transcriptomics and coexpression data, we performed RT-qPCR on selected genes in anthers from preanthesis flowers of myb99-1 and wild-type plants. Altogether, down-regulation in transcript levels was confirmed for various genes participating in phenylpropanoid metabolism, including PAL1, PAL4, 4CL2, CSE, UGT72B1, and UGT73B2, or primary metabolism, namely TK2, as well as in lipid metabolism, LAP5 and LAP6. Notably, the transcripts of MYB21 and MYB24 were almost undetectable in myb99 anthers (Fig. 4). In addition, transcripts of PHOSPHOFRUCTOKINASEα2 (PFPα2), PFPβ1, RIBULOSE-PHOSPHATE 3-EPIMERASE (RP3E), 3DQS, EPSPS, 4CL3, CCR3, CHS, F3H, and OMT1 did not show any significant change and are likely not directly regulated by MYB99 (Supplemental Fig. S7).
Figure 4.
myb99 knockout mutation leads to down-regulation in expression of genes in primary and phenylpropanoid metabolic pathways. RT-qPCR of anthers collected from preanthesis flowers of the wild type (WT) and myb99-1 mutants is shown. Down-regulation of TK2, PAL1, PAL4, 4CL2, LAP5, LAP6, CSE, UGT72B1, and UGT73B2 metabolic genes was detected in myb99-1. Significant decrease in expression was also measured for the transcripts of MYB21 and MYB24. Shown are means and se of four biological replicates. Asterisks denote statistically significant differences from the wild type calculated using Student’s t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Recombinant UGT73B2 and UGT79B6 Generate the Archetypal Arabidopsis Anther Diglycosylated Flavonols
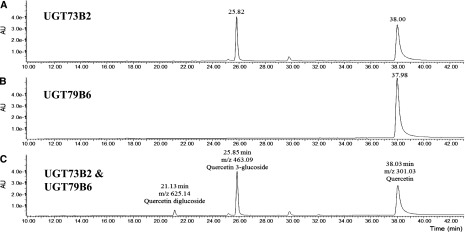
A previous report showed that UGT79B6 catalyzes the terminal glycosylation of the archetypal flavonols produced in Arabidopsis anthers (Yonekura-Sakakibara et al., 2014). These pollen-specific flavonols in fact undergo two consecutive glycosylation steps. First, a Glc is added to the third position of the flavonoid C-ring, resulting in the production of flavonol 3-O-glycoside intermediates. The second or terminal Glc is then added to the sugar moiety attached to the flavonoid aglycons, resulting in the production of diglycosylated flavonols. According to transcriptomic data previously obtained from ms1 mutants, UGT78D2 was suspected to catalyze the first glycosylation step in Arabidopsis anthers (Yonekura-Sakakibara et al., 2014). Our study here suggested that a different protein, UGT73B2, is associated with pollen flavonol glycosylation and is possibly a downstream target of MYB99. To evaluate the activity of UGT73B2 and UGT79B6 in the production of pollen diglycosylated flavonols, we expressed the two enzymes in Escherichia coli cells and examined their activity (either individual or when coincubated) with either kaempferol or quercetin and UDP-Glc. While the UGT73B2 protein catalyzed the conversion of kaempferol or quercetin to their respective glycosylated products (i.e. quercetin 3-glucoside), the UGT79B6 protein showed no activity toward these flavonols (Fig. 5, A and B). As expected, UGT73B2 favored the primary glycosylation at the 3-hydroxyl position, which was in agreement with the occurrence of flavonol-3-diglucosides in the Arabidopsis pollen. Importantly, coincubation of the two UGTs (i.e. UGT73B2 and UGT79B6) with either kaempferol or quercetin (and UDP-Glc) resulted in the production of diglycosylated flavonols, identical to the ones found in Arabidopsis pollen (Fig. 5C; Supplemental Fig. S8). This supported the hypothesis that these two enzymes sequentially catalyze the primary and secondary glycosylations of flavonols in the Arabidopsis anthers.
Figure 5.
Production of pollen-specific flavonols by the consecutive reactions of UGT73B2 and UGT79B6. A, The glycosylation of quercetin on the 3-hydroxyl group by the catalytic activity of UGT73B2 was confirmed using a quercetin 3-glucoside standard. B, UGT79B6, as expected, did not show any activity toward quercetin. C, The combined activity of UGT73B2 and UGT79B6 resulted in the production of quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides. A.U, Arbitrary units; m/z, mass-to-charge ratio.
MYB21 and MYB24 Affect Phenylpropanoid Metabolism and Regulation in Anthers
To examine the possible functional link between MYB99, MYB21, and MYB24, we examined the previously reported mutant plants myb21-5, myb24-5, and myb21-5/myb24-5. myb21-5 possesses a null mutation in the coding sequence of the first exon of the gene, causing an early stop codon, while in myb24-5 (SALK_038872), a T-DNA insertion was localized to the first intron of the gene (Reeves et al., 2012). Both myb21-5 and myb24-5 single mutants display semi male sterility with few viable seeds. In the myb21/myb24 double mutant, a more severe phenotype was observed, which led to complete sterility, as reported before (Song et al., 2011). We subsequently carried out untargeted metabolic analysis of myb21-5, myb24-5, myb21-5/myb24-5, and wild-type anthers (dissected from preanthesis flowers). We detected 109, 186, and 46 mass signals that were significantly decreased when compared with those in wild-type anthers in myb21-5, myb24-5, and myb21/myb24, respectively. These mass signals corresponded to 23, 31, and 20 putatively identified and annotated metabolites that were decreased in the anthers of myb21-5, myb24-5, and myb21-5/myb24-5, respectively, compared with the wild type (Table 1). We also detected a decrease in 11 unknown compounds that could not be annotated, four of which were also decreased in myb99-1 anthers (unknowns 5, 7, 9, and 10 in Supplemental Data Set S2). Much like in the anthers of myb99-1, the myb21-5, myb24-5, and myb21-5/myb24-5 mutant lines displayed impaired phenylpropanoid-associated metabolism. We detected a decrease of five, 13, and two flavonol glycosides in myb21-5, myb24-5, and myb21-5/myb24-5, respectively, compared with the wild type (Table 1). The pollen-specific flavonols, kaempferol and quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides, failed to accumulate in myb24-5 anthers, while no significant changes from the wild type could be detected in the levels of these metabolites in the anthers of myb21-5 and myb21-5/myb24-5 (Table 1), suggesting that MYB21 might not be required for the production of these pollen-specific flavonols. Furthermore, levels of 17 HCAAs were significantly decreased in anthers of myb21-5, myb24-5, and myb21-5/myb24-5, which corresponded to the reductions detected in the myb99-1 mutant. Targeted metabolite analysis showed that the anthers of myb24-5 and myb21/myb24 contained significantly reduced levels of naringenin chalcone (Table 1). Unexpectedly, this was not the case in the anthers of myb21-5, which displayed more than a 2-fold enrichment of this metabolite when compared with the wild type.
To examine if MYB21 and MYB24 are involved in the transcriptional regulation of phenylpropanoid pathway genes during pollen wall development, we performed RT-qPCR on anthers collected from preanthesis flowers of myb21-5, myb24-5, myb21-5/myb24-5, and the wild type. Notably, a decrease in the transcript levels of multiple phenylpropanoid metabolic genes was detected in the three mutant genotypes compared with the wild type, which corresponded with the observed metabolic changes. Transcripts of PAL4, LAP5, UGT73B2, and UGT79B6 were reduced in isolated anthers of myb21-5 compared with the wild type (Fig. 6). In myb24-5, the transcripts of TK2, PAL1, PAL4, CSE, and UGT79B6 were decreased, and in the myb21-5/myb24-5 double mutant, we detected decreases in TK2, PAL4, 4CL2, CSE, UGT73B2, and UGT79B6. The transcripts of PAL4 and UGT79B6 were commonly decreased in the three mutant backgrounds compared with the wild type, while the MYB99 transcript level was significantly increased in these mutants (i.e. myb21-5, myb24-5, and myb21-5/myb24-5), with up to 9-fold enrichment in the double mutant (Fig. 6), suggesting that MYB21 and MYB24 negatively regulate the expression of MYB99 in the tapetum.
Figure 6.
myb21-5, myb24-5, and myb21-24 knockout mutants are down-regulated in the expression of genes from primary and phenylpropanoid metabolic pathways. RT-qPCR of anthers collected from preanthesis flowers is shown. Down-regulation of TK2, PAL4, UGT73B2, and UGT79B6 was detected in the anthers of the mutant lines as well as a significant increase in expression of MYB99 transcripts. Shown are means and se of four biological replicates. Asterisks denote statistically significant differences from the wild type (WT) calculated using Student’s t test: *, P < 0.05 and **, P < 0.01.
MYB99, MYB21, and MYB24 Can Activate Promoters of Phenylpropanoid-Associated Genes and of TK2 in Primary Metabolism
We applied a transient expression, dual-luciferase assay system in Nicotiana benthamiana leaves (Hellens et al., 2005) to measure promoter activation by MYB99, MYB21, and MYB24. The MYB21, MYB24, and MYB99 coding sequences were expressed under the control of the Arabidopsis UBIQUITIN1 promoter (UBQpro:MYB21, UBQpro:MYB24, and UBQpro:MYB99) and coinfiltrated with another vector containing the putative promoter sequence of the tested genes fused to the LUCIFERASE (LUC) reporter (Hellens et al., 2005). We then measured the LUC and RENNILASE (REN) luminescence ratio (i.e. LUC/REN ratio) in infiltrated leaves. To assess any basal activation of putative promoters, an empty vector was used in each coinfiltration experiment as a negative control. The results showed that MYB99 was able to activate promoters of TK2, the three isoforms of the PAL gene (PAL1, PAL2, and PAL4), the two LAP genes (LAP5 and LAP6), and UGT79B6 (Fig. 7A). Conversely, MYB99 was not able to activate the promoters of 4CL2, CSE, UGT72B1, and UGT73B2. MYB21 expression activated the promoters of TK2, PAL4, and UGT73B2, while MYB24 activated the promoters of PAL1, PAL2, UGT73B2, and UGT79B6 (Supplemental Fig. S9). Noticeably, the UGT72B1 promoter could not be activated by any of the MYB proteins and is probably not regulated by them.
Figure 7.
MYB99 activates promoters of structural genes and regulators of the phenylpropanoid pathway. A, Putative promoters of genes from primary and phenylpropanoid metabolic pathways coinfiltrated with a vector containing MYB99 under the regulation of the ubiquitin promoter. B, Coinfiltration of MYB21, MYB24, and MYB99 with their putative promoters. Background promoter activity was assayed by coinfiltration with an empty vector of the same type. Shown are means and se of six biological replicates. Asterisks denote statistically significant differences from the wild type calculated using Student’s t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
We subsequently tested the activation of MYB99, MYB21, and MYB24 promoters by each of these transcription factors and detected a complex interaction network, in which each of these genes can affect the transcription of the other in a coordinated manner. MYB99 activated the promoter of MYB24, which in turn activated that of MYB21. Interestingly, MYB21 repressed the MYB99 promoter-driven expression while activating that of MYB24 (Fig. 7B). These results were in agreement with the up-regulation of MYB99 in the anthers of myb21-5, myb24-5, and myb21-5/24-5 and the down-regulation of MYB21 and MYB24 in myb99-1 anthers.
To examine if MYB99, MYB21, and MYB24 can coactivate the promoter of a single gene, we performed coinfiltration transient expression assays with all possible combinations of the three transcription factors and tested their activity with the PAL2 or PAL4 promoter (Supplemental Fig. S10). Notably, the PAL2 promoter, which was strongly activated by either MYB24 or MYB99 (over 10-fold enrichment compared with background), was not activated when MYB24 and MYB99 were coexpressed. Similarly, the PAL4 promoter, which was strongly activated by either MYB21 or MYB99, was only weakly activated (less than 3-fold enrichment compared with background) when these two transcription factors were coexpressed. Any other combination of the three transcription factors resulted in very weak and insignificant activation levels of the PAL2 or PAL4 promoters. As our experiments were performed in N. benthamiana, additional factors might be missing that facilitate activation by combinations of these transcription factors in this heterologous plant system. For example, MYB21 and MYB24 were shown to form a bHLH-MYB transcription complex in anthers that can be activated by jasmonate release (Qi et al., 2015). To compare these results with previous activation assays, the PAL2 promoter was retested for activation by a single transcription factor. As expected, the PAL2 promoter was activated by either MYB99 or MYB24 but not by MYB21 (Fig. 7; Supplemental Fig. S9).
DISCUSSION
A Similar Triad of R2R3-MYB Transcription Factors in Arabidopsis and Petunia Regulates Different Branches of the Phenylpropanoid Pathway
In the course of this study, we showed that MYB99 is required for normal pollen development in Arabidopsis. Decrease in pollen viability was evident in myb99, which was accompanied by a distorted pollen wall structure and irregularities in pollen shape. Interestingly, these deformed pollen grains seemed to aggregate in the anther locule rather than adhere to the stigma. This phenotype is analogous to the one observed in the Arabidopsis lap mutants, producing distorted pollen grains that remain tightly attached to the anthers. The two LAP genes, LAP5 and LAP6, encode anther-specific enzymes that take part in the biosynthesis of triketide and tetraketide α-pyrones, important lipid components of sporopollenin. Although the two proteins exhibit structural similarities to CHS that catalyzes the conversion of p-coumaroyl-CoA to naringenin chalcone, their involvement in the phenylpropanoid pathway was not established. Previous studies showed that the pollen exine of lap mutants is deformed and its anthers are depleted from both chalcones and flavonol glycosides (Dobritsa et al., 2010), which is similar to myb99 (Table 1). These phenotypic similarities between lap mutants and myb99 could indicate a possible role for MYB99 in regulating the production of sporopollenin-related lipid components and not merely phenylpropanoids. This assumption was further corroborated by decreased expression of LAP5 and LAP6 in myb99 anthers and experiments showing activation of the LAP5 and LAP6 promoters by MYB99 and not by MYB21 and MYB24, placing the two LAP genes under MYB99 control.
We also observed impairments in stamen filament growth in myb99 mutants and in MYB99-overexpressing plants, which in both cases appeared to be shorter then wild-type plants. While fertility in myb99 was not affected by this, the shorter filament size likely caused the sterility observed in MYB99-overexpressing plants; this phenotype was also reported before for myb21 mutants (Mandaokar et al., 2006), suggesting a functional link between these two genes in regulating stamen filament growth in addition to their role in controlling tryphine phenylpropanoid composition suggested here.
We discovered that two previously characterized R2R3-MYB transcription factors, namely MYB21 and MYB24, shown to control jasmonate-regulated stamen growth (Reeves et al., 2012), were also required for pollen wall development. We found that MYB21, MYB24, and MYB99 are coexpressed in dissected young anthers (stages 8 and 9) of wild-type and mutant plants. While MYB99 was reported to be predominantly expressed in tapetal cells (Alves-Ferreira et al., 2007), the expression of MYB21 and MYB24 was reported in stamen filaments and also in anthers (Reeves et al., 2012), but their expression in the tapetum was not examined. It is therefore important to learn more about the cell-specific expression of these two genes in order to understand if they can directly regulate MYB99 expression in the tapetum.
We examined if these three genes can effectively activate the expression of one another by performing promoter activation assays and measuring their expression in the anthers of their respective mutant plants. We have deduced a putative model explaining the relationship between these regulators (Fig. 8). While MYB99 expression is negatively regulated by MYB21, it activates the promoter of MYB24. MYB21 and MYB24 appear to activate the promoters of one another. In support of this model, we detected a significant decrease in MYB21 and MYB24 transcript levels in the anthers of myb99 and increased expression of MYB99 in the anthers of myb21, myb24, and myb21/24.
Figure 8.
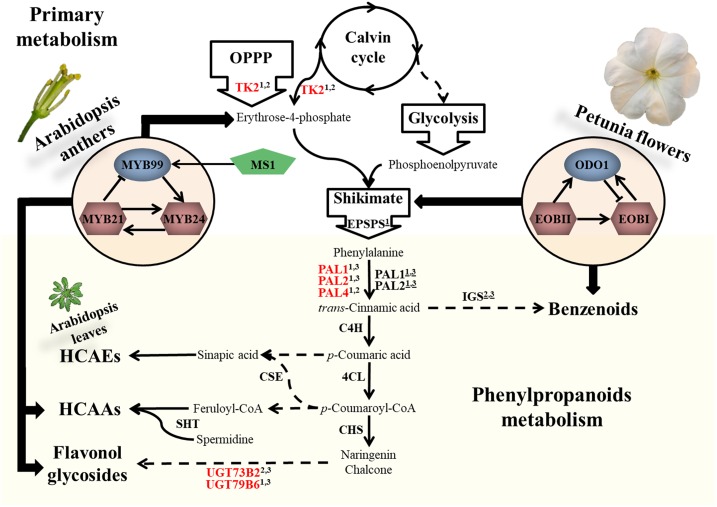
Scheme depicting the regulation of phenylpropanoid metabolism in Arabidopsis anthers and petunia flowers. Solid arrows represent one enzymatic reaction, and dashed arrows represent multiple reactions. Down-regulated genes in myb99 anthers are marked in red, and gene promoters that were tested positive in activation assays are marked with superscript 1, 2, or 3 for MYB99, MYB21, or MYB24, respectively. Underlined superscripts 1, 2, or 3 represent petunia orthologs that were shown to be regulated by ODO1, EOBI, or EOBII, respectively. IGS, Isoeugenol synthase; SHT, spermidine hydroxycinnamoyl transferase.
Furthermore, myb99 anthers displayed reduced levels of HCAAs and flavonol glycosides, which corresponded with decreased expression of MYB21 and MYB24 in this mutant. On the other hand, myb21/myb24 anthers exhibited impaired HCAA production with only a minimal impact on flavonol glycoside metabolism, which corresponded with the increased MYB99 expression in this mutant. This suggested that increased MYB99 expression partially rescued phenylpropanoid production in myb21/myb24 anthers. While MYB99 seems to directly affect flavonol glycoside levels in the tapetum, MYB21 and MYB24 are probably more important for the production of HCAAs. Noticeably, increased MYB99 expression in myb21/24 anthers did not rescue plant sterility or the decreased expression of phenylpropanoid network genes observed in this mutant (Fig. 6), excluding the expression of PAL1, which displayed transcript levels similar to those in the wild type. It also cannot be ruled out that the MYB99 expression changes were a result of MYB21 and MYB24 deficiency outside the tapetum domain.
The three MYB factors appear to act together in a coordinated network in which each regulator affects the expression of the other. It is therefore difficult to predict transcriptional and metabolic changes that mutations in these genes exert on anther phenylpropanoid metabolism and gene expression. Interestingly, unlike the results obtained from promoter activation experiments using a single transcription factor (Fig. 7; Supplemental Fig. S9), the coexpression of MYB99, MYB21, and MYB24 in N. benthamiana leaves did not activate the PAL2 and PAL4 promoters (Supplemental Fig. S10), suggesting that these three transcription factors do not coactivate target genes.
The relationship between MYB99, MYB21, and MYB24 resembles the regulatory network controlling benzenoid production in petunia floral scent (Spitzer-Rimon et al., 2012). First, the two networks are phylogenetically related; as mentioned above, the Arabidopsis MYB99 protein is related to the ODO1 regulatory factor from petunia. The Arabidopsis MYB21 and MYB24 and their petunia orthologs EOBI and EOBII are part of a different subclade in the MYB family. Second, both networks trigger the production of metabolites from the phenylpropanoid pathway by simultaneously regulating genes in primary and phenylpropanoid metabolism (Fig. 8). Although some of the target genes differ between petunia and Arabidopsis, both networks commonly regulate PAL1 and PAL2. Lastly, ODO1, EOBI, and EOBII affect the expression of one another to activate the biosynthesis of scent compounds (Verdonk et al., 2005; Van Moerkercke et al., 2011; Spitzer-Rimon et al., 2012). Similarly, the three Arabidopsis transcription factors function in a coordinated manner to instigate the biosynthesis of other classes of phenylpropanoids in anthers (i.e. flavonol glycosides and HCAAs), ultimately affecting tryphine composition and pollen viability.
The Arabidopsis MYB Triad Controls the Exclusive Production of Tapetal Diglycosylated Flavonols and HCAAs for Coating Pollen
The occurrence of flavonols in pollen has been reported in several plant species (Ross et al., 2005; Arráez-Román et al., 2007; Moreira et al., 2008; Tao et al., 2011; Li et al., 2012). Yet, thus far, an anther-specific transcription factor that mediates the production of pollen coat flavonols was not described. In this study, we have shown that the MYB99 transcription factor, together with MYB21 and MYB24, are required for the biosynthesis of the two major diglycosylated flavonols in Arabidopsis pollen. As previously suggested (Stracke et al., 2010), we identified a transcriptional regulatory mechanism for flavonol biosynthesis in anthers that is controlled by MYB99 triad members. Moreover, these transcription factors coregulate a suite of metabolic genes that take part in controlling upstream precursor pathways, core phenylpropanoid metabolism, as well as flavonol glycoside and HCAA biosynthesis in the anther.
Down-regulation of PAL4, 4CL2, and CSE transcripts was evident in the anthers of myb99 and myb21/24. In the phenylpropanoid metabolic pathway, common enzymatic reactions lead to the biosynthesis of lignin, flavonols, HCAEs, and HCAAs. The first of these reactions is catalyzed by PAL, which converts the amino acid Phe into cinnamic acid, and the last reaction is catalyzed by 4CL, which generates p-coumaroyl-CoA from p-coumaric acid (Fig. 8). The consecutive enzymatic reactions of CSE and 4CL mediate the conversion of p-coumaroyl-CoA to caffeoyl-CoA in the biosynthesis pathway leading to lignin and HCAEs but not toward the biosynthesis of flavonol glycosides or HCAAs (Vanholme et al., 2013). Transient activation assays performed with the putative promoter sequences of these genes (i.e. PAL4, 4CL2, and CSE) showed that the PAL4 promoter was activated by MYB99 and MYB21. The promoters of both 4CL2 and CSE did not show any activation in our experiments, indicating that these two genes are probably not directly regulated by these MYB factors, or alternatively, their expression requires the involvement of other transcription factors. Since both 4CL2 and CSE were characterized in the past with relation to lignin production in Arabidopsis and sterility was never reported in mutant plants of these genes, we cannot be certain regarding the role of these enzymes in the production of tapetum flavonols or HCAAs. As for PAL4, according to our results, it is highly likely that it takes part in an anther-specific pathway and is important for the production of pollen phenylpropanoids. Moreover, PAL4 expression in the anther is controlled by MYB99 and MYB21.
We also detected down-regulation in PAL1 transcripts in myb99 anthers and a small but significant decrease in PAL1 expression in myb24 anthers. Promoter activation experiments confirmed that both MYB99 and MYB24 were able to activate the promoter of this gene while MYB21 could not, indicating that PAL1 expression in the anther is probably regulated by MYB99 and MYB24. As for PAL2, we could not confirm any decrease in transcription in any of the mutant backgrounds, but surprisingly, both MYB99 and MYB24 strongly activated the promoter of this gene. This suggests that PAL2 expression in anthers could be regulated by MYB99 and MYB24 but probably involves other transcription factors as well. The role of PAL1 and PAL2 genes in pollen development coincides with previous reports on the pal1/pal2 double mutant that did not exhibit any gross morphological differences other than reduced pollen production and varying degrees of male sterility (Rohde et al., 2004; Huang et al., 2010). In the same study (Huang et al., 2010), the pal1/pal2/pal4 triple mutant had a more severe phenotype and was completely sterile, confirming our finding of the importance of PAL1, PAL2, and PAL4 to pollen development.
The UGT73B2 protein was previously characterized as a kaempferol 3-O-UDP-Glc transferase (Kim et al., 2006), although its specific substrates and in vivo function were never described. As for UGT79B6, it was shown to act in glycosylation of kaempferol and quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside in Arabidopsis tapetal cells (Yonekura-Sakakibara et al., 2014). The involvement of these two proteins in the production of pollen diglycosylated flavonols was confirmed by determining their in vitro enzyme activity with quercetin and kaempferol as substrates. While UGT73B2 catalyzed the primary glycosylation of kaempferol or quercetin at the third hydroxy group, the consecutive glycosylation was executed by UGT79B6, which was reported earlier to act as a glycosyl-glycosyl transferase (Yonekura-Sakakibara et al., 2014). These assays resulted in the in vitro production of diglycosylated flavonols identical to the ones prevalent in Arabidopsis pollen, thus implicating the combined activity of UGT73B2 and UGT79B6 in the production of the typical Arabidopsis pollen flavonols.
The expression of UGT73B2 was down-regulated in myb99, myb21, and myb21/24 anthers compared with wild-type anthers, and promoter activation assays showed that both MYB21 and MYB24 were able to activate its promoter region. As for UGT79B6, its expression remained unchanged in myb99 anthers but was down in both myb21 and myb24 single mutants, as well as in the myb21/24 double mutant, compared with wild-type anthers. Subsequent promoter activation experiments revealed that the UGT79B6 promoter could be activated by either MYB99 or MYB24. Taken together with the gene expression and metabolic data obtained from myb99 and myb24 anthers in which the two major pollen diglycosylated flavonols were decreased, it is highly likely that UGT73B2 expression is controlled by MYB21 and MYB24, while the expression of UGT79B6 is controlled by MYB24 and MYB99.
The significance of MYB99 to phenylpropanoid metabolism was further demonstrated in Arabidopsis MYB99-overexpressing plants. Overexpression led to the accumulation of p-coumaric acid as well as HCAEs in leaves. We also detected an increase in a diglycosylated quercetin isomer that is found in Arabidopsis leaves but not in anthers. The biosynthetic pathway for HCAE and flavonol glycoside production shares common enzymatic steps that only diverge when p-coumaric acid is converted to either sinapic acid or p-coumaroyl-CoA (Fig. 8). It is therefore likely that MYB99 can drive metabolic flux into these two pathways possibly by enrichment in precursor levels that can lead to the production of both products. Interestingly, p-coumaric acid also contributes to sporopollenin biosynthesis by forming ester and ether linkages to in-chain hydroxy lauric acids (Morant et al., 2007). Although we did not examine the chemical composition of sporopollenin in myb99 mutants, it is tempting to propose that by regulating PAL gene expression in the tapetum, MYB99 contributes to the accumulation of p-coumaric acid monomers that are ultimately deposited as sporopollenin material. Yet, the relation between MYB99 and sporopollenin biosynthesis requires further investigation.
HCAAs have been previously reported to be localized to pollen walls of various plant species (Meurer et al., 1988), likely as part of the pollen tryphine material, yet, their role in pollen development remains elusive. The BAHD acyltransferase, SHT, catalyzes the conjugation of hydroxycinnamoyl-CoA esters to spermidine in Arabidopsis tapetal cells (Grienenberger et al., 2009). In Arabidopsis flowers, SHT expression in the tapetum is limited to the early stages of flower development during sporopollenin biosynthesis and pollen coat formation. A decrease in the HCAA content of sht pollen affected pollen autofluorescence, indicative of these metabolites’ involvement in tryphine composition. Deficiency in at least 14 different HCAAs was evidenced in young anthers of myb99, myb21, myb24, and myb21/24. This corresponded with a decrease in expression of the core phenylpropanoid structural genes PAL1 and PAL4 (see above). Previous studies reported that ms1 floral buds are devoid of HCAAs, and more recently MS1 was shown to directly bind to the SHT promoter (Yu, 2015). We therefore suggest that by regulating core phenylpropanoid metabolism, MYB99, MYB21, and MYB24 work together with MS1 in regulating the production of HCAAs in Arabidopsis anthers.
The MYB Triad Controls Precursor Supply from Primary Metabolism to the Phenylpropanoid Pathway by Regulating TK2 Gene Expression
The biosynthesis of specialized metabolites in plants relies on a constant carbon supply that meets production demands. Therefore, transcriptional regulation of specialized metabolites is likely to be associated with the activation of primary, precursor pathways (Aharoni and Galili, 2011). The most recent example to this idea was demonstrated by heterologous expression of the Arabidopsis flavonol biosynthesis regulator MYB12 in tomato, which led to an enhanced supply of carbon, energy, and reducing power from primary metabolism. It appears that promoters of genes from the glycolysis, shikimate, and phenylpropanoid pathways are directly regulated by MYB12 in order to coordinate metabolic flux toward the biosynthesis of flavonols and HCAEs in fruit (Zhang et al., 2015).
Here, we found that overexpression of MYB99 in Arabidopsis leaves activates the expression of primary metabolism genes associated with glycolysis (ENO, G6PD, PFP, and PGK), the OPPP (PGD, TK, and RP3E), and the Calvin cycle (TK). We also detected the up-regulation of genes from the intermediate shikimate pathway (3DQS, DHQ-SDH, and EPSPS) that directs primary metabolites toward the biosynthesis of aromatic amino acids. Since all phenylpropanoids are ultimately derived from Phe, it is crucial to maintain metabolic flux in order to secure precursor supply and availability of this amino acid. Out of the primary metabolism-related genes examined in this work, we confirmed the down-regulation of TK2 transcript levels in the anthers of myb99, myb24, and the myb21/24 mutants. Concurring with these results, coexpression analysis of TK2 in Arabidopsis revealed that it is highly coregulated with multiple genes from the core phenylpropanoid pathway, further indicating its association with this pathway. The significance of TK activity for phenylpropanoid metabolism was previously demonstrated in tobacco plants (Henkes et al., 2001). A small decrease in TK activity affected the flux of primary metabolites through the shikimate pathway while dramatically decreasing phenylpropanoid metabolism. In plants, TK catalyzes reversible reactions in the Calvin cycle and oxidative pentose phosphate pathway, and by doing so it directly regulates the levels of erythrose-4-phosphate, one of the substrates required for the shikimate pathway (Fig. 8). Erythrose-4-phosphate together with phosphoenolpyruvate (a metabolic product of glycolysis) are further metabolized in the shikimate pathway, in which they are converted to the three aromatic amino acids. Lastly, promoter activation assays showed that both MYB21 and MYB99 were able to activate the promoter of TK2. We therefore suggest that the MYB99-MYB21-MYB24 triad of transcription factors regulates the expression of TK2 in anthers, subsequently driving metabolic flux from primary metabolism, through the shikimate pathway, toward phenylpropanoid biosynthesis for tryphine material production.
Following this study, we propose that additional plant species utilize a similar transcriptional regulatory triad to instigate the biosynthesis of specific metabolites in various branches of the phenylpropanoid pathway. Apart from petunia, another example for this conserved triad can be found in snapdragon flowers, in which protein orthologs of MYB99 (Tamagnone et al., 1998) as well as MYB21 and MYB24 (Moyano et al., 1996) were shown to regulate genes in the phenylpropanoid pathway associated with flavonols and lignin production. Yet, the coregulation of these three snapdragon protein orthologs was not demonstrated to date. The discovery of the MYB99-MYB21-MYB24 triad is thus an interesting but, at this stage, infrequent example of how plants utilize a common set of transcriptional factors, coregulating each other, to control the biosynthesis of different branches of a large species-wide pathway of specialized metabolism.
MATERIALS AND METHODS
Plant Growth and Material
Arabidopsis (Arabidopsis thaliana) plants were grown in climate rooms (22°C, 16/8 h of light/dark). The myb99 (ecotype Columbia) mutants were obtained from the European Arabidopsis Stock Center (http://arabidopsis.info/; stock IDs SALK_052877C and SALK_003193C), and the myb21-5, myb24-5, and myb21/24 mutants was kindly provided by Jason W. Reed from the University of North Carolina. T-DNA insertions for MYB99 SALK mutants were identified using oligonucleotides designed by the iSECT tool (Signal T-DNA Express Web site), and myb21-5, myb24-5, and myb21/24 mutation/insertion were identified by PCR as described (Reeves et al., 2012). Arabidopsis plants expressing the MYB99-HA-tag gene and plants expressing MYB99 under the control of the Cauliflower mosaic virus 35S promoter were generated by the floral dip method (Clough and Bent, 1998). Transformant seedlings were selected on petri dishes containing Murashige and Skoog medium (purchased from Duchefa) with kanamycin and then transferred to soil. For metabolic and transcriptomics analyses, leaf tissue was harvested from 18-d-old plants and anthers (stages 8 and 9; Sanders et al., 1999) dissected from preanthesis flowers of ∼4-week-old plants; 100 and 30 anthers were used (per biological replicate) for RNA and metabolite extractions, respectively. Flower buds in floral stages 10 to 12 were collected (Smyth et al., 1990) for protein and metabolite extraction of myb99-1/HA-tag plants. Plant material was immediately frozen in liquid nitrogen and kept at −80°C until processed.
Phylogenetic Analysis
Arabidopsis R2R3-MYB factor full protein sequence was acquired from the PLAZA Web site (http://bioinformatics.psb.ugent.be/plaza/), which provides an online platform to perform evolutionary analyses and data mining within the green plant lineage. The full protein sequences of the various R2R3-MYB factors together with the petunia (Petunia hybrida) proteins ODO1, EOBI, and EOBII were aligned using the default parameters of the MUSCLE algorithm, and the nonconserved regions in the N and C termini were then removed in an unbiased manner before realignment. Multiple sequence alignment was then processed into a maximum-likelihood phylogenetic tree with the MEGA software version 7.0 (Tamura et al., 2013), with bootstrapping of 1,000 iterations for validation.
Cloning and Construction of Vectors
Reactions requiring proofreading were performed with Phusion High-Fidelity DNA Polymerase (New England Biolabs), and all other reactions were performed with the GoTaq green mix (Promega) and the respective oligonucleotides (Supplemental Data Set S1). The detailed cloning procedure for the various vectors used in this work can be found in Supplemental Methods S1.
Protein Separation and Immunoblotting
Frozen floral bud powder was extracted for 10 min at 100°C in 60 mm Tris-HCl, pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 0.005% (w/v) Bromphenol Blue, and 0.7 m β-mercaptoethanol, cooled, and centrifuged for 5 min at 14,000g. Protein concentration was determined in the supernatant; 60-µg protein aliquots were separated by SDS-PAGE, blotted, and transferred. Membranes were reversibly stained with Coomassie Blue to confirm transfer. The polyclonal antibody against HA-tag was obtained from Santa Cruz.
Gene Expression Analysis Using RT-qPCR
For floral buds and leaves, RNA extraction was performed with the TRIzol method (Sigma-Aldrich), and for anthers, it was performed using the Qiagen RNeasy Micro Kit. RNA quantity and purity were assessed with a NanoDrop ND-1000 spectrophotometer and with agarose gel electrophoresis. All RNA samples were treated with DNase I (Sigma). Complementary DNA libraries were prepared using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Four biological replicates were sampled for each experiment. Plants were grown together and sampled at the same time of the day, within several days. RT-PCR conditions are specified in Supplemental Methods S2.
Electron Microscopy Analysis and Alexander Staining
For SEM analysis, we collected inflorescences from 5-week-old wild-type and mutant plants. SEM was immediately performed using an FEI XL30 FEG ESEM device, working in the wet mode. Three biological replicates were sampled from anthers of myb99-1, myb99-2, and wild-type plants and scored for pollen viability according to Alexander (1969). The percentage of aborted pollen was calculated from a single biological replicate obtained from myb99-1.
Microarray Analysis
Two biological replicates were sampled from leaves of wild-type and MYB99-ox1 plants. Extracted RNA was then amplified and labeled using the standard Affymetrix protocol and hybridized to Affymetrix ATH1 GeneChips according to the manufacturer’s guidelines (Mueller et al., 2008). Statistical analysis of transcriptome data was carried out using Partek Genome Suite software (www.partek.com). Data preprocessing and normalization were performed using the Robust Microarray Averaging algorithm (Irizarry et al., 2003). Batch effects between the replicates were not found. Differentially expressed genes were identified using ANOVA according to false discovery rate < 0.05 and at least a 2-fold change between the genotypes (Supplemental Data Set S3).
RNA Sequencing Analysis
Libraries for Illumina high-throughput strand-specific RNA sequencing were prepared as follows. Two biological replicates were sampled from myb99-1 and the wild type and a single biological replicate was sampled from MYB99-ox1. Two micrograms of total RNA from each sample was used for preparation of RNA sequencing libraries using the method described (Zhong et al., 2011) with minor modifications. Libraries were then pooled and sequenced with an Illumina HiSeq2000 instrument, and reads were aligned to the TAIR10 version of the Arabidopsis genome (www.arabidopsis.org) using TOPHAT version 2.0.10 (Rozen and Skaletsky, 2000). SAM files were generated using the SAMtools version 0.1.19 (Li et al., 2009). Gene quantification was done using HTseq-count (Anders et al., 2015), and normalization of read counts and differential expression analysis were performed with DESeq2 (Auer and Doerge, 2010) as described in the DESeq2 vignette.
Metabolite Analysis
Anthers (stages 8 and 9) collected from closed buds (n = 5 per genotype, 30 per replicate) were homogenized and extracted in 50 µL of 80% methanol and 0.1% (v/v) formic acid. For leaf and floral bud analysis (n = 4 per genotype), 100 mg of ground sample was extracted in 300 µL of 80% methanol and 0.1% (v/v) formic acid. Anther leaf and floral bud samples were subsequently cleaned through a 0.22-µm filter, sonicated for 20 min at room temperature, and then injected into an ultra-HPLC device coupled to a quadrupole time-of-flight mass spectrometer. The injection conditions were as previously described (Mintz-Oron et al., 2012). Samples were injected in the positive and negative ionization modes, and preprocessing was performed independently for each mode. Accurate mass, retention time, elemental composition, dual-energy fragments obtained from tandem mass spectrometry daughter ion scan, UV spectra, and references used for the identification of each metabolite are presented in Supplemental Data Set S2. Specific information on data analysis can be found in Supplemental Methods S3.
Generation and Purification of Glucosyltransferase Recombinant Enzymes and Glucosyltransferase Assays
Procedures used for cloning, protein expression, and protein isolation of UGT73B2 and UGT29B6 as well as detailed glycosyltransferase assay conditions are as described in Supplemental Methods S4.
Dual-Luciferase Assay
Nicotiana benthamiana plants were grown in climate rooms (22°C, 16/8 h of light/dark). Plants were grown until they had six leaves and then infiltrated with Agrobacterium tumefaciens GV3101. Plants were maintained in the climate rooms and, after 4 to 5 d, 1-cm discs were collected from the fourth and fifth leaves of each plant. Six biological replicates with their respective negative controls were used per assay. The experiment was performed as previously described (Hellens et al., 2005) with minor changes detailed in Supplemental Methods S5.
Accession Numbers
Sequence data from this article can be found in TAIR under the following accession numbers: MYB99 (AT5G62320), MYB21 (AT3G27810), MYB24 (AT5G40350), MYB20 (AT1G66230), MYB40 (AT5G14340), MYB42 (AT4G12350), MYB43 (AT5G16600), MYB85 (AT4G22680), MS1 (AT5G22260), ENO1 (AT1G74030), G6PD6 (AT5G40760), PFPβ1 (AT1G12000), PFPα2 (AT1G76550), PFPα1 (AT1G20950), PGK (AT1G79550), PGD2 (AT3G02360), TK2 (AT2G45290), RP3E (AT1G63290), 3DQS (AT5G66120), DHQ-SDH (AT3G06350), EPSPS (AT1G48860), EMB1144 (AT1G48850), PAL1 (AT2G37040), PAL2 (AT3G53260), PAL4 (AT3G10340), C4H (AT2G30490), 4CL2 (AT3G21240), 4CL3 (AT1G65060), CCR1 (AT1G15950), CCR3 (AT5G14700), CCR4 (AT2G23910), CHS (AT5G13930), LAP5 (AT4G34850), LAP6 (AT1G02050), CSE (AT1G52760), F3H (AT3G51240), OMT1 (AT5G54160), CCoAOMT1 (AT4G34050), UGT72B1 (AT4G01070), UGT73B2 (AT4G34135), UGT79B6 (AT5G54010), and SHT (AT2G19070). Sequence data for the petunia proteins used for multiple sequence alignments (Fig. 1) can be found in the GenBank/EMBL data libraries under the following accession numbers: ODO1 (Q50EX6), EOBI (AGC00814), and EOBII (EU360893).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The motif for interaction with bHLH proteins is not present in the MYB99-subclade amino acid sequences.
Supplemental Figure S2. RT-qPCR of MYB99 mutants and overexpressing plants.
Supplemental Figure S3. Rescue of pollen-specific flavonol glycoside production in myb99 by MYB99 overexpresssion.
Supplemental Figure S4. Staining for ectopic lignification in MYB99-overexpressing plants.
Supplemental Figure S5. Expression profile of UGT genes.
Supplemental Figure S6. Coexpression analysis of the first and second isoforms of the TK gene in Arabidopsis.
Supplemental Figure S7. Gene transcripts with no significant changes in myb99 compared with the wild type.
Supplemental Figure S8. Mass spectra profiles of pollen-specific flavonol quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside.
Supplemental Figure S9. Examination of MYB24 and MYB21 transcription factor capacity to activate promoters of candidate genes through transient expression assays.
Supplemental Figure S10. Coinfiltration of MYB21, MYB24, and MYB99 inhibits promoter activation of the PAL2 and PAL4 genes.
Supplemental Table S1. Fold change values for metabolites identified by comparative nontargeted profiling.
Supplemental Data Set S1. List of oligonucleotides used in this study.
Supplemental Data Set S2. Liquid chromatography-mass spectrometry data of putatively identified metabolites.
Supplemental Data Set S3. Microarray results obtained from leaves of wild-type and MYB99-ox1 plants.
Supplemental Data Set S4. DESeq analysis of anthers collected from wild-type, myb99-1, and MYB99-ox1 plants.
Supplemental Methods S1. Cloning and construction of vectors (van Engelen et al., 1995; Sarrion-Perdigones et al., 2013).
Supplemental Methods S2. Gene expression analysis using RT-qPCR (Livak and Schmittgen, 2001; Pellino et al., 2011).
Supplemental Methods S3. Metabolite data analysis (Hochberg and Benjamini, 1990; Smith et al., 2006; Mintz-Oron et al., 2008; Handrick et al., 2010).
Supplemental Methods S4. Generation and purification of glucosyltransferase recombinant enzymes and glucosyltransferase assay conditions.
Supplemental Methods S5. Dual-luciferase assay.
Acknowledgments
We thank Jason Reed for providing the myb21, myb24, and myb21/24 mutants; Sergey Malitsky for assistance with liquid chromatography-mass spectrometry analysis; Efrat Weithorn for help with analyzing the transcriptomic data; Sagit Meir, Nir Shachaf, and Jedrzej Szymanski for help with XCMS and data analysis; Wanqi Liang and Jianxin Shi for their critical comments and input on rice; and Samuel Bocobza for providing the tomato UBQ10 promoter sequence.
Footnotes
Articles can be viewed without a subscription.
References
- Aharoni A, Galili G (2011) Metabolic engineering of the plant primary-secondary metabolism interface. Curr Opin Biotechnol 22: 239–244 [DOI] [PubMed] [Google Scholar]
- Ahlers F, Lambert J, Wiermann R (1999) Structural elements of sporopollenin from the pollen of Torreya californica Torr. (Gymnospermae): Using the H-1-NMR technique. Z Naturforsch C 54: 492–495 [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Arráez-Román D, Zurek G, Bässmann C, Almaraz-Abarca N, Quirantes R, Segura-Carretero A, Fernández-Gutiérrez A (2007) Identification of phenolic compounds from pollen extracts using capillary electrophoresis-electrospray time-of-flight mass spectrometry. Anal Bioanal Chem 389: 1909–1917 [DOI] [PubMed] [Google Scholar]
- Auer PL, Doerge RW (2010) Statistical design and analysis of RNA sequencing data. Genetics 185: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Bubert H, Lambert J, Steuernagel S, Ahlers F, Wiermann R (2002) Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Z Naturforsch C 57: 1035–1041 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg C, van Ohlen M, Handrick V, Vogt T (2012a) The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta 236: 51–61 [DOI] [PubMed] [Google Scholar]
- Fellenberg C, Ziegler J, Handrick V, Vogt T (2012b) Polyamine homeostasis in wild type and phenolamide deficient Arabidopsis thaliana stamens. Front Plant Sci 3: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser WT, Scott AC, Forbes AES, Glasspool IJ, Plotnick RE, Kenig F, Lomax BH (2012) Evolutionary stasis of sporopollenin biochemistry revealed by unaltered Pennsylvanian spores. New Phytol 196: 397–401 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Handrick V, Vogt T, Frolov A (2010) Profiling of hydroxycinnamic acid amides in Arabidopsis thaliana pollen by tandem mass spectrometry. Anal Bioanal Chem 398: 2789–2801 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M (2001) A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9: 811–818 [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen D, Olbrich A, Knüfer J, Krüger A, Hoppert M, Polle A, Fulda M (2011) Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J 68: 715–726 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang Z, Cao J (2013) Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol (Stuttg) 15: 249–263 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim BG, Ko JH, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Molecular cloning, expression, and characterization of a flavonoid glycosyltransferase from Arabidopsis thaliana. Plant Sci 170: 897–903 [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Li C, Wang X, Ran L, Tian Q, Fan D, Luo K (2015a) PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol 56: 2436–2446 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Suen DF, Huang CY, Kung SY, Huang AHC (2012) The maize tapetum employs diverse mechanisms to synthesize and store proteins and flavonoids and transfer them to the pollen surface. Plant Physiol 158: 1548–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kim JI, Pysh L, Chapple C (2015b) Four isoforms of Arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol 169: 2409–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Huang XX, Li Q, Cao Y, Bao Y, Meng XF, Li YJ, Fu C, Hou BK (2016) UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J 88: 26–42 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Meurer B, Wray V, Wiermann R, Strack D (1988) Hydroxycinnamic acid-spermidine amides from pollen of Alnus glutinosa, Betula verrucosa and Pterocarya fraxinifolia. Phytochemistry 27: 839–843 [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T (2012) Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci USA 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K, Fujiwara T, Iki T, Kuroda K, Yamashita K, Tamura M, Fujisawa Y, Watanabe A (2014) Transcriptome sequencing and profiling of expressed genes in cambial zone and differentiating xylem of Japanese cedar (Cryptomeria japonica). BMC Genomics 15: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L, Dias LG, Pereira JA, Estevinho L (2008) Antioxidant properties, total phenols and pollen analysis of propolis samples from Portugal. Food Chem Toxicol 46: 3482–3485 [DOI] [PubMed] [Google Scholar]
- Moyano E, Martínez-Garcia JF, Martin C (1996) Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell 8: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzac I, Wang J, Anzellotti D, Zhang H, Ibrahim RK (2000) Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch Biochem Biophys 375: 385–388 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen HA, Makaroff CA (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L) Heynh ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Pellino M, Sharbel TF, Mau M, Amiteye S, Corral JM (2011) Selection of reference genes for quantitative real-time PCR expression studies of microdissected reproductive tissues in apomictic and sexual Boechera. BMC Res Notes 4: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27: 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Grienenberger E, Douglas CJ (2015) The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113: 170–182 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejón JD, Delalande F, Schaeffer-Reiss C, Alché JD, Rodríguez-García MI, Van Dorsselaer A, Castro AJ (2016) The pollen coat proteome: At the cutting edge of plant reproduction. Proteomes 4: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM (2005) Comparison of lignin deposition in three ectopic lignification mutants. New Phytol 168: 123–140 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SA, ElSohly MA, Sultana GNN, Mehmedic Z, Hossain CF, Chandra S (2005) Flavonoid glycosides and cannabinoids from the pollen of Cannabis sativa L. Phytochem Anal 16: 45–48 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palací J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D (2013) GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162: 1618–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ. (1994) Pollen exine: The sporopollenin enigma and the physics of pattern. In Scott RJ, Stead MA, eds, Molecular and Cellular Aspects of Plant Reproduction. Cambridge University Press, Cambridge, UK, pp 49–81 [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Shin B, Choi G, Yi H, Yang S, Cho I, Kim J, Lee S, Paek NC, Kim JH, Song PS, et al. (2002) AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J 30: 23–32 [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G (2006) XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78: 779–787 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, Cna’ani A, Ben Zvi MM, Masci T, Edelbaum O, Yu Y, Shklarman E, Ovadis M, et al. (2012) The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24: 5089–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemans P, Lepot K, Marshall CP, Le Hérissé A, Javaux EJ (2010) FTIR characterisation of the chemical composition of Silurian miospores (cryptospores and trilete spores) from Gotland, Sweden. Rev Palaeobot Palynol 162: 577–590 [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188: 985–1000 [DOI] [PubMed] [Google Scholar]
- Sun YJ, Hord CLH, Chen CB, Ma H (2007) Regulation of Arabidopsis early anther development by putative cell-cell signaling molecules and transcriptional regulators. J Integr Plant Biol 49: 60–68 [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Yang N, Duan JA, Wu D, Guo J, Tang Y, Qian D, Zhu Z (2011) Simultaneous determination of eleven major flavonoids in the pollen of Typha angustifolia by HPLC-PDA-MS. Phytochem Anal 22: 455–461 [DOI] [PubMed] [Google Scholar]
- Uimari A, Strommer J (1997) Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J 12: 1273–1284 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Haring MA, Schuurink RC (2011) The transcription factor EMISSION OF BENZENOIDS II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J 67: 917–928 [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J (2007) Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol 144: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA (2007a) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Li JG, Pei M, Gu H, Chen ZL, Qu LJ (2007b) Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep 26: 219–228 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Nakabayashi R, Sugawara S, Tohge T, Ito T, Koyanagi M, Kitajima M, Takayama H, Saito K (2014) A flavonoid 3-O-glucoside:2′′-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J 79: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. (2015) Identification and characterisation of MS1 putative interacting proteins and regulatory targets in Arabidopsis. PhD thesis. University of Nottingham, Nottingham, United Kingdom [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
- Zhu J, Lou Y, Xu X, Yang ZN (2011) A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol 53: 892–900 [DOI] [PubMed] [Google Scholar]