Abstract
Cfr is a radical S-adenosylmethionine (RS) methylase that appends methyl groups to C8 and C2 of adenosine 2503 in 23S ribosomal RNA. Methylation of C8 confers resistance to several classes of antibiotics that bind in or near the peptidyl transferase center of the bacterial ribosome, including the synthetic antibiotic linezolid. The Cfr reaction requires the action of five conserved cysteines, three of which ligate a required [4Fe–4S] cluster cofactor. The two remaining cysteines play a more intricate role in the reaction, one of which (Cys338) becoming transiently methylated during catalysis. The function of the second (Cys105) has not been rigorously established; however, in the related RlmN reaction, it (Cys118) initiates resolution of a key protein-nucleic acid cross-linked intermediate by abstracting the proton from the carbon center (C2) undergoing methylation. We previously proposed that, unlike RlmN, Cfr would utilize a polyprotic base during resolution of the protein-nucleic acid cross-linked intermediate during C8 methylation, and, like RlmN, use a monoprotic base during C2 methylation. We based this proposal on the fact that solvent hydrons could exchange into the product during C8 methylation, but not during C2 methylation. Herein, we show that Cys105 of Cfr has a similar function to that of Cys118 of RlmN while methylating C8 of A2503, and provide evidence for one molecule of water that is in close contact with it, which provides the exchangeable protons during catalysis.
INTRODUCTION
Antibiotic resistance is a growing problem that threatens to hark back an era of rampant bacterial infection similar to that before the discovery and use of penicillin. The Centers for Disease Control estimates that 23,000 people die each year just in the United States because of antibiotic resistant infections.1 More than forty percent of all clinically employed antibiotics target the ribosome.2, 3 A common strategy used by bacteria to circumvent the action of these antibiotics is to modify ribosomal RNA (rRNA) and/or ribosomal amino acid residues to impede antibiotic binding. One such modification is methylation of C8 of adenosine 2503 (A2503) of 23S rRNA, which resides ultimately at the entrance to the nascent peptide exit channel of the ribosome. This modification, installed by Cfr, hinders the binding of multiple classes of antibiotics that target the peptidyltransferase center, including phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A, the macrolides josamycin and spiramycin, and even the synthetic oxazolidinone linezolid.4, 5 In addition to methylating C8 of A2503, Cfr can also methylate C2 of the same nucleotide; however, C8 methylation is heavily favored and may even be required before C2 methylation. Cfr is homologous to the ubiquitous bacterial RNA methylase RlmN, which exclusively methylates C2 of the exact same nucleotide of rRNA as well as C2 of adenosine 37 in several transfer RNAs (tRNAs).6
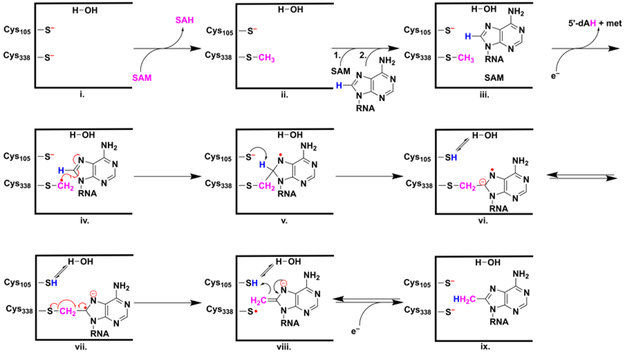
Cfr and RlmN are members of the radical S-adenosylmethionine (SAM) superfamily of enzymes, which all house an essential [4Fe-4S] cluster that participates in the reductive cleavage of SAM to methionine and a 5’-deoxyadenosyl 5’-radical (5’-dA•). The 5’-dA• usually initiates turnover by abstracting a target substrate hydrogen atom.7 Recent studies on RlmN and Cfr have shown that the first step in their mechanisms of catalysis is the transfer of a methyl group from SAM to an absolutely conserved cysteine (C338 in Cfr and C355 in RlmN), producing a methylcysteinyl residue (mCys).8 Upon release of S-adenosylhomocysteine (SAH), a second molecule of SAM binds to the enzyme and is converted into a 5’-dA•. The 5’-dA• abstracts a hydrogen atom from the mCys residue, with the resulting methylene radical adding to the carbon center undergoing methylation (C8 in Cfr and C2 in RlmN) to yield a protein–RNA crosslink containing an unpaired electron on the nucleotide base.9-11 A general base then abstracts the proton from the nascent sp3-hybridized C2 or C8 carbon center to initiate resolution of the crosslink, leading to a thiyl radical and an enamine (Scheme 1). Reduction of the thiyl radical and tautomerization of the enamine yields the final product.10, 12
Scheme 1.
Mechanistic proposal for catalysis by Cfr.
In addition to C338 (C355 in RlmN), Cfr contains another absolutely conserved cysteinyl residue, C105 (C118 in RlmN), which has been shown to gate resolution of the protein–RNA crosslink. When it is changed to alanyl or seryl, the protein–RNA crosslink persists.12 It was originally proposed that C105 is required for formation of a disulfide bond with C338 concomitant with release of the methylated product from the protein–RNA crosslink (Figure S1).13, 14 However, recent EPR and crystallographic studies of a trapped RlmN C118A-tRNA covalent complex clearly show that C118 is the base that abstracts the C2 proton in the RlmN reaction.10, 12
The identity of the active-site base that mediates C8 deprotonation has been uncertain as a result of studies examining the fate of the C2 or C8 proton during turnover.11, 14 In one study, RlmN and Cfr reactions were performed with a 342-nucleotide fragment of 23S rRNA containing deuterium at C2 of all adenosine nucleotides. MS analysis of the 2-methyladenosine (m2A) products showed full retention of the substrate-derived C2 deuteron in reactions containing RlmN and ~95% retention of the C2 deuteron in reactions containing Cfr, which was interpreted as evidence of a hydride shift from C2 or C8 to the nascent methyl moiety during the reaction.11 In another study, Cfr was overproduced in an E. coli methionine auxotroph that was cultured in media supplemented with [methyl-d3]-methionine. In vivo, [methyl-d3]-methionine is converted to [methyl-d3]-SAM, which affords a [methyl-d3]-mCys residue on Cfr. When Cfr [methyl-d3]-mCys was reacted with a 7-nucleotide (7mer) rRNA fragment containing A2503, an envelope of isotopic products was observed, indicating solvent–hydron exchange during methylation and arguing against a hydride-shift mechanism.14 It was proposed that solvent-hydron exchange during the Cfr-mediated production of 8-methyladenosine (m8A) might be due to the action of a polyprotic base.14 These results led to the suggestion that Cfr uses different bases when methylating C2 versus C8. Herein, we show via deuterium-transfer experiments that analogously to RlmN, C105 of Cfr, and not a polyprotic amino acid, is the required base both for C2 and for C8 methylation, and that exchange is mediated most likely by one tightly bound water molecule during C8 methylation.
Materials and Methods
Materials.
Vent polymerase and Antarctic phosphatase were purchased from New England Biolabs (Ipswich, MA). Nuclease P1 from Penicillium citrinum was purchased from Sigma-Aldrich (St. Louis, MO). All oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) and used as received. Talon cobalt-resin was purchased from Clontech (Mountain View, CA). PD-10 pre-poured gel-filtration columns were purchased from GE Biosciences (Piscataway, NJ). S-Adenosylmethionine (SAM) and S-adenosyl[methyl-d3]methionine (d3-methyl-SAM) were synthesized enzymatically and purified as described previously.15 2H8-adenosine 5’-triphosphate (90% D) was purchased from Cambridge Isotopes (Andover, MA). RNA substrates (155mer, deu155mer, 16mer, deu16mer) were prepared using run-off transcription as previously described.9 E. coli flavodoxin and flavodoxin reductase were generated as previously described.16
Production of Apo Cfrwt, Cfrwt, and CfrC105A.
Overexpression and purification of apo Cfrwt, holo Cfrwt, and Cfr C105A was performed as previously described.8, 9, 12 Apo Cfr refers to the enzyme lacking its [4Fe–4S] cluster as well as a methylated Cys355 residue. Holo Cfr refers to the enzyme that is isolated with its [4Fe–4S] cluster intact and with a methylated Cys355 residue.
Production of CfrK79Q, CfrE91A, and CfrR327Q.
Cfr variants were generated using the Stratagene QuikChange II kit (Agilent Technologies) with the primers shown in Table S1. However, Pfu polymerase was substituted with Vent polymerase in the PCR reaction. CfrK79Q and CfrR327Q were overproduced and purified in their holo forms as previously described.8, 9, 12, 14, 16 CfrE91A was overproduced and purified in its apo form.
Reconstitution of Apo Cfrwt and Apo CfrE91A:
Reconstitution of apo Cfrwt and apo CfrE91A was performed as previously described, with the exception that a 20-fold excess of iron and sulfide, and a 2000-fold excess of DTT were used in the reconstitution reaction.8, 9
In vitro Transcription of the 155mer and deu155mer RNA Substrates:
The 155mer RNA substrate was transcribed in vitro as previously described with minor modifications.9, 12 Triton and spermidine were withheld from the reaction and the MgCl2 concentration used in the reactions was optimized for each new stock of DNA template used. 2H8-adenosine 5’-triphosphate (90% D) was used instead of ATP to produce the 155mer with deuterium at all positions in adenosine (deu155mer).
Synthesis and Purification of the 7mer RNA Substrate:
The 7mer RNA substrate, consisting of E. coli 23S rRNA bases 2500-UCGAUGU-2506, was obtained from Dharmacon Inc. (GE Healthcare). The substrate was deprotected using Dharmacon’s published standard protocol, lyophilized to dryness, and resuspended in deoxygenated water. The concentration was determined using the supplied extinction coefficient of 69,900 M−1cm−1.
Generation of the 16mer RNA Template (nucleotides 2494 to 2509 of E. coli 23S RNA):
The 16mer DNA template was generated through annealing of a forward (5’-cggaaattaatacgactcactataggcacctcgatgtcgg-3’) and a reverse (3’-mCmCgacatcgaggtgcctatagtgagtcgtattaatttccg-5’) primer, where the underlined portions are the T7 promoter regions and where mC stands for 2’-methoxycytosine. A solution (100 μL) containing 5 nmol of each primer was incubated at 100 °C for 1 min and then allowed to cool to room temperature over 40 min. The resulting template was used in in vitro transcription reactions to generate the 16mer RNA substrate as previously described.9 Transcription reactions contained the following in a final volume of 10 mL: 30 mM Tris–HCl, pH 8.4, 26 mM MgCl, 10 ng/μL DNA template, 20 mM DTT, 4 mM each NTP and ~4 mg of T7 RNA polymerase. The reaction was incubated at 37 °C for 1 h and then quenched by adding EDTA to a final concentration of 50 mM. The reaction was exchanged into water using a PD-10 gel-filtration column. The eluate containing RNA was concentrated using an Amicon Centricon with a YM-3 membrane. An extinction coefficient of 186,000 M−1 cm−1 was used to determine the concentration. The extinction coefficient was calculated using “Oligo Calc: Oligonucleotide Properties Calculator” (basic.northwestern.edu/biotools/oligocalc.html). 2H8-adenosine 5’-triphosphate (90% D) was used instead of ATP to produce the 16mer with deuterium at all positions in adenosine (deu16mer).
Cfr Variant Turnover Assays:
Assays contained in a final volume of 100 μL: 20 μM Cfr variant, 100 mM EPPS, pH 8, 10 mM MgCl2, 2 mM SAM, and 100 μM 155mer RNA substrate. The reactions were initiated by adding dithionite to a final concentration of 2 mM. Aliquots were removed at designated times and added to a quench solution containing 100 mM H2SO4 and 100 μM tryptophan (internal standard, IS). The samples were neutralized with 20 μL 2 × P1 nuclease buffer (250 mM sodium acetate, pH 6.0, 45 mM NaCl and 4 mM ZnCl2). The RNA was digested to its individual nucleotides by incubating the neutralized samples overnight at 37 °C with 0.25 units of P1 nuclease and 5 U of Antarctic phosphatase. The samples were centrifuged for 15 min to remove any precipitate, and the supernatants were analyzed as previously described.9, 12, 14 Each assay was performed in duplicate. The m2,8A standard was synthesized as previously described.17
Multiple Turnover Assays with Cfr wt and the 155mer RNA Substrate:
Assays contained the following in a final volume of 100 μL: 5 μM Cfr wt, 100 mM EPPS, pH 8, 10 mM MgCl2, 100 μM 155mer, 2 mM SAM, and either 50 μM flavodoxin, 25 μM flavodoxin reductase, and 2 mM NADPH or 2 mM dithionite. Aliquots were removed at designated times and quenched in a solution of 100 mM H2SO4 and 100 μM tryptophan (IS). The samples were digested and analyzed as described above. Each assay was performed in triplicate.
Assays with Cfr wt and the 7mer or 16mer RNA Substrates:
Assays contained the following in a final volume of 100 μL: 100 μM Cfr wt, 100 mM, EPPS pH 8, 10 mM MgCl2, 2 mM SAM, 200 μM 7mer or 16mer, and 2 mM dithionite or 100 μM flavodoxin, 100 μM flavodoxin reductase and 2 mM NADPH. Reactions were initiated by adding NADPH or dithionite, and aliquots were removed at designated times and added to a quench solution of 100 mM H2SO4 and 100 μM tryptophan (IS). The samples were prepared for analysis by mass spectrometry as described for assays with the 155mer RNA substrate. These assays were performed in triplicate.
Neutral-Loss Mass Spectrometry for Analysis of Digested RNA:
Neutral-loss mass spectrometry (NLMS) was used to analyze the isotopic distribution of the Cfr reaction products as described previously.14 A neutral loss of 132 (natural abundance ribose) or 138 (perdeuterated ribose) was used for the assays described herein.
Preparation of CfrD2O for Use in D2O Exchange Assays:
50 mL of Cfr S200 buffer prepared in H2O (20 mM HEPES, pH 7.1 (pD 7.5), 500 mM KCl, 5 mM MgCl2, 25% glycerol) was twice dried down and resuspended in 30 mL D2O and then dried down again. The dried buffer was subsequently introduced into the anaerobic chamber and diluted to a final volume of 50 mL with D2O. Solid DTT was then added to a final concentration of 5 mM, generating S200D2O buffer. Apo Cfrwt→rcn (50 to 100 μL) was diluted to 4 mL with S200D2O buffer and concentrated to 200 μL in a 10,000 MWCO 4 mL centricon. The concentrated enzyme was diluted with 2 mL of D2O S200 buffer and concentrated again to 200 μL. A Bradford assay was performed to determine the concentration of the protein. The equivalent steps were performed using H2O S200 buffer for CfrH2O to be used in parallel assays with CfrD2O.
Preparation of 155merD2O:
The 155mer RNA substrate was diluted to a final volume of 4 mL with 100% anaerobic D2O and concentrated to 200 μL in a 10,000 MWCO 4 mL centricon. The 155mer was further diluted with an additional 2 mL of D2O, concentrated to 200 μL and used in D2O exchange assays as described below.
D2O Exchange Assays to Observe Deuterium Exchange from Solvent to Product.
CfrD2O and CfrH2O were used in assays to observe deuterium exchange from solvent into the methylated product. Aliquots (0.5 mL) of reaction mixtures containing 100 mM EPPS, pH 7.6 (pD 8), 10 mM MgCl2, and 200 μM 16mer or 7mer RNA substrate (see above for 155mer preparation in D2O) were lyophilized and resuspended in 0.5 mL of D2O. SAM, 155merD2O, and enzyme were added to final concentrations of 2 mM, 100 μM and 10 μM (155mer) or 100 μM (16mer and 7mer). Reactions were initiated by adding 2 mM dithionite, resulting in a 95% final percentage of D2O. Aliquots were removed at designated times and added to a quench solution of 100 mM H2SO4 and 100 μM tryptophan (IS). Samples were digested and analyzed using NLMS. A control, in which reactions were diluted in H2O, was performed in parallel.
Assays to Examine Deuterium Exchange from Perdeuterated RNA to Solvent:
Reactions contained the following in a final volume of 100 μL: apo Cfrwt→rcn (10 μM for deu155mer, 100 μM for deu16mer), 100 mM EPPS, pH 8, 10 mM MgCl2, 2 mM SAM, RNA substrate (100 μM of deu155mer, or 200 μM of deu16mer), 50 μM flavodoxin, 25 μM flavodoxin reductase, and 2 mM NADPH (FFR system). Dithionite (2 mM) was substituted for the FFR reducing system in reactions with the deu16mer substrate. All reaction products were analyzed using NLMS.
Assays to Examine Deuterium Exchange from SAM Derived Methyl Group to Solvent:
Reactions contained the following in a final volume of 100 μL: apo Cfrwt→rcn (10 μM for 155mer, 100 μM for 16mer and 7mer), 100 mM EPPS, pH 8, 10 mM MgCl2, 2 mM [methyl-d3]-SAM, RNA substrate (100 μM of 155mer, or 200 μM of 16mer or 7mer), 50 μM flavodoxin, 25 μM flavodoxin reductase, and 2 mM NADPH. Dithionite (2 mM) was substituted for the FFR reducing system in reactions with the 16mer and 7mer. At designated times, 10 μL aliquots were removed and the reaction was quenched by mixing each aliquot with 10 μL of a solution containing 100 mM H2SO4 and 100 μM tryptophan (IS). Samples were digested and analyzed using NLMS. A control reaction was performed using unlabeled SAM to determine an apparent kinetic isotope effect for [methyl-d3]-SAM. All assays were performed in triplicate.
RESULTS
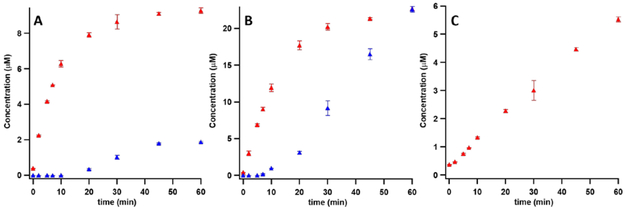
In our initial studies to identify the key base in the Cfr reaction, we focused on conserved polyprotic amino acid residues that are predicted to be in the active site of Cfr based on a homology model of the protein generated from the structure of RlmN with bound SAM (Figure S2).18-20 One such strictly conserved residue is K79.14 However, when a K79Q variant was constructed and used in Cfr reactions, it catalyzed production of m8A with a kcat of 3.34 × 10−2 ± 5.2 × 10−5 min−1 and m2,8A with a kcat of 2.64 × 10−3 ± 4.2 × 10−6 min−1 (Figure 1, Panel A). Given that the wild-type (wt) enzyme exhibits a kcat of 2.44 × 10−1 ± 5.6 × 10−2 min−1, it is unlikely that this amino acid is the targeted base in the reaction. Changes to other potential general bases (Glu91 and Arg327) gave similar results, ruling those out as well (Figure 1). The inability to find a conserved polyprotic amino acid residue in Cfr that causes a substantial loss in activity when changed to an amino acid with a nonacidic functional group led to a re-examination of the solvent–hydron exchange reported previously.12
Figure 1:
Time-dependent formation of m8A (red) and m2,8A (blue) by Cfr K79Q (A), Cfr E91A (B) and Cfr R327Q (C) in assays containing 20 μM Cfr variant, 100 μM 155mer RNA substrate, and 2 mM SAM. In all assays, production of m8A was observed, ruling out these conserved residues as active site bases. Error bars represent the mean ± SD of two replicates.
In the experiments herein, Cfr is overproduced in its apo form, as previously described, and then reconstituted with iron and sulfide to regenerate its required iron-sulfur (Fe-S) cluster.8, 9, 12 This form of the protein is designated apo Cfrwt→rcn, (Figure S3). Cfr prepared in this manner lacks the mCys residue, which allows us to probe the exchange of deuterium from the methyl moiety of SAM during catalysis by employing [methyl-d3]-SAM in our reactions.
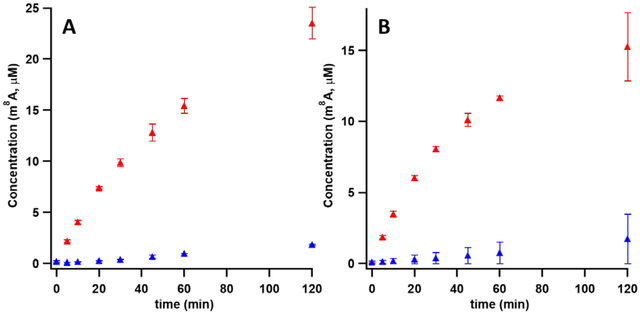
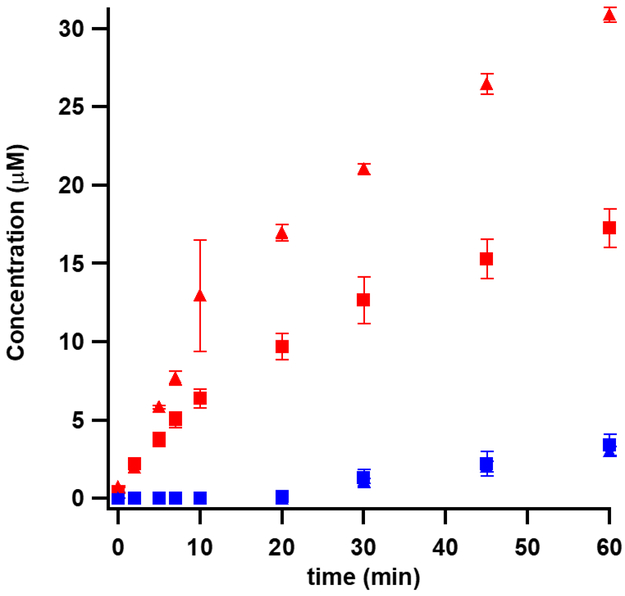
In initial experiments, the effect of substrate size on the extent of solvent-hydron exchange was assessed by using three different lengths of RNA: a 7mer spanning E. coli 23S rRNA bases 2500-2506; a 16mer spanning E. coli 23S rRNA bases 2494-2509; and a 155mer spanning E. coli 23S rRNA bases 2454-2608. In the presence of 200 μM 7mer or 16mer, 100 μM Cfr produces 25 μM and 15 μM m8A product in 120 min, respectively. Methyl-2,8-adenosine (m2,8A) is not observed with the 7mer, while the 16mer affords production of m2,8A, but it is below the limit of quantification (Figure 2). Moreover, only minute amounts of product is observed with these smaller substrates when the flavodoxin reducing system (flavodoxin, flavodoxin reductase, NADPH) is replaced with dithionite as the required reductant. By contrast, in the presence of 100 μM 155mer, 5 μM Cfr supports multiple turnovers in 60 min when using either the flavodoxin reducing system (7 turnovers) or dithionite (4 turnovers) as the electron source. Using this substrate, production of both m8A and m2,8A is observed (Figure 3).
Figure 2:
Reactions comparing the production of methylated product with the 7mer (A) or 16mer (B) RNA substrates by apo Cfrwt→rcn when using dithionite (red) or flavodoxin/flavodoxin reductase (blue) as reductants. The reactions contained 100 μM Cfr, 200 μM 7mer or 16mer RNA substrate, and 2 mM SAM. With both the small substrates, production of m8A is better when using dithionite as a reductant. Error bars represent the mean ± SD of three replicates.
Figure 3:
Formation of m8A (red) and m2,8A (blue) by Cfr wt using dithionite (squares) or the flavodoxin/flavodoxin reductase (FFR) reducing system (triangles). The reactions contained 5 μM Cfr wt, 2 mM SAM, and 100 μM 155mer RNA substrate. The FFR reducing system supports better turnover of the 155mer RNA substrate than dithionite. Error bars represent the mean ± SD of three replicates.
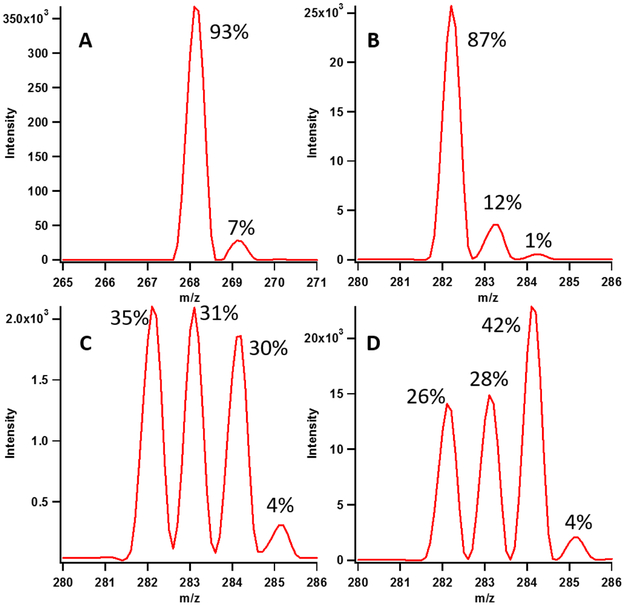
To assess whether a polyprotic amino acid is the targeted base during C8 methylation by Cfr, reaction mixtures were reconstituted in 95% D2O to determine the extent of deuterium exchange into the m8A product. Shown in Figure 4 (panel A) is an adenosine standard, which exhibits a parent ion [M+H]+ at 268.1 m/z and an [M+H]+ +1 ion at 269.1 m/z (7% of that of the parent ion) due to contributions of heavier isotopes in the molecule (predominantly 13C). Upon turnover in the presence of the 155mer substrate, the peak at 268.1 m/z shifts to 282.1, which is consistent with the addition of a methyl moiety (+14 Da) to C8 of A2503. The peak at 269.1 m/z shifts to 283.1, and now represents 12% of the magnitude of the parent ion (Figure 4, panel B). This observation suggests that when Cfr methylates C8 in the presence of the 155mer substrate, only a small amount of solvent hydrons exchange into the product, which is inconsistent with a polyprotic amino acid as the base shown in intermediate v of Scheme 1. In the case of a polyprotic amino acid (e.g. a lysyl residue), up to 66% of the product would be expected to contain a deuterium atom exchanged into the nascent methyl moiety. Production of m2,8A is also observed (m/z 296.1), and this product contains a further 5% increase in deuterium content (m/z 297.1), indicating that a very small amount of solvent deuterium exchanges into the C2 methyl moiety of m2,8A during Cfr-mediated methylation of m8A (Figure S4, Panel A).
Figure 4:
Representative isotopic abundance of substrate adenosine (A) and m8A (B-D) from assays performed in the presence of D2O with the 155mer (B), 16mer (C) and 7mer (D) RNA substrates after two hours. In panel B, only 5% of deuterium is observed to exchange in with the 155mer. In contrast, the 16mer and 7mer allow for greater incorporation of one and two deuterons into the product. Only a maximum of two deuterons gets incorporated into the product with the 16mer and 7mer. Isotopic abundance variation for assays performed in the presence of D2O is presented in Table S2.
When similar reactions are performed with the 16mer and 7mer substrates, extensive exchange of deuterium into the m8A product is observed. Reactions containing the 16mer substrate result in an approximate ratio of 1:1:1 of d0-, d1-, and d2-containing products (Figure 4, Panel C), while reactions containing the 7mer substrate result in an even larger incorporation of solvent deuterium in the product (26% d0, 28% d1 and 42% d2, Figure 4, Panel D). Importantly, the observed peak at m/z 285 for the reactions with the 16mer and 7mer substrates (4%) is consistent with the natural isotopic abundance of 13C in the product. Therefore, it appears that Cfr does not catalyze exchange of all three hydrogens of the installed methyl group. Given the unlikeliness of a polyprotic amino acid and the observation that only up to two solvent-derived hydrons are exchanged into the methyl moiety of the m8A product, the simplest explanation for the observed exchange is the presence of an isolated reservoir of protons in the active site, which is most likely an anchored or trapped water molecule that is in association with the general base. In addition, the above results suggest that the smaller substrates allow greater solvent exchange with the active site base, most likely by slowing down one or more of the steps following proton abstraction, including tautomerization and protonation of the enamine intermediate. Given the low rate of exchange observed with the 155mer RNA substrate, it is unlikely that water itself is the base.
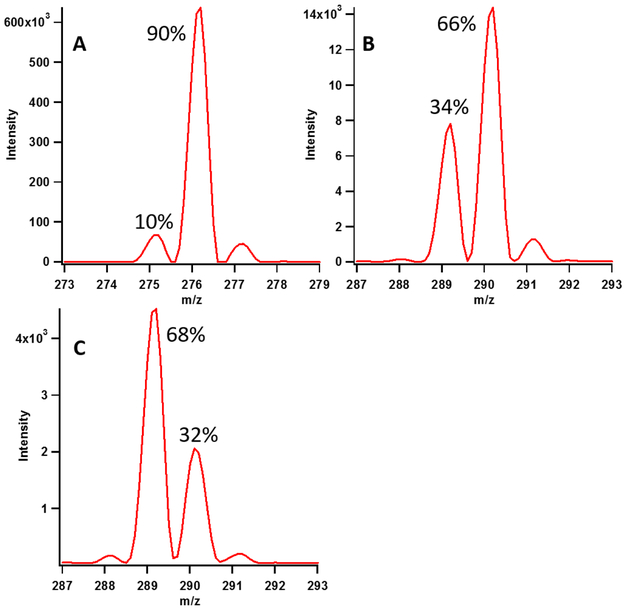
A subsequent set of complementary experiments were conducted with RNA substrates containing deuterium at C2 and C8 of all adenosines to quantitatively assess the transfer of deuterium from the substrate into the nascent methyl moiety. One substrate, termed the deu155mer, was generated by in vitro transcription of the 155mer in the presence of [U-2H]ATP. An adenosine moiety labeled in this fashion displays an m/z of 276.1, as shown in Figure 5 (Panel A); however, about 10% of the substrate is missing one deuterium atom (m/z 275.1), which is likely at C8 of the molecule. Previous studies investigating the acidity of purine carbon acids showed that isotopic exchange takes place at C8 under neutral conditions ~2000 times faster than at C2.21 In the RlmN reaction, the C2 hydrogen is returned to the methyl moiety of the product without exchange with solvent hydrons. However, an analysis of the exchange of the C8 hydrogen with Cfr has not been conducted. To assess the fate of the C8 hydrogen during C8 methylation by Cfr, apo Cfrwt→rcn was incubated with SAM, reductant, and either the deu155mer or the analogous 16mer (deu16mer). In the presence of the deu155mer, the m8A produced displays a major peak at m/z 290 (66%), which corresponds to a methylated product in which the C8 deuterium is retained (Figure 5, Panel B). Also observed is a peak at m/z 289, which is present at 34%. This peak corresponds to an m8A product in which the C8 deuterium is not returned to the nascent methyl moiety, indicating solvent exchange before return. Production of m2,8A that contains a similar deuterium content as the m8A product is observed with the deu155mer, suggesting that methylation at C2 takes places with significantly less deuterium exchange than methylation at C8 (Figure S4, Panel B).
Figure 5:
Representative isotopic distribution of substrate adenosine (A) and m8A (B and C) from assays performed with deu155mer (B) and deu16mer (C) after 120 minutes. Panel B shows the majority peak at m/z 290, which is consistent with the C8 deuteron being returned to the methyl group. Panel C shows that the majority species is m/z 289, which is consistent with the C8 deuteron not being returned to the methylated product. Isotopic abundance variation for assays performed with deuRNA substrates is presented in Table S3.
The exchange observed during C8 methylation supports the hypothesis that hydrons on a tightly bound water molecule are in facile exchange with the protonated target base. When the reaction is performed with the deu16mer, a majority species containing no deuterium in the methyl group (m/z 289, 68%) is observed (Figure 5, Panel C). The larger exchange of the C8 deuteron with the deu16mer as compared to the deu155mer (58% vs. 24% when correcting for the percentage of deuterium in the starting substrate) supports our hypothesis that the smaller substrates likely slow down a step subsequent to deprotonation at C8. Time-dependent analysis of exchange during production of m8A with the deu16mer shows that the extent of deuterium exchange does not vary with respect to time, suggesting that at the completion of a reaction cycle (including product release), no further exchange takes place (Figure S5).
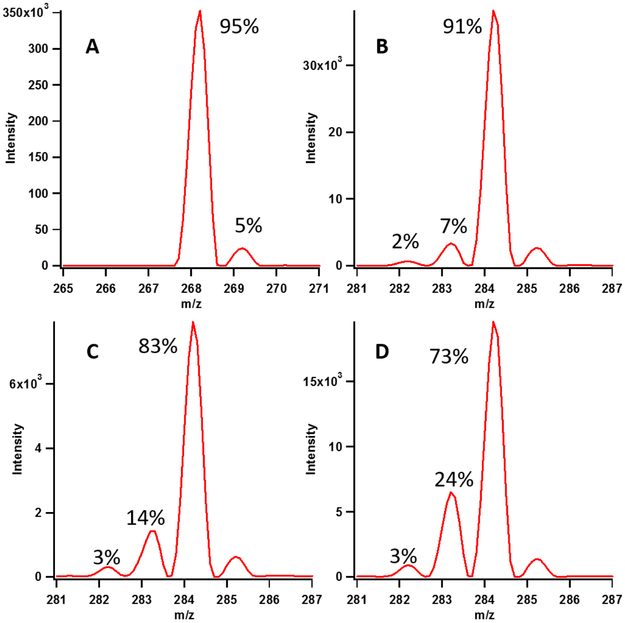
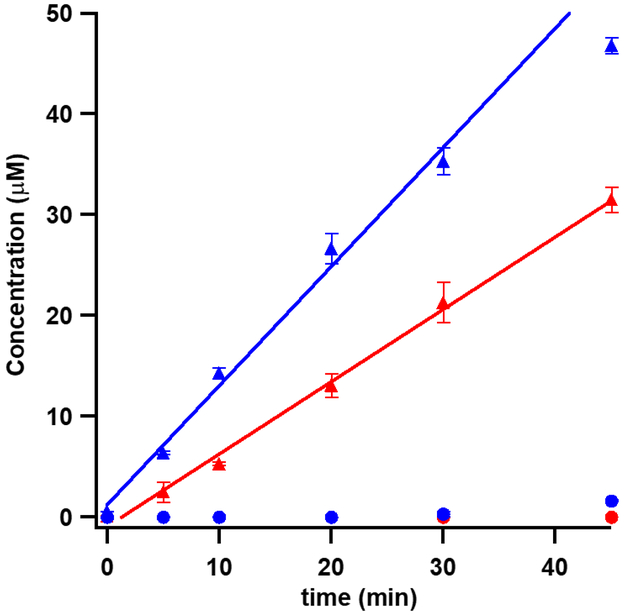
Finally, we assessed the exchangeability of deuterium incorporated from [methyl-d3]-SAM in an effort to examine deuterium exchange during the enamine/imine tautomerization step (Scheme 1). When using the 155mer as the substrate, the m8A product displays a majority species containing two deuterium atoms (m/z 284, 91%), with a small percentage of product containing one (7%) or no (2%) deuterium atoms (Figure 6, Panel B). These results are consistent with the 5’-dA• abstracting a deuterium atom from the mCys residue without further significant loss of deuterium from the C1 moiety.14 The loss of a small amount of SAM-derived deuterium is best explained by a reversible enamine:imine tautomerization. Kinetic analysis of the production of m8A with [methyl-d3]-SAM showed an apparent kinetic isotope effect on Vmax of the overall reaction of 1.65 ± 0.17 (Figure 7). We observed production of m2,8A when using [methyl-d3]-SAM that exchanged out a small amount of SAM-derived deuterons during methylation at C2, consistent with less exchange occurring during methylation of C2 than C8 (Figure S4). The 16mer and 7mer were also used in reactions with [methyl-d3]-SAM. As before, the smaller substrates led to an increase in the amount of deuterium exchange (Figure 6).
Figure 6:
Representative isotopic distribution of substrate adenosine (A) and product m8A (B-D) from assays performed with [methyl-d3]-SAM, apo Cfrwt→rcn, and 155mer (B), 16mer (C) and 7mer (D) after 120 minutes. In all product spectra the majority species is d2-containing m8A (m/z 284) with smaller amounts of d1 and d0-containing products. Additionally, with decreasing substrate size, greater loss of deuterium is observed. Isotopic abundance variation for assays performed with [methyl-d3]-SAM is presented in Table S4.
Figure 7:
Reactions comparing the rate of production of m8A (closed triangles) and m2,8A (closed circles) by apo Cfrwt→rcn when using unlabeled SAM (blue) or [methyl-d3]-SAM (red). The reactions contained 10 μM apo Cfrwt→rcn, 100 μM 155mer RNA substrate, and 2 mM methyl-d3-SAM or SAM. From the ratio of the slopes, an apparent kinetic isotope effect of 1.65 ± 0.17 can be extracted. Error bars represent the mean ± SD of three replicates.
DISCUSSION
In this work, we studied the exchange of hydrons out of various substrates in the Cfr reaction as well as the exchange of solvent hydrons into the Cfr product during turnover. In contrast to a previous suggestion, we find no evidence for a polyprotic base that mediates exchange in the reaction. This conclusion is based on the inability to identify an amino acid in the active site of a Cfr structural model that could act as a polyprotic base and that affords a poorly active catalyst upon its substitution. It is also based on the finding that Cfr-mediated exchange of solvent hydrons into the product of the reaction is not as extensive as would be predicted for a polyprotic base when using a good substrate (155mer), given that the product should contain approximately 33% of solvent hydrons. Our results are more consistent with a monoprotic base — most likely Cys105 — that is in close association with only one molecule of water. Moreover, the relatively high rate of exchange of deuterium out of a good substrate (155mer) containing a C8 deuterium atom, but a relatively low exchange of deuterium from D2O into the methylated product, is consistent with cysteine acting as the base. The relatively low fractionation factor associated with cysteines (~0.4-0.5) would favor an intermediate in step vi of Scheme 1 that contains hydrogen on Cys105 rather than deuterium.22 Therefore, exchange in from D2O would be disfavored, while exchange out from a substrate containing a C8 deuteron would be favored; although, structural perturbations of the protein in the presence of D2O could also affect the extent of deuterium exchange.
Our finding that up to two solvent hydrons can exchange into the methyl group of the product necessitates — based on the mechanism shown in Scheme 1 — that there be a second step in which exchange can take place. We propose that this step is the imine-enamine tautomerization shown between intermediates viii and ix in Scheme 1, which is shown as being reversible. Consistent with this suggestion is the exchange of deuterium out of the methylated product that is observed when using [methyl-d3]-SAM in reactions. According to Scheme 1, this exchange can take place essentially after turnover is completed but before the substrate leaves the active site. Our observation that exchange into the methyl moiety of the m8A product is not enhanced upon longer incubation times when the deu16mer as a substrate (Figure S5) suggests that, upon dissociation of the product of the reaction, it does not rebind in a manner that allows further modification to C8. Presumably, C8 methylation effects a conformational change in A2503 that causes it to bind with its C2 carbon in proximity with Cfr’s catalytic machinery. In addition, the low exchange of deuterium out of the methyl group of the product when [methyl-d3]-SAM is used in reactions containing a good substrate is consistent with this exchange being mediated by a cysteinyl group. Our suggestion that Cys105 is the base that mediates exchange when Cfr is acting on C8 of A2503 is consistent with our studies of formation and decay of the protein-RNA cross-linked intermediate. When the C105A variant is used in EPR studies, the paramagnetic cross-linked intermediate forms but does not decay during the time span of the experiment.12 The inability to observe significant exchange in the RlmN-catalyzed reaction might suggest that a tightly bound water molecule is not present adjacent to the catalytic base in the RlmN active site, perhaps due to the larger ring structure on which RlmN operates as compared to that on which Cfr operates. Nevertheless, the exchange observed in the Cfr reaction is important, because it rules out a mechanism involving a hydride shift.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the Huck Institute for Life Sciences Genomics Core for DNA sequencing.
Funding Sources
This work was supported by NIH GM-122595 to S. J. B, who is also an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- m2A
2-methyladenosine
- m8A
8-methyladenosine
- 5’-dA
5’-deoxyadenosine
- EPR
Electron Paramagnetic Resonance
- MS
mass spectrometry or mass spectrum
- (m2,8A)
methyl-2,8-Adenosine
- mCys
methylcysteine residue
- PTC
peptidyl transferase center
- RS
radical SAM
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
Footnotes
Supporting Information.
Table S1-S4 and Figures S1-S5 are available free of charge on the ACS Publications website at DOI: http://pubs.acs.org
REFERENCES
- (1).CDC. (2013) Antibiotic Resistance Threats in the United States, U.S. Department of Health and Human Services, https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed Feb 16, 2017).
- (2).Poehlsgaard J, and Douthwaite S (2005) The bacterial ribosome as a target for antibiotics, Nat. Rev. Microbiol 3, 870–881. [DOI] [PubMed] [Google Scholar]
- (3).Steitz TA (2010) From the structure and function of the ribosome to new antibiotics (Nobel Lecture), Angew. Chem. Int. Ed 49, 4381–4398. [DOI] [PubMed] [Google Scholar]
- (4).Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, and Vester B (2006) The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics, Antimicrob. Agents Chemother 50, 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Smith LK, and Mankin AS (2008) Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors, Antimicrob. Agents Chemother 52, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Benítez-Páez A, Villarroya M, and Armengod M-E (2012) The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy, RNA 18, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, and Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods, Nucleic Acids Res. 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Grove TL, Radle MI, Krebs C, and Booker SJ (2011) Cfr and RlmN contain a single [4Fe–4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation, J. Am. Chem. Soc 133, 19586–19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Grove TL, Livada J, Schwalm EL, Green MT, Booker SJ, and Silakov A (2013) A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr, Nat. Chem. Biol 9, 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schwalm EL, Grove TL, Booker SJ, and Boal AK (2016) Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA, Science 352, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yan F, and Fujimori DG (2011) RNA methylation by radical SAM enzyme RlmN and Cfr proceeds via methylene transfer and hydride shift, Proc. Natl. Acad. Sci. U S A 108, 3930–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Silakov A, Grove TL, Radle MI, Bauerle MR, Green MT, Rosenzweig AC, Boal AK, and Booker SJ (2014) Characterization of a cross-linked protein-nucleic acid substrate radical in the reaction catalyzed by RlmN, J. Am. Chem. Soc 136, 8221–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, and Rosenzweig AC (2011) Structural basis for methyl transfer by a radical SAM enzyme, Science 332, 1089–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, and Booker SJ (2011) A radically different mechanism for S-adenosylmethionine-dependent methyltansferases, Science 332, 604–607. [DOI] [PubMed] [Google Scholar]
- (15).Iwig DF, and Booker SJ (2004) Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine, Biochemistry 43, 13496–13509. [DOI] [PubMed] [Google Scholar]
- (16).Lanz ND, Grove TL, Gogonea CB, Lee KH, Krebs C, and Booker SJ (2012) RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins, Methods Enzymol. 516, 125–152. [DOI] [PubMed] [Google Scholar]
- (17).Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, and Fujimori DG (2010) RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA, J. Am. Chem. Soc 132, 3953–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Roy A, Kucukural A, and Zhang Y (2010) I-TASSER: a unified platform for automated protein sturcutre and function prediction, Nature Protocols 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015) The I-TASSER Suite: Protein structure and fuction prediction, Nature Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang Y (2008) I-TASSER server for protein 3D structure prediction, BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Elvidge JA, Jones JR, O’Brien C, and Evans EA (1971) Isotopic hydrogen exchange in purine, adenine, adenosine, and benzimidazole, Chem. Commun, 394–395. [Google Scholar]
- (22).Schowen RL (1972) Mechanistic deductions from solvent isotope effects, Prog. Phys. Org. Chem 9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.