Abstract
L-carnitine is a candidate therapeutic for the treatment of septic shock, a condition that carries a ≥40% mortality. Responsiveness to L-carnitine may hinge on unique metabolic profiles that are not evident from the clinical phenotype. To define these profiles, we performed an untargeted metabolomic analysis of serum from 21 male sepsis patients enrolled in a placebo-controlled L-carnitine clinical trial. Although treatment with L-carnitine is known to induce changes in the sepsis metabolome, we found a distinct set of metabolites that differentiated 1-year survivors from non-survivors. Following feature alignment, we employed a new and innovative data reduction strategy followed by false discovery correction, and identified 63 metabolites that differentiated carnitine-treated 1-year survivors versus non-survivors. Following identification by MS/MS and database search, several metabolite markers of vascular inflammation were determined to be prominently elevated in the carnitine-treated non-survivor cohort, including fibrinopeptide A, allysine, and histamine. While preliminary, these results corroborate that metabolic profiles may be useful to differentiate L-carnitine treatment responsiveness. Furthermore, these data show that the metabolic signature of L-carnitine-treated non-survivors is associated with a severity of illness (e.g., vascular inflammation) that is not routinely clinically detected.
Keywords: liquid chromatography-mass spectroscopy, pharmacometabolomics, vascular inflammation
Graphical Abstract
Introduction
Sepsis remains a significant hazard to human health and its most severe form, septic shock, carries a mortality rate of ≥ 40%.1 Sepsis can be described as a “metabolic crisis” as it is associated with alterations in global metabolism as evidenced by disruption in pathways that lead to alterations in serum amino acid and free fatty acid levels.2–5 In addition, there is evidence that sepsis induces a modest carnitine deficiency and that non-survivors have perturbations in carnitine processing.4, 6 Carnitine is critically important for energy homeostasis because it is required for the transport of long-chain fatty acids into the mitochondria that are needed for acyl-CoA generation and beta-oxidation. These mechanisms have served as the basis for testing the efficacy of supplemental L-carnitine as a therapeutic for septic shock.
We have previously demonstrated the therapeutic value of supplemental L-carnitine as a treatment strategy for human patients with vasopressor-dependent septic shock.7 Importantly, we have discerned that the pre-treatment metabolome may identify phenotypes of septic shock patients that are responsive to L-carnitine supplementation. In addition, we introduced the concept that L-carnitine provokes sepsis metabolism (as an oral glucose tolerance test during pregnancy uncovers latent susceptibility to diabetes mellitus), since the post-treatment carnitine and acylcarnitine concentrations distinguished septic shock survivors and non-survivors.8 However, with the exception of the acylcarnitines, there was only a modestly detectable metabolic response (by quantitative nuclear magnetic resonance spectroscopy) to L-carnitine, and the pre-treatment differentiation was primarily due to differences in the concentration of ketone bodies.6 Given the known metabolic derangements of sepsis and the expected downstream metabolic consequences of L-carnitine supplementation, we aimed to further define the metabolic response to L-carnitine and to investigate differences in the metabolome of 1-year septic shock survivors and non-survivors. The ultimate goal of this investigation was to better understand how the metabolic changes of sepsis relate to patient-centered outcomes, and the potential role of provoked metabolic testing to elicit unique metabolic signatures that relate to those outcomes. To accomplish these objectives, we employed untargeted LC-MS-based metabolomics and developed a novel data reduction approach to study serum samples acquired from patients enrolled in a phase I L-carnitine clinical trial. We hypothesized that a metabolic signature of carnitine treatment would distinguish septic shock survivors from non-survivors.
Materials and Methods
Clinical trial details
Serum samples were collected from subjects enrolled in a clinical trial of L-carnitine for the treatment of vasopressor-dependent septic shock.7 The clinical trial was IRB approved, FDA regulated (IND #107,086) and registered on clinicaltrial.gov (NCT01193777); all subjects or their surrogates provided informed consent prior to enrollment. Details related to the clinical trial such as inclusion and exclusion criteria, L-carnitine administration parameters and serum collection details have been previously described.8 While the trial was conducted prior to release of the sepsis-3 definitions,1, due to a priori inclusion criteria related to the study hypothesis all patients met criteria for the new definition of septic shock. Although the primary outcome of the clinical trial was safety, mortality was assessed. Carnitine-treated patients had a better 1-year mortality rate than placebo-treated patients (56% versus 75%; p=0.06).9 For the purposes of this study, interpretation of untargeted metabolomics data with respect to 1-year survival was limited to male subjects (n=21) as detailed in the results section.
Sample collection and extraction
Blood samples were collected via an indwelling IV or arterial catheter before, 24h after and 48h after the initiation of a 12h L-carnitine (12 g) or saline (control) infusion. Blood samples were processed to generate serum as previously described.6 Serum was stored at −80°C until processing for metabolomic analysis. To prepare serum for metabolomics, samples were thawed on ice and 50 uL aliquots of were transferred to polypropylene microcentrifuge tubes. A quality control (QC) sample was prepared by pooling 20 serum samples selected at random from the batch and was thereafter treated identically to other samples. The serum was vortexed with 200 uL of organic solvent (1:1:1 methanol:acetonitrile:acetone) containing a mixture of internal standard compounds (supporting information: Table S1, internal standards). After 5 minutes on ice, samples were centrifuged (15,000 × g, 4°C, 10 min). Supernatant (200 μL) was transferred to clean tubes and dried under a gentle stream of nitrogen gas at room temperature. The samples were then reconstituted in 9:1 water:methanol (50 μL) and were transferred to autosampler vials for untargeted metabolomics assay.
Untargeted LC-MS metabolomics
Samples were analyzed on an Agilent 1200LC/ 6530 qTOF MS (Santa Clara, CA) using a reversed-phase liquid chromatography (RPLC) method similar to that previously described.10 Briefly, 10 μL of reconstituted sample was injected on a Waters Acquity HSS T3 column, 2.1 mm inner diameter × 100 mm length, 1.8 μ particle diameter. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The column was eluted using a 20-minute gradient from 2 to 75% B, followed by a 7-minute wash at 98%B and a 10-minute re-equilibration at 2% B (total run time 37 min, flow rate 0.44 mL/min). Each sample was analyzed twice; once in positive ion mode and once in negative ion mode. At the beginning of the batch and after every 8th biological sample, a QC sample was assayed. MS instrument parameters were as follows: gas temperature 325°C, drying gas flow 10L/min, nebulizer 45psig, sheath gas temperature 400°C, sheath gas flow 12L/min, capillary voltage 4000V/3500V (positive ion/negative ion), acquisition mass range m/z 50–1000, acquisition rate 2 spectra/sec, reference mass correction enabled.
Feature detection and annotation
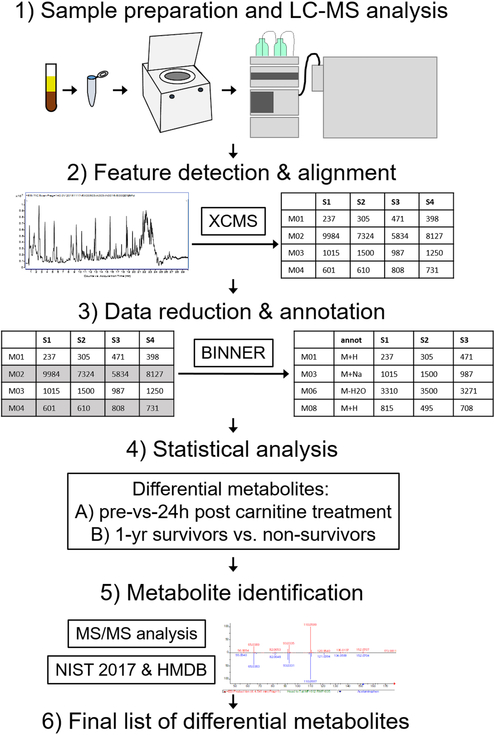
Major steps in the data analysis workflow for this study are visually illustrated in Figure 1. Instrument vendor format LC-MS data files were converted to generic mzXML format using msconvert, a component of the ProteoWizard software suite.11 Untargeted feature detection and alignment were performed using XCMS12–14, which operates in the R environment for statistical computing15, using default parameters. Detected features were putatively annotated using a novel in-house mass spectral data annotation tool, Binner (https://binner.med.umich.edu/). Untargeted metabolomics studies generate a large number of features many of which have been shown to be redundant, containing contaminants, artifacts, and degeneracies.16 Presence of these features complicates compound identification and leads to inflated false discovery rates in downstream statistical analysis. Our new tool first groups features into bins using retention time, then computes correlation of feature intensities across all samples, followed by hierarchical clustering to subdivide bins into smaller clusters. Additionally, Binner computes a matrix of mass differences on a per-bin basis and compares these to the list of known adducts and neutral losses. The application of Binner resulted in 25.3% and 36.5% data reduction in the positive and negative mode data, respectively. Following feature annotation using Binner, QC sample runs were assessed and corrected for peak area intensity drift using LOESS fitting17 using a macro-enabled Excel workbook, MetaboDrift, as previously described.18 The resulting data set can be found at: http://www.metabolomicsworkbench.org/.
Figure 1:
Schematic of the sample analysis and data processing workflow. Serum samples were extracted with solvent and subjected to LC-MS analysis (step 1). Features were detected and aligned in the resulting data using XCMS (step 2). Data were then processed using data reduction and annotation software Binner, which enabled removal of redundant features including isotopes, fragments and minor adducts while annotating “primary” features with putative ion type (M+H, M+Na, etc) (step 3). Remaining features were subject to univariate statistical analysis with FDR correction to detect differential features between pre and 24h post carnitine samples, as well as between 1-year sepsis survivors and non-survivors (step 4). Differential features were subject to MS/MS analysis and database search to assign metabolite identity when possible or provide putative annotation when not (step 5), resulting in the final lists of differential metabolites included in this manuscript.
Statistical analysis
Following drift correction, intensity values of detected features were median normalized and log transformed using generalized log transformation (glog 2). Statistical comparison of features (T0h, T24h and T48h) of carnitine-treated 1-year survivors and non-survivors and placebo-treated 1-year survivors and non-survivors were compared using generalized estimation equation (R studio, version 1.1.46; geepack with the function geeglm).19 The resulting p values of each analysis were corrected for false discovery using the method of Storey, et al.20 To prioritize features for identification, they were ranked in ascending order and a false discovery corrected p value of 0.05 was used as a cutoff. To assess the acute influence of carnitine-treatment on the metabolome, a Student’s t-test was used to compare the T0h and T24h timepoints in carnitine-treated patients. Demographic data were compared using a Mann-Whitney U test; a p value of ≤ 0.05 was considered significant.
Database search, MS/MS analysis and feature identification
The lists of features differentiating carnitine- and placebo-treated survivor and non-survivor groups were subjected to the workflow to identify as many metabolites as possible. All features annotated as isotopes (M+1, M+2, etc.) were removed from the data set. Features that Binner annotated as in-source fragments or lower-abundance adducts of a higher-abundance “primary” feature were visually examined as spectra and extracted ion chromatograms using MassHunter Qualitative Analysis software. If the peak shape of a putative adduct or fragment closely matched that of the primary feature to which it was assigned, it was removed from the list of differential features. The m/z values of remaining features were compared with a local library of approximately 200 known metabolites commonly detected in human plasma using our LC-MS method. Features matching m/z and retention time of one of these standards were annotated as identified according to Metabolite Standards Initiative (MSI) Level 1 criteria.21
All differential features not identified using authentic standards were subject to MS/MS analysis. The QC sample was analyzed using identical LC conditions as the initial LC-MS analysis with the instrument set to acquire targeted MS/MS data on the unidentified ions in question. MS/MS parameters were as follows: precursor ion isolation width 1.3 m/z, quadrupole MS/MS mass range m/z 50–1700, acquisition rate 2 spectra/sec, collision energy 20. The resulting MS/MS spectra of unknowns were searched against the NIST 2017 MS/MS library using NIST MS Search software, version 2.3 (Gaithersburg, MD). Features with a high forward- or reverse-score match (>700) and good visual alignment of multiple MS/MS peaks with an accurate-mass reference spectrum were considered identified to MSI level 1 criteria, and were further annotated as having been identified by MS/MS spectral match. Finally, differential features not identified using authentic standards or MS/MS analysis were searched by m/z using the Human Metabolome Database (HMDB; http://www.hmdb.ca)22–23 specifying the ion mode in which they were detected, unknown adduct type and 20ppm mass accuracy. Features with m/z values matching metabolites previously reported to be detected in human serum or plasma (based on the HMDB entry) were putatively annotated (MSI level 2) in the data.
Results
Subject characteristics
A total of 31 subjects (21 male, 10 female) participated in the original study of L-carnitine administration in sepsis7; their demographic and clinical characteristics have been previously reported.8 However, since there was only one 1-year survivor among the females, and because of our prior observation of distinct responses of the sexes to carnitine administration6, we elected to perform an untargeted metabolomics analysis of only the male subjects enrolled in this clinical trial to decrease heterogeneity given the relatively small sample size. Subject characteristics for these male subjects are provided in Table 1. One year mortality for carnitine- and placebo-treated males was 45% and 57%, respectively. Follow-up studies of our current findings in a larger cohort that will be better powered to assess differences in drug response by sex is currently underway in our laboratory.
TABLE I:
Baseline Demographics and Clinical Characteristics of Male Septic Shock 1-year Survivors and Non-survivors
| Variable | Survivors (n=10) | Non-survivors (n=11) | p value |
|---|---|---|---|
| Age (IQR) | 60.5 (43–66) | 68 (60–76) | 0.07 |
| Race, n (%) | |||
| African American | 3 (30) | 10 (90) | 0.31 |
| Caucasian | 7 (70) | 1 (10) | 0.31 |
| Comorbidities, n (%) | |||
| Coronary artery disease | 3 (30) | 3 (27) | >0.99 |
| Cerebrovascular accident | 3 (30) | 0 (0) | 0.09 |
| Hypertension | 7 (70) | 7 (64) | >0.99 |
| Type I DM | 2 (20) | 3 (27) | >0.99 |
| Type II DM | 2 (20) | 1 (9) | 0.59 |
| Chronic renal insufficiency | 3 (30) | 2 (18) | 0.64 |
| Malignancy | 1 (10) | 3 (27) | 0.59 |
| T0 Total SOFA score (IQR) | 11.5 (8–14) | 13 (6.5–14.5) | 0.98 |
IQR, interquartile range; DM, diabetes mellitus; SOFA, Sequential Organ Failure Assessment score; age and SOFA values are median.
Impact of L-carnitine on the serum metabolome of septic shock patients
Untargeted LC-MS data were first analyzed to study the acute effect of L-carnitine administration on the serum metabolome of male septic shock patients, irrespective of their short- or long-term survival status. To this end, we compared the metabolome of subjects before and T24h after the initiation of L-carnitine supplementation. We also performed the same T0h vs T24h comparison of the metabolomics data from the placebo group. In the carnitine-treated patients, 179 features (91 positive ion mode, 88 negative ion mode) differentiated 0 from 24h time points based on an uncorrected p-value <0.05 (supporting information: Table S2, differential (unadjusted p <0.05) compounds between 0h and 24h timepoints in carnitine-treated subjects); but only 5 positive ion-mode features and no negative ion-mode features had a FDR-corrected p-value <0.05 (Table S2). In placebo-treated subjects, no features were differential T0h from T24h time points following FDR correction (data not shown).
Putative identification of FDR-corrected differential features in carnitine-treated subjects was attempted using a local metabolite library, MS/MS analysis and HMDB database searching as described in the methods. The top 3 differential features were attributed to L-carnitine (M+H, M+Na, and 2M+H adducts); the remaining two features could not readily be identified. Although detailed identification efforts were not attempted on features that did not achieve FDR-adjusted significance, among positive ion mode features with uncorrected p<0.05, 21 of the 91 features could be readily assigned as acylcarnitine M+H ions; preliminary examination suggested that many of the remaining features were isotopes, adducts, or fragments of these species. These results corroborate the findings of our prior targeted metabolomics study which demonstrated that post-carnitine treatment levels of acylcarnitines differentiated septic shock survival.6 However, reliable detection of systemic alterations to metabolism induced by L-carnitine supplementation, beyond the formation of acylcarnitines, will necessitate additional studies in larger cohorts; these efforts are currently underway in our laboratory.
Specific metabolites differentiate L-carnitine and placebo treated septic shock survivors from non-survivors
Next, the untargeted metabolomics data were used to assess the differences between the serum metabolome of 1-year carnitine- and placebo- treated sepsis survivors and non-survivors. After the removal of redundant and spurious features identified by Binner, positive and negative mode data contained 2823 and 2615 features, respectively. These were subjected to statisical analyis as descrtibed in the Methods section. In carnitine- treated patients, 63 features had FDR ≤ 5% (supporting information: Table S4, differentiating metabolites of carnitine-treated survivors vs nonsurvivors); no negative ion-mode features had a FDR-corrected p-value <0.05. In placebo-treated subjects there were 88 features that differentiated survivors from survivors with FDR <0.05 (supporting information:Table S5, differentiating metabolites of placebo-treated survivors vs nonsurvivors).
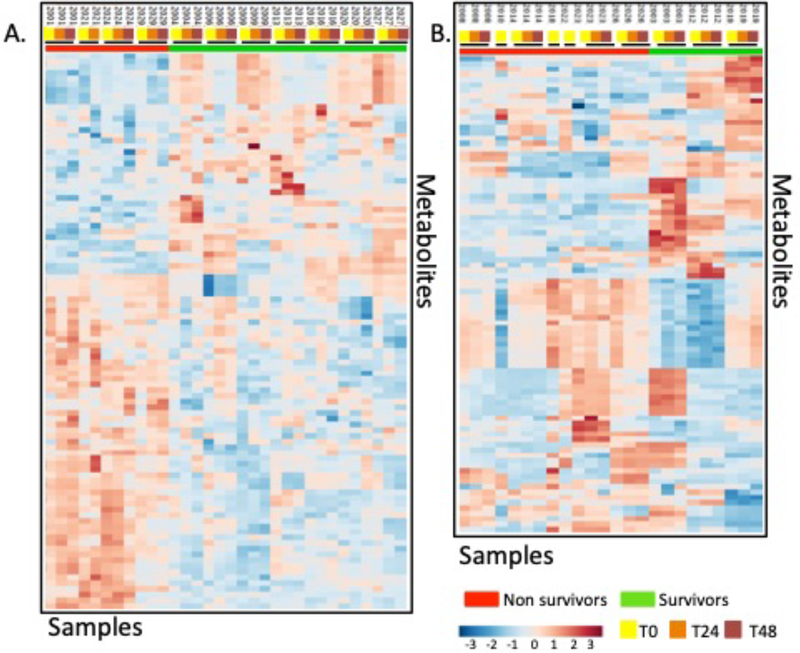
Due to the known differences in sepsis severity between the carnitine-treated and placebo-treated groups (Total sequential organ failure assessment (SOFA) score, Table II; supporting information: Table S3, SOFA scores, placebo- and carnitine-treated patients)24 the statistical comparisons of carnitine- and placebo-treated survivors versus non-survivors were performed separately. Figures 2A and 2B show the heatmaps for the features with an FDR ≤ 10% from carnitine and placebo-treated groups, respectively. There are noticable metabolic differences between carnitine-treated survivors and non-survivors, however the placebo group appears to be more heterogeneous. Notably, in most cases, the samples obtained from the same subject at T0h, T24h and T48h are more similar to each other than to other samples at the same timepoint.
TABLE II:
Baseline Demographics and Clinical Characteristics of Carnitine and Placebo-treated Male Septic Shock Patients
| Variable | Carnitine-treated (n=11) | Placebo-treated (n=10) | p value |
|---|---|---|---|
| Age (IQR) | 62 (51–74) | 66.5 (55–70) | 0.54 |
| Race, n (%) | |||
| African American | 2 (18) | 2 (20) | >0.99 |
| Caucasian | 9 (82) | 8 (80) | >0.99 |
| Comorbidities, n (%) | |||
| Coronary artery disease | 4 (36) | 2 (20) | 0.64 |
| Cerebrovascular accident | 1 (30) | 2 (20) | 0.59 |
| Hypertension | 7 (64) | 7 (70) | >0.99 |
| Type I DM | 1 (9) | 4 (40) | 0.15 |
| Type II DM | 2 (18) | 1 (10) | >0.99 |
| Chronic renal insufficiency | 3 (27) | 2 (20) | >0.99 |
| Malignancy | 1 (9) | 3 (30) | 0.31 |
| T0 Total SOFA score (IQR) | 14 (9–14) | 8 (6–12) | 0.02 |
IQR, interquartile range; DM, diabetes mellitus; SOFA, Sequential Organ Failure Assessment score; age and SOFA values are median.
Figure 2.
Heatmaps of the metabolite features (FDR<0.1) of (A) carnitine-treated and (B) placebo-treated male septic shock patients. The black bars indicate samples from the same patient.
To carry out feature identification the top ranking differential features were searched against a local metabolite library and then subjected to targeted LC-MS/MS analysis. To establish a high-confidence identification (MSI Level 1) we required either a high-quality MS/MS spectral match or high-confidence accurate mass and retention time match from an authentic standard previously analyzed using the same LC-MS method. All features not meeting these criteria but with an accurate-mass match (within 20 ppm) to an endogenous metabolite previously detected in human serum (based on data reported at the human metabolome database; http://www.hmdb.ca/) were considered either putatively annotated (MSI level 2); remaining unidentified features were considered unknown (MSI level 4). Table 3 contains the list of confidently-identified (MSI Level 1) metabolites determined to differentiate carnitine-treated survivors and non-survivors, along with the m/z value, retention time, observed adduct type and FDR-corrected p-value for each feature.
TABLE III:
Identified Metabolites Differentiating Carnitine-treated 1-year Survivors from Non-survivors
| HMDB ID | Adduct (observed) | m/z (observed) | RT (min) | FDR-adjusted p value | |
|---|---|---|---|---|---|
| Endogenous | |||||
| N-Acetyl-L-phenylalanine | 0000512 | M+H | 163.0355 | 3.37 | 0.0000 |
| Adipoyl-L-carnitine | 0028747 | M+H | 290.1436 | 2.48 | 0.0001 |
| Fibrinopeptide A | C0952* | M+H | 768.8494 | 9.12 | 0.0005 |
| N-(3-acetamidopropyl)pyrrolidin-2-one | 0061384 | 185.128028 | 3.8417 | 0.0008 | |
| N,N-dimethylguanosine | 0004824 | M+H | 312.1302 | 3.56 | 0.0078 |
| Phenylalanyl-tyrosine | 0029007 | M+H | 329.153524 | 21.445 | 0.0105 |
| Glucosamine | 0001514 | M+H | 180.0875 | 3.560 | 0.0171 |
| isoleucyl-proline / leucyl proline | 0028908/0011175 | 229.154547 | 1.3083 | 0.0171 | |
| Allysine | 0001263 | M+Na | 168.063352 | 1.7559 | 0.0187 |
| N-Methyl-phenylalanine | 0029224 | M+H | 142.9403 | 0.89 | 0.0218 |
| Methoxytryptophol | 0001896 | M+H | 192.104478 | 5.0838 | 0.0254 |
| Histamine | 0000870 | M+Na | 134.0687 | 3.165 | 0.0376 |
| Pantetheine | 0003426 | M+H | 279.1389 | 7.931 | 0.0475 |
| Exogenous/drug-related |
HMDB ID: human metabolomics database entry number (http://www.hmdb.ca/).
KEGG ID, no HMDB ID available.
All features were identified either by MS/MS spectral match with NIST 2017 MS/MS library or by accurate mass and retention time match with authentic standard in local metabolite library.
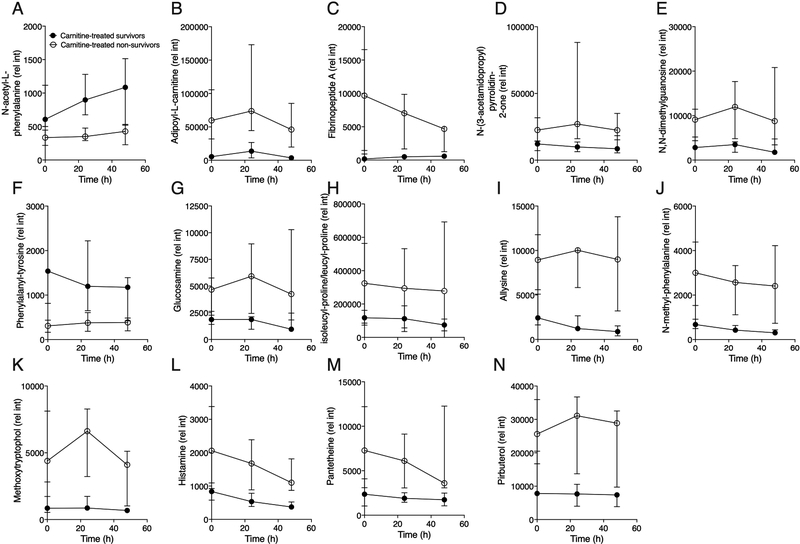
The relative abundance of these features over the three timepoints of the study (T0h, T24h and T48h) is visually illustrated in the graphs shown in Figure 3 (endogenous and exogenous metabolites in carnitine-treated subjects). Additional putatively annotated or unidentified differential features are listed in (supporting information: Tables S4, differentiating metabolites of carnitine-treated survivors vs nonsurvivors and S5, differentiating metabolites of placebo-treated survivors and non-survivors).
Figure 3.
Temporal profiles of the metabolites that differentiated carnitine-treated non-survivors and survivors (FDR <0.05). These included endogenous (A-M) and an exogenous metabolites (N). With the exception of (A) N-acetyl-L-phenylalanine and (F) phenylalanyltyrosine, all endogenous metabolites were higher in carnitine-treated non-survivors. Data are the median (IQR) of 7 male carnitine-treated survivors and 4 male carnitine-treated non-survivors at all time points except T48h (non-survivors, n=3).
Discussion
Using untargeted LC-MS metabolomics with a novel annotation approach (Binner) to assess global changes in the serum induced by supplemental L-carnitine in patients with septic shock, we made the novel findings that: 1) L-carnitine administration appears to induce few acute metabolic changes aside from increased levels of acylcarnitines (T0h-T24h); and 2) L-carnitine treated non-survivors in this study, although clinically similar to survivors, were metabolically distinct from them, suggesting that this phenotype has a poor prognosis and may be non-responsive to intervention with L-carnitine supplementation; and 3) that overall, there is broad metabolic heterogeneity in patients with septic shock. We have previously shown that this group also had substantially higher concentrations of carnitine and acylcarnitines following L-carnitine administration compared with survivors.6
The distinct metabolic phenotype of L-carnitine treated non-survivors is illustrated in Figure 3 which shows that the majority of the metabolites that differentiated 1-year survival were elevated in non-survivors, suggesting these compounds may be by-products of more severe metabolic derangement occurring in these individuals. Most metabolites that differentiated L-carnitine-treated survivors from non-survivors appeared to be elevated at the T0h (pre-treatment) timepoint, suggesting that they may be associated with sepsis survival independent of carnitine treatment. Importantly, while carnitine-treated patients were clinically distinct from placebo-treated patients (by total SOFA score, Table II), all carnitine-treated patients were clinically similar (supporting information: Table S3, SOFA scores, placebo- and carnitine-treated patients).
The distinct metabolic phenotype of carnitine-treated non-survivors is attributable to elevations in several endogenous metabolites that have previously been reported to be associated with inflammation and, in aggregate, portray a pro-inflammatory-vascular injury state in L-carnitine-treated non-survivors. Fibrinopeptide A (Figure 3C), a biomarker of activation of the coagulation pathway, has previously been associated with vascular inflammation, which is closely tied to lung injury and sepsis.25–26 Additional metabolites detected as elevated in carnitine-treated sepsis non-survivors include glucosamine (HMDB0001514, Figure 3G), an endogenous amino-monosaccharide which is actively produced during tissue injury, tissue repair and wound healing which may increase in the blood as a consequence of inflammation27, histamine (HMDB0000870, Figure 3L), which increases vascular inflammation and has been shown to be casually associated with sepsis28, and allysine (HMDB0001263, Figure 3I), a by-product of lysine metabolism that participates in the biosynthesis and cross-linking of collagen and elastin, which are known to arise at sites of inflammation.29 In addition to this inflammatory signal, non-survivors had higher levels of N-methyl phenylalanine (HMDB0029224; Figure 3J) which is a constituent of bacterial pili, the structures that protrude from bacteria and anchor them to other cells.30
One drug-related metabolite, pirbuterol (HMDB0015407, Figure 3N) was also elevated in non-survivors. Although detailed records of drugs administered in the course of treatment of these patients were not available, it is conceivable that greater renal dysfunction (as suggested by the higher renal SOFA score) in non-survivors may have influenced drug clearance as well as that non-survivors may have received higher doses and/or more frequent drug dosing. In future studies, collecting more detailed drug treatment records for all subjects might help clarify the origins of these metabolites.
Compared to the number of endogenous metabolites that had elevated concentrations two metabolites were elevated in sepsis survivors. Phenylalanyl-tyrosine (Figure 3F; HMDB0029007) is a dipeptide that consists of phenylalanine and tyrosine both of which are precursors of catecholamines. We have previously reported that serum tyrosine concentration is elevated in carnitine-treated survivors.8 Levels of N-acetyl-L-phenylalanine (Figure 3A; HMDB0000512), a metabolite most often associated phenylketonuria, it has also been shown to be elevated in liver dysfunction.31 Carnitine-treated survivors had higher liver SOFA scores (supporting information: Table S3, SOFA scores, placebo- and carnitine-treated patients) that may explain this finding. However, even though N-acetyl-L-phenylalanine was more abundant in the serum of survivors, our findings do not indicate that they are associated with a poor sepsis prognosis.
We acknowledge several limitations of our study. The most notable, as discussed above, is the limited number of subjects, which was constrained by the size of the original clinical trial.14 The phase I clinical trial was small and we further reduced it by removing the female participants. This is because we and others have shown that peak blood carnitine concentrations following intravenous supplementation may be influenced by sex.6, 32 Furthermore, in our cohort, there was only one female carnitine-treated 1-year survivor which would further the variance already known to exist in the sepsis metabolome. Nevertheless, while small, this group of patients is unique and critically important to furthering understanding the mechanistic underpinnings of L-carnitine’s therapeutic response. Secondly, untargeted metabolomics produces rich data consisting of thousands of features of biological origin, but is limited to relative quantitation and requires a complex data analysis and compound identification workflow. A major aspect of this complexity originates from the fact that a large portion of the features detected in untargeted metabolomics data have been shown to be redundant, containing contaminants, artifacts, and degeneracies.16 The high prevalence of such degenerate features leads to inflated false discovery rates, which may particularly affect the outcome of studies with a limited sample size. Our novel feature annotation and data reduction approach, Binner, reflects an advancement in the removal of redundancies from the data to ensure reported features reflect unique biological metabolites. Future studies including analysis of differential metabolites using targeted approaches that enable robust quantitation will be important to validate and extend the impact of our present findings.
Conclusions
Using untargeted LC-MS metabolomics, we identified a metabolically distinct phenotype of septic shock that is associated with poor long-term prognosis of survival, non-responsiveness to L-carnitine supplementation, and that was not readily clinically distinguished. Importantly, to the best of our knowledge, this is a unique phenotype that has not been previously described using metabolomics.33 To date, the primary biochemical signature of sepsis has been attributed to energy disruption. With further development, quantitation of the metabolites we detected may become useful for predicting disease outcome or potential effectiveness of L-carnitine or other therapeutic agents. Additionally, these metabolites provide direct insight into the extent of metabolic derangement which occurs in septic shock, focusing attention on vascular inflammation as a factor that could affect long-term survival and potential drug-responsiveness of sepsis patients.
Supplementary Material
Acknowledgements:
This work was supported by a metabolomics supplement to a grant from the National Institute of General Medical Sciences (NIGMS; R01GM103799 to AEJ), R01GM111400 to KAS, K23GM113041 to MAP and R03CA211817 to AK. Research reported in this publication was supported by Core Services supported by the National Institutes of Health (NIH) under award numbers U24 DK097153 (MRC2), U2CES026553 (MCHEAR), U24DK112342 (MiCAS) and U2CES030164 (MCIDC). The clinical trial was supported by a grant from the American Heart Association (10POST3560001) and the Cannon Foundation (SRG10–004). The content is solely the responsibility of the authors and does not necessarily present the official views of the NIGMS, NIDDK, the National Institute of Environmental Health Sciences or the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION:
The following supporting information is available free of charge at ACS website http://pubs.acs.org
Table S1. Internal standards included in extraction solvent.
Table S2. Potentially differential (unadjusted p <0.05) compounds between 0h and 24h timepoints in carnitine-treated subjects.
Table S3. Baseline (T0) Sequential Organ Failure Assessment (SOFA) Scores of Carnitine and Placebo-treated Male Septic Shock 1-year Survivors and Non-survivors.
Table S4: Differential (FDR-corrected p <0.10) compounds between 1-year sepsis survivors and non-survivors in carnitine-treated subjects.
Table S5. Differential (FDR-corrected p <0.10) compounds between 1-year sepsis survivors and non-survivors in placebo-treated subjects.
References
- 1.Singer M; Deutschman CS; Seymour CW; Shankar-Hari M; Annane D; Bauer M; Bellomo R; Bernard GR; Chiche JD; Coopersmith CM; Hotchkiss RS; Levy MM; Marshall JC; Martin GS; Opal SM; Rubenfeld GD; van der Poll T; Vincent JL; Angus DC, The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315 (8), 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mogensen KM; Lasky-Su J; Rogers AJ; Baron RM; Fredenburgh LE; Rawn J; Robinson MK; Massarro A; Choi AM; Christopher KB, Metabolites Associated With Malnutrition in the Intensive Care Unit Are Also Associated With 28-Day Mortality. JPEN J Parenter Enteral Nutr 2017, 41 (2), 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrario M; Cambiaghi A; Brunelli L; Giordano S; Caironi P; Guatteri L; Raimondi F; Gattinoni L; Latini R; Masson S; Ristagno G; Pastorelli R, Mortality prediction in patients with severe septic shock: a pilot study using a target metabolomics approach. Sci Rep 2016, 6, 20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langley RJ; Tsalik EL; van Velkinburgh JC; Glickman SW; Rice BJ; Wang C; Chen B; Carin L; Suarez A; Mohney RP; Freeman DH; Wang M; You J; Wulff J; Thompson JW; Moseley MA; Reisinger S; Edmonds BT; Grinnell B; Nelson DR; Dinwiddie DL; Miller NA; Saunders CJ; Soden SS; Rogers AJ; Gazourian L; Fredenburgh LE; Massaro AF; Baron RM; Choi AM; Corey GR; Ginsburg GS; Cairns CB; Otero RM; Fowler VG Jr.; Rivers EP; Woods CW; Kingsmore SF, An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013, 5 (195), 195ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neugebauer S; Giamarellos-Bourboulis EJ; Pelekanou A; Marioli A; Baziaka F; Tsangaris I; Bauer M; Kiehntopf M, Metabolite Profiles in Sepsis: Developing Prognostic Tools Based on the Type of Infection. Crit Care Med 2016, 44 (9), 1649–62. [DOI] [PubMed] [Google Scholar]
- 6.Puskarich MA; Evans CR; Karnovsky A; Das AK; Jones AE; Stringer KA, Septic Shock Nonsurvivors Have Persistently Elevated Acylcarnitines Following Carnitine Supplementation. Shock 2018, 49 (4), 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puskarich MA; Kline JA; Krabill V; Claremont H; Jones AE, Preliminary safety and efficacy of L-carnitine infusion for the treatment of vasopressor-dependent septic shock: a randomized control trial. JPEN J Parenter Enteral Nutr 2014, 38 (6), 736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puskarich MA; Finkel MA; Karnovsky A; Jones AE; Trexel J; Harris BN; Stringer KA, Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Annals of the American Thoracic Society 2015, 12 (1), 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puskarich MA; Kline JA; Krabill V; Claremont H; Jones AE, Preliminary Safety and Efficacy of L-carnitine Infusion for the Treatment of Vasopressor-Dependent Septic Shock: A Randomized Control Trial. JPEN J Parenter Enteral Nutr 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans CR; Karnovsky A; Kovach MA; Standiford TJ; Burant CF; Stringer KA, Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. Journal of proteome research 2014, 13 (2), 640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers MC; Maclean B; Burke R; Amodei D; Ruderman DL; Neumann S; Gatto L; Fischer B; Pratt B; Egertson J; Hoff K; Kessner D; Tasman N; Shulman N; Frewen B; Baker TA; Brusniak M-Y; Paulse C; Creasy D; Flashner L; Kani K; Moulding C; Seymour SL; Nuwaysir LM; Lefebvre B; Kuhlmann F; Roark J; Rainer P; Detlev S; Hemenway T; Huhmer A; Langridge J; Connolly B; Chadick T; Holly K; Eckels J; Deutsch EW; Moritz RL; Katz JE; Agus DB; MacCoss M; Tabb DL; Mallick P, A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnology 2012, 30, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith CA; Want EJ; O’Maille G; Abagyan R; Siuzdak G, XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006, 78 (3), 779–87. [DOI] [PubMed] [Google Scholar]
- 13.Tautenhahn R; Patti GJ; Rinehart D; Siuzdak G, XCMS Online: a web-based platform to process untargeted metabolomic data. Anal Chem 2012, 84 (11), 5035–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryce J; Boschi-Pinto C; Shibuya K; Black RE; Group WCHER, WHO estimates of the causes of death in children. Lancet 2005, 365 (9465), 1147–52. [DOI] [PubMed] [Google Scholar]
- 15.R Core Team R: A language and environment for statistical computing, R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- 16.Mahieu NG; Patti GJ, Systems-Level Annotation of a Metabolomics Data Set Reduces 25000 Features to Fewer than 1000 Unique Metabolites. Analytical chemistry 2017, 89 (19), 10397–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn WB; Broadhurst D; Begley P; Zelena E; Francis-McIntyre S; Anderson N; Brown M; Knowles JD; Halsall A; Haselden JN; Nicholls AW; Wilson ID; Kell DB; Goodacre R, Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature protocols 2011, 6 (7), 1060–83. [DOI] [PubMed] [Google Scholar]
- 18.Thonusin C; IglayReger HB; Soni T; Rothberg AE; Burant CF; Evans CR, Evaluation of intensity drift correction strategies using MetaboDrift, a normalization tool for multi-batch metabolomics data. Journal of chromatography. A 2017, 1523, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.RStudio: Integrated Development for R. . http://www.rstudio.com/.
- 20.Storey JD, A direct approach to false discovery rates. J Roy Stat Soc B 2002, 64, 479–498. [Google Scholar]
- 21.Sumner LW; Amberg A; Barrett D; Beale MH; Beger R; Daykin CA; Fan TW; Fiehn O; Goodacre R; Griffin JL; Hankemeier T; Hardy N; Harnly J; Higashi R; Kopka J; Lane AN; Lindon JC; Marriott P; Nicholls AW; Reily MD; Thaden JJ; Viant MR, Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics : Official journal of the Metabolomic Society 2007, 3 (3), 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS, Current progress in computational metabolomics. Briefings in Bioinformatics 2007, 8 (5), 279–293. [DOI] [PubMed] [Google Scholar]
- 23.Wishart DS; Jewison T; Guo AC; Wilson M; Knox C; Liu Y; Djoumbou Y; Mandal R; Aziat F; Dong E; Bouatra S; Sinelnikov I; Arndt D; Xia J; Liu P; Yallou F; Bjorndahl T; Perez-Pineiro R; Eisner R; Allen F; Neveu V; Greiner R; Scalbert A, HMDB 3.0--The Human Metabolome Database in 2013. Nucleic acids research 2013, 41 (Database issue), D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincent JL; Moreno R; Takala J; Willatts S; De Mendonca A; Bruining H; Reinhart CK; Suter PM; Thijs LG, The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive care medicine 1996, 22 (7), 707–10. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe E; Smith DM Jr.; Delcarpio JB; Sun J; Smart FW; Van Meter CH Jr.; Claycomb WC, Cardiomyocyte transplantation in a porcine myocardial infarction model. Cell transplantation 1998, 7 (3), 239–46. [DOI] [PubMed] [Google Scholar]
- 26.Gando S; Nanzaki S; Sasaki S; Aoi K; Kemmotsu O, Activation of the extrinsic coagulation pathway in patients with severe sepsis and septic shock. Crit Care Med 1998, 26 (12), 2005–9. [DOI] [PubMed] [Google Scholar]
- 27.Chuang KH; Peng YC; Chien HY; Lu ML; Du HI; Wu YL, Attenuation of LPS-induced lung inflammation by glucosamine in rats. American journal of respiratory cell and molecular biology 2013, 49 (6), 1110–9. [DOI] [PubMed] [Google Scholar]
- 28.Neugebauer E; Lorenz W; Rixen D; Stinner B; Sauer S; Dietz W, Histamine release in sepsis: a prospective, controlled, clinical study. Crit Care Med 1996, 24 (10), 1670–7. [DOI] [PubMed] [Google Scholar]
- 29.Eyre DR; Paz MA; Gallop PM, Cross-linking in collagen and elastin. Annual review of biochemistry 1984, 53, 717–48. [DOI] [PubMed] [Google Scholar]
- 30.Strom MS; Lory S, Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. The Journal of biological chemistry 1991, 266 (3), 1656–64. [PubMed] [Google Scholar]
- 31.Wang X; Lv H; Zhang A; Sun W; Liu L; Wang P; Wu Z; Zou D; Sun H, Metabolite profiling and pathway analysis of acute hepatitis rats by UPLC-ESI MS combined with pattern recognition methods. Liver international : official journal of the International Association for the Study of the Liver 2014, 34 (5), 759–70. [DOI] [PubMed] [Google Scholar]
- 32.Reuter SE; Evans AM, Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet 2012, 51 (9), 553–72. [DOI] [PubMed] [Google Scholar]
- 33.Langley RJ; Wong HR, Early Diagnosis of Sepsis: Is an Integrated Omics Approach the Way Forward? Mol Diagn Ther 2017, 21 (5), 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.