Abstract
Lipoyl synthase (LipA in bacteria) is a radical S-adenosylmethionine (SAM) enzyme that catalyzes the second step of the de novo biosynthesis of the lipoyl cofactor: the insertion of sulfur at C6 and C8 of a pendant octanoyl chain. In addition to the [4Fe–4S] cluster that is characteristic of the radical SAM (RS) enzymes, LipA contains a second [4Fe–4S] cluster that, though controversial, has been the proposed to be degraded during turnover to supply the inserted sulfur atoms. A consequence of this proposed role is that the destruction of its iron-sulfur cluster renders the enzyme in an inactive state. Recently, it was shown that Escherichia coli proteins NfuA or IscU can confer catalytic properties to E. coli LipA in vitro. In this article, we present methods for characterizing LipA and analyzing its activity in vitro, and provide strategies to monitor the pathway for the regeneration of LipA’s auxiliary cluster by E. coli iron-sulfur carrier protein NfuA.
Keywords: S-adenosylmethionine, radical, lipoic acid, iron-sulfur cluster, protein maturation
1. Introduction
Lipoic acid is an eight carbon straight-chain fatty acid containing sulfur atoms at C6 and C8 (Reed, et al., 1951; Reed, et al., 1953). It plays a central role in multienzyme complexes involved in the oxidative decarboxylation of α-keto acids, the breakdown of glycine, and the degradation of acetoin in certain types of bacteria (Reed, 1974). In this capacity, it functions as a redox-active cofactor due to its ability to convert between states containing free thiol groups and a dithiolane ring and as a shuttle to move reaction intermediates between successive active sites in multienzyme complexes. In its role as a cofactor, lipoic acid is covalently attached through an amide linkage to a specific lysyl residue of a lipoyl carrier protein (LCP), one of the subunits of the multienzyme complexes (Cronan, 2016).
Early studies identified the pathway for the de novo biosynthesis of the lipoyl cofactor in E. coli and outlined the roles of two dedicated enzymes: an octanoyltransferase, LipB, and a radical S-adenosylmethionine (SAM) enzyme, LipA (Miller, et al., 2000; Zhao, et al., 2003; Cronan, 2016). In an offshoot from fatty acid biosynthesis, LipB transfers an octanoyl group from an acyl carrier protein to an LCP, which generates the substrate for LipA. LipA subsequently inserts sulfur atoms at C6 and C8 to generate the lipoyl cofactor. The inability to synthesize the lipoyl cofactor is embryonic lethal in mice, and defects in its biosynthesis result in disastrous biochemical outcomes in humans, such as seizures, developmental delays, and death (Yi, et al., 2005; Landgraf, et al., 2016). Further, lipoic acid supplementation has been shown to be therapeutic for a wide range of clinical ailments associated with oxidative stress, including diabetes and Alzheimer’s disease (Holmquist, et al., 2007; Borcea, et al., 1999).
There are several challenges associated with the biosynthesis of the lipoyl cofactor, despite its structural simplicity. The first obstacle is removal of hydrogens from C6 and C8 of the octanoyl chain, which LipA overcomes through the use of radical chemistry. As a radical SAM (RS) enzyme, LipA coordinates a [4Fe–4S]2+ cluster that is absolutely required for its function. In its reduced [4Fe–4S]+ form, the iron-sulfur (Fe/S) cluster can inject an electron into the sulfonium of SAM, inducing its cleavage to L-methionine, and a 5′-deoxyadenosyl 5′-radical (5′-dA•). This high energy radical species is a potent oxidant capable of cleaving C-H bonds displaying homolytic bond-dissociation energies of 98 kcal/mol or greater.
The second outstanding question is the source of the sulfur used for the sulfur insertion. In addition to the [4Fe–4S] cluster common to all RS enzymes, LipA coordinates a second ‘auxiliary’ cluster that appears to be the source of the inserted sulfur atoms (Lanz and Booker, 2015). As such, the [4Fe–4S] auxiliary cluster is destroyed during catalysis, which is supported by an overwhelming amount of biochemical evidence (Douglas, et al., 2006; Lanz, et al., 2014; McGlaughlin, et al., 2016; Cicchillo, et al., 2004, Harmer, et al., 2014, Cicchillo, et al, 2005). For example, LipA can only undergo up to one turnover in vitro in the absence of exogenously added sulfur. Further, when LipA is isolated from E. coli cultures using 34S as the sole sulfur source and then subsequently used in in vitro assays, the lipoyl product contains almost exclusively 34S at C6 and C8 (Cicchillo and Booker, 2005).
Due to the sacrificial role of its auxiliary cluster, LipA typically catalyzes less than one equivalent of product formation in in vitro assays. Recently, the E. coli [4Fe–4S] cluster carrier protein NfuA was shown to regenerate LipA’s auxiliary cluster to support catalysis in vitro (McCarthy and Booker, 2017). Remarkably, it accomplishes this feat at a rate that does not limit the rate of catalysis. In the absence of NfuA, the scaffold protein IscU can fulfill a similar role. Herein, we describe methods for isolating and characterizing LipA and monitoring the regeneration of its auxiliary cluster in vitro. This chapter aims to complement other previously published Methods in Enzymology papers that highlight the characterization of RS enzymes that contain multiple [Fe–S] clusters (Lanz, et al., 2012) and RS enzymes that contain cobalamin cofactors (Blaszczyk, Wang, & Booker, 2017), with additional insight into analysis of Fe–S cluster regeneration in vitro in RS enzymes that consume their clusters during catalysis.
2. Overproduction and Isolation of E. coli LipA and E. coli NfuA
The generation of LipA protein of high purity and activity has been notoriously difficult due to issues of solubility and the air sensitive nature of its two [4Fe–4S] clusters. Initial characterizations resulted in protein that had poor activity most likely due to incomplete cofactor incorporation which is essential for the enzyme’s activity. With regard to NfuA, the E. coli protein had been typically isolated aerobically and then subjected to a chemical reconstitution procedure to insert its iron-sulfur (Fe/S) cluster, leaving it unclear as to what the in vivo cluster configuration is (Angelini, et al., 2008; Py, et al., 2012; Boutigny, et al., 2013). Following the methods detailed herein, we have generated robust amounts of LipA and NfuA proteins that are >90% homogenous. Further, the proteins contain intact [4Fe–4S] clusters in its as-isolated state without the need for chemical reconstitution allowing for extensive biochemical analysis.
2.1. Gene cloning and expression strategy
The genes encoding LipA and NfuA were amplified from E. coli genomic DNA (strain W3110) using the Polymerase Chain Reaction (PCR) and inserted into the pET28a vector using the NdeI/ EcoRI restriction sites and HindIII/XhoI restriction sites, respectively (Cicchillo et al., 2004, McCarthy and Booker, 2017). The resulting constructs allowed for overexpression of the lipA and nfuA genes under the control of the strong T7 promoter. Further, these constructs conferred kanamycin resistance for selection in vivo and encoded an N-terminal hexahistidine (His6) tag, which provided a simple purification strategy. A second plasmid, pDB1282, encodes the isc operon from Azotobacter vinelandii and confers ampicillin resistance, making it compatible for coexpression with the pET28a-lipA and pET28a-nfuA constructs. E. coli BL21(DE3) cells, which are commonly used with the pET systems, were co-transformed with either the pET28a-lipA or pET28a-nfuA construct and pDB1282 for gene expression.
E. coli LipA and E. coli NfuA are overproduced in E. coli cultured in M9 minimal medium supplemented with glucose as a carbon source and ammonium chloride as a nitrogen source, and cultures are grown with limited aeration. During the overproduction, iron is not typically added to the cultures until gene expression is induced at ~OD600=0.6. We have found that in some cases, iron starvation can induce cells to release siderophores, which have an extremely high affinity for iron and can increase iron-sulfur cluster incorporation in some proteins, such as LipA.
2.2. Protocol for expression of the lipA and nfuA genes
Co-transform E.coli BL21(DE3) cells with pDB1282 and either pET28a-lipA or pET28a-nfuA using standard protocols. Use the antibiotic markers within the plasmids to select for colonies containing both pDB1282 and pET28a-lipA or pET28a-nfuA constructs by plating transformants on Luria-Bertani (LB) plates containing kanamycin (50 μg/mL) and ampicillin (100 μg/mL).
Select a single colony and inoculate 200 mL LB media containing kanamycin (50 μg/mL) and ampicillin (100 μg/mL). Incubate at 37 ºC with shaking at 250 rpm for ~14 h. Simultaneously, place four 6 L flasks containing 4 L M9 minimal media in a shaker overnight at 37 ºC in order to warm culture media.
Immediately prior to inoculation of large flasks, add kanamycin (50 μg/mL) and ampicillin (100 μg/mL).
Inoculate each 6 L flask with 10 mL of the 200 mL overnight LB culture and incubate the cultures at 37 ºC with shaking at 180 rpm.
Once the cells reach an OD600=0.3 (~5 h), add 0.2% (w/) L-arabinose to induce expression of genes on pDB1282. Take a 1 mL sample of each flask directly before arabinose induction (pre-induction sample) for SDS-PAGE analysis.
At an OD600=0.6, place the flasks in an ice bath for ~1 h and add FeCl3 to a final concentration of 50 μM. Adjust the temperature of the incubator to 18 ºC. Take a sample of each flask (post-arabinose sample) for SDS-PAGE analysis, normalizing the number of cells to the pre-induction sample.
Induce gene expression by adding 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and allow expression to proceed for ~18 h at 18 ºC with shaking at 180 rpm.
The following day, take a sample of each flask (post-IPTG sample) for SDS-PAGE analysis, normalizing the number of cells to the post-arabinose sample. Harvest the bacterial cells by centrifugation at 7,500 × g for 12 min. Flash-freeze the resulting cellular pellet (~3 g/L) in liquid nitrogen and store in liquid nitrogen until further use.
Analyze the overproduction of the desired protein by SDS-PAGE (12.5%) using the samples collected during the growth (pre-induction, post-arabinose induction, post-IPTG induction).
2.3. Purification of E. coli LipA and NfuA
Isolating proteins that contain Fe/S clusters is challenging due to their oxygen sensitive nature. Oxygen contamination can lead to the degradation of the clusters, resulting in proteins with poor stability and enzymatic activity. As such, careful handling of the protein during all purification steps is vital to ensure that the quality of the protein does not suffer because of oxidative damage. Ideally, the isolation and characterization of Fe/S cluster-containing proteins should be performed in an anaerobic chamber. The Fe/S clusters coordinated by LipA and NfuA are especially sensitive to oxygen, so maintaining an anaerobic environment is essential. LipA is also sensitive to repeated freeze-thaw cycles, so the purification should be carried out to completion without interruption using chilled buffers, and the protein should be stored in small aliquots. The general procedure for isolating Fe–S cluster-containing proteins has been previously described in detail (Lanz et al., 2012).
Autoclave plastic centrifuge tubes and cycle into anaerobic chamber while hot. Allow the tubes to equilibrate in the chamber several days before purification.
In the anaerobic chamber, prepare 250 mL of each of the buffers needed for the purification using oxygen-free water: Lysis Buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 20 mM imidazole, 10 mM 2-mercaptoethanol), Wash Buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 45 mM imidazole, 10 mM 2-mercaptoethanol, 10% glycerol), Elution Buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 500 mM imidazole, 10 mM 2-mercaptoethanol, 10% glycerol) and Storage Buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 5 mM dithiothreitol, 20% glycerol).
Resuspend the cell paste (~60 g) in 150 mL Lysis Buffer supplemented with lysozyme (1 mg/mL) and DNAse I (0.1 mg/mL) in a metal sonication cup. Once the solution is homogeneous, allow it to stir for 30 min at room temperature and then add one EDTA-free Sigmafast protease inhibitor tablet.
Place the sonication cup in an ice–water bath on a stir-plate and slowly stir.
Lyse the cells by sonic disruption using a Fisher 550 Sonic Dismembrator by sonicating for 1 min at 30% output (setting of 7) followed by resting for 8 min to allow for temperature re-equilibration. Repeat for a total of five cycles. The cell lysate should become noticeably darker as the cells burst, releasing Fe–S containing proteins into solution.
Following sonication, pour the cell lysate into the plastic centrifuge tubes that were previously autoclaved and allowed to equilibrate in the anaerobic chamber. Seal the juncture between the cap and tube tightly with vinyl tape to avoid oxygen contamination.
Remove the tightly sealed tubes from the glovebox and centrifuge at 45,000 × g for 1 h at 4 ºC.
Cycle the centrifuge tubes back into the anaerobic glove box and pour the supernatant into a 250 mL flask. Take a 50 μL sample of the supernatant and scrape a small amount of the pellet into an Eppendorf tube for SDS-PAGE analysis.
Equilibrate a column containing 50 mL Ni-NTA resin with 100 mL chilled Lysis Buffer.
After equilibration, load the supernatant onto the column. Collect the flow-through in a separate container and take a 50 μL sample for SDS-PAGE analysis.
Wash the column with 100 mL chilled Wash Buffer and take a 50 μL sample for SDS-PAGE analysis. Repeat the wash step and collect another 50 μL sample for SDS-PAGE analysis.
Elute the desired protein using chilled Elution Buffer. Allow ~15 mL of Elution Buffer to flow over the column before collecting the protein, which is when the dark brown colored band will begin to elute. Stop collecting once the color of the eluate returns to clear.
Concentrate the protein in an Amicon stir-cell fitted with a YM-10 membrane (10 kDa molecular weight cutoff) in the anaerobic glovebox. It is vital to keep the stirring apparatus on ice throughout the entire concentration step.
Once the protein has been concentrated to ~2.5 mL, exchange it into chilled Storage buffer using a PD-10 column. Collect 3.5 mL and aliquot into Eppendorf tubes.
Take a 5 μL aliquot of the final purified protein for SDS-PAGE analysis.
Remove the protein aliquots from the anaerobic chamber and immediately flash-freeze in liquid nitrogen. Store the protein aliquots in liquid nitrogen until use, avoiding freezing and thawing, which leads to activity loss.
Analyze the purification by SDS-PAGE (12.5%) using the samples collected of the cellular pellet, supernatant, flow-through, wash, and final protein.
3. Initial Characterization of As-isolated Protein
Following the isolation, routine characterizations should be performed to determine the yield and to obtain a qualitative estimate of the cofactor incorporation in the protein. These routine analyses provide a straightforward method to monitor consistency among enzyme preparations. Protein concentration is determined via the Bradford method (Bradford, 1976) using bovine serum albumin (fraction V) as a standard. However, because the Bradford method is biased by amino acid composition, a correction factor should be obtained through amino acid analysis to account for differences in amino acid sequence. This step was especially important in the analysis of LipA, which has a correction factor of nearly 1.5. In this case, its amino acid composition significantly changes the protein concentration empirically determined by the Bradford method, which has further implications in determining its activity and the stoichiometry of its Fe–S cluster cofactors. In addition to protein concentration, colorimetric iron and sulfide analyses should be performed (Beinert, 1978; Beinert, 1983). It is important to note that these methods simply estimate the amount of iron and sulfide in solution; they do not provide evidence for the biologically relevant form of the cofactor or its coordination environment. For a more advanced understanding, Mössbauer and EPR spectroscopies should be used, as detailed in other methods papers (Pandelia, et al., 2015; Lanz, et al., 2012).
3.1. Protocol for obtaining amino acid correction factor
Contact a facility capable of performing amino acid analysis, such as the University of California-Davis proteomics facility.
Ensure that the protein you wish to analyze is >95% homogenous.
Using a NICK column (GE healthcare) exchange 100 μL of the protein (~40 mg/mL) into 10 mM NaOH in order to remove the Fe–S cluster.
Perform three Bradford assays each with an independent standard curve to determine the protein concentration.
Average the three values together, and record as the concentration determined via the Bradford method.
Carefully place 100 μL of the protein in three Corning Pyrex test tubes (10 × 75 mM) and seal the top with parafilm.
Use a needle to add a hole to the top of each of the glass tubes and lyophilize the protein samples in vacuo.
Ship to the proteomics facility in order to determine the true concentration of the sample based on its amino acid composition.
Average the three values of the concentration determined by amino acid analysis.
Generate the amino acid correction factor by dividing the concentration determined by amino acid analysis by the concentration determined by the Bradford method.
Apply this ratio in every subsequent Bradford analysis performed on the protein to ensure an exact determination of the concentration.
3.2. Colorimetric Iron Analysis Protocol
Prepare the following solutions: Reagent A (1.35 g SDS, 450 μL saturated iron-free ammonium acetate, in a final volume of 30 mL dH20); Reagent B (270 mg ascorbic acid, 9 mg sodium meta-bisulfite, 450 μL saturated iron-free ammonium acetate, in a final volume of 6 mL dH20); and Reagent C (18 mg ferene in a final volume of 1 mL dH20).
In the anaerobic chamber, dilute the protein to 10 μM in 50 mM NaOH.
Aliquot into four 300 μL samples and remove the samples from the anaerobic chamber.
Prepare a 100 μM iron standard stock using Claritas PPT iron standard (Fisher Scientific).
Dilute the 100 μM iron standard to the following concentrations in a final volume of 300 μL: 0 μM, 1.66 μM, 3.33 μM, 16.6 μM, 33.3 μM, 50 μM, 66.6 μM, 83.3 μM, and 100 μM).
Add 300 μL of Reagent A to each sample and standard and mix.
Add 300 μL Reagent B to each sample and standard and mix.
Incubate the samples and standards in a 37 ºC water bath for 15 min.
Add 15 μL of Reagent C to each sample and standard. Mix and then centrifuge at 14,000 × g for 5 min.
Measure the absorbances of the standards at 592 nm using the first standard (0 μM) as a blank.
Measure the absorbances of each of the four samples.
Plot the concentration versus absorbance of each of the standards and use the linear equation to calculate the concentrations of each of the four samples.
Average the concentration of iron in each of the four samples and normalize to the protein concentration in order to determine the amount of iron per protein.
3.3. Colorimetric Sulfide Analysis Protocol
Prepare the following solutions anaerobically: 1% zinc acetate (w/v%); 3 M NaOH; 0.2% dimethyl-4-phenylenediamine (DMPD) in 5 N HCl; 23 mM FeCl3 in 1.2 N HCl; and 200 μM Na2S).
Complete steps 3-10 anaerobically.
Dilute the protein to 10 μM in 50 mM NaOH (prepared in deoxygenated water) to a final volume of 1000 μL.
Aliquot 100 μL of the 10 μM protein solution into five Eppendorf tubes.
Create a standard curve using the 200 μM Na2S solution by making 100 μL standards at the following concentrations: 0 μM, 10 μM, 20 μM, 30 μM, 40 μM, 50 μM, 60 μM, and 80 μM.
Add 300 μL of the 1% zinc acetate solution to each of the standards and samples.
Add 15 μL of 3 M NaOH to each of the standards and samples. Mix by inversion.
Let stand for 2 h to allow zinc sulfide to form and settle to the bottom of the tubes.
Underlay the standards and samples with 75 μL of the 0.2% DMPD solution.
Quickly add 2 μL of 23 mM FeCl3 to each sample and mix by inversion.
Remove standards and samples from the anaerobic chamber and centrifuge at 14,000 × g for 15 min,
Remove 400 μL of each standard and sample and place in unused Eppendorf tubes.
Add 600 μL 2 mM EPPS, pH 8.0, to each standard and sample.
Let stand for 30 min at room temperature and then measure the absorbance at 666 nm.
Plot the concentration versus the absorbance from each standard. Use the linear equation to determine the concentration of sulfide in each sample and normalize the sulfide to the protein concentration to determine the amount of sulfide per protein.
4. Methods for Isotopically Labelling Iron-sulfur Clusters to Monitor Cluster Transfer
4.1. Protocol for the Expression of the nfuA Gene in Apo Form
Introduce the pET28a-nfuA construct into BL21(DE3) cells using standard transformation procedures. Spread cells on an LB plate supplemented with 50 μg/mL kanamycin.
The following day, use a single colony to inoculate 200 mL LB media in a 250 mL flask supplemented with 50 μg/mL kanamycin. Incubate the starter culture at 37 ºC with shaking at 250 rpm for ~14 h. Place 4 6 L flasks containing 4 L M9 minimal media in the shaker overnight at 37 ºC to warm the culture media.
Prior to inoculation of the large flasks, add 50 μg/mL kanamycin and 400 μL 10,000× micronutrient mix (3 nM (NH4)2Mo7O24, 30 nM CoCl2•6H20, 400 nM H3BO3, 10 nM CuSO4•5H2O, 80 nM MnCl2•4H20, and 10 nM ZnSO4•7H20).
Use 20 mL of the overnight starter culture to inoculate each of the four large flasks.
At an OD600=0.3, supplement cultures with 200 mL amino acid mixture (alanine (800 μM), arginine (400 μM), asparagine (400 μM), aspartic acid (400 μM), cysteine (100 μM), glutamic acid (600 μM), glutamine (600 μM), glycine (800 μM), histidine (200 μM), isoleucine (400 μM), leucine (800 μM), lysine (400 μM), methionine (200 μM), phenylalanine (400 μM), proline (400 μM), serine (10 mM), threonine (400 μM), tryptophan (100 μM), tyrosine (200 μM), and valine (600 μM). Take a 1 mL sample (uninduced sample) for SDS-PAGE analysis.
At an OD600=0.6, add 1,10-phenanthroline to each of the flasks to a final concentration of 75 μM and allow the flasks to shake for an additional 15 min.
Place the flasks in an ice-water bath for ~1 h. Turn the temperature of the incubator to 18 ºC.
Induce overexpression of the desired gene by adding 200 μM IPTG and allow expression to take place at 18 ºC with shaking at 180 rpm for ~18 h.
On the following day, take a sample (post-IPTG induction) normalized to the number of cells in the uninduced sample for SDS-PAGE analysis.
Harvest the cells by centrifugation at 7,500 × g for 12 min. Flash-freeze the resulting cell pellet (~2g/L) in liquid nitrogen and store in liquid nitrogen until further use.
Isolate the apo protein using the exact method described for the holo protein.
Analyze the overproduction of the protein by SDS-PAGE analysis (12.5%) using the samples collected.
4.2. Synthesis of 34S-Labeled Sulfide from 34S-Labeled Elemental Sulfur
Perform all steps under a constant flow of nitrogen.
Cool a two-neck round-bottomed flask containing a magnetic stir bar to −78 ºC in a dry ice/acetone bath.
Add 0.5 g elemental 34S-labeled elemental sulfur to the cooled flask and pass ammonia gas into the flask until approximately 75 mL liquid ammonia forms.
Carefully and slowly add a two-fold molar excess of sodium metal to the reaction and allow it to stir for 30 min.
Remove the reaction from the dry ice/acetone bath and allow it to warm to room temperature.
Evaporate the ammonia under a stream of nitrogen gas to yield a white precipitate.
Add isopropanol until the blue color dissipates, indicating that the excess sodium metal is quenched.
Dissolve the white precipitate in 15 mL H20.
Remove the solvent using a rotary evaporator to yield a solid containing both NaOH and 34S-labeled sodium sulfide (Na234S).
Dissolve the solid in a minimum amount of water and quantify the sulfide concentration using the colorimetric sulfide analysis protocol detailed above.
4.3. Reconstitution of Apo NfuA using Na234S
Perform all reconstitution steps under strictly anaerobic conditions.
Dilute Apo NfuA to 100 μM in reconstitution buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 5 mM DTT, and 20% glycerol) in a total volume of 20 mL.
Add DTT (5 mM final concentration) in three increments every 20 min. Incubate the solution on ice for 1 h.
Slowly, add FeCl3 (2 mM final concentration) every five min a total of five times. Incubate the solution for an additional 30 min.
Add the Na234S (2 mM final concentration) in five increments every 30 min. Check the pH of the Na234S solution before adding and neutralize the NaOH left from the synthesis procedure if necessary. Incubate the reaction on ice overnight.
The following day, remove protein precipitation by centrifuging the reconstituted protein at 14,000 × g for 15 min.
Concentrate the solution to ~2 mL.
Remove aggregates from the reconstitution procedure by size-exclusion chromatography using a HiPrep 16/60 Sephacryl HR S-200 column (GE Health Sciences) equilibrated in storage buffer (flow-rate of 0.5 mL/min) and connected to an ÅKTA protein liquid chromatography system (GE Health Sciences) housed in an anaerobic chamber.
Collect the fractions containing the reconstituted protein and concentrate to ~2 mL. Flash-freeze in liquid nitrogen until further use. Re-determine the protein concentration using the Bradford method and perform an iron and sulfide analyses on the reconstituted protein.
5. Strategies for Analyzing Protein Complex Formation
Various methods exist to measure protein-protein interactions; however, it is often difficult to characterize interactions that are transient in nature. A non-exhaustive list of methods includes co-immunoprecipitation, isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and analytical molecular-sieve chromatography. The benefit of using co-immunoprecipitation is the ability to pull-down protein complexes in vivo that are biologically relevant. However, while it is possible to pull protein complexes out of the cell, the use a cross-linking reagent is often needed to capture weak interactors. Both ITC and SPR are enticing options because they can quantitatively determine the Kd associated with the binding event in protein-protein interactions or with protein-small molecule interactions. One downfall associated with ITC is the need for large amounts of material for analysis.
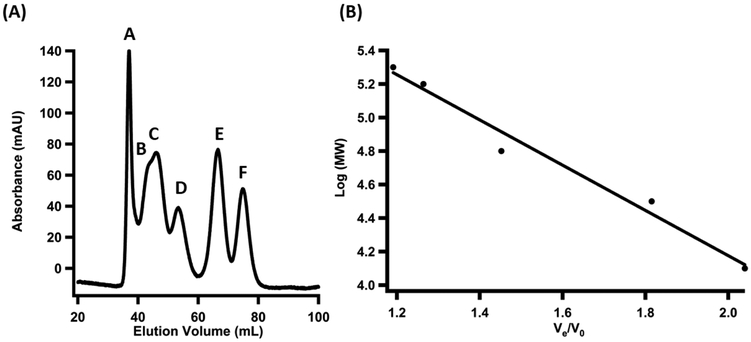
Qualitatively, molecular-sieve chromatography can be used to determine if in vitro mixtures of proteins co-elute as a complex rather than as individual components. A standard curve can be generated using a suite of standards in order to estimate the molecular weight of the complexes. This method is inferior to ITC and SPR in the sense that it only provides a qualitative assessment of whether the proteins form a complex. However, it takes very small amounts of material and can be performed in a single day. Similar to co-immunoprecipitation, this method does not capture transient interactions. Following its completion, other methods can be used for a more quantitative assessment of the binding event.
5.1. Protocol for performing analytical molecular sieve chromatography
Equilibrate a HiPrep 16/60 Sephacryl HR S-200 column (GE Healthcare) connected to an ÅKTA protein liquid chromatography system (GE Health Sciences) housed in an anaerobic chamber with buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 5 mM DTT, and 15% glycerol).
It is essential to ensure that the glovebox is fully devoid of oxygen contamination (<1 ppm O2) and to add fresh reductant (5 mM DTT) to buffer directly before use to prevent disulfide formation between proteins.
Prepare a 500 μL sample in buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 5 mM DTT, and 15% glycerol) containing the following proteins as standards: Cytochrome C (0.3 mg/mL), Carbonic Anhydrase (0.3 mg/mL), Bovine Serum Albumin (0.80 mg/mL), Alcohol Dehydrogenase (0.65 mg/mL), β-Amylase (0.75 mg/mL), and Blue Dextran (0.40 mg/mL).
Load the mixture containing the standards onto the column and flow at a rate of 0.5 mL/min until all of the standards elute. Record the elution volume of the Blue Dextran, which indicates the experimentally determined void volume (V0), and the elution volumes (Ve) of each of the additional standards, which will be used to generate a standard curve.
Dilute the LipA protein to make a 500 μL sample at a concentration of 100 μM. Load the sample onto the column and elute at a rate of 0.5 mL/min. Record its elution volume.
Dilute the NfuA protein to make a 500 μL sample with a concentration of 100 μM. Load the sample onto the column and elute at a rate of 0.5 mL/min. Record its elution volume.
Make a 500 μL sample mixture containing 100 μM LipA protein and 100 μM NfuA protein. Incubate at room temperature for 5 min and then load onto the column and elute at a rate of 0.5 mL/min. Collect protein containing fractions and record the elution volumes of each peak observed in the chromatogram.
Concentrate the fractions corresponding to each protein peak observed in the chromatogram. Analyze by SDS-PAGE (12.5%) to confirm the identity of the proteins in each fraction.
Generate a standard curve of the Log(molecular weight) versus Ve/V0 for each of the standards.
Divide the experimentally determined elution volume of each of the peaks in the chromatograms of each sample by the void volume of the column (Ve/V0).
Use the linear equation from the standard curve to solve for the Log(molecular weight) of the individual proteins as well as the protein complexes.
6. Monitoring Iron-sulfur Cluster Transfer Using Quantitative Analysis
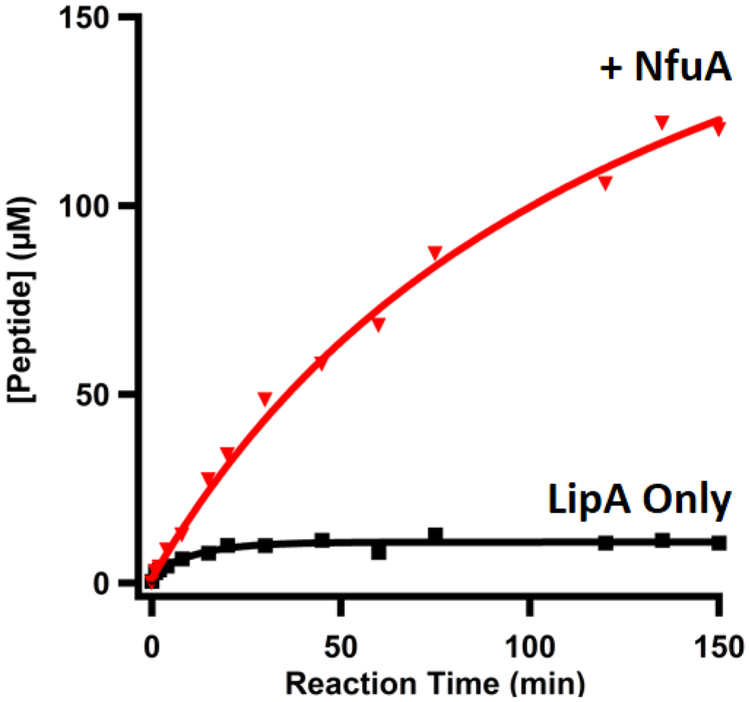
A strategy for accurately monitoring lipoyl cofactor production in vitro was not established until the early 2000s (Miller, et al., 2000). This strategy was based on an indirect assay in which activation of apo-pyruvate dehydrogenase by lipoylation was coupled to the reduction of NAD+, which could be monitored spectrophotometrically. Through this method, the rate of NADH formation was directly dependent on the amount of lipoylated pyruvate dehydrogenase. Since that time, more direct methods have been developed to quantitatively monitor the formation of the monothiolated intermediate and lipoyl product using liquid-chromatography-mass spectrometry (LC-MS), which is described below. This method allows the activity of LipA to be quantified and the effect of NfuA on LipA’s activity to be assessed. Additionally, by using NfuA with isotopically labeled clusters, as detailed above, the transfer of an iron-sulfur cluster can be directly monitored. Further, including a competitive chelator such as citrate in the reaction can serve to ensure that the cluster transfer from NfuA to LipA is direct, as opposed to a diffusive mechanism wherein LipA obtains a cluster from solution (Ding, et al., 2004).
6.1. Sample preparation for analysis of reaction products by liquid-chromatography-mass spectrometry (LC-MS) to assess the effect of NfuA on LipA activity
Reactions are performed anaerobically. Reactions are typically prepared in a total volume of 200 μL and contain the following: buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 10% glycerol), 600 μM octanoyl peptide substrate (Glu-Ser-Val-[N6-octanoyl]Lys-Ala-Ala-Ser-Asp), 25 μM LipA, 1 μM S-adenosylhomocysteine (SAH) nucleosidase (to prevent product inhibition), and 2 mM SAM. An additional and identical reaction, but containing 400 μM NfuA, is also prepared. Importantly, DTT has been shown to be capable of non-physiological iron-sulfur cluster transfer and should not be used in assay buffers (Vranish, et al., 2015).
A 500 μL quench solution containing 100 mM H2SO4, and 20 μM AtsA (peptide internal standard, Pro-Met-Ser-Ala-Pro-Ala-Arg-Ser-Met) is prepared, and 15 μL is aliquoted into 11 Eppendorf tubes.
Initiate the reaction by adding the low-potential reductant. In LipA/NfuA assays, 2 mM dithionite is typically used. The initiation of the two reactions (with and without NfuA) can be staggered by 30 s in order to perform them simultaneously.
At the specified time points (t=0, 1, 2, 4, 8, 15, 30, 60, 90, 120, and 150 min), add 15 μL of the reaction into 15 μL of the quench solution.
After the reaction is complete, remove the reaction tubes from the anaerobic chamber and centrifuge for 30 min at 14,000 × g.
Avoiding any precipitation, pipette 25 μL of each time point into an appropriate autosampler vial for LC-MS analysis.
6.2. Sample preparation for analysis of reaction products in the presence of a competitive chelator or exogenous 34S-labeled sulfide to ensure direct cluster transfer from NfuA to LipA
Prior to performing activity assays with LipA and NfuA, incubate each protein individually with a five-fold excess sodium citrate. Assess whether the citrate has a significant effect on the Fe–S clusters of LipA and NfuA by obtaining a UV-visible spectrum of the protein before and after a 1.5 h incubation with citrate.
Perform a LipA only control and a LipA/NfuA activity assay exactly as described previously.
Set up two additional reactions containing both LipA and NfuA. In one reaction, add a five-fold excess of citrate. In the other reaction, add Na234S to a final concentration of 1 mM.
When analyzing data from the LipA/NfuA reaction with citrate, assess whether the addition of this chelator inhibits product formation by LipA. Citrate can chelate metals from solution and will inhibit multiple turnover of LipA if LipA’s auxiliary cluster is regenerated chemically from iron and sulfide in solution after each turnover. The absence of an effect is consistent with a direct cluster transfer from NfuA through protein-protein interactions.
As a second method (or if citrate chelates the cluster out of the protein of interest), analyze whether 34S from exogenous Na234S included in the reaction mixture is observed in the lipoyl product. The observation of product that contains sulfur at natural abundance suggests that the attached sulfur atom did not come from solution.
6.3. Protocol for the quantification of reaction products by LC-MS
Prepare 50 μL of a 500 μM standard in deoxygenated water that contains buffer (50 mM HEPES, pH 7.5, 300 mM KCl, and 10% glycerol), 50 mM H2SO4, 2 mM TCEP, 20 μM AtsA internal standard, 500 μM octanoyl peptide, 500 μM 6-thio-octanoyl peptide, and 500 μM lipoyl peptide. The peptides used for quantification (octanoyl, 6-thio-octanoyl, lipoyl, AtsA) are >98% pure and can be purchased from ProImmune Custom Peptides.
Prepare 300 μL of a dilution mix containing the following: buffer (50 mM HEPES, pH 7.5, 300 mM KCl, and 10% glycerol), 50 mM H2SO4, 2 mM TCEP, and 20 μM AtsA internal standard.
Aliquot 25 μL of the dilution mix into eleven Eppendorf tubes.
Perform a 1:2 serial dilution of the 500 μM standard by mixing 25 μL of the standard with 25 μL of the dilution mix.
Continue serial diluting the standards by a factor of 2 to generate standards for quantification (500 μM, 250 μM, 125 μM, 62.5 μM, 31 μM, 16 μM, 8 μM, 4 μM, 2 μM, 1 μM, 0.5 μM, and 0.25 μM).
Analyze the standards and reaction mixtures by LC-MS (Agilent Technologies 1200 system coupled to an Agilent Technologies 6410 QQQ mass spectrometer) using an Agilent Technologies Zorbax Extend-C18 column Rapid Resolution HT (4.6 mm × 50 mm, 1.8 μm particle size) equilibrated in 98% Solvent A (0.1% formic acid, pH 2.6) and 2% Solvent B (100% acetonitrile). Detect the compounds using electrospray ionization in positive mode (ESI+) with the following parameters: 340 ºC nitrogen gas temperature, 9.0 L/min flow-rate, 40 PSI nebulizer pressure, and 4000 V capillary pressure. Apply a gradient of 2-23% Solvent B from 0.8 min to 3.5 min and maintain at 23% Solvent B until 8 min. Return to 2% Solvent B from 8 min to 10 min and then allow the column to re-equilibrate for 2 min before the next injection. Maintain a flow-rate of 0.4 mL/min throughout the entire method.
Use the associated MassHunter program to quantify the decay of the octanoyl peptide substrate and formation of the 6-thio-octanoyl peptide intermediate and lipoyl product.
Conclusions
LipA catalyzes the attachment of sulfur atoms to an unactivated octanoyl chain in the biosynthesis of the lipoyl cofactor, an essential compound used in various multienzyme complexes in Nature and implicated in human health and disease. Refined techniques for the overproduction and isolation of Fe–S containing proteins allowed for the isolation of homogenous LipA with high activity for robust mechanistic study, although the enzyme only catalyzes a single turnover due its inactivation upon the destruction of its auxiliary cluster. Recent work has identified a pathway for reforming the auxiliary cluster to allow for multiple turnovers. Biochemical methods to study the molecular details of cofactor repair and/or reinsertion in proteins that sacrifice their Fe–S clusters will help fill outstanding gaps in our understanding of the mechanism of protein maturation.
Figure 1.
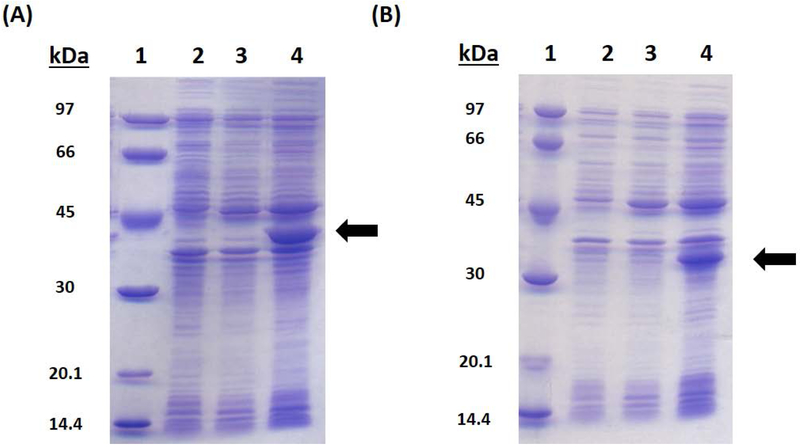
SDS-PAGE analysis of (A) E. coli LipA overproduction and (B) E. coli NfuA overproduction. The lanes are as follows: (1) Molecular Weight Ladder (2) Uninduced cells (3) Arabinose-induced cells (4) IPTG-induced cells. The arrows point to the protein of interest.
Figure 2.
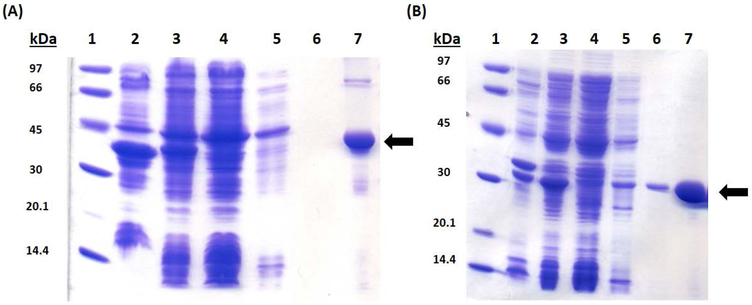
SDS-PAGE analysis of (A) E. coli LipA isolation and (B) E. coli NfuA isolation. The lanes are as follows: (1) Molecular Weight Ladder (2) Pellet (3) Supernatant (4) Flow-through (5) Wash 1 (6) Wash 2 (7) Pure protein. The arrows point to the protein of interest.
Figure 3.
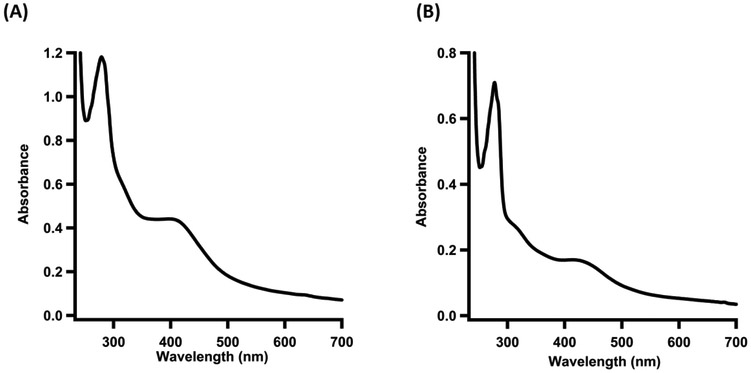
UV-visible spectrum of (A) As-isolated E. coli LipA and (B) As-isolated E. coli NfuA.
Figure 4.
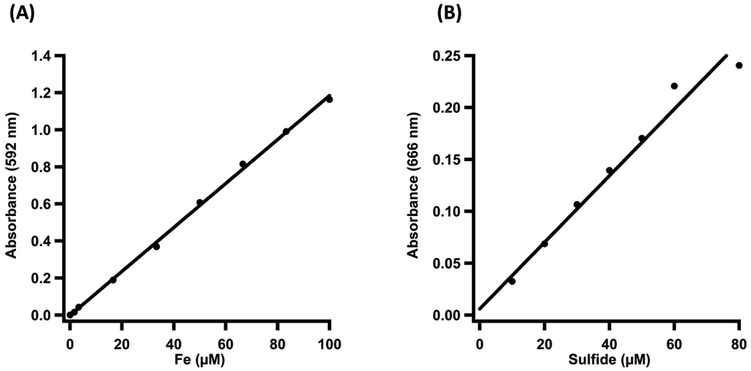
(A) Iron Analysis Standard Curve (B) Sulfide Analysis Standard Curve.
Figure 5.
Molecular Sieve Chromatography. (A) Chromatogram of the suite of standards used to generate the standard curve as follows: (A) Blue Dextran, 2000 kDa; (B) β-Amylose, 200 kDa; (C) Alcohol Dehydrogenase, 150 kDa; (D) Bovine Serum Albumin, 66 kDa; (E) Carbonic Anhydrase, 29 kDa; (F) Cytochrome C, 12.4 kDa.
(B) Standard curve generated from the elution volume of each of the standards corrected for the void volume of the column versus the log molecular weight of each of the standards.
Figure 6.
Time-dependent formation of the monothiolated intermediate and lipoyl product in the absence (15 μM LipA, shown in black) and presence (400 μM NfuA, shown in red) of NfuA.
Table 1.
Parameters for LC-MS/MS analysis of unlabeled reaction intermediate and product.
| Compound | Parent Ion | Fragmentator Voltage | Product Ion 1 | Product Ion 2 |
|---|---|---|---|---|
| Octanoyl Peptide | 932.5 | 240 | 552.3 (37) | 210.1 (51) |
| 6-thio-octanoyl Peptide | 964.5 | 240 | 584.3 (37) | 242.1 (51) |
| Lipoyl Peptide | 996.5 | 240 | 616.4 (37) | 274.1 (51) |
| AtsA Peptide (IS) | 474.4 | 135 | 229.1 (12) | 153.1 (26) |
| Tryptophan (IS) | 188.0 | 130 | 146.1 (10) | 118.0 (21) |
Respective collision energies are in parentheses
Table 2.
Parameters for LC-MS/MS analysis of 34S-labeled reaction intermediate and product.
| Compound | Parent Ion |
Fragmentation Voltage | Product Ion 1 | Product Ion 2 |
|---|---|---|---|---|
| Octanoyl Peptide | 932.5 | 240 | 552.3 (37) | 210.1 (51) |
| 32S-6-thio-octanoyl Peptide | 964.5 | 240 | 584.3 (37) | 242.1 (51) |
| 34S-6-thio-octanoyl Peptide | 966.5 | 240 | 586.3 (37) | 244.1 (51) |
| 32S/32S Lipoyl Peptide | 996.5 | 240 | 616.4 (37) | 274.1 (51) |
| 32S/34S Lipoyl Peptide | 998.5 | 240 | 618.4 (37) | 276.1 (51) |
| 34S/34S-Lipoyl Peptide | 1000.5 | 240 | 620.4 (37) | 278.1 (51) |
| AtsA Peptide (IS) | 474.4 | 135 | 229.1 (12) | 153.1 (26) |
| Tryptophan (IS) | 188 | 130 | 146.1 (10) | 118.0 (21) |
Respective collision energies are in parentheses
Acknowledgements
This work was supported by the National Institutes of Health grant GM122595 (S.J.B.) and NSF grant MCB-1158486 (S.J.B.). S.J.B. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Reed LJ, Busk BGD, Gunsalus IC, & Schnakenberg GHF (1951) Crystalline a-Lipoic Acid: A Catalytic Agent Associated with Pyruvate Dehydrogenase. Science 114:93. [DOI] [PubMed] [Google Scholar]
- 2.Reed LJ, et al. (1953) Isolation, characterization and structure of α-lipoic acid. J. Am. Chem. Soc 75:1267–1270. [Google Scholar]
- 3.Cronan JE (2016) Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol. Mol. Biol. Rev 80:429–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed L (1974) Multienzyme complexes. Acc. Chem. Res 7:40–46. [Google Scholar]
- 5.Miller JR, et al. (2000) Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39:15166–15178. [DOI] [PubMed] [Google Scholar]
- 6.Zhao S, Miller JR, Jiang Y, Marletta MA, & Cronan JE Jr. (2003) Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem. Biol 10:1293–1302. [DOI] [PubMed] [Google Scholar]
- 7.Yi X & Maeda N (2005) Endogenous production of lipoic acid is essential for mouse development. Mol. Cell. Biol 25:8387–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landgraf BJ, McCarthy EL, & Booker SJ (2016) Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annual Review of Biochemistry 85(1):485–514. [DOI] [PubMed] [Google Scholar]
- 9.Frey PA, and Booker SJ (2001) Radical mechanisms of S-adenosylmethionine-dependent enzymes. Adv Protein Chem 58, 1–45 [DOI] [PubMed] [Google Scholar]
- 10.Lanz ND & Booker SJ (2015) Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochimica et biophysica acta 1853(6):1316–1334. [DOI] [PubMed] [Google Scholar]
- 11.Douglas P, Kriek M, Bryant P, & Roach PL (2006) Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew. Chem 118:5321–5323. [DOI] [PubMed] [Google Scholar]
- 12.Lanz ND, et al. (2014) Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase. Biochemistry 53:4557–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin MI, et al. (2016) Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc. Natl. Acad. Sci. U S A 113:9446–9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cicchillo RM, et al. (2004) Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry 43:6378–6386. [DOI] [PubMed] [Google Scholar]
- 15.Cicchillo RM, et al. (2004) Escherichia coli lipoyl synthase binds two distinct [4Fe–4S] clusters per polypeptide. Biochemistry 43:11770–11781. [DOI] [PubMed] [Google Scholar]
- 16.Harmer JE, et al. (2014) Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J 464:123–133. [DOI] [PubMed] [Google Scholar]
- 17.Cicchillo RM & Booker SJ (2005) Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J. Am. Chem. Soc 127:2860–2861. [DOI] [PubMed] [Google Scholar]
- 18.McCarthy EL & Booker SJ (2017) Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. 358(6361):373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanz ND, et al. (2012) Chapter Seven - RlmN and AtsB as Models for the Overproduction and Characterization of Radical SAM Proteins Methods in Enzymology, ed Hopwood DA (Academic Press; ), Vol 516, pp 125–152. [DOI] [PubMed] [Google Scholar]
- 20.Blaszczyk AJ, Wang RX, & Booker SJ (2017) TsrM as a Model for Purifying and Characterizing Cobalamin-Dependent Radical S-Adenosylmethionine Methylases. Methods in enzymology 595:303–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelini S, et al. (2008) NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem 283(20):14084–14091. [DOI] [PubMed] [Google Scholar]
- 22.Py B, et al. (2012) Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Molecular microbiology 86(1):155–171. [DOI] [PubMed] [Google Scholar]
- 23.Boutigny S, et al. (2013) Physical and functional interactions of a monothiol glutaredoxin and an iron sulfur cluster carrier protein with the sulfur-donating radical S-adenosyl-L-methionine enzyme MiaB. The Journal of biological chemistry 288(20):14200–14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 25.Beinert H (1983) Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Analytical Biochemistry 131(2):373–378. [DOI] [PubMed] [Google Scholar]
- 26.Beinert H (1978) Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 54:435–445. [DOI] [PubMed] [Google Scholar]
- 27.Pandelia ME, Lanz ND, Booker SJ, & Krebs C (2015). Mossbauer spectroscopy of Fe/S proteins. Biochim Biophys Acta, 1853(6), 1395–1405. doi: 10.1016/j.bbamcr.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 28.Snell EE (1992) Microorganisms in vitamin and biofactor research. Journal of nutritional science and vitaminology Spec No:34-39.29. [DOI] [PubMed] [Google Scholar]
- 29.Snell EE (1993) From bacterial nutrition to enzyme structure: a personal odyssey. Annual review of biochemistry 62:1–27. [DOI] [PubMed] [Google Scholar]
- 30.Holmquist L, et al. (2007) Lipoic acid as a novel treatment for Alzheimer’s disease and related dementias. Pharmacology & therapeutics 113(1):154–164. [DOI] [PubMed] [Google Scholar]
- 31.Borcea V, et al. (1999) alpha-Lipoic acid decreases oxidative stress even in diabetic patients with poor glycemic control and albuminuria. Free radical biology & medicine 26(11-12):1495–1500. [DOI] [PubMed] [Google Scholar]
- 32.Ding H, Clark RJ, & Ding B (2004) IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J. Biol. Chem. 279:37499–37504. [DOI] [PubMed] [Google Scholar]
- 33.Vranish JN, Russell WK, Yu LE, Cox RM, Russell DH, and Barondeau DP (2015). Fluorescent probes for tracking the transfer of iron-sulfur cluster and other metal cofactors in biosynthetic reaction pathways. Journal of the American Chemical Society, 137 (1), 390–8. [DOI] [PMC free article] [PubMed] [Google Scholar]