Abstract
Pseudoenzymes generally lack detectable catalytic activity despite adopting the overall protein fold of their catalytically competent counterparts, indeed ‘pseudo’ family members seem to be incorporated in all enzyme classes. The small GTPase enzymes are important signaling proteins, and recent studies have identified a number of new family members with non-canonical residues within the catalytic cleft, termed pseudoGTPases. To illustrate recent discoveries in the field we use the p190RhoGAP proteins as an example. p190RhoGAP proteins (ARHGAP5 and ARHGAP35) are the most abundant GAPs for the Rho family of small GTPases. These are key regulators of Rho signaling in processes such as cell migration, adhesion and cytokinesis. Structural biology has complemented and guided biochemical analyses for these proteins and has allowed discovery of two cryptic pseudoGTPase domains, and the re-classification of a third, previously identified, GTPase-fold domain as a pseudoGTPase. The three domains within p190RhoGAP proteins illustrate the diversity of this rapidly expanding pseudoGTPase group.
Keywords: pseudoGTPase, pseudoenzyme, pseudokinase, small GTPase, p190RhoGAP, Rho signaling
Introduction
Pseudoenzymes are defined as members of an enzyme class which contain mutations in the highly conserved canonical catalytic residues found in enzyme counterparts [1, 2]. The loss of conserved residues can render these proteins catalytically deficient, or conversely can be associated with a redesign of the catalytic cleft to utilize non-canonical residues for enzymatic processes [3]. The pseudoenzymes continue to emerge as functionally important members of their respective enzymatic families, and have roles as regulators and modifiers of a large number of signaling pathways [4]. Pseudoenzymes have been described in the protein kinase, phosphatase, protease, GTPase and other families [3, 5], and are thought to encompass approximately 10% of each enzyme fold [2, 6]. For the small GTPase family a number of pseudoGTPases have been identified over the last four or five years [7–9]. Consequently there is a growing group of annotated pseudoGTPases within the fold. We recently identified multiple pseudoGTPase domains in the p190RhoGAP proteins, which are important regulators of RhoA signaling [9, 10]. In this mini-review we discuss the discovery and characterization of these multiple pseudoGTPase domains and summarize the current state of the pseudoGTPase group using the p190RhoGAP proteins as examples.
The small GTPase domain
Proteins belonging to the Ras superfamily of small GTPases act as bimodal molecular switches that translate extracellular stimuli to intracellular responses in myriad cellular pathways [11]. They are guanine nucleotide-binding proteins whose signaling mode is determined by their nucleotide-bound state: when bound to GTP, small GTPases assume an ON/active conformation and bind downstream effector proteins, conversely, when bound to GDP, small GTPases assume the OFF/inactive conformation. These enzymes require additional regulatory proteins to achieve biologically functional GTP/GDP cycling [12]; the otherwise slow exchange of bound GDP for GTP is facilitated by guanine nucleotide exchange factors (GEF), and their slow intrinsic enzymatic activity requires a GTPase activating protein (GAP) to promote hydrolysis. Some GTPases are further negatively regulated by guanine nucleotide disassociation inhibitors (GDI) which bind and extract the GTPase from the plasma membrane [12]. A range of inputs therefore control the downstream signaling of canonical small GTPases (Figure 1A).
Figure 1. Schematic of canonical GTPase cycling and classes of pseudoGTPase.
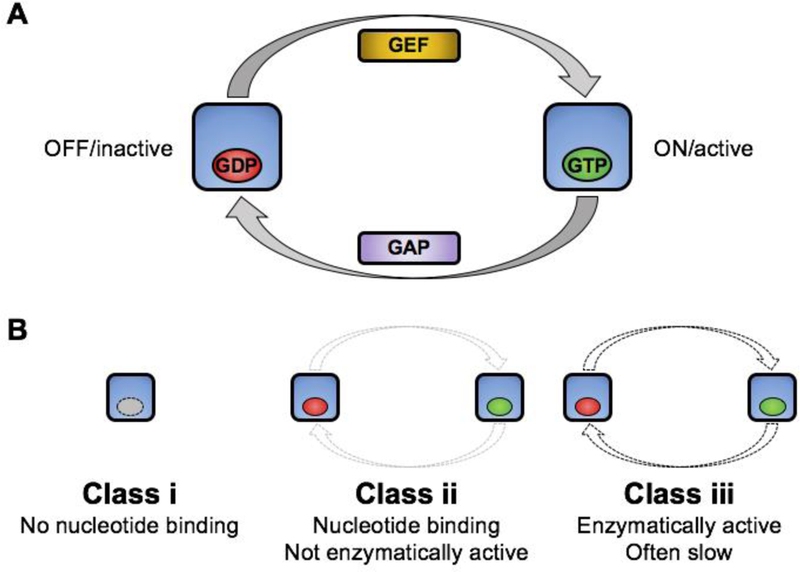
A) GTPase cycling is illustrated with OFF/inactive GTPase shown on left bound to GDP (red) and ON/active GTPase shown in right bound to GTP (green). Guanine nucleotide exchange factors (GEF) and GTPase activating proteins (GAP) are indicated. B) Classes of pseudoGTPase. Class (i) has no nucleotide binding activity and therefore no catalytic activity, class (ii) binds nucleotide but with no activity (*under standard catalytic conditions), and class (iii) binds nucleotide and is catalytically active. Color scheme as in A.
Small GTPases are typically single-domain proteins (e.g. H-Ras) but can also exist within large multidomain proteins (e.g. the GGAPs [13]). This domain is usually between 20 and 25 kDa in molecular weight, and adopts a well conserved three-dimensional Rossmann-type fold, a common structural motif occurring in nucleotide-binding proteins that consists of alternating beta strands and alpha helical segments which form a sandwich [14]. A set of nearly invariant sequence motifs are present, and these are termed the “G motifs”: G1 (P-loop), G2 (Switch I), G3 (Switch II), G4 and G5 [15–17] (Figure 2A). Together, the G motifs are responsible for binding nucleotide and catalyzing GTP hydrolysis to GDP (Figure 2B). Specifically, G1 (phosphate-binding P-loop, GxxxxGKS/T) accommodates the phosphates of GTP/GDP. G2 (Switch I) contains an invariant Thr residue that contacts the GTP γ-phosphate and Mg2+. Similarly G3 (Switch II, DxxGQ/H/T), contains aspartic acid and glycine residues which coordinate the γ-phosphate and Mg2+. The G3 Gln residue is a key enzymatic residue that is highly conserved: mutation of this residue (Q61) in Ras leads to loss of GTPase activity [18] and freezes Ras in the GTP-bound ON/active state leading to cellular transformation. Together, G2 and G3 undergo large conformational changes during nucleotide cycling and are sometimes termed the “γ-phosphate sensors” [19]. When the small GTPase is bound to GTP, the G2 (Switch I) motif is also the major binding site for downstream effector proteins. G4 (N/TKxD) and G5 (SAR/K) contact the guanine base directly and dictate nucleotide binding specificity (i.e. prevent non-guanine base nucleotides from binding) [15]. Conservation of the G motifs in active GTPases has been well catalogued and the critical GTP-binding and catalytic residues are established [17] (Figure 2). The pseudoGTPases, by definition, are the subset of GTPases that sequentially diverge at one or more of these conserved G motifs while maintaining a canonical Ras-like fold [4].
Figure 2. G motif conservation in canonical GTPases and p190RhoGAP.
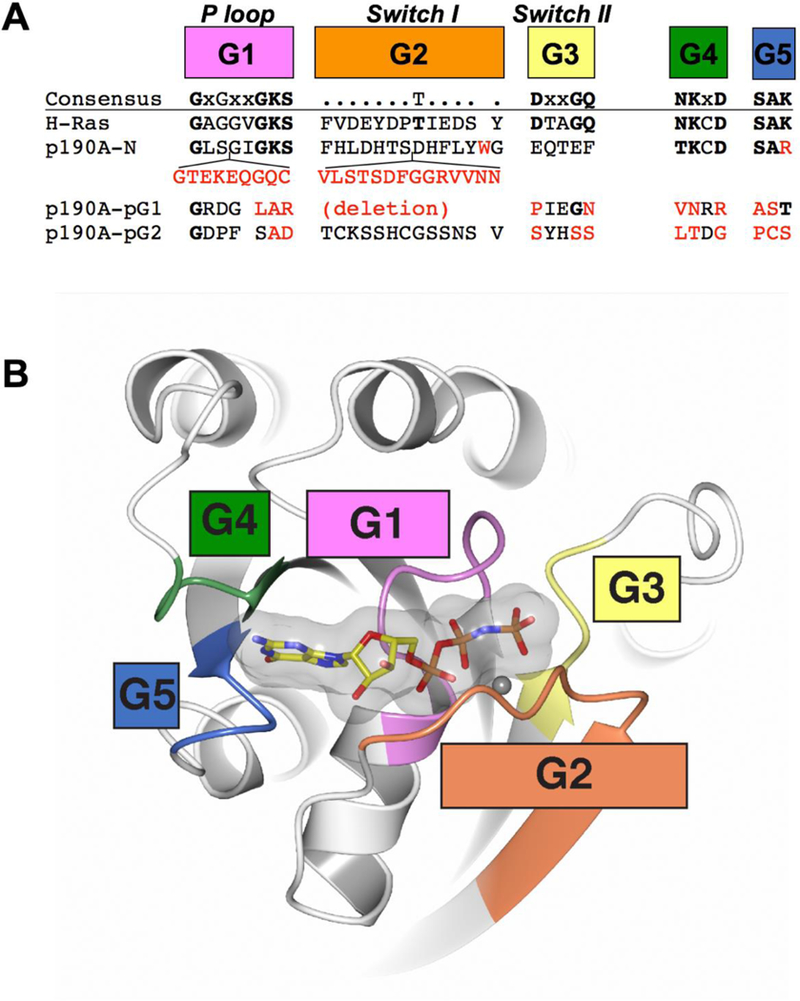
A) Active small GTPases contain five conserved G motifs termed G1 through G5. Structure-based sequences for the G motifs of H-Ras, p190RhoGAP-A N-terminal GTPase domain (p190A-N) and p190RhoGAP-A pG1 pseudoGTPase domain (p190A-pG1) are shown. Predicted sequences for the G motifs of p190RhoGAP-A pG2 pseudoGTPase (p190A-pG2) domain are also shown. B) Ribbon diagram of H-Ras bound to a nonhydrolyzable GTP analog (PDB ID: 5p21 [54]). G motifs are colored as in part A.
Identification of pseudoGTPases
The pseudokinases are the archetypal pseudoenzymes. Identification of the full kinome in 2002 by Manning and colleagues clearly highlighted that the family contains both conventional active kinases and a subset that lack one or more of the consensus sequences known to mediate phosphoryl transfer [20]. The disruption of conserved sequence presumably indicated that most of these were catalytically inactive, and by 2006 the term ‘pseudokinase’ was used to define these ~10% of the kinome [21], however, biochemical and structural analyses have shown that some pseudokinases retain catalytic competency [22–28]. The extensive studies of pseudokinases illustrate that pseudoenzymes, enzymes mutated in the conserved catalytic residues, are not required to be catalytically inactive; indeed these proteins are rich in their variety of enzymatic activities ranging from those that are unable to bind nucleotide [29–33] to those that are essentially normal kinases [22, 30].
In contrast to the bioinformatics-driven identification of the entire pseudokinase group, the pseudoGTPases have emerged in a more piecemeal manner and over a longer time. Although the Rnd (Rnd1, Rnd2 and Rnd3) [34, 35] and RGK (Rad, Rem1, Rem2, Gem/Kir) groups were easily identified as small GTPases despite mutations in their G motifs [36–40], the pseudoGTPase group also includes ‘cryptic’ GTPase domains which have very low sequence identity compared to active GTPases and harbor completely divergent G motif sequences. These cryptic GTPase domains can be elusive in sequence-based bioinformatics searches, but upon crystal structure determination have been revealed to adopt the canonical GTPase-like fold. Cryptic GTPase domains have been found in CENP-M, fungal dynein LIC, and the p190RhoGAP proteins [7–9], and their collective identification raised the possibility that a larger number of cryptic pseudoGTPases have yet to be identified. Consequently, similar to the pseudokinases, the pseudoGTPases contain catalytically inactive (e.g. the Rnd group) and catalytically active (e.g. the RGK group) members, but contrasting the pseudokinases also contain divergent family members that are structurally similar but sequentially completely degraded in their catalytic motifs (Figure 2). The use of structural biology has been key for discovery of the cryptic domains, and below we discuss the identification and analysis of the multiple pseudoGTPase domains within the p190RhoGAP proteins.
The p190RhoGAP proteins
The p190RhoGAP orthologs, p190RhoGAP-A and -B, are the most abundant GAPs for the Rho family of small GTPases [41], and are important regulators of Rho signaling processes including cell migration, adhesion and cytokinesis [42–44]. These are large (approximately 1500 amino acid) multidomain proteins whose domain structure is conserved between the two proteins and comprises a GTPase fold at the N-termini (termed the N-GTPase) followed by four FF domains, and a GAP domain that is located at the C-termini [45–47] (Figure 3). The region between the FF and GAP domains was originally termed the ‘middle domain’, and was expected to be structureless [48], however, recent studies have discovered previously unidentified pseudoenzyme components within the domain structure of p190RhoGAP proteins (see below).
Figure 3. Updated domain organization of p190RhoGAP proteins.
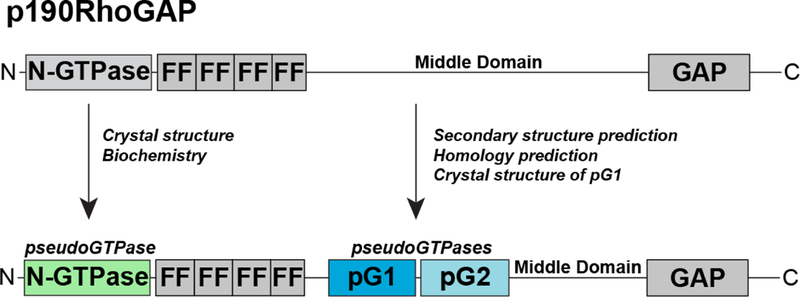
(Top) previous domain assignment of p190RhoGAP proteins. N-GTPase indicates N-terminal GTPase domain, FF indicates FF domains 1 through to 4, and GAP indicates C-terminal GAP domain. The middle domain is indicated, and the location of the newly identified pseudoGTPase domains, pG1 and pG2 are shown.
The N-GTPase domain of p190RhoGAP proteins
The N-GTPase domain of p190RhoGAP proteins was first identified in both p190RhoGAP-A and -B by sequence similarity with the Ras-like small GTPases [45, 46]. These early sequence-based discoveries also showed unusual regions within the N-GTPase domain, particularly the presence of long inserts [45, 46]. Biochemical studies of the N-GTPase domain yielded varying results with respect to GTP binding and/or GTPase activities [49–51], and the biochemical function of this domain remained unclear until a recent study determined the co-crystal structure of N-GTPase with GTP and conducted detailed biochemical and mutagenesis analyses [10]. This work has shown that the N-GTPase domain of p190RhoGAP proteins is, in fact, a pseudoGTPase domain that binds nucleotide but lacks catalytic activity.
The crystal structure of p190RhoGAP-A N-GTPase reveals numerous differences between N-GTPase and canonical small GTPases that likely contribute to a lack of hydrolytic activity in N-GTPase. Most notably, insert sequences which have not been observed in other small GTPases disrupt the G1, G2 (Switch I) and G3 (Switch II) motifs (Figure 2). Furthermore, the invariant Thr residue in G2 (Switch I) and the catalytic Gln residue from G3 (Switch II) are not conserved. These structurally defined sequence differences indicate extensive modifications around the nucleotide binding cleft. This is further supported by analysis of the coordination of the GTP phosphates, where γ-phosphate binding and coordination of Mg2+ involve additional unique contacts from N-GTPase that are not observed in typical GTPases [10]. In contrast, high conservation of the G4 and G5 motifs are consistent with maintained GTP binding specificity, and this is observed in the crystal structure. Biochemical analyses show that GTP bound to N-GTPase does not undergo hydrolysis to GDP, that GTP does not exchange readily, and that mutation of GTP-contacting residues leads to destabilization of the protein. The summation of these biochemical and structural studies together suggests that GTP binds tightly to the p190RhoGAP N-GTPase domain, but that the role of GTP binding is not to act as a conformational regulator, but rather to function as a stabilizer intrinsic to maintaining the fold of the domain.
Identification of two new pseudoGTPase domains within p190RhoGAP
The region between the FF and GAP domains of p190RhoGAP proteins (Figure 3) was originally termed the ‘middle domain’, and was expected to be structureless [48]. This region of approximately 700 residues had not, however, been fully analyzed since the late 1990s. We conducted sequence analysis and discovered an extensive region of strongly predicted secondary structure encompassing approximately 400 residues [9]. Secondary structure-based fold homology detection suggested that this region contained two GTPase-like domains which we termed pG1 and pG2, but also indicated low sequence identity of less than 20% compared to any small GTPase domain. We therefore determined the X-ray crystal structure of the first of these domains. The crystal structure conclusively showed that the pG1 domain is a GTPase-like fold with very strong structural similarity to Ras-like small GTPases. Despite this similarity, the crystal structure also showed that there are extensive differences in the nucleotide binding pocket that preclude nucleotide binding; each of the five G motifs are divergent from canonical small GTPases (Figure 2). Specifically, the sequence of G1 (phosphate-binding P-loop), G3 (Switch II), G4 and G5 motifs are completely degraded, and the G2 motif (Switch I) is deleted. These sequence changes contribute to an altered nucleotide binding pocket in which the phosphate binding cavity is sterically occluded and the binding site for the guanine base is changed. We biochemically confirmed the inability to bind nucleotide. The second of the predicted GTPase-like domains has so far proven intractable for structural studies, however, homology prediction and sequence alignment suggest that similar to pG1, all five G motifs are divergent from canonical small GTPases. These studies therefore strongly suggest that p190RhoGAP proteins contain two cryptic GTPase domains that are degraded in their nucleotide binding site, but maintain the small GTPase fold.
PseudoGTPase domains in p190RhoGAP are evolutionarily conserved
The p190RhoGAP proteins are found throughout evolution including ancestral eukaryotic species like sponge (A. queenslandica). Vertebrates contain two p190RhoGAP proteins (p190RhoGAP-A and -B with a roughly 50% sequence identity), while invertebrates contain a single p190RhoGAP protein. Sequence analysis among these p190RhoGAP proteins reveals that the N-GTPase, pG1, and pG2 pseudoGTPase domains are evolutionarily conserved; specifically, strong conservation of the G motif sequences throughout evolution supports that these domains arose as pseudoGTPase domains, rather than ones that lost activity over time. This raises the interesting possibility that the small GTPase fold emerged during evolution to fill both catalytic and noncatalytic/scaffolding, and supports that noncatalytic pseudoGTPase domains contribute necessary functions separate from catalytic ones.
p190RhoGAP proteins contain multiple pseudoGTPase domains of different classes
Pseudokinases can be classified into subgroups based on ATP binding and phosphorylation activity [30]. In a similar manner we propose that the pseudoGTPases can also be classified according to GTP binding and hydrolysis activity. These include those that class (i) have no nucleotide binding activity (and therefore no catalytic activity), class (ii) bind nucleotide but with no activity (under standard assay conditions), and class (iii) bind nucleotide and are catalytically active [9] (Figure 1B). Remarkably, our work indicates that p190RhoGAP proteins contain pseudoGTPase domains that belong to both class (i) and class (ii).
The newly identified p190RhoGAP pG1 (and likely pG2) domains (Figure 3) are class (i) pseudoGTPases. These lack nucleotide binding, and comparison of p190RhoGAP pG1 and pG2 with previously identified class (i) pseudoGTPase domains (CENP-M and dynein LIC [7, 8]) confirms the common feature of a lack of conserved G motif residues. The structures of each of these pseudoGTPase domains reveal that the nucleotide binding site is occluded and thus provides a common structural theme for this class of pseudoGTPase. Likewise, the p190RhoGAP N-GTPase domain (Figure 3) has now been shown to be a nucleotide binding catalytically inactive pseudoGTPase [10]. This falls into the class (ii) category, and places this domain alongside the Rnd family of small GTPases (Rnd1, Rnd2 and Rnd3/RhoE) which bind GTP constitutive but lack hydrolysis activity [34, 35]. To date, no class (ii) pseudoGTPase that binds GDP constitutively has been identified. Similar to p190RhoGAP N-GTPase, the Rnd family maintain selectivity for GTP by conservation of G4 and G5 motifs, but are mutated in the G1, G2 and G3 motifs to varying extents. The p190RhoGAP proteins do not contain class (iii) pseudoGTPase domains, defined by the RGK family (Rad, Rem1, Rem2, Gem/Kir) which are degraded in their G1, G2 and G3 motifs but maintain catalytic activity, although reportedly with slow kinetics in some cases [40, 52, 53].
Concluding remarks
The identification of three new pseudoGTPase domains within the p190RhoGAP proteins allows a better understanding of this type of pseudoenzyme. Notably, discovery of the cryptic pseudoGTPase domains pG1 and pG2 in p190RhoGAP raises the likelihood that other pseudoGTPase domains have yet to be identified, and highlights that robust bioinformatics combining sequence and structural homology analyses may be necessary to identify the full pseudoGTPase complement.
Taken together, the varying catalytic activities of pseudoGTPases along with their G motif conservation suggests general classification rules. Cryptic pseudoGTPases, those with both very low sequence identity to active GTPases and lacking the consensus G motifs, may comprise much of the class (i) pseudoGTPases. In contrast, the class (ii) pseudoGTPases are perturbed in the G1, G2 and/or G3 motifs which disrupts catalysis, but contain conserved G4 and G5 motifs to allow binding of the guanine base. Class (iii) pseudoGTPases have the highest conservation within the G motifs, and maintain catalytic activity by relying on novel nucleotide coordinations which are best described by crystal structures. Future discoveries of pseudoGTPase domains will help refine these activity guidelines to allow a more comprehensive understanding of this type of pseudoenzyme.
Acknowledgments
Funding
National Institutes of Health grants R01NS086078 and R01GM102262 funded this work.
Abbreviations
- GEF
guanine nucleotide exchange factor
- GAP
GTPase activating protein
- GDI
guanine nucleotide disassociation inhibitor
- GGAP
GTP-binding and GTPase activating proteins
- GTP
guanosine triphosphate
- GDP
guanosine diphosphate
- CENP-M
Centromere protein M
- LIC
light intermediate chain
- pG1
pseudoGTPase-1
- pG2
pseudoGTPase-2
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Todd AE, Orengo CA, and Thornton JM, Sequence and structural differences between enzyme and nonenzyme homologs. Structure, 2002. 10(10): p. 1435–51. [DOI] [PubMed] [Google Scholar]
- 2.Pils B and Schultz J, Inactive enzyme-homologues find new function in regulatory processes. J Mol Biol, 2004. 340(3): p. 399–404. [DOI] [PubMed] [Google Scholar]
- 3.Eyers PA and Murphy JM, The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol, 2016. 14(1): p. 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy JM, Mace PD, and Eyers PA, Live and let die: insights into pseudoenzyme mechanisms from structure. Curr Opin Struct Biol, 2017. 47: p. 95–104. [DOI] [PubMed] [Google Scholar]
- 5.Reiterer V, Eyers PA, and Farhan H, Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol, 2014. 24(9): p. 489–505. [DOI] [PubMed] [Google Scholar]
- 6.Murphy JM, Farhan H, and Eyers PA, Bio-Zombie: the rise of pseudoenzymes in biology. Biochem Soc Trans, 2017. 45(2): p. 537–544. [DOI] [PubMed] [Google Scholar]
- 7.Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, et al. , The pseudo GTPase CENP-M drives human kinetochore assembly. Elife, 2014. 3: p. e02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroeder CM, Ostrem JM, Hertz NT, and Vale RD, A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. Elife, 2014. 3: p. e03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiegler AL and Boggon TJ, p190RhoGAP proteins contain pseudoGTPase domains. Nat Commun, 2017. 8(1): p. 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stiegler AL and Boggon TJ, The N-terminal GTPase domain of p190RhoGAP proteins is a pseudoGTPase. Structure, 2018: p. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wennerberg K, Rossman KL, and Der CJ, The Ras superfamily at a glance. J Cell Sci, 2005. 118(Pt 5): p. 843–6. [DOI] [PubMed] [Google Scholar]
- 12.Cherfils J and Zeghouf M, Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev, 2013. 93(1): p. 269–309. [DOI] [PubMed] [Google Scholar]
- 13.Xia C, Ma W, Stafford LJ, Liu C, Gong L, Martin JF, et al. , GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol Cell Biol, 2003. 23(7): p. 2476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanukoglu I, Proteopedia: Rossmann fold: A beta-alpha-beta fold at dinucleotide binding sites. Biochem Mol Biol Educ, 2015. 43(3): p. 206–9. [DOI] [PubMed] [Google Scholar]
- 15.Bourne HR, Sanders DA, and McCormick F, The GTPase superfamily: conserved structure and molecular mechanism. Nature, 1991. 349(6305): p. 117–27. [DOI] [PubMed] [Google Scholar]
- 16.Mishra AK and Lambright DG, Invited review: Small GTPases and their GAPs. Biopolymers, 2016. 105(8): p. 431–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetter IR and Wittinghofer A, The guanine nucleotide-binding switch in three dimensions. Science, 2001. 294(5545): p. 1299–304. [DOI] [PubMed] [Google Scholar]
- 18.Der CJ, Finkel T, and Cooper GM, Biological and biochemical properties of human rasH genes mutated at codon 61. Cell, 1986. 44(1): p. 167–76. [DOI] [PubMed] [Google Scholar]
- 19.Splingard A, Menetrey J, Perderiset M, Cicolari J, Regazzoni K, Hamoudi F, et al. , Biochemical and structural characterization of the gem GTPase. J Biol Chem, 2007. 282(3): p. 1905–15. [DOI] [PubMed] [Google Scholar]
- 20.Manning G, Whyte DB, Martinez R, Hunter T, and Sudarsanam S, The protein kinase complement of the human genome. Science, 2002. 298(5600): p. 1912–34. [DOI] [PubMed] [Google Scholar]
- 21.Boudeau J, Miranda-Saavedra D, Barton GJ, and Alessi DR, Emerging roles of pseudokinases. Trends Cell Biol, 2006. 16(9): p. 443–52. [DOI] [PubMed] [Google Scholar]
- 22.Min X, Lee BH, Cobb MH, and Goldsmith EJ, Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure, 2004. 12(7): p. 1303–11. [DOI] [PubMed] [Google Scholar]
- 23.Shi F, Telesco SE, Liu Y, Radhakrishnan R, and Lemmon MA, ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A, 2010. 107(17): p. 7692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee K, Sharma M, Jahn R, Wahl MC, and Sudhof TC, Evolution of CASK into a Mg2+-sensitive kinase. Sci Signal, 2010. 3(119): p. ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. , The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol, 2011. 18(9): p. 971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, and Hubbard SR, Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol, 2012. 19(8): p. 754–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey FP, Byrne DP, Oruganty K, Eyers CE, Novotny CJ, Shokat KM, et al. , The Tribbles 2 (TRB2) pseudokinase binds to ATP and autophosphorylates in a metal-independent manner. Biochem J, 2015. 467(1): p. 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eyers PA, Keeshan K, and Kannan N, Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol, 2017. 27(4): p. 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, and Manning G, Structure of the Pseudokinase VRK3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site. Structure, 2009. 17(1): p. 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, et al. , A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J, 2014. 457(2): p. 323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel O, Griffin MDW, Panjikar S, Dai W, Ma X, Chan H, et al. , Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association. Nat Commun, 2017. 8(1): p. 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha BH and Boggon TJ, The crystal structure of pseudokinase PEAK1 (Sugen kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization. J Biol Chem, 2018. 293(5): p. 1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecointre C, Simon V, Kerneur C, Allemand F, Fournet A, Montarras I, et al. , Dimerization of the Pragmin Pseudo-Kinase Regulates Protein Tyrosine Phosphorylation. Structure, 2018. 26(4): p. 545–554 e4. [DOI] [PubMed] [Google Scholar]
- 34.Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, and Settleman J, Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol, 1996. 16(6): p. 2689–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, and Chardin P, A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol, 1998. 141(1): p. 187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynet C and Kahn CR, Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science, 1993. 262(5138): p. 1441–4. [DOI] [PubMed] [Google Scholar]
- 37.Cohen L, Mohr R, Chen YY, Huang M, Kato R, Dorin D, et al. , Transcriptional activation of a ras-like gene (kir) by oncogenic tyrosine kinases. Proc Natl Acad Sci U S A, 1994. 91(26): p. 12448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, and Kelly K, Gem: an induced, immediate early protein belonging to the Ras family. Science, 1994. 265(5169): p. 241–4. [DOI] [PubMed] [Google Scholar]
- 39.Finlin BS and Andres DA, Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J Biol Chem, 1997. 272(35): p. 21982–8. [DOI] [PubMed] [Google Scholar]
- 40.Finlin BS, Shao H, Kadono-Okuda K, Guo N, and Andres DA, Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem J, 2000. 347 Pt 1: p. 223–31. [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent S and Settleman J, Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur J Cell Biol, 1999. 78(8): p. 539–48. [DOI] [PubMed] [Google Scholar]
- 42.Arthur WT and Burridge K, RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell, 2001. 12(9): p. 2711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang JH, Gill S, Settleman J, and Parsons SJ, c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J Cell Biol, 1995. 130(2): p. 355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manchinelly SA, Miller JA, Su L, Miyake T, Palmer L, Mikawa M, et al. , Mitotic down-regulation of p190RhoGAP is required for the successful completion of cytokinesis. J Biol Chem, 2010. 285(35): p. 26923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Settleman J, Narasimhan V, Foster LC, and Weinberg RA, Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell, 1992. 69(3): p. 539–49. [DOI] [PubMed] [Google Scholar]
- 46.Burbelo PD, Miyamoto S, Utani A, Brill S, Yamada KM, Hall A, et al. , p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J Biol Chem, 1995. 270(52): p. 30919–26. [DOI] [PubMed] [Google Scholar]
- 47.Bedford MT and Leder P, The FF domain: a novel motif that often accompanies WW domains. Trends Biochem Sci, 1999. 24(7): p. 264–5. [DOI] [PubMed] [Google Scholar]
- 48.Roof RW, Haskell MD, Dukes BD, Sherman N, Kinter M, and Parsons SJ, Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol Cell Biol, 1998. 18(12): p. 7052–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster R, Hu KQ, Shaywitz DA, and Settleman J, p190 RhoGAP, the major RasGAP-associated protein, binds GTP directly. Mol Cell Biol, 1994. 14(11): p. 7173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatsis N, Lannigan DA, and Macara IG, The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J Biol Chem, 1998. 273(51): p. 34631–8. [DOI] [PubMed] [Google Scholar]
- 51.Roof RW, Dukes BD, Chang JH, and Parsons SJ, Phosphorylation of the p190 RhoGAP N-terminal domain by c-Src results in a loss of GTP binding activity. FEBS Lett, 2000. 472(1): p. 117–21. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J, Reynet C, Caldwell JS, and Kahn CR, Characterization of Rad, a new member of Ras/GTPase superfamily, and its regulation by a unique GTPase-activating protein (GAP)-like activity. J Biol Chem, 1995. 270(9): p. 4805–12. [DOI] [PubMed] [Google Scholar]
- 53.Opatowsky Y, Sasson Y, Shaked I, Ward Y, Chomsky-Hecht O, Litvak Y, et al. , Structure-function studies of the G-domain from human gem, a novel small G-protein. FEBS Lett, 2006. 580(25): p. 5959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, and Wittinghofer A, Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J, 1990. 9(8): p. 2351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


