SUMMARY
While current immune checkpoint therapy (ICT) mainly targets lymphoid cells, it is associated with a broader remodeling of the tumor micro-environment. Here, using complementary forms of high dimensional profiling, we define differences across all hematopoietic cells from syngeneic mouse tumors during unrestrained tumor growth or effective ICT. Unbiased assessment of gene expression of tumor infiltrating cells by single cell RNA sequencing (scRNAseq) and longitudinal assessment of cellular protein expression by mass cytometry (CyTOF) revealed significant remodeling of both the lymphoid and myeloid intratumoral compartments. Surprisingly, we observed multiple subpopulations of monocytes/macrophages, distinguishable by the markers CD206, CX3CR1, CD1d and iNOS, that change over time during ICT in a manner partially dependent on IFNγ. Our data support the hypothesis that this macrophage polarization/activation results from effects on circulatory monocytes and early macrophages entering tumors, rather than on pre-polarized mature intratumoral macrophages.
Graphical Abstract

In Brief:
Comprehensive changes in the tumor microenvironment during successful immune checkpoint therapy are profiled, implicating a key role for polarization of infiltrating macrophages in the anti-tumor immune milieu
INTRODUCTION
Work over the last two decades has shown that the immune system has a profound influence on cancer induction and outgrowth (Balkwill et al., 2005; Schreiber et al., 2011). One advance in our understanding of how tumors form in immunocompetent individuals has been the demonstration that immunity can paradoxically inhibit tumor outgrowth while also shaping tumor immunogenicity by a process known as cancer immunoediting (Schreiber et al., 2011; Shankaran et al., 2001). As a consequence of this process, immunologically sculpted cancer cells with reduced immunogenicity expand and establish an immunosuppressive tumor microenvironment that compromises the host-protective actions of tumor immunity (Schreiber et al., 2011). This immunosuppression is mediated, at least in part, through expression on T cells of two immune checkpoints: Cytotoxic T-Lymphocyte Associated Antigen-4 (CTLA-4) and Programmed Death-1 (PD-1) (Freeman et al., 2000; Leach et al., 1996). Monoclonal antibody (mAb) targeting of CTLA-4 or PD-1 can inhibit these checkpoints and unleash the immune system’s ability to destroy established tumors in both mice and some cancer patients (Sharma and Allison, 2015; Topalian et al., 2015).
To date, the approach to understand the mechanism of immune checkpoint therapy (ICT) has focused on T cells because they are the direct targets of anti-CTLA-4 or anti-PD-1. Accumulating evidence suggests that these mAbs function through distinct, non-redundant mechanisms (Gubin et al., 2014; Parry et al., 2005; Wei et al., 2017). We and others have studied these effects by monitoring the actions of ICT antibodies on T cells directed against tumor-specific mutant neoantigens that are major targets for immune responses reactivated by ICT (Gubin et al., 2015; Schumacher and Schreiber, 2015). We previously showed that anti-CTLA-4 and/or anti-PD-1 induced both overlapping as well as distinct changes in tumor-specific CD8+ T cells (Fehlings et al., 2017; Gubin et al., 2014). This work supports the concept that anti-CTLA-4 enhances T cell priming against tumor antigens, anti-PD-1 enhances metabolism and effector function of tumor-specific T cells while both antibodies together induce a maximal anti-tumor effector phenotype. Recent work from the Allison group provided further evidence that anti-CTLA-4 and anti-PD-1-exerted their anti-tumor effects via distinct lymphocyte-dependent mechanisms in both preclinical models and human cancer patients (Wei et al., 2017). Despite these insights, many aspects remain unknown about ICT-induced tumor regression, including potential changes that ICT may induce in myeloid cells that often constitute a significant cellular fraction of the tumor microenvironment (Franklin et al., 2014) and that can either facilitate or inhibit immune control of cancer.
Herein, we use two complementary forms of high dimensional profiling—scRNASeq (Dixit et al., 2016) and CyTOF (Spitzer and Nolan, 2016)—to assess transcriptional and functional changes of multiple immune cell populations infiltrating syngeneic tumors of mice treated with either control mAb (where tumor progression occurs) or different ICT mAb (where tumor rejection occurs). We find dramatic changes in the infiltrating cell populations associated with different ICT leading to therapeutic tumor rejection. We additionally identify similarities and differences in the various hematopoietic cell populations that are detected as a consequence of targeting the CTLA-4 and/or PD-1 checkpoints. We find a particularly striking influence of different ICTs on intratumoral monocytes/macrophages leading to a high degree of plasticity and complexity within this cell population. Our data suggest that the changes in intratumoral myeloid cell populations stem from a functional branch point effecting infiltrating circulatory monocytes and not via reprogramming of pre-polarized pro-tumor versus anti-tumor macrophages. Overall, this study reveals heretofore unrecognized dynamic cellular changes occurring during ICT thereby providing a view into the process at an unprecedented molecular and cellular depth.
RESULTS
Isolation of Tumor Infiltrating Cells for High Dimensional Profiling
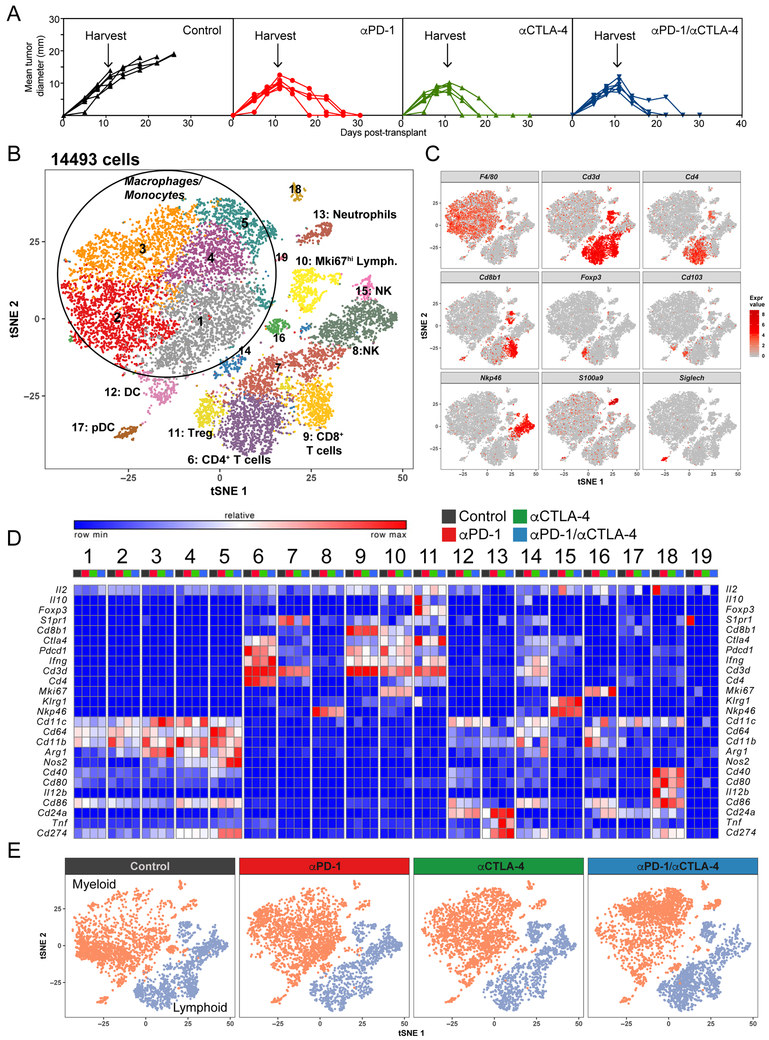
In this study, we used the T3 murine methylcholanthrene (MCA)-induced sarcoma line that we had previously characterized with respect to its mutational and neoantigen landscapes, immunogenicity, susceptibility to ICT and in vivo response kinetics (Gubin et al., 2014; Matsushita et al., 2012). T3 sarcoma cells form progressively growing tumors in naive syngeneic wild type mice that are uniformly rejected following therapeutic administration of anti-CTLA-4 or anti-PD-1 either alone or together (anti-PD-1/anti-CTLA-4) (Figure 1A). We also previously demonstrated that the optimal day to study hematopoietic cell infiltration into T3 tumors was day 11 after implantation of 1 × 106 T3 cells regardless of whether tumor-bearing mice were treated with control mAb (and thus experienced unrestrained tumor expansion) or ICT mAb (leading to complete tumor rejection). We therefore established T3 tumors in four groups of naive mice followed by treatment of each group on days 3, 6 and 9 with either control mAb, anti-PD-1, anti-CTLA-4 or anti-PD-1/anti-CTLA-4. Tumors were harvested on day 11, disaggregated, and their immune cell content analyzed either by scRNAseq using a 10x genomics pipeline or, following staining with panels of heavy-metal labeled mAbs, by CyTOF (Figure S1A). Consistent with our previous findings (Fehlings et al., 2017; Gubin et al., 2014), the number and proportion of CD45+ cells in T3 tumors increased in all ICT groups compared to tumors in control mAb treated mice (Figure S1B).
Figure 1. Identification of Intratumoral Cell Populations by scRNAseq.
(A) T3 tumor growth in mice treated with different ICTs. (B) tSNE plot of intratumoral cells from all groups merged. (C) tSNE plot of infiltrating cells displaying marker gene expression. (D) Heat map displaying normalized expression of select genes in each cell cluster. (E) tSNE plots of infiltrating lymphoid and myeloid cells.
Identification of Tumor Infiltrating Immune Cells Using scRNAseq
The scRNAseq analysis of sorted CD45+ tumor infiltrating cells yielded data for three to four thousand cells per condition with a coverage of ~100,000 reads per cell (Figure S1C). To define the subpopulation structure of the tumor infiltrating immune cells, we computationally pooled data from the four treatment groups representing ~15,000 cells total and used graph-based clustering to identify transcriptional clusters consisting of individual cell types (Figure 1B). By comparison with the ImmGen database and assessing known cell type markers, this resulted in elaboration of 7 distinct lymphoid clusters, 5 distinct monocyte/macrophage clusters, and smaller clusters of neutrophils, conventional dendritic cells (cDC) and plasmacytoid DCs (pDC) (Figure 1B, 1C, 1D and S2). Cellular identities were confirmed by flow cytometry (Figure S1D). We were surprised by the unexpected complexity and dynamics of the intratumoral monocyte/macrophage compartment in response to ICT (Figure 1E).
Identification of ICT-Induced Changes in Tumor Infiltrating Lymphocytes Using scRNAseq and CyTOF
We knew that ICT-induced rejection of T3 requires both CD4+ and CD8+ T cells and had identified some of the changes that occur within these cellular compartments upon ICT (Fehlings et al., 2017; Gubin et al., 2014). To increase resolution and more accurately define the lymphoid subpopulations identified by scRNAseq, we computationally separated lymphoid cells (~5000 cells total for all four treatments) and reanalyzed the data at higher granularity (Figure 2A). This approach yielded 12 distinct lymphoid subpopulations broadly defined by the distribution of classical marker genes (Figures 2B and S3A). Clusters for scRNAseq are annotated as “XXX_s#” where XXX represents the cell type, s the cluster as defined by scRNAseq and # the specific cluster. In addition, using a panel of metal-labeled mAbs and peptide-MHC-I (pMHC-I) tetramers, we assessed expression of 37 selected surface or intracellular markers and tumor antigen-specific T cell receptors (TCR) (Figures S3B and S3C) by CyTOF on CD45+ cells from disaggregated T3 tumors to independently identify the major quantitative and qualitative changes in protein expression that occur in lymphoid cells as a consequence of ICT. Clusters for CyTOF are annotated as “XXX_c#” where XXX represents the cell type, c the cluster as defined by CyTOF and # the specific cluster. Using this combinatorial approach, it became evident that each ICT induced changes in subpopulation proportions, their transcriptional profiles and their protein expression.
Figure 2. ICT Remodels Tumor Infiltrating Lymphocytes.
(A) tSNE scRNAseq plot from merged treatment data of exclusively intratumoral lymphocytes. (B) tSNE scRNAseq plot of lymphocytes displaying select marker gene expression. (C) tSNE scRNAseq plots with annotated clusters of intratumoral lymphocytes. (D) Percentage of cells in individual clusters by condition. (E) Histograms of Foxp3 expression in Tregs. Cells were gated on live CD45+ CD4+ Foxp3+ CD25+ cells. Histograms are representative from 3 individual mice per treatment condition and represent 2 independent experiments. (F) Heat map of GSEA identifying pathway enrichment by cluster. (G) Percentage of cells in Mki67hi by condition and tSNE plots displaying Cd4 or Cd8 expression.
Regulatory T cells
It is known that intratumoral regulatory T cells (Tregs) in the mouse express FoxP3, CD4 and both PD-1 and CTLA-4 and can suppress anti-tumor immunity (Tanaka and Sakaguchi, 2017). It is also known that the 9D9 anti-mouse CTLA-4 mAb used in the current study can partially deplete intratumoral Tregs in vivo (Selby et al., 2013; Simpson et al., 2013). Consistent with reports by others, the number and percent of Tregs decreased substantially in all ICT groups compared to Tregs infiltrating progressively growing tumors in mice treated with control mAb as assessed by scRNAseq and flow cytometry (Figures 2C, 2D and S1D). The greatest decrease occurred in the anti-CTLA-4 treated group. Tregs remaining after therapy displayed transcriptomic differences to the depleted population, including reductions in Il10 and foxp3 gene expression (Figure S3A), suggesting their reduced ability to suppress anti-tumor responses on a per cell basis. Similar changes in Foxp3 were also observed when the remaining cells were analyzed by flow cytometry (Figure 2E). Levels of Foxo1, sell, Gzmb, Klrg1 and Icos were also decreased in Tregs that remained after treatment with anti-CTLA-4 either alone or in combination with anti-PD-1 (Figures 1D and S3A).
CyTOF analysis revealed two Treg subpopulations (Treg_c1 and Treg_c2) (Figure 3A, 3B and S3B). Treg_c2 was distinguishable from Treg_c1 by its exclusive expression of KLRG1 and higher expression of CTLA-4, ICOS, GITR, VISTA and CD25 (Figure 3C). Following anti-CTLA-4 (+/− anti-PD-1) therapy, the percentage of lymphocytes in Treg_c2 was reduced nearly 9-fold (Figure 3D). In addition, expression on a per cell basis of several functional markers such as CTLA-4, VISTA, CD39, ICOS and OX40 was reduced in Treg_c2 with ICT as compared to Treg_c2 from tumors in control mAb treated mice (Figure 3C). Anti-PD-1 monotherapy also induced a significant reduction in the percent of lymphocytes in Treg_c2 but to a lower extent. The percentage of cells in Treg_c1 was only significantly reduced in tumors from mice treated with anti-CTLA-4 (5.9%-control mAb versus 2.6%-anti-CTLA-4) (Figure 3D). In addition, ICT reduced expression of Foxp3 and CD25 in Treg_c1 (Figure 3C). In sum, ICT effected both quantitative and qualitative changes in intratumoral Tregs, with the most significant reduction in Treg numbers and functional markers occurring when anti-CTLA-4 was used.
Figure 3. Characterization of Intratumoral T Cell Populations by CyTOF.
(A) tSNE plots of lymphoid cells identifying clustering strategy for T cell clusters. (B) tSNE plots of intratumoral CD4+ T cells by treatment condition annotated with distinct clusters. (C) Heat map displaying normalized expression of select genes in each cluster. (D) Percentage of cells in each clusters by condition. (E) tSNE plots of intratumoral CD8+ T cells (neoantigen-specific are highlighted) by treatment condition. (F) Percentage of cells in individual clusters by condition. (G) Heat map displaying normalized expression of select genes in each neoantigen-specific cluster. For C and F, each dot represents 2 pooled mice harvested and stained independently (N=5). Bar indicates mean percent ± SD of either (C) total lymphoid or (F) total CD8+ cells. Representative data from 3-4 independent experiments. *p<0.05, **p<0.01, ***p<0.001, Dunnett’s multiple comparison (DMC).
CD4+ T cells
Single Cell RNAseq revealed three distinct clusters of FoxP3− CD4+ T cells (CD4_s1, CD4_s2 and CD4_s3) (Figure 2C). Both CD4_s1 and CD4_s2 expressed high levels of Cd4 and functional markers such as Pdcd1 (PD-1), Csf1 and Ifng (Figures 2B and S3A). CD4_s3 expressed intermediate levels of the Cd4 transcript and few other functional markers. While both CD4_s1 and CD4_s2 appeared to be activated, CD4_s2 expressed higher levels of genes upregulated as a consequence of T cell activation such as Pdcd1, Lag3, Ctla4, Icos and Ifng. Gene Set Enrichment Analysis (GSEA) revealed that CD4_s2 displayed upregulation of pathways associated with hypoxia and costimulation by the CD28 family and signaling via NF-κB, TCR, IL-2/STAT5 and MAPK (Figure 2F). ICT treatment induced a reduction of cells in CD4_s1 and increased the number of cells in CD4_s2 and CD4_s3, with the largest shift occurring following anti-PD-1/anti-CTLA-4 therapy (Figure 2D). ICT not only increased the percent of T cells within CD4_s2 but also altered gene expression on a per cell basis since these cells showed decreased CD200, Pdcd1 and Lag3 expression and increased Ifng and Ctla4 expression following ICT (Figure S3A).
CyTOF analysis revealed the presence of multiple subpopulations of CD4+ T cells (Figures 3A and 3B). The CD4_c2 CyTOF cluster phenotypically resembled the CD4_s2 scRNAseq cluster since it expressed the highest levels of CD4, TIM-3, PD-1, CTLA-4, LAG-3, CD137 and ICOS (Figure 3C). In addition, cells in CD4_c2 also expressed Tbet, GITR, VISTA and CD39. CD4_c2 comprised only 3% of the lymphoid infiltrate in progressively growing tumors from control mAb treated mice but increased more than 2-fold following anti-PD-1 or anti-CTLA-4 and 4-fold with combination ICT (Figure 3D). In contrast, all ICT groups showed decreases in CD4_c3, whose component cells expressed high levels of PD-1 but lacked TIM-3, LAG-3 and VISTA. Anti-CTLA-4 treatment, either alone or together with anti-PD-1, increased the percentage of cells in three CD4+ T cell clusters (CD4_c1, CD4_c6 and CD4_c8) that express the proliferation marker Ki67, with the most dramatic increase occurring in CD4_c1 [3.9%-control mAb; 12.3%-anti-CTLA-4; 17.7%-anti-PD-1/anti-CTLA-4] (Figures 3C and 3D). The remaining CD4+ T cell clusters (CD4_c4, CD4_c5 and CD4_c7) differed little following ICT. In sum, these data show that anti-CTLA-4 (and to a lesser extent anti-PD-1) ICT induces a shift in the intratumoral CD4+ T cell population towards one that is more proliferative, more activated, and expresses high levels of IFNγ.
CD8+ T cells
Single cell RNAseq revealed two distinct subsets of CD8+ T cells (CD8_s1 and CD8_s2) (Figure 2C). When compared to one another, CD8_s1 expressed higher levels of genes associated with T cell activation and/or exhaustion (e.g., Havcr2 (TIM-3), Lag3, Pdcd1) (Wherry and Kurachi, 2015) as well as Serpinb9 and Gzmb (Figure S3A). GSEA revealed that CD8_s1 displayed upregulation in pathways associated with TCR signaling and oxidative phosphorylation (OxPhos) (Figure 2F). In contrast, CD8_s2 showed increased expression of Cxcr3, Ccl5, Ifit3, Ifit3b and lacked expression of Havcr2 (Figure S3A). Following ICT, proportions of cells in CD8_s2 increased while the number of cells in CD8_s1 decreased (Figure 2D). Gene expression within each cluster also changed with ICT: CD8_s1 cells showed decreased expression of Pdcd1 while anti-CTLA-4 but not anti-PD-1 increased expression of Gzmb and Ccl5 (Figure S3A). Expression of Klrg1 was increased in both CD8 clusters upon ICT. These findings indicate ICT changes the intratumoral CD8+ T cell population from one showing characteristics of exhaustion/dysfunction to one showing signs of reactivation.
A limitation of the scRNAseq approach is that it does not distinguish between tumor-specific versus non-specific CD8+ T cells. Therefore, we included pMHC-I tetramers in our CyTOF lymphoid panel that were specific for TCRs that recognize the two major neoantigens expressed by T3 cells—mutant (m) Lama4 and mAlg8. By gating on tetramer+ CD8+ T cells we found that intratumoral mLama4-specific CD8+ T cells were present at a higher frequency than mAlg8-specific CD8+ T cells under all treatment conditions (Figure S4A), a result consistent with our previous report (Gubin et al., 2014). Anti-CTLA-4 induced decreases in expression of TIM-3, LAG-3 and PD-1 and increases in expression of granzyme B in tumor neoantigen-specific CD8+ T cells (Figure S4B), while anti-PD-1 therapy decreased expression of LAG-3 and PD-1. These results recapitulate our prior findings using flow cytometry (Gubin et al., 2014).
To better capture the neoantigen-specific CD8+ T cell heterogeneity, we computationally excluded non-CD8+ T cells and re-clustered tetramer+ and tetramer− CD8+ T cells as shown in Figure 3A. [See tSNE plot and heat map of all CD8+ T cells (Figures S4C and S4D).] Upon re-clustering, four individual mLama4-specific CD8+ T cell subpopulations (CD8_c1, CD8_c2, CD8_c3 and CD8_c4) were identified (Figure 3E). The largest cluster (CD8_c1) of tumor neoantigen-specific CD8+ T cells in tumors from control mAb treated mice represented 35% of the total CD8+ T cell infiltrate and was characterized by the highest expression of markers of exhaustion/dysfunction (e.g., CD27, TIM-3, GITR, LAG-3, CD39, CTLA-4 and PD-1) (Figures 3F and 3G). CD8_c1 was reduced 2.3-, 3.1-, and 3.8-fold in tumors from mice treated with anti-PD-1, anti-CTLA-4, or anti-PD-1/anti-CTLA-4, respectively. Whereas tumor neoantigen-specific CD8+ T cells in CD8_c2 represented 5.5% of total CD8+ T cells in growing tumors from control mAb treated mice, their density increased to 20.8% and 16.1% in tumors from anti-CTLA-4 and anti-PD-1/anti-CTLA-4 treated mice, respectively. Similarly, the percentage of neoantigen-specific CD8+ T cells in CD8_c3 rose from 1.7% in control mAb treated mice to 10.2% in anti-CTLA-4- and 9.6% in anti-PD-1/anti-CTLA-4-treated mice. CD8_c3 expressed the highest number of KLRG1+ CD8+ T cells. No effects of anti-PD-1 monotherapy were observed on either CD8_c2 or CD8_c3. Cells in CD8_c4 expressed the lowest amounts of CTLA-4, granzyme B, CD39 and TIM-3 but expression of these markers increased following either anti-PD-1 or anti-CTLA-4 monotherapies (Figure 3G). CD8+ T cells specific for mAlg8 were only found in CD8_c5, presumably as a consequence of their limited numbers. Nearly all neoantigen-specific CD8+ T cells expressed Ki67 (Figure S4E). Together these data show that anti-CTLA-4 and/or anti-PD-1 have both similar and distinct activating effects on tumor-infiltrating CD8+ T cells that are most evident in antigen-specific CD8+ T cells.
NK and γδ T cells
Single cell RNAseq identified three distinct clusters of NK Cells (NK_s1, NK_s2 and NK_s3) (Figure 2C). NK_s2 selectively expressed Klra3, Klra8 and Klra9 (Figure S3A). NK_s3 selectively expressed Gzmd, Gzme, Gzmg, Ifitm1, Ifitm2, and the highest levels of Klrg1, which increased even more upon ICT. All three NK cell clusters displayed increased Gzmb expression upon anti-PD-1 or anti-CTLA-4 treatment, with the highest expression occurring in NK_s3. CyTOF identified six distinct clusters of NK cells (NK_c6, NK_c7, NK_c11, NK_c14, NK_c15 and NK_c17) (Figure S3B and S5A) and showed that anti-CTLA-4 induced marked upregulation of Granzyme B in KLRG1+ NK cells that phenotypically resemble the NK_s3 population detected by scRNAseq (Figures S3C). These data suggest ICT increases proliferation and maturation of NK cells as indicated by their KLRG1 expression (Huntington et al., 2007).
Single cell RNAseq also identified a cluster (Tγδ_s) positive for Cd3e, Klra5, Cd7 and Xcl, and negative for both Cd4 and Cd8 that consisted largely of γδ T cells (Figures S3A, S5B and S5C). The presence and fraction of γδ T cells was also observed using flow cytometry (Figures S1D and S5D). ICT had minimal effects on γδ T cells at day 11 post-tumor transplantation.
Additional changes occurring in the lymphoid compartment
Interestingly, several of the scRNAseq clusters were identified more on the basis of their functional markers rather than their known cellular subtype. Specifically, Mki67hi cells, identified using unsupervised clustering, contained a mixture of CD4+ T cells, CD8+ T cells, Tregs and NK cells based on their profound cell proliferation signature (Figures 2B, 2C and 2F). The cellular components of this cluster underwent major changes upon ICT (Figure 2G). In progressively growing tumors from mice treated with control mAb, this cluster was comprised predominantly of CD8+ T cells and NK cells. However, in response to anti-CTLA-4 therapy (+/− anti-PD-1) the numbers of Mki67hi CD4+ T cells dramatically increased indicating a major impact of anti-CTLA-4 on the proliferative capacity of CD4+ T cells. This cluster also showed striking expression patterns of genes linked to T cell proliferation: Ezh2; Ung, Hmmr, Cenpa and Mcm5 (Figure S5E).
Another functionally defined cluster contained particularly activated T cells (Tact) and was comprised of both CD4+ and CD8+ T cells (Figures 2B and 2C). All cells within this cluster were enriched in TCR activation gene signatures (Figure S5F) and had the highest Ifng gene expression among all populations (Figure S3A). Whereas the quantity of cells occupying Tact did not substantially change upon ICT (Figure 2D), its component cells underwent dramatic transcriptional changes showing increased expression of Ifng and decreased expression of Pdcd1 and Lag3 (Figure S3A).
Overall, these findings highlight the integrated changes occurring in the intratumoral lymphocyte compartment following ICT that include (a) a dramatic reduction in Treg frequency and presumed function; (b) NF-κB- and IFNγ-driven remodeling of CD4+ and CD8+ T cells leading to increased expression of an anti-tumor effector gene signature (e.g., Ifng); (c) alterations in markers of T cell function/activation; and (d) an overall increase in effector T cell expansion with increased numbers of proliferating CD4+ T cells within tumors.
Remodeling of Intratumoral Monocytes/Macrophages by ICT
Intratumoral monocytes/macrophages constitute the largest cluster in the scRNAseq tSNE plots and undergo a striking remodeling following ICT (Figure 1E). Monocytes/macrophages from progressively growing tumors in control mAb treated mice expressed high levels ofMrc1 (CD206)—a marker frequently associated with anti-inflammatory, immunosuppressive macrophages (Figure 4A) (Biswas and Mantovani, 2010; Murray et al., 2014; Qian and Pollard, 2010). In contrast, ICT-induced accumulation of myeloid cells that expressed high levels of Nos2 (iNOS)—a marker associated with classical IFNγ activated, pro-inflammatory macrophages expressing anti-tumor activity. The altered expression of CD206 and iNOS was also observed at the protein level as revealed by flow cytometry (Figure 4B). We therefore looked more closely at the subpopulation structure defined by scRNAseq in Figure 1B. Five major monocyte/macrophage subpopulations were identified by unbiased clustering that were associated with tumor progression or regression (Figure 4C). Specifically, monocytes/macrophages in progressively growing tumors from control mAb treated mice almost exclusively populated clusters Mac_s1 and Mac_s2 (Figure 4D). Cells in Mac_s1 were characterized by the absence of classical macrophage maturation markers such as Mertk and expressed the highest levels of Ccr2, Il1r2 and Itga4, consistent with a monocyte phenotype (Figures 4E and S2). The percentage of cells within this subpopulation increased with anti-PD-1 or anti-CTLA-4 treatment but, interestingly, decreased with combination ICT (Figure 4D). In contrast, Mac_s2 cells were characterized as having a distinctly anti-inflammatory gene expression profile and expressed genes encoding Mrc1, Ccl2 and most strikingly, Cx3cr1 (Figure 4E). Mac_s2 cells also expressed signatures of active OxPhos and respiratory election transport (but not enhanced glycolysis) (Figure 4F). In progressively growing tumors, 30% of the CD45+ tumor infiltrating cells resided in Mac_s2 but were reduced almost three-fold (to 7-12%) in tumors following ICT (Figure 4D). This cluster also expressed high levels of Mertk following ICT (Figure 4E). Conversely, residual expression of Ccr2 in Mac_s2 cells decreased upon ICT.
Figure 4. ICT Remodels Intratumoral Monocytes/Macrophages.
(A) tSNE plots from scRNAseq highlighting Mrc1 and Nos2 expressing cells. (B) iNOS and CD206 expression represented as mean fluorescent intensity with mean ± SEM (**p<0.01, ***p<0.005, unpaired t test). (C) tSNE plots with annotated clusters of intratumoral monocytes/macrophages. (D) Percentage of cells in individual clusters by condition. (E) Heat map displaying normalized expression of select genes in each monocyte/macrophage cluster. (F) Heat map of GSEA identifying pathway enrichment by cluster. Representative data (B) from 3 individual mice per treatment condition and represent 3 independent experiments.
Macrophages in the Mac_s3, Mac_s4 and Mac_s5 clusters represented together less than 10% of the CD45+ tumor infiltrating cells from growing tumors in control mAb treated mice but increased dramatically upon ICT, achieving maximal levels in the combination ICT group (Figure 4D). GSEA showed that Mac_s3 displayed reduced transcriptional signatures of active OxPhos and respiratory electron transport compared to Mac_s2 (Figure 4F). Mac_s4 expressed high levels of Cd1d1, intermediate levels of Nos2 and Cd274 (PD-L1) and little or no expression of Mrc1 and Cx3cr1 (Figure 4E). GSEA revealed that Mac_s4 cells displayed pathway enrichment in NF-κB signaling, inflammatory responses and hypoxia (Figure 4F). These cells also appeared to be the most metabolically active, because they not only display strong enrichment in OxPhos and respiratory electron transport signatures but also expressed strong signatures of glycolysis. Cells in Mac_s5 expressed the highest level of Nos2 and Cd274 (Figure 4E) and displayed strong enrichment of other genes indicative of IFNγ response and NF-κB signaling. Although both Mac_s4 and Mac_s5 exhibited overlapping characteristics, they could be differentiated from one another by their mutually exclusive expression of Cd1d1 and IFN response signatures, respectively. Mac_s5 displayed characteristics of classically activated, monocyte-derived macrophages based on the IFNγ and NF-κB signature identified by GSEA, the strong expression of Nos2, and the down regulation of Ccr2. Although Mertk was not expressed in cells within Mac_s5, a recent report revealed that Mertk expression is epigenetically silenced in macrophages exposed to IFNγ (Qiao et al., 2016).
These data support the concept that in tumors undergoing ICT, monocytes/macrophages receive numerous signals and commit to a number of distinct response programs, some of which are induced by the combinatorial engagement of the IFNγ and NF-κB signaling pathways.
In sum, scRNAseq analysis identified 5 major genes that combinatorially define the 5 monocyte/macrophage subpopulations: Ccr2-predominantly in Mac_s1; Cx3crl exclusive to Mac_s2; Mrc1 shared by Mac_s2 and Mac_s3; Cd1d1 exclusive to Mac_s4 and Nos2, most highly expressed in Mac_s5 macrophages and to a lesser extent in a subset of Mac_s4 cells (Figure 4E).
We further scrutinized these findings using a myeloid-focused CyTOF panel (Figure S6). CyTOF identified multiple populations of monocytes/macrophages, cDC including DC1 and pDC (Figures 5A and 5B) but, in contrast to scRNAseq, also revealed substantial numbers of neutrophils and eosinophils among the infiltrating cells (Figures S6A and S6B). Using markers identified by scRNAseq that defined the monocyte/macrophage subpopulations responsive to ICT, CyTOF identified similar changes at the protein level for these transcripts (Figure S6C). Specifically, CX3CR1+CD206+ macrophages were observed in progressively growing tumors in mice treated with control mAb but were dramatically reduced in tumors from mice treated with anti-PD-1 and/or anti-CTLA-4. Conversely, whereas limited numbers of macrophages in progressively growing tumors expressed iNOS, they dramatically increased in number following ICT. As also observed using scRNAseq, CD1d was expressed in both iNOS+ as well as iNOS− macrophages.
Figure 5. Characterization of Intratumoral Myeloid Cell Populations by CyTOF.
(A) CyTOF tSNE plot of total tumor infiltrating CD45+ cells. Identification of individual populations is overlaid. (B) tSNE plots shown as the density of equal numbers of CD45+ viable cells per treatment group. (C) tSNE plots of annotated intratumoral monocytes/macrophages. (D) Percentage of cells in individual monocyte/macrophage clusters. (E) Heat map displaying normalized expression of selected genes in each cluster. (F) Percentage of cells in individual monocyte/macrophage clusters. For (D) and (F), each dot represents 2 pooled mice harvested and stained independently (N=5). Bar indicates mean percent ± SD of CD45+ cells upon different ICT. Representative data from 3-4 independent experiments. *p<0.05, **p<0.01, ***p<0.001, DMC.
CyTOF analysis revealed the presence of 8 subpopulations of monocytes/macrophages (Mac_c1-Mac_c8) (Figure 5C) with certain subpopulations predominating in progressively growing tumors of mice treated with control mAb. The most evident of these was Mac_c3 representing 26% of the CD45+ tumor infiltrate (Figure 5D). Mac_c3 cells expressed CX3CR1, CD206, IL-4Rα, VCAM-1 and CD64 and resembled Mac_s2 cells both phenotypically and quantitatively (Figures 5D and 5E). This cluster was noticeably diminished following treatment with anti-PD-1 and/or anti-CTLA-4. Concomitant with the loss of the Mac_c3 cluster, we observed an increased cellular density of iNOS+ macrophages in Mac_c4. This cluster displayed properties of both Mac_s4 and Mac_s5. In the absence of ICT, only 2% of the CD45+ tumor infiltrate was represented within the iNOS+ Mac_c4 cluster. In contrast, this cluster more than doubled (5.5%) in tumors from mice treated with anti-PD-1, and increased 5-7-fold following treatment with anti-CTLA-4 either alone (10.8%) or in combination with anti-PD-1 (14.7%).
CyTOF also identified other subpopulations of monocytes/macrophages including two clusters, Mac_c6 and Mac_c7, of CCR2+, CD11b+ Ly6C+, Ly6G− cells that phenotypically resembled Mac_s1 monocytes (Figure 5E). When compared to Mac_c6, Mac_c7 expressed higher levels of Ly6C and CCR2 but lacked expression of CD64 that was heterogeneously expressed on Mac_c6. These data suggest that Mac_c7 is the most immature monocyte that, when exposed to IFNγ begins to differentiate to a macrophage by upregulating Fc receptors and downregulating Ly6C and Ccr2. Mac_c6 plus Mac_c7 together represented 12.1% of the CD45+ tumor infiltrate in tumors from control mAb treated mice (Figure 5F). CyTOF analysis did not reveal differences in the cellularity of Mac_c6 or Mac_c7 after either anti-PD-1 or anti-CTLA-4 monotherapy but after combination therapy the cellular density of both clusters was reduced to 6.0% of the CD45+ infiltrate. This decrease was also seen by scRNAseq for Mac_s1 following combination ICT (Figure 4D).
The cells within the other Mac_c clusters were MerTK+ macrophages displaying markers of differing levels of activation (Figure 5E). Cells in Mac_c1 and Mac_c5 expressed CD206, PD-L2 and CD64 but lacked expression of CX3CR1 and could be differentiated from one another by the higher expression of MerTk, CD38 and IL-4Rα in Mac_c1 versus higher expression of CCR7 and CD63 in Mac_c5. Mac_c1 and Mac_c5 represented 9.1% and 7.3%, respectively, of the CD45+ cells in tumors from control mAb treated mice (Figure 5F). The cellular density of these two clusters increased significantly in tumors from mice treated with anti-PD-1, but not with anti-CTLA-4 (+/−anti-PD-1). Although the percent of cells in Mac_c1 and Mac_c5 did not change with anti-CTLA-4 treatment, the expression of several markers were altered following ICT including reduced expression of F4/80 and IL-4Rα on a per cell basis (Figure 5E).
Temporal Dynamics of Intratumoral Monocyte/Macrophage Compartment Remodeling
Whereas the integrated analysis of transcriptional and proteomic data established the major monocyte/macrophage subpopulations that comprise the intratumoral myeloid compartment and demonstrated the remodeling of this compartment upon ICT, it provided few insights into the origins of the cells that populate the individual clusters. To gain insight into the temporal dynamics of the observed remodeling, we analyzed scRNAseq data using Monocle2 (Trapnell et al., 2014), which suggested that the starting point corresponds to cells within the Mac_s1 cluster (monocytes), which then assume one of two fates: either they become macrophages within Mac_s2 or occupy Mac_s4 and Mac_s5 (Figures 6A and 6B). This analysis also revealed the complexity of cells within Mac_s3, indicating that it could be a composite of two subclusters, one of which is closer to Mac_s2, while the other one is closer to Mac_s4 and Mac_s5. The pseudotime-organized sequence of “differentiation/activation” events suggests that neither CX3CR1+ macrophages nor iNOS+ macrophages are present early in a tumor but rather accumulate as the tumor microenvironment is shaped by treatment of tumor bearing mice with control versus ICT mAbs. This leads to a computationally based hypothesis of an ICT-induced branch point in the fate of intratumoral myeloid cells.
Figure 6. Restructuring of Intratumoral Monocytes/Macrophages Revealed by Longitudinal Analyses.
(A) tSNE scRNAseq plot with corresponding analysis of monocyte/macrophage data by Monocle2. (B) Monocyte/macrophage clusters from scRNAseq overlaid on Monocle2 pseudotime plot. (C) CyTOF tSNE plot (all time points harvested combined and displayed) with macrophage/monocytes clusters annotated. (D) Heat map displaying normalized expression of markers in each monocyte/macrophage cluster. (E) tSNE plots of total CD45+ cells overlaid with individual markers per treatment group over time. (F) Kinetics of Mac_c3 (CX3CR1+ CD206+) or Mac_c8 (iNOS+). (G) Monocle2 analysis of CyTOF time course. (H) Individual markers overlaid on Monocle2 pseudotime plot. Representative data from 2-3 independent experiments.
To validate this hypothesis, we performed longitudinal studies of the polarization process by establishing matched cohorts of tumor bearing mice that were then treated with either (a) control mAb or (b) anti-PD-1/anti-CTLA-4 administered every 3 days post-tumor transplant. Matched sets of tumors residing in mice for 5, 6, 7, 8, 9, 10, 11 or 12 days were harvested on the same day and analyzed using the myeloid cell CyTOF panel (Figures 6C, 6D, S7A, S7B and S7C). Detailed clustering analysis of the longitudinal CyTOF experiments produced clusters that were generally consistent with both the static CyTOF and scRNAseq analyses. Because the longitudinal experiments were performed independently of the static analyses, the numbering of the respective CyTOF clusters are different. Specifically, in the experiment presented in Figure 6, Mac_c9 contained CCR2+ Ly6C+ monocytes and was comparable to Mac_s1; Mac_c3 contained CX3CR1+ CD206+ iNOS− macrophages and was comparable to Mac_s2; Mac_c4 and Mac_c5 contained CX3CR1− CD206+ iNOS− macrophages and was comparable to Mac_s3; and Mac_c8 contained iNOS+ macrophages and was comparable to Mac_s4 and Mac_s5. Following treatment of tumor bearing mice with either control mAb or ICT, CCR2+ monocytes (Mac_c9 cells) were detected in tumors as early as the first time point sampled (day 5) and represented 14% and 13% of intratumoral CD45+ cells, respectively (Figures 6E and S7B). At this time point, neither CX3CR1+ CD206+ iNOS− Mac_c3 cells nor CX3CR1− CD206− iNOS+ Mac_C8 cells were detected in the tumors. At later time points the percent of Mac_c9 cells decreased in both the control mAb and ICT groups. In progressively growing tumors from mice treated with control mAb, CX3CR1+ CD206+ iNOS− Mac_c3 macrophages began to accumulate on day 9, at which time 14% of the CD45+ cells were contained in this cluster and subsequently increased reaching 41% by day 12 (Figures 6E, 6F and S7B). In contrast, macrophages from tumor bearing mice treated with anti-PD-1/anti-CTLA-4 displayed a CX3CR1− CD206− iNOS+ Mac_c8 phenotype that noticeably increased starting at day 8 and then remained stable thereafter at 11-14% of the CD45+ cells. This longitudinal analysis thus revealed that individual clusters of monocytes/macrophages displayed distinctive kinetics of formation and/or disappearance.
We additionally noted the presence of two clusters of CX3CR1− CD206+ iNOS− cells (Mac_c13 and Mac_c15) that were observed after either control or ICT treatment only in tumors harvested on day 5 where they represented ~30% and 20%, respectively, of the tumor infiltrate (Figures 6D, 6E and S7B). The presence of these two populations diminished rapidly and was not detectable post day 6, explaining why we never saw them in the static CyTOF analyses performed on day 11 tumors. However, starting at day 6-7, two other CX3CR1− CD206+ iNOS− myeloid cell populations (Mac_c4 and Mac_c5) became detectable.
Figures 6G and 6H suggest that CCR2+ Ly6c+ Mac_c9 cells experience the same differentiation/activation branch point to become either CX3CR1+ CD206+ iNOS− or CX3CR1− CD206− iNOS+ cells over time following their exposure to influences in the tumor microenvironment arising as a consequence of treatment with control versus ICT mAbs. Overall, these data confirm the existence of an ICT-induced branch point occurring 7-9 days post-tumor cell transplantation when the myeloid compartment in tumors undergoes remodeling leading to formation of either a tumor-promoting or anti-tumor microenvironment.
IFNγ is a Key Macrophage Branch Point Mediator
The appearance of intratumoral iNOS+ macrophages on day 7 with ICT (Figure 6E) corresponds to the nearly simultaneous arrival in the tumor of neoantigen-specific T cells (Gubin et al., 2014). Since iNOS induction in macrophages is known to require IFNγ plus an NF-κB inducing stimulus, we hypothesized that IFNγ from ICT-reinvigorated T cells drives the monocyte/macrophage branch point towards development of activated iNOS+ macrophages. To test this hypothesis, we established parallel cohorts of T3 tumor bearing mice treated on days 3, 6, and 9 with either control mAb or anti-PD-1/anti-CTLA-4 and subsequently injected the latter cohort with either irrelevant hamster mAb or neutralizing hamster anti-mouse IFNγ on day 7 or 9. We specifically chose these time points to ensure that IFNγ had been induced in the tumor microenvironment. All tumors were harvested at day 11 and iNOS+-, CX3CR1+ CD206+-, and CX3CR1− CD206+-macrophages were quantitated by CyTOF analysis (Figure 7A). As expected, iNOS+ macrophages constituted only 1.2% of CD45+ cells in tumors from mice treated with control mAb followed by irrelevant hamster mAb. This percentage increased 11.4-fold to 13.7% of CD45+ cells upon treatment of tumor bearing mice with anti-PD-1/anti-CTLA-4 followed by irrelevant hamster mAb. However, injection of neutralizing anti-IFNγ on day 7 into tumor bearing mice treated with anti-PD-1/anti-CTLA-4 inhibited the increased occurrence iNOS+ macrophages by 7.3-fold. The inhibition of iNOS+ macrophages was reduced by 2.2-fold when anti-IFNγ addition was delayed to day 9, where macrophages would have encountered transient exposure IFNγ. Thus, IFNγ production as a consequence of ICT dependent reinvigoration of T cells positively drives polarization of newly arrived monocytes to become iNOS positive macrophages.
Figure 7. IFNγ-Dependent iNOS+ Monocyte/Macrophages Branching and Comparison to Human Macrophages.
(A) Percentage of intratumoral monocytes/macrophages under different treatment conditions. (B) Comparison of mouse tumor infiltrating macrophage clusters with human macrophages under differing stimulation conditions (GSE46903) by principle component analysis (top) or represented as fold change enrichment (bottom). (C) tSNE Projection for the human monocyte/macrophage GSE46903 dataset.
In contrast, ICT nearly completely ablated the appearance of CX3CR1+ CD206+ macrophages in T3 tumors (from 33.1% with control mAb to 1.3% with ICT, a 24.9-fold reduction). However, in this case, the inhibition was not reversed when anti-IFNγ was added on either day 7 or 9. Interestingly, the percent of CX3CR1− CD206+ macrophages remained unchanged when tumor bearing mice were treated with anti-PD-1/anti-CTLA-4 and showed only a modest increase (approximately 1.7-fold) when anti-IFNγ was added on either day 7 or day 9. Thus, the macrophage branch point induced by ICT is a consequence of at least two influences: whereas IFNγ is required for induction of iNOS+ macrophages, another, currently unknown moiety that presumably arises as a consequence of ICT, ablates induction of CX3CR1+ CD206+ macrophages.
Linking Our Findings to Human Monocytes/Macrophages
We lastly asked whether the populations of monocytes/macrophages we had identified in progressively growing or ultimately rejecting mouse tumors had corresponding human counterparts by comparing transcriptional profiles of the 5 clusters of monocytes/macrophages identified by scRNAseq analysis to data from a study that comprehensively profiled human monocytes and macrophages under differing stimulation conditions (Figure 7B). This study contained 299 profiles that encompass 32 different stimulation conditions. We reanalyzed these data and organized the 299 profiles as individual dots on a tSNE plot (Figure 7C). We then computed enrichment of individual signatures of each tumor-associated monocyte/macrophage subtype defined by our scRNAseq analysis within this human dataset. The Mac_s1 signature was distinctly upregulated in unstimulated human monocytes, thus demonstrating similarities between our tumor infiltrating murine monocytes and human blood monocytes. Cells within our Mac_s2 and Mac_s3 clusters displayed distinct similarities with human macrophages that were activated under anti-inflammatory conditions (glucocorticoids, PGE2, etc.). Conversely, cells within our Mac_s4 and Mac_s5 clusters showed strong enrichment for genes in human macrophages stimulated under pro-inflammatory conditions, including various combinations of IFNγ, TNFα and LPS, with Mac_s5 (activated, iNOS+ macrophages) displaying the strongest similarity to IFNγ activated human macrophages. Overall, this analysis establishes that the monocytes/macrophages in mouse tumors have human counterparts.
In sum, integration of scRNAseq and CyTOF data provide compelling support to the hypothesis that ICT leads to a change within the tumor microenvironment that favors the generation IFNγ-activated, highly glycolytic, pro-inflammatory macrophages that can express anti-tumor potential while suppressing induction of anti-inflammatory, immunosuppressive macrophages that facilitate tumor growth.
DISCUSSION
In this study, we employed two different high dimensional analysis approaches to profile immune cells infiltrating T3 MCA sarcomas either growing progressively in mice treated with control mAb or destined to undergo complete immunologic rejection as a consequence of ICT. Using a combination of scRNAseq and CyTOF we identified qualitative and quantitative changes that occur in both lymphoid and myeloid cell populations. The combinatorial use of scRNAseq and CyTOF has revealed unique advantages to each method plus the advantage of using the methodologies together. Whereas each approach uses unsupervised clustering to segregate different populations, scRNAseq is completely unbiased as it analyzes expression of 1000-4000 or more genes. This is in contrast to CyTOF that employs ~40 markers that are pre-selected based on a priori knowledge. In addition, scRNAseq permits transcriptomic analysis/comparisons between individual infiltrating cell populations including the use of GSEA and comparisons to human datasets. However, scRNAseq suffers from the caveat that mRNA and protein expression do not always directly correlate. It is therefore significant that we confirmed and extended our key findings made with scRNAseq using CyTOF as a measure of protein expression. The current study not only provides additional insights into the mechanisms of action of anti-PD-1 or anti-CTLA-4 monotherapies but also provides insights into why the combination of both mAbs induces particularly strong anti-tumor responses. Thus, we can compare and contrast the molecular and cellular changes that occur as a consequence of ICT and link them to the induction of unequivocal clinical outcomes.
The current study confirms and extends two recent studies that used mass cytometry to profile changes in tumor infiltrating T cells following ICT (Fehlings et al, Wei et al). Importantly, the major findings on the lymphocyte directed effects of ICT made in both previous studies are recapitulated in the current study, thus demonstrating the generality of the findings between different tumor types and with some exceptions between mouse and human. However, a major difference between the current and previous studies is that we also present effects of ICT on the intratumoral myeloid compartment thereby providing a more comprehensive view of the cellular dynamics of ICT-induced anti-tumor responses.
The current study makes several key observations: (1) ICT induces the expansion and/or contraction of specific subsets of tumor infiltrating myeloid as well as lymphoid cells; (2) in the mouse, anti-CTLA-4 plays a major role in reducing the number of intratumoral Tregs with the highest immunosuppressive potential and enhancing proliferation and IFNγ production by intratumoral non-Treg CD4+ T cells; (3) anti-CTLA-4 or anti-PD-1 monotherapy reduces the exhaustion/dysfunctional phenotype of tumor-specific CD8+ T cells but by different mechanisms; (4) targeting both PD-1 and CTLA-4 can produce unique effects not seen in monotherapy; (5) the tumor associated monocyte/macrophage compartment is highly complex and dynamic; (6) intratumoral myeloid cell subpopulations display the spectrum of activation states ranging from a predominantly anti-inflammatory phenotype in progressively growing tumors to a predominantly proinflammatory phenotype in tumors that will reject in ICT treated mice; (7) the activation state of intratumoral macrophages is influenced by the effects that cytokines in the tumor microenvironment have on circulatory monocytes as they enter the tumor and differentiate into macrophages; (8) IFNγ neutralization inhibits tumor rejection and induction of activated iNOS+ proinflammatory macrophages in tumors from ICT treated mice; and (9) macrophage subpopulations identified in our mouse tumor model have human equivalents.
In conclusion, the results of this study provide new insights into the transcriptional, molecular and functional changes that occur within major intratumoral immune cell populations following successful cancer immunotherapy. These insights strongly support the argument for the need to consider targeting/engaging both innate (e.g., macrophages) as well as adaptive (CD4+ and CD8+ T cells) immunity to improve the efficacy of cancer immunotherapy. They also demonstrate the utility of using high dimensional profiling approaches to not only help identify optimal combinations among the plethora of immuno-oncology drugs currently available but also in identifying potential novel biomarkers of response to make cancer immunotherapy more effective, more specific and safer than the therapies available to us today.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robert D. Schreiber (rdschreiber@wustl.edu).
Mice
Wild type mice were purchased from Taconic Farms. All in vivo experiments used 8–12 week old male, 129S6 background mice (to match the sex and strain of the tumors) housed in our specific pathogen-free animal facility. All studies were performed in accordance with procedures approved by the AAALAC accredited Animal Studies Committee of Washington University in St. Louis.
Tumor transplantation
The T3 MCA-induced sarcoma line used in this study was previously generated as described (Shankaran et al.) and was banked as low-passage tumor cells. Tumor cells derived from frozen stocks and propagated in vitro in RPMI media (Hyclone) supplemented with 10% FCS (Hyclone) were washed extensively, resuspended at a density of 6.67 × 106 cells/ml in endotoxin-free PBS and then 150 μl injected (1 ×106 cells) subcutaneously into the flanks of recipient mice. Tumor cells were >90% viable at the time of injection as assessed by trypan blue exclusion. Tumor growth was quantified by caliper measurements and expressed as the average of two perpendicular diameters.
Immune checkpoint therapy
Mice were treated intraperitoneally with 200 μg anti-PD-1 (RMP1-14) or anti-CTLA-4 (9D9), used alone or in combination on days 3, 6, and 9 post-tumor transplant. For controls, mice were injected with 200 μg of rat IgG2a isotype control antibody. Antibodies were diluted in PBS prior to injection for a total volume of 0.5 ml per mouse. For longitudinal analyses by CyTOF, 2 groups of mice were injected 5, 6, 7, 8, 9, 10, 11, or 12 days prior to harvest. One group was treated with control antibody and the other was treated with anti-PD-1 and anti-CTLA-4. The treatment schedule was the same as used for the single time point experiments, beginning 3 days after tumor injection and continuing every 3 days until harvest.
In vivo IFNγ neutralization
Mice were treated intraperitoneally with 250 μg anti-IFNγ (H22) or control anti-bacterial glutathione S-transferase (PIP) on either day 7 or day 9 post-tumor transplant.
Antibodies for in vivo use
Anti-PD-1 (rat IgG2a clone RMP1-14) and anti-CTLA-4 (murine IgG2b clone 9D9) antibodies were purchased from Leinco Technologies and BioXcell, respectively. Isotype control antibody (rat IgG2b) was purchased from Leinco Technologies. Neutralizing anti-IFNγ (hamster IgG clone H22) was purchased from Leinco Technologies. Anti-bacterial glutathione S-transferase (hamster IgG clone PIP) was used as an isotype control and was purchased from Leinco Technologies.
Tumor harvest
Established tumors were excised from mice, minced and treated with 1 mg/ml type IA collagenase (Sigma) in HBSS (Hyclone) for 1 h at 37 °C. Cells were washed thrice. Red blood cells were lysed using ACK lysis buffer (150mM NH4Cl 10 mM KHCO3 0.1 mM EDTA) made in house. To remove aggregates and clumps, cells were filtered through a 40-micron strainer.
Fluorescence-activated cell sorting and conventional flow cytometry
For sorting of intratumoral CD45+ cells, single cell suspensions of digested tumor were stained for 5 minutes at room temperature with 500 ng of Fc block (anti-CD16/32) and then stained with anti-CD45-PerCP/Cy5.5 (Biolegend) (1:200) plus Zombie NIR Viability dye (Biolegend) (1:500) in 100 μl of staining buffer (PBS with 2% FCS) for 30 minutes at 4 °C. Sorting of live CD45+ cells was performed on a BD FACSAria II (BD Biosciences). The purity of the sorted cells for single cell RNA sequencing was greater than 96% as assessed during post-sort cellular analysis. For comprehensive flow cytometry analysis of cell subsets in tumors, single cell suspensions of digested tumor were as above (see STAR methods table for antibody clones and dilutions). For FoxP3 staining of tumor infiltrating lymphocytes, cells were isolated, washed, stained with surface antibodies for 20 minutes, and then fixed and permeabilized using the Foxp3 transcription factor staining kit (ThermoFisher Scientific). Cells were then stained with anti-Foxp3-FITC (ebioscience) (1:100) for 30 minutes at 4 °C. All flow cytometry was performed on the LSR Fortessa X-20 (BD Biosciences) and analyzed using FlowJo software (TreeStar).
Single Cell RNA Sequencing
Single cell RNA sequencing library generation
Droplet-based 3’ end massively parallel single-cell RNA sequencing (scRNAseq) was performed by encapsulating sorted live CD45+ tumor infiltrating cells into droplets and libraries were prepared using Chromium Single Cell 3’ Reagent Kits v1 according to manufacturer’s protocol (10x Genomics). The generated scRNAseq libraries were sequenced using an Illumina HiSeq2500.
Single cell RNA sequencing alignment, barcode assignment and UMI counting
The Cell Ranger Single-Cell Software Suite available at https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome was used to perform sample demultiplexing, barcode processing and single-cell 3’ counting. Cellranger mkfastq was used to demultiplex raw base call files from the HiSeq4000 sequencer, into sample-specific fastq files. Files were demultiplexed with 97%+ perfect barcode match, and 82%+ q30 reads. Afterwards, fastq files for each sample were processed with Cellranger count was used to align samples to mm10 genome, filter and quantify. For each sample the recovered-cells parameter was specified as 10,000 cells that we expected to recover for each individual library. The resulting analysis files for each sample were aggregated using the cellranger aggr pipeline, which performed a between-sample normalization step and merged four samples into one. Samples by default were subsampled to have equal number of confidently mapped reads per cell.
Preprocessing analysis with Seurat package.
The Seurat pipeline was applied to combined datasets(Butler et al., 2018). Genes that were expressed in less than 3 cells and cells that expressed less than 400 and more than 3500 genes, were excluded. Data was normalized with a scale factor of 104. Latent variable - number of UMI’s - were regressed out using a negative binomial model (function RegressOut). Most variable genes were detected by the MeanVarPlot function and used for subsequent analysis. Principle component analysis (PCA) was performed on about 3,000 genes with PCA function. A tSNE dimensional reduction was performed on the scaled matrix (with most variable genes only) using first 40 PCA components to obtain a two-dimensional representation of the cell states. For clustering, we used the function FindClusters that implements SNN (shared nearest neighbor) modularity optimization based clustering algorithm on 40 PCA components with resolution 0.5 - 1.5, leading to 13-24 clusters. A resolution of 1 was chosen for the analysis.
Dimensionality reduction and clustering
First, the most variable genes were detected using the MeanVarPlot function. PCA was then performed on about 3000 genes with PCA function. Cells are represented with tSNE (t-distributed Stochastic Neighbor Embedding). We applied RunTSNE function on normalized data, using first 40 PCA components. For clustering, we used function FindClusters that implements SNN (shared nearest neighbor) modularity optimization based clustering algorithm on 40 PCA components with resolution 1.
Identification of cluster-specific genes and marker-based classification
To identify marker genes, the FindAllMarkers function was used with likelihood-ratio test for single cell gene expression. For each cluster, only genes that were expressed in more than 25% of cells with at least 0.25 fold difference were considered. To characterize clusters, we used ImmGen, GeneQuery and Enrichr. To perform pathway analysis, we compared clusters inside cohorts (e.g., T cells, macrophages) with different parameters (0 fold and threshold of at least 10% of cells, expressing this gene inside cluster). For heat map representation, mean expression of markers inside each clusters was used.
Reanalysis of available RNA sequencing datasets
All RNA sequencing datasets were obtained from GEO and processed with GENE-E software. If needed, data was quantile and log2 normalized. Plots were created in R with ggplot2 package.
Lymphoid population analysis
To examine lymphoid cells, clusters expressing Cd3e and/or Nrc1 (Nkp46) were extracted from aggregated samples. Most variable genes, PCA, tSNE, clustering (resolution 1.8 on 40 first PCAs) and marker selection analysis were performed as described above.
Analysis of human macrophages
Expression data was retrieved from GEO with GSE46903 ID. For analysis, we selected only monocyte and macrophage samples. According to the dataset description, peripheral blood monocytes were stimulated with GM-CSF or M-CSF, and subsequently stimulated with different compounds. We excluded all M-CSF-derived macrophages and non-72 hour time points. M0-GM-CSF 72 hour samples were used as baseline. We used limma package to perform differential expression in order to understand gene changes upon treatments compared to baseline. For each comparison, gene lists were obtained and ranked by t-statistics. To compare human macrophages under different stimuli with mouse monocytes/macrophages from our scRNAseq data, we compared genes, upregulated or downregulated inside each scRNAseq cluster with genes, upregulated or downregulated in each human monocyte/macrophage treatment condition. The goal was to get a list of signed scores that will represent the level of concordance between treatments and each cluster.
For each cluster, we created two gene sets: 100 upregulated and downregulated genes. GSEA (with fgsea package) was performed using this gene set against ranked gene lists from treatment comparisons. To obtain a score, we subtracted the normalized enrichment score (NES) of downregulated genes from NES of upregulated genes. To further investigate the correlation (or lack thereof) between clusters and treatments, we visualized genes associated with each cluster in human macrophages. We reduced the dimensions of macrophage datasets with Rtsne package and plotted each sample in two dimensions using the ggplot2 package. Samples were then highlighted using Z-scored expression profiles of cluster-associated genes.
CyTOF
Production of Exchanged Peptide-MHC Class I Monomers
Peptide-MHC Class I monomers refolded with an ultraviolet light-cleavable conditional ligand were prepared in-house as described (Gubin et al., 2014). Briefly, recombinant H-2Kb heavy chain and human β2 microglobulin light chain were produced in Escherichia coli as inclusion bodies and refolded in the presence of a UV-cleavable peptide (SIINFE-J-L, where J represents 3-amino-3-(2-nitro) phenyl-propionic acid (Peptide 2.0). Monomers were captured by anion exchange (HiTrap Q HP, GE), biotinylated, and purified by gel filtration FPLC. Ultraviolet light-induced ligand exchange with mLama4 and mAlg8 peptides was performed as described (Gubin et al., 2014).
Tetramerization of Peptide-MHC Class I Monomers with Metal-Bound Streptavidin
Streptavidin modified to contain six free cysteines separated by flexible linkers was produced in-house and coupled to heavy metals as previously described, with the exception that Eu-153, Tb-159, Dy-161, and Er-166 (Fluidigm) were used(Fehlings et al., 2017). For tetramerization, each streptavidin-metal conjugate was incubated with exchanged peptide-MHC monomers at a 1:4 molar ratio and quenched with biotin before staining as described (Fehlings et al., 2017).
Mass Cytometry Antibodies
Antibody clones and suppliers are listed in Methods Table. For antibody conjugations done in-house, 100 μg of carrier-free antibody was coupled to metal-labeled X8 polymer according to the manufacturer’s instructions (Fluidigm).
Myeloid Panel Staining
Following collagenase digestion and disaggregation of tumors into a single cell suspension as previously described (Gubin et al., 2014), four million cells per sample were transferred into 96 well plates, washed once with CyFACS (metal-free PBS + 4% FCS + 2 mM EDTA) and blocked with 40 μg/mL each of rat IgG2a (Leinco), mouse IgG2a (Leinco), and Armenian hamster IgG (Leinco), together with 10 μg/mL 2.4G2 antibody (BD Biosciences) for 5 minutes in a final volume of 50 μL. A master mix of titrated amounts of metal-labeled antibodies was then added in a 50 μL volume and incubated at 37 °C for 15 minutes followed by a 45-minute incubation at room temperature. Cells were washed twice in CyFACS and stained with a titrated amount of metal-labeled anti-PE secondary antibody before being washed twice in PBS and stained with 5 μM cisplatin (Sigma) for 3 minutes on ice for the discrimination of dead cells. Following one wash in CyFACS, cells were incubated in Cytofix (BD Biosciences) for 30 minutes on ice and subsequently washed in permeabilization buffer (BD Biosciences) prior to intracellular staining with a titrated master mix of metal-labeled antibodies at room temperature for 30 minutes. The cells were then washed and fixed a second time in 4% paraformaldehyde in PBS (Electron Microscopy Sciences) at 4° C at least overnight. Each sample was then barcoded with a unique combination of palladium metal barcodes using the manufacturer’s instructions (Fluidigm). Following barcoding, cells were pooled together and incubated overnight in 2% paraformaldehyde in PBS containing 62.5 nM Iridium nucleic acid intercalator (Fluidigm). The cells were then washed once with PBS, once with de-ionized water and then diluted in water containing 10% EQ Calibration Beads (Fluidigm) at 1 million cells per ml before acquisition on a CyTOF2 mass cytometer (Fluidigm).
Lymphoid Panel Staining
Single cell suspensions of tumor digests were washed in CyFACS and stained with 2 μg/mL anti-CD64 APC (BioLegend) for 20 minutes at 4° C. After washing twice, cells were incubated with anti-APC coated microbeads (Miltenyi) according to the manufacturer’s instructions prior to negative selection using LS columns (Miltenyi). Four million cells per sample were then transferred into 96 well plates and blocked with 40 μg/mL each of rat IgG2a (Leinco), mouse IgG2a (Leinco), and Armenian hamster IgG (Leinco) together with 10 μg/mL 2.4G2 antibody (BD Biosciences) in a final volume of 30 μL CyFACS containing 0.1% sodium azide (Sigma) for 5 minutes. Cells were then stained with double-coded metal-labeled MHC Class I tetramers at a final concentration of 10 μg/mL each for 15 minutes at 37° C. A master mix of titrated amounts of metal-labeled antibodies was then added in a 50 μL volume and incubated at 37°C for 15 minutes followed by a 45 minute incubation at room temperature. Cells were washed twice in PBS and stained with 5 μM cisplatin (Sigma) for 3 min on ice for the discrimination of dead cells prior to a wash in CyFACS and permeabilization and fixation using Foxp3/Transcription Factor Staining Buffer (ThermoFisher) for 30 minutes on ice. Cells were washed twice in permeabilization buffer (ThermoFisher) and then for intracellular staining cells were incubated with a titrated panel of antibodies in permeabilization buffer for 30 minutes at room temperature. Cells were then washed twice in CyFACS and fixed a second time in 4% paraformaldehyde in PBS (Electron Microscopy Sciences) at 4°C at least overnight. Each sample was then barcoded with a unique combination of palladium metal barcodes using the manufacturer’s instructions (Fluidigm). Following barcoding, cells were incubated overnight in 2% paraformaldehyde in PBS containing 62.5 nM Iridium nucleic acid intercalator (Fluidigm). The cells were then washed once with PBS, once with de-ionized water and then diluted in water containing 10% EQ Calibration Beads (Fluidigm) at 1 million cells per ml before acquisition on a CyTOF2 mass cytometer (Fluidigm).
CyTOF Data Analysis
Mass cytometry data were randomized with the Fluidigm acquisition software version 6.0.626 and normalized with the Matlab normalizer from Finck et al. (v.7.14.0.739 run in MATLAB R2012a) (Finck et al.). Sample barcodes were resolved using a single cell debarcoder tool (Zunder et al.). Individual samples were manually gated using Cytobank to exclude normalization beads, cell debris, dead cells, and doublets for the identification of CD45+ Cisplatin-negative cells for further downstream analyses. For dimensionality reduction analysis the viSNE tool (Amir el et al.) was used in Cytobank to apply the Barnes-Hut implementation of the t-SNE algorithm (van der Maaten, 2014) . Separate viSNE plots were generated for all CD45+ cells in the myeloid panel, and for subsets of total lymphoid cells (defined as TCRβ+ and/or NKp46 (Nrc1)+ cells) or CD8+ T cells (defined as TCRβ+ CD90.2+ CD8+) from the lymphoid panel. Data were then exported into the Matlab implementation of Phenograph (Levine et al., 2015) and transformed using a cofactor of ten for subsequent clustering analysis. Visualization of clusters identified by Phenograph was done using the R package ggplot2. Heat maps of relative marker expression were generated in GENE-E from the Broad Institute (https://software.broadinstitute.org/GENE-E/).
Monocle Analysis of scRNAseq and CyTOF Data
Pseudo-temporal analysis of macrophage populations was performed using the Monocle R package (Trapnell et al., 2014). For scRNAseq, genes expressed in more that 50 cells were ranked based on differential analysis between clusters, and 1000 of the top genes were used for analysis. For CYTOF dataset all relevant proteins were used. Dimensionality reduction was done using the DDRTree method and cells ordered with the orderCells function.
Statistical Analyses
Dunnett’s multiple comparison tests were used for comparison of individual T cell cluster frequencies in anti- PD-1, anti-CTLA-4, or combination treatment versus that of the isotype antibody control group. Student’s t-tests were used for analysis of conventional flow cytometry data.
Data and available
Single cell RNAseq data is publicly available through GEO: GSE119352.
Supplementary Material
Supplemental Figure 1. CD45+ Cellular Infiltration in T3 Tumors for Single Cell Analysis and scRNAseq Alignment and Statistics (related to Figure 1 and 2).
(A) Experimental setup. Cohorts of T3 tumor bearing mice were treated with control mAb, anti-PD-1, anti-CTLA-4 or anti-PD-1/anti-CTLA-4 on days 3, 6, 9. Tumors were harvested on day 11 post-tumor transplant, digested and either subjected to CyTOF analysis or stained for CD45 and sorted. Sorted CD45+ cells were then subjected to scRNAseq using a 10x Genomics pipeline. For scRNAseq tumors from 5 mice from each treatment condition were pooled before sorting. For CyTOF analysis, each treatment condition contained 10 tumor bearing mice, with tumors from 2 pooled mice harvested and stained independently (N=5). (B) Percentage of CD45+ cells in T3 tumors across treatment conditions. (C) Quality control of alignment and summary of statistics from CellRanger. (D) Percentage of intratumoral CD45+ cell types. Staining and gating strategies performed as previously described (Noguchi et al., 2017). Data in (B) and (C) indicate mean percent ± SEM (5 mice per condition). *<0.05, **p<0.01, ***p<0.005, unpaired t test. Representative data from at least 3 independent experiments.
Supplemental Figure 2. Identification of scRNAseq Cellular Clusters by Comparison with ImmGen Data (related to Figure 1 and 2)
For each cluster, 7-15 of the most upregulated genes were used as marker genes. Mean expression of marker genes inside ImmGen-defined populations represent value of enrichment inside the population for each cluster (Heng et al., 2008).
Supplemental Figure 3. Identification of Tumor Infiltrating Lymphocyte Populations by scRNAseq and CyTOF (related to Figure 2 and 3)
(A) Heat map from scRNAseq displaying normalized expression of select genes in each lymphocyte cluster. (B) tSNE CyTOF plot generated in viSNE of subsampled lymphoid [TCRβ+ and/or NKp46 (Nrc1)+] events. Subsets identified with PhenoGraph clustering are overlaid. (C) Heat map from CyTOF displaying normalized expression of panel markers in each individual cell cluster.
Supplemental Figure 4. Identification of Neoantigen-Specific CD8+ T Cells By CyTOF (related to Figure 3)
(A) Percentages of total intratumoral CD8+ T cells specific for either mLama4 or mAlg8 as assessed by MHC class I H-2Kb tetramers loaded with mLama4 or mAlg8 peptide and labeled with a two-metal combination. Bar indicates mean percent ± SD of CD8+ cells upon different ICT. Each dot represents 2 pooled mice harvested and stained independently (N=5). Representative data from 3-4 independent experiments. (B) Median expression of TIM-3, LAG-3, or Granzyme B in intratumoral CD8+ T cells specific for mLama4, mAlg8, or neither neoantigen. Tetramer positive or negative CD8+ T cells were manually gated. Plotted as median expression from intratumoral CD8+ T cells from 2 pooled mice harvested and stained independently (N=5) ± SEM. Representative data from 3-4 independent experiments. (C) Total tumor infiltrating CD8+ cells segregated as a tSNE plot by viSNE and assessed as discrete clusters using Phenograph. (D) Heat map of mean expression intensity for each marker tested in the CD8+ T cell population. (E) Total CD8+ T cells segregated by tSNE are plotted and expression of Ki67 is highlighted with neoantigen-specific clusters indicated. *p<0.05, **p<0.01, ***p<0.001, NS, not significant, Dunnett’s multiple comparison.
Supplemental Figure 5. Identification of Intratumoral NK Cells and γδ T Cells (related to Figure 2 and 3)
(A) Frequency of cells defined in CyTOF clusters of NK cells. Each dot represents tumors from 2 pooled mice harvested and stained independently (N=5). Bar indicates mean percent ± SD of lymphoid [TCRβ+ and/or NKp46 (Nrc1)+] cells upon different ICT. Representative data from 3-4 independent experiments. (B) tSNE scRNAseq plots of lymphoid cells with select gene (s) highlighted by color. (C) Z-score of normalized expression of selected genes across ImmGen datasets. (D) Flow cytometry plots displaying γδTCR+ and CD3ε+ cells after gating on CD45+ CD4− CD8− Nkp46−. Representative data from 3 independent experiments. (E) tSNE scRNAseq plots with select gene (s) highlighted by color. (F) T_act cluster resembles activated or stimulated T cells by GSEA. *p<0.05, **p<0.01, ***p<0.001, Dunnett’s multiple comparison.
Supplemental Figure 6. Intratumoral CD45+ Cells Revealed by the Myeloid CyTOF Panel (related to Figure 5)
(A) Total CD45+ cells segregated as a tSNE plot by viSNE with 37 markers and individual clusters identified using PhenoGraph. Identity of individual populations are shown. (B) Frequency of cells defined in clusters of neutrophils and eosinophils as a percentage of total viable CD45+ cells. Bar indicates mean percent ± SD of CD45+ cells upon different ICT. Each dot represents 2 pooled mice harvested and stained independently (N=5) (C) Heat map showing mean expression intensity for each marker. *p<0.05, **p<0.01, ***p<0.001, Dunnett’s multiple comparison.
Supplemental Figure 7. Longitudinal Analysis of Intratumoral CD45+ Cells Revealed by CyTOF Myeloid Panel (related to Figure 6)
(A) Total CD45+ cells segregated as a tSNE plot by viSNE with 37 markers and individual clusters identified using PhenoGraph. (B) Heat map displaying the percentage (of intratumoral CD45+ cells) of monocytes/macrophages by cluster, day, and treatment. (C) Heat map showing mean expression intensity for each marker per treatment group and day analyzed. Representative data from 2-3 independent experiments.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CTLA-4 (clone 9D9) | BioXCell | Cat# BP0164 |
| Anti-PD-1 (clone RMP1-14) | Leinco | Cat# P372 |
| Anti-CD16/32 (clone 2.4G2) | BD Biosciences | Cat#553141 |
| Mouse IgG2a (clone HKSP84) | Leinco | Cat# I-118 |
| Rat IgG2a (clone 1-1) | Leinco | Cat# I-1177 |
| Hamster IgG (clone PIP) | Leinco | Cat# I-140 |
| Anti-mouse CD45 (clone 30-F11) | Fluidigm | Cat# 3089005B |
| Anti-mouse CD90.2 (clone 30-H12) | BioLegend | Cat# 105302 |
| Anti-mouse CD44 (clone IM7) | BioLegend | Cat # 103002 |
| Anti-mouse TCRb (clone H57-597 ) | BioLegend | Cat # 109202 |
| Anti-mouse CD24 (clone M1/69) | BioLegend | Cat # 101802 |
| Anti-mouse CD19 (clone 6D5) | BioLegend | Cat # 115502 |
| Anti-mouse CD278 (clone C398.4A) | BioLegend | Cat #313502 |
| Anti-mouse CD127 (clone A7R34) | BioLegend | Cat # 135002 |
| Anti-mouse CXCR3 (clone CXCR3-173) | BioLegend | Cat # 126517 |
| Anti-mouse CD335 (clone 29A1.4) | BioLegend | Cat# 137602 |
| Anti-mouse CD150 (clone TC15-12F12.2) | BioLegend | Cat# 115902 |
| Anti-mouse T-bet (clone 4B10) | BioLegend | Cat# 644825 |
| Anti-mouse VISTA (clone MIH63) | BioLegend | Cat# 150202 |
| Anti-mouse Tim-3 (clone RMT3-23) | BioLegend | Cat# 119702 |
| Anti-mouse CTLA-4 (clone UC10-4B9) | BioLegend | Cat# 106302 |
| Anti-mouse Ly6A/E (clone D7) | BioLegend | Cat# 108102 |
| Anti-mouse CD38 (clone 90) | BioLegend | Cat# 102702 |
| Anti-mouse Granzyme B (clone GB11) | BioLegend | Cat# 92229 |
| Anti-mouse Fas (clone SA367H8) | BioLegend | Cat# 152602 |
| Anti-mouse I-A/I-E (clone M5/114.15.2) | BioLegend | Cat# 107602 |
| Anti-mouse CD1d (clone 1B1) | BioLegend | Cat# 123504 |
| Anti-mouse F4/80 (clone BM8) | BioLegend | Cat# 123102 |
| Anti-mouse VCAM1 (clone 429 (MVCAM.A) | BioLegend | Cat# 105702 |
| Anti-mouse CX3CR1 (clone SA011F11) | BioLegend | Cat# 149002 |
| Anti-mouse CD43 (clone S11) | BioLegend | Cat# 143202 |
| Anti-mouse CD103 (clone 2E7) | BioLegend | Cat# 121402 |
| Anti-mouse XCR-1 (clone ZET) | BioLegend | Cat# 148202 |
| Anti-mouse CD63 (clone NVG-2) | BioLegend | Cat# 143902 |
| Anti-mouse CD206 (clone C068C2) | BioLegend | Cat# 141702 |
| Anti-mouse CD64 (clone X54-5/7.1) | BioLegend | Cat# 139302 |
| Anti-mouse CD124 (clone I015F8) | BioLegend | Cat# 144802 |
| Anti-mouse VISTA (clone MIH63) | BioLegend | Cat# 150202 |
| Anti-mouse SIRPa (clone P84) | BioLegend | Cat# 144002 |
| Anti-PE (clone PE001) | BioLegend | Cat# 408102 |
| Anti-mouse CD40 (clone 3/23) | BioLegend | Cat# 124610 |
| Anti-mouse CD8a (clone 53-6.7) | Leinco | Cat# C375 |
| Anti-mouse CD4 (clone GK1.5) | Leinco | Cat# C1333 |
| Anti-mouse CD62L (clone MEL-14 ) | Leinco | Cat# C2118 |
| Anti-mouse KLRG-1 (clone 2F1) | Leinco | Cat# K155 |
| Anti-mouse CD25 (clone PC61) | Leinco | Cat # C1194 |
| Anti-mouse Lag-3 (clone C9B7W) | Leinco | Cat# C2248 |
| Anti-mouse Ly6G (clone 1A8) | Leinco | Cat# L280 |
| Anti-mouse CD11b (clone M1/70) | Leinco | Cat# C377 |
| Anti-mouse CD45R/B220 (clone RA3-6B2) | Leinco | Cat# C383 |
| Anti-mouse PD-L2 (clone TY25) | Leinco | Cat# C2292 |
| Anti-mouse CD80 (clone 16-10A1) | Leinco | Cat# C2045 |
| Anti-mouse PD-L1 (clone 10F.9G2) | Leinco | Cat# P363 |
| Anti-mouse Ly6C (clone HK1.4) | Leinco | Cat# L136 |
| Anti-mouse CD11c (clone N418) | Leinco | Cat# C2119 |
| Anti-mouse CD86 (clone GL1) | Leinco | Cat# C2158 |
| Anti-mouse Ki67 (clone B56) | BD Biosciences | Cat # 556003 |
| Anti-mouse Siglec F (clone E50-2440) | BD Biosciences | Cat# 552125 |
| Anti-mouse GITR (clone DTA-1) | Thermo Fisher | Cat# 16-5874-83 |
| Anti-mouse OX-40 (clone OX-86) | Thermo Fisher | Cat# 16-1341-85 |
| Anti-mouse CD39 (clone 24DMS1) | Thermo Fisher | Cat# 16-0391-83 |
| Anti-mouse CD27 (clone LG.7F9) | Thermo Fisher | Cat# 14-0271-85 |
| Anti-mouse EOMES (clone dan11mag) | Thermo Fisher | Cat# 14-4875-82 |
| Anti-mouse PD-1 (clone RMP1-30) | Thermo Fisher | Cat# 14-9981-85 |
| Anti-mouse CD137 (clone 17B5) | Thermo Fisher | Cat# 16-1371-85 |
| Anti-mouse FoxP3 (clone FJK-16s) | Thermo Fisher | Cat# 14-5773-82 |
| Anti-mouse BST2/PDCA-1 (clone 927) | Thermo Fisher | Cat# 16-3172-85 |
| Anti-mouse MerTK (clone DS5MMER) | Thermo Fisher | Cat# 14-5751-82 |
| Anti-mouse CD83 (clone Michel-17) | Thermo Fisher | Cat# 14-0831-82 |
| Anti-mouse CCR7 (clone 4B12) | Thermo Fisher | Cat# 16-1971-85 |
| Anti-mouse Nos2 (clone CXNFT) | Thermo Fisher | Cat# 14-5920-82 |
| Anti-mouse CCR2 (clone 475301 ) | RnD Systems | Cat# MAB55381-100 |
| Anti-mouse CD103 FITC (clone 2E7) (1:100 dilution) | BioLegend | Cat# 121419 |
| Anti-mouse CD11b PerCP/Cy5.5 (clone M1/70) (1:100 dilution) | BioLegend | Cat# 101227 |
| Anti-mouse CD45 BV605 (clone 30-F11) (1:800 dilution) | BioLegend | Cat# 103139 |
| Anti-mouse CD45 PE/Cy7 (clone 30-F11) (1:200 dilution) | BioLegend | Cat# 103113 |
| Anti-mouse CD45 PerCP/Cy5.5 (clone 30-F11) (1:200 dilution) | BioLegend | Cat# 103131 |
| Anti-mouse I-A/I-E BV650 (clone M5/114.15.2) (1:2000 dilution) | BioLegend | Cat# 107641 |
| Anti-mouse CD24 BV711 (clone M1/69) (1:1000 dilution) | BD Bioscience | Cat# 563450 |
| Anti-mouse CD11c BV786 (clone HL3) (1:100 dilution) | BD Bioscience | Cat# 563735 |
| Anti-mouse F4/80 BUV395 (clone T45-2342) (1:400 dilution) | BD Bioscience | Cat# 565614 |
| Anti-mouse CD64 APC (clone X54-5/7.1) (1:400 dilution) | BioLegend | Cat# 139305 |
| Anti-mouse CD64 BV421 (clone X54-5/7.1) (1:400 dilution) | BioLegend | Cat# 139309 |
| Anti-mouse Ly6G Alexa Fluor 700 (clone 1A8) (1:200 dilution) | BD Bioscience | Cat# 561236 |
| Anti-mouse PDCA-1 BV650 (clone 927) (1:200 dilution) | Biolegend | Cat# 127019 |
| Anti-mouse B220 BUV395 (clone RA3-6B2) (1:200 dilution) | BD Bioscience | Cat# 563793 |
| Anti-mouse SiglecF Alexa Fluor 647 (clone E50-2440) (1:200 dilution) | BD Bioscience | Cat# 562680 |
| Anti-mouse FceR1 PE/Cy7 (clone MAR-1) (1:200 dilution) | Biolegend | Cat# 134317 |
| Anti-mouse CD117 (c-KIT) FITC (clone ACK2) (1:200 dilution) | Biolegend | Cat# 135115 |
| Anti-mouse NKp46 FITC (clone 29A1.4) (1:200 dilution) | Biolegend | Cat# 137605 |
| Anti-mouse CD19 BV650 (clone 1D3) (1:200 dilution) | BD Bioscience | Cat# 563235 |
| Anti-mouse CD4 BV711 (clone RM4-5) (1:200 dilution) | Biolegend | Cat# 100549 |
| Anti-mouse CD8a BV786 (clone 53-6.7) (1:200 dilution) | BD Bioscience | Cat# 563332 |
| Anti-mouse CD8b PerCP/Cy5.5 (clone YTS156.7.7) (1:200 dilution) | Biolegend | Cat# 126609 |
| Anti-mouse CD3e APC (clone 145-2C11) (1:200 dilution) | Biolegend | Cat# 100311 |
| Anti-mouse gdTCR PE/Cy7 (clone GL3) (1:200 dilution) | Biolegend | Cat# 118123 |
| Anti-mouse CD25 APC (clone 3C7) (1:200 dilution) | Biolegend | Cat# 101909 |
| Anti-mouse Foxp3 FITC (clone FJK-16s) (1:100 dilution) | Thermo Fisher | Cat# 11-5773-80 |
| Anti-mouse iNOS/Nos2 PE (clone CXNFT) (1:200 dilution) | Thermo Fisher | Cat# 12-5920-80 |
| Anti-mouse CD206 APC (clone C068C2) (1:200 dilution) | Biolegend | Cat# 141707 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RPMI-1640 | HyClone | Cat# SH30096 |
| Defined fetal bovine serum | HyClone | Cat# SH30070 |
| Mutant Lama4 peptide, sequence VGFNFRTL | Peptide 2.0 | Custom order |
| Mutant Alg8 peptide, sequence ITYTWTRL | Peptide 2.0 | Custom order |
| Recombinant H-2Kb | Gubin et al | Purified in-house |
| Recombinant streptavidin | Fehlings et al | Purified in house |
| Iridium | Fluidigm | Cat# 201192A |
| Cisplatin | Fluidigm | Cat# 201064 |
| Bismuth(III) nitrate pentahydrate | Sigma-Aldrich | Cat# 254150 |
| Nitric acid, 70% | Sigma-Aldrich | Cat# 225711 |
| EQ 4-element beads | Fluidigm | Cat# 201078 |
| Antibody stabilizer | Candor | Cat# 131 125 |
| tris-(2-carboxyethyl) phosphine | Thermo Fisher | Cat# 77720 |
| 16% paraformaldehyde | EMS Sciences | Cat# 15710 |
| Collagenase Type IA | Sigma-Aldrich | Cat# C9891 |
| Critical Commercial Assays | ||
| Cytofix | BD Biosciences | Cat# 554655 |
| Perm/Wash | BD Biosciences | Cat# 554723 |
| Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher | Cat# 00-5523-00 |
| X8 Antibody Labeling Kit | Fluidigm | N/A (metal specific) |
| DN3 Antibody Labeling Kit | Fluidigm | N/A (metal specific) |
| 20-Plex Pd Barcoding Kit | Fluidigm | Cat# 201060 |
| Deposited Data | ||
| CD45+ scRNAseq | This paper | Repository ID: GSE119352 [Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/)] |
| T3 checkpoint therapy × 5 replicates (CyTOF) | This paper | Repository ID: FR-FCM-ZYPM [FLOW Repository (https://flowrepository.org)] |
| T3 checkpoint therapy: time course, days 5-12 (CyTOF) | This paper | Repository ID: FR-FCM-ZYPN [FLOW Repository (https://flowrepository.org)] |
| T3 checkpoint therapy: anti-IFNγ (CyTOF) | This paper | Repository ID: FR-FCM-ZYPX [FLOW Repository (https://flowrepository.org)] |
| Experimental Models: Cell Lines | ||
| T3 mouse MCA sarcoma line | Generated in house. | N/A |
| Experimental Models: Organisms/Strains | ||
| Mus musculus (mouse)/129S6 strain | Taconic | 129SVE-M |
| Software and Algorithms | ||
| MATLAB R2017b | https://www.mathworks.com/ | N/A |
| Cyt | http://www.c2b2.columbia.edu/danapeerlab/html/cyt.html | N/A |
| Cell Ranger 1.3.1 | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest | N/A |
| Seurat 1.4.0 | https://satijalab.org/seurat/;RRID:SCR_007322 | N/A |
| Monocle 2.6.4 | http://cole-trapnell-lab.github.io/monocle-release/ | N/A |
| ggplot2 2.2.1 | https://ggplot2.tidyverse.org/index.html | N/A |
| Rtsne 0.13 | https://github.com/jkrijthe/Rtsne | N/A |
| ImmGen | https://www.immgen.org | N/A |
Highlights:
High dimensional analyses of successful ICT in tumor bearing mice
ICT induces changes in intratumoral myeloid and lymphoid cells
Tumor associated monocytes/macrophages display complex cytokine-driven phenotypes
Different cytokines act on tumor infiltrating monocytes to drive macrophage polarization
ACKNOWLEDGEMENTS
The authors thank all members of the Schreiber and Artyomov labs for helpful discussions and technical support. We especially thank K. Zaitsev (Artyomov lab) for computational discussions. We thank E. Unanue and G. Randolph (Washington University) for helpful comments and criticisms. We would also like to thank L. Yang and C. Wilson for assistance with MHC multimer development and staining. We thank E. Lantelme and D. Brinja at the Flow Cytometry Core for technical support with FACS cell sorting. This work was supported by grants to R.D. Schreiber and subcontract to E.W. Newell from the NCI (RO1 CA190700). This work was also supported by grants to R.D. Schreiber from the Parker Institute for Cancer Immunotherapy, the Cancer Research Institute, Janssen Pharmaceutical Company of Johnson and Johnson, the Prostate Cancer Foundation, and by a Stand Up To Cancer—Lustgarten Foundation Pancreatic Cancer Convergence Dream Team Translational Research Grant. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. M.M. Gubin was supported by a postdoctoral training grant (Irvington Postdoctoral Fellowship) from the Cancer Research Institute. E. Esaulova is partially supported by a grant (08-08) from the Government of Russian Federation. J.P. Ward is supported by the Paul Calabresi Career Development Award in Clinical Oncology (PCACO) K12 (4K12CA167540-05). Aspects of studies including mass cytometry were performed with assistance by the Immunomonitoring Laboratory (IML) which is supported by the Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs and the Alvin J. Siteman Comprehensive Cancer Center that, in turn, is supported by NCI Cancer Center Support Grant P30CA91842.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
E.W.N is a board director and shareholder of immunoSCAPE Pte.Ltd. M.F is Director, Scientific Affairs and shareholder of immunoSCAPE Pte. Ltd. R.D.S. is a cofounder, scientific advisory board member, stockholder and royalty recipient of Jounce Therapeutics and Neon Therapeutics and is a scientific advisory board member for BioLegend, Codiak Biosciences, Constellation Pharmaceuticals, Lytix Biopharma and NGM Biopharmaceuticals.
REFERENCES
- Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, and Pe’er D (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F, Charles KA, and Mantovani A (2005). Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 7, 211–217. [DOI] [PubMed] [Google Scholar]
- Biswas SK, and Mantovani A (2010). Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology 11, 889–896. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866 e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlings M, Simoni Y, Penny HL, Becht E, Loh CY, Gubin MM, Ward JP, Wong SC, Schreiber RD, and Newell EW (2017). Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8(+) T cells. Nat Commun 8, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, and Bendall SC (2013). Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, and Li MO (2014). The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. (2000). Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 192, 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Artyomov MN, Mardis ER, and Schreiber RD (2015). Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 125, 3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, et al. (2014). Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW, and Immunological Genome Project, C. (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, Smyth MJ, Tarlinton DM, and Nutt SL (2007). NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol 178, 4764–4770. [DOI] [PubMed] [Google Scholar]
- Leach DR, Krummel MF, and Allison JP (1996). Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. (2012). Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. (2014). Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, Selby MJ, Graziano RF, Mardis ER, Korman AJ, et al. (2017). Temporally Distinct PD-L1 Expression by Tumor and Host Cells Contributes to Immune Escape. Cancer Immunol Res 5, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, Linsley PS, Thompson CB, and Riley JL (2005). CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol 25, 9543–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, and Pollard JW (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Kang K, Giannopoulou E, Fang C, and Ivashkiv LB (2016). IFN-gamma Induces Histone 3 Lysine 27 Trimethylation in a Small Subset of Promoters to Stably Silence Gene Expression in Human Macrophages. Cell Rep 16, 3121–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, and Smyth MJ (2011). Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, and Schreiber RD (2015). Neoantigens in cancer immunotherapy. Science 348, 69–74. [DOI] [PubMed] [Google Scholar]
- Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, and Korman AJ (2013). Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res 1, 32–42. [DOI] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, and Schreiber RD (2001). IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410, 1107–1111. [DOI] [PubMed] [Google Scholar]
- Sharma P, and Allison JP (2015). The future of immune checkpoint therapy. Science 348, 56–61. [DOI] [PubMed] [Google Scholar]
- Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, et al. (2013). Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 210, 1695–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, and Nolan GP (2016). Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, and Sakaguchi S (2017). Regulatory T cells in cancer immunotherapy. Cell Res 27, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, and Pardoll DM (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L (2014). Accelerating t-SNE using Tree-Based Algorithms. J Mach Learn Res 15, 3221–3245. [Google Scholar]
- Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, et al. (2017). Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell 170, 1120–1133 e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, and Kurachi M (2015). Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir el AD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. (2015). Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 10, 316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. CD45+ Cellular Infiltration in T3 Tumors for Single Cell Analysis and scRNAseq Alignment and Statistics (related to Figure 1 and 2).
(A) Experimental setup. Cohorts of T3 tumor bearing mice were treated with control mAb, anti-PD-1, anti-CTLA-4 or anti-PD-1/anti-CTLA-4 on days 3, 6, 9. Tumors were harvested on day 11 post-tumor transplant, digested and either subjected to CyTOF analysis or stained for CD45 and sorted. Sorted CD45+ cells were then subjected to scRNAseq using a 10x Genomics pipeline. For scRNAseq tumors from 5 mice from each treatment condition were pooled before sorting. For CyTOF analysis, each treatment condition contained 10 tumor bearing mice, with tumors from 2 pooled mice harvested and stained independently (N=5). (B) Percentage of CD45+ cells in T3 tumors across treatment conditions. (C) Quality control of alignment and summary of statistics from CellRanger. (D) Percentage of intratumoral CD45+ cell types. Staining and gating strategies performed as previously described (Noguchi et al., 2017). Data in (B) and (C) indicate mean percent ± SEM (5 mice per condition). *<0.05, **p<0.01, ***p<0.005, unpaired t test. Representative data from at least 3 independent experiments.
Supplemental Figure 2. Identification of scRNAseq Cellular Clusters by Comparison with ImmGen Data (related to Figure 1 and 2)
For each cluster, 7-15 of the most upregulated genes were used as marker genes. Mean expression of marker genes inside ImmGen-defined populations represent value of enrichment inside the population for each cluster (Heng et al., 2008).
Supplemental Figure 3. Identification of Tumor Infiltrating Lymphocyte Populations by scRNAseq and CyTOF (related to Figure 2 and 3)
(A) Heat map from scRNAseq displaying normalized expression of select genes in each lymphocyte cluster. (B) tSNE CyTOF plot generated in viSNE of subsampled lymphoid [TCRβ+ and/or NKp46 (Nrc1)+] events. Subsets identified with PhenoGraph clustering are overlaid. (C) Heat map from CyTOF displaying normalized expression of panel markers in each individual cell cluster.
Supplemental Figure 4. Identification of Neoantigen-Specific CD8+ T Cells By CyTOF (related to Figure 3)
(A) Percentages of total intratumoral CD8+ T cells specific for either mLama4 or mAlg8 as assessed by MHC class I H-2Kb tetramers loaded with mLama4 or mAlg8 peptide and labeled with a two-metal combination. Bar indicates mean percent ± SD of CD8+ cells upon different ICT. Each dot represents 2 pooled mice harvested and stained independently (N=5). Representative data from 3-4 independent experiments. (B) Median expression of TIM-3, LAG-3, or Granzyme B in intratumoral CD8+ T cells specific for mLama4, mAlg8, or neither neoantigen. Tetramer positive or negative CD8+ T cells were manually gated. Plotted as median expression from intratumoral CD8+ T cells from 2 pooled mice harvested and stained independently (N=5) ± SEM. Representative data from 3-4 independent experiments. (C) Total tumor infiltrating CD8+ cells segregated as a tSNE plot by viSNE and assessed as discrete clusters using Phenograph. (D) Heat map of mean expression intensity for each marker tested in the CD8+ T cell population. (E) Total CD8+ T cells segregated by tSNE are plotted and expression of Ki67 is highlighted with neoantigen-specific clusters indicated. *p<0.05, **p<0.01, ***p<0.001, NS, not significant, Dunnett’s multiple comparison.
Supplemental Figure 5. Identification of Intratumoral NK Cells and γδ T Cells (related to Figure 2 and 3)
(A) Frequency of cells defined in CyTOF clusters of NK cells. Each dot represents tumors from 2 pooled mice harvested and stained independently (N=5). Bar indicates mean percent ± SD of lymphoid [TCRβ+ and/or NKp46 (Nrc1)+] cells upon different ICT. Representative data from 3-4 independent experiments. (B) tSNE scRNAseq plots of lymphoid cells with select gene (s) highlighted by color. (C) Z-score of normalized expression of selected genes across ImmGen datasets. (D) Flow cytometry plots displaying γδTCR+ and CD3ε+ cells after gating on CD45+ CD4− CD8− Nkp46−. Representative data from 3 independent experiments. (E) tSNE scRNAseq plots with select gene (s) highlighted by color. (F) T_act cluster resembles activated or stimulated T cells by GSEA. *p<0.05, **p<0.01, ***p<0.001, Dunnett’s multiple comparison.
Supplemental Figure 6. Intratumoral CD45+ Cells Revealed by the Myeloid CyTOF Panel (related to Figure 5)
(A) Total CD45+ cells segregated as a tSNE plot by viSNE with 37 markers and individual clusters identified using PhenoGraph. Identity of individual populations are shown. (B) Frequency of cells defined in clusters of neutrophils and eosinophils as a percentage of total viable CD45+ cells. Bar indicates mean percent ± SD of CD45+ cells upon different ICT. Each dot represents 2 pooled mice harvested and stained independently (N=5) (C) Heat map showing mean expression intensity for each marker. *p<0.05, **p<0.01, ***p<0.001, Dunnett’s multiple comparison.
Supplemental Figure 7. Longitudinal Analysis of Intratumoral CD45+ Cells Revealed by CyTOF Myeloid Panel (related to Figure 6)
(A) Total CD45+ cells segregated as a tSNE plot by viSNE with 37 markers and individual clusters identified using PhenoGraph. (B) Heat map displaying the percentage (of intratumoral CD45+ cells) of monocytes/macrophages by cluster, day, and treatment. (C) Heat map showing mean expression intensity for each marker per treatment group and day analyzed. Representative data from 2-3 independent experiments.