Abstract
Background
Grass glucuronoarabinoxylan (GAX) substitutions can inhibit enzymatic degradation and are involved in the interaction of xylan with cell wall cellulose and lignin, factors which contribute to the recalcitrance of biomass to saccharification. Therefore, identification of xylan characteristics central to biomass biorefining improvement is essential. However, the task of assessing biomass quality is complicated and is often hindered by the lack of a reference for a given crop.
Results
In this study, we created a reference library, expressed in glucose units, of Miscanthus sinensis GAX stem and leaf oligosaccharides, using DNA sequencer-Assisted Saccharide analysis in high throughput (DASH), supported by liquid chromatography (LC), nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Our analysis of a number of grass species highlighted variations in substitution type and frequency of stem and leaf GAX. In miscanthus, for example, the β-Xylp-(1 → 2)-α-Araf-(1 → 3) side chain is more abundant in leaf than stem.
Conclusions
The reference library allows fast identification and comparison of GAX structures from different plants and tissues. Ultimately, this reference library can be used in directing biomass selection and improving biorefining.
Electronic supplementary material
The online version of this article (10.1186/s13068-019-1451-6) contains supplementary material, which is available to authorized users.
Keywords: Grass xylan, Bioenergy, Tissue variation, Species variation, Xylan branching, DASH
Background
Plant xylan polysaccharides have attracted attention due to their numerous applications not only in the papermaking, baking and food industries but also in respect to bioenergy production. Branched xylan is the main hemicellulose in many crops. This xylan is made of a linear chain of β-(1 → 4)-linked xylopyranosyl (Xylp) residues, which can be substituted by α-(1 → 2)-linked (4-O-methyl-)glucuronic acid ([Me]GlcA) and acetylation at the O-2 and/or O-3 positions [1]. In monocots, such as grasses, and also in gymnosperms, xylan is additionally modified by α-(1 → 3)-linked l-arabinofuranosyl residues (Araf). The Araf residues may be further substituted at O-2 with an Araf or a Xylp residue [2]. Cereal grain endosperm contains neutral arabinoxylan (AX), which is monosubstituted with α-(1 → 3)-linked Araf residues or di-substituted with α-(1 → 2)-linked and α-(1 → 3)-linked Araf residues [1, 3]. Araf residues of both glucuronoarabinoxylan (GAX) and endosperm AX may be esterified with a feruloyl (Fer)- or coumaryl group at O-5 position [4, 5]. Feruloylation has been implicated in cross-linking of different xylan chains and in cross-linking to lignin [6, 7]. Feruloylated Araf structures can be further substituted with β-(1 → 2)-linked Xylp groups or additional sugars [8, 9]. Corn bran xylan was found to contain α-(1 → 2)-linked l-Galp on the Xylp residue of the Fer-Araf-Xylp oligomeric structure [10–12], and this was recently found to be present in other cereal grains as well [13, 14]. Among the ferulate-containing xylan side-chain variants, 5-O-Fer-Araf structure appears most abundant, followed by the Xylp-[5-O-Fer]-Araf structure [8]. Grass GAX and AX is acetylated but to a lesser extent than dicot glucuronoxylan. However, in addition to acetyl groups being added to the backbone Xylp residues, the Araf substituents can carry acetyl groups at O-2 [15].
Most of the energy in the lignocellulosic biomass is locked within the secondary cell walls in the form of cellulose and xylan, which form a dense matrix with lignin [16]. Lignocellulosic plant cell wall recalcitrance is a barrier to cost-effective cellulosic biofuel production. Cell wall recalcitrance in regards to xylan is influenced by various factors: heavy substitution of xylan, which can impair the action of hydrolytic enzymes; specific branching points, which can serve as cross-linking sites with lignin and the pattern of branching, which can affect the interaction of xylan with cellulose [17]. The importance of xylan in recalcitrance is illustrated by the finding that removal of xylan in switchgrass resulted in materials that achieved nearly 100% glucose yields in subsequent enzymatic hydrolysis [18].
In order to improve the biomass of bioenergy crops and optimise the biorefining processes, information on the structure of xylan, and its variation, is crucial. However, little is known about the variability of xylan structure of grasses in, e.g. different tissues like stems and leaves. In addition, analysis methods of xylooligosaccharides have to be fast, accurate and robust. The majority of detailed structural information on xylan in grasses is based on analysis of cereal grain xylans such as corn cob, oat spelt, barley husks or wheat endosperm often using LC, NMR and MS [19–23]. More recently, structures of xylan of lignified tissues were analysed in grasses [24–26]. Oligosaccharides with different degrees of polymerisation (DP), glycosidic linkages and saccharide composition can be resolved with DNA sequencer-Assisted Saccharide analysis High throughput (DASH) which can analyse simultaneously 96 samples by capillary electrophoresis [27]. Our study provides the detailed structural characterisation of xylan oligosaccharides from Miscanthus sinensis stem cell walls hydrolysed with xylan-specific glycosyl hydrolases (GH) from the families 10 and 11. This information was used to generate a DASH reference library of GAX oligosaccharides with their corresponding glucose units (GU) as mobility standards. This library allows the fast and quantitative comparison of GAX oligosaccharide structures using the high-throughput method DASH, as shown by the comparison of oligosaccharide profiles of leaf and grass GAX from different grasses. Relative quantification of side chains can be achieved as exemplified by the more detailed analysis of miscanthus stem and leaf GAX.
Results
Development of a GAX oligosaccharide library from Miscanthus sinensis stem
In order to use DASH as a high-throughput method to characterise GAX structures and achieve relative quantification of different xylan substitutions, a GAX oligosaccharide library had to be created to serve as a standard for structural analysis. To develop this library, Miscanthus sinensis stem GAX was hydrolysed with endo-β-xylanases GH10 and GH11, respectively. Most reported xylanases classify into these two families, which are described to have slightly different substrate specificities. Briefly, GH10 xylanases are more capable of hydrolysing adjacent to substitutions of the xylan backbone, while GH11 xylanases preferably act on relatively unsubstituted parts of xylan [28, 29].
To describe the various hydrolysis products we use the heteroxylan naming system suggested by Faure et al. [30] with the exception of xylooligosaccharide standards which are described as X1–X6 corresponding to xylose, xylobiose, xylotriose, xylotetraose, xylopentaose and xylohexaose.
The DASH capillary electropherograms were first aligned using the internal mobility migration markers mixed with each sample to eliminate variation between capillaries [27]. The GAX oligosaccharide library was compiled by assigning the characterised oligosaccharide structures to DASH peaks based on their migration in DASH. The migration of the oligosaccharides was compared to the migration of dextran standards to provide migration information in a method we adapted here from liquid chromatography [31]. By providing the migration of the xylooligosaccharides in GU, the identification is more robust to any changes in capillary sequencer variation. Comparison of oligosaccharide GU migration therefore allows the fast and reliable annotation of GAX structures in unknown samples to all DASH users.
To characterise the detailed structure of GAX oligosaccharides, we separated the hydrolysis products by SEC and all fractions were analysed by DASH. Based on the result of the DASH analysis, SEC fractions were selected in which the respective oligosaccharides were highly abundant. Part of these fractions was subjected to secondary enzymatic hydrolysis followed by DASH analysis. The other part was used to separate possible structural isomers by Hydrophilic Interactions Liquid Chromatography (HILIC) followed by off-line Matrix-Assisted laser Desorption Ionisation (MALDI)-Mass Spectrometry. The oligosaccharides were then subjected to high-energy MALDI Collision-Induced Dissociation (CID) for detailed structural analysis. Ultimately, DASH peaks were matched to oligosaccharide structures characterised with HILIC–MALDI–MS/MS CID by combining data on peak abundance and enzyme sensitivity from different SEC fractions.
Based on the previously characterised composition of grass GAX [1], in the MALDI-CID spectra we assign uronic acid substitutions as GlcA and furanosyl pentosyl substitutions as Araf on a 1,4-linked Xylp backbone. Given that no GlcA residues on xylan have been reported to carry a methyl modification on the O-6, we assign methyl group modifications at O-4. In addition, all oligosaccharides characterised here are generated by GH10 or GH11 endo-β-1,4 xylanases and therefore the non-reducing end backbone Xylp residues cannot be modified at the O-4. Glycosidic bond and cross-ring product ions are labelled according to the nomenclature of Domon and Costello [32]. The D, E, G and V ions are labelled according to previously established nomenclature [33–35]. We used sequential digests of the oligosaccharides with enzymes to gain further insights into the structure and to confirm the results of the MALDI-CID: arabinofuranosidases GH51 and GH62 hydrolyse single Araf substitutions but not Xylp [36], which helps to distinguish the nature of pentosyl side chains; glucuronidases GH67 and GH115 both remove [Me]GlcA substitutions. However, GH67 can only remove terminal GlcA from the non-reducing end, whereas GH115 preferably acts on substitutions of internal regions although it will also cleave GlcA from non-reducing terminal Xylp residues albeit at a slower rate [37, 38].
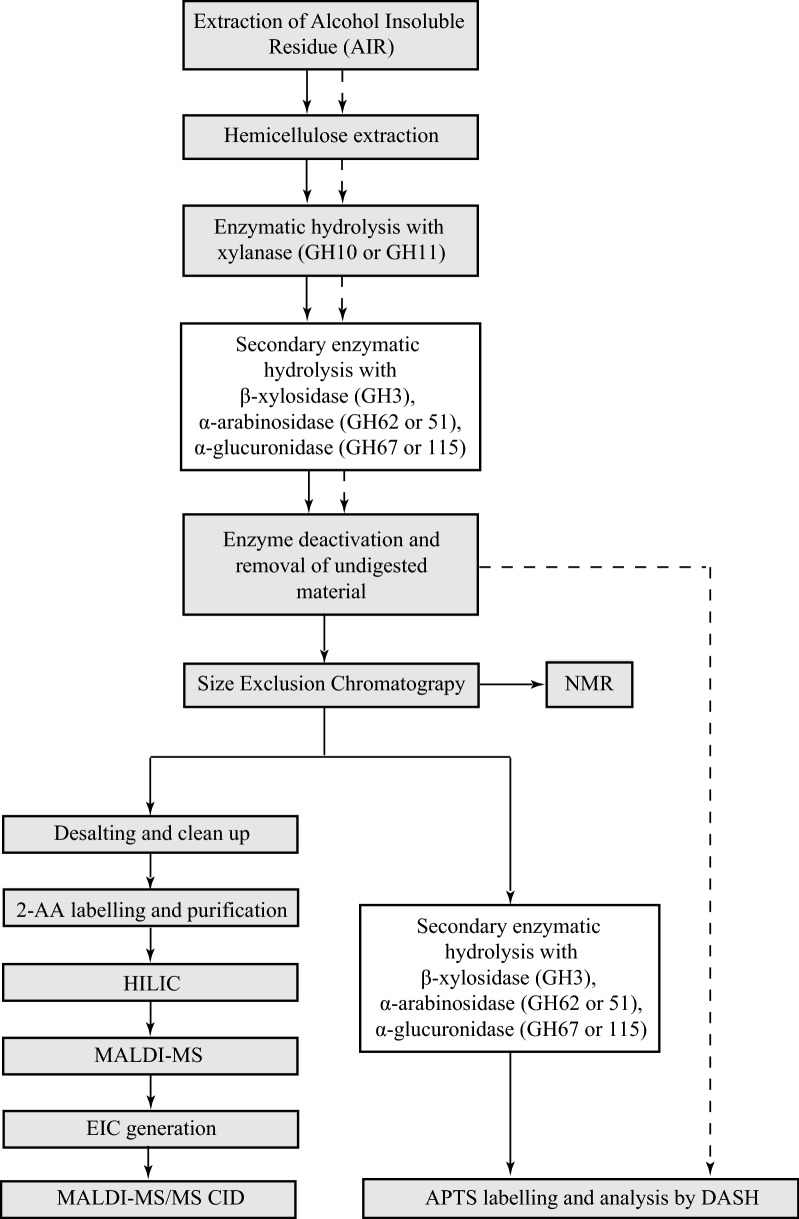
An overview of the analysis is depicted in Fig. 1.
Fig. 1.
Overview of the technical procedure used to characterise GAX structures. Creation of the GAX oligosaccharide library from Miscanthus sinensis including extensive structural analysis (solid lines), generation of standard DASH profiles prior to SEC (dashed lines). White filled boxes indicate optional steps
GAX oligosaccharide profiles of xylanase GH10 and GH11 hydrolysis of Miscanthus sinensis
In brief, a standard DASH profile was generated by APTS labelling of GH10 and GH11 hydrolysis products of Miscanthus sinensis stem alcohol-insoluble residue (AIR), respectively, and analysis by DASH (Fig. 2).
Fig. 2.
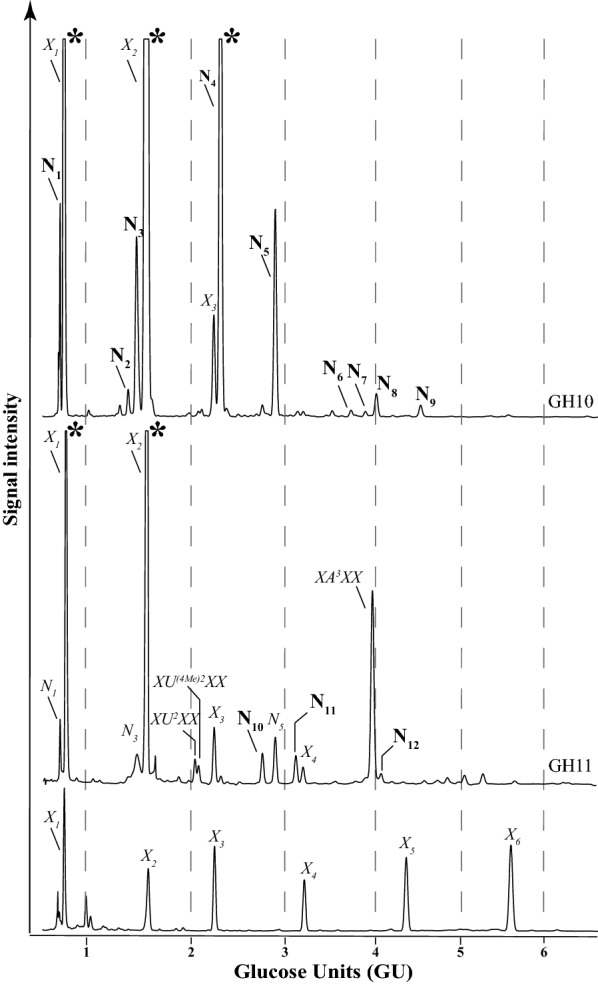
DASH profile of GAX oligosaccharides after hydrolysis with xylanases prior to SEC separation. GH10 (top panel), GH11 (middle panel), β-(1 → 4)-xylooligosaccharide standards X1-X6 (bottom panel). In addition, xylooligosaccharides XU2XX, XU(4Me)2XX and XA3XX as well as unknown peaks N1-N12 are labelled. Oligosaccharide migration is expressed in glucose units (GU). Asterisks (*) mark off-scale peaks
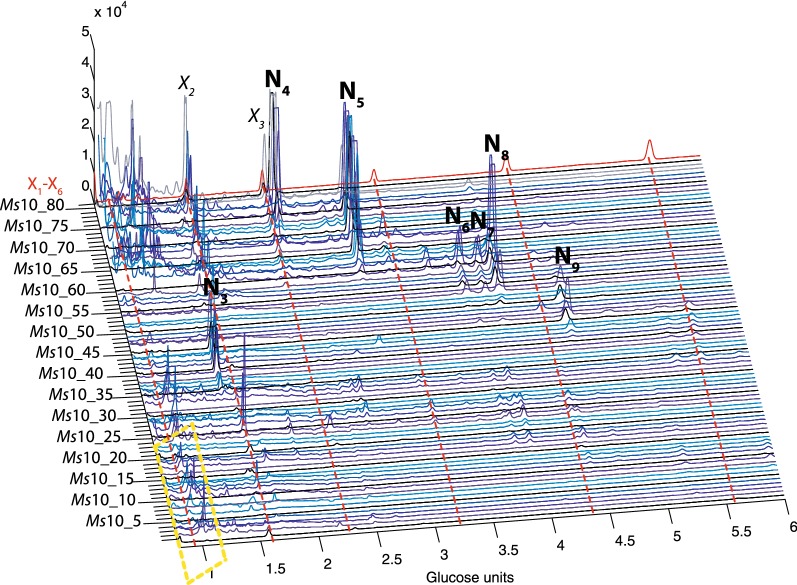
In order to partially separate the oligosaccharides for further structural analysis, the hydrolysis products were subjected to Size Exclusion Chromatography (SEC). Eighty Miscanthus sinensis GH10 and GH11 oligosaccharide SEC fractions (Ms10_1–80 and Ms11_1–80) were collected and labelled with APTS and analysed by DASH, revealing separation of a number of oligosaccharides by SEC (Fig. 3 and Additional file 1: Figure S1) in comparison to the standard DASH profile prior to SEC (Fig. 2). Figure 3 and Additional file 1: Figure S1 also show the power of DASH to study a large number of oligosaccharide mixtures, to aid interpretation of the SEC separation and selection of fractions of interest.
Fig. 3.
DASH profile of SEC fractions from GH10 hydrolysis of miscanthus stem. 80 SEC fractions were separated and analysed by DASH (blue and grey traces), xylooligosaccharide standards X1–X6 (red trace). Area marked with yellow dashed box corresponds to background noise. Unknown peaks N3–N9 are labelled
Some oligosaccharides separated by DASH eluted at the same time as β-(1 → 4)-xylooligosaccharide (X1–X6) standards; other oligosaccharides (namely XU2XX, XU(4Me)2XX and XA3XX) were previously structurally characterised [27–29]. The identity of their DASH peaks was confirmed by MALDI-CID (data not shown). Remaining unknown peaks were labelled N1–12 (Fig. 2). The detailed results of the structural analysis of each oligosaccharide are described below. Table 1 summarises the GAX oligosaccharide structures generated by GH10 and GH11 hydrolysis and their corresponding GU. Table 2 summarises the sensitivity/resistance to enzymatic hydrolysis of GAX oligosaccharides and Additional file 1: Figure S2 shows the sensitivity/resistance of selected oligosaccharides to enzymatic hydrolysis.
Table 1.
Structures of GAX oligosaccharides and their migration positions by DASH expressed in glucose units (GU)
| Nomenclature | Structure | Unknown structure | MALDI-CID or NMR figure | Glucose units |
|---|---|---|---|---|
| U(4Me)2X | 4-O-Me-α-GlcpA-(1 → 2)-β-Xylp-(1 → 4)-Xylp | N1 | S3 | 0.72 |
| X | Xylp | 0.80 | ||
| U2XX | α-GlcpA-(1 → 2)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N2 | 1.40 | |
| U(4Me)2XX | 4-O-Me-α-GlcpA-(1 → 2)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N3 | S4 | 1.48 |
| X2 | β-Xylp-(1 → 4)-Xylp | 1.57 | ||
| XU 2 XX | β-Xylp-(1 → 4)-[α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 2.03 | ||
| XU (4Me)2 XX | β-Xylp-(1 → 4)-[4-O-Me-α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 2.07 | ||
| X3 | β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 2.24 | ||
| A3X | α-Araf-(1 → 3)-β-Xylp-(1 → 4)-Xylp | N4 | S5 | 2.31 |
| A3U(4Me)2XX | α-Araf-(1 → 3)-β-Xylp-(1 → 4)-[4-O-Me-α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N10 | S8 | 2.74 |
| XA3X | β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-Xylp | N5 | S6 | 2.87 |
| B2,3U(4Me)2XX or D2,3U(4Me)2XX |
Araf-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-[α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-[α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp |
N11 | S9 | 3.09 |
| D2,3X | β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-Xylp | M1 | S12 | 3.12 |
| X4 | β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 3.17 | ||
| A3A3X | α-Araf-(1 → 3)-β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-Xylp | N6 (Z1) | 5A | 3.70 |
| A3XXX | α-Araf-(1 → 3)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N7 (Z2) | 5B | 3.85 |
| XA 3 XX | β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 3.92 | ||
| D2,3XX | β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N8 (Z3) | 5C 5D |
3.97 |
| XA3XU(4Me)2XX | β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-[4-O-Me-α-GlcpA-(1 → 2)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | N12 | S10 | 4.01 |
| X5 | β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 4.32 | ||
| XA3A3X | β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-[α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-Xylp | N9 | S7 | 4.47 |
| XD2,3XX | β-Xylp-(1 → 4)-[β-Xylp-(1 → 2)-α-Araf-(1 → 3)]-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | M2 | S11 | 4.83 |
| X6 | β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-Xylp | 5.54 |
Table 2.
Enzymatic analysis of GAX oligosaccharides
| Oligosaccharide | Unknown structure | GH10 product | GH11 product | Enzyme sensitivity | ||||
|---|---|---|---|---|---|---|---|---|
| CgGH3 xylosidase | PcGH51 α-arabinosidase | PaGH62 α-arabinosidase | CjGH67 α-glucuronidase | BoGH115 α-glucuronidase | ||||
| U(4Me)2X | N1 | ●● | ● | − | − | − | − | + |
| U2XX | N2 | ● | − | − | − | + | + | |
| U(4Me)2XX | N3 | ●● | ● | − | − | − | + | + |
| A3X | N4 | ●● | − | + | + | NT | − | |
| XU 2 XX | − | ● | − | − | − | + | + | |
| XU (4Me)2 XX | − | ● | − | − | − | + | + | |
| XA3X | N5 | ●● | ●● | NT | + | + | NT | − |
| A3A3X | N6 (Z1) | ● | NT | + | − | NT | − | |
| A3XXX | N7 (Z2) | ● | NT | + | + | NT | NT | |
| XA 3 XX | − | ●● | − | NT | + | NT | − | |
| D2,3XX | N8 (Z3) | ● | + | − | − | NT | − | |
| XA3A3X | N9 | ● | − | + | − | NT | NT | |
| A3U(4Me)2XX | N10 | ● | NT | + | − | NT | − | |
| B2,3U(4Me)2XX or D2,3U(4Me)2XX |
N11 | ● | − | − | NT | NT | − | |
| XA3XU(4Me)2XX | N12 | ● | NT | + | + | NT | + | |
| D2,3X | M1 | ● | + | − | − | NT | − | |
| XD2,3XX | M2 | ● | + | − | − | NT | − | |
The dots indicate, which xylanase produces the oligosaccharide; one dot (●) indicates minor products while two dots (●●) indicate major products. Oligosaccharides sensitive to GH enzymes are marked with a plus (+), those resistant with a minus (−); NT not tested. Oligosaccharides XU2XX, X(4Me)2XX and XA3XX have been previously structurally characterised [27–29]
Structural characterisation of the oligosaccharides N1–N9 from the xylanase GH10 digest of Miscanthus sinensis
After GH10 hydrolysis, three oligosaccharides co-eluted with X1, X2 and X3 of the xylooligosaccharide standards in DASH. We identified nine additional peaks from oligosaccharides of unknown structure which were named N1–N9 (Fig. 2). The [Me]GlcA modified hydrolysis products mainly accumulated in earlier SEC fractions independent of their molecular size (Fig. 3, fractions Ms10_24–46) because of the negative charge of the Bio-Gel P2, which results in the exclusion of uronic acids [39].
The oligosaccharide N1 in the DASH trace was analysed from the unfractionated GH10 miscanthus stem hydrolysis products (oligosaccharide mix prior to SEC fractionation). The [M + Na]+ ion at m/z 616.0 corresponds to a Pent2 structure modified with one MeGlcA. The MALDI-CID reveals the structure of N1 as U(4Me)2X (Additional file 1: Figure S3).
The Y2 ion (m/z 426.0) shows there are two Xylp residues in the backbone. The presence of the significant Y1 ion (m/z 294.1) indicates that the reducing-end Xylp is unsubstituted. The 0,2X1 ion (m/z 523.9) indicates that the non-reducing-end Xylp is modified at the O-2 with a MeGlcA, while the presence of the V3 product ion (m/z 539.9) indicates that the GlcA is modified with a methyl group at the O-4. Consistent with this, N1 oligosaccharide was sensitive to GH115 glucuronidase digestion, although resistant to GH67 hydrolysis (Table 2 and Additional file 1: Figure S2A) due to it not being an ideal substrate for the GH67 enzyme [37].
The oligosaccharide N2 was in low abundance in the DASH trace and MALDI-CID showed it is the unmethylated counterpart of the N3 oligosaccharide (data not shown).
The oligosaccharide N3 in the DASH trace was analysed from fraction Ms10_40. The [M + Na]+ ion at m/z 748.1 corresponds to a Pent3 structure modified with one MeGlcA. The MALDI-CID reveals the structure of N3 as U(4Me)2XX (Additional file 1: Figure S4): The series of Y and cross-ring 1,5X ions gives crucial sequence information showing the positioning of the [Me]GlcA on the xylan backbone. The cross-ring 0,2X2 ion (m/z 655.9) indicates that the non-reducing-end Xylp is modified at the O-2 with a methylated uronic acid. The non-reducing end E2 (m/z 461.0) product ion confirms this linkage assignment and the presence of V4 (m/z 671.9) product ion indicates that the GlcA is modified with a methyl group at the O-4. The N3 oligosaccharide was susceptible to digestion with GH67 and GH115 glucuronidase (Table 2 and Additional file 1: Figure S2A), with the GH67-sensitivity confirming that the GlcA substitution is at the non-reducing end.
The oligosaccharide N4 in the DASH trace was analysed from fraction Ms10_75. The [M + Na]+ ion at m/z 558.1 corresponds to a Pent3 structure, but it does not co-migrate with Xyl3 in DASH, suggesting it is a substituted Xyl2. The MALDI-CID reveals the structure of N4 as A3X (Additional file 1: Figure S5): A major Y1 ion (m/z 294.1) is indicative of an unsubstituted reducing-end Xylp. The presence of the 0,2X1 ion (m/z 336.1) and the E2 product ion (m/z 271.1) indicates that the second Xylp from the reducing-end is unsubstituted at the O-2. The presence of the G3 ion (m/z 510.7) indicates the pentose substitution is Araf. Consistent with this, N4 oligosaccharide was sensitive to GH62 and GH51 arabinosidase digestions (Table 2).
The oligosaccharide N5 in the DASH trace was analysed from fraction Ms10_65. The [M + Na]+ ion at m/z 690.1 corresponds to a Pent4 structure, but it does not co-migrate with Xyl4 in DASH suggesting it is a substituted Xyl3. The MALDI-CID reveals the structure of N5 as XA3X (Additional file 1: Figure S6).
The 1,5X ion shows that the 2-AA-derivatised reducing-end Xylp is not modified. The cross-ring 0,2X1 ion (m/z 336.1) indicates that the O-2 in the middle Xylp is also not substituted. This is supported by the presence of the E2 (m/z 403.1) product ion, while the presence of the W2 sugar lactone ion (m/z 424.1) [29] indicates that the middle Xylp is substituted at O-3 with a pentose residue. The presence of the G3α ion (m/z 642.0) indicates the substitution is Araf. Consistent with this, the N5 oligosaccharide was sensitive to GH62 and GH51 arabinosidase digestions (Table 2).
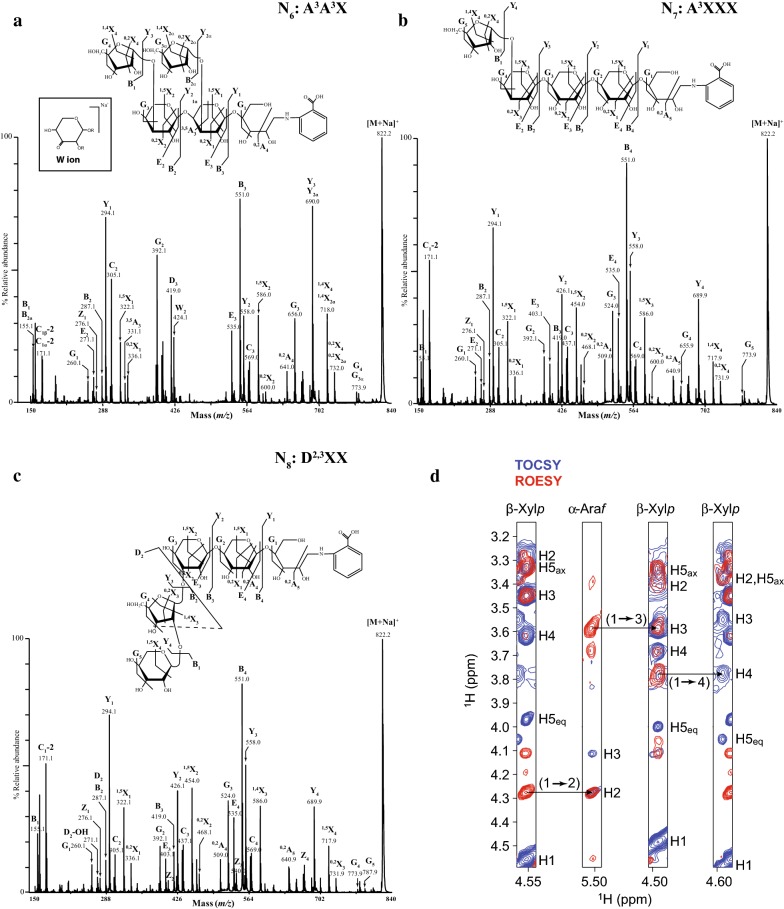
The oligosaccharides N6–N8 in the DASH trace were analysed from fraction Ms10_55 (Fig. 4a, the structure N9 was more abundant in fraction Ms10_45 and corresponds to a Pent6 oligosaccharide). N6–N8 oligosaccharides had the same m/z 822.0 [M + Na]+ corresponding to Pent5 structural isomers (Fig. 4b). The extracted ion chromatogram (EIC) from off-line HILIC-MALDI-CID mass spectrometry of 2-AA labelled SEC fractions showed that Ms10_55, contained three structural isomers of Pent5 observed as [M + Na]+ at m/z 822.0 (Z1–Z3, Fig. 4c).
Fig. 4.
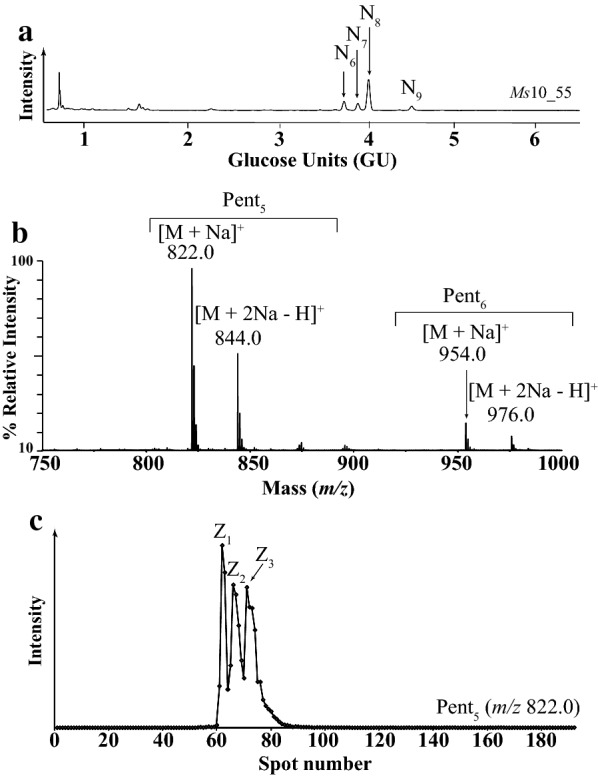
Characterisation of oligosaccharides in SEC fraction Ms10_55. a DASH capillary electropherograms showing four unknown oligosaccharides N6, N7, N8 and N9. b MALDI-ToF–MS of the 2-AA labelled oligosaccharides, showing a major sodiated and doubly sodiated ion corresponding to Pent5 (m/z 822.0, m/z 844.0, respectively) and a minor ion corresponding to Pent6 (m/z 954.0 and 976.0, respectively). c HILIC separation of structural isomers of m/z 822.0 [M + Na]+ followed by off-line-MALDI-ToF–MS, results in the EIC showing three structural isomers Z1, Z2 and Z3, respectively
Arabinofuranosidase digestion of the oligosaccharides followed by DASH and HILIC-MALDI showed that the oligosaccharides Z1, Z2 and Z3 correspond to N6, N7 and N8 (Table 2).
The MS/MS spectra for N6, N7 and N8 oligosaccharides are shown in Fig. 5.
Fig. 5.
High-energy MALDI-CID spectra of oligosaccharides in SEC fraction Ms10_55. a Z1 structure (N6 in DASH) A3A3X. Inset: proposed chemical structure for W product ion [29]; b Z2 structure (N7 in DASH) A3XXX; c Z3 structure (N8 in DASH) D2,3XX. d NMR analysis of the Z3 structure. H-1 strip plots from 2D 1H-1H TOCSY (blue) and ROESY (red) spectra showing the NOE connectivity arising from the β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-β-Xylp glycosidic linkages
The presence of the G ions (m/z 773.9) in the N6, N7 and N8 spectra indicates Araf substitution. The presence of the 1,5X1 cross-ring ion showed that the 2-AA labelled reducing-end Xylp is not substituted in any of the three structural isomers. The significant Y2, 1,5X2 and 0,2X2 ions (m/z 426.1, 454.0 and 468.1, respectively) in the spectra of N7 and N8 indicate that the second Xylp residue from the reducing end is also not substituted. In contrast, the absence of these ions from the N6 spectrum and the concomitant presence of the W2 (m/z 424.1) sugar lactone ion indicate that in this isomer the second Xylp residue from the reducing-end is substituted at O-3. The presence of the G3 product ion at m/z 656.0 in the spectrum of N6 indicates that the third Xylp residue from the reducing end is also substituted at the O-3 with Araf. This is confirmed by the presence of the E2 (m/z 271.1) product ion which indicates that the third Xylp from the reducing end is not substituted at O-2. Hence the N6 structure was identified as A3A3X. Consistent with this, the N6 oligosaccharide was sensitive to GH51 arabinosidase digestion. Possibly, due to steric hindrance of Araf substitutions on two consecutive Xylp residues, N6 was resistant to GH62 hydrolysis (Table 2).
The presence of the series of B, Y and 1,5X ions differing by 132 Da in the MALDI-CID spectra of the N7 and N8 structural isomers, indicates the linear sequence of pentoses. Based on the MALDI-CID spectra alone the precise structure of N7 and N8 oligosaccharides could not be identified. However, N7 was found to be sensitive to both GH62 and GH51 arabinofuranosidases, while N8 was resistant to both enzymes. Taking the MALDI-CID data and the information from enzyme sensitivity together, N7 was identified as A3XXX. N8 was further analysed by Nuclear Magnetic Resonance (NMR) to gain further insight into the detailed structure of this oligosaccharide (Fig. 5d and Additional file 2: Table S1).
Chemical-shift assignments were obtained using 2D 1H-1H TOCSY (TOtal Correlated SpectroscopY) and ROESY (Rotating-frame Overhauser Effect SpectroscopY) alongside 2D 13C HSQC (Heteronuclear Single Quantum Correlation) and HSQC-TOCSY experiments. The non-reducing-end Xylp residue was readily identified and the chemical shifts of the H-1 and C-1 were consistent with a β configuration. The Xylp (1 → 2) linkage to α-Araf was apparent from the intense NOE from Xylp H-1 to Araf H-2 taken together with the downfield shift of Araf C-2 characteristic of a glycosidic bond. The Araf-(1 → 3)-Xylp and Xylp-(1 → 4)-Xylp links were also confirmed by a combination of NOEs and the downfield 13C shifts of the linked carbon. H3 and H4 assignments of β-Xylp were apparent from relative TOCSY and NOESY intensities of the cross-peak connecting to H1 (both were stronger for H3). The reducing-end Xylp could not be identified due to peak broadening and overlap (the peaks for the reducing-end-adjacent Xylp are also significantly broadened). Our chemical-shift assignments are in accordance with previous 1H and 13C NMR analysis of a feruloylated D2,3X oligosaccharide from shoots of wiregrass (Cynodon dactylon) [40], except the nuclei involved in the feruloyl linkage. Therefore, structure N8 was identified as D2,3XX. In addition, this structure was found to be sensitive to a GH3 β-xylosidase (CgGH3) that cleaves terminal Xylp residues from β-Xylp-(1 → 2)-Araf-(1 → 3)-Xylp structures, confirming the assignment (Table 2).
The oligosaccharide N9 in the DASH trace was analysed from fraction Ms10_45. The [M + Na]+ ion at m/z 954.3 corresponds to a Pent6 structure. The MALDI-CID reveals the structure of N9 as XA3A3X (Additional file 1: Figure S7).
The 1,5X1 (m/z 322.1) and the Y1 (m/z 294.2) ions show that the reducing-end Xylp residue is unsubstituted. The reducing-end G2 (m/z 392.2) and G3 (m/z 656.1) product ions show that the second and third Xylp residues from the reducing end are substituted at O-3. The presence of the G3α/G4β ion (m/z 906.4, loss of 48 Da) indicate the presence of at least one terminal Araf. GH51 arabinosidase hydrolysis resulted in an oligosaccharide co-migrating with xylotetraose (X4; data not shown), confirming that both pentose substitutions are Araf residues. Similar to N6 oligosaccharide, N9 was resistant to GH62 arabinosidase hydrolysis (Table 2 and Additional file 1: Figure S2C).
Structural characterisation of the oligosaccharides N10, N11 and N12 from the xylanase GH11 digest of Miscanthus sinensis
After GH11 hydrolysis, four oligosaccharides co-eluted with X1, X2, X3 and X4 of the xylooligosaccharide standards; three oligosaccharides were structurally characterised with high-energy MALDI-CID and were identical to the previously characterised xylooligosaccharides: XU2XX and XU(4Me)2XX and XA3XX. Furthermore, three oligosaccharides, N1, N3 and N5, were identified based on their GU unit assigned from the GH10 library. However, we also identified three additional peaks from oligosaccharides of unknown structure which were named N10–N12. For the detailed structural characterisation of these oligosaccharides, the GH11 oligosaccharide mixture was loaded on a SEC column (Additional file 1: Figure S1 fractions Ms11_01 to Ms11_80) and unknown structures were consequently analysed by high-energy MALDI-CID.
The oligosaccharide N10 in the DASH trace was analysed from fraction Ms11_05. The [M + Na]+ ion at m/z 1012.8 corresponds to a Pent5 structure modified with one MeGlcA. The MALDI-CID reveals the structure of N10 as A3U(4Me)2XX (Additional file 1: Figure S8): The presence of the Y1 (m/z 294.3) and Y2 (m/z 426.3) ions indicate that the reducing end and the adjacent Xylp are unsubstituted. The E3 ion (m/z 403.4) and H3 (m/z 435.3) sugar lactone indicate that the third Xylp from the reducing end is modified at O-2 with a methylated glucuronic acid, while the V3α product ion (m/z 936.3) indicates that the GlcA is modified with a methyl group at O-4. The series of Y3 (m/z 748.3) and Y4 (m/z 880.2) indicate that two pentosyl residues are present at the non-reducing end while the absence of the non-reducing end 3,5A2 ion (m/z 199.0) indicates that the non-reducing end Xylp is substituted by a pentose. Finally the presence of the G4 reducing-end ion (m/z 846.3) is indicative of a substitution at O-3 of the reducing-end Xylp residue. The N10 oligosaccharide was found sensitive to GH51 hydrolysis indicating that the pentosyl modification on the fourth Xylp residue from the reducing end is an Araf residue (Table 2).
The oligosaccharide N11 in the DASH trace was analysed from fraction Ms11_70. The [M + Na]+ ion at m/z 1144.8 corresponds to a Pent6 structure modified with one MeGlcA. The MALDI-CID combined with enzymatic hydrolysis was not conclusive but indicates two putative structures of N11 as B2,3U(4Me)2XX or D2,3U(4Me)2XX (Additional file 1: Figure S9).
The presence of the Y1 (m/z 294.3) and Y2 (m/z 426.3) ions indicated that the reducing end and the adjacent Xylp are unsubstituted. The E4 ion (m/z 535.4) and H4 (m/z 567.3) sugar lactone indicate that the third Xylp from the reducing end is modified at O-2 with a MeGlcA. This assignment is also confirmed by the presence of the G3 reducing-end ion (m/z 714.3). The V3α product ion (m/z 1068.3) indicated that the GlcA was modified with a methyl group at the O-4 position. The presence of a series of Y ions, Y4 (m/z 880.2) and Y5 (m/z 1012.2), indicates that the reducing-end sugar sequence of this oligosaccharide is a linear pentose chain. However, the presence of G4 ion (m/z 846.3) indicated that the fourth Xylp residue from the reducing end is modified at O-3. Furthermore the reducing-end 0,2X4 ion (m/z 1054.3) is indicative of a modification at O-2 of the fifth pentosyl group from the reducing end and therefore pointing to a structure similar to the D2,3XX (Fig. 5c). N11 oligosaccharide was resistant to CgGH3 xylosidase which would be in accordance with the B2,3U(4Me)2XX assignment or could be explained by steric hindrance by MeGlcA modification on the adjacent Xylp residue in the case of D2,3U(4Me)2XX assignment (Table 2).
The oligosaccharide N12 in the DASH trace was analysed from fraction Ms11_05. The [M + Na]+ ion at m/z 1276.9 corresponds to a Pent7 structure modified with one MeGlcA. The MALDI-CID combined with enzymatic hydrolysis reveals the structure of N12 as XA3XU(4Me)2XX (Additional file 1: Figure S10).
The series of Y ions (Y1, Y2, Y3, Y4 and Y5; m/z 294.4, m/z 426.4, m/z 748.3, m/z 880.3 and m/z 1144.2, respectively) indicate that the oligosaccharide is modified on the third Xylp residue from the reducing end with [Me]GlcA and also on the fifth Xylp residue from the reducing end with a pentosyl group. The presence of H3 (m/z 699.4) sugar lactone confirms that the third Xylp residue is modified by a [Me]GlcA and indicates that this modification is at O-2. Additionally, the presence of G5 (m/z 978.3) reducing-end ions indicates that the fifth Xylp residue is modified at O-3. The V3α product ion (m/z 1200.4) indicates that the GlcA was modified with a methyl group at O-4. This oligosaccharide was found sensitive to GH51 and GH62 hydrolysis, indicating that the pentosyl modification on the fifth Xylp residue from the reducing end is an Araf residue, and to GH115 hydrolysis confirming that the uronic acid modification on the third Xylp from the reducing end is a [Me]GlcA (Table 2 and Additional file 1: Figure S2A).
Comparative structural analysis of GAX oligosaccharides derived from different grasses and different tissues
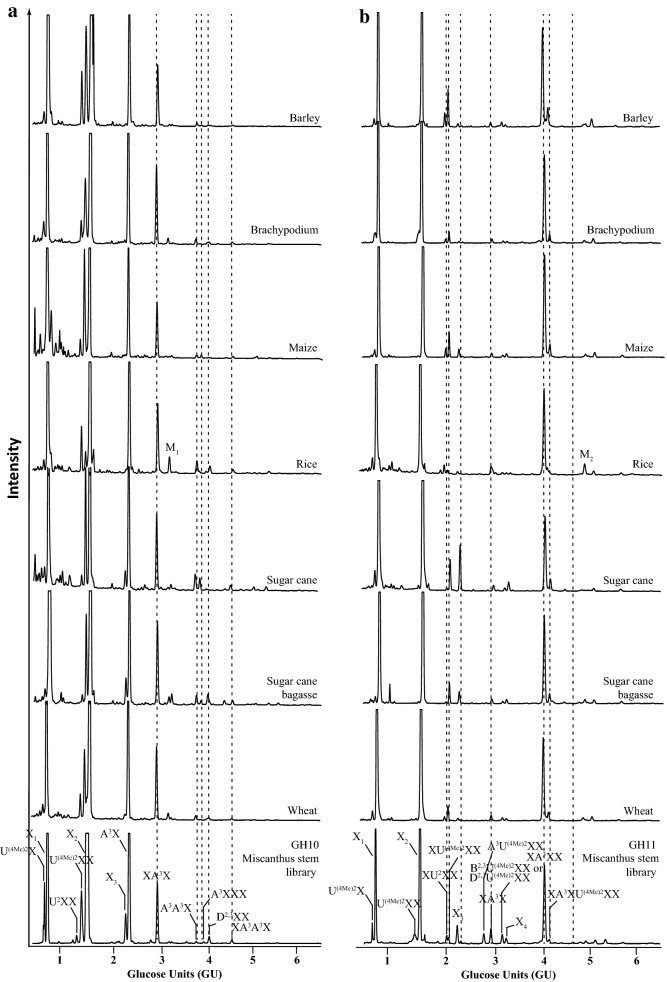
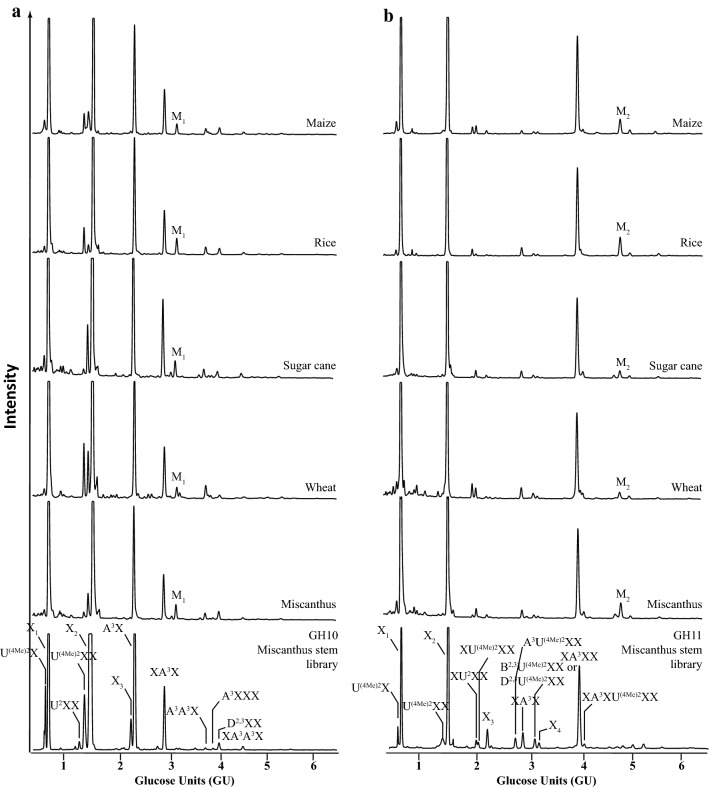
The developed reference library was used to characterise variation of GAX in agriculturally important grasses in different aerial tissues. We analysed the xylooligosaccharide products of GH10 and GH11 hydrolysis from stems and leaves of brachypodium, maize, rice, sugar cane, wheat, leaves of miscanthus and stems of barley as well as sugar cane bagasse (Figs. 6 and 7).
Fig. 6.
Comparison of DASH profiles of GAX oligosaccharides from stem of various grasses. a GH10 hydrolysis products; b GH11 hydrolysis products. Respective Miscanthus oligosaccharide library (bottom panel). Note the presence of two unknown peaks M1 and M2 in the rice DASH profile
Fig. 7.
Comparison of DASH profiles of GAX oligosaccharides from leaves of various grasses. a GH10 hydrolysis products; b GH11 hydrolysis products. Respective Miscanthus oligosaccharide library (bottom panel). Note the abundance of the two unknown peaks M1 and M2 in profile of leaf material
The DASH profiles of the xylooligosaccharides of the GH10 and the GH11 digests of all grasses analysed look very similar and nearly all peaks can be annotated based on the GU of their oligosaccharides. The main peaks are X1, X2, A3X and XA3X, respectively. Overall, arabinosylation appears more abundant than glucuronidation and the latter seems more variable in abundance across grasses. In addition, the ratio of methylated and unmethylated GlcA seems to widely differ in grasses with rice GAX being rather unmethylated, whereas major proportion of GlcA in, e.g. miscanthus and sugar cane GAX is methylated. There is also a slight variance in abundance of the more complex oligosaccharides. Moreover, we can detect additional peaks: The most characteristic peaks, M1 and M2, are present in the DASH profiles of leaf GAX from all grasses analysed, but very rare in GAX of stem with the exception of rice stem GAX, in which M1 and M2 are substantial.
Structural characterisation of the oligosaccharides M1 and M2 from leaf GAX
In order to identify the unknown oligosaccharides M1 and M2, maize leaf GH10 and rice leaf GH11 oligosaccharide products, respectively, were separated by SEC. The suitable SEC fractions (Zm10_65 for structure M1 and Os11_60 for structure M2) were further analysed by MALDI-ToF/ToF Mass Spectrometry, structural isomers were separated by HILIC and structurally characterised by high-energy MALDI-CID (Additional file 1: Figure S11).
The oligosaccharide M1 in the DASH trace was analysed from fraction Zm10_65 (Additional file 1: Figure S11A). The unknown oligosaccharide had an m/z 690.00 [M + Na]+, corresponding to a Pent4 structure (Additional file 1: Figure S11B). It did not co-migrate with Xyl4 in DASH, suggesting it is a substituted xylooligosaccharide. Off-line HILIC-MALDI-MS separated the two structural isomers with m/z 690.00 (M1 and XA3X, Additional file 1: Figure S11C) and the respective MALDI-CID spectra revealed the structure of M1 as D2,3X (Additional file 1: Figure S12): The presence of a series of Y ions (Y1, Y2 and Y3; m/z 294.3, 426.3 and 558.3, respectively) and the presence of 1,4X2 and 1,5X3 ions (m/z 454.3 and 586.3, respectively) indicate that the M1 is a linear DP4 pentose chain. The presence, however, of the G3 product ion (m/z 642.4, loss of 48 Da) indicates the existence of an Araf residue in the structure. In addition, the presence of the G4 product ion (m/z 656.5) gives evidence of a terminal Xylp residue on the structure. Finally, the presence of the 0,2X2 cross-ring ion (m/z 600.3) suggests that a pentose is 2-linked on the penultimate Xylp residue. The assignment of M1 as D2,3X was confirmed by the finding that the structure is resistant to the hydrolysis with GH62 arabinosidase, GH67 and GH115 glucuronidases, but sensitive to CgGH3 β-xylosidase hydrolysis (Table 2 and Additional file 1: Figure S2D).
The oligosaccharide M2 in the DASH trace was analysed from fraction Os11_60 (Additional file 1: Figure S11D). The unknown oligosaccharide had an m/z 954.5 [M + Na]+, corresponding to a Pent6 structure (Additional file 1: Figure S11E). It did not co-migrate with Xyl6 in DASH, suggesting it is a substituted xylooligosaccharide. Off-line HILIC-MALDI-CID revealed that this was the only structural isomer present in the sample (Additional file 1: Figure S11F). The MALDI-CID revealed the structure of M2 as XD2,3XX (Additional file 1: Figure S13): The significant Y1 ion (m/z 294.4) and the 1,5X1 ion (m/z 322.4) indicate that the 2-AA labelled Xylp residue is not substituted. The Y2 and Y3 ions (m/z 426.4 and 690.4, respectively) show that the second Xylp is unsubstituted but that the third Xylp residue from the reducing end is modified with two pentose residues. The reducing-end G3 (m/z 524.5) product ion and the concomitant presence of the W3 (m/z 556.4) sugar lactone indicate that the substitution on the third Xylp from the reducing end is at O-3. This assignment is further verified by the presence of the D2 (m/z 271.4) product ion. The existence of the G4 (m/z 906.5) product ion indicates the presence of an Araf substitution. Finally, the presence of the 3,5A2 non-reducing end cross-ring fragment ion indicates the presence of an unsubstituted Xylp at the non-reducing end. The assignment of M2 as XD2,3XX was confirmed by the finding that the structure is resistant to the hydrolysis with GH62 arabinosidase, GH67 and GH115 glucuronidases, but sensitive to CgGH3 β-xylosidase hydrolysis (Table 2 and Additional file 1: Figure S2D).
Relative quantitative differences in oligosaccharide structure and abundance in Miscanthus sinensis stem and leaf GAX
By DASH the relative quantity of reducing-end labelled oligosaccharides within a sample can be determined by the relative fluorescence intensity of the corresponding electropherogram peaks [27]. To analyse the difference in GAX structure in stems and leaves in greater detail, we quantified the frequency of substitution of all characterised library oligosaccharides in the GH10 and GH11 digests of three biological replicates of miscanthus using the DASHboard software (Fig. 8). The substitution frequency was calculated after normalisation of values by comparing the abundance of side chains to the Xylp residues in the backbone.
Fig. 8.
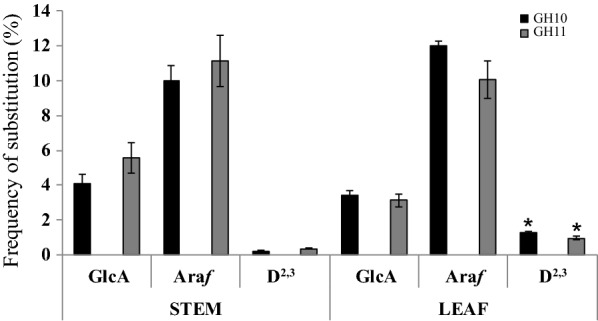
Quantification of substitution frequency of miscanthus stem and leaf GAX. All characterised GH10 and GH11 hydrolysis products were analysed by DASH using the DASHboard software for quantification. The substitution frequency was calculated after normalisation of values by comparing the abundance side chain to the Xylp residues in the backbone. Error bars represent mean ± SD (n = 3). A significant difference was observed in the D2,3 substitution frequency of the GH10 digest (p = 0.002) and of the GH11 digest (p = 0.2) between stems and leaves and between the GH10 and G11 digest in leaves only (p = 0.01) using the paired t-test for two-tailed distribution, marked with asterisk
First of all, the overall substitution frequency of xylan with [Me]GlcA, Araf and D2,3 calculated from the two different digests with GH10 and GH11, respectively, is consistent with only a minor difference for the D2,3 structure in leaves, showing a slightly higher frequency in the GH10 digest only (1.3% versus 1%, p value 0.01).
In both, stems and leaves of miscanthus, arabinosylation of xylan is more frequent than glucuronidation, ranging between 10 and 12% of Araf substitutions versus 3–6% of [Me]GlcA substitutions. However, there is a significant difference between tissues regarding the amount of D2,3 substitution both in the GH10 (p value 0.002) and the GH11 (p value 0.02) digests. In leaves the D2,3 substitution frequency of the GH10 and GH11 digest is 1.3% and 1%, respectively, whereas in stem it is down to 0.2% and 0.4%, respectively. Concomitantly, with the presence of the additional D2,3 structure (D2,3X and XD2,3XX) in leaf xylan, our data show that the frequency of the D2,3 structure is increased in leaves.
Discussion
In this study, we used a combination of SEC, DASH, HILIC and MALDI-CID to elucidate the detailed structure of grass GAX oligosaccharides released by GH10 and GH11 xylanases. Based on these data, we developed a DASH reference library of structurally identified oligosaccharides and expressed their migration in GU. It has been reported previously that DASH can separate oligosaccharides with minor structural differences like methylation of GlcA [27]. Here, we show that DASH can successfully resolve structural isomers as demonstrated by the separation of the three DP5 oligosaccharides (m/z 822.0). Few approaches allow for wider screening of cell wall polysaccharide structures. Microarray-based glycan-profiling techniques [41, 42], for example, integrate the sequential extraction of glycans with the generation of microarrays, which are probed with monoclonal antibodies (mAbs) or carbohydrate-binding molecules (CBMs) with specificities for cell wall components. Although Microarray-based glycan profiling is a powerful technique offering high-throughput analysis of a wider variety of cell wall polymers, it is limited by the availability of mAbs and CBMs and their epitope specificity. A non-destructive high-throughput method for the compositional analysis of plant cell wall polymers is Fourier Transformed Mid-Infrared (FT-IR) spectroscopy [43]. This approach can provide structural information about substitution nature and frequency of cereal arabinoxylan [44]. The DASH reference library generated here allows the fast and robust comparison of a large number of GAX samples (96 samples in 50 min), providing detailed structural and quantitative information on biomass structural variation. Information gathered from DASH analysis will ultimately greatly facilitate the biorefining selection process of appropriate biomass.
We analysed the GAX structure of a number of different grass species (miscanthus, barley, brachypodium, maize, rice, sugar cane and wheat). The DASH profiles of all grasses analysed and the quantification data of xylan substitution of Miscanthus sinensis showed that Araf side chains are more frequent than [Me]GlcA substitutions, which is consistent with sugar composition analysis of wheat straw [26, 45] and MS and NMR analysis of miscanthus, rice and brachypodium of the entire actively growing aerial portions [26]. Unlike cereal grain AX where 2-linked Araf residues are abundant [1, 46–48], we did not detect any such substitutions, which is consistent with earlier reports on the structure of grass GAX in lignified tissues [24, 49, 50]. The overall xylan structure of the different grass species and of the different tissues is remarkably conserved, which is in line with earlier studies on grass xylan structure [51]. However, differences between species and tissues are detectable. Interestingly, the xylan structure of rice stems appears more similar to the xylan structure in leaves than to other xylan stem structures, forming an exception in the grasses analysed, perhaps because of the immature developmental stage of the culms collected. Plants exploit a number of variations in the xylan structure that were not studied in this work, e.g. the pattern of substitutions along the xylan backbone, feruloylation and coumaroylation of Araf and acetylation of backbone Xylp. These could be studied by exploitation of additional carbohydrate active enzymes and generation of additional standards in the DASH mobility library.
The most characteristic tissue-specific differences were identified in the form of the two oligosaccharides D2,3X and XD2,3XX, which are (apart from in rice) scarce in stem xylan but abundant in leaf xylan. Quantification of the overall frequency of the D2,3 side chain in miscanthus suggests that it is significantly more abundant in leaves than in stem. The D2,3 structure has been linked to feruloylation of xylan, and hence cross-linking and reduced digestibility [8]. Interestingly, the lignin amount and composition also differs in cell walls of miscanthus stems versus leaves [52]. Therefore xylan structural changes in different tissues might reflect specific levels and positioning of cross-linking, which can be a way of adjusting polymer structures to different functional requirements of the cell wall depending on its particular composition. A disaccharide side chain on xylan has been reported in sorghum [53] and switchgrass [54] although not detected by Bowman et al. [24] in a similar xylan analysis of switchgrass. The presence of this disaccharide side chain cannot be excluded although with our approach all disaccharide side-chain modifications positively identified were β-Xylp-(1 → 2)-α-Araf-(1 → 3) structures. The only exception was the GH11 product, putatively assigned as D2,3U(4Me)2XX, which was found resistant to CgGH3 β-xylosidase. Resistance to CgGH3 β-xylosidase could either be due to steric hindrance of the enzyme or could indicate the presence of an Araf-(1 → 2)-α-Araf-(1 → 3) side chain on this GH11 hydrolysis product (B2,3U(4Me)2XX). However, the Araf-(1 → 2)-Araf-(1 → 3)- adjacent to a Xylp modified by a GlcA residue would be in a different substitution context to the Mazumder et al. [54] and Verbruggen et al. [48] reported oligosaccharides. If the latter is true then we could hypothesise that GH10 and GH11 xylanases hydrolyse slightly different parts of GAX. It is also, however, possible that the Araf-(1 → 2)-α-Araf-(1 → 3) side chain is not present in the tissues and plant species analysed here, or that it is present in amounts below the detection level.
It has been reported that the degree of methylation of GlcA on xylan varies between grass species and that in miscanthus methylation of xylan is more predominant than in, for example, rice or Brachypodium [49, 50]. This finding is consistent with our data on miscanthus xylan.
Some qualitative differences of oligosaccharides reflect enzyme specificity and distinct tolerance of the xylanases GH10 and GH11 to substitutions [28], for example, A3X from GH10 and XA3XX from GH11. However, some oligosaccharide such as A3U(4Me)2XX, D2,3U(4Me)2XX (or B2,3U(4Me)2XX) and XA3XU(4Me)2XX, although only minor products, were only detectable in GH11 digestion and might indicate that GH10 and GH11 act on different domains of grass xylan as they resemble different substitution patterns. The fact that the level of 3-linked Araf and [Me]GlcA substitutions is remarkably similar independent of the xylanase used does not necessarily support this hypothesis. Surprisingly, some of these minor digestion products of GH11 harbour substitutions at the non-reducing end of the oligosaccharide, which are not predicted as products and might be the result of either using the enzymes in excess or that the GH11 enzyme preparation used in this study was contaminated with small amounts of either GH10 xylanase or β-xylosidase. Alternatively, these oligosaccharides could derive from the non-reducing end of the xylan polymer and would be produced by a single cleavage at the reducing end of the oligosaccharide.
Conclusions
As characterised here in grass stems and leaves from several species, mainly three GAX sugar side chains, 3-linked Araf, 2-linked GlcA/MeGlcA and 3-linked Xylp-(1 → 2)-Araf, are utilised to decorate the xylan molecule. Our GAX oligosaccharide reference library of DASH mobilities, developed in this study, provides a means to study structural aspects of xylan and might help to shed light on how structural changes of xylan correlate with the interaction of this polysaccharide with other cell wall components, how this influences its biological function, its mechanical properties and recalcitrance of the cell wall. Furthermore it provides a high-throughput quantitative method for the selection of suitable lignocellulosic biomass and tailoring of biorefining processes.
Methods
Plant material
The plant materials used in this study were fresh material from Miscanthus sinensis (miscanthus; Wageningen University, Netherlands, Luisa Trindade & Oene Dolstra), Hordeum vulgare (barley; University of Dundee, Claire Halpin), Brachypodium distachyon (Brachypodium; (grown at University of Cambridge greenhouse), Zea mays (maize; University of Cambridge, Paul Dupree), Oryza sativa (rice; grown at University of Cambridge greenhouse), Saccharum officinarum (sugar cane; EMBRAPA-Brazil, Christiane Farinas), sugar cane bagasse (University of Sao Paulo-Brazil, Marcos Buckeridge) and Triticum aestivum (wheat; University of Nottingham, Greg Tucker).
Extraction of alcohol-insoluble residue (AIR)
Plant stems and leaves were harvested, submerged in 96% (v/v) ethanol and boiled at 70 °C for 30 min to inactivate enzymes. Following homogenisation using a ball mixer mill (Glen Creston), the pellet was collected by centrifugation (4000 x g for 15 min) and was washed with 100% (v/v) ethanol, twice with chloroform:methanol (2:1), followed by successive washes with 65% (v/v), 80% (v/v) and 100% (v/v) ethanol. The remaining pellet of AIR was air dried. Aqueous suspensions (0.5 mg/ml; at 21 °C) of AIR were prepared using a glass homogeniser and kept for further analysis.
Hemicellulose extraction
Hemicelluloses were extracted by treating AIR preparations (2 g for Size exclusion chromatography (SEC) fractionation; 50 μg for small-scale digestions) with a small volume of 4 M NaOH (5 ml for SEC fractions; 20 μl for small-scale digestions) for 1 h at room temperature before the pH was adjusted to about pH 6.0 with 1 N HCl; 50 mM ammonium acetate buffer pH 6.0 was added (200 ml for SEC fractionation; 0.5 ml for small-scale digestions). Note: Alkali treatments results in the removal of acetylation and feruloylation.
Enzymatic hydrolysis and enzymes
Glycoside hydrolases of different Carbohydrate Active enZYme families were used in this study: GH10 endo-β-1,4-xylanase CjGH10A from Cellvibrio japonicus [55]; GH11 endo-β-1,4-xylanase NpGH11A from Neocallimastix patriciarum [56]; GH67 α-glucuronidase CjGH67 from Cellvibrio japonicus [37, 57] and GH115 α-glucuronidase BoGH115 from Bacteroides ovatus [38, 58]; GH62 α-arabinofuranosidase PaGH62 from Penicillium aurantiogriseum [59], a gift from Novozymes; GH51 α-arabinofuranosidase PcGH51 from Pseudomonas cellulose [36]; GH3 xylosidase CgGH3 from Chaetomium globosum (NS39127) and GH3 β-1,4 xylosidase TrGH3 from Trichoderma reesei [60] were both gifts from Novozymes. All enzymes were added at a final concentration of 2 μM and incubated at 21 °C under constant shaking for 24 h. Enzymatic hydrolysis progression was monitored by Polysaccharide Analysis Using Carbohydrate Gel Electrophoresis [61] and if necessary, the enzyme amount was adjusted to ensure complete hydrolysis.
Enzyme deactivation and removal of undigested material
Enzymes were then deactivated by boiling for 30 min undigested cell wall material was removed by centrifugation. In case of SEC, centrifugation was followed by filtration (Whatman 45 μm). Samples were then dried in a centrifugal evaporator.
Size exclusion chromatography
Dried hydrolysed cell wall materials (corresponding to 2 g of AIR) were resuspended in 2 ml distilled water. Sample (2 ml) was applied onto the column and eluted with distilled water. SEC was performed on a gravity-driven Bio-Gel P-2 column (190 × 2.5 cm, BioRad) as previously described [62]. 2 ml fractions were collected, concentrated to 100 μl in a centrifugal evaporator and 10 μl were analysed by DASH as described below. A total of 80 SEC fractions were collected for each grass species and xylanase hydrolysis analysed. The SEC fraction naming system includes information on the species (Ms: Miscanthus sinensis, Zm: Zea mays and Os: Oryza sativa), the xylanase (10: GH10 and 11: GH11) and the fraction number (_01 to _80). The fractions with xylooligosaccharides of interest were subjected to secondary enzymatic hydrolysis or dried, desalted, reductively aminated with 2-anthranilic acid (2-AA), purified by HILIC and structurally characterised by high-energy MALDI-CID.
APTS labelling and analysis by DASH
The derivatisation of oligosaccharides with 8-aminopyrene-1,4,6-trisulfonate (APTS) was performed according to previously developed protocols [27]. A set of 7 fluorophore (DY-481XL-NHS ester)-labelled amino acids and peptides was used as electrophoretic mobility standards (Asp–Asp–Asp–Asp; Asp–Asp–Asp; Glu–Glu; Cysteic acid; l-2-Aminoadipic acid; Glycine; Gly–Gly–Gly) to align the electropherograms. These electrophoretic mobility standards were mixed with each sample prior to DASH separation serving as internal mobility markers. DASH data generated by the DNA sequencer were processed with the DASHboard software [27]. Control experiments without the substrates were performed under the same conditions in order to identify any non-specific compounds in the enzymes or labelling reagents. An APTS-derivatised dextran ladder (0.1 M TFA hydrolysis at 100 °C for 2 h; 50 μg μl−1 dextran in 200 μl TFA solution) was simultaneously separated by DASH and was used to provide the GU migration positions.
Desalting and clean up for HILIC separation
Xylooligosaccharides from SEC fractions were desalted using HyperSep Hypercarb cartridges (Thermo-Hypersil-Keystone, Runcorn, Cheshire, UK) as previously described [29]. Oligosaccharides were lyophilised and then derivatised with 2-aminobenzoic acid (2-AA) as described below.
Reductive amination and purification for HILIC separation
SEC-purified xylooligosaccharides were reductively aminated with 2-AA (Sigma) and then purified from the reductive amination reagents using a Glyko Clean S cartridge (Prozyme, San Leandro, CA) as previously described [63].
HILIC-MALDI-MS and MALDI-MS/MS CID analysis
Capillary HILIC was carried out using an LC-Packings Ultimate system (Dionex, CA, USA) equipped with an amide-80 column (300 μm × 25 cm; 3 μm particle size; Dionex) as previously described [63]. Briefly, the LC system was used to generate the gradient that flowed at 3 μl min−1. Solvent A was 50 mm ammonium formate adjusted to pH 4.4 with formic acid. Solvent B was 5% solvent A in acetonitrile. The labelled oligosaccharides dissolved in 95% acetonitrile were loaded onto the column (20 μl) and eluted with increasing aqueous concentrations. The following gradient conditions were applied: 0 min, 5% solvent A, 95% solvent B; 6 min, 25% solvent A, 75% solvent B; 86 min, 45% solvent A, 55% solvent B. The system operated at ambient temperature. The column eluent passed through a capillary UV detector (set at 254 nm) to the MALDI sample spotter. For HILIC-MALDI-ToF/ToF Mass Spectrometry, a Probot sample fraction system (Dionex) was employed for automated spotting of the HPLC eluent onto a MALDI target at 20 s intervals. After air drying, the sample spots were overlaid with 0.5 μl 2,5-DHB matrix (1 mg ml−1 in 50% aqueous methanol) and analysed by MALDI-ToF/ToF–MS on an AB-Sciex 4700. The MS spectra were obtained in automatic mode with an average 1500 laser shots/spectrum (mass range 400–2500 Da). The oligosaccharide molecular ions [M + Na]+ were identified in the MALDI data and their HILIC elution positions were determined by carrying out an extracted ion chromatogram (EIC). High-energy MALDI-CID spectra were acquired with an average 10,000 laser shots/spectrum, using a high collision energy (1 kV). The oligosaccharide ions were allowed to collide in the CID cell with argon at a pressure of 2 × 10−6 Torr.
NMR analysis
Saponified miscanthus AIR was hydrolysed with GH10 xylanase, followed by GH115 xylan glucuronidase, GH51 arabinofuranosidase and TrGH3 β-1,4-xylanase using enzyme hydrolysis conditions described above. The resulting N8 oligosaccharide was then isolated by SEC as described above. Consequently, SEC fractions containing the N8 oligosaccharide were pooled, solubilised in 0.6 ml D2O and analysed by NMR.
NMR spectra were recorded at 298 K with a Bruker AVANCE III spectrometer operating at 600 MHz equipped with a TCI CryoProbe. Two-dimensional 1H-1H TOCSY, ROESY, 13C HSQC and HSQC-TOCSY experiments were performed, using established methods [64]; the mixing times were 70 ms and 200 ms for the TOCSY and ROESY experiments, respectively. Chemical shifts were measured relative to internal acetone (δH = 2.225, δC = 31.07 ppm). Data were processed using the Azara suite of programs (v. 2.8, copyright 1993–2014, Wayne Boucher and Department of Biochemistry, University of Cambridge, unpublished) and chemical-shift assignment was performed using Analysis v2.2 [65].
Oligosaccharide naming system
The various hydrolysis products are named according to the Faure et al. [30] naming system. This naming system utilises a single letter code where the uppercase letters identify the substituents of the main xylan chain. The letter “A” is attributed to single Araf substitution, the letter “U” is used for single GlcA substitution and the letter “X” for unsubstituted Xylp residues. The superscript numbers indicate substitution linkage position on Xylp. Information of side-chain modifications is included in the superscript part of the name, for example, “Me” for methylation. Finally, further substitutions of the side chains receive a new letter assignment, for example, the Araf-(1 → 2)-α-Araf-(1 → 3) side chain has been designated the “B2,3” character and the β-Xylp-(1 → 2)-α-Araf-(1 → 3) substitution has been designated the “D2,3” character [30].
DASHboard software and substitution frequency quantitation
Data generated by the DNA sequencer were processed in DASHboard software [27] which was developed to complement the profiling technique and perform tasks such as visualisation of data, alignment of electropherograms, peak area quantification and export to Excel for further analysis.
Relative quantitation of substitution frequency was calculated after normalisation of values by comparing the abundance of side chains to the Xylp residues in the backbone. Value normalisation allowed the accurate peak area calculation by DASH software because electropherograms from higher dilution were utilised for the peak area calculation of highly abundant mono- and oligosaccharides (such as xylose and xylobiose) and the electropherograms of same samples but of lower dilution were utilised for peak area calculation of less abundant oligosaccharides. The peak area ratio between each of the xylanase oligosaccharide products and a reference oligosaccharide (XA3X and XA3XX for GH10 and GH11 digestions, respectively) resulted in the normalised values for each oligosaccharide. The total backbone Xylp present in the digested xylan was calculated as the sum of the relative quantity of each of these digestion products multiplied by the number of Xylp residues present in each structure (total backbone Xylp). For each specific side chain, side-chain substitution was calculated as the sum of the relative quantity of each of these side chains (Araf, [Me]GlcA and D2,3) multiplied by the number of side chains present in each structure (side-chain substitution). Hence, side-chain substitution frequency was calculated as the ratio between (side-chain substitution) and (total backbone Xylp).
Additional files
Additional file 2: Table S1. 1H and 13C NMR assignments of β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp, at 25 °C in D2O.
Authors’ contributions
TT designed the experimental part and carried out the analysis of xylooligosaccharides. MS and CF supported the analysis. KS performed the NMR analysis. DVR updated the DASHboard software. TT, NA and PD analysed the data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Tina Theys and Dr. Rita Marques for technical support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article and its additional files.
Consent for publication
All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Funding
The work was funded by the BBSRC (BSBEC BIOMASS BB/G016216/1 and BB/K005537/1) and the European Community’s Seventh Framework Programme SUNLIBB (FP7/2007–2013) under the Grant Agreement No. 251132 to P.D. The Cambridge NMR facility infrastructure is funded by the BBSRC and the Wellcome Trust. We are grateful for plant material from Aberystwyth, Rothamsted and Wageningen University, NL.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DASH
DNA sequencer-Assisted Saccharide analysis in High throughput
- Araf
arabinofuranose/arabinofuranosyl
- Xylp
xylopyranose/xylopyranosyl
- [Me]GlcA
[methyl]glucuronic acid/glucuronyl
- (G)AX
(glucurono)arabinoxylan
- AIR
alcohol-insoluble residue
- GH
glycosyl hydrolase
- GU
glucose units
References
- 1.Ebringerová A, Heinze T. Xylan and xylan derivatives—biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun. 2000;21:542–556. [Google Scholar]
- 2.Saulnier L, Vigouroux J, Thibault J-F. Isolation and partial characterization of feruloylated oligosaccharides from maize bran. Carbohydr Res. 1995;272(2):241–253. doi: 10.1016/0008-6215(95)00053-v. [DOI] [PubMed] [Google Scholar]
- 3.Tian L, Gruppen H, Schols HA. Characterization of (Glucurono)arabinoxylans from oats using enzymatic fingerprinting. J Agric Food Chem. 2015;63(50):10822–10830. doi: 10.1021/acs.jafc.5b04419. [DOI] [PubMed] [Google Scholar]
- 4.Hatfield RD, Rancour DM, Marita JM. Grass cell walls: a story of cross-linking. Front Plant Sci. 2017;7:2056. doi: 10.3389/fpls.2016.02056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller-Harvey I, Hartley RD, Harris PJ, Curzon EH. Linkage of p-coumaroyl and feruloyl groups to cell-wall polysaccharides of barley straw. Carbohydr Res. 1986;148(1):71–85. [Google Scholar]
- 6.de Buanafina OMM. Feruloylation in grasses: current and future perspectives. Mol Plant. 2009;2(5):861–872. doi: 10.1093/mp/ssp067. [DOI] [PubMed] [Google Scholar]
- 7.Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127(2):111–127. [Google Scholar]
- 8.Wende G, Fry SC. 2-O-β-d-xylopyranosyl-(5-O-feruloyl)-l-arabinose, a widespread component of grass cell walls. Phytochemistry. 1997;44(6):1019–1030. [Google Scholar]
- 9.Chiniquy D, Sharma V, Schultink A, Baidoo EE, Rautengarten C, Cheng K, Carroll A, Ulvskov P, Harholt J, Keasling JD, et al. XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc Natl Acad Sci USA. 2012;109(42):17117–17122. doi: 10.1073/pnas.1202079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appeldoorn MM, de Waard P, Kabel MA, Gruppen H, Schols HA. Enzyme resistant feruloylated xylooligomer analogues from thermochemically treated corn fiber contain large side chains, ethyl glycosides and novel sites of acetylation. Carbohydr Res. 2013;381:33–42. doi: 10.1016/j.carres.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Allerdings E, Ralph J, Steinhart H, Bunzel M. Isolation and structural identification of complex feruloylated heteroxylan side-chains from maize bran. Phytochemistry. 2006;67(12):1276–1286. doi: 10.1016/j.phytochem.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Appeldoorn MM, Kabel MA, Van Eylen D, Gruppen H, Schols HA. Characterization of oligomeric xylan structures from corn fiber resistant to pretreatment and simultaneous saccharification and fermentation. J Agric Food Chem. 2010;58(21):11294–11301. doi: 10.1021/jf102849x. [DOI] [PubMed] [Google Scholar]
- 13.Coelho E, Rocha MAM, Moreira ASP, Domingues MRM, Coimbra MA. Revisiting the structural features of arabinoxylans from brewers’ spent grain. Carbohydr Polymers. 2016;139:167–176. doi: 10.1016/j.carbpol.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Schendel RR, Meyer MR, Bunzel M. Quantitative profiling of feruloylated arabinoxylan side-chains from graminaceous cell walls. Front Plant Sci. 2016;6:1249. doi: 10.3389/fpls.2015.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii T. Acetylation at O-2 of arabinofuranose residues in feruloylated arabinoxylan from bamboo shoot cell-walls. Phytochemistry. 1991;30(7):2317–2320. doi: 10.1016/0031-9422(91)83639-3. [DOI] [PubMed] [Google Scholar]
- 16.McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Curr Opin Plant Biol. 2008;11(3):314–320. doi: 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Simmons TJ, Mortimer JC, Bernardinelli OD, Pöppler A-C, Brown SP, deAzevedo ER, Dupree R, Dupree P. Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat Commun. 2016;7:13902. doi: 10.1038/ncomms13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeMartini JD, Pattathil S, Miller JS, Li HJ, Hahn MG, Wyman CE. Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energy Environ Sci. 2013;6(3):898–909. [Google Scholar]
- 19.Hoije A, Sandstrom C, Roubroeks JP, Andersson R, Gohil S, Gatenholm P. Evidence of the presence of 2-O-β-d-xylopyranosyl-α-l-arabinofuranose side chains in barley husk arabinoxylan. Carbohydr Res. 2006;341(18):2959–2966. doi: 10.1016/j.carres.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Marcotuli I, Hsieh YSY, Lahnstein J, Yap K, Burton RA, Blanco A, Fincher GB, Gadaleta A. Structural variation and content of arabinoxylans in endosperm and bran of durum wheat (Triticum turgidum L.) J Agric Food Chem. 2016;64(14):2883–2892. doi: 10.1021/acs.jafc.6b00103. [DOI] [PubMed] [Google Scholar]
- 21.Pastell H, Virkki L, Harju E, Tuomainen P, Tenkanen M. Presence of α-linked 2-O-β-d-xylopyranosyl-α-l-arabinofuranosyl side chains in cereal arabinoxylans. Carbohydr Res. 2009;344(18):2480–2488. doi: 10.1016/j.carres.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Toole G, Wilson R, Parker M, Wellner N, Wheeler T, Shewry P, Mills E. The effect of environment on endosperm cell-wall development in Triticum aestivum during grain filling: an infrared spectroscopic imaging study. Planta. 2007;225(6):1393–1403. doi: 10.1007/s00425-006-0448-0. [DOI] [PubMed] [Google Scholar]
- 23.Ordaz-Ortiz JJ, Devaux M-F, Saulnier L. Classification of wheat varieties based on structural features of arabinoxylans as revealed by endoxylanase treatment of flour and grain. J Agric Food Chem. 2005;53(21):8349–8356. doi: 10.1021/jf050755v. [DOI] [PubMed] [Google Scholar]
- 24.Bowman MJ, Dien BS, Vermillion KE, Mertens JA. Structural characterization of (1 → 2)-β-xylose-(1 → 3)-α-arabinose-containing oligosaccharide products of extracted switchgrass (Panicum virgatum, L.) xylan after exhaustive enzymatic treatment with α-arabinofuranosidase and β-endo-xylanase. Carbohydr Res. 2014;398:63–71. doi: 10.1016/j.carres.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni A, Pena MJ, Avci U, Mazumder K, Urbanowicz B, Pattathil S, Yin Y, O’Neill MA, Roberts AW, Hahn MG, et al. The ability of land plants to synthesize glucuronoxylans predates the evolution of tracheophytes. Glycobiology. 2011;22(3):1–15. doi: 10.1093/glycob/cwr117. [DOI] [PubMed] [Google Scholar]
- 26.Peña MJ, Kulkarni AR, Backe J, Boyd M, O’Neill MA, York WS. Structural diversity of xylans in the cell walls of monocots. Planta. 2016;244(3):589–606. doi: 10.1007/s00425-016-2527-1. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Jackson P, Rubtsov D, Faria-Blanc N, Mortimer J, Turner S, Krogh K, Johansen K, Dupree P. Development and application of a high throughput carbohydrate profiling technique for analyzing plant cell wall polysaccharides and carbohydrate active enzymes. Biotechnol Biofuels. 2013;6(1):94. doi: 10.1186/1754-6834-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biely P, Vrsanská M, Tenkanen M, Kluepfel D. Endo-[beta]-1,4-xylanase families: differences in catalytic properties. J Biotechnol. 1997;57(1–3):151–166. doi: 10.1016/s0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- 29.Maslen SL, Goubet F, Adam A, Dupree P, Stephens E. Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydr Res. 2007;342(5):724–735. doi: 10.1016/j.carres.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Faure R, Courtin CM, Delcour JA, Dumon C, Faulds CB, Fincher GB, Fort S, Fry SC, Halila S, Kabel MA, et al. A brief and informationally rich naming system for oligosaccharide motifs of heteroxylans found in plant cell walls. Aust J Chem. 2009;62(6):533–537. [Google Scholar]
- 31.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal Biochem. 1996;240(2):210–226. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 32.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5(4):397–409. [Google Scholar]
- 33.Chai W, Piskarev V, Lawson AM. Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal Chem. 2001;73(3):651–657. doi: 10.1021/ac0010126. [DOI] [PubMed] [Google Scholar]
- 34.Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P. GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 2013;74(3):423–434. doi: 10.1111/tpj.12135. [DOI] [PubMed] [Google Scholar]
- 35.Spina E, Sturiale L, Romeo D, Impallomeni G, Garozzo D, Waidelich D, Glueckmann M. New fragmentation mechanisms in matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry of carbohydrates. Rapid Commun Mass Spectrom. 2004;18(4):392–398. doi: 10.1002/rcm.1350. [DOI] [PubMed] [Google Scholar]
- 36.Beylot MH, Emami K, McKie VA, Gilbert HJ, Pell G. Pseudomonas cellulosa expresses a single membrane-bound glycoside hydrolase family 51 arabinofuranosidase. Biochem J. 2001;358(3):599–605. doi: 10.1042/0264-6021:3580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurizzo D, Nagy T, Gilbert HJ, Davies GJ. The structural basis for catalysis and specificity of the pseudomonas cellulosa α-glucuronidase, GlcA67A. Structure. 2002;10(4):547–556. doi: 10.1016/s0969-2126(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 38.Rogowski A, Basle A, Farinas CS, Solovyova A, Mortimer JC, Dupree P, Gilbert HJ, Bolam DN. Evidence that GH115 α-glucuronidase activity, which is required to degrade plant biomass, is dependent on conformational flexibility. J Biol Chem. 2014;289(1):53–64. doi: 10.1074/jbc.M113.525295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thibault J-F. Separation of a-d-galacturonic acid oligomers by chromatography on polyacrylamide gel. J Chromatogr A. 1980;194(3):315–322. [Google Scholar]
- 40.Himmelsbach D, Hartley RD, Borneman W, Poppe L, Van Halbeek H. Structure of a feruloylated arabinoxylan tetrasaccharide that contains the β-d-Xylp-(1 → 2)-α-l-Araf element by1H and13C NMR spectrocopy. Magn Reson Chem. 1994;32(3):158–165. [Google Scholar]
- 41.Moller I, Marcus SE, Haeger A, Verhertbruggen Y, Verhoef R, Schols H, Ulvskov P, Mikkelsen D, Jr, Knox JP, Willats W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconjugate J. 2008;25(1):37–48. doi: 10.1007/s10719-007-9059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Costa RM, Pattathil S, Avci U, Lee SJ, Hazen SP, Winters A, Hahn MG, Bosch M. A cell wall reference profile for Miscanthus bioenergy crops highlights compositional and structural variations associated with development and organ origin. New Phytol. 2017;213(4):1710–1725. doi: 10.1111/nph.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badhan A, Wang Y, McAllister TA. Analysis of complex carbohydrate composition in plant cell wall using Fourier Transformed Mid-Infrared Spectroscopy (FT-IR) In: Abbott DW, van Lammerts Bueren A, editors. Protein-carbohydrate interactions: methods and protocols. New York: Springer; 2017. pp. 209–214. [DOI] [PubMed] [Google Scholar]
- 44.Robert P, Marquis M, Barron C, Guillon F, Saulnier L. FT-IR investigation of cell wall polysaccharides from cereal grains. Arabinoxylan infrared assignment. J Agric Food Chem. 2005;53(18):7014–7018. doi: 10.1021/jf051145y. [DOI] [PubMed] [Google Scholar]
- 45.Sun X-F, Sun RC, Fowler P, Baird MS. Extraction and characterization of original lignin and hemicelluloses from wheat straw. J Agric Food Chem. 2005;53(4):860–870. doi: 10.1021/jf040456q. [DOI] [PubMed] [Google Scholar]
- 46.Bengtsson S, Aman P, Andersson RE. Structural studies on water-soluble arabinoxylans in rye grain using enzymatic hydrolysis. Carbohydr Polymers. 1992;17(4):277–284. [Google Scholar]
- 47.Gruppen H, Kormelink FJM, Voragen AGJ. Water-unextractable cell wall material from wheat flour. 3. A structural model for arabinoxylans. J Cereal Sci. 1993;18(2):111–128. [Google Scholar]
- 48.Verbruggen MA, Beldman G, Voragen AGJ. Enzymic degradation of sorghum glucuronoarabinoxylans leading to tentative structures. Carbohydr Res. 1998;306(1–2):275–282. doi: 10.1016/s0008-6215(97)10065-9. [DOI] [PubMed] [Google Scholar]
- 49.Pena MJ, Kulkarni AR, Backe J, Boyd M, O’Neill MA, York WS. Structural diversity of xylans in the cell walls of monocots. Planta. 2016;244:589–606. doi: 10.1007/s00425-016-2527-1. [DOI] [PubMed] [Google Scholar]
- 50.Kulkarni A, Pattathil S, Hahn MG, York WS, O’Neill MA. Comparison of arabinoxylan structure in bioenergy and model grasses. Ind Biotechnol. 2012;8(4):222–229. [Google Scholar]
- 51.Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT. Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol. 2015;170(1):234–249. doi: 10.1104/pp.15.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Costa RMF, Lee SJ, Allison GG, Hazen SP, Winters A, Bosch M. Genotype, development and tissue-derived variation of cell-wall properties in the lignocellulosic energy crop Miscanthus. Ann Bot. 2014;114(6):1265–1277. doi: 10.1093/aob/mcu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verbruggen MA, Spronk BA, Schols HA, Beldman G, Voragen AGJ, Thomas JR, Kamerling JP, Vliegenthart JFG. Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr Res. 1998;306(1):265–274. doi: 10.1016/s0008-6215(97)10064-7. [DOI] [PubMed] [Google Scholar]
- 54.Mazumder K, York WS. Structural analysis of arabinoxylans isolated from ball-milled switchgrass biomass. Carbohydr Res. 2010;345(15):2183–2193. doi: 10.1016/j.carres.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 55.Charnock SJ, Spurway TD, Xie H, Beylot M-H, Virden R, Warren RAJ, Hazlewood GP, Gilbert HJ. The topology of the substrate binding clefts of glycosyl hydrolase family 10 xylanases are not conserved. J Biol Chem. 1998;273(48):32187–32199. doi: 10.1074/jbc.273.48.32187. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert HJ, Hazlewood GP, Laurie JI, Orpin CG, Xue GP. Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol. 1992;6(15):2065–2072. doi: 10.1111/j.1365-2958.1992.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 57.Nagy T, Nurizzo D, Davies GJ, Biely P, Lakey JH, Bolam DN, Gilbert HJ. The α-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J Biol Chem. 2003;278(22):20286–20292. doi: 10.1074/jbc.M302205200. [DOI] [PubMed] [Google Scholar]
- 58.Ryabova O, Vršanská M, Kaneko S, van Zyl WH, Biely P. A novel family of hemicellulolytic α-glucuronidase. FEBS Lett. 2009;583(9):1457–1462. doi: 10.1016/j.febslet.2009.03.057. [DOI] [PubMed] [Google Scholar]
- 59.Peng W, Pedersen NR, Pettersson D, Ekloff JM, Nymand-Grarup S, Palmen LG, Monrad RN, Spodsberg N, Stringer MA, Blom C et al. Compositions comprising polypeptides having xylanase activity and polypeptides having arabinofuranosidase activity. 2017. US 2017/0335302 A1.
- 60.Margolles-Clark E, Tenkanen M, Nakari-Setälä T, Penttilä M. Cloning of genes encoding alpha-l-arabinofuranosidase and beta-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62(10):3840–3846. doi: 10.1128/aem.62.10.3840-3846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goubet F, Jackson P, Deery MJ, Dupree P. Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem. 2002;300(1):53–68. doi: 10.1006/abio.2001.5444. [DOI] [PubMed] [Google Scholar]
- 62.Tryfona T, Theys TE, Wagner T, Stott K, Keegstra K, Dupree P. Characterisation of FUT4 and FUT6 β-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases arabidopsis root growth salt sensitivity. PLoS ONE. 2014;9(3):e93291. doi: 10.1371/journal.pone.0093291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tryfona T, Stephens E. Analysis of carbohydrates on proteins by offline normal-phase liquid chromatography MALDI-ToF/ToF-MS/MS. Meth Mol Biol. 2010;658(2):137–151. doi: 10.1007/978-1-60761-780-8_8. [DOI] [PubMed] [Google Scholar]
- 64.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR spectroscopy: principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- 65.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Bioinform. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2: Table S1. 1H and 13C NMR assignments of β-Xylp-(1 → 2)-α-Araf-(1 → 3)-β-Xylp-(1 → 4)-β-Xylp-(1 → 4)-β-Xylp, at 25 °C in D2O.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article and its additional files.