Abstract
Background
Lysine crotonylation, as a novel evolutionarily conserved type of post-translational modifications, is ubiquitous and essential in cell biology. However, its functions in tea plants are largely unknown, and the full functions of lysine crotonylated proteins of tea plants in nitrogen absorption and assimilation remains unclear. Our study attempts to describe the global profiling of nonhistone lysine crotonylation in tea leaves and to explore how ammonium (NH4+) triggers the response mechanism of lysine crotonylome in tea plants.
Results
Here, we performed the global analysis of crotonylome in tea leaves under NH4+ deficiency/resupply using high-resolution LC-MS/MS coupled with highly sensitive immune-antibody. A total of 2288 lysine crotonylation sites on 971 proteins were identified, of which contained in 15 types of crotonylated motifs. Most of crotonylated proteins were located in chloroplast (37%) and cytoplasm (33%). Compared with NH4+ deficiency, 120 and 151 crotonylated proteins were significantly changed at 3 h and 3 days of NH4+ resupply, respectively. Bioinformatics analysis showed that differentially expressed crotonylated proteins participated in diverse biological processes such as photosynthesis (PsbO, PsbP, PsbQ, Pbs27, PsaN, PsaF, FNR and ATPase), carbon fixation (rbcs, rbcl, TK, ALDO, PGK and PRK) and amino acid metabolism (SGAT, GGAT2, SHMT4 and GDC), suggesting that lysine crotonylation played important roles in these processes. Moreover, the protein-protein interaction analysis revealed that the interactions of identified crotonylated proteins diversely involved in photosynthesis, carbon fixation and amino acid metabolism. Interestingly, a large number of enzymes were crotonylated, such as Rubisco, TK, SGAT and GGAT, and their activities and crotonylation levels changed significantly by sensing ammonium, indicating a potential function of crotonylation in the regulation of enzyme activities.
Conclusions
The results indicated that the crotonylated proteins had a profound influence on metabolic process of tea leaves in response to NH4+ deficiency/resupply, which mainly involved in diverse aspects of primary metabolic processes by sensing NH4+, especially in photosynthesis, carbon fixation and amino acid metabolism. The data might serve as important resources for exploring the roles of lysine crotonylation in N metabolism of tea plants. Data were available via ProteomeXchange with identifier PXD011610.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5716-z) contains supplementary material, which is available to authorized users.
Keywords: Camellia sinensis L., Lysine crotonylation, Ammonium deficiency/resupply, Primary metabolism, Enzymatic activity
Background
Ammonium (NH4+) and nitrate (NO3−) are the major sources of nitrogen (N) in higher plants. As a kind of beverage leafy crop, tea plant (Camellia sinensis L.) preferred NH4+ over NO3− [1, 2]. Compared with the NH4+ supply, the contents of chlorophyll and biomass were more lower in tea plants when NO3− was supplied as the sole nitrogen source [3]. Compared with NO3− supply, the biosynthesis of free amino acids and catechins was more effective in tea plants under NH4+ supply, resulting from expression of N transporter genes [4]. NH4+ deficiency could lead to clearly reduce the accumulation of amino acid, while NH4+ supply could improve the status of carbohydrate in tea plants [1, 4]. Moreover, previous study in our laboratory showed that the levels of lysine acetylation in tea leaves changed dynamically under NH4+ resupply. And lysine acetylated proteins might regulate photosynthesis, glycolysis and secondary metabolism in tea leaves after NH4+ resupply [5]. However, the global profiling of lysine crotonylated proteins in tea plants in response to NH4+ resupply remains largely unknown.
Post-translational modifications (PTMs) of histone were well-known for its critical roles in cellular pathways, which could change the physicochemical properties of proteins and affect their activity and stability [6, 7]. As a novel evolutionarily conserved type of PTMs, the histone crotonylation level could be regulated by cellular concentration of crotonyl-CoA [8]. The previous reports demonstrated that a subset of genes could be differentially regulated by histone crotonylation, and selective histone decrotonylation could repress the global transcription of mouse embryonic stem cells to some extent [9]. Recently, the global profiling of crotonylation has been reported in tobacco and rice [10, 11], which revealed that crotonylation was correlated with signal transduction and cellular physiology. However, there have not been systematically reported about the dynamic viewing of nonhistone lysine crotonylation responding to N, especially ammonium. In order to explore whether the crotonylation is connected with NH4+ resupply and to obtain a comprehensive characterization of lysine crotonylation in tea leaves, we adopted an integrated system using the peptide prefractionation, immunoaffinity enrichment, and coupling with highly sensitive mass spectrometry combined with affinity purification analysis. We examined the crotonylated proteins in tea leaves under NH4+ deficiency/resupply and analyzed the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI) of these proteins. This research not only greatly extended the list of crotonylated proteins in tea plants but also paved the way for exploring the roles of crotonylated proteins in the utilization of N in tea plants.
Methods
Plant materials and growth conditions
The 1-year-old seedlings of the tea plants cultivar ‘QN3’ planted in pots at the greenhouse of the Tea Research Institute, Qingdao Agricultural University in Shandong Province of China (36°19′ N, 120°23′ E, 54.88 m above the sea level), were used to Hydroponics. For hydroponics, the tea plants were suspended in a hydroponic nutrient solution (Additional file 1). The growth conditions were set as follows: temperature, 25 ± 1 °C /15 ± 1 °C (14 h day/10 h night); lighting, 260–280 μM•m− 2•s− 1 photon flux densities; and humidity, 75 ± 5% relative humidity. After hydroponic seedlings were grown in the greenhouse for two weeks to recover, which used to perform the NH4+ deficiency/resupply experiments. Firstly, hydroponic seedlings were transferred to NH4+ deficiency nutrient solution ((NH4)2SO4 deleted) for 14 days (tea plants of NH4+ deficiency). Then, these seedlings were retransferred to NH4+ resupply nutrient solution ((NH4)2SO4 added) for 3 days. Finally, the third and/or fourth mature leaves from the terminal bud were sampled at NH4+ deficiency (DN), 3 h (3hN) and 3 days (3dN) of NH4+ resupply. All samples were quickly frozen in liquid nitrogen and stored at − 80 °C for further study. Three biological replicates were performed for each sampling time point.
Physiological determinations
For physiological experiments, more than ten plants were harvested and pooled for each treatment group at DN, 3hN and 3dN. And the leaves were collected three times as biological replicates. The nitrogen content (NC) of tea leaves was measured after Kjeldahl digestion. The chlorophyll content (CC) of samples was measured as described by Lichtenthaler et al [12]. The measurement of obtain maximum photochemical quantum yield of PS II (Fv/Fm) referred to Zheng et al. [13]. Samples were analyzed for the contents of free amino acids (AA), and measurement followed the Sate Standard of China for tea content determination recorded as: GB 8314–87.
Western blot
The samples of tea leaves that in been grown in the presence of DN, 3hN and 3dN, were performed by western blotting (WB) analysis based on a previously described method [14]. For western blot, protein was diluted with SDS loading buffer, and 30 μg protein of each sample was separated by 12% SDS-PAGE and electro-blotted onto PVDF. First antibody and second antibody were probed using Anti-crotonyllysine Antibody (PTM-502, PTM Biolabs, Hangzhou, China) in the 1:1000 dilution and HRP AffiniPure Goat Anti-Rabbit IgG (31,430, Thermo Fisher Scientific, Waltham, USA) in 1:10000 dilution, respectively.
Protein extraction and digestion
The protein extraction and digestion of the samples were following by previous method [15]. And then the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM NH4HCO3 to urea concentration less than 2 M. Finally, trypsin (Promega, Madison, WI, USA) was added at 1:50 trypsin-to-protein mass ratio for 12 h and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion. And the more detailed information was shown in Additional file 2.
HPLC fractionation and affinity enrichment
For global proteome analysis, the tryptic peptides were fractionated into fractions by high pH reverse-phase HPLC using Agilent 300 Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). Briefly, peptides were first separated with a gradient of 8 to 32% acetonitrile (pH 9.0, Fisher Chemical) over 60 min into 60 fractions. Then, the peptides were combined into four fractions and dried by vacuum centrifuging. 200 μg peptides were used for HPLC fractionation in this process.
To enrich crotonylated peptides, tryptic peptides dissolved in NETN buffer (100 Mm NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0) were incubated with pre-washed antibody beads (PTM-402, PTM Biolabs, Hangzhou, China) at 4 °C overnight with gentle shaking. Then the beads were washed four times with NETN buffer and twice with H2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid (Sigma, Saint Louis, USA). Finally, the eluted fractions were combined and vacuum-dried. For LC-MS/MS analysis, the resulting peptides were desalted with C18 ZipTips (Millipore, Saint Louis, USA) according to the manufacturer’s instructions, followed by LC-MS/MS analysis. In this process, 1.5 mg peptides were used for each affinity enrichment.
LC-MS/MS analysis
The tryptic peptides were dissolved in solvent A (water on 0.1% formic acid) and directly loaded onto a home-made reversed-phase analytical column (15-cm length, 75 μm i.d.). For global proteome analysis, the gradient of was comprised of an increase from 6 to 23% solvent B (0.1% formic acid in 90% acetonitrile) over 40 min, 23 to 35% in 14 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific, Waltham, USA). For crotonylome analysis, the gradient of was comprised of an increase from 7 to 25% solvent B (0.1% formic acid in 90% acetonitrile) over 38 min, 25 to 40% in 14 min and climbing to 80% in 4 min then holding at 80% for the last 4 min, all at a constant flow rate of 700 nL/min on an EASY-nLC 1000 UPLC system.
The peptides were subjected to NSI (neutral spray ionization) source followed by tandem mass spectrometry (MS/MS) in Q Exactive™ Plus (Thermo Fisher Scientific, Waltham, USA) coupled online to the UPLC. The electrospray voltage applied was 2.0 Kv for crotonylome analysis or 2.1 Kv for global proteome analysis. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using NCE setting as 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. In order to improve the efficiency of mass spectrometry, automatic gain control (AGC) was set at 5E4, signal threshold was set at 10000 ions/s for crotonylome analysis or 20,000 ions/s for global proteome analysis, the maximum injection time was set at 200 ms for crotonylome analysis or 100 ms for global proteome analysis, and the dynamic exclusion time of tandem mass spectrometry was set at 15 s for crotonylome analysis or 30 s for global proteome analysis.
Database search and bioinformatic analysis
For protein quantification of global proteome and crotonylome, the resulting MS/MS data were processed using MaxQuant search engine (v.1.5.2.8). Tandem mass spectra were searched against Camellia sinensis database (36,951 sequences, http://www.plantkingdomgdb.com/tea_tree/) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 4 missing cleavages for lysine crotonylome, and 2 missing cleavages for global proteome. The mass tolerance for precursor ions was set as 20 ppm in First search and 5 ppm in Main search, and the mass tolerance for fragment ions was set as 0.02 Da. For global proteome analysis, carbamidomethyl on Cys was specified as fixed modification, oxidation on Met and acetylation on the protein N-terminus was specified as variable modifications. For crotonylome analysis, carbamidomethyl on Cys for fixed modification and oxidation on Met, crotonylation on lysine and acetylation on the protein N-terminus for variable modifications. And then, Label-free quantification method was LFQ, FDR was adjusted to < 1% and minimum score for peptides or modified peptides were set > 40.
Soft motif-x (http://motif-x.med.harvard.edu/) was used to analyze the model of sequences with amino acids in specific positions of modify-21-mers (10 amino acids upstream and downstream of the site) in all protein sequences. The subcellular localization was performed by wolfpsort (http://www.genscript.com/wolf-psort.html). The GO annotation and enrichment analyses were done by UniProt-GOA database (http://www.ebi.ac.uk/GOA/). KEGG database was adopted for the enrichment of pathways by functional annotation tool of DAVID against the background of Camellia sinensis. To perform a PPI network analysis, the STRING database (http://string-db.org/) was used and then functional protein interaction networks were visualized by using Cytoscape (v.3.7.0).
Enzyme assays
The activity of Rubisco, TK, PRK, SGAT, GGAT and SHMT were measured at DN, 3hN and 3dN in tea leaves, using ELISA kit from Jiangsu Meimian industrial Co. Ltd. The enzyme activity was tested using ELISA kit from Jiangsu Meimian industrial Co. Ltd. in 50 mg sample of each enzyme. Finally, the reaction was terminated by the addition of a sulphuric acid solution and the color change was measured spectrophotometrically at a wavelength of 450 nm. Each treatment was designed with three replicates randomly. The more detail information was shown in Additional file 3.
Results
Physiological characterization of tea leaves after NH4+ resupply
To describe the major physiological changes of tea plants in varying NH4+ resupply, we measured the NC, CC, Fv/Fm and AA under NH4+ deficiency/resupply (Additional file 4: Figure S1). We found that NC, CC, Fv/Fm and AA at DN were all lower than those of NH4+ resupply, and they showed upward trends with the NH4+ resupply prolonged (3hN and 3dN). These results indicated that NH4+ could affect the physiological property of tea leaves.
WB analysis of tea leaves after NH4+ resupply
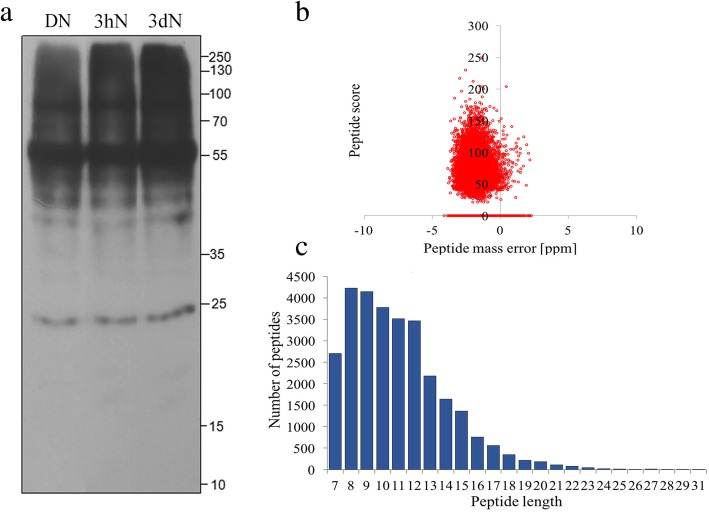
In order to test whether there was crotonylation in proteins of tea leaves after NH4+ resupply, we performed SDS-PAGE and WB analysis. As a quantitative control in three samples, the results of SDS-PAGE showed that the amount of proteins added to the three samples was consistent (Additional file 4: Figure S2). And then, the results of WB showed that proteins in tea leaves were widely crotonylated, and crotonylation levels showed changes between each time point (Fig. 1a), suggesting that lysine crotonylation underwent significantly dynamic changes in response to NH4+ resupply.
Fig. 1.
The WB analysis and proteome-wide identification of lysine crotonylation sites in tea leaves. a WB analysis of the total protein content of tea leaves showing duplicates at three time-points after NH4+ resupply. b The mass error distributions of crotonylation profiles. c The peptide length distributions of crotonylation profiles
Detection of lysine crotonylome and global proteome in tea leaves
In this study, we combined antibody against crotonylated lysine, LC − MS/MS and intensive bioinformatics for comprehensive study of lysine crotonylome in tea leaves upon NH4+ treatment. In order to validate the MS data, we firstly checked the mass error of the identified peptides. The distribution of mass errors was near zero, and most errors were less than 4 ppm, indicating the accuracy of the MS data (Fig. 1b). The lengths of most identified peptides were in the range of 7 to 17 amino acids, which were consistent with the property of tryptic peptides, indicating that the sample preparation achieved a reasonable standard (Fig. 1c). Each treatment was designed with three biological replicates. As a result, a total of 2260 modified peptides were identified on 971 proteins, containing 2288 crotonylation sites (Additional file 5). Among of them, the majority of crotonylated proteins (698, 71.9%) identified contained only one or two crotonylation sites (Additional file 4: Figure S3). Fifty representative LC-MS/MS spectra of crotonylated peptides were shown in Additional file 6. Furthermore, global proteome data for normalization were also collected with identified 5312 protein groups (Additional file 7).
Functional classification and motif analysis of the lysine crotonylome in tea leaves
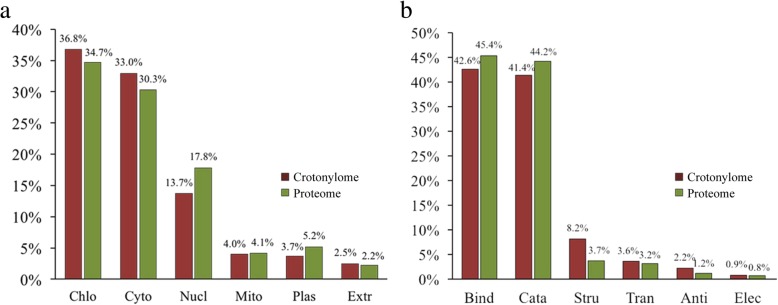
To better understand the function of proteins in crotonylome and global proteome, we conducted their functional classification. The results of subcellular localization revealed that a majority of crotonylated proteins were mainly located in chloroplast (36.8%), cytoplasm (33.0%) and nucleus (13.7%), suggesting that the crotonylated proteins distributed broadly in tea leaves (Additional file 8: Fig. 2a). The subcellular localization of global proteome showed that the proteins were mainly located in chloroplast (34.7%), cytoplasm (30.3%) and nucleus (17.8%). Then, the analysis of molecular functions showed that crotonylated proteins and global proteins presented a similar pattern, and the vast majority of proteins participated in binding and catalytic activity (Additional file 8: Fig. 2b). And the percentage of crotonylome and global proteome in each category was also very close. Based on the results, we found that the subcellular localization and molecular functions between the crotonylome and global proteome have no significant difference.
Fig. 2.
Functional classification of lysine crotonylome compared to global proteome. a Subcellular localization of lysine crotonylome compared to global proteome. b Molecular function of lysine crotonylome compared to global proteome. Abbreviation: Chlo for chloroplast, Cyto for cytoplasm, Nucl for nucleus, Mito for mitochondria, Plas for plasma membrane, Extr for extracellular, Bind for binding, Cata for catalytic activity, Stru for structural molecule activity, Tran for transporter activity, Anti for antioxidant activity, and Elec for electron carrier activity
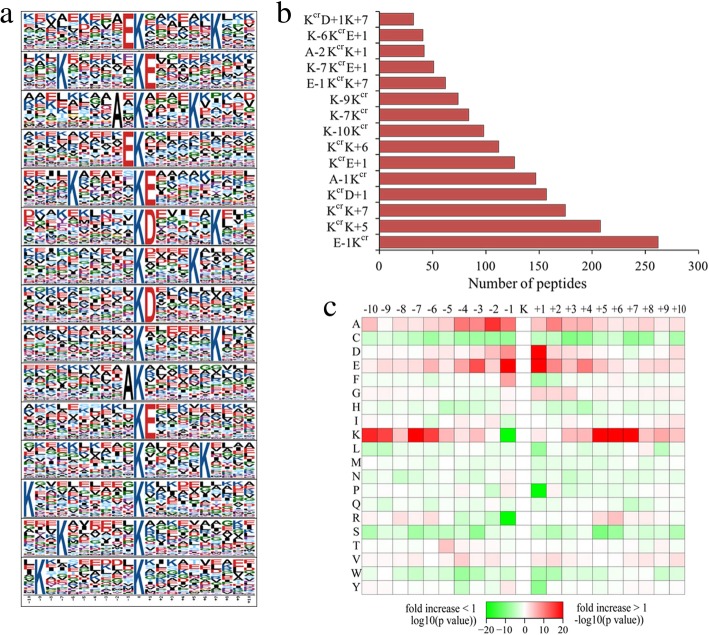
To determine whether there were any specific amino acid biases adjacent to crotonylation sites, we investigated the sequence of all identified crotonylated peptides with the motif-x program (Additional file 9). As a result, the amino acid sequences were classified into 15 conserved motifs, including E-1Kcr, KcrD + 1 K + 7, KcrK + 5, KcrD + 7, A-1Kcr, KcrE+ 1/+ 6, K-10/− 9/−7Kcr, E-1KcrK + 7, K-7/−6KcrE+ 1, A-2KcrK + 1 (Fig. 3a). Among them, E-1Kcr, KcrD + 1, A-1Kcr, KcrD + 1 and KcrE+ 1 have been reported as crotonylation motifs [10, 16], while the others were first reported in our study. In addition, we found that the abundances of E-1Kcr, E-1Kcr, KcrK + 7 and KcrD + 1 were comparatively higher than the other 11 conserved motifs (Fig. 3b). In accordance with these findings, crotonylation was preferred on lysine residues that adjacent to alanine, glutamate and lysine (Fig. 3c).
Fig. 3.
Motif analysis of lysine crotonylated peptides. a Crotonylated sequence motifs and conservation of crotonylation sites. The 0 position K refers to the crotonylation sites. b Number of identified peptides containing crotonylation in each motif. c Heatmap of the amino acid compositions of the crotonylation sites showing the frequency of the different of amino acids around the crotonylation. “+ 1” and “-1” represent the position around the crotonylation
The crotonylated proteins in tea leaves after NH4+ resupply
From these, we quantified 2164 crotonylation sites on 945 proteins, and then normalized the quantitative lysine crotonylome with the data of the global proteome (Additional file 10). Compared with the NH4+ deficiency, 232 crotonylation sites on 183 proteins were up-regulated and 72 crotonylation sites on 63 proteins were down-regulated at 3hN based on a fold-change threshold > 1.5, whereas 218 crotonylation sites on 172 proteins were up-regulated and 57 crotonylation sites on 52 proteins were down-regulated at 3dN (Additional file 11). Furthermore, 39 crotonylation sites on 33 proteins were up-regulated and 71 crotonylation sites on 50 proteins were down-regulated at 3dN/3hN. These results proved that lysine crotonylation in tea leaves could directly respond to NH4+ resupply.
Design of Venn diagram and functional analysis of lysine crotonylated proteins
To deeply investigate the cellular processes regulated by crotonylation in tea leaves at 3 h and 3d of NH4+ resupply, a Venn diagram of differentially expressed crotonylated proteins (DCPs) was structured (Additional file 4: Figure S4, Additional file 12). We found that 121 DCPs were specifically observed at 3hN/DN, and 143 DCPs were specifically observed at 3dN/DN. Moreover, 94 DCPs were identical in two conditions. These results proved that the expression of crotonylated proteins was specific at different time points of NH4+ resupply.
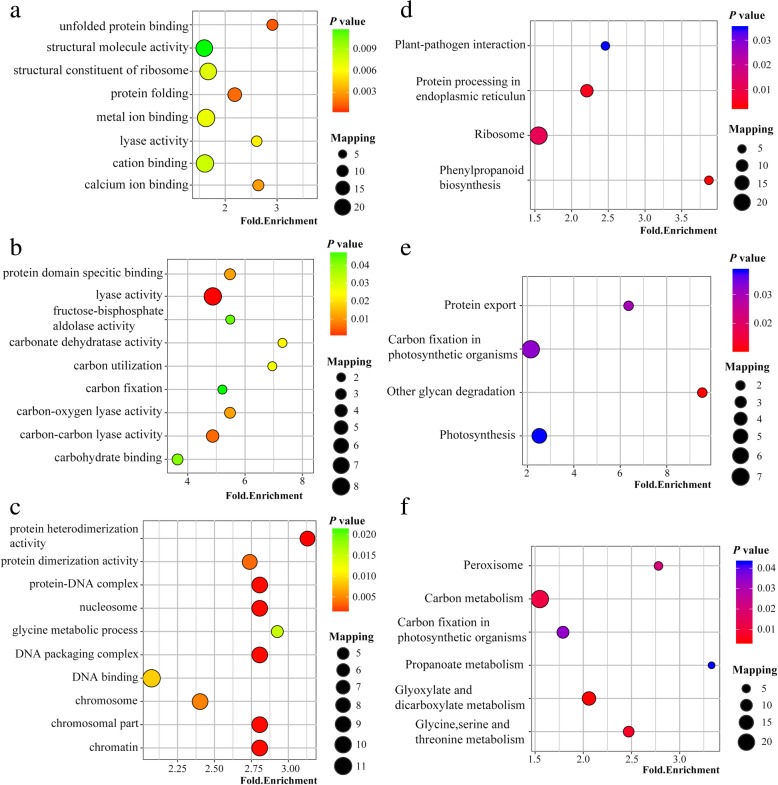
To explore the relevant biological functions and pathways, we performed GO and KEGG enrichment analysis based on the data of Venn analysis (Additional files 13 to 14). The results showed that GO terms of DCPs were enriched exclusively at 3hN/DN, such as structural molecule activity, metal ion binding, cation binding, protein folding and structural constituent of ribosome (Fig. 4a). The GO terms of common DCPs in 3hN/DN and 3dN/DN were similar and both included terms associated with lyase activity, carbon-carbon lyase activity, fructose bisphosphate aldolase activity, carbohydrate binding and carbon fixation (Fig. 4b). Moreover, GO terms of DCPs were enriched exclusively at 3dN/DN, such as protein heterodimerization activity, nucleosome, DNA packaging complex, chromatin and glycine metabolic process (Fig. 4c). These results implied that crotonylated proteins in tea leaves plays an important role in these processes by sensing NH4+ nutrition.
Fig. 4.
Enrichment analysis of DCPs in tea leaves after NH4+ resupply. a GO enrichment analysis of specific DCPs at 3hN/DN. b GO enrichment analysis of common DCPs at 3hN/DN and3dN/DN. c GO enrichment analysis of specific DCPs at 3dN/DN. d KEGG pathway enrichment analysis of specific DCPs at 3hN/DN. e KEGG pathway enrichment analysis of common DCPs at 3hN/DN and3dN/DN. f KEGG pathway enrichment analysis of specific DCPs at 3dN/DN
KEGG pathway analysis showed that 3hN/DN-specific DCPs were enriched to ribosome, protein processing in endoplasmic reticulum, phenylpropanoid biosynthesis and plant−pathogen interaction (Fig. 4d). Furthermore, common DCPs modulated at 3hN/DN and 3dN/DN were enriched to similar pathways, including photosynthesis, carbon fixation in photosynthetic organisms, protein export and other glycan degradation (Fig. 4e). In addition, many 3dN/DN-specific DCPs were enriched to carbon metabolism, glyoxylate and dicarboxylate metabolism, peroxisome, glycine, serine and threonine metabolism (Fig. 4f). In total, crotonylated proteins in tea leaves after NH4+ resupply was involved in photosynthesis, carbon metabolism, glyoxylate and dicarboxylate metabolism, ribosome, glycine, serine and threonine metabolism.
The analysis of interaction network in lysine crotonylated proteins
To deeply understand the interactions of crotonylated proteins, we constructed the PPI networks for DCPs (Additional file 15). The results showed that 86 specific DCPs were closely connected at 3hN/DN, which mapped to the protein interaction database (Fig. 5a). There was a strong interaction between crotonylated proteins involved in photosynthesis or biosynthesis of amino acids. In this network, 28 crotonylated proteins were identified with the node degree over 10, such as endoplasmin homolog and ALDO. Furthermore, 66 common DCPs were closely connected between 3hN/DN and 3dN/DN (Fig. 5b). There was a close interaction between crotonylated proteins involved in photosynthesis, carbon metabolism and biosynthesis of amino acids. Of which 16 crotonylated proteins were identified with the node degree over 10, such as FNR, PsbO and PsaN. In addition, 70 specific DCPs were closely connected at 3dN/DN (Fig. 5c). There was a close interaction between crotonylated proteins involved in carbon fixation or glycine, serine and threonine metabolism. Thereinto, 21 crotonylated proteins were identified with the node degree over 10, such as SBPase, SHMT, γ-ATPase and PRK. These results can be clearly seen that crotonylated proteins had a close interaction in many primary metabolic processes of tea leaves under NH4+ deficiency/resupply.
Fig. 5.
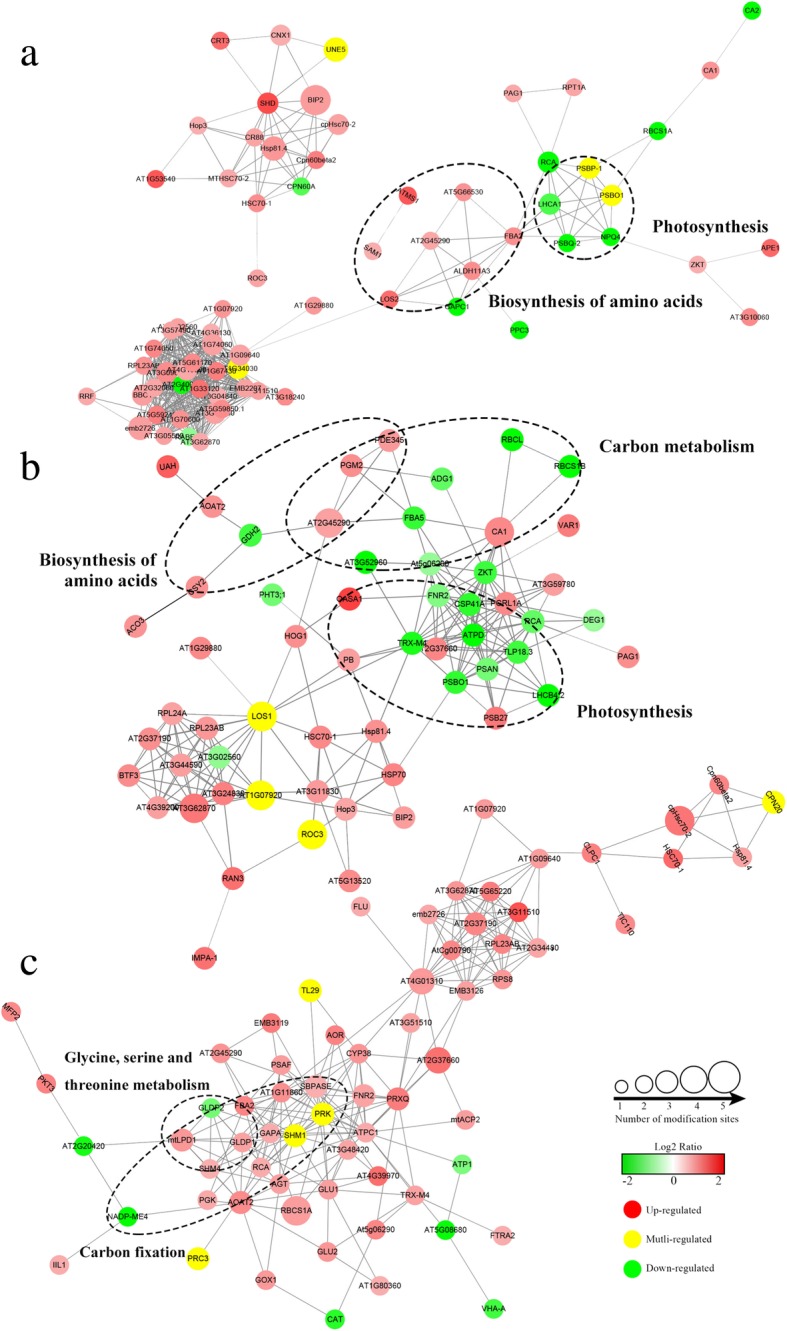
PPI network of DCPs after NH4+ resupply. a The PPI of specific DCPs at 3hN/DN. b The PPI of common DCPs at 3hN/DN and3dN/DN, c The PPI of specific DCPs at 3dN/DN
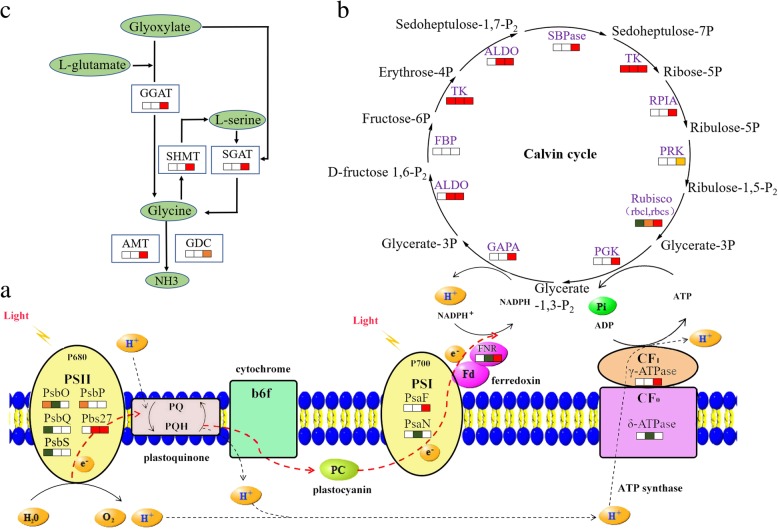
The changes of crotonylation sites on crotonylated proteins involved in photosynthesis
To deeply investigate the photosynthesis regulated by lysine crotonylation, we summarized the crotonylated proteins in tea leaves under NH4+ deficiency/resupply (Fig. 6a, Additional file 16). We found that there were 6 crotonylation sites on 4 specific crotonylated proteins changed obviously at 3hN/DN, containing K270 (up-regulated) and K144 (down-regulated) on PsbO, K119 (down-regulated) and k247 (up-regulated) on PsbP, K147 (down-regulated) on PsbQ and K185 (down-regulated) on PsbS. Meanwhile, there were 5 crotonylation sites on 5 common crotonylated proteins changed significantly between 3hN/DN and 3dN/DN, containing K159 (down-regulated) on PsaN, K121 (down-regulated) on PsbO, K121 (up-regulated) on Pbs27, K238 (down-regulated) on FNR and K117 (down-regulated) on δ-ATPase. Among them, K121 on PsbO and K117 on δ-ATPase decreased more than 3 times and 2 time from 3hN to 3dN, respectively. Furthermore, at 3dN/DN, there were 6 crotonylation sites on 4 specific crotonylated proteins changed significantly, including K124 (up-regulated) on PsaF, K116 (up-regulated) and K169 (up-regulated) on Pbs27, K81 (up-regulated) and K203 (up-regulated) on FNR and K194 (up-regulated) on γ-ATPase.
Fig. 6.
DEPs in involved in primary metabolic processes after NH4+ resupply. a photosynthesis. b Calvin cycle. c glycine, serine and threonine metabolism. The rectangle was divided into three equal parts (the left of rectangle represented specific DCPs at 3hN/DN; the middle of rectangle represented common DCPs at 3hN/DN and 3dN/DN; the right of rectangle represented specific DCPs at 3dN/DN). The color in the rectangle represents the crotonylated proteins were regulated after NH4+ resupply (red indicated up-regulation; yellow indicates mutli-regulation; green indicated down-regulation)
The changes of crotonylation sites on crotonylated proteins involved in carbon fixation
To check into the carbon fixation in photosynthetic organisms regulated by lysine crotonylation, we mainly analyzed the crotonylated proteins of Calvin cycle in tea leaves under NH4+ deficiency/resupply (Fig. 6b, Additional file 17). We found that there were 2 crotonylation sites on 2 specific crotonylated proteins marked change at 3hN/DN, containing K138 (down-regulated) on rbcs and K314 (up-regulated) on TK. Meanwhile, there were 7 crotonylation sites on 4 common crotonylated proteins changed significantly between 3hN/DN and 3dN/DN, such as K190 (up-regulated) on rbcl, K84 (down-regulated) on rbcs, K172 (up-regulated) and K4 (up-regulated) on ALDO, K133 (up-regulated) and K710 (up-regulated) on TK. Furthermore, at 3dN/DN, there were 15 crotonylation sites on 9 specific crotonylated proteins changed highly, including K63 (up-regulated), K110 (up-regulated), K144 (up-regulated) and K160 (up-regulated) on rbcs, K480 (up-regulated) on TK, K207 (up-regulated) and K233 (down-regulated) on PRK, K125 (up-regulated) on PGK, K212 (up-regulated) on RPIA, K80 (up-regulated) on ALDO, K108 (up-regulated) and K305 (up-regulated) on SBPase. Among them, K207 on PRK, K212 on RPIA, K63 and K144 on rbcs were up-regulated from 3 h to 3d of NH4+ resupply.
The changes of crotonylation sites on crotonylated proteins involved in amino acid metabolism
To deeply research the amino acid metabolism regulated by lysine crotonylation, especially glycine, serine and threonine (Fig. 6c, Additional file 18), we characterized the crotonylated proteins in tea leaves under NH4+ deficiency/resupply. Interestingly, there was no significant change in crotonylated proteins at 3hN/DN. However, there were 12 crotonylation sites on 7 specific crotonylated proteins changed dramatically at 3dN/DN, such as K52 (up-regulated) on SGAT, K441 (up-regulated) on SHMT4, K4 (up-regulated), K35 (up-regulated) and K357 (up-regulated) on GGAT2, K478 (up-regulated) and K265 (down-regulated) on GDC, K146 (up-regulated) and (up-regulated) K201 on LPD1. And K146 on LPD1 also was up-regulated from 3hN to 3dN.
The changes of enzymatic activity and protein contents of DCPs
To investigate whether the activities of DCPs were changed after NH4+ resupply, we measured the activity of six DCPs involved in Calvin cycle, serine and glycine metabolism, respectively (Additional file 19). Among them, Rubisco (rbcl and rbcs were subunits of Rubisco), TK and PRK were the key enzymes for CO2 fixation, reduction of 3-phosphoglycerate and ribulose-1,5-bisphosphate (RUBP) regeneration in Calvin cycle, respectively. Meanwhile, SGAT, GGAT and SHMT were directly involved in serine and glycine synthesis. As a result, the activity of Rubisco, TK and PRK were significantly enhanced after NH4+ resupply. And the activity of SGAT, GGAT and SHMT were specifically increased at 3dN. Besides, we found that the expression levels of some proteins had no significant change (F > 1.5, P < 0.05) after NH4+ resupply which were dramatically expressed at crotonylation level involved in Calvin cycle and serine and glycine synthesis (Additional file 20).
Discussion
As a kind of evergreen crop, tea plants highly prefer to NH4+. However, the lysine crotonylation of proteins in tea leaves responding to NH4+ nutrition has not been studied. Therefore, we investigated the global profiling of lysine crotonylation in tea leaves under NH4+ deficiency/resupply. A total of 2288 high-confident crotonylation sites on 971 proteins were identified, which greatly expanded the catalog of crotonylated proteins in plants. With the quantitative lysine crotonylome data normalized to the data of the global proteome, we noticed that hundreds of lysine residues and crotonylated proteins changed significantly during NH4+ resupply. These DCPs were associated with primary metabolism, including photosynthesis, carbon fixation and amino acid metabolism. The PPI network analysis also indicated that a wide range of protein interactions involved in these biological processes was likely modulated by crotonylation.
DCPs participated in photosynthesis after NH4+ resupply
Photosynthesis could convert light energy into chemical energy to synthesize NADPH and ATP. Previous research showed that total photosynthetic performances were positively correlated with N content [17]. N deficient leaves damaged capacity for electron transport, thus limiting ATP synthesis and RuBP regeneration [18, 19]. In our study, we noticed that NC and Fv/Fm showed upward trends after NH4+ resupply. Moreover, large proportions of crotonylated proteins related to photosynthesis were significantly changed after NH4+ resupply (Fig. 6a), mainly including PsbO, PsbP, PsbQ, Pbs27, PsaN, PsaF, FNR, γ-ATPase and δ-ATPase, suggesting that lysine crotonylation could play active roles in response to NH4+ resupply.
PS II was a protein complexes that converted light energy into the electrochemical potential energy required to split water into H+, electrons, and molecular oxygen, which was dynamic and constantly underwent assembly, disassembly and repair [20, 21]. However, the rate of repairment and destruction of PS II was unbalanced under various stress conditions, like N deficiency [22, 23]. PsbO, PsbP and PsbQ were extrinsic proteins of PS II complexes, which were indispensable for photosynthetic oxygen evolution and essential for the regulation and stabilization of PS II in higher plants [24, 25]. However, the information about PsbO, PsbP and PsbQ in tea plants were very limited. In this research, the crotonylated PsbO (K121, K144 and K270), PsbP (K119 and K247) and PsbQ (K147) were significantly changed at crotonylation level after NH4+ resupply. So, our results could provide references for exploring the functions of related proteins and improving the stability of PS II in tea leaves after NH4+ resupply. In addition, lipoprotein Pbs27 was involved in assembly of the water-splitting site of PS II and the turnover of the complex [26]. In our research, crotonylated Pbs27 was significantly up-regulated at K121 and K116, K121 and K169 at 3hN/DN and 3dN/DN, respectively. This suggested that the crotonylated Psb27 might play a positive function in repair of PS II after NH4+ resupply. Thus, considering the dynamic changes of crotonylated proteins and the upward trends of Fv/Fm after NH4+ resupply, we speculate that NH4+ could influence the crotonylation levels of proteins contained in PS II, thus modulating the activity of PS II.
As a sunlight energy converter, the regulation of PS I cyclic electron transport has been considered necessary for photosynthesis and plant growth. N deficiency preferentially affected the activity of PS I, which displayed lower efficiencies for electron flow from intermediate carriers to final PS I acceptors [26]. In PS I, PsaN and PsaF were essential intermediates for electron transferring from plastocyanin to the oxidized reaction centre of P700+. In the present study, crotonylated PsaN (K159) was significantly down-regulated at 3hN/DN and 3dN/DN, while crotonylated PsaF (K124) up-regulated only at 3dN/DN. In addition, we found that crotonylated FNR (K81 and K203), γ-ATPase synthase (K195) and δ-ATPase (K117) were notably changed at 3dN/DN. And K117 on δ-ATPase decreased more than 2 time from 3hN to 3dN. It was reported that FNR had been implicated in cyclic electron transfer around PS I, which catalyzed the last of the light reactions by transferring electrons from ferredoxin to NADP+, and was essential for efficient photosynthetic performance [17, 27]. As subunits of ATP synthase, γ-ATPase and δ-ATPase were participated in creating the energy storage molecule ATP, which could facilitate electron transport in both PS I and PS II [28, 29]. From the above, we speculate that lysine crotonylation played important roles in electron transport of PS I in tea leaves after NH4+ resupply, and specific expression of crotonylated FNR, γ-ATPase and δ-ATPase could have a significant effect on promoting photosynthetic electron transport.
DCPs involved in the carbon fixation after NH4+ resupply
Under abiotic stress, particularly in nitrogen nutrition changes, plants gradually adjust the mechanisms of carbon metabolism. Several papers described that N deficiency exacerbated the limitations of photosynthesis performance by reducing the capacity of key enzymes involved in carbon fixation [30, 31]. The Calvin cycle is the primary pathway of carbon fixation. Previous study showed that the transcription levels of genes in the Calvin cycle had distinctly changed when N deficiency seedlings received N [32]. In this work, the almost all enzymes in the Calvin cycle were specifically expressed at crotonylation level after NH4+ resupply (Fig. 6b), such as rbcs, rbcl, TK, ALDO, PGK and PRK, suggesting that lysine crotonylation participated in the regulation of carbon fixation in tea leaves under NH4+ resupply.
Rubisco catalyzes the carboxylation of RuBP, which is the first step of the Calvin cycle. Previous studies showed that the content of Rubisco was strictly controlled to maintain C/N balance, and the N source provided by Rubisco degradation under N deficiency [30]. N deficiency reduced the activity of Rubisco in Arabidopsis and Kentucky Bluegrass, while the activity of Rubisco increased with increasing N content [33, 34]. The results from the current paper indicated that the activity of Rubisco increased after NH4+ resupply, crotonylated rbcs (K63, K110, K144 and K160) and rbcl (K190) changed significantly. However, the expressions of rbcs and rbcl showed no significant changes at protein level after NH4+ resupply. So, we speculate that lysine crotonylation was closely related to the activity of Rubisco, and crotonylated Rubisco had a role in regulating the carboxylation of RuBP and the balance of C/N after NH4+ resupply.
TK centrally locates in the Calvin cycle where it catalyzes the generation of ribose 5-phosphate and erythrose 4-phosphate. Up to now, the crotonylated TK has not been reported in plants. In this research, the activity of TK and the abundance of crotonylated TK were increased after NH4+ resupply. Because TK was important to maintaining RuBP regeneration and aromatic amino acids synthesis [35, 36], crotonylated TK might be contributing to the regulation of these processes in tea leaves after NH4+ resupply.
PRK catalyzes the phosphorylation of D-Ribulose 5-phosphate into RuBP, which is the last step of the Calvin cycle. The activity of PRK often determined the metabolic rate in organisms for which carbon fixation was key to survival [37]. Low N reduced the activity and content of PRK in transgenic tobacco, thus limiting photosynthesis and biomass production [38]. In this study, K207 on PRK was increased after NH4+ resupply. The crotonylated PRK (K207 and K233) was significantly changed at 3dN, and the activity of PRK was increased under NH4+ deficiency/resupply. As a consequence, we deduce that the crotonylated PRK played key roles in RuBP regeneration after NH4+ resupply.
DCPs involved in the amino acid metabolism after NH4+ resupply
The high levels of amino acids principally contributed to mellowness and freshness in tea [39]. NH4+ supply was found to effectively enhance the biosynthesis of catechins and free amino acids in tea leaves [1]. Our present study showed that the content of free amino acids was significantly increased after NH4+ resupply, and large amounts of enzymes participated in glycine, serine and threonine metabolism were specifically changed at crotonylation level after 3dN, not at 3hN (Fig. 6c), suggesting that lysine crotonylation could selectively participate in the amino acid metabolism under NH4+ deficiency/resupply.
GGAT and SGAT can function cooperatively in the production of glycine. Previous research showed that the activity of GGAT was sharply down-regulated and the activity of SGAT was little affected in rice under N deficiency. The substrate glutamate was accumulated while downstream products of glycine and serine were significantly reduced, indicated that the glycine formation from glyoxylate relies sensitively on GGAT than SGAT [40]. However, the response mechanism of SGAT and GGAT at crotonylation level have not been reported. In this research, we found that the activity of GGAT and SGAT were increased at 3dN, and crotonylated GGAT (K4, K35 and K367) and SGAT (K52) were significantly up-regulated, while GGAT and SGAT were not significantly changed at protein level after NH4+ resupply. We contemplate that lysine crotonylation might participate in regulating the activity of GGAT and SGAT after NH4+ resupply, but it still needs further investigation. In addition, according to previous reports, the improvement of GGAT activity could enhance the efficiency of nitrate and ammonium assimilation [41]. Thus, the crotonylated GGAT might plays important roles in N metabolism and glycine formation in tea leaves after NH4+ resupply.
SHMT is an essential player in the serine homeostasis, which catalyzes the reversible serine to glycine conversion. In microalgae, the SHMT might aid in maintaining cellular homeostasis during later stages of N deficiency [42]. In Agrostis stolonifera, the abundance of SHMT decreased with N deficiency, affecting the biosynthesis of amino acids and lipids [43]. In this study, the abundance of SHMT showed no remarkable change under NH4+ deficiency/resupply, while the activity of SHMT and the abundance of crotonylated SHMT4 (SHMT subunit) were significantly increased at 3dN/DN. We have reason to deduce that the crotonylated SHMT4 might affect the activity of SHMT and then consequently regulate the amino acids homeostasis in tea leaves after NH4+ resupply.
Conclusion
Above all, lysine crotonylation in tea leaves subjected to NH4+ deficiency/resupply was described as a pioneering study of crop. We quantified 2288 crotonylation sites on 971 proteins in tea leaves. After NH4+resupply, the crotonylated proteins played key roles in primary metabolic processes, especially in photosynthesis, carbon fixation and amino acid metabolism in tea leaves. Furthermore, the results of analysis implied that lysine crotonylation was associated with activities of certain enzymes by sensing NH4+. The ultimate function of the crotonylated proteins requires further thoroughly research by site-specific crotonylation antibodies. The information obtained from the lysine crotonylation could be helpful in exploring metabolic mechanism of proteins of tea plants in response to N.
Additional files
The nutrient solution formula. (XLSX 10 kb)
The detailed description of protein extraction and trypsin digestion. (DOCX 18 kb)
The methods of enzyme activity determination. (DOCX 49 kb)
Figure S1. The physiological analyses of tea leaves after NH4+ resupply. Figure S2. SDS-PAGE of three samples under NH4+ deficiency/resupply. Figure S3. The number of crotonylation sites identified per protein. Figure S4. The Venn diagram analysis of DCPs at 3 h and 3d of NH4+ resupply. (DOCX 343 kb)
The information of lysine crotonylome. (XLSX 3355 kb)
Fifty representative LC-MS/MS spectra of crotonylated peptides. (DOCX 8024 kb)
The information of global proteome. (XLSX 1932 kb)
The information of subcellular localization. (XLSX 326 kb)
The information of motif-X. (XLSX 505 kb)
The data of normalization. (XLSX 717 kb)
The information of DCPs. (XLSX 219 kb)
The information of Venn diagram. (XLSX 76 kb)
The information of GO enrichment analysis of DCPs. (XLSX 18 kb)
The information of KEGG enrichment analysis of DCPs. (XLSX 14 kb)
The information of PPI analysis of DCPs. (XLSX 45 kb)
The information of DCPs involved in photosynthesis. (XLSX 12 kb)
The information of DCPs involved in carbon fixation. (XLSX 14 kb)
The information of DCPs involved in amino acid metabolism. (XLSX 11 kb)
The enzyme activities of DCPs after NH4+ resupply. (DOCX 183 kb)
The information of proteins was concerned with DCPs. (XLSX 11 kb)
Acknowledgements
We would like to thank all authors for valuable discussions. We also would like to thank PTM Biolabs (Hangzhou, China) for providing the data of proteomic.
Funding
This work was supported by the Significant Application Projects of Agriculture Technology Innovation in Shandong Province, the Technology System of Modern Agricultural Industry in Shandong Province (SDAIT-19-01) and the Special Foundation for Distinguished Taishan Scholar of Shangdong Province (No.ts201712057), National Natural Science Foundation of China (31800588). The funds played no role in study design, data analysis, and manuscript preparation.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository with the dataset identifier PXD011610. If any further data is required, it will be available from the corresponding author on reasonable requests.
Abbreviations
- ALDO
Fructose-bisphosphate aldolase
- FNR
Ferredoxin--NADP reductase
- GDC
Glycine dehydrogenase
- GGAT2
Glutamate-glyoxylate aminotransferase 2-like
- LPD1
Dihydrolipoyl dehydrogenase 1
- Pbs27
Photosystem II Pbs27 protein
- PGK
Phosphoglycerate kinase
- PRK
Phosphoribulokinase
- PsaF
Photosystem I reaction center subunit III
- PsaN
Photosystem I reaction center subunit N
- PsbO
Oxygen-evolving enhancer protein 1
- PsbP
Oxygen-evolving enhancer protein 2
- PsbQ
Oxygen-evolving enhancer protein 3–2
- PsbS
Photosystem II 22 kDa protein
- rbcl
Ribulose bisphosphate carboxylase large chain
- rbcs
Ribulose bisphosphate carboxylase small chain
- RPIA
Ribose-5-phosphate isomerase 3
- Rubisco
Ribulose bisphosphate carboxylase oxygenase
- RuBP
Ribulose-1,5-bisphosphate
- SBPase
Sedoheptulose-1,7-bisphosphatase
- SGAT
Serine-glyoxylate aminotransferase
- SHMT4
Serine hydroxymethyltransferase 4
- TK
Transketolase
- γ-ATPase
ATP synthase gamma chain
- δ-ATPase
ATP synthase delta chain
Author’s contributions
JS and CQ, carried out the experiment, collected and organized data and wrote the manuscript. WQ, participated in designing the experiment and directed the study. YW, reviewed the manuscript. LS, helped organize data. YL, helped do the experiment. ZD, corresponding author, raised the hypothesis underlying this work, designed the experiment, and helped organize the manuscript structure. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable. The tea plants used in this study were provided by Tea Research Institute of Qingdao Agricultural University. It does not require ethical approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianhao Sun, Email: sunjianhaotea@163.com.
Chen Qiu, Email: qiuchentea@163.com.
Wenjun Qian, Email: qau-WenjunQian@qau.edu.cn.
Yu Wang, Email: wangyutea@163.com.
Litao Sun, Email: slttea@163.com.
Yusheng Li, Email: yusheng8207@163.com.
Zhaotang Ding, Email: dzttea@163.com.
References
- 1.Huang H, Yao Q, Xia E, Gao L. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J Agric Food Chem. 2018;66(37):9828–9838. doi: 10.1021/acs.jafc.8b01995. [DOI] [PubMed] [Google Scholar]
- 2.Yang YY, Li XH, Ratcliffe RG, Ruan JY. Characterization of ammonium and nitrate uptake and assimilation in roots of tea plants. Russ J Plant Physiol. 2013;60(1):91–99. doi: 10.1134/S1021443712060180. [DOI] [Google Scholar]
- 3.Kai F, Dongmei F, Zhaotang D, Yanhua S, Xiaochang W. Cs-miR156 is involved in the nitrogen form regulation of catechins accumulation in tea plant (Camellia sinensis L.) Plant Physiol Biochem Ppb. 2015;97:350–360. doi: 10.1016/j.plaphy.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Liu M-Y, Burgos A, Zhang Q, Tang D, Shi Y, Ma L, Yi X, Ruan J. Analyses of transcriptome profiles and selected metabolites unravel the metabolic response to NH4+ and NO3− as signaling molecules in tea plant ( Camellia sinensis L.) Sci Hortic. 2017;218:293–303. doi: 10.1016/j.scienta.2017.02.036. [DOI] [Google Scholar]
- 5.Jiang J, Gai Z, Wang Y, Fan K, Sun L, Wang H, Ding Z. Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genomics. 2018;19(1):840. doi: 10.1186/s12864-018-5250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159(2):458. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H, Shu L, Garcia BA, Yingming Z. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115(6):2376–2418. doi: 10.1021/acs.chemrev.5b00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Wang DL, Zhao Y. Quantitative Crotonylome analysis expands the roles of p300 in the regulation of lysine Crotonylation pathway. Proteomics. 2018;18(15):e1700230. doi: 10.1002/pmic.201700230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, Liu X, Chen J, Gao S, Liu L, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27(7):898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H, Liu X, Li F, Li W, Zhang J, Xiao Z, Shen L, Li Y, Wang F, Yang J. First comprehensive proteome analysis of lysine crotonylation in seedling leaves of Nicotiana tabacum. Sci Rep. 2017;7(1):3013. doi: 10.1038/s41598-017-03369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Xue C, Fang Y, Chen G, Peng X, Zhou Y, Chen C, Liu G, Gu M, Wang K, et al. Global involvement of lysine Crotonylation in protein modification and transcription regulation in Rice. Mol Cell Proteomics. 2018;17(10):1922–1936. doi: 10.1074/mcp.RA118.000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148(1):350–382. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- 13.Zheng C, Wang Y, Ding Z, Zhao L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front Plant Sci. 2016;7:1858. doi: 10.3389/fpls.2016.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Andres O, Sanchez-Niño MD, Cannata-Ortiz P, Ruiz-Ortega M, Egido J, Ortiz A, Sanz AB. Histone lysine crotonylation during acute kidney injury in mice. Dis Model Mech. 2016;9(6):633–645. doi: 10.1242/dmm.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Li W, Wang C, Fan P, Cao L, Wu Z, Wang F. Ultradeep lysine Crotonylome reveals the Crotonylation enhancement on both histones and nonhistone proteins by SAHA treatment. J Proteome Res. 2017;16(10):3664. doi: 10.1021/acs.jproteome.7b00380. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Yu H, Liu Y, Liu X, Zhang Y, Bu C, Yuan S, Chen Z, Xie G, Li W. Chromodomain protein CDYL acts as a Crotonyl-CoA hydratase to regulate histone Crotonylation and spermatogenesis. Mol Cell. 2017;67(5):853–866. doi: 10.1016/j.molcel.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Nikiforou C, Manetas Y. Inherent nitrogen deficiency in Pistacia lentiscus preferentially affects photosystem I: a seasonal field study. Funct Plant Biol. 2011;38(11):848–855. doi: 10.1071/FP11040. [DOI] [PubMed] [Google Scholar]
- 18.Rui DU, Zhang HH, Tian Y, Sun GY. Effects of different nitrogen forms on photosynthetic Characteristcs of PSII energy distribution in tobacco leaves. Hubei Agric Sci. 2014;34(31):659–662. [Google Scholar]
- 19.Mu X, Chen Q, Chen F, Yuan L, Mi G. Within-leaf nitrogen allocation in adaptation to low nitrogen supply in maize during grain-filling stage. Front Plant Sci. 2016;7:699. [DOI] [PMC free article] [PubMed]
- 20.Nelson N, Yocum CF. Structure and function of photosystems I and II. Annu Rev Plant Biol. 2006;57(57):521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- 21.Nickelsen J, Rengstl B. Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol. 2013;64(64):609–635. doi: 10.1146/annurev-arplant-050312-120124. [DOI] [PubMed] [Google Scholar]
- 22.Kolber Z, Zehr J, Falkowski P. Effects of growth irradiance and nitrogen limitation on photosynthetic energy conversion in photosystem II. Plant Physiol. 1988;88(3):923–929. doi: 10.1104/pp.88.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa T, Sonoike K. Effects of bleaching by nitrogen deficiency on the quantum yield of photosystem II in Synechocystis sp. PCC 6803 revealed by chlorophyll fluorescence measurements. Plant Cell Physiol. 2016;57(3):558–567. doi: 10.1093/pcp/pcw010. [DOI] [PubMed] [Google Scholar]
- 24.Barber J, Nield J, Morris EP, Zheleva D, Hankamer B. The structure, function and dynamics of photosystem two. Physiol Plant. 2010;100(4):817–827. doi: 10.1111/j.1399-3054.1997.tb00008.x. [DOI] [Google Scholar]
- 25.Bricker TM, Frankel LK. Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant photosystem II: a critical analysis. J Photochem Photobiol B Biol. 2011;104(1–2):165–178. doi: 10.1016/j.jphotobiol.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Nowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, Rögner M. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18(11):3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumit VI, Essigke T, Cortez N, Ullmann GM. Mechanistic insights into ferredoxin–NADP(H) reductase catalysis involving the conserved glutamate in the active site. J Mol Biol. 2010;397(3):814–825. doi: 10.1016/j.jmb.2010.01.063. [DOI] [PubMed] [Google Scholar]
- 28.Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimataariga Y, Hase T. Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP+ reductase. Nat Struct Biol. 2001;8(2):117–121. doi: 10.1038/84097. [DOI] [PubMed] [Google Scholar]
- 29.Yamori W, Takahashi S, Makino A, Price GD, Badger MR, Von CS. The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 2011;155(2):956–962. doi: 10.1104/pp.110.168435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rott M, Martins NF, Thiele W, Lein W, Bock R, Kramer DM, Schöttler MA. ATP synthase repression in tobacco restricts photosynthetic Electron transport, CO2 assimilation, and plant growth by Overacidification of the thylakoid lumen. Plant Cell. 2011;23(1):304–321. doi: 10.1105/tpc.110.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honoki R, Ono S, Oikawa A, Saito K, Masuda S. Significance of accumulation of the alarmone (p) ppGpp in chloroplasts for controlling photosynthesis and metabolite balance during nitrogen starvation in Arabidopsis. Photosynth Res. 2017;135(1–3):1–10. doi: 10.1007/s11120-017-0402-y. [DOI] [PubMed] [Google Scholar]
- 32.Zong YZ, Shangguan ZP. Nitrogen deficiency limited the improvement of photosynthesis in maize by elevated CO2 under drought. J Integr Agric. 2014;13(1):73–81. doi: 10.1016/S2095-3119(13)60349-4. [DOI] [Google Scholar]
- 33.Scheible W-R, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136(1):2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song H, Yan QW, Tu B, Chen YJ, Zhang L, Liu W, Chen X, Xie FC, Qin LG. Effects of water and nitrogen interaction on chlorophyll fluorescence parameters and RuBisCO activity in Kentucky Bluegrass. Chin J Grassland. 2017;39(05):31–38. [Google Scholar]
- 35.Lin ZH, Zhong QS, Chen CS, Ruan QC, Chen ZH, You XM. Carbon dioxide assimilation and photosynthetic electron transport of tea leaves under nitrogen deficiency. Bot Stud. 2016;57(1):37. doi: 10.1186/s40529-016-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. A small decrease of plastid Transketolase activity in antisense tobacco Transformants has dramatic effects on photosynthesis and Phenylpropanoid metabolism. Plant Cell. 2001;13(3):535–551. doi: 10.1105/tpc.13.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miziorko HM. Phosphoribulokinase: current perspectives on the structure/function basis for regulation and catalysis. Adv Enzymol Relat Area Mol Biol. 2000;74:95–127. doi: 10.1002/9780470123201.ch3. [DOI] [PubMed] [Google Scholar]
- 38.Banks FM, Driscoll SP, Parry MA, Lawlor DW, Knight JS, Gray JC, Paul MJ. Decrease in phosphoribulokinase activity by antisense RNA in transgenic tobacco. Relationship between photosynthesis, growth, and allocation at different nitrogen levels. Plant Physiol. 1999;119(3):1125–1136. doi: 10.1104/pp.119.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan J, Haerdter R, Gerendás J. Impact of nitrogen supply on carbon/nitrogen allocation: a case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Biol. 2010;12(5):724–734. doi: 10.1111/j.1438-8677.2009.00288.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Mao X, Ou J, Ye N, Zhang J, Peng X. Distinct photorespiratory reactions are preferentially catalyzed by glutamate:glyoxylate and serine:glyoxylate aminotransferases in rice. J Photochem Photobiol B Biol. 2015;142(142C):110–117. doi: 10.1016/j.jphotobiol.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Igarashi D, Tsuchida H, Miyao M, Ohsumi C. Glutamate:Glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiol. 2006;142(3):901–910. doi: 10.1104/pp.106.085514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai V, Muthuraj M, Gandhi MN, Das D, Srivastava S. Real-time iTRAQ-based proteome profiling revealed the central metabolism involved in nitrogen starvation induced lipid accumulation in microalgae. Sci Rep. 2017;7:45732. doi: 10.1038/srep45732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Jiang Z, Huang B. Nitrogen deficiency-induced protein changes in immature and mature leaves of creeping bentgrass. J Am Soc Hortic Sci Am Soc Hortic Sci. 2011;136(6):399–407. doi: 10.21273/JASHS.136.6.399. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The nutrient solution formula. (XLSX 10 kb)
The detailed description of protein extraction and trypsin digestion. (DOCX 18 kb)
The methods of enzyme activity determination. (DOCX 49 kb)
Figure S1. The physiological analyses of tea leaves after NH4+ resupply. Figure S2. SDS-PAGE of three samples under NH4+ deficiency/resupply. Figure S3. The number of crotonylation sites identified per protein. Figure S4. The Venn diagram analysis of DCPs at 3 h and 3d of NH4+ resupply. (DOCX 343 kb)
The information of lysine crotonylome. (XLSX 3355 kb)
Fifty representative LC-MS/MS spectra of crotonylated peptides. (DOCX 8024 kb)
The information of global proteome. (XLSX 1932 kb)
The information of subcellular localization. (XLSX 326 kb)
The information of motif-X. (XLSX 505 kb)
The data of normalization. (XLSX 717 kb)
The information of DCPs. (XLSX 219 kb)
The information of Venn diagram. (XLSX 76 kb)
The information of GO enrichment analysis of DCPs. (XLSX 18 kb)
The information of KEGG enrichment analysis of DCPs. (XLSX 14 kb)
The information of PPI analysis of DCPs. (XLSX 45 kb)
The information of DCPs involved in photosynthesis. (XLSX 12 kb)
The information of DCPs involved in carbon fixation. (XLSX 14 kb)
The information of DCPs involved in amino acid metabolism. (XLSX 11 kb)
The enzyme activities of DCPs after NH4+ resupply. (DOCX 183 kb)
The information of proteins was concerned with DCPs. (XLSX 11 kb)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium via the PRIDE partner repository with the dataset identifier PXD011610. If any further data is required, it will be available from the corresponding author on reasonable requests.