Abstract
MicroRNAs (miRNAs), a subgroup of small noncoding RNAs, play critical roles in tumor growth and metastasis. Accumulating evidence shows that the dysregulation of miRNAs is associated with the progression of hepatocellular carcinoma (HCC). However, the molecular mechanism by which miR-942-3p contributes to HCC remains undocumented. The association between miR-942-3p expression and the clinicopathological characteristics in HCC patients was analyzed by The Cancer Genome Atlas data set. The targets of miR-942-3p were identified by bioinformatic analysis and dual luciferase report assay. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and Transwell assays were performed to assess the functional role of miR-942-3p in HCC cells. Consequently, we found that miR-942-3p expression level was elevated in HCC tissues and cell lines as compared with the normal tissues and was associated with the pathological stage and tumor node metastasis (TNM) stage, acting as an independent prognostic factor of poor survival in patients with HCC. Ectopic expression of miR-942-3p enhanced the proliferation and invasive potential of HCC cells, but inhibition of miR-942-3p expression had the opposite effects. Mannose-binding lectin 2 (MBL2) was further identified as a direct target of miR-942-3p and possessed a negative correlation with miR-942-3p expression and unfavorable survival in patients with HCC. Restoration of MBL2 inhibited the progression of HCC cells and attenuated the tumor-promoting effects induced by miR-942-3p. In conclusion, miR-942-3p may act as an oncogenic factor in HCC cells by targeting MBL2 and provide a potential marker for patients with HCC.
Keywords: miR-942-3p, proliferation, invasion, MBL2, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) ranks as the second leading cause of cancer-related death and its incidence rate is gradually rising annually in the world.1 In spite of the great advances in the therapeutic strategies of HCC, such as hepatectomy, ablation, and chemotherapy, the prognostic outcomes of the patients are still unfavorable owing to its unlimited proliferation and distant metastasis.2 It is well known that the initiation and progression of HCC is associated with multiple genetic mutations as well as the activation of oncogenes or inactivation of antioncogenes.3 Therefore, identification of the promising therapeutic targets may provide insights into the early detection and treatment of HCC.
MicroRNAs (miRNAs), a subgroup of small noncoding RNAs, function by binding to the 3′-untranslated region (3′-UTR) of their target messenger RNAs (mRNAs).4 Increasing data show that the dysregulation of miRNAs is associated with the progression of HCC.5-7 MiRNAs act as oncogenes8-10 or tumor suppressors,11-13 involved in the pathogenesis of HCC. But, little knowledge is known about the role of miR-942 in cancers. Some studies show that miR-942 has an increased expression in Huntington disease and influences its progression14 and Kaposi’s sarcoma-associated herpes virus lytic replication.15 Moreover, miR-942 expression is upregulated in metastatic renal cell carcinoma (MRCC)16,17 and lung adenocarcinoma18 and predicts poor survival in patients with MRCC.16,17 miR-942 facilitates MRCC cell migration17 and cancer stem cell–like traits in esophageal carcinoma via activation of the Wnt/β-catenin pathway19 and induces cell apoptosis in cancers.20,21 These studies suggest that miR-942 acts an oncogenic factor in cancer cells.
However, the functional role of miR-942 in HCC remains unclear. In the present study, we found that increased expression of miR-942-3p was associated with the pathological stage and TNM stage, acting as an independent prognostic factor of poor survival in patients with HCC. Reexpression of miR-942-3p promoted the proliferation and invasive potential of HCC cells by targeting mannose-binding lectin 2 (MBL2) and might be a potential prognostic marker for HCC.
Materials and Methods
Clinical Data
The data including 276 cases of patients with HCC, overall survival (OS) time and status, recurrence time and status, and miR-942-3p and MBL2 expression levels were downloaded from The Cancer Genome Atlas (TCGA) Liver Cancer database (https://genome-cancer.ucsc.edu).
Identification of the Target Genes of miR-942-3p
The target genes of miR-942-3p were identified by using the prediction tool TargetScanHuman7.1 (http://www.targetscan.org/vert_71/), and according to the cumulative weighted text score, 14 target genes of miR-942-3p were selected for further investigation.
Cell Culture
Normal liver tissue and HCC cell lines (Huh7, SMMC-7721, SK-hep-1, HepG2, LO2, and Huh6) were purchased from Chinese Academy of Sciences Cell Bank and were cultured in Dulbecco Modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction
The total RNA of HCC cells was extracted by using TRIzol. Reverse transcription was performed by universal RT-PCR kit (M-MLV) and complementary DNA amplification using the SYBR Green Master Mix kit (Takara, Otsu, Japan). Total RNA for miRNAs was isolated using a High Pure miRNA isolation kit (Roche) and reverse transcription-polymerase chain reaction (RT-PCR) using a TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Shanghai, China). Data were analyzed using the comparative Ct method (2−△△Ct). Three experiments were performed for each clone. The primers were listed in Supplementary Table S1.
Western Blot Analysis
LO2 and HepG2 cell lines were harvested and extracted using lysis buffer. Cell extracts were boiled in loading buffer and equal amounts of cell extracts were separated on 15% sodium dodecyl sulfate -polyacrylamide gel electrophoresis (SDS-PAGE) gels. Separated protein bands were transferred into polyvinylidene fluoride membranes. The primary antibodies against MBL2 (24207-1-AP, rabbit polyclonal antibody; Proteintech, Rosemont, Illinois) and β-actin (ab16039, rabbit polyclonal antibody; Abcam, Cambridge, Massachusetts) were diluted at a ratio of 1:1000 according to the instructions and incubated overnight at 4°C. The specific details were described as previously reported.11
Luciferase Reporter Assay
LO2 and HepG2 cell lines were seeded into 96-well plates and were cotransfected with a mixture of 60 ng of luciferase, 6 ng of pRL-CMV Renilla luciferase reporter, and miR-942-3p mimic or inhibitor. After 48 hours of incubation, the luciferase activities were measured with a dual-luciferase reporter assay (Promega, Madison, Wisconsin).
Plasmid, siRNAs, and miR-942-3p Mimic and Inhibitor
Plasmid-mediated pcDNA3.1-MBL2 (MBL2), small interfering RNA (siRNA) targeting MBL2 (si-MBL2), miR-942-3p mimic and inhibitor, and the control vectors were purchased from Genepharma (Shanghai, China). LO2 and HepG2 cell lines were planted in six-well plates 24 hours prior to si-MBL2, pcDNA3.1-MBL2, miR-942-3p mimic or inhibitor transfection with 50% to 70% confluence and then were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, California), according to the manufacturer’s instructions.
Cell Viability and Invasion Assays
Cell viability was analyzed by using the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, and cell invasive potential was conducted by using Transwell assay. The specific details were described as previously reported.11
Statistical Analysis
Statistical analyses were conducted by SPSS 20.0 (IBM, SPSS, Chicago, Illinois) and GraphPad Prism. Student t test, χ2 test, and analysis of variance were used to evaluate the statistical significance for the comparisons of the groups. Pearson correlation coefficient analysis was used to analyze the correlations of miR-942-3p with its target genes in HCC tissues. The OS and recurrence curves were analyzed with the Kaplan-Meier and log-rank test. Univariate or multivariate analysis was performed by using a Cox proportional hazards regression model. P < .05 was considered statistically significant.
Results
Upregulation of miR-942-3p Expression Was Associated With Poor Survival in Patients With HCC
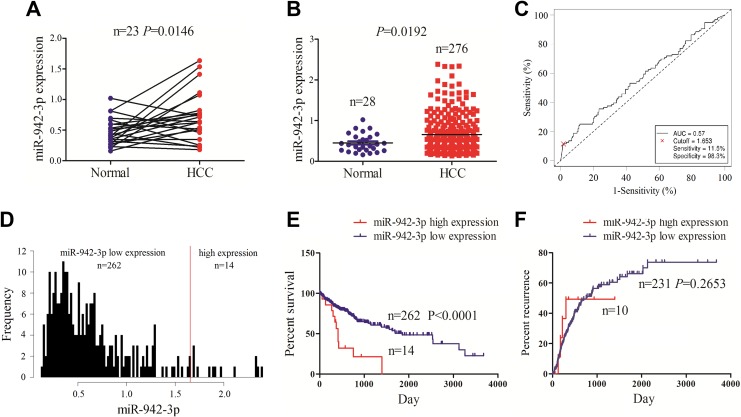
Out results showed that miR-942-3p expression level was increased in paired (Figure 1A) and unpaired HCC tissues (Figure 1B) in comparison with the adjacent normal tissues by using TCGA data set. According to the OS time, survival status, and miR-942-3p expression level, we obtained a cutoff value of miR-942-3p in HCC tissues (Figure 1C) and divided the patients into 2 groups: high miR-942-3p expression and low miR-942-3p expression (Figure 1D). We further analyzed the association between miR-942-3p expression and the clinicopathological parameters in patients with HCC and found that high expression of miR-942-3p was associated with the pathological stage (P = .047) and TNM stage (P = .037), but had no association with other factors (each P > .05; Table 1). Kaplan-Meier analysis showed that the patients with high miR-942-3p expression displayed a poorer survival (Figure 1E), but had no difference in tumor recurrence (Figure 1F), as compared to those with low miR-942-3p expression. Univariate and multivariate Cox regression analyses revealed that high miR-942-3p expression was an independent prognostic factor of poor survival in patients with HCC (Table 2).
Figure 1.
The expression of miR-942-3p was associated with poor survival in patients with hepatocellular carcinoma (HCC). A and B, The Cancer Genome Atlas (TCGA) analysis showed that the expression level of miR-942-3p was increased in paired and unpaired HCC tissues as compared with the normal tissues. C, Receiver operating characteristic (ROC) curve was used to obtain a cutoff value of miR-942-3p in patients with HCC. D, Patients with HCC were divided into high or low miR-942-3p expression group according to the cutoff value. E and F, Kaplan-Meier analysis demonstrated that the patients with high miR-942-3p expression displayed a poorer survival but had no difference in tumor recurrence as compared with those with low miR-942-3p expression in patients with HCC.
Table 1.
The Association of miR-942-3p Expression With Clinicopathologic Characteristics in Patients With HCC.
| Variables | Cases (n) | miR-942-3p | P Value | |
|---|---|---|---|---|
| High | Low | |||
| Total | 260 | 14 | 246 | |
| Age (years) | ||||
| ≥60 | 136 | 6 | 130 | |
| <60 | 124 | 8 | 116 | .325 |
| Gender | ||||
| Male | 173 | 10 | 163 | |
| Female | 87 | 4 | 83 | .469 |
| Pathological stage | ||||
| Ⅰ/Ⅱ | 191 | 7 | 184 | |
| Ⅲ/Ⅳ | 69 | 7 | 62 | .047 |
| TNM stage | ||||
| T1/T2 | 194 | 7 | 187 | |
| T3/T4 | 66 | 7 | 59 | .037 |
| Lymphatic invasion | ||||
| Negative | 191 | 11 | 180 | |
| Positive | 69 | 3 | 66 | .465 |
| Distant metastasis | ||||
| Negative | 206 | 11 | 195 | |
| Positive | 54 | 3 | 51 | .585 |
TNM: tumor node metastasis.
Table 2.
Cox Regression Analysis of miR-942-3p Expression as Survival Predictor.
| Variables | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | ||
|---|---|---|---|---|
| RR (95% CI) | P Value | RR (95% CI) | P Value | |
| Age (years) | ||||
| ≥ 60 vs <60 | 1.192 (0.775-1.833) | .423 | NA | NA |
| Gender | ||||
| Male vs Female | 0.922 (0.593-1.434) | .718 | NA | NA |
| Pathological stage | ||||
| Ⅲ/Ⅳ vs Ⅰ/Ⅱ | 2.919 (1.892-4.530) | <.0001 | 1.689 (0.232-12.318) | .605 |
| TNM stage | ||||
| T3 + T4 vs T1 + T2 | 2.967 (1.920-4.584) | <.0001 | 1.654 (0.225-12.174) | .621 |
| Lymphatic invasion | ||||
| Positive vs negative | 1.109 (0.675-1.824) | .683 | NA | NA |
| Distant metastasis | ||||
| Positive vs negative | 1.289 (0.778-2.135) | .324 | NA | NA |
| miR-942-3p expression | ||||
| High vs low | 3.774 (1.981-7.191) | <.0001 | 3.127 (1.629-6.002) | .001 |
NA: not analyzed; TNM: tumor node metastasis.
MiR-942-3p Promoted the Proliferation and Invasion of HCC Cells
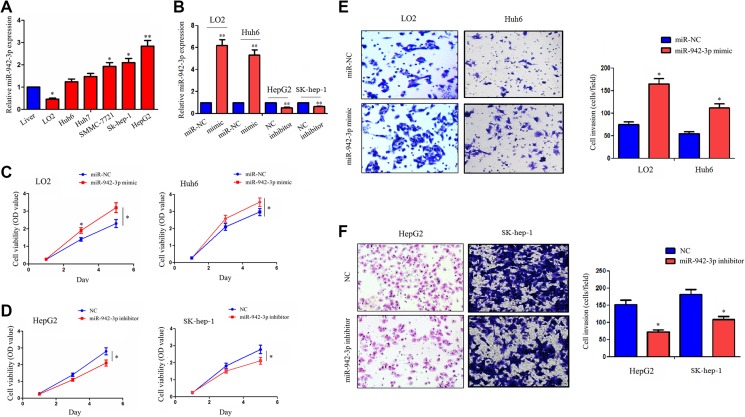
We detected the expression levels of miR-942-3p in different HCC cell lines by quantitative polymerase chain reaction (qPCR) analysis, indicating that miR-942-3p had a lower expression level in LO2 and Huh6 cell lines but a higher expression level in HepG2 and SK-hep-1 cell lines, as compared with the normal liver tissue (Figure 2A). The transfection efficiency of miR-942-3p mimic in LO2 and Huh6 cell lines and miR-942-3p inhibitor in HepG2 and SK-hep-1 cell lines were respectively validated by qPCR analysis (Figure 2B). To investigate the function of miR-942-3p in HCC cells, we performed the MTT and Transwell assays and found that miR-942-3p mimic increased the cell viability (Figure 2C) and invasive potential (Figure 2E) in LO2 and Huh6 cell lines, while miR-942-3p inhibitor had the opposite effects in HepG2 and SK-hep-1 cell lines, as compared with the control group (Figure 2D, F).
Figure 2.
miR-942-3p promoted the proliferation and invasion of hepatocellular carcinoma (HCC) cells. A, Quantitative polymerase chain reaction (qPCR) analysis of the expression levels of miR-942-3p in different HCC cell lines and normal liver tissue. B, The qPCR assessment of the transfection efficiency of miR-942-3p mimic or inhibitor in LO2 or HepG2 cell line. C and D, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that miR-942-3p mimic promoted the cell viability in LO2 and Huh6 cell lines, but its inhibitor inhibited the cell viability in HepG2 and SK-hep-1 cell lines as compared with the control group. E and F, Transwell assay showed that miR-942-3p mimic promoted the invasive potential in LO2 and Huh6 cell lines, but its inhibitor weakened the invasive potential in HepG2 and SK-hep-1 cell lines as compared with the control group. Data were given as mean ± standard error of the mean of 3 experiments. *P < .05; **P < .01.
Mannose-Binding Lectin 2 Was Identified to Have a Negative Correlation With miR-942-3p Expression in Patients With HCC
According to the cumulative weighted text scores, we used the prediction tool TargetScanHuman7.1 to identify14 target genes of miR-942-3p and detected their expression levels in paired HCC tissues (n = 23), which indicated that 5 genes (MBL2, ROS4, GNG12, SASH1, and SOS2) had a significantly decreased expression in HCC tissues (Figure 3A), and only MBL2 displayed a negative correlation with miR-942-3p expression in HCC tissues (Figure 3B, C). We further obtained a cutoff value of MBL2 in HCC tissues (Figure 3D) and divided the patients into high and low MBL expression groups (Figure 3E). We found that MBL2 expression was associated with the pathological stage (P = .001) and TNM stage (P = .004), but had no association with other factors (each P > .05, Supplementary Table S2). Kaplan-Meier analysis demonstrated that the patients with high MBL2 expression exhibited a better survival but had no difference in tumor recurrence as compared with those with low MBL2 expression (Figure 3F). Multivariate Cox regression analyses revealed that MBL2 expression was not an independent prognostic factor of OS in patients with HCC (Supplementary Table S3).
Figure 3.
MiR-942-3p had a negative correlation with mannose-binding lectin 2 (MBL2) expression in patients with hepatocellular carcinoma (HCC). A, The Cancer Genome Atlas (TCGA) analysis of the expression levels of 14 target genes of miR-942-3p in paired HCC tissues (n = 23). B and C, Pearson correlation analysis revealed that miR-942-3p had a negative correlation with MBL2 expression rather than other target genes in HCC tissues. D, Receiver operating characteristic (ROC) curve was used to determine a cutoff value of MBL2 in patients with HCC. E, Patients with HCC were divided into high or low MBL2 expression group according to the cutoff value. F, Kaplan-Meier analysis demonstrated that the patients with high MBL2 expression displayed a better survival but had no difference in tumor recurrence as compared with those with low MBL2 expression in patients with HCC.
Mannose-Binding Lectin 2 Was Validated as a Direct Target of miR-942-3p in HCC Cells
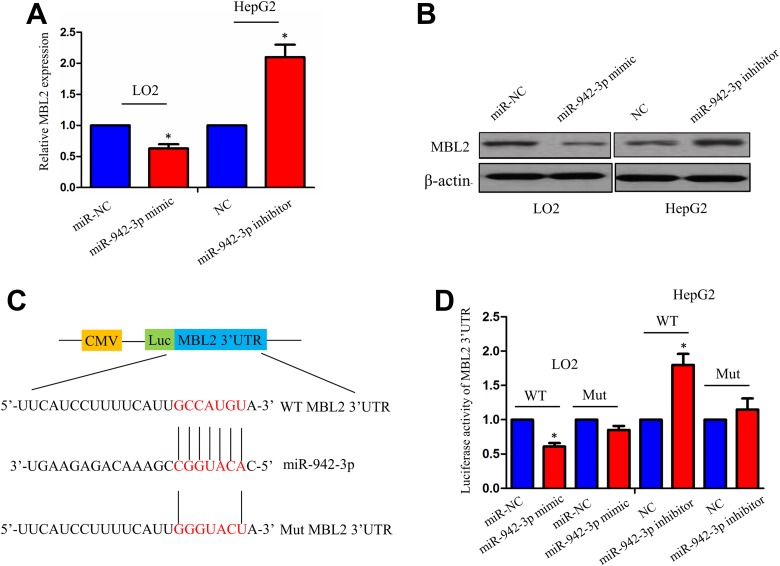
MR-942-3p mimic and inhibitor were respectively transfected into LO2 and HepG2 cell lines. After transfection for 48 hours, qPCR and Western blot analysis indicated that miR-942-3p mimic dramatically decreased the mRNA and protein levels of MBL2 in LO2 cells, but miR-942-3p inhibitor increased their expression levels in HepG2 cells (Figure 4A, B). Furthermore, luciferase reporter vector containing the wild-type (WT) or mutant (Mut) 3′-UTR of MBL2 (Figure 4C) was cotransfected with miR-942-3p mimic or inhibitor in LO2 or HepG2 cell line. The results showed that the luciferase activity of WT 3′-UTR of MBL2 was decreased by miR-942-3p mimic in LO2 cells but increased by miR-942-3p inhibitor in HepG2 cells (Figure 4D). However, the luciferase activity of Mut 3′-UTR of MBL2 was not affected by miR-942-3p mimic or inhibitor in HCC cells (Figure 4D).
Figure 4.
Identification of mannose-binding lectin 2 (MBL2) as a direct target of miR-942-3p in hepatocellular carcinoma (HCC) cells. A and B, Quantitative polymerase chain reaction (qPCR) and Western blot analysis of the effects of miR-942-3p mimic or inhibitor on the messenger RNA (mRNA) and protein levels of MBL2 in LO2 or HepG2 cell line. C, The binding sites of miR-942-3p with wild-type (WT) or mutant (Mut) 3′-untranslated region (3′-UTR) of MBL2. D, The luciferase activities of WT or Mut 3′-UTR of MBL2 after cotransfection with miR-942-3p mimic or inhibitor and WT or Mut 3′-UTR of MBL2 in HCC cells. Data were given as mean ± standard error of the mean of 3 experiments. *P< .05.
Mannose-Binding Lectin 2 Reversed the Tumor-Promoting Effects of miR-942-3p in HCC Cells
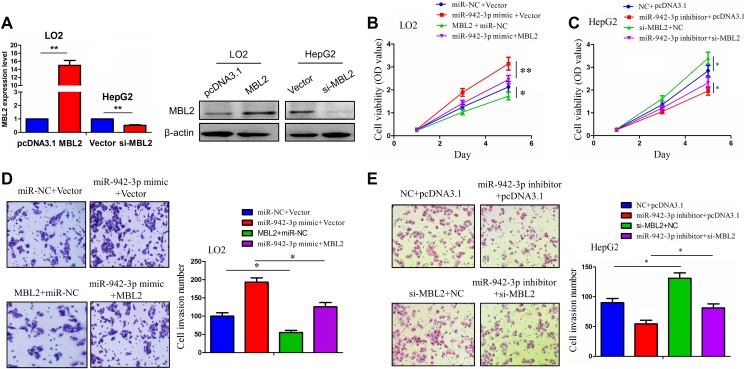
To verify the functional association between miR-942-3p and MBL2 expression in HCC cells, we conducted the MTT and Transwell assays. The transfection efficiency of MBL2 plasmid in LO2 cells or si-MBL2 in HepG2 cells was determined by qPCR and Western blot analysis (Figure 5A). Then, we found that MBL2 overexpression inhibited the cell viability and invasive potential and reversed the tumor-promoting effects induced by miR-942-3p in LO2 cells (Figure 5B, D), while knockdown of MBL2 enhanced cell viability and invasive potential and reversed the tumor suppressive effects caused by miR-942-3p inhibitor in HepG2 cells (Figure 5C, E).
Figure 5.
Mannose-binding lectin 2 (MBL2) reversed the tumor-promoting effects of miR-942-3p in hepatocellular carcinoma (HCC) cells. A, Quantitative polymerase chain reaction (qPCR) and Western blot analysis of the transfection efficiency of MBL2 plasmid or si-MBL2 in LO2 or HepG2 cell line. B and C, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) analysis of the cell viability after cotransfection with miR-942-3p mimic and MBL2 plasmid or miR-942-3pinhibitor and si-MBL2 in HCC cells. D and E, Transwell analysis of the cell invasive potential after cotransfection of miR-942-3p mimic and MBL2 plasmid or miR-942-3p inhibitor and si-MBL2 in HCC cells. Data were given as mean ± standard error of the mean of 3 experiments. *P< .05; **P< .01.
Discussion
Some studies have shown that miR-942-3p expression is increased in multiple malignancies16-18 and is associated with poor prognosis in patients with MRCC.16,17 Interestingly, it has been reported that activation of ERK pathway can downregulate the miRNAs through phosphorylating exportin-522 and Pin1 impairs the biogenesis of miRNAs by mediating conformation change of XPO5 in HCC,23 indicating that these pathways may explain the downregulation of miRNA expression in HCC, but miR-942-3p is not regulated by these pathways.22,23 The clinical significance of miR-942-3p in HCC is also unclear. In our study, miR-942-3p expression was found upregulated and associated with the pathological and TNM stages in patients with HCC. Multivariate Cox regression analysis revealed miR-942-3p as an independent prognostic factor of poor survival in patients with HCC. These findings indicated that miR-942-3p might be a potential biomarker of HCC.
Functionally, some studies have shown that miR-942-3p promotes cell proliferation and invasion and induces cell apoptosis in several cancers.19-21 We also assessed the role of miR-942-3p in HCC cells and found that miR-942-3p mimic enhanced cell proliferation and invasion, whereas its inhibitor had opposite effects in HCC cells. Our results suggested that miR-942-3p might act as an oncogenic factor in HCC cells.
Furthermore, we identified the target genes of miR-942-3p and found that MBL2 was downregulated and had a negative correlation with miR-942-3p expression in HCC tissues. Low-risk genotype of MBL2 is associated with increased risk of bacterial infections in children with B acute lymphoblastic leukemia.24 The serum levels of MBL2 are decreased in colon and lung cancers and exhibit a negative correlation with miR-27a expression and an increased cancer risk.25,26 The polymorphism of MBL2 gene is associated with the risk of gastric cancer.27 In our study, high MBL2 expression was associated with pathological and TNM stages and a favorable survival in patients with HCC. Mannose-binding lectin 2 activates the classical apoptosis signaling and associates with breast cancer following the treatment of doxorubicin and cyclophosphamide.28 We further confirmed that miR-942-3p downregulated MBL2 expression by binding to its 3′-UTR, and MBL2 inhibited the proliferation and invasion of HCC cells and reversed the tumor-promoting effects of miR-942-3p. These findings indicated that miR-942-3p might promote the proliferation and invasion of HCC cells by targeting MBL2.
Conclusions
Our findings demonstrated that miR-942-3p expression was upregulated in HCC tissues and acted as an independent prognostic factor of poor survival in patients with HCC. Further investigations showed that miR-942-3p promoted the proliferation and invasion of HCC cells by targeting MBL2 and might be a potential prognostic marker for HCC.
Supplemental Material
Supplementary_Tables_(3) for MiR-942-3p Promotes the Proliferation and Invasion of Hepatocellular Carcinoma Cells by Targeting MBL2 by Chun-Yang Xu, Jun-Feng Dong, Zi-Qi Chen, Guo-Shan Ding and Zhi-Ren Fu in Cancer Control
Footnotes
Authors’ Note: Chun-Yang Xu, Jun-Feng Dong, and Zi-Qi Chen contributed equally to this article. The present study was approved by the Ethics Committee and Protection of Human Subjects Committee of Changzheng Hospital (No. 2017 1324 B463), and we confirmed that all patients gave informed consent.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Natural Science Foundation of China (Grant No. 81671576 and 81470900), Shanghai Sailing Program (No. 17YF1425500), and Shanghai Municipal Health and Family Planning Commission Scientific Research Project (No. 201640274).
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. [DOI] [PubMed] [Google Scholar]
- 3. Lee JS. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 2015;21(3):220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babski J, Maier LK, Heyer R, et al. Small regulatory RNAs in Archaea. RNA Biol. 2014;11(5):484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu T, Zhang X, Sha K, Liu X, Zhang L, Wang B. miR-709 up-regulated in hepatocellular carcinoma, promotes proliferation and invasion by targeting GPC5. Cell Prolif. 2015;48(3):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Yang C, Xu Y, Cheng F, et al. miR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/β-catenin signaling through targeting BCL9. Cell Death Dis. 2017;8(8):e2999. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Xu B, Xu T, Liu H, Min Q, Wang S, Song Q. MiR-490-5p suppresses cell proliferation and invasion by targeting BUB1 in hepatocellular carcinoma cells. Pharmacology. 2017;100(5-6):269–282. [DOI] [PubMed] [Google Scholar]
- 8. Han G, Zhang L, Ni X, et al. MicroRNA-873 promotes cell proliferation, migration, and invasion by directly targeting TSLC1 in hepatocellular carcinoma. Cell Physiol Biochem. 2018;46(6):2261–2270. [DOI] [PubMed] [Google Scholar]
- 9. Huang L, Cai JL, Huang PZ, et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996–2008. [PMC free article] [PubMed] [Google Scholar]
- 10. Jia B, Tan L, Jin Z, Jiao Y, Fu Y, Liu Y. MiR-892a promotes hepatocellular carcinoma cells proliferation and invasion through targeting CD226. J Cell Biochem.2017;118(6):1489–1496. [DOI] [PubMed] [Google Scholar]
- 11. Chen SY, Ma DN, Chen QD, et al. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer. 2017;8(4):617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin Y, Chen W, Liu B, et al. MiR-200c Inhibits the tumor progression of glioma via targeting moesin. Theranostics. 2017;7(6):1663–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang Z, Wang X, Xu X, et al. MicroRNA-608 inhibits proliferation of bladder cancer via AKT/FOXO3a signaling pathway. Mol Cancer. 2017;16(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Díez-Planelles C, Sánchez-Lozano P, Crespo MC, et al. Circulating microRNAs in Huntington’s disease: emerging mediators in metabolic impairment. Pharmacol Res. 2016;108:102–110. [DOI] [PubMed] [Google Scholar]
- 15. Yan Q, Shen C, Qin J, et al. HIV-1 Vpr inhibits Kaposi’s sarcoma-associated herpesvirus lytic replication by inducing microRNA miR-942-5p and activating NF-κB signaling. J Virol. 2016;90(19):8739–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kovacova J, Juracek J, Poprach A, et al. Candidate microRNA biomarkers of therapeutic response to sunitinib in metastatic renal cell carcinoma: a validation study in patients with extremely good and poor response. Anticancer Res. 2018;38(5):2961–2965. [DOI] [PubMed] [Google Scholar]
- 17. Prior C, Perez-Gracia JL, Garcia-Donas J, et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS One. 2014;9(1):e86263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PLoS One. 7(9):e46045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ge C, Wu S, Wang W, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6(13):10964–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu N, Zuo C, Wang X, et al. miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget. 2014;5(13):4959–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H. MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One. 2014;9(4):e94501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun HL, Cui R, Zhou J, et al. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell. 2016;30(5):723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J, Pu W, Sun HL, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018;25(9):1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pana ZD, Samarah F, Papi R, et al. Mannose binding lectin and ficolin-2 polymorphisms are associated with increased risk for bacterial infections in children with B acute lymphoblastic leukemia. Pediatr Blood Cancer. 2014;61(6):1017–1022. [DOI] [PubMed] [Google Scholar]
- 25. Zanetti KA, Haznadar M, Welsh JA, et al. 3’-UTR and functional secretor haplotypes in mannose-binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012;72(6):1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pine SR, Mechanic LE, Ambs S, et al. Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J Natl Cancer Inst. 2007;99(18):1401–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang FY, Tahara T, Arisawa T, et al. Mannan-binding lectin (MBL) polymorphism and gastric cancer risk in Japanese population. Dig Dis Sci. 2008;53(11):2904–2908. [DOI] [PubMed] [Google Scholar]
- 28. Jamieson D, Sunter N, Muro S, et al. Pharmacogenetic association of MBL2 and CD95 polymorphisms with grade 3 infection following adjuvant therapy for breast cancer with doxorubicin and cyclophosphamide. Eur J Cancer. 2017;71:15–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Tables_(3) for MiR-942-3p Promotes the Proliferation and Invasion of Hepatocellular Carcinoma Cells by Targeting MBL2 by Chun-Yang Xu, Jun-Feng Dong, Zi-Qi Chen, Guo-Shan Ding and Zhi-Ren Fu in Cancer Control