Abstract
The major sensor kinase controlling the initiation of development in Bacillus subtilis, KinA, functions by activating the phosphorelay signal-transduction system in response to as yet unknown signal ligands. KinA contains, within its amino-terminal signal-sensing region, three PAS domains that, in other proteins, are known to be involved in sensing changes in oxygen concentration and redox potential among other functions. The most amino-terminal PAS domain, PAS-A, was found to bind ATP and catalyze exchange of phosphate between ATP and nucleoside diphosphates. A cysteine-to-alanine mutation in PAS-A increased the affinity for ATP 5-fold, decreased the exchange reaction 2-fold, and stimulated KinA-dependent sporulation. A model for the role of ATP and the exchange reaction in the PAS domain in sensor kinase signal transduction is presented in which the free energy of nucleotide hydrolysis drives the conformational changes that activate or deactivate the sensor kinase in response to signal ligand binding.
For bacteria, the ability to sense and adapt rapidly to a wide variety of environmental conditions is the key to persistence and survival. Two-component signal-transduction systems are integral in a bacterium's adaptive response to cellular and environmental change and mediate the activation or repression of a large repertoire of genes and pathways in response to specific internal and external signals (1). Two-component systems are composed of a signal ligand-responsive histidine protein kinase (sensor kinase) and a response regulator transcription factor. Signals specific for each pathway are sensed and interpreted by the sensor kinase and result in an ATP-dependent autophosphorylation reaction to a histidine residue. The phosphoryl group subsequently is transferred to an aspartate residue of the response regulator, resulting in the activation of its transcriptional properties. The mechanism by which sensor kinases sense and interpret signals is fundamental to the understanding of the roles of two-component systems in the complex cellular processes required for bacterial development, virulence, and pathogenicity (2).
Recently, a large family of proteins has been identified (3) that contains cytosolic signaling modules known as PAS domains, a name coined from the proteins in which these domains originally were recognized (Drosophila Period protein, vertebrate aryl hydrocarbon nuclear translocator, and Drosophila Single-minded protein). PAS domains are found in proteins from all three kingdoms of life, where they monitor changes in overall energy level of the cell, redox potential, oxygen, and light and also mediate protein/protein interactions (3). In bacteria, they are found almost exclusively in sensor kinases of two-component systems.
In some cases, the sensing properties of PAS domains are determined by the associated signal ligands. The ligands for a number of prokaryotic PAS domains have been identified and include FAD (4), heme (5), 4-hydroxycinnamyl (6), and 2Fe–2S centers (7). The conserved fold of PAS domains suggests that similar signaling strategies may be used in all PAS domain-containing proteins, but the specificity of signaling of each pathway likely is to be mediated by the associated signal ligand.
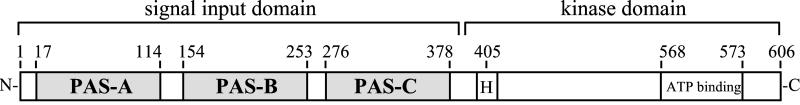
In response to nutrient depletion, the Gram-positive bacterium Bacillus subtilis initiates a complex developmental process that results in the production of a resistant endospore. Initiation of sporulation in B. subtilis is controlled by an expanded, two-component signal-transduction system termed a phosphorelay (8). Multiple sensor kinases are involved in the initiation of development in B. subtilis (9), but the sensor kinase responsible for the majority of phosphate input into the phosphorelay is KinA (10). KinA is a soluble cytoplasmic protein that is active as a dimer and is composed of an amino-terminal signal-input domain and a carboxyl-terminal kinase domain (Fig. 1). The signal-input domain contains three PAS domains, PAS-A, PAS-B, and PAS-C, which presumably regulate the activity of the kinase in response to sporulation-inducing signals.
Figure 1.
Domain organization of KinA. The PAS domains of the amino-terminal signal-input domain and His-405 and ATP-binding site of the kinase domain are indicated. Amino acid residue numbers are also indicated.
Recently, it has been shown that the PAS-A domain of KinA is essential for the activity of this sensor kinase and that PAS-B/PAS-C are involved in homodimerization with PAS-B/PAS-C of the other subunit of the dimer (11). This report describes studies to elucidate the nature of the PAS-A domain signal ligand(s) that are likely to be involved in the regulation of the initiation of development in B. subtilis by means of the activation of phosphorelay sensor kinase A.
Materials and Methods
Purification of the PAS-A Domain of KinA.
The PAS-A domain of KinA spans amino acids 17–114 (Fig. 1). To study the properties of the isolated domain, an expression vector was constructed to allow PAS-A to be purified from Escherichia coli. A DNA fragment corresponding to PAS-A was amplified from the chromosome of B. subtilis JH642 by using PCR with oligonucleotide primers KinA5′Nco (5′-GGATTCCATGGAACAGGATACGCAGCATGTTAAAC-3′) and KinAN3′Xho (5′-CGTTATCTCGAGAGTCGATTCCGGGCTTGCAGG-3′) and cloned into the NcoI and XhoI restriction sites of pET28a (Novagen), producing a carboxyl-terminal His-tag. The resulting construct was confirmed by DNA sequencing.
The PAS-A domain was expressed in E. coli BL21 (DE3) (Novagen) and purified by nickel-affinity chromatography by using His-bind resin (Novagen) according to the manufacturer's instructions. After extensive washing with 20 mM and 50 mM imidazole in buffer A (25 mM Tris⋅Cl, pH 8.0/10 mM KCl), the PAS-A domain was eluted with buffer A containing 100 mM imidazole, dialyzed extensively against buffer A, and concentrated with a Centriprep-10 column (Amicon). When this procedure was used, PAS-A was purified to >99% homogeneity, as determined by SDS/PAGE.
Direct Photoaffinity Labeling of PAS-A with [α-32P]ATP.
Direct photo-cross-linking of nucleotides to proteins is a widely used technique for the investigation of nucleotide-binding sites (12, 13). The PAS-A domain was photoaffinity-labeled with [α-32P]ATP in buffer B {50 mM K-EPPS [4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid], pH 8.5/20 mM MgCl2/0.1 mM EDTA/5% (vol/vol) glycerol} as described (11). PAS-A (30 μM) was mixed with 40 μCi (1 μCi = 37 kBq) of [α-32P]ATP diluted to a final concentration of 2 μM in unlabeled ATP, and the reaction mixture (50 μl) was incubated on ice in the dark for 15 min. A sample (25 μl) was removed and exposed to UV light for 30 min on ice while the remainder was kept in the dark. UV light was generated by two 15-W General Electric G15T8 UV lamps at a distance of 3 cm from the sample. The samples then were subjected to SDS/PAGE on 15% (wt/vol) acrylamide gels by using the Tricine buffer system (14) and autoradiography. When required, the amount of labeled protein was quantified by phosphorimaging (Molecular Dynamics PhosphorImager SF).
Phosphorylation of PAS-A and KinA-C.
Phosphorylation reactions were carried out at 25°C in buffer B. PAS-A (75 μM) was phosphorylated with 5 μCi of [γ-32P]ATP or [γ-32P]GTP diluted to a final concentration of 100 μM in unlabeled ATP or GTP, respectively, for 180 min. KinA-C (5 μM), the isolated kinase domain of KinA, was phosphorylated for 60 min with 5 μCi of [γ-32P]ATP diluted to a final concentration of 100 μM in unlabeled ATP. The reactions were stopped by the addition of SDS/PAGE sample buffer, and labeled proteins were visualized by SDS/PAGE on 15% (wt/vol) acrylamide gels in the Tricine buffer system and subsequent autoradiography.
Binding of ATP to the PAS-A Domain.
The dissociation constant of ATP binding to PAS-A was determined by using the ultrafiltration technique (15, 16). PAS-A (20 μM) was incubated for 180 min at 25°C in 50 mM K-EPPS, pH 8.5/20 mM MgCl2/100 mM KCl, with increasing concentrations of ATP containing [α-32P]ATP at constant specific activity. The amount of ATP bound to PAS-A or remaining free in solution was determined by using Microcon-10 filtration units (Amicon), with subsequent liquid scintillation counting corrected for nonspecific binding of label.
Acid/Base Stability of PAS-A∼P.
The nature of the phosphorylated amino acid in PAS-A was investigated by determining the pH stability of phospho-amino acid linkage. PAS-A and KinA-C were phosphorylated with [γ-32P]ATP as described above and separated by SDS/PAGE. Gel slices containing PAS-A∼P and KinA-C∼P were excised and treated with 0.1 M HCl/0.1 M Hepes buffer, pH 7.0, or 1.0 M NaOH for 2 h at 55°C as described (8) and analyzed by autoradiography.
Catalytic Activity of PAS-A.
Reactions were carried out in 50 mM K-EPPS, pH 8.5/20 mM MgCl2. PAS-A (100 μM) was mixed with 0.5 μCi of α-32P- or γ-32P-labeled ATP, diluted to a final concentration of 1 mM with unlabeled ATP in the presence of 1 mM unlabeled ADP or GDP. The reactions were incubated at 25°C for 180 min, and reaction products were analyzed by PEI-cellulose TLC, with 0.75 M KH2PO4, pH 3.75, as the solvent. The TLC plates were air-dried and subjected to autoradiography and phosphorimaging.
Mutant Construction.
A kinA DNA fragment including the promoter and signal-input domain (amino acids 1–382) was cloned into the SacI and PstI sites of pJM105A (17) to produce plasmid pES1. This plasmid was used as the template for in vitro site-directed mutagenesis with the Muta-Gene kit (Bio-Rad) to introduce a point mutation into kinA, resulting in the replacement of the cysteine residue at position 75 by alanine. Plasmids based on pES1 integrate into the chromosome of B. subtilis by a single (Campbell-type) crossover event between plasmid- and chromosomally encoded kinA sequences, resulting in one truncated and one intact copy of kinA. pES1 and pES1-C75A were used to transform B. subtilis JH19980 [trpC2, phe-1, kinB (10)], and transformants were selected on Schaeffer's agar plates containing 6 μg/ml chloramphenicol at 37°C.
Sporulation Assays.
The sporulation efficiency of B. subtilis strains was determined by measuring the relative proportion of chloroform-resistant spores in liquid cultures, which were grown under sporulating conditions as described (18).
Results
PAS-A Is an ATP-Binding Domain.
It has been shown previously that the isolated kinase domain of KinA (KinA-C, Fig. 1) binds and hydrolyzes ATP, albeit at a much reduced level compared with the complete kinase (11). Surprisingly, in vitro studies with various isolated domains of KinA revealed that the PAS-A domain also could be labeled by [γ-32P]ATP. Because the binding and hydrolysis of ATP was thought to be an exclusive function of the kinase domain, this phenomenon was investigated further.
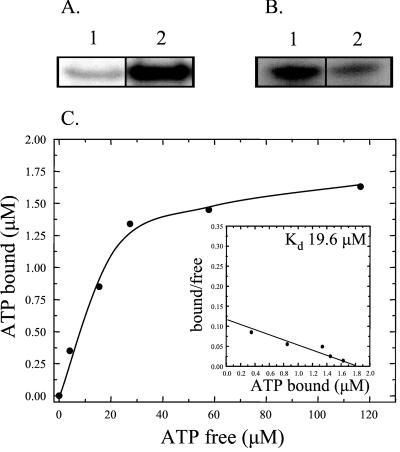
Direct photo-cross-linking of nucleotides to proteins has been used extensively for the identification of nucleotide-binding sites in proteins (12, 13). In the absence of UV irradiation, [α-32P]ATP remained tightly associated with PAS-A after electrophoresis (Fig. 2A, lane 1), and the binding was increased more than 10-fold after exposure to UV (Fig. 2A, lane 2) as a consequence of photoactivation of the purine ring of ATP. UV cross-linking of [α-32P]ATP to PAS-A was unaffected by the presence of reducing agents such as DTT (data not shown). PAS-A also becomes labeled in the presence of [γ-32P]ATP (Fig. 2B, lane 1) and [γ-32P]GTP (Fig. 2B, lane 2). ATP appears to act as a more efficient phosphodonor than GTP, resulting in an ≈5-fold increase in the amount of labeled PAS-A in the presence of [γ-32P]ATP; however, this simply may represent increased binding.
Figure 2.
Binding and phosphorylation of PAS-A by ATP. (A) PAS-A was incubated with [α-32P]ATP and kept in the dark (lane 1) or exposed to UV light (lane 2), followed by SDS/PAGE with subsequent autoradiography. (B) Phosphorylation of PAS-A with [γ-32P]ATP (lane 1) or [γ-32P]GTP (lane 2). Labeled protein was visualized by autoradiography after SDS/PAGE. (C) Binding curve of [α-32P]ATP binding to PAS-A was obtained by nonlinear regression analysis of the raw data followed by Scatchard plot transformation of the data (Inset).
To get an indication of the relative affinity of binding, the dissociation constant (Kd) of the interaction between PAS-A and [α-32P]ATP was determined. The binding curve obtained (Fig. 2C) is consistent with a single ATP molecule binding to one site on PAS-A. Further, the Kd of the PAS-A/ATP interaction was calculated from the Scatchard plot of the binding data to be 19.6 μM (Fig. 2C Inset).
These data, combined with the observation that a water-soluble compound with an absorbance spectrum identical to that of ATP can be extracted from purified PAS-A (data not shown), demonstrate that ATP is the likely ligand for the PAS-A domain of KinA and that this domain represents a nucleotide-binding domain. Given the established role of PAS domains as sensing modules, these observations raise the possibility that binding of ATP and/or phosphorylation of PAS-A influence the activation of KinA.
Acid/Base Stability of PAS-A∼P.
Because it is possible to get an idea of the type of amino acid phosphorylated by determining the pH stability of the linkage between the amino acid residue and the phosphoryl group, the pH stability of the putative PAS-A∼P was compared with that of the phosphohistidine of KinA-C.
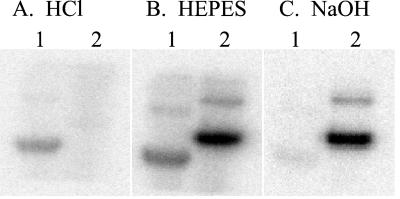
The phosphoryl moiety of PAS-A∼P was stable after treatment with HCl (Fig. 3A, lane 1), whereas the phosphohistidine of KinA-C∼P was labile under acidic conditions (Fig. 3A, lane 2), indicating that phosphorylation of PAS-A does not result in a phosphoramidate such as phosphohistidine (19, 20). In contrast, the phosphohistidine of KinA-C was stable after treatment with NaOH (Fig. 3C, lane 2), whereas the phosphoryl group of PAS-A∼P was labile under basic conditions (Fig. 3C, lane 1). The pH stability profile is characteristic of O-linked phosphates such as phosphoserine or phosphothreonine (21), suggesting that PAS-A is phosphorylated on a serine or threonine residue. Both PAS-A∼P and KinA-C∼P were stable in Hepes buffer at pH 7.0 (Fig. 3B).
Figure 3.
Acid and base stability of PAS-A∼P. PAS-A (lane 1) and KinA-C (lane 2) were phosphorylated with [γ-32P]ATP and separated by SDS/PAGE. Gels were treated with 0.1 M HCl (A), 0.1 M Hepes, pH 7.0 (B), or 1 M NaOH (C) at 55°C and analyzed by autoradiography.
PAS-A Is a Catalytic Domain with a Nucleoside Diphosphate Kinase-Like Activity.
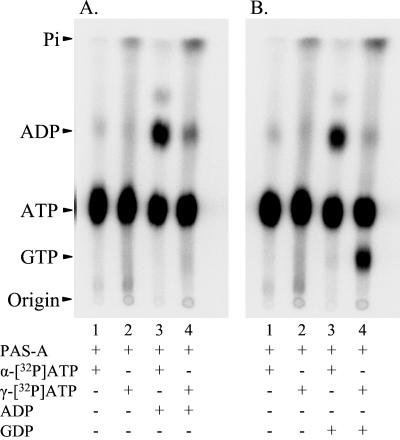
The PAS-A domain of KinA becomes phosphorylated by [γ-32P]ATP and, to a lesser extent, by [γ-32P]GTP (Fig. 2B), indicating that this domain may possess catalytic activity. The apparent enzymatic activity of PAS-A was investigated further by assaying the reaction products on TLC. In the presence of unlabeled ADP and [α-32P]ATP, PAS-A catalyzed the formation of [α-32P]ADP (Fig. 4A, lane 3). This could come about if the γ-phosphate group was transferred from [α-32P]ATP to ADP to form unlabeled ATP, which would not be detected in this experiment, or if PAS-A acted as a phosphatase of ATP. When [γ-32P]ATP was used as the phosphodonor in the presence of unlabeled ADP, the amount of labeled ADP produced was much lower, and little if any increase in labeled inorganic phosphate was detected (Fig. 4A, lane 4), eliminating the possibility that PAS-A was acting as a phosphatase. Whether PAS-A was responsible for the low amount of labeled ADP formed (Fig. 4A, lane 4) or whether this represents contamination of the [γ-32P]ATP by label in the α or β positions could not be determined. Regardless, PAS-A appears to be catalyzing the exchange of the phosphate between ATP and ADP.
Figure 4.
Catalytic activity of PAS-A. (A) [α-32P]ATP (lanes 1 and 3) or [γ-32P]ATP (lanes 2 and 4) was used as phosphodonor and mixed with PAS-A in the absence (lanes 1 and 2) or the presence (lanes 3 and 4) of unlabeled ADP. (B) [α-32P]ATP (lanes 1 and 3) or [γ-32P]ATP (lanes 2 and 4) was used as phosphodonor and mixed with PAS-A in the absence (lanes 1 and 2) or the presence (lanes 3 and 4) of unlabeled GDP. In all cases, the reaction products were analyzed by TLC and autoradiography. The identity of the nucleotides was confirmed by comparison with nonradioactive standards run on the same TLC plate. Pi, inorganic phosphate.
The exchange activity of PAS-A was more obvious when the reaction was stimulated by unlabeled GDP, as GTP and ATP separate on the TLC. Unlabeled GDP addition resulted in the production of [α-32P]ADP and [γ-32P]GTP when [α-32P]ATP or [γ-32P]ATP was used as the phosphodonor, respectively (Fig. 4B, lanes 3 and 4). However, GTP was much less efficient as a phosphodonor than ATP (data not shown). The phosphorylated enzyme intermediates of PAS-A are also evident from the increase in labeled protein that remained at the origin of the TLC plate when [γ-32P]ATP or [γ-32P]GTP was used as phosphodonor (Fig. 4 A and B, lanes 2 and 4).
The reactions described here are consistent with PAS-A possessing a nucleoside diphosphate kinase (NDPK)-like activity capable of catalyzing the exchange of the terminal phosphate of ATP or GTP to ADP or GDP. Several experiments were undertaken to ensure that this activity was a property of PAS-A and not a contaminating protein. Although the PAS-A preparation used was essentially pure by SDS/PAGE criteria, the preparation was subjected to nondenaturing gel electrophoresis followed by excision of the PAS-A band and electroelution. The eluted protein retained the activity. Similarly, further purification by DEAE-Sepharose and Affi-Gel blue chromatography did not separate the activity from the PAS-A protein.
The KinA C75A Mutation Increases in Vivo Sporulation Efficiency.
As part of a parallel project to determine the function of cysteine residues in the activity of KinA, we constructed an integrative plasmid, pES1-C75A, encoding a mutant KinA in which the cysteine residue at amino acid position 75 was replaced by alanine. This plasmid and pES1 encoding wild-type KinA were used to transform a B. subtilis strain deficient in KinB (JH19980) to determine whether a sporulation phenotype would arise as a consequence of the KinA C75A mutation. Strains lacking KinA and KinB have a sporulation frequency at least five orders of magnitude less than a strain lacking only KinB. Initially, sporulation phenotypes were identified by plating the strains on Schaeffer's agar plates to induce sporulation. Under these conditions, the strain harboring KinA C75A exhibited a hypersporulating phenotype compared with the isogenic strain encoding wild-type KinA.
Sporulation efficiency of the strains in liquid culture was measured by determining the proportion of chloroform-resistant spores in cultures of B. subtilis grown under sporulation-inducing conditions. Compared with B. subtilis JH642, inactivation of kinB reduced the sporulation efficiency from 61% to 30% in JH19980∷pES1 (Table 1). The KinA C75A substitution increased sporulation efficiency in a kinB background from 30% to 48% (Table 1). If plasmid pES1-C75A was integrated into a strain prototrophic for KinB, the transformants were observed to accumulate sporulation-negative suppressor mutations at a relatively high frequency. This phenotype is a well established consequence of inappropriate timing of sporulation, where the initiation of sporulation during growth selects for secondary suppressors blocked in sporulation (22), and it indicates that the C75A mutation results in a more active KinA.
Table 1.
Sporulation efficiency of B. subtilis strains
| Strain | Relevant genotype | % Sporulation |
|---|---|---|
| JH642 (wild type) | trpC2, phe-1 | 61 ± 7.5 |
| JH19980∷pES1 | trpC2, phe-1, kinB | 30 ± 0.6 |
| JH19980∷pES1-C75A | trpC2, phe-1, kinB, kinA C75A | 48 ± 1.5 |
The results of at least three independent experiments are shown.
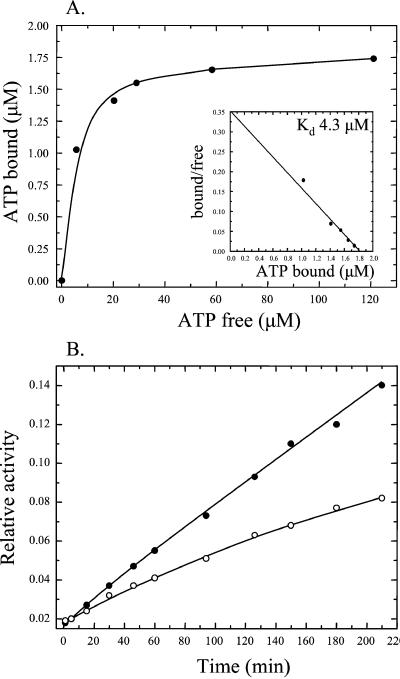
To determine whether the C75A substitution influences the ATP binding and catalytic activity of PAS-A, this domain was amplified from the chromosome of B. subtilis JH19980∷pES1-C75A by PCR and cloned into pET28a, and the protein was purified as described above. Purified PAS-A C75A bound ATP with higher affinity (Kd = 4.3 μM) than the wild-type PAS domain, as revealed by an ≈5-fold reduction in the Kd of the PAS-A C75A/ATP interaction compared with the wild-type (Fig. 5A). Furthermore, the catalytic activity of PAS-A C75A was assayed by determining the relative amount of [α-32P]ADP produced from [α-32P]ATP and unlabeled ADP with time, as a consequence of the transfer of the γ-phosphate of [α-32P]ATP to unlabeled ADP. Compared with wild-type PAS-A, the rate of production of [α-32P]ADP catalyzed by PAS-A C75A was reduced by more than 2-fold (Fig. 5B). Thus, the KinA C75A mutation increases both the sporulation efficiency in vivo and the ATP binding of the PAS-A domain by ≈5-fold, whereas the NDPK-like activity of the domain is reduced.
Figure 5.
ATP binding and catalytic activity of PAS-A C75A. (A) The binding curve of [α-32P]ATP binding to PAS-A C75A was obtained by nonlinear regression analysis of the raw data followed by Scatchard plot transformation (Inset). (B) Catalytic activity of PAS-A. Wild-type PAS-A (100 μM, ●) or PAS-A C75A (100 μM, ○) was incubated with 1 mM [α-32P]ATP in the presence of 1 mM unlabeled ADP, and samples were removed at time intervals and the reaction was stopped by freezing in a dry ice/ethanol bath. Reaction products were analyzed by TLC and quantified by phosphorimaging. The relative amount of [α-32P]ADP formed with time is expressed as the ratio of [α-32P]ADP to [α-32P]ATP.
Discussion
PAS domains are highly conserved signaling modules found in proteins from all three kingdoms of life. Of the proteins harboring PAS domains encoded in the genome of B. subtilis, only one is not a sensor kinase, and two sensor kinases, KinA and KinE (another sporulation-related sensor kinase), possess more than one PAS domain, suggesting that these sensing modules are important for the biological function of sensor kinases and for the initiation of development in this organism. Although the exact role of each PAS domain in KinA-mediated signal transduction is unclear, previous mutational studies have demonstrated the importance of the most amino-terminal PAS domain, PAS-A, for the activity of KinA (11).
The present data demonstrate that the isolated PAS-A domain of KinA binds ATP with relatively high affinity. The dissociation constant of the PAS-A/ATP interaction was calculated to be ≈20 μM, the same order of magnitude as the Km for ATP (74 μM) of the complete sensor kinase (23). PAS-A can be phosphorylated by [γ-32P]ATP (and, to a lesser extent, by [γ-32P]GTP) and catalyzes the transfer of the γ-phosphate group of ATP to ADP or GDP. The exchange reaction of PAS-A appears similar to that of NDPK, an enzyme that catalyzes the synthesis of nucleoside triphosphates from the corresponding nucleoside diphosphates via a phosphohistidine enzyme intermediate.
NDPK is an important cellular enzyme that monitors and maintains nucleotide pools and has been implicated in a number of regulatory processes, including signal transduction and development (24). Furthermore, NDPK-like activity has been observed in a number of proteins that are not classical NDPK enzymes (25–28). Commonly, NDPKs are phosphorylated as a reaction intermediate on a conserved histidine residue. In contrast, the phosphoryl group of PAS-A∼P is acid-stable and base-labile, indicating that the phosphoenzyme intermediate of PAS-A is not phosphohistidine but is more likely to be an O-linked phosphate such as phosphoserine or phosphothreonine. Mitochondrial NDPK is phosphorylated on a serine residue (29), and it is possible that the exchange reaction catalyzed by PAS-A occurs via a phosphoserine intermediate. However, it is not clear whether the phosphorylation of PAS-A is part of the KinA reaction mechanism, and the possibility that phosphorylation may be due to a spurious reaction of the isolated domain with no physiological significance cannot be ruled out.
These data raise several questions as to the biological significance of the ATP-binding and NDPK-like activity of PAS-A. It is possible that this inherent, NDPK-like activity is part of a mechanism to sense the cellular pool of ATP (and possibly other nucleotides). Although PAS-A does become labeled by [γ-32P]GTP, the phosphorylation of KinA by [γ-32P]ATP is not affected by GDP or GTP (data not shown), indicating that PAS-A is not sensing changes in guanine nucleotide pools. The likely structural changes induced as a consequence of ATP binding and phosphorylation might regulate the autophosphorylation of the kinase domain of KinA and the input of phosphate into the phosphorelay. Simple binding of ATP to PAS-A could constitute a signal regulating the activity of KinA, with the observed NDPK-like activity of PAS-A being part of a turnover or desensitizing reaction. One of the established roles of PAS domains is to monitor changes in the energy status and redox potential of the cell, which, in essence, is measuring the capacity to produce ATP. Although it is not difficult to imagine scenarios connecting cellular energy potential and development, ATP binding does not constitute compelling evidence for such a role.
A B. subtilis strain carrying a mutant KinA in which the cysteine residue at position 75 in PAS-A was replaced by alanine showed increased sporulation efficiency. The effect of the C75A mutation on sporulation is modest in liquid culture. However, on solid media, the mutant strain clearly sporulates earlier, as evidenced by colony morphology, and sporulates inappropriately as the mutant strain segregates sporulation-negative secondary mutations visible as papillae to overcome this problem. In both of these regards, the mutant strain resembles strains with sof mutations of Spo0A (22) and Rap phosphatase mutations (30), each of which increases the tendency to sporulate during growth.
As a consequence of the C75A mutation, the Kd of ATP binding by the isolated PAS-A C75A domain was decreased and its NDPK-like activity was reduced to less than 50% of the wild-type level. Thus, it appears that it is the ATP-bound form of the PAS domain that is more active in sporulation, and the loss of NDPK activity, which may serve to turn over ATP, prevents the mutant PAS-A domain from returning to the non-ATP-bound, presumably inactive state. The C75A mutation may have an effect on the interaction of the PAS-A domain with itself, other domains of KinA, or a putative effector molecule. Although PAS-A in the wild-type protein is monomeric (11), the C75A mutation may induce the dimerization properties that PAS domains are known for. Perhaps dimerization of PAS-A is a prerequisite to KinA activity. The folding of KinA might alter the kinetics of ATP binding and phosphorylation and either increase or decrease the effects of the C75A mutation.
It is counterintuitive to correlate high ATP concentrations, an energy-sufficient state, with the initiation of sporulation that is associated with nutrient deprivation, when cells become limited in their capacity to produce ATP. Further, the Kd of ATP binding (≈20 μM) is well below the concentration of the cellular ATP pool (1–3 mM). Thus, it seems more likely that ATP is not the ligand being sensed by the sensor kinase KinA. ATP may have a role in transmitting the state, plus or minus ligand, of the signal-sensing domain of the sensor kinase to the autokinase domain. Signal recognition by sensor kinases involves noncovalent-bonding interactions that drive the interconversion of conformational states, resulting in ATP hydrolysis and autophosphorylation. There exist many examples of proteins such as actin or G proteins, recently termed energases, that transduce Gibbs free energy of nucleotide hydrolysis to useful work (31). Perhaps the ATP bound to the PAS-A domain is quasiequivalent to the GTP of G proteins and serves as a mediator of the state of ligand binding by driving the interconversion of the conformational states of KinA. Signal ligands might act to either promote or prevent ATP binding or ATP hydrolysis, making it possible for different signals to activate or inactivate KinA.
KinA contains three PAS domains in its sensing domain, and the experiments described in this study have concentrated on the most amino-terminal PAS domain. Conceivably, each of the KinA PAS domains could have a different function and/or sense different parameters. It will be of interest to determine the nature of the PAS-B and PAS-C signal ligands, if any exist, and to determine whether the PAS domains act independently or cooperatively to influence the activity of KinA. The data described here extend the currently limited inventory of known PAS domain ligands and may have important, broad implications for PAS domain-mediated signal sensing in all organisms.
Acknowledgments
We thank Emil Schultz for constructing the C75A mutation. This work was supported, in part, by Grant GM19416 from the National Institute of General Medical Sciences, National Institutes of Health, U.S. Public Health Service. This is manuscript 14327-MEM from The Scripps Research Institute.
Abbreviation
- NDPK
nucleoside diphosphate kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Barrett J F, Hoch J A. Antimicrob Agents Chemother. 1998;42:1529–1536. doi: 10.1128/aac.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor B L, Zhulin I B. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikov S I, Barnes L A, Gitin Y, Parkinson J S. Proc Natl Acad Sci USA. 2000;97:5830–5835. doi: 10.1073/pnas.100118697. . (First Published May 16, 2000; 10.1073/pnas.100118697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monson E K, Ditta G S, Helinski D R. J Biol Chem. 1995;270:5243–5250. doi: 10.1074/jbc.270.10.5243. [DOI] [PubMed] [Google Scholar]
- 6.Pellequer J-L, Wagner-Smith K A, Kay S A, Getzoff E D. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 8.Burbulys D, Trach K A, Hoch J A. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 9.Jiang M, Shao W, Perego M, Hoch J A. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 10.Trach K A, Hoch J A. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Fabret C, Kanamaru K, Stephenson K, Dartois V, Perego M, Hoch J A. J Bacteriol. 2001;183:2795–2802. doi: 10.1128/JB.183.9.2795-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagan B, Knowles J R. Methods Enzymol. 1977;46:64–115. [Google Scholar]
- 13.Knorre D G, Vlassov V V. Affinity Modification of Biopolymers. Boca Raton, FL: CRC; 1989. pp. 60–64. [Google Scholar]
- 14.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 15.Kamberov E S, Atkinson M R, Ninfa A J. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 16.Jiang P, Peliska J A, Ninfa A J. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 17.Perego M. In: Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 615–624. [Google Scholar]
- 18.Tzeng Y-L, Hoch J A. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 19.Fujitaki J M, Smith R. Methods Enzymol. 1984;107:23–26. doi: 10.1016/0076-6879(84)07004-x. [DOI] [PubMed] [Google Scholar]
- 20.Hultquist D E. Biochim Biophys Acta. 1968;153:329–340. doi: 10.1016/0005-2728(68)90078-9. [DOI] [PubMed] [Google Scholar]
- 21.Martensen T M. Methods Enzymol. 1984;107:3–23. doi: 10.1016/0076-6879(84)07003-8. [DOI] [PubMed] [Google Scholar]
- 22.Spiegelman G, Perego M, Day J, Trach K A, Hoch J A. J Bacteriol. 1990;172:5011–5019. doi: 10.1128/jb.172.9.5011-5019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimshaw C E, Huang S, Hanstein C G, Strauch M A, Burbulys D, Wang L, Hoch J A, Whiteley J M. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- 24.Lu Q, Park H, Egger L A, Inouye M. J Biol Chem. 1996;271:32886–32893. doi: 10.1074/jbc.271.51.32886. [DOI] [PubMed] [Google Scholar]
- 25.Hiromura M, Yano M, Mori H, Inoue M, Kido H. J Biol Chem. 1998;273:5435–5438. doi: 10.1074/jbc.273.10.5435. [DOI] [PubMed] [Google Scholar]
- 26.Kapatral V, Bina X, Chakrabarty A M. J Bacteriol. 2000;182:1333–1339. doi: 10.1128/jb.182.5.1333-1339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzeng C M, Kornberg A. J Biol Chem. 2000;275:3977–3983. doi: 10.1074/jbc.275.6.3977. [DOI] [PubMed] [Google Scholar]
- 28.Ishige K, Noguchi T. Biochem Biophys Res Commun. 2001;281:821–826. doi: 10.1006/bbrc.2001.4415. [DOI] [PubMed] [Google Scholar]
- 29.Struglics A, Hakansson G. Eur J Biochem. 1999;262:765–773. doi: 10.1046/j.1432-1327.1999.00432.x. [DOI] [PubMed] [Google Scholar]
- 30.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 31.Purucker M. Trends Biochem Sci. 2001;26:417–421. doi: 10.1016/s0968-0004(01)01880-1. [DOI] [PubMed] [Google Scholar]