Abstract
Protein chemical cross-linking combined with mass spectrometry has become an important technique for the analysis of protein structure and protein−protein interactions. A variety of cross-linkers are well developed, but reliable, rapid, and user-friendly tools for large-scale analysis of cross-linked proteins are still in need. Here we report MetaMorpheusXL, a new search module within the MetaMorpheus software suite that identifies both MS-cleavable and noncleavable cross-linked peptides in MS data. MetaMorpheusXL identifies MS-cleavable cross-linked peptides with an ion-indexing algorithm, which enables an efficient large database search. The identification does not require the presence of signature fragment ions, an advantage compared with similar programs such as XlinkX. One complication associated with the need for signature ions from cleavable cross-linkers such as DSSO (disuccinimidyl sulfoxide) is the requirement for multiple fragmentation types and energy combinations, which is not necessary for MetaMorpheusXL. The ability to perform proteome-wide analysis is another advantage of MetaMorpheusXL compared with programs such as MeroX and DXMSMS. MetaMorpheusXL is also faster than other currently available MS-cleavable cross-link search software programs. It is imbedded in MetaMorpheus, an open-source and freely available software suite that provides a reliable, fast, user-friendly graphical user interface that is readily accessible to researchers.
Keywords: proteomics, MS-cleavable chemical cross-link, protein−protein interactions, ion index
Graphical Abstract
INTRODUCTION
Cross-linking mass spectrometry (XL−MS) has been widely used to determine protein structure and identify protein− protein interactions.1–5 Protein chemical cross-linking uses a small-molecule bridge to form a covalent bond between two proximal amino acids, with the cross-linker’s length determining the distance at which the cross-link can form. This bond-length restriction can be used to characterize protein structure or protein−protein interactions. Compared with other protein structure determination methods (X-ray crystallography, nuclear magnetic resonance, and cryo-electron microscopy) that depend on challenging sample preparation procedures, protein structure determination using chemical cross-linking is much more straightforward. Cross-linked proteins are enzymatically digested (e.g., with trypsin) into peptides and analyzed by LC−MS/MS. Wang et al.6 successfully employed XL−MS in conjunction with cryo-electron microscopy and computational modeling to fully resolve dynamic structures of the human 26S proteasome. In addition, they detected dynamic states of the proteasome subunits Rpn1, Rpn6, and Rpt6 and identified several new proteasome-interacting proteins. Chen et al.7 used XL−MS to interpret the architecture of yeast RNA polymerase−TFIIF complex (TFIIF is a transcription initiation factor). Despite these successes, the XL−MS search itself is still in need of significant improvement.
One limitation of some cross-link search programs (e.g., X! Link8 and Xlink-Identifier9) is that they employ a database of all possible theoretical peptide dimers. The number of theoretical peptide−peptide combinations increases quadratically with database size; if a protein database contains n peptides, then the possible number of peptide−peptide combinations is n(n + 1)/2, which defines the search space.2 In general, this type of search algorithm is inefficient and not feasible for large databases because of the significant time and computational power required and the greater chance of incorrect false-positive identifications.10
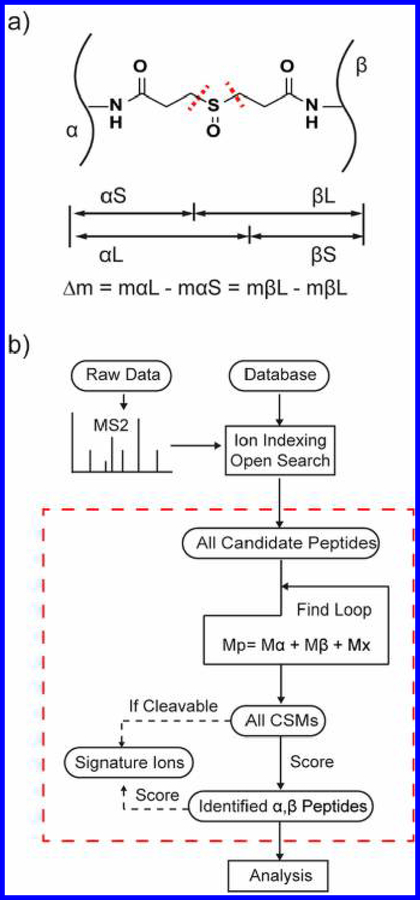
MS-cleavable cross-linkers such as DSSO can make the job of peptide identification more straightforward compared with noncleavable cross-linkers. When fragmented, MS-cleavable cross-linked peptides yield two pairs of signature ions (αS/βL and αL/βS, where α (alpha) and β (beta) refer to the two cross-linked peptides and S and L refer to the “short” and “long” pieces of the fragmented cross-linker molecule) (Figure 1a). These ion pairs have a signature mass difference between them, which is used to help identify cross-linked peptides. The signature ions allow the search algorithm to distinguish the ion series associated with each of the individual cross-linked peptides.11 MS-cleavable cross-linked peptides also typically generate more fragment ions than peptides connected with a noncleavable cross-linker, thereby improving identification.12
Figure 1.
MetaMorpheusXL for MS-cleavable cross-link search. (a) Four signature fragment ions are generated from DSSO cross-linked peptides (α and β peptides) using CID/HCD (CID: collision-induced dissociation/ HCD: higher-energy collisional dissociation). If the cross-linker is cleaved at the left of the sulfoxide moiety, then it will generate the αS and βL fragments; if the cross-linker is cleaved at the right, then it will generate the αL and βS fragments. The signature mass difference (Δm) between αL and αS, βL, and βS is the same; Δm is 31.97 Da for DSSO. (b) Workflow of MetaMorpheusXL (detailed explanation in the Methods section). In the “Find Loop”, the candidate PSMs are matched pairwise to attempt to satisfy the equation: Mprecursor = Malpha + Mbeta + Mcross‑linker. (M, mass; x, cross-linker).
Many programs have been designed for noncleavable cross-link studies (for example xQuest,3 pLink,13 Protein Prospector,14 Xi,15 ECL,16 and Kojak17), which separately search the alpha and beta peptides by treating one of them as a modification on the other. These programs are limited, however, to noncleavable cross-linkers.
MeroX18 and DXMSMS19 were developed to be able to search cleavable cross-links by searching theoretical dimers. MeroX avoids searching all theoretical dimers by using a DiBond algorithm18 to reduce the number of dimer candidates, but it is still limited to only be able to search small databases. In 2015, Liu et al.4 developed a search strategy (XlinkX 1.0) that searches fragmentation spectra for these signature ion pairs. The XlinkX 1.0 algorithm first finds the masses of each of the two cross-linked peptides by identifying all four signature ion peaks, followed by a standard fragment-based search to determine their sequences. However, because of the lack of signature fragment ions in many experimental spectra, this strategy leaves many cross-linked peptides unidentified. In response to these difficulties, researchers have had to use complicated, optimized fragmentation methods to maximize the chances of observing all four signature ions. Since then, the developers of XlinkX have improved their program to avoid requiring the presence of all four signature ions in a spectrum.12 However, the approach implemented in XlinkX 2.0 still relies on the detection of at least one signature ion of high intensity. Another problem with requiring the presence of signature ions is that some cross-linkers with bond strengths comparable to peptide amide bonds (such as DSBU, disuccinimidyl dibutyric urea) make the generation of signature ions difficult.
The present work describes a novel search program for the detection of both MS-cleavable and noncleavable cross-linked peptides, and it is the first software program reported with both capabilities. The search strategy has been implemented in the computer program MetaMorpheus,20 which has a user-friendly graphical user interface (GUI). Novel cross-linker molecules are easily added if desired. A fragment-ion index scheme makes the search computationally efficient.17,20–22 Additionally, the MetaMorpheus software suite contains multiple other functions useful to proteomics researchers such as traditional bottom-up search algorithms, mass calibration,22 post-translational modification (PTM) discovery,22,23 and label-free quantification.20
METHODS
Cross-Link Search Algorithm
MetaMorpheusXL identifies alpha and beta cross-linked peptides with an ion-indexed open search algorithm (as outlined in Figure 1b). First, all fragment ions are indexed according to their m/z prior to searching to increase the speed of the search.20,21 (See the Supporting Information for a description of the indexing algorithm.) Then, fragmentation spectra are searched against a target and decoy database24 (decoys are generated by reversing sequences of each target protein) with an unlimited precursor mass tolerance (an “open-mass” search21), and all candidate peptides for each spectrum are held in memory. Second, all candidate peptides for each spectrum from the first step are paired in an attempt to find a combined mass (the two peptides plus the cross-linker mass) that matches the precursor ion mass. If such a mass match is found, that peptide pairing is considered a CSM (cross-linked peptide spectrum match) candidate. The next step is scoring these CSMs, and that depends on whether the cross-linker is MS-cleavable. If cleavable, the algorithm searches the spectrum for any signature fragment ions that could arise from the peptide pair. All possible cross-link site pairs for one CSM are considered during the generation of theoretical fragment ions; the pairing with the most fragment ion matches between theoretical and experimental is considered to emanate from correct cross-linking. This information thus informs the position of the cross-link within the pair. After attempting to match all theoretical ions from the peptide pair, the score of a CSM is the summed count of both peptides’ observed fragment ions plus any signature fragment ions if the cross-linker was MS-cleavable. Next, the CSMs are ranked by score. The false discovery rate (FDR) is estimated using the target-decoy strategy.24 (See the Supporting Information for a description of MetaMorpheusXL’s FDR estimation.) Candidate CSMs that meet a suitable FDR threshold (e.g., 1%) are considered an identified pair of cross-linked alpha and beta peptides. MetaMorpheusXL’s output is a tab-delimited text file, which is readable by Percolator,17,25 a semisupervised learning program, to potentially increase the number of target CSMs below a desired FDR. MetaMorpheusXL also formats its output into pepXML, enabling easy visualization of cross-link peptide search results with publicly available software such as ProXL,26 Kojak Spectrum Viewer, and TransProteomicPipeline (TPP).27
Sample Preparation
BSA (bovine serum albumin; 1 μg/μL; Sigma) was dissolved in PBS (phosphate-buffered saline) buffer. Ribosomes (13.3 μM; NEB) were diluted to 1 μg/μL with HEPES buffer. Freshly prepared 50 mM MS-cleavable cross-linker DSSO (disuccinimidyl sulfoxide, Thermo Scientific) dissolved in DMSO was added to a final concentration of 1 mM. After incubating at RT for 60 min, the reaction was quenched by adding Tris buffer to 40 mM. The samples were digested with a modified eFASP procedure as described.28 In brief, the cross-link reaction samples were washed with 8 M urea, 0.1% DCA using a 30 kDa cutoff Ultrafree filter (Millipore). The samples were reduced with 20 mM DTT for 30 min, alkylated with 20 mM iodoacetamide for 60 min, and digested with 1 μg trypsin per 40 μg protein overnight at 37 °C. The peptide digests were dried in vacuo, resuspended in 0.1% TFA, and desalted with C18 OMIX ZipTip (Agilent). The final peptides were dissolved in 95:5 H2O/ACN with 0.2% formic acid.
Mass Spectrometry
Samples (∼2 μg protein each injection) were analyzed via HPLC (NanoAcquity, Waters)-ESI−MS/MS (Q Exactive HF, ThermoFisher Scientific). The HPLC separation employed a 15 cm × 365 μm fused silica capillary microcolumn packed with 3 μm diameter, 100 Å pore size C18 beads (Magic C18; Bruker), with an emitter tip pulled to ∼2 μm using a laser puller (Sutter Instruments). Peptides were loaded on-column at a flow-rate of 400 nL/min for 30 min, then eluted over 120 min at a flow rate of 300 nL/min with a gradient from 5 to 35% acetonitrile in 0.1% formic acid. The gradient was then ramped to 70% acetonitrile in 0.1% formic acid over 5 min and held for 5 min, then reduced to 2% acetonitrile in 0.1% formic acid over 5 min and held for 15 min. Full-mass profile scans were performed in the Orbitrap between 375 and 1500 m/z at a resolution of 120 000, followed by MS/MS HCD (higher energy collisional dissociation) scans of the 10 highest intensity parent ions with z > 2 at 30 CE (relative collision energy) and 15 000 resolution, with a mass range starting at 100 m/z. Dynamic exclusion was enabled with an exclusion window of 30 s.
Analysis of MS/MS Spectra
Single protein data of DSSO cross-linked BSA, DSSO cross-linked E. coli ribosome, and BS3 (bis(sulfosuccinimidyl)-suberate, a noncleavable cross-linker) cross-linked protein complex data of yeast Pol II (ProteomeXchange Data set Identifier PXD004749) were analyzed.7 After data-dependent acquisition, tandem mass spectral data were first calibrated using MetaMorpheus’ mass calibration function.22 (See the Supporting Information for a description of the calibration algorithm.) The generated .mzML files were searched by MetaMorpheusXL (MetaMorpheus version 0.0.237). The small database used for DSSO cross-linked E. coli ribosome contained the 52 known protein sequences of ribosomal complex and another 41 protein sequences for proteins known to interact with the E. coli ribosome. We also searched the ribosome data against the complete E. coli proteome database, which contains 4443 proteins. The search took ∼6.4 min and resulted in 35% fewer CSMs below 1% FDR than the CSMs identified from searching against the small database containing 93 proteins. Detailed results are shown in Supplementary Figure S-2b. The database used for BS3 cross-linked yeast Pol II contained the 12 protein sequences of the Pol II complex. The DSSO data were also searched with XlinkX 2.0 and the BS3 data with Kojak 1.5 for comparison. The distances between each lysine−lysine pair of identified cross-linked peptides from E. coli ribosome and yeast Pol II were further validated by mapping to known structures with a custom Python script.
RESULTS AND DISCUSSION
MetaMorpheusXL Does Not Require the Observation of Signature Ions
MetaMorpheusXL identifies alpha and beta peptides based on peptide fragment ions in an “open-mass” search; the observation of signature ions is not required for its MS-cleavable cross-link search. This is an advantage of MetaMorpheusXL over XlinkX, which requires high-intensity signature ions to be present in the spectrum. Other software programs such as MeroX and DXMSMS also do not require the observation of signature ions, but these programs lack the ability to search large databases.
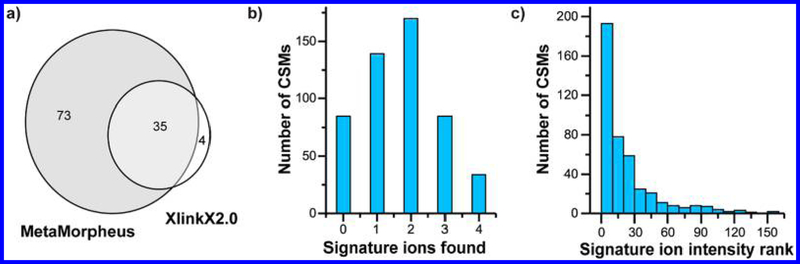
To evaluate MetaMorpheusXL’s performance for MS-cleavable cross-link searches, we first analyzed DSSO cross-linked BSA data. At 1% FDR, 513 CSMs were identified by MetaMorpheusXL, which correspond to 108 unique cross-linked peptide pairs; XlinkX 2.0 identified 104 CSMs with 39 unique cross-linked peptide pairs, 35 of which had also been found in MetaMorpheusXL (Figure 2a). MetaMorpheusXL found five times as many CSMs (three times as many unique cross-linked peptide pairs) as XlinkX 2.0 in 1/10th the computational time (SI, Figure S-2a).
Figure 2.
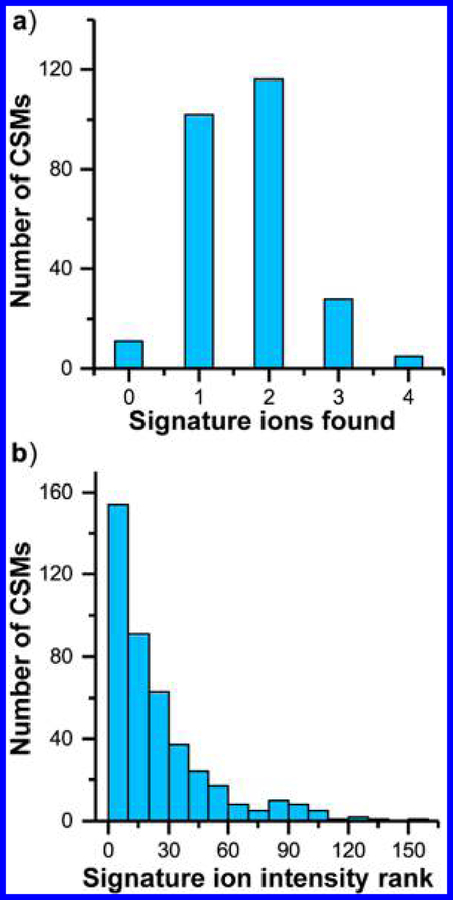
MS-cleavable cross-link search of BSA. (a) Comparison of identified unique cross-linked peptide pairs of BSA by MetaMorpheusXL and XlinkX 2.0. (b) Distribution of number of signature ions observed in the MS2 spectra from identified CSMs. (c) Relationship between number of CSMs and the most intense signature ions’ intensity ranks.
Of the 513 CSMs identified by MetaMorpheusXL, 34 contained all 4 signature ions, 85 contained 3 signature ions, 170 contained 2 signature ions, 139 contained 1 signature ion, and 85 contained no signature ions (Figure 2b). A majority of CSMs thus had fewer than four signature ions. CSMs containing zero signature ions were not detected by XlinkX 2.0, but many were detected by MetaMorpheusXL, as the peptide fragment ions provided sufficient information for characterization.
We also examined the intensity distribution of the signature ions. XlinkX 2.0 requires one signature ion to be among the most intense fragment peaks for each CSM. However, the intensities of signature ions depend on the type of cross-linker, MS instrumentation, and acquisition parameters. With the MS method we used for DSSO cross-linked BSA (HCD with 30 CE), we found that although roughly 50% of the most intense signature ions are among the top 15 most intense peaks in a spectrum, ∼25% are not in the top 30 peaks and 2.5% are not in the top 100 (Figure 2c). Because MetaMorpheusXL is not dependent on matching signature ions first, it provides a wider variety of choices for cross-linkers and acquisition parameters compared with other algorithms.
DSSO Cross-Linked Ribosome Analysis
We also generated DSSO cross-linked E. coli ribosomal complex data to further validate that MetaMorpheusXL could be used for protein complexes. The E. coli ribosome contains 52 proteins, and a wealth of detailed structural data exists to assist in validating the search results. Experimental replications were not performed here. In addition, no sample enrichments or prefractionation steps were employed.
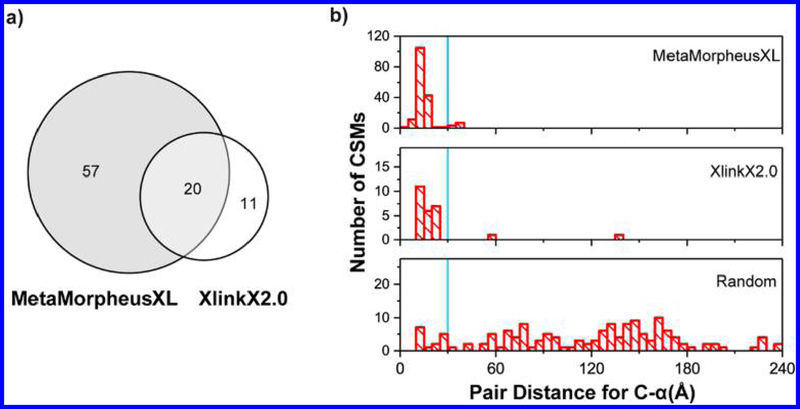
At 1% FDR, MetaMorpheusXL found 262 CSMs including 46 inter- and 216 intra-CSMs (inter: cross-linked peptides from different proteins; intra: cross-linked peptides from a single protein); XlinkX 2.0 identified 49 CSMs including 28 inter- and 21 intra-CSMs. The pepXML output of the ribosome cross-links was visualized by ProXL26 (SI, Figure S-3). MetaMorpheusXL detected five times more CSMs than XlinkX 2.0, similar to the results for DSSO cross-linked BSA data. In total, MetaMorpheusXL identified 77 unique cross-linked peptide pairs, while XlinkX 2.0 identified 31 (Figure 3a).
Figure 3.
Analysis of DSSO cross-linked ribosome. (a) Comparison of identified unique cross-linked peptide pairs of E. coli ribosome by MetaMorpheusXL and XlinkX 2.0. (b) Cα−Cα distance distribution for experimentally observed lysine−lysine pairs from MetaMorpheusXL, XlinkX 2.0, and a random distribution. The blue lines denote the 30 Å cutoff.
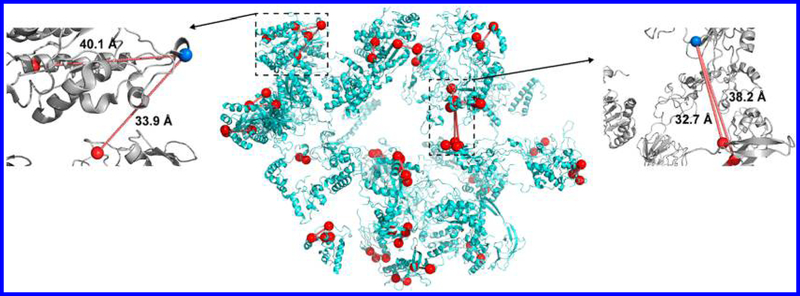
We further investigated the results by mapping the identified cross-link residues to the known structure of the ribosomal complex. Distances between the Cα carbon of cross-linked residues were expected to be within 30 Å based on the cross-linker spacer arm length and structural flexibility considerations. In total, 171 of the 262 CSMs identified by MetaMorpheusXL could be mapped to the E. coli ribosome structure (PDB: 3jcd). Four unique cross-linked peptide pairs (from 11 CSMs) had distances greater than the 30 Å restriction; the distances of the rest of the CSMs fell within the expected 30 Å (Figures 3b and 4). Of the four unique cross-linked peptide pairs, two of them (32.7 and 33.9 Å) were very close to 30 Å. The other two contained an intra-cross-link with distance of 40.1 Å and an inter-cross-link with distance of 38.2 Å. The structure suggested that these four cross-links may occur due to flexibility in the protein structure (Figure 4). Additionally, because they were identified multiple times and shared cross-linked residues with other cross-linked peptides, they are likely to be true cross-linked peptides. For the results produced by XlinkX 2.0, 21 of the 31 unique cross-linked peptide pairs could be mapped to the ribosome structure, 2 had distances larger than the 30 Å restriction, and the large distances 56.9 and 139 Å are not likely to be transient cross-links. More intra-CSMs could be detected than inter-CSMs, consistent with the known structure of the ribosome (the 52 proteins of the ribosome being dispersed around the rRNA).
Figure 4.
Structure of DSSO cross-linked E. coli ribosome (PDB: 3jcd) with cross-linked lysine residues and distances shown. Cross-links are identified using MetaMorpheusXL at a 1% FDR cutoff. The whole E. coli ribosome structure is shown in the middle; red lines indicate Cα−Cα distances between the cross-linked lysine (marked as red spheres). Four cross-link outliers are also shown: at left panel with an inter-cross-link with Cα−Cα distance of 40.1 Å on P0A7V0(36) and P0A7V0(58) and an intra-cross-link with Cα−Cα distance of 33.9 Å on P0A7V0(36) and P0A7W7(69); at right panel with an intra-cross-link with Cα−Cα distance of 38.2 Å on P0A7L0(141) and P0A7N9(58) and another intra-cross-link with Cα−Cα distance of 32.7 Å on P0A7L0(141) and P0A7N9(9).
We further analyzed the distribution of number of signature ions observed from identified CSMs and the relationship between the number of CSMs and intensity ranks of the most intense signature ions from the MetaMorpheusXL results. Similar to the intensity distribution of signature ions for BSA, the majority of CSMs lack two or three signature ions, and ∼5% of CSMs have no signature ion matches (Figure 5a). The rank distribution of the most intense signature ions also followed a similar pattern to that obtained for BSA: 75% of the most intense signature ions were fou nd in the most intense 30 peaks of a spectrum (Figure 5b).
Figure 5.
Validation of signature ions from DSSO cross-linked E. coli ribosome. (a) Distribution of number of signature ions observed in the MS2 spectra from identified CSMs. (b) Relationship between number of CSMs and the most intense signature ions’ intensity ranks.
Because MetaMorpheusXL uses fragment-ion indexing prior to searching, it was faster than XlinkX 2.0 for both the BSA data and the ribosomal complex data (SI, Figure S-2a).
Noncleavable Cross-Link Search with MetaMorpheusXL
MetaMorpheusXL can also be used for the identification of peptides with noncleavable cross-linkers. A high-quality data set of yeast Pol II complex acquired by Chen et al. was used for this study.7 The data set contains four raw files from peptide digests of the 12 subunits of yeast Pol II (523 kDa) cross-linked with cross-linker BS3.
MetaMorpheusXL identified five types of PSMs in this data set: intraprotein cross-links, interprotein cross-links, loop-links, dead-end-links, and single-peptide PSMs. MetaMorpheusXL identified 2075 PSMs (peptide spectrum matches) with 1% FDR from the yeast Pol II data. About 76% of the PSMs were single-peptide PSMs, which is consistent with other cross-linking studies, and ∼11% of the PSMs were loop-links (cross-linked residues are in the same peptide) or dead-end-links (cross-linker reacted with one residue). In total, 277 CSMs were identified (168 intra-CSMs and 109 inter-CSMs), including 97 unique cross-linked peptide pairs.
The search speed of MetaMorpheusXL for noncleavable cross-links was also assessed. Kojak is one of the most efficient software tools for cross-link studies. The algorithms of Kojak and MetaMorpheusXL yielded comparable search times (data not shown).
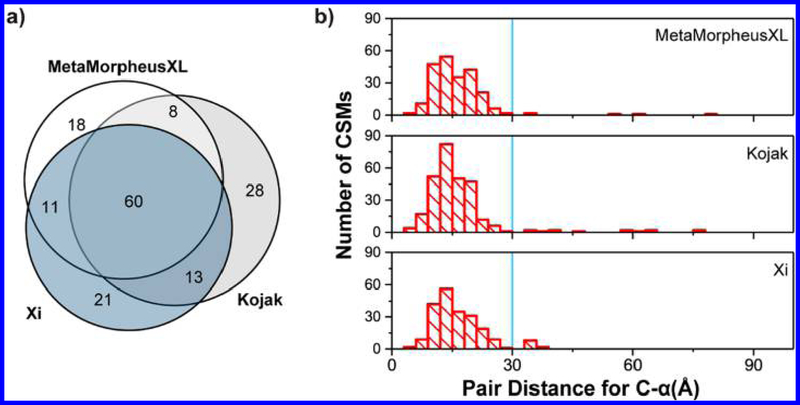
To assess the cross-links identified with MetaMorpheusXL, we analyzed the yeast Pol II data in parallel with Kojak and compared it with the original result from the paper that used a program named Xi.7 Kojak identified 315 CSMs (109 unique cross-linked peptide pairs) at 1% FDR, the paper reported 287 CSMs (105 unique cross-linked peptide pairs) with high confidence, and MetaMorpheusXL identified 277 CSMs (97 unique cross-linked peptide pairs). Among the unique cross-linked peptide pairs identified by all three programs, 60 were common to all three analyses (Figure 6a).
Figure 6.
Analysis of noncleavable cross-linked yeast Pol II complex results. (a) Comparison of identified unique cross-linked peptide pairs of yeast Pol II complex by Kojak, Xi, and MetaMorpheusXL. (b) Cα−Cα distance distribution for experimentally observed lysine−lysine pairs from MetaMorpheusXL, Kojak, and Xi. The blue lines denote the 30 Å cutoff.
We then validated the cross-linked residues by comparison with the published crystal structure data (PDB: 1wcm, Figure 6b). In the MetaMorpheusXL result, 227 of all identified CSMs fell in regions consistent with the published structure, whereas 5 (2.2%) were not within the 30 Å cutoff. The level of structural agreement was better than that for Kojak (13 of 284, 4.6% not within cutoff) or Xi (11 of 214, 5.1% were not within cutoff).
CONCLUSIONS
MetaMorpheusXL is a new search algorithm designed for large-scale studies of chemically cross-linked peptides. The approach increases the number of identified MS-cleavable cross-linked peptides compared with existing software. The algorithm is readily compatible with any cross-linker and will benefit developers and researchers who want to test the performance of different cross-linkers. MetaMorpheusXL has publicly available source code (https://github.com/smith-chem-wisc/MetaMorpheus). The software is user-friendly, and the search results can easily be pipelined to downstream software (e.g., Percolator et al.).
Supplementary Material
ACKNOWLEDGMENTS
We thank Brian L. Frey, who helped edit the manuscript. We thank Anthony J. Cesnik and Leah V. Schaffer, who also contributed daily input and guidance to the improvement of MetaMorpheus. We thank Prof. Lingjun Li’s lab for supplying EThcD (Electron-Transfer/Higher-Energy Collision Dissociation) fragmented cross-linked BSA data. This work was supported by grant U24CA199347 from the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acs.jproteo-me.8b00141.
Supplementary Figure S-1. Example explanation of MetaMorpheusXL’s workflow. Supplementary Figure S-2. Computation time comparison between MetaMorpheusXL and XlinkX 2.0. Supplementary Figure S-3. Circle plot from ProXL displays cross-links of DSSO cross-linked ribosome proteins. Supplementary Figure S-4. Examples of annotated spectra from E. coli ribosome data with different numbers of identified signature ions (4 to 0) and from different score ranges (high to low). Supplementary Figure S-5. Identification of intraprotein cross-links composed from consecutive sequences. Table- S1. Parameter usage for MetaMorpheusXL. (PDF)
Notes
The authors declare no competing financial interest.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository29 with the data set identifier PXD009000.
REFERENCES
- (1).Liko I; Allison TM; Hopper JT; Robinson CV Mass spectrometry guided structural biology. Curr. Opin. Struct. Biol 2016, 40, 136−144. [DOI] [PubMed] [Google Scholar]
- (2).Barysz H; Malmström J Development of large-scale cross-linking mass spectrometry. Mol. Cell. Proteomics 2017, mcp.R116.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rinner O; Seebacher J; Walzthoeni T; Mueller L; Beck M; Schmidt A; Mueller M; Aebersold R Identification of cross-linked peptides from large sequence databases. Nat. Methods 2008, 5 (4), 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Liu F; Rijkers DTS; Post H; Heck AJR Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 2015, 12 (12), 1179−1184. [DOI] [PubMed] [Google Scholar]
- (5).Yu C; Huang L Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology. Anal. Chem 2018, 90 (1), 144−165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang X; Cimermancic P; Yu C; Schweitzer A; Chopra N; Engel JL; Greenberg C; Huszagh AS; Beck F; Sakata E; et al. Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 2017, 16 (5), 840−854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chen ZA; Jawhari A; Fischer L; Buchen C; Tahir S; Kamenski T; Rasmussen M; Lariviere L; Bukowski-Wills J-C; Nilges M; et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J 2010, 29 (4), 717−726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lee YJ; Lackner LL; Nunnari JM; Phinney BS Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res 2007, 6 (10), 3908−3917. [DOI] [PubMed] [Google Scholar]
- (9).Du X; Chowdhury SM; Manes NP; Wu S; Mayer MU; Adkins JN; Anderson GA; Smith RD Xlink-identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J. Proteome Res 2011, 10 (3), 923−931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ji C; Li S; Reilly JP; Radivojac P; Tang H XLSearch: A probabilistic database search algorithm for identifying cross-linked peptides. J. Proteome Res 2016, 15 (6), 1830−1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kao A; Chiu C; Vellucci D; Yang Y; Patel VR; Guan S; Randall A; Baldi P; Rychnovsky SD; Huang L Development of a Novel Cross-linking Strategy for Fast and Accurate Identification of Cross-linked Peptides of Protein Complexes. Mol. Cell. Proteomics 2011, 10 (1), M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Liu F; Lössl P; Scheltema R; Viner R; Heck AJR Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun 2017, 8, 15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yang B; Wu Y-J; Zhu M; Fan S-B; Lin J; Zhang K; Li S; Chi H; Li Y-X; Chen H-F; et al. Identification of cross-linked peptides from complex samples. Nat. Methods 2012, 9 (9), 904−906. [DOI] [PubMed] [Google Scholar]
- (14).Trnka MJ; Baker PR; Robinson PJJ; Burlingame AL; Chalkley RJ Matching Cross-linked Peptide Spectra: Only as Good as the Worse Identification. Mol. Cell. Proteomics 2014, 13 (2), 420−434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fischer L; Chen ZA; Rappsilber J Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers. J. Proteomics 2013, 88, 120−128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yu F; Li N; Yu W ECL: an exhaustive search tool for the identification of cross-linked peptides using whole database. BMC Bioinf 2016, 17 (1), 1−8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hoopmann MR; Zelter A; Johnson RS; Riffle M; Maccoss MJ; Davis TN; Moritz RL Kojak: Efficient analysis of chemically cross-linked protein complexes. J. Proteome Res 2015, 14 (5), 2190−2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Götze M; Pettelkau J; Fritzsche R; Ihling CH; Schäfer M; Sinz A Automated assignment of MS/MS cleavable cross-links in protein 3d-structure analysis. J. Am. Soc. Mass Spectrom 2015, 26 (1), 83−97. [DOI] [PubMed] [Google Scholar]
- (19).Petrotchenko EV; Borchers CH ICC-CLASS: Isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinf 2010, 11 (1), 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Millikin RJ; Solntsev SK; Shortreed MR; Smith LM Ultrafast Peptide Label-Free Quantification with FlashLFQ. J. Proteome Res 2018, 17 (1), 386−391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kong AT; Leprevost FV; Avtonomov DM; Mellacheruvu D; Nesvizhskii AI MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry−based proteomics. Nat. Methods 2017, 14 (5), 513−520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Solntsev SK; Shortreed MR; Frey BL; Smith LM Enhanced Global Post-translational Modification Discovery with MetaMorpheus. J. Proteome Res 2018, 17 (5), 1844−1851. [DOI] [PubMed] [Google Scholar]
- (23).Cesnik AJ; Shortreed MR; Sheynkman GM; Frey BL; Smith LM Human Proteomic Variation Revealed by Combining RNA-Seq Proteogenomics and Global Post-Translational Modification (G-PTM) Search Strategy. J. Proteome Res 2016, 15 (3), 800−808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4 (3), 207−214. [DOI] [PubMed] [Google Scholar]
- (25).Käll L; Canterbury JD; Weston J; Noble WS; MacCoss MJ Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4 (11), 923−925. [DOI] [PubMed] [Google Scholar]
- (26).Riffle M; Jaschob D; Zelter A; Davis TN ProXL (protein cross-linking database): A platform for analysis, visualization, and sharing of protein cross-linking mass spectrometry data. J. Proteome Res 2016, 15 (8), 2863−2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hoopmann MR; Mendoza L; Deutsch EW; Shteynberg D; Moritz RL An Open Data Format for Visualization and Analysis of Cross-Linked Mass Spectrometry Results. J. Am. Soc. Mass Spectrom 2016, 27 (11), 1728−1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Erde J; Loo RRO; Loo JA Improving proteome coverage and sample recovery with enhanced FASP (eFASP) for quantitative proteomic experiments. Methods Mol. Biol 2017, 1550, 11−18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Vizcaíno JA; Côté RG; Csordas A; Dianes JA; Fabregat A; Foster JM; Griss J; Alpi E; Birim M; Contell J; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 2012, 41 (D1), D1063−D1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.