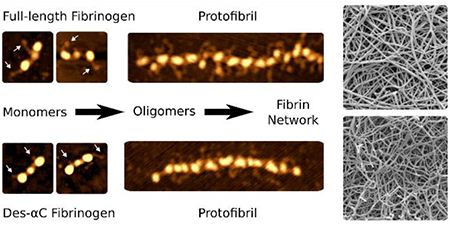
Abstract
The flexible C-terminal parts of fibrinogen’s Aα chains named the αC regions have been shown to play a role in fibrin self-assembly, although many aspects of their structure and functions remain unknown. To examine the involvement of the αC regions in the early stages of fibrin formation, we used high-resolution atomic force microscopy to image fibrinogen and oligomeric fibrin. Plasma-purified full-length human fibrinogen or des-αC fibrinogen lacking most of the αC regions, untreated or treated with thrombin, were imaged. Up to 80% of the potentially existing αC regions were visualized and quantified; they were highly heterogeneous in their length and configurations. Conversion of fibrinogen to fibrin was accompanied by an increase of the incidence and length of the αC regions as well as transitions from more compact conformations, such as a globule on a string, to extended and more flexible offshoots. Concurrent dynamic turbidimetry, confocal microscopy, and scanning electron microscopy revealed that trimming of the αC regions slowed down fibrin formation, which correlated with longer protofibrils, thinner fibers, and a denser network. No structural distinctions, except for the incidence of the αC regions, were revealed in laterally aggregated protofibrils made of the full-length or des-αC fibrinogens, suggesting a pure kinetic effect of the αC regions on the fibrin architecture. This work provides a structural molecular basis for the promoting role of the αC regions in the early stages of fibrin self-assembly and reveals this stage of fibrin formation as a potential therapeutic target to modulate the structure and mechanical properties of blood clots.
Graphical Abstract
High-resolution atomic force microscopy imaging reveals the role of fibrinogen αC regions in the early stages of fibrin self-assembly.
Introduction
Fibrinogen (Fg) is a soluble blood plasma protein that can be proteolytically converted to polymeric fibrin, the structural and mechanical scaffold of hemostatic clots and vessel-obstructing pathological thrombi. The 340-kDa Fg molecule consists of three pairs of polypeptide chains, designated Aα, Bβ and γ, arranged into an elongated structure 45nm in length and 2–5nm in diameter (Fig.S1).1–3
The C-terminal portions of the Aα chains, called the αC regions, are ≈400-residues long, mostly disordered and highly flexible.4 Based on many studies, the αC regions do the following: 1) mediate intermolecular interactions during fibrin self-assembly,5–10 2) modulate the structure and mechanical properties of fibrin,9,11–14 3) serve as a substrate for fibrin cross-linking by factor XIIIa,5,8,15–18 4) contain binding sites for plasminogen, tPA, α2-antiplasmin, fibronectin19–22 and adhesive cellular receptors,23–25 5) increase susceptibility of fibrin to lysis,12 although their cross-linking by factor XIIIa decreases the fibrinolysis rate,26 6) provide cleavage sites for plasmin,27 7) determine hydrodynamic properties of Fg in solution,28,29 and 8) mediate adhesion of Fg to artificial surfaces.30,31 The physiological importance of the Fg’s αC regions is confirmed by reported dysfibrinogenemias (Fgs Dusart, Caracas II, Marburg, Milano III, Lincoln, and Otago) in which truncation or mutation of the αC regions predisposes to thrombosis or bleeding.4 Therefore, studies on the structure and functions of the αC regions have been of great biological and medical importance.
In the absence of the three-dimensional atomic structure of the αC regions32,33, the current view on their structural organization is based on sequence analysis, differential scanning calorimetry, and transmission electron microscopy.5,7,34–36 It is generally accepted that in human Fg each αC region (Aα221–610) consists of two structurally distinct portions: the C-terminal portion (Aα392–610), called the αC-domain, contains a compact folded domain, while the N-terminal portion (Aα221–391), called αC-connector, forms a flexible tether linking the αC-domain to the bulk of the molecule. The only direct microscopic observation of the αC regions was done by transmission electron microscopy of rotary-shadowed Fg. It revealed an additional (fourth) globule near the central part of the molecule corresponding to the αC regions interacting with each other,5,36 but the αC connector regions were not visualized.
The goal of our work was to get new structural information on the αC regions in Fg and its derivatives and, by combining the structural and functional approaches, to study the involvement of the αC regions in the early stages of fibrin self-assembly.
To obtain structural characteristics of the αC regions, we visualized fibrinogen and fibrin using atomic force microscopy (AFM), which has become a valuable tool to study protein molecules.37–41 Recently, we improved the AFM-based visualization of fibrin(ogen) using a modified graphite surface as a substrate for protein adsorption.42 This method enabled us to obtain single-molecule images with a resolution comparable to or better than that of transmission electron microscopy, which is necessary for visualization of the Fg αC regions. To get functional characteristics of the αC regions, we used dynamic turbidity, laser scanning confocal microscopy, and scanning electron microscopy.
Here we provide a molecular structural basis for the role of the unstructured Fg’s αC regions in fibrin self-assembly based on the single-molecule imaging of the full-length Fg and Fg subfraction I-9 with truncated αC regions (des-αC Fg). The novelty of the work is based on the precise quantification of the αC regions’ structure, which was only possible due to development of the remarkably high-resolution AFM imaging, and gives important information about its functions. We analyze the incidence, contour length and morphology of the αC regions in two Fg variants and their derivatives, as well as fibrin polymerization kinetics, and final clot structure.
Methods
Handling and characterization of the full-length fibrinogen:
Protein samples were prepared from lyophilized human Fg (HYPHEN Biomed, France). The powder was dissolved in water and dialyzed against 20 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl. After centrifugation at 20,000g and 4°C for 15 minutes to remove all micro-precipitates, the fibrinogen (Fg) concentration was determined by absorbance at λ = 280 nm using an extinction coefficient of 1.51 for 1 mg/ml in a 1-cm cuvette. All the full-length Fg preparations were 93–95% pure, as determined using 10% SDS-PAGE in reducing conditions (Fig. S2) and 98% clottable by thrombin, indicating that Fg was fully functional.
Purification and characterization of human des-αC Fg subfraction:
We used a thrombin-coagulable catabolic subfraction Fg I-9 (currently termed des-αC Fg) – a population of Fg molecules without the αC regions or without a part of the αC regions comprising a minor part of circulating plasma Fg.43 The procedure used for isolation of fraction I-9 from pooled human plasma43 is briefly described below. Following two consecutive saturated Glycine precipitations at 5°C, cold insoluble material was removed, fraction I-2 was harvested at −2°C, by 8% ethanol precipitation. Fraction I-5 was subsequently precipitated by increasing the ethanol concentration to 16%. Fraction I-5 (amounting to 5–10% of Fg in plasma) was dissolved in 0.28 M NaCl, 0.1 M sodium phosphate, pH 6.4, to 0.1–0.15% protein concentration. The solution was cooled to −2°C, ethanol was slowly added to 8% concentration, and the precipitate, fraction I-6, was harvested by centrifugation at −2°C. The remaining soluble fibrinogen was precipitated by raising the ethanol concentration to 16%, and redissolved in 0.1 M NaCl, 0.01 M sodium phosphate, pH 6.4. Addition of Glycine to 2.1 M precipitated fraction I-8. Fg remaining in the supernatant, fraction I-9, was precipitated by adding ammonium sulfate to 33% saturation. Final isolates were dialyzed and stored in 0.3M NaCl at −70°C.
The des-αC Fg subfraction lacks the C-terminal portions of its Aα chains and contains the Aα chain core remnants with intact N-terminus44 ranged from 46.5 kDa to 22.6 kDa (or ~66% to 32% of the mass of intact Aα chain) (Fig. S2). As estimated from these measurements, the subfraction lacks virtually all of the αC-domains (Aα392–610) and variable lengths of the αC-connector (Aα221–391). Evaluated by tryptic peptide maps44 and SDS-PAGE (Fig. S2), the Bβ and γ chains of des-αC Fg were intact. The concentration of des-αC Fg was determined by absorbance at λ = 280 nm using an extinction coefficient of 1.6 for 1 mg/ml in a 1-cm cuvette, as for recombinant Aα251 Fg without the αC regions. As a matter of fact, larger quantities of plasma-purified Fg I-9 are available compared to recombinant Fg Aα251, allowing the variety of different experiments performed in this study.
Sample preparations for AFM:
Fibrinogen samples were prepared by dilution of the initial protein solution to 1–3 μg/ml concentration with a 20 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl and 2 mM CaCl2. To make samples ready for imaging, typically 3 μl of the diluted solution was applied on a substrate surface and kept for 5–15 s. A 150-μl volume drop of fresh milli-Q water was then carefully placed above the sample for 10 s and then removed with a flow of air making the surface dry for imaging in air.
For fibrin formation, the initial protein solution was first diluted to the desired concentration in the same buffer. To produce short fibrin oligomers and individual protofibrils, 0.15 mg/ml Fg was mixed with 0.05 U/ml thrombin (final concentration), incubated for 5 minutes at room temperature, then diluted 100-fold to 1.5 μg/ml concentration with the buffer and immediately used for AFM sample preparation as described above. To produce long fibrin protofibrils and their lateral aggregates, 0.02 mg/ml Fg was mixed with 0.05 U/ml thrombin, incubated for 6–12 minutes at room temperature and used for AFM sample preparation without further dilution.
All AFM samples were performed at least in triplicate.
Substrate selection for AFM:
To perform high-resolution AFM of Fg and fibrin, we used a modified hydrophilized graphite as a substrate for sample preparation. Highly oriented pyrolytic graphite (HOPG) was rendered hydrophilic with an amphiphilic graphite modifier (GM) as described elsewhere45,42. Because results of AFM imaging strongly depend on the properties of the substrate, we tested and compared 3 other substrates: clean freshly cleaved mica, mica modified with APTES, and glow-discharged glass (Supplementary Section 1). However, GM-HOPG was the only substrate that allowed reproducible visualization of the Fg’s αC regions. AFM images of full-length Fg molecules adsorbed on the substrates other than GM-HOPG are shown in the Fig. S3.
Acquisition and processing of AFM images:
AFM imaging was performed using a MFP-3D microscope (Asylum Research – Oxford Instruments, USA) or an Ntegra Prima microscope (NT-Mdt, Russia) in a tapping mode with a typical scan rate of 0.8 Hz. Images were taken in air using sharpened silicon cantilevers SSS-SEIHR (Nanosensors, Germany) with a guaranteed tip radius <5 nm or homemade super-sharp cantilevers with a tip radius about 1 nm.46 FemtoScan Online software (http://www.femtoscanonline.com) was used to filter and analyze the AFM data.
Dynamic turbidity measurements:
To follow and quantify the kinetics of fibrin formation from different Fg variants, dynamic turbidimetry was used. Fibrin self-assembly was initiated by adding 0.05 U/ml thrombin (final) to a solution containing 0.02 mg/ml Fg in a buffer (20 mM Tris-HCl, 150 mM NaCl, and 2 mM CaCl2, pH 7.4). Turbidity measurements were carried out at 350 nm using a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific, USA) at room temperature. The following parameters were extracted from turbidimetric curves: the lag phase, which corresponds to formation of fibrin oligomers and protofibrils; average slope of rise in optical density, which correlates with the rate of protofibril lateral aggregation; and the final optical density, which reflects the amount of protein and averaged fiber diameter in the clot (Fig. S4).
Laser scanning confocal microscopy:
To prepare fibrin clots for confocal microscopy, 1.5 mg/ml full-length Fg (4.4 μM) or des-αC Fg (5 μM; estimated molar concentrations of des-αC Fg are based on an average molecular weight (Mw = 300 kDa) between recombinant Fg Aα251 completely lacking the αC regions, Mw = 260 kDa47 and the full-length Fg (340 kDa)) in a 20 mM Tris-HCl buffer, pH 7.4, 150 mM NaCl, 2 mM CaCl2 containing 0.06 mg/ml (4%) Alexa-488 labeled Fg (Invitrogen, Thermo Fisher Scientific, USA) was mixed with thrombin and incubated for 2 hours at room temperature in a cover glass bottomed chamber. To keep the same Fg/thrombin molar ratio (≈2000/1) with correction for the lower molecular weight of the des-αC Fg, we added 0.22 U/ml (2.2 nM) thrombin (final concentration) to the full-length Fg and 0.25 U/ml (2.5 nM) thrombin to des-αC Fg. Images were taken in a Zeiss LSM 880 microscope using a C-Apochromat 40x water immersion objective lens (NA 1.2) and a PMT detector. An argon laser was used for excitation at 488 nm. Confocal stacks of 15 μm depth were taken several micrometers from the glass surface. The experiments were performed in triplicate.
Fiji48 and Imaris 8.3.1 (Bitplane – Oxford Instruments, Switzerland) software were used to present and analyze confocal images. To determine the branch point density in fibrin networks with the Imaris Filament Tracer software, identical tracing parameters were used for all images; short loops (<2 μm) and branches (<1.5 μm) were excluded from the analysis. Imaris Filament Tracer calculates a number of branch points corresponding to each node of the network based on a number of branches exiting the node (see schematic examples in Fig. S5). Visual evaluation of tracing results showed that the algorithm gives a fair estimation of a total number of branch points per image for both networks.
Scanning electron microscopy:
1 mg/ml full-length Fg (2.9 μM) or des-αC Fg (3.3 μM) in a 20 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl and 2 mM CaCl2 was mixed with 0.15 U/ml (1.5 nM) or 0.17 U/ml (1.7 nM) thrombin (respective final concentrations) and incubated for 1 hour at room temperature in plexiglass microdialysis cells. Specimens were washed 3 times with a low ionic strength 50 mM sodium cacodylate-HCl buffer (pH 7.4) for 10 minutes, fixed overnight in 2% glutaraldehyde, then washed 3 times and dehydrated with a series of graded concentrations of ethanol (30–100vol%) in the cacodylate buffer. The dehydrated samples were then rinsed with increasing concentrations of hexametihyldisilazane (HMDS) in ethanol (50–100 vol%). Finally, the specimens were left to dry under a fume hood in 100% HMDS. The dried samples were mounted, sputter-coated with gold-palladium in a Sputter Coating Unit E5100 (Polaron Equipment, UK) at 2.2 kV and 20 mA for 1.5 min, and examined in a FEI Quanta 250 FEG scanning electron microscope (FEI, USA). Several fields of each clot were taken at different magnifications before choosing the areas that were characteristic of the entire clot. The experiments were performed in triplicate.
Statistical analysis:
Statistical analysis was performed in R49 using the package Psych.50 The Chi-square test was used for nominal scale data, Welch’s unequal variances two-sample t-test and non-parametric Wilcoxon rank sum test were used for ordinal scale data.
Results
Molecular structure of the full-length and des-αC Fgs.
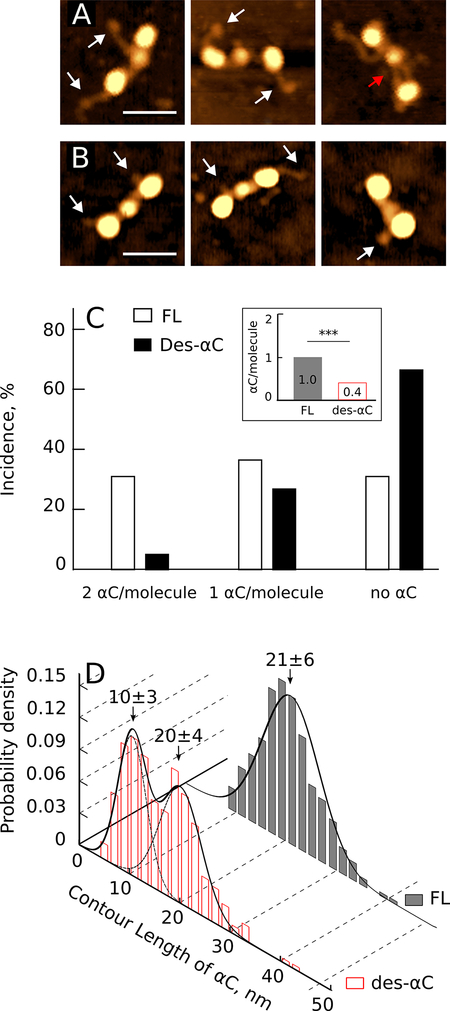
An individual Fg molecule adsorbed on the GM-graphite substrate was visualized in AFM as a linear sequence of three distinct globular regions connected by thin rods representing the α-helical coiled coils (Fig. 1A,B). The central globular region was smaller than the lateral regions with an average height of 1.8±0.3nm (n=125), while two identical lateral globular regions were higher with an average height of 2.9±0.3nm (n=250). The structural differences between the full-length Fg (Fig. 1A) and des-αC Fg (Fig. 1B) were the distinct incidence and length of the filamentous protrusions that represented the αC regions. Most of the full-length Fg molecules had one or two long strands sticking out from the lateral globules or near them (Fig. 1A), while des-αC Fg typically had only short stubs or no visible overhangs (Fig. 1B). In the full-length Fg, some of these protrusions had a small single globule on their end, which could represent a compact αC-domain linked to the bulk molecule via a flexible αC-connector.4 In the AFM images, both the end globule and thin αC-connector were often observed, in contrast to transmission electron microscopy images of Fg, where the globular regions were observed without the connector regions. Some αC regions were connected to the central globule of the Fg molecule (Fig. 1A, red arrow), as observed earlier with transmission electron microscopy of Fg.5 Quantitatively, both αC regions were seen in 32% of the full-length Fg molecules, only one was observed in 37%, and no αC-related protrusions were revealed in 31% of the full-length Fg molecules (Fig. 1C). This was quite distinct from des-αC Fg, of which only 6% of molecules exposed two protrusions, 27% had one short filament, and no extensions were seen in 67% of the des-αC Fg molecules (Fig. 1C). A chi-square test showed a significant difference in the proportion of molecules with two, one, or no αC regions in the full-length and des-αC Fgs (p<0.001, n=400 for each sample). On average, only 0.4 αC regions per molecule were visualized in des-αC Fg compared to 1 αC region per monomer in full-length Fg (Fig. 1C, inset).
figure 1.
incidence and contour length distribution of the αc regions in the full-length and des-αc fgs. a: afm images of individual full-length fg molecules with their αc regions exposed. white arrows point to the globular structures at the free ends of the αc regions, a red arrow tags the αc region connected to the central globule of a fg molecule. magnification bar = 30 nm. b: des-αc fg molecules with partially truncated αc regions shown by white arrows. magnification bar = 30 nm. c: percentage of the full-length fg (fl) and des-αc fg molecules with one, two or no visible αc regions. an inset shows the average number of visible αc regions per one fg monomer (***p<0.001). d: histograms of the αc regions’ contour lengths in the full-length (fl) and des-αc fg.
The contour length of the αC regions in the AFM images was also different in the full-length and des-αC Fg, consistent with SDS-PAGE (Fig.S2). The peak contour length of the αC-related protrusions in the full-length Fg was 21±6nm (n=280) with a unimodal distribution, while in des-αC Fg the distribution was bimodal with two maxima at 20±4nm and 10±3nm (n=280), showing the heterogeneity due to variable truncation of the Aα chains (Fig. 1D).
In summary, single-molecule AFM imaging confirmed that Fg’s αC regions could be visualized and this enabled us to see morphological distinctions between the full-length and des-αC Fg molecules that differed in the incidence and contour length of the αC regions. Moreover, in the full-length Fg we could identify compact terminal αC-domains linked to the bulk molecule via flexible αC connectors, thus confirming the proposed structure of the αC regions based on transmission electron microscopy,5 differential scanning calorimetry,33 and NMR spectroscopy.32
Structure of fibrin oligomers formed from the full-length and des-αC Fgs.
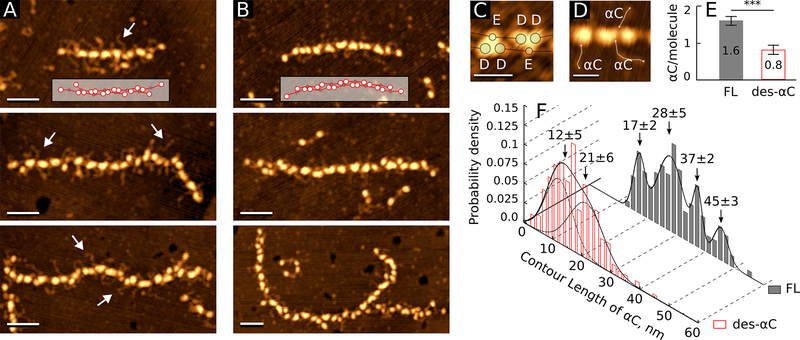
To see if the αC regions are involved in the early stages of fibrin self-assembly, we compared the structures of fibrin oligomers formed from the full-length and des-αC Fg variants by thrombin cleavage. We identified fibrin oligomers in the AFM images as elongated double-stranded structures containing periodically (≈22.5nm) spaced oval or “heart-shaped” structures (Fig. 2A,B). Each of these structures within a fibrin oligomer corresponded to a single D-E-D complex that holds together two strands of the oligomer. The variability of this shape was likely due to protofibril twisting and the resulting non-uniform spatial orientation of the D-E-D complex on the substrate. The longer oligomers were more curved, but otherwise the structural characteristics of two-stranded fibrin oligomers did not depend on the degree of polymerization.
figure 2.
incidence and contour length distribution of the αc regions in fibrin oligomers formed from full-length and des-αc fgs. a: individual fibrin oligomers formed from the full-length fg. an inset shows how fibrin monomers are lined up in the protofibril. white arrows point to the αc regions magnification bars = 50 nm. b: fibrin oligomers formed from the des-αc fg. an inset shows how monomers are lined up in the protofibril. magnification bars = 50 nm. c: two enlarged “heart-shaped” particles showing the dispositions of the globular parts of d and e regions. magnification bars = 20 nm. d: protofibril fragment; the lines show how the contour length of the αc regions was measured. magnification bars = 20 nm. e: the average number of visible αc regions per one monomer in fibrin oligomers formed from the full-length (n=40) and des-αc fgs (n=40) variants (***p<0.001). f: histograms of the αc regions’ contour lengths in fibrin oligomers formed from the full-length fg (n=140) and des-αc fg (n=246).
The average height of the whole D-E-D complexes was 3.6±0.3nm (n=150). The heart-shaped structures were present as tri-nodular complexes made from globular parts of two D regions and one E region based on its symmetry (Fig. 2C). The distances between the geometric centers of D or E globules in the D-E or D-D structures were equal to 6.8±1.2nm (n=100) and 10.1±1.7nm (n=50), respectively.
To evaluate the possible role of the αC regions in fibrin polymerization, we determined the number of the αC regions extending from two-stranded short fibrin oligomers or longer protofibrils relative to the number of monomeric molecules forming each oligomer (Fig. 2D,E). On average, in the oligomers made from the full-length Fg, we could identify 1.6 αC regions per monomer, while in the oligomers made from des-αC Fg, only 0.8 αC regions per fibrin monomer were observed. Thus, in the oligomers the incidence or “visibility” of the αC regions increased substantially compared to Fg monomers (Figs. 1C, inset, and 2E). However, the number of αC regions was still significantly smaller in the des-αC-oligomers than in the oligomers made from the full-length Fg (p<0.001, Welch’s two-sample t-test, n=40 for both samples). Remarkably, the bimodal distribution and the average contour length of the truncated αC regions in des-αC fibrin oligomers were not significantly different from those in des-αC Fg molecules (Wilcoxon rank sum test p=0.6319, nm=194, no=246) (compare Figs. 2F and 1D). In contrast, the αC regions’ contour length in fibrin oligomers made from the full-length Fg was significantly longer than that in the full-length Fg monomers (Wilcoxon rank sum test p<0.001 nm=165, no=140) and had a broad multimodal distribution (Fig. 2F), reflecting a conformational change in the full-length αC regions happening upon conversion from Fg to fibrin.
Thus, for both Fg variants more αC regions were seen in fibrin oligomers than in the corresponding Fg monomers; however, in the full-length Fg the αC regions’ contour length increased after conversion to oligomeric fibrin, while in the des-αC Fg the contour length of shortened αC regions did not change upon fibrin oligomerization.
Structural basis for the αC region-dependent variations in the kinetics of fibrin polymerization.
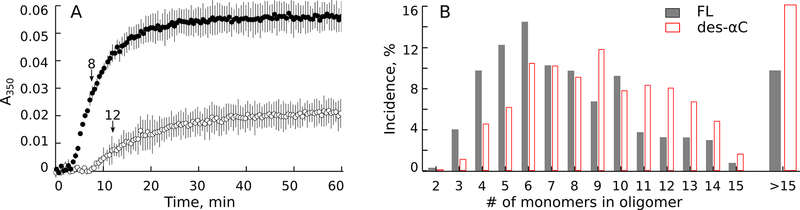
Average turbidity curves for clotting of the full-length and des-αC Fgs (Fig. 3A) revealed a big difference in the kinetics of fibrin formation. In des-αC Fg vs. full-length Fg the average lag time was longer and comprised 8.2±0.5min and 3.7±0.2min, respectively (p<0.001, unpaired t-test). The rate of protofibril lateral aggregation determined by the slope of the curve was smaller in des-αC Fg vs. full-length Fg, equal to (11±3)×10−4a.u./min and (55±6)×10−4a.u./min, respectively (p<0.001, unpaired t-test). These kinetic differences suggest that without the αC regions protofibril formation is delayed and their lateral aggregation is slowed down. In addition, the maximal optical density of “des-αC clots” was 2.5 times lower than that of the “full-length clots”, suggesting a dramatic effect of the αC regions on the final structure of fibrin.
figure 3.
kinetics of clotting of the full-length and des-αc fgs. a: average turbidity curves of the full-length (●) and des-αc (○) fgs at 0.02 mg/ml concentration activated with 0.05 u/ml thrombin (n=3 for each sample). fibrin formation was followed as a change in turbidity at λ=350 nm with time. arrows show the time points at which the samples were extracted and prepared for afm shown in fig. s5. b: length distributions of fibrin oligomers measured from the afm images presented as the number of monomers comprising each oligomer (degree of polymerization). the histograms were made at the time points shown in a: 8 min after the initiation of clotting of the full-length fg (n=400), and 12 min after the initiation of clotting of the des-αc fg (n=400). structures containing >15 fibrin monomers were infrequent, so they were combined in the histogram.
To provide a structural basis for the observed differences in polymerization kinetics, intermediate fibrin structures formed from des-αC and full-length Fgs were imaged and compared. The samples were taken at or near the time points corresponding to the middle of the ascending parts of turbidimetry curves, i.e. at or near the gelation point determined in a parallel experiment by eye as the clotting time. AFM showed that at this time point a mixture of fibrin monomers, oligomers, aggregating protofibrils, and fibers was present in the samples (Fig.S6). We measured contour lengths of individual double-stranded oligomers found in the images and converted their lengths to the degree of polymerization for each oligomer using the formula where n is the number of monomers (degree of polymerization), L is the measured contour length of an oligomer in nanometers, and 22.5nm corresponds to ½-length of fibrin monomers that are half-staggered. Then we calculated the relative numbers of oligomeric structures with various degrees of polymerization (Fig. 3B). In des-αC fibrin, oligomers built of more than 9–10 monomers were more frequent compared to the structures made from the full-length Fg. On the contrary, in the fibrin oligomers made from the full-length Fg, the fraction of shorter structures (3–6 monomers) was bigger compared to the fibrin oligomers formed from the des-αC Fg. These distinctions were consistent at the time points within 6–12min on the polymerization curve at which we interrupted fibrin formation and visualized intermediate fibrin structures using a relatively narrow range of thrombin concentrations varying from 0.04 U/ml to 0.06 U/ml. Aggregated protofibrils formed from the full-length and des-αC Fgs had very similar backbone structures, yet with different numbers of αC regions near and between the strands (Figs.S6B and S6D).
Thus, the dynamic turbidimetry correlated with AFM imaging revealed a significant effect of the αC regions on fibrin polymerization kinetics. The results of AFM indicate that these differences in kinetics are associated with longer intermediate fibrin oligomers made from the des-αC Fg compared to those made from the full-length Fg, suggesting a larger threshold length of des-αC-protofibrils at which they can undergo lateral aggregation.
Structure of hydrated fibrin clots with the full-length and truncated αC regions.
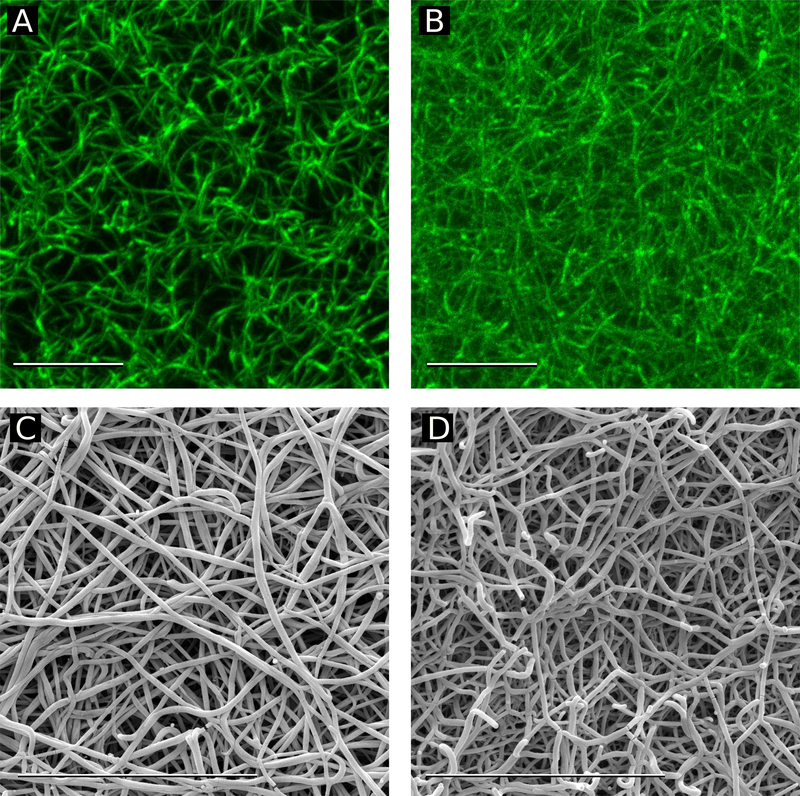
The lower maximum turbidity of the des-αC clots (Fig. 3A) suggests that fibrin fibers formed from des-αC Fg are thinner than fibers formed from full-length Fg.51 To confirm this hypothesis, the final structures of the two types of fully hydrated mature fibrin clots were studied with confocal microscopy. Full-length and des-αC fibrin networks looked very different with a substantially higher density of fibrin fibers, many more branch points, and thinner fibers in the clots made from des-αC Fg (Fig. 4A, B).
figure 4.
fluorescent confocal and scanning electron microscopy of fibrin clots made from the full-length and des-αc fgs at the same fg concentration and fg/thrombin molar ratio. confocal microscopy images of a: a fibrin clot made from full-length fg, b: a fibrin clot made from des-αc fg. images are the maximum intensity projections of 15-μm z-stacks, image size 35.4x35.4 μm2. electron microscopy images of c: a fibrin clot made from full-length fg. d: a fibrin clot made from des-αc fg. magnification bars = 10 μm.
To estimate the network porosity, the number of branch points per volume (branch point density) was measured using the Imaris Filament Tracer image analyzing tool. With confocal microscopy we analyzed 4 clots made from the full-length Fg and 3 clots made from des-αC Fg; the results obtained were reproducible. For each clot, 20 randomly selected images were examined, and an average branch point density was determined in each clot. The mean density of branch points was 5.7±2.2 per 100μm3 in the clots made from the full-length Fg and 11.1±2.4 per 100μm3 in the clots made from des-αC Fg (p=0.0344, Welch’s two-sample t-test, nFL=4, nI-9=3), confirming the visual impression of a much higher density of branch points of the des-αC fibrin network. With the same protein mass, a higher network density implies that individual fibers are thinner, which is in line with the reduced lateral aggregation of protofibrils formed from des-αC Fg and smaller turbidity of the des-αC fibrin clot (Fig. 3A).
Diameter of fibers in fibrin clots made from the full-length and des-αC Fgs.
Scanning electron microscopy of fibrin clots showed that the fibrin network made of the full-length Fg had thicker fibers than that made of the des-αC Fg (Fig. 4C, D). The average diameter of the full-length fibrin fibers was 119±25nm, while the des-αC fibrin fibers were 83±18 nm in diameter (p< 0.01, Welch’s two-sample t-test, n=400 for both samples). This result is consistent with the lower maximum turbidity of the des-αC clots (Fig. 3A) and with a higher number of branch points in these clots revealed by confocal microscopy. It could also be noted that the des-αC clots appear to have more fiber ends than the clots prepared from full-length Fg, which is another sign of impaired polymerization (Fig. 4C, D). Together all three methods (dynamic turbidimery, confocal microscopy and scanning electron microscopy) showed that in the absence of the αC regions fibrin clots were much denser because they were formed of thinner fibers arranged into a highly branched and less porous network.
Discussion
Physiological relevance of des-αC Fg variants.
One of the general approaches to study the αC regions is using Fg variants where the αC regions are partially or fully absent, such as proteolytic fragment X produced by plasmin cleavage of Fg5,7,52 or recombinant Fg Aα251 in which the C-termini of the Aα chains are deleted at position 251.12,47 Here, for comparison with the full-length Fg, we used a natural catabolic Fg subfraction, des-αC Fg (also named Fg I-9), purified from normal human plasma,43,44 which has never been characterized structurally before. Des-αC Fg is produced naturally in vivo by enzymatic digestion of the C-terminal parts of Fg Aα chains.43,52–57 It belongs to 30% of the total clottable Fg comprising partially truncated Fg with variable parts of the αC regions, while the full-length Fg with intact αC regions normally comprise about 70%.58
Although the role of Fg catabolic derivatives is poorly understood, based on in vitro studies of proteolytic fragment X it has been suggested that truncated Fg variants interfere with coagulation process and have mild anticoagulant activity.59 Another study has shown increased levels of Fg mRNA and elevated plasma Fg concentration in animals after intraperitoneal injection of fragment X, suggesting a signaling role of the truncated Fg derivatives.60 These and other data emphasize the importance and physiological relevance of studies on the natural Fg variants with partially cleaved Aα chains. Additionally, larger quantities of plasma-purified des-αC Fg are available compared to recombinant Fg Aα251, allowing the variety of different experiments performed in this study.
What is the real length of the αC regions?
The distribution of αC region’s contour lengths in the full-length Fg was broad and skewed towards larger values with a maximum at 21±6 nm (Fig. 1D). Remarkably, this peak length is much shorter than the theoretical full contour length of 148 nm, considering that the αC regions contain 390 residues with the backbone length of one residue to be equal to 0.38 nm.61 This discrepancy could be interpreted in three possible ways. 1) As discussed above, many of the αC regions in the full-length Fg are proteolytically truncated. 2) There is evidence that the long αC regions are likely partially folded4, so their visible length could be much less than the theoretical full contour length. 3) The αC regions could be only partially exposed outwards because of their interaction with the bulk of the molecule. The proposed explanations are not mutually exclusive, and our experimental data indirectly support all of them.
Remarkably, conversion of the full-length Fg to oligomeric fibrin is followed by exposure of the αC regions that are more than 2-fold longer than in the initial monomers, reaching 45–50 nm vs. ~20 nm, respectively (Figs. 1D and 2F). On the contrary, the lengths of the αC regions in des-αC Fg monomers and des-αC fibrin oligomers are identical and smaller (Figs. 1D and 2FE), suggesting the lack of any cryptic portions exposed in des-αC fibrin. This “recovery” of the αC regions in the full-length fibrin versus Fg supports the assumption that the αC regions either undergo conformational unfolding/elongation or dissociate from the compact core of Fg upon conversion to fibrin, which is not mutually exclusive.
The incidence of the αC regions deviates from the theory.
Based on the literature, analysis of the incidence of the αC regions in molecular images of Fg is complicated by variability of sample preparation procedures and imaging techniques. In contrast, the appearance and incidence of the αC regions in fibrin oligomers and monomers is more robust and there is good agreement between the results of our work and that reported by others. Specifically, the incidence of the αC regions in the full-length fibrin oligomers of 1.6 αC/molecule observed in our study (Fig. 2E) is almost equal to 1.5 αC/molecule in the full-length fibrin monomers shown by Veklich et al.5 Our results are also in line with the data obtained by SDS-PAGE analysis by Mosesson and coworkers44 who estimated the number of Aα chains containing at least some of the αC regions to be equal to ≈1.6 αC/molecule for the full-length Fg and 0.7 αC/molecule for des-αC Fg. Both values match our data: an average number of the αC regions in the full-length and des-αC fibrin oligomers were equal to 1.6 αC regions/molecule and 0.8 αC regions/molecule, respectively (Fig. 2ED). The coinciding results on the incidence of the αC regions in the three independent studies suggest that there is about 20% of the αC regions in Fg preparations that are either cleaved off or buried in the molecule, thus remaining unexposed and perhaps non-functional.
Variable structural forms of the αC regions.
The appearance of the αC regions in the full-length Fg is diverse and can be roughly segregated into the following types: 1) αC regions connected to the central region of the molecule (Fig. 1A, red arrow); 2) prominent string-like αC regions with a small globule on the end (Fig. 1A, white arrows); 3) prominent and relatively long curved or bent αC regions (Fig. 2A, white arrows). The first and second types were more typical for Fg, while the third type was more characteristic of fibrin oligomers. This diversity confirms a high structural flexibility of the αC regions and suggests that more compact conformations, such as a globule on a string, dominate in Fg, while extended and more flexible conformations prevail in fibrin. For comparison, in des-αC Fg and fibrin, where virtually all the αC-domains are missing, the shapes of the αC regions were more uniform, usually showing up as straight or bent short strings, likely representing the αC-connectors. The results strongly suggest that conformational transitions of the αC regions from a more compact to more elongated and extended state is a part of the overall structural transformation of Fg to fibrin.
The kinetic mechanism for the effects of the αC regions on fibrin formation and structure.
Experiments with the proteins lacking all (recombinant Fg Aα251) or almost all (Fg fragment X) αC regions have shown that the αC regions enhance fibrin self-assembly, most likely via promoting protofibrils’ lateral aggregation.5,47 Our data (Figs. 3–4) agree with these studies (Table S1) showing unambiguously that without the αC regions, fibrin polymerization is delayed (longer lag period and slower fibrin formation) and the fibrin network has thinner fibers with more branch points and lower maximum turbidity (with a partial exception for fragment X). It is conceivable that the αC regions act in three ways: 1) accelerate formation of protofibrils; 2) modulate (shorten) the threshold length needed for their lateral aggregation; 3) enhance the lateral aggregation of protofibrils.
To discriminate between these possibilities, we used the AFM characterization of intermediate fibrin oligomers formed at the rise of the turbidity curve (Fig. 3A). It was found that the αC regions reduce the threshold length of protofibrils at which they begin to aggregate laterally. At the same time, a careful examination of laterally aggregated double protofibrils did not reveal any structural signs of impaired lateral aggregation in the absence of the αC regions; aggregated protofibrils formed from the full-length and des-αC Fgs had very similar backbone structures, yet with a different number of αC regions on and between the protofibrils (Figs.S6B and S6D).
Thus, the αC regions accelerate fibrin self-assembly by reducing the threshold length of protofibrils, thus speeding up their lateral aggregation as simulated previously.51 Therefore, the effect of the αC regions on the thickness of fibrin fibers and network density is likely purely kinetic without affecting the structure of aggregating protofibrils.
Conclusions
In summary, our results confirmed that after conversion of Fg to fibrin, the αC regions become exposed, undergo partial unfolding and/or conformational elongation and their contour length increases substantially, perhaps enabling them to mediate homomeric intermolecular interactions during early stages of fibrin self-assembly. However, the effects of the αC regions on fibrin formation have a kinetic nature due to reduction of the threshold length of protofibrils, leading to accelerated lateral aggregation. The final structure and macroscopic properties of fibrin clots are affected by kinetics of the earliest stages of fibrin self-assembly. Therefore, these early steps comprise a good potential therapeutic target to modulate the structure and mechanical properties of blood clots.
Supplementary Material
Acknowledgements
This work was supported by NIH grant UO1-HL116330, NSF grant DMR 1505662, scholar award from American Society of Hematology, and by the Program for Competitive Growth at Kazan Federal University. We thank Dr. Artem Zhmurov for the image of Fg structure (Fig.S1).
Notes and references
- 1.Kollman JM, Pandi L, Sawaya MR, Riley M and Doolittle RF, Biochemistry, 2009, 48, 3877–3886. [DOI] [PubMed] [Google Scholar]
- 2.Spraggon G, Everse SJ and Doolittle RF, Nature, 1997, 389, 455–462. [DOI] [PubMed] [Google Scholar]
- 3.Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL and Medved L, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weisel JW and Medved L, Ann. N. Y. Acad. Sci, 2001, 936, 312–327. [DOI] [PubMed] [Google Scholar]
- 5.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W and Weisel JW, J. Biol. Chem, 1993, 268, 13577–13585. [PubMed] [Google Scholar]
- 6.Tsurupa G, Pechik I, Litvinov RI, Hantgan RR, Tjandra N, Weisel JW and Medved L, Biochemistry, 2012, 51, 2526–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorkun OV, Veklich YI, Medved’ LV, Henschen AH and Weisel JW, Biochemistry, 1994, 33, 6986–6997. [DOI] [PubMed] [Google Scholar]
- 8.Litvinov RI, Yakovlev S, Tsurupa G, V Gorkun O, V Medved L and Weisel JW, Biochemistry, 2007, 46, 9133–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson NE, Ding F, Bucay I, O’Brien ET, Gorkun OV, Superfine R, Lord ST, Dokholyan NV and Falvo MR, Biophys. J, 2013, 104, 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ping L, Huang L, Cardinali B, Profumo A, V Gorkun O and Lord ST, Biochemistry, 2011, 50, 9066–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan EA, Mockros LF, Weisel JW and Lorand L, Biophys. J, 1999, 77, 2813–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collet J-P, Moen JL, Veklich YI, V Gorkun O, Lord ST, Montalescot G and Weisel JW, Blood, 2005, 106, 3824–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falvo MR, Millard D, O’Brien ET, Superfine R and Lord ST, J. Thromb. Haemost, 2008, 6, 1991–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helms CC, Ariens RAS, Uitte De Willige S, Standeven KF and Guthold M, Biophys. J, 2012, 102, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouapi KN, Bell JD, Smith KA, Ariens RAS, Philippou H and Maurer MC, Blood, 2016, 127, 2241–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrnes JR, Duval C, Wang Y, Hansen CE, Ahn B, Mooberry MJ, Clark MA, Johnsen JM, Lord ST, Lam WA, Meijers JCM, Ni H, Ariëns RAS and Wolberg AS, Blood, 2015, 126, 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.V Matsuka Y, V Medved L, Migliorini MM and Ingham KC, Biochemistry, 1996, 35, 5810–5816. [DOI] [PubMed] [Google Scholar]
- 18.Tsurupa G, Mahid A, Veklich Y, Weisel JW and Medved L, Biochemistry, 2011, 50, 8028–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsurupa G and Medved L, Biochemistry, 2001, 40, 801–808. [DOI] [PubMed] [Google Scholar]
- 20.Mosher DF, J. Biol. Chem, 1975, 250, 6614–6621. [PubMed] [Google Scholar]
- 21.Tamaki T and Aoki N, Biochim. Biophys. Acta, 1981, 661, 280–286. [DOI] [PubMed] [Google Scholar]
- 22.Sakata Y and Aoki N, J. Clin. Invest, 1980, 65, 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JW, Ruggeri ZM, Kunicki TJ and Cheresh DA, J. Biol. Chem, 1990, 265, 12267–12271. [PubMed] [Google Scholar]
- 24.Suehiro K, Mizuguchi J, Nishiyama K, Iwanaga S, Farrell DH and Ohtaki S, J. Biochem, 2000, 128, 705–710. [DOI] [PubMed] [Google Scholar]
- 25.Litvinov RI, Farrell DH, Weisel JW and Bennett JS, J. Biol. Chem, 2016, 291, 7858–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duval C, Allan P, Connell SDA, Ridger VC, Philippou H and Ariens RAS, Thromb. Haemost, 2014, 111, 842–850. [DOI] [PubMed] [Google Scholar]
- 27.V Pizzo S, Schwartz ML, Hill RL and Mckee PA, J. Biol. Chem, 1972, 247, 636–645. [PubMed] [Google Scholar]
- 28.Raynal B, Cardinali B, Grimbergen J, Profumo A, Lord ST, England P and Rocco M, Thromb. Res, 2013, 132, e48–e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardinali B, Profumo A, Aprile A, Byron O, Morris G, Harding SE, Stafford WF and Rocco M, Arch. Biochem. Biophys, 2010, 493, 157–168. [DOI] [PubMed] [Google Scholar]
- 30.Yermolenko IS, V Gorkun O, Fuhrmann A, Podolnikova NP, Lishko VK, Oshkadyerov SP, Lord ST, Ros R and Ugarova TP, J. Biol. Chem, 2012, 287, 41979–41990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo J, Rafailovich MH, Medved L, Tsurupa G, Kudryk BJ, Liu Y, Galanakis DK, Bohdan J, Liu Y and Galanakis DK, J. Thromb. Haemost, 2010, 8, 2727–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burton R, Tsurupa G, Medved L and Tjandra N, Biochemistry, 2006, 45, 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsurupa G, Tsonev L and Medved L, Biochemistry, 2002, 41, 6449–6459. [DOI] [PubMed] [Google Scholar]
- 34.Doolittle RF, Watt KWK, Cottrell BA, Strong DD and Riley M, Nature, 1979, 280, 464–468. [DOI] [PubMed] [Google Scholar]
- 35.Medved’ LV, Gorkun OV and Privalov PL, FEBS Lett, 1983, 160, 291–295. [DOI] [PubMed] [Google Scholar]
- 36.Erickson HP and Fowler WE, Ann. N. Y. Acad. Sci, 1983, 408, 146–163. [DOI] [PubMed] [Google Scholar]
- 37.Yermolenko IS, Lishko VK, Ugarova TP and Magonov SN, Biomacromolecules, 2011, 12, 370–379. [DOI] [PubMed] [Google Scholar]
- 38.Sheng S, Gao Y, Khromov A, V Somlyo A, Somlyo AP and Shao Z, J. Biol. Chem, 2003, 278, 39892–39896. [DOI] [PubMed] [Google Scholar]
- 39.Kodera N, Yamamoto D, Ishikawa R and Ando T, Nature, 2010, 468, 72–76. [DOI] [PubMed] [Google Scholar]
- 40.Miyagi A, Ando T and Lyubchenko YL, Biochemistry, 2011, 50, 7901–7908. [DOI] [PubMed] [Google Scholar]
- 41.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M and Bustamante C, Nat. Struct. Mol. Biol, 2011, 18, 1394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Protopopova AD, Barinov NA, Zavyalova EG, Kopylov AM, Sergienko VI and Klinov DV, J. Thromb. Haemost, 2015, 13, 570–579. [DOI] [PubMed] [Google Scholar]
- 43.Mosesson MW and Sherry S, Biochemistry, 1966, 5, 2829–2835. [DOI] [PubMed] [Google Scholar]
- 44.Mosesson MW, Finlayson JS, Umfleet RA and Galanakis D, J. Biol. Chem, 1972, 247, 5210–5219. [PubMed] [Google Scholar]
- 45.Klinov D, Dwir B, Kapon E, Borovok N, Molotsky T and Kotlyar A, Nanotechnology, 2007, 18, 225102. [Google Scholar]
- 46.V Klinov D, V Lagutina I, V Prokhorov V, Neretina T, Khil PP, Lebedev YB, Cherny DI, V Demin V and Sverdlov ED, Nucleic Acids Res, 1998, 26, 4603–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorkun OV, Henschen-Edman AH, Ping LF and Lord ST, Biochemistry, 1998, 37, 15434–15441. [DOI] [PubMed] [Google Scholar]
- 48.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A, Nat. Methods, 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Core Team, 2016.
- 50.W. Revelle, 2016.
- 51.Weisel JW and Nagaswami C, Biophys. J, 1992, 7, 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marder VJ, Shulman NR and Carroll WR, J. Biol. Chem, 1969, 244, 2111–2119. [PubMed] [Google Scholar]
- 53.Sherman LA, Mosesson MW and Sherry S, Biochemistry, 1969, 8, 1515–1523. [DOI] [PubMed] [Google Scholar]
- 54.Gron B, Bennick A, Nieuwenhuizen W, Bjornsen S and Brosstad F, Thromb. Res, 1988, 52, 413–424. [DOI] [PubMed] [Google Scholar]
- 55.Mosesson MW, Galanakis DK and Finlayson JS, J. Biol. Chem, 1974, 249, 4656–4664. [PubMed] [Google Scholar]
- 56.Sherman LA, Fletcher AP and Sherry S, J. Lab. Clin. Med, 1969, 73, 574–583. [PubMed] [Google Scholar]
- 57.Holm B, Nilsen DWT and Godal HC, Thromb. Res, 1986, 41, 879–884. [DOI] [PubMed] [Google Scholar]
- 58.Holm B and Godal HC, Thromb. Res, 1984, 35, 279–290. [DOI] [PubMed] [Google Scholar]
- 59.Marder VJ and Shulman NR, J. Biol. Chem, 1969, 244, 2120–2125. [PubMed] [Google Scholar]
- 60.H. M. Princen, H. J. Moshage, J. J. Emeis, H. J. Haard, W. Nieuwenhuizen and S. H. Yap, Thromb. Haemost, 1985, 53, 212–215. [PubMed] [Google Scholar]
- 61.Zhmurov A, Brown AEX, Litvinov RI, Dima RI, Weisel JW and Barsegov V, Structure, 2011, 19, 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.