Abstract
Receptor tyrosine kinases (RTKs) control a wide range of developmental processes, from the first stages of embryogenesis to postnatal growth and neurocognitive development in the adult. A significant share of our knowledge about RTKs comes from genetic screens in model organisms, which provided numerous examples demonstrating how specific cell fates and morphologies are abolished when RTK activation is either abrogated or significantly reduced. Aberrant activation of such pathways has also been recognized in many forms of cancer. More recently, studies of human developmental syndromes established that excessive activation of RTKs and their downstream signaling effectors, most notably the Ras signaling pathway, can also lead to structural and functional defects. Given that both insufficient and excessive pathway activation can lead to abnormalities, mechanistic analysis of developmental RTK signaling must address quantitative questions about its regulation and function. Patterning events controlled by the RTK Torso in the early Drosophila embryo are well-suited for this purpose. This mini review summarizes current state of knowledge about Torso-dependent Ras activation and discusses its potential to serve as a quantitative model for studying the general principles of Ras signaling in development and disease.
Keywords: Drosophila, Torso, Ras, Patterning, Cancer, RASopathies
1. Introduction
Receptor tyrosine kinases (RTKs) are critically involved in controlling a wide range of developmental processes in organisms from planaria to humans (Rink et al., 2011; Rogers et al., 2017; Toyoda et al., 2010). In most of the studied systems, RTKs are activated by locally produced ligands that establish patterns of gene expression, which are in turn converted to specific morphogenetic outcomes. While the overall picture of tissue patterning by transient RTK activation is well established, even the very basic questions about the relevant parameters of signaling transients are poorly understood. For instance, it is not clear what parameter(s), e.g. duration, maximum amplitude, or integrated signal, dictate diverse patterning outcomes. Furthermore, if the critical parameters are identified, what is the range of parameter values that may give normal outcomes? In other words, what are the “necessary and sufficient” signaling conditions required for proper development? We propose that the Torso RTK signaling in the early Drosophila embryo (Furriols and Casanova, 2003) is an excellent model system for quantitative studies aimed at addressing some of the questions proposed above. Torso as an experimental system is attractive because of the simplicity of its anatomy, the possibility of conducting quantitative studies of fixed and live embryos, and its amenability to sophisticated genetic perturbations. In this review, we briefly summarize this signaling pathway, some of the outstanding questions about the mechanistic and functional properties of this pathway, and how this system can be harnessed for applied studies of diseases.
Torso signaling is initiated during the 2nd hour of embryonic development, when nine sequential divisions of the zygotic nucleus establish a layer of nuclei under the common plasma membrane (Foe and Alberts, 1983). After four more divisions, this layer is transformed into an epithelium which begins to morph into the 3D structures of future organs. Torso signaling patterns the anterior and posterior termini of the embryo: these regions give rise to the nonsegmented structures of the future larva. The formation of these structures, which include the most anterior parts of the head and the posterior spiracle, depends on Torso (Casanova and Struhl, 1989; Furriols and Casanova, 2003). This RTK is translated from a uniformly distributed maternal transcript and is activated by Trunk, a diffusible ligand which is processed into its active form only at the embryonic poles (Fig. 1A,B) (Amarnath et al., 2017; Jenni et al., 2015; Johnson et al., 2013; Sprenger and Nüsslein-Volhard, 1992). A combination of a uniformly expressed receptor and locally produced ligand is a common scenario for developmental RTK signaling (Nonomura et al., 2013; Toyoda et al., 2010), and results in localized activation of the highly conserved Ras signaling cascade, leading to the dual phosphorylation and activation of the extracellular signal regulated kinase (ERK) (Fig. 1A,B) (Gabay et al., 1997), an enzyme with numerous intracellular substrates and functions (Futran et al., 2013; Kim et al., 2014).
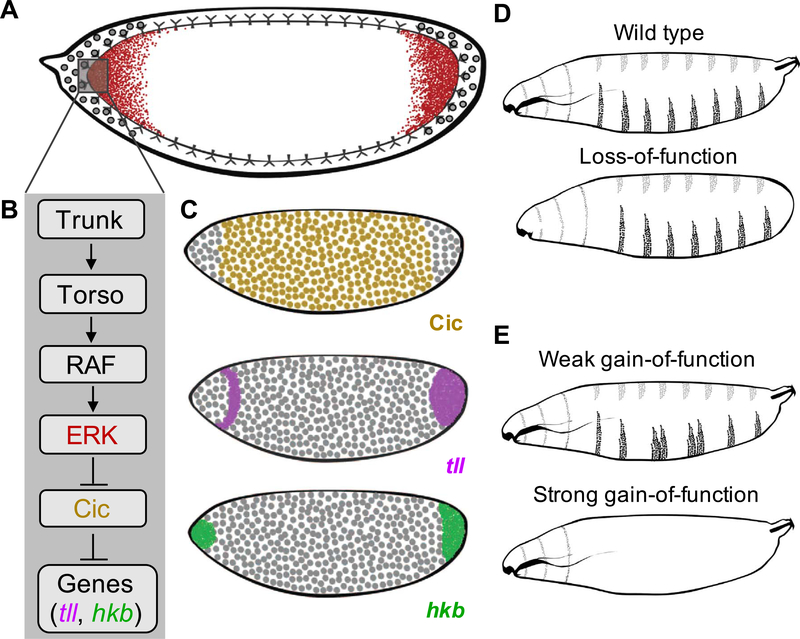
Fig. 1.
Torso-mediated ERK activation in the early Drosophila embryo. (A) A cartoon of the RTK Torso driven ERK activation (red) in the early fly embryo. (B) Schematic of the Torso pathway. (C) Active ERK induces expression of terminal genes such as tailless (magenta) and huckebein (green) by antagonizing Capicua-mediated repression (yellow). (D, E) Schematics representing the larval cuticle phenotype for wild type (D), loss-of-function (LOF) (D), and gain-of-function (GOF) backgrounds (E) in the Torso pathway.
Dually phosphorylated ERK (dpERK) transmits Torso activation through transcriptional repressor Capicua (Cic) (Fig. 1B,C) (Ajuria et al., 2011; Jiménez et al., 2000). Cic is translated from a uniformly distributed transcript and localizes to the blastoderm nuclei. Following Torso activation, ERK-dependent phosphorylation of Cic at the poles first rapidly antagonizes its repressor function and then causes its nuclear export and cytoplasmic degradation (Grimm et al., 2012). As a consequence, Cic represses its targets in the middle of the embryo, but not at the poles, where several genes are de-repressed in response to Torso activation (Jiménez et al., 2000). Among these genes are transcription factors tailless (tll) and huckebein (hkb), which are essential for specifying the terminal structures (Fig. 1C) (Jiménez et al., 2000). In the absence of Torso activation, these genes are not expressed, and the terminal structures are lost (Fig. 1D) (Schupbach and Wieschaus, 1986; Strecker et al., 1989). On the other hand, when Torso signaling is not restricted to the poles the terminal structures are essentially not affected, but the segmented pattern of the larva is disrupted, reflecting loss of Cic-dependent repression in the middle of the embryo (Fig. E) (Klingler et al., 1988; Strecker et al., 1989). Thus, quantitative control of Torso is essential for proper patterning: lack of activation results in the loss of the terminal structures, but ectopic activation causes defects in segmentation.
Patterning of the terminal structures displays many of the key features associated with developmental RTK-signaling: a pulse of RTK activation establishes 2D patterns of gene expression that leads to 3D tissue morphogenesis. Since its discovery more than three decades ago (Nüsslein-Volhard et al., 1987; Schupbach and Wieschaus, 1986), we have developed a comprehensive understanding of the Torso pathway from the genes and molecules involved to the resulting phenotypes, thus providing a useful basis for developing novel quantitative frameworks. Work done in this system continues to discover important mechanisms with both fundamental and applied implications. Below, we cover some of these studies, and discuss how we can continue to learn about and from the system.
2. Some open questions in terminal patterning
Which features of ERK activation at the poles are important for normal terminal patterning? Since ERK signaling in this system works by relief of repression, it appears that the levels of ERK activation would only need to cross some threshold value, after which point signaling dynamics should not affect the patterning outcomes. This scenario is consistent with the results of recent optogenetic studies, which established that prolonging the duration of ERK activation at the poles causes no detectable defects in terminal patterning (Johnson et al., 2017). This robustness is in striking contrast to what is observed in the middle of the embryo, where even low levels of transient ERK activation can be lethal. In fact, defects in segmentation can be caused by levels of signaling that are as low as 10% of ERK activation at the poles, suggesting that the threshold needed for the induction of the terminal fates is also significantly lower that than the wild type signal provided by Torso activation (Goyal et al., 2017a; Johnson et al., 2017). We argue that this design makes the terminal system robust with respect to variations in the strength of Torso signaling.
How many active ERK molecules correspond to the threshold? In thinking about this question, it is important to realize that ERK molecules in vivo can exist in at least four distinct phosphorylation states (Fig. 2A) (Canagarajah et al., 1997; Payne et al., 1991). These four states are generated by the joint actions of MEK1, a kinase which phosphorylates the tyrosine and threonine residues within the activation loop of ERK, and several phosphatases that dephosphorylate these sites. The dually phosphorylated ERK is the most active of these forms, but the other two forms also have partial enzymatic activity. Depending on parameters, such as the amounts and the nature of the ERK phosphatases, the ERK activation network can generate a broad range of the relative amounts of the four states, including the ones where the monophosphorylated states are the most abundant (Iwamoto et al., 2016; Rubinstein et al., 2016). Thus, estimating the number of active ERK molecules at the threshold is a challenging task. As a first step in addressing this challenge, it is important to measure the absolute levels of core components of the ERK activation circuit, including ERK itself and the relevant phosphatases. The phosphatases responsible for ERK dephosphorylation in the embryo are yet to be determined. One plausible candidate is the dual specificity phosphatase MKP3, which can dephosphorylate both sites within the activation loop of ERK. However, knockdown of MKP3 causes only a slight increase in ERK phosphorylation at the poles, suggesting that other phosphatases are involved (Kim et al., 2014).
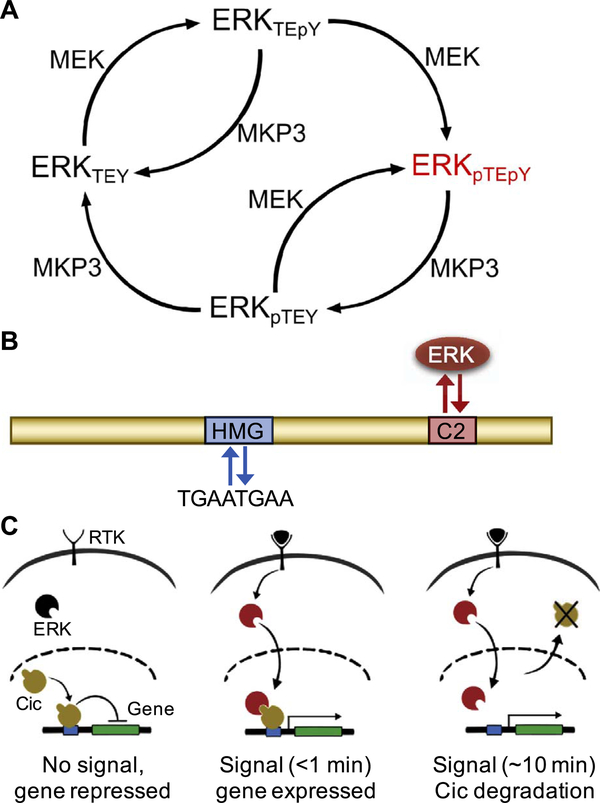
Fig. 2.
Active, dually phosphorylated ERK antagonizes Cic repressor activity. (A) Reaction network depicting the regulation of ERK phosphorylation state by kinase MEK and phosphatase MPK3. Active, dually phosphorylated ERK is depicted in red. (B) Schematic of the Cic protein (yellow) including its major functional domains, high mobility group (HMG) box for DNA binding (blue), and the C2 motif, which functions as a docking site for ERK (red). (C) In response to ligand-induced RTK activation, ERK (black) is phosphorylated; active ERK (red) is translocated to the nucleus, resulting in fast (< 1 min) derepression of the target gene, and cytoplasmic export (~10 min) and subsequent degradation of Cic (yellow).
The next set of questions is related to the transcriptional interpretation of ERK activation. A number of studies focused on the ERK-dependent control of Cic, leading to a detailed understanding of the binding interface of the two proteins (Fig. 2B) (Astigarraga et al., 2007; Futran et al., 2015; Jiménez et al., 2012). Interestingly, this interaction is greatly enhanced when ERK is phosphorylated, which may prevent spurious ERK/Cic binding and interference with the repressor function of Cic in the absence of RTK signaling. While the functional significance of the ERK/Cic interaction is firmly established, how this interaction leads to the relief of transcriptional repression by Cic remains unclear. Live imaging of embryos expressing the fluorescently-tagged Cic established that signaling by Torso results in the nuclear export and cytoplasmic degradation of Cic, suggesting that this can be the mechanism for the ERK-dependent gene derepression (Fig. 2C) (Grimm et al., 2012). On the other hand, studies of the later phase of ERK activation in the fly embryo revealed that relief of transcriptional repression is observed before a significant reduction of Cic levels in the nucleus (Fig. 2C) (Lim et al., 2013). In one of the possible mechanisms that can reconcile these observations, ERK can counteract gene repression by Cic while Cic is still bound to chromatin. In this case, the system would not have to “wait” for Cic to dissociate from the chromatin and would therefore respond as soon as Cic is phosphorylated. This raises the question about the processes leading to the first encounter between active ERK and Cic. We propose that active ERK can first bind to chromatin and then search for Cic that is engaged at the enhancers of its target genes. This possibility is suggested by studies reporting that ERK can bind to DNA, either on its own, or recruited by DNA bound proteins, including ERK substrates (Göke et al., 2013; Hu et al., 2009; Yue et al., 2017).
In addition to Cic, ERK phosphorylates three other transcription factors in the early embryo. The global transcriptional co-repressor Groucho (Gro) is one of these additional substrates (Cinnamon et al., 2008; Helman et al., 2011). Gro plays a critical role in the terminal patterning system. Indeed, loss of Gro phenocopies loss of Cic and is essential for the Cic-dependent repression in the blastoderm (Cinnamon et al., 2008). In contrast to Cic, which is eventually degraded, Gro phosphorylation at the poles can be detected long after the transient pulse of ERK signaling. Furthermore, targeted expression of the phosphomimetic form of Gro, mimicking its constitutive phosphorylation, interferes with the Cic-dependent repression. Based on this observation, it has been proposed that phosphorylation of Gro provides a long-term memory in the interpretation of transient ERK signaling, enabling the expression of the ERK- target genes when the levels of Cic are re-established (Helman et al., 2011).
The other two substrates are the anterior patterning determinants Bicoid (Bcd) (Janody et al., 2000; Ronchi et al., 1993) and Hunchback (Hb) (Kim et al., 2010). Both of these proteins are readily phosphorylated by ERK in vitro and can interfere with the ERK-dependent downregulation of Cic in the embryo (Kim et al., 2010). Specifically, the extent of Cic downregulation at the anterior pole is increased in the absence of Bcd and Hb. This interference has been interpreted as direct competition for active ERK and proposed to fine-tune the expression borders of the ERK-target genes (Kim et al., 2013, 2010). At this point, it is unclear whether Cic, Gro, Bcd, and Hb are the only ERK substrates in the embryo. Based on comparison with other ERK-dependent events, most notably the C. elegans germline development (Arur et al., 2009; Futran et al., 2013), it is likely that other substrates will be identified in the future, leading to a more complete view of ERK signaling in the embryo.
3. A platform for studies of pathogenic mutations
Given the ubiquitous role of the Ras pathway in organismal development, it is not surprising that deregulated Ras signaling has been associated with multiple human diseases (Hanahan and Weinberg, 2011; Rauen, 2013; Shaikh et al., 2000). Somatic mutations in the pathway have long been recognized as key drivers of tumorigenesis (Hanahan and Weinberg, 2011). More recently, autosomal dominant mutations have been discovered in the germline and associated with a large class of human developmental abnormalities, known as the RASopathies (Jindal et al., 2015; Rauen, 2013). These mutations arise mainly during spermatogenesis and provide competitive advantage to developing sperm cells with the de novo mutations (Giannoulatou et al., 2013; Maher et al., 2016). The affected individuals display a spectrum of phenotypes, including congenital heart defects, craniofacial dysmporhisms, stunted growth, and learning disabilities. Remarkably, even though the pathogenic mutations are present in all cells of the organism, defects are observed only in certain tissues and organs (Rauen, 2013). Patterning and morphogenesis apparently proceed normally in some tissues, despite Ras signaling playing a critical role in their development. The origins of this phenotypic specificity have remained unclear.
The relative simplicity of Ras signaling in Drosophila, with a single isoform for all pathway components, and the fact that Drosophila Ras signals almost exclusively through the ERK pathway, makes it an attractive system for the in-depth analysis of mutations from the RASopathies. Our group has recently analyzed a panel of mutations in MEK1 (Goyal et al., 2017a; Jindal et al., 2017a, 2017b) (Dsor1 in Drosophila, henceforth referred to as MEK, since Drosophila has only one homologue of MEK), an enzyme that directly phosphorylates and activates ERK, the terminal enzyme in the Ras/ERK pathway (Fig. 3A) (Seger et al., 1992). Given the fact that MEK is a highly regulated enzyme, pathogenic mutations can affect multiple processes involved in MEK regulation and function (Bromberg-White et al., 2012; Nikolaev et al., 2011; Rodriguez-Viciana et al., 2006). Studies with purified components suggest that mutant MEK variants have partial enzymatic activity, even in the absence of their phosphorylation by RAF, an upstream kinase in the ERK pathway (Fig. 3A) (Jindal et al., 2017a). This constitutive activity is further enhanced when MEK is phosphorylated by Raf, generating a state that is just as active as the phosphorylated wild type protein. Importantly, the activating phosphorylation of the mutant MEK variants proceeds at a faster rate. These studies reveal changes in at least three reactions involved in MEK regulation and function (Jindal et al., 2017a), illustrating the complexity of the effects of the pathogenic mutations, even at the level of a single protein.
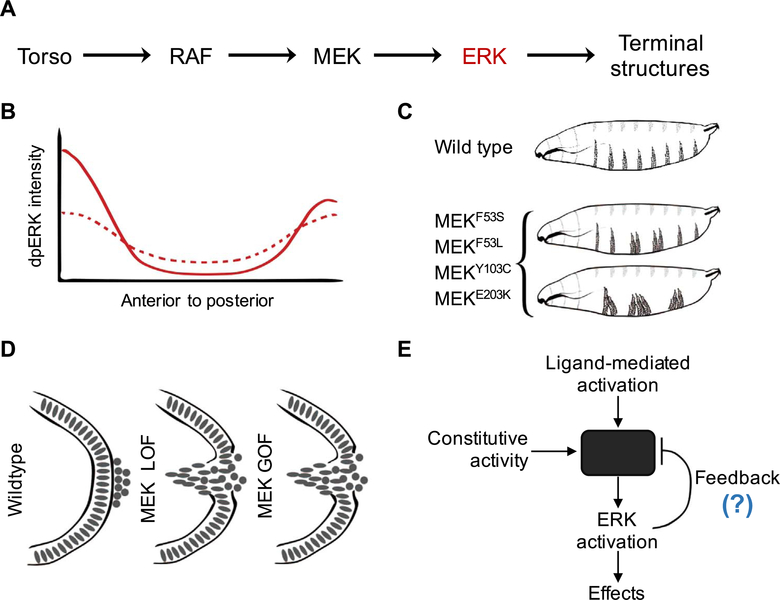
Fig. 3.
Quantitative studies in the early embryo allow identification of divergent effects of pathogenic mutations. (A) Simplified schematic of Torso RTK signaling in the early Drosophila embryo. (B) Schematic representation of the spatial profile of active, dually phosphorylated ERK (dpERK) for wild-type (solid) and pathogenic MEK variants (dotted) in the early embryo. (C) Schematics representing the larval cuticle phenotype for wild type and various gain-of-function (GOF) MEK mutants. (D) Pole-hole phenotype as a readout of the pathway function for MEK loss-of-function (LOF) (via MEK RNAi), and MEK GOF (via GOF mutations F53S, F53L, Y130C, and E203K). (E) Proposed negative-feedback based model to explain the divergent effects of activating mutations on ERK activation
How do these effects play out in vivo, where MEK is subject to other levels of control, including the deactivating phosphatases and additional binding partners? It turned out that the answer to this question depends on cellular context: these mutations cause ectopic pathway activation in cells that are not exposed to extracellular signals and, at the same time, desensitize the pathway to endogenous inputs (Fig. 3B) (Goyal et al., 2017a). These results come from studies in which a targeted gene expression system was used to uniformly express either wild type MEK or mutant variants in the early embryo. Note that the central region of the embryo and the embryonic poles, provide an opportunity to simultaneously study how the same mutation affects the ERK pathway in the basal and activated states. Since all of these mutations cause RAF-independent phosphorylation of ERK in vitro, it was expected that they would cause ectopic activation in the middle of the embryo, which was indeed the case. Surprisingly, the activating mutations resulted in significant reduction of ERK activation at the poles where Torso signaling is normally active, yet, in the presence of the MEK mutations, the normal peak of ERK activity was significantly decreased. Both of these effects are functionally significant: ectopic activation in the middle of the embryo disrupts segmentation (Fig. 3C), while reduced activation at the poles leads to defects in the terminal structures (Fig. 3D).
Thus, activating mutations can have very different effects in individual cells, even during the same stage of development. While the molecular mechanisms underlying this paradoxical effect are yet to be determined, mathematical modeling suggests that it can be explained by a negative feedback loop (Fig. 3E). The activating mutations can induce the expression of a negative regulator of RTK signaling, making the pathway less sensitive to ligand-induced activation at the poles. The importance of the negative feedback mechanisms is becoming increasingly appreciated in studies of human cancers, where negative feedbacks have been proposed to cause the activating effects of pharmacological inhibitors of the Ras signaling (Caunt et al., 2015). Specifically, the presence of activating somatic mutations triggers the expression of negative regulators of RTK signal transduction, such as ERK phosphatases. The expression of these components is lost in response to drug treatment, causing paradoxical increase of pathway activation after an initial decrease following drug administration (Caunt et al., 2015; Friday et al., 2008). We argue that the inhibitory effects of the activating mutations from the RASopathies is another manifestation of the same effect.
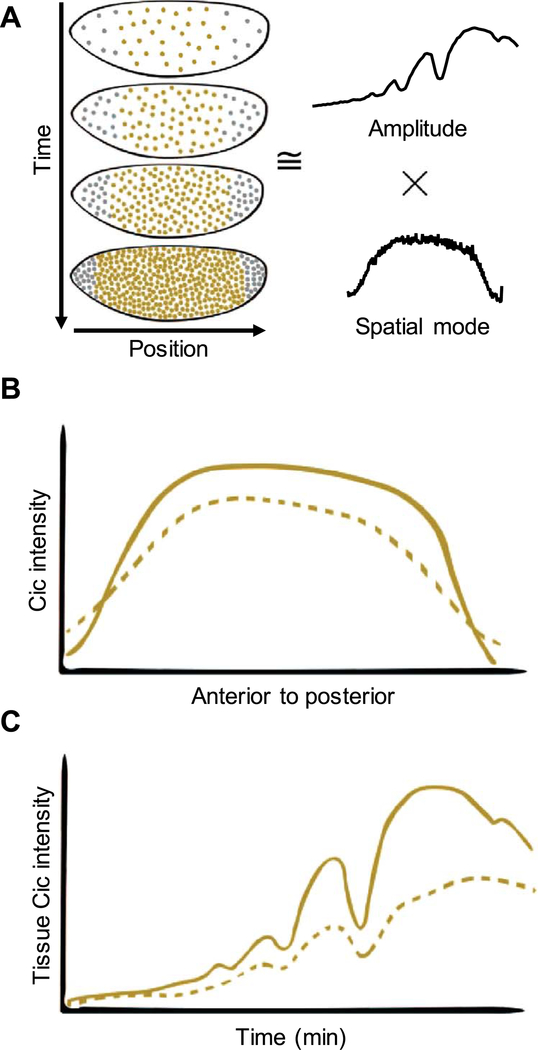
In addition to the larval cuticle and the spatial pattern of ERK activation in fixed embryos, studies of the activating mutations can use the spatiotemporal pattern of the nuclear levels of Cic, the main sensor of ERK signaling in the embryo. We found that this pattern of Cic can be accurately represented by a product of a single function of the anterior-to-posterior distance along the embryo and a time-dependent amplitude (Fig. 4A) (Goyal et al., 2017b). The spatial component reflects the joint effects of the spatially uniform synthesis of Cic and its downregulation in response to Torso signaling at the poles (Fig. 4B). The temporal component reflects the dynamics of nuclear cleavages (Fig. 4C). The quantitative characteristics of both of these functions provide another set of quantifiable readouts for the studies of the activating mutations (Fig. 4B,C). The main advantage of this approach is that it characterizes signaling dynamics in live embryos. Thus, the experimental approaches established by the fundamental studies of the terminal system can be used for designing sensitive assays for the mechanistic analysis of pathogenic mutations. Note that while functional analysis of mutations from the RASopathies is pursued in multiple experimental models, the blastoderm stage embryo offers unrivalled opportunities for statistical comparison of multiple sequence variants.
Fig. 4.
Capicua dynamics as a functional assay to analyze the Ras pathway mutations. (A) Low dimensional approximation of Cic dynamics in terms of temporal amplitude and primary spatial mode. (B) Schematic representation of the spatial profiles of Cic intensity for wild type (solid) and MEK mutants (dotted) across the embryo length. (C) Schematic representation of the dynamics of the Cic amplitudes for wild type (solid) and MEK mutants (dotted). Each visible peak corresponds to mitosis in the respective nuclear cycle.
4. Concluding remarks
Studies of the Torso pathway started as a part of a comprehensive program for understanding genetic control of Drosophila embryogenesis (Schupbach and Wieschaus, 1986). At this point, the key players involved in the Ras-dependent control of the terminal structures have been identified, but the system continues to provide fundamental insights into the mechanisms of Ras signaling and serves as a convenient testing ground for investigating the effects of pathogenic mutations. Using the rapidly improving mass-spectrometry techniques (Presler et al., 2017; Wühr et al., 2015; Yang et al., 2016), it is now possible to identify all of the binding partners of the core components of the terminal signaling cascade, including ERK and Cic. Furthermore, the recently developed optogenetic techniques (Johnson et al., 2017) enable the investigation of the dynamic changes of these interactomes at different levels and at different durations of Ras signaling. These studies may shed light on the molecular mechanisms by which Ras signaling relieves transcriptional repression by Cic, a process that has been documented in human diseases (Forés et al., 2017; Okimoto et al., 2016; Tan et al., 2018; Tanaka et al., 2017) and can provide a new target for drug development in multiple therapeutic areas, including cancer.
Another class of systems-level approaches to Torso signaling relies on the signal-dependent changes of Cic binding to chromatin. A recently study used this approach to identify a new mechanism for modulation the transcriptional repression by Cic (Papagianni et al., 2018). Specifically, the genomic targets of Cic in the blastoderm could be assigned to two broad classes based on the strength of Cic binding sites in their regulatory DNA regions. The regulatory regions with the affinity sites, such as the enhancers of tll and hkb, readily recruit Cic on their own and are expressed only in cells that receive Ras signaling. On the other hand, Cic recruitment to the regulatory regions with the low affinity sites requires a co-factor, which must be present in the nucleus and have binding sites adjacent to those of Cic. This co-factor turned out to be Dorsal (Dl), a NF-kb transcription factor that plays a cardinal role in the dorsoventral patterning of the embryo. The nuclear import of Dl is possible only in the ventral part of the blastoderm. As a consequence, Cic is recruited to the regulatory regions with weak sites only in nuclei with significant levels of Dl (Papagianni et al., 2018). This results in the ventral repression of several important regulators of dorsoventral patterning, genes such as zen and dpp. This elegant mechanism allows one repressor to simultaneously act downstream of two different signaling pathways. In the future, it will be important to investigate whether Ras-independent mechanisms are used in other developmental processes that rely on the modulation of gene repression by Cic, both in Drosophila and in other organisms.
Finally, when it comes to using the terminal system as platform for studies of pathogenic mutations, the next challenge is to analyze their effects in heterozygous conditions. For instance, the effects of mutations from the RASopathies have been analyzed in conditions where the levels of the activating variants of MEK were at least 10-fold higher than the levels of the wild type protein (Goyal et al., 2017a), which is very different from the 1:1 ratio in individuals with the RASopathies. In the future, the heterozygous backgrounds can be engineered using the rapidly developing techniques for genome editing (Gratz et al., 2013; Port et al., 2014; Ren et al., 2013). This should enable rigorous tests of several of the proposed molecular mechanisms for the effects of these mutations. For instance, the phosphorylation independent enzymatic activity of mutant MEK variants may give rise to ectopic ERK signaling only when MEK is overexpressed, but not under heterozygous conditions, where it can be counteracted by the deactivating phosphatases. When combined with the increasingly quantitative assays, including a system for parallel live imaging of multiple embryos, these studies should allow us to study developmental Ras signaling at an unprecedented resolution and understand how it is affected by a wide range of genetic and pharmacological perturbations.
Acknowledgements
We thank members of the Shvartsman and Schüpbach laboratories for comments and suggestions. We also thank Granton Jindal for providing critical feedback. This work was supported by NIH grants R01GM086537 (Y.G. and S.Y.S.) and R01GM077620 (T.S.). Y.G. would also like to thank the Schmidt Science Fellows program, in partnership with the Rhodes Trust for their support.
References
- Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, Andreu MJ, Helman A, González-crespo S, Paroush Z, Courey AJ, Jiménez G, 2011. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 924, 915–924. 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarnath S, Stevens LM, Stein DS, 2017. Reconstitution of Torso signaling in cultured cells suggests a role for both Trunk and Torso-like in receptor activation. Development 144, 677–686. 10.1242/dev.146076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T, 2009. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl. Acad. Sci. USA 106, 4776–4781. 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Grossman R, Díaz-Delfín J, Caelles C, Paroush Z, Jiménez G, 2007. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 26, 668–677. 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-White JL, Andersen NJ, Duesbery NS, 2012. MEK genomics in development and disease. Brief. Funct. Genom 11, 300–310. 10.1093/bfgp/els022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ, 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90, 859–869. [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G, 1989. Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev. 3, 2025–2038. [DOI] [PubMed] [Google Scholar]
- Caunt CJ, Sale MJ, Smith PD, Cook SJ, 2015. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat. Rev. Cancer 15, 577–592. 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- Cinnamon E, Helman A, Ben-Haroush Schyr R, Orian A, Jimenez G, Paroush Z, 2008. Multiple RTK pathways downregulate Groucho-mediated repression in Drosophila embryogenesis. Development 135, 829–837. 10.1242/dev.015206. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM, 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci 61, 31–70. [DOI] [PubMed] [Google Scholar]
- Forés M, Simón-Carrasco L, Ajuria L, Samper N, González-Crespo S, Drosten M, Barbacid M, Jiménez G, 2017. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 13, e1006622 10.1371/journal.pgen.1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, Adjei AA, 2008. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 68, 6145–6153. 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- Furriols M, Casanova J, 2003. In and out of Torso RTK signalling. EMBO J. 22, 1947–1952. 10.1093/emboj/cdg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futran AS, Kyin S, Shvartsman SY, Link AJ, 2015. Mapping the binding interface of ERK and transcriptional repressor Capicua using photocrosslinking. Proc. Natl. Acad. Sci. USA 112, 8590–8595. 10.1073/pnas.1501373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futran AS, Link AJ, Seger R, Shvartsman SY, 2013. ERK as a model for systems biology of enzyme kinetics in cells. Curr. Biol 23, R972–R979. 10.1016/j.cub.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ, 1997. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124, 3535–3541. [DOI] [PubMed] [Google Scholar]
- Giannoulatou E, McVean G, Taylor IB, McGowan SJ, Maher GJ, Iqbal Z, Pfeifer SP, Turner I, Burkitt Wright EMM, Shorto J, Itani A, Turner K, Gregory L, Buck D, Rajpert-De Meyts E, Looijenga LHJ, Kerr B, Wilkie AOM, Goriely A, 2013. Contributions of intrinsic mutation rate and selfish selection to levels of de novo HRAS mutations in the paternal germline. Proc. Natl. Acad. Sci. USA 110, 20152–20157. 10.1073/pnas.1311381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Chan Y-S, Yan J, Vingron M, Ng H-H, 2013. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol. Cell 50, 844–855. 10.1016/j.molcel.2013.04.030. [DOI] [PubMed] [Google Scholar]
- Goyal Y, Jindal GA, Pelliccia JL, Yamaya K, Yeung E, Futran AS, Burdine RD, Schüpbach T, Shvartsman SY, 2017a. Divergent effects of intrinsically active MEK variants on developmental Ras signaling. Nat. Genet 10.1038/ng.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal Y, Levario TJ, Mattingly HH, Holmes S, Shvartsman SY, Lu H, 2017b. Parallel imaging of Drosophila embryos for quantitative analysis of genetic perturbations of the Ras pathway. Dis. Model. Mech 10, 923–929. 10.1242/dmm.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM, 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm O, Sanchez Zini V, Kim Y, Casanova J, Shvartsman SY, Wieschaus E, 2012. Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139, 3962–3968. 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA, 2011. Hallmarks of cancer: the next generation. Cell 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Helman A, Cinnamon E, Mezuman S, Hayouka Z, Von Ohlen T, Orian A, Jiménez G, Paroush Z, 2011. Phosphorylation of groucho mediates RTK feedback inhibition and prolonged pathway target gene expression. Curr. Biol 21, 1102–1110. 10.1016/J.CUB.2011.05.043. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho H, Woodard C, Wang H, Jeong J-S, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H, 2009. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139, 610–622. 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto N, DAlessandro LA, Depner S, Hahn B, Kramer BA, Lucarelli P, Vlasov A, Stepath M, Bohm ME, Deharde D, Damm G, Seehofer D, Lehmann WD, Klingmuller U, Schilling M, 2016. Context-specific flow through the MEK/ERK module produces cell- and ligand-specific patterns of ERK single and double phosphorylation. Sci. Signal 9 10.1126/scisignal.aab1967, (ra13–ra13). [DOI] [PubMed] [Google Scholar]
- Janody F, Sturny R, Catala F, Desplan C, Dostatni N, 2000. Phosphorylation of bicoid on MAP-kinase sites: contribution to its interaction with the torso pathway. Development 127, 279–289. [DOI] [PubMed] [Google Scholar]
- Jenni S, Goyal Y, von Grotthuss M, Shvartsman SY, Klein DE, 2015. Structural basis of neurohormone perception by the receptor tyrosine kinase torso. Mol. Cell 60, 941–952. 10.1016/j.molcel.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Jiménez G, Guichet A, Ephrussi A, Casanova J, 2000. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 14, 224–231. 10.1101/GAD.14.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Shvartsman SY, Paroush Z, 2012. The Capicua repressor–a general sensor of RTK signaling in development and disease. J. Cell Sci 125, 1383–1391. 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Burdine RD, Rauen KA, Shvartsman SY, 2015. RASopathies: unraveling mechanisms with animal models. Dis. Model. Mech 8, 769–782. 10.1242/dmm.020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Humphreys JM, Yeung E, Tian K, Patterson VL, He H, Burdine RD, Shvartsman SY, Goldsmith EJ, 2017a. How activating mutations affect MEK1 regulation and function. J. Biol. Chem 292, 18814–18820. 10.1074/jbc.C117.806067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Yamaya K, Futran AS, Kountouridis I, Balgobin CA, Schüpbach T, Burdine RD, Shvartsman SY, 2017b. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc. Natl. Acad. Sci. USA 114, 510–515. 10.1073/pnas.1615651114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, Goyal Y, Pannucci NL, Schüpbach T, Shvartsman SY, Toettcher JE, 2017. The spatiotemporal limits of developmental Erk signaling. Dev. Cell 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TK, Crossman T, Foote KA, Henstridge MA, Saligari MJ, Forbes Beadle L, Herr A, Whisstock JC, Warr CG, 2013. Torso-like functions independently of Torso to regulate Drosophila growth and developmental timing. Proc. Natl. Acad. Sci. USA 110, 14688–14692. 10.1073/pnas.1309780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Coppey M, Grossman R, Ajuria L, Jiménez G, Paroush Z, Shvartsman SY, 2010. MAPK substrate competition integrates patterning signals in the Drosophila embryo. Curr. Biol 20, 446–451. 10.1016/j.cub.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Iagovitina A, Ishihara K, Fitzgerald KM, Deplancke B, Papatsenko D, Shvartsman SY, 2013. Context-dependent transcriptional interpretation of mitogen activated protein kinase signaling in the Drosophila embryo. Chaos 23, 25105 10.1063/1.4808157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Paroush Z, Nairz K, Hafen E, Jimenez G, Shvartsman SY, 2014. Substrate-dependent control of MAPK phosphorylation in vivo. Mol. Syst. Biol 7, 467 10.1038/msb.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler M, Erdélyi M, Szabad J, Nüsslein-Volhard C, 1988. Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature 335, 275–277. 10.1038/335275a0. [DOI] [PubMed] [Google Scholar]
- Lim B, Samper N, Lu H, Rushlow C, Jiménez G, Shvartsman SY, 2013. Kinetics of gene derepression by ERK signaling. Proc. Natl. Acad. Sci. USA 110, 10330–10335. 10.1073/pnas.1303635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher GJ, McGowan SJ, Giannoulatou E, Verrill C, Goriely A, Wilkie AOM, 2016. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc. Natl. Acad. Sci. USA 113, 2454–2459. 10.1073/pnas.1521325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, Michielin O, Muehlethaler K, Speiser D, Beckmann JS, Xenarios I, Halazonetis TD, Jongeneel CV, Stevenson BJ, Antonarakis SE, 2011. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet 44, 133–139. 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Yamaguchi Y, Hamachi M, Koike M, Uchiyama Y, Nakazato K, Mochizuki A, Sakaue-Sawano A, Miyawaki A, Yoshida H, Kuida K, Miura M, 2013. Local apoptosis modulates early mammalian brain development through the elimination of morphogen-producing cells. Dev. Cell 27, 621–634. 10.1016/j.devcel.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Frohnhöfer HG, Lehmann R, 1987. Determination of anteroposterior polarity in Drosophila. Science 238, 1675–1681. 10.1126/SCIENCE.3686007. [DOI] [PubMed] [Google Scholar]
- Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, Asthana S, Hrustanovic G, Flanagan J, Tulpule A, Blakely CM, Haringsma HJ, Simmons AD, Gowen K, Suh J, Miller VA, Ali S, Schuler M, Bivona TG, 2016. Inactivation of Capicua drives cancer metastasis. Nat. Genet 49, 87–96. 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni A, Forés M, Shao W, He S, Koenecke N, Andreu MJ, Samper N, Paroush Z, González-Crespo S, Zeitlinger J, Jiménez G, 2018. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proc. Natl. Acad. Sci. USA 115, 1807–1812. 10.1073/pnas.1713930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DM, Rossomando AJ, Martino P, Erickson AK, Her JH, Shabanowitz J, Hunt DF, Weber MJ, Sturgill TW, 1991. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase). EMBO J. 10, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen H-M, Lee T, Bullock SL, 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111, E2967–E2976. 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presler M, Van Itallie E, Klein AM, Kunz R, Coughlin ML, Peshkin L, Gygi SP, Wühr M, Kirschner MW, 2017. Proteomics of phosphorylation and protein dynamics during fertilization and meiotic exit in the Xenopus egg. Proc. Natl. Acad. Sci. USA, 201709207. 10.1073/pnas.1709207114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA, 2013. The RASopathies. Annu. Rev. Genom. Hum. Genet 14, 355–369. 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu L-P, Yang Z, Mao D, Sun L, Wu Q, Ji J-Y, Xi J, Mohr SE, Xu J, Perrimon N, Ni J-Q, 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110, 19012–19017. 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink JC, Vu HT-K, Sánchez Alvarado A, 2011. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780. 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA, 2006. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311, 1287–1290. 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- Rogers WA, Goyal Y, Yamaya K, Shvartsman SY, Levine MS, 2017. Uncoupling neurogenic gene networks in the Drosophila embryo. Genes Dev. 31, 634–638. 10.1101/gad.297150.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi E, Treisman J, Dostatni N, Struhl G, Desplan C, 1993. Down-regulation of the Drosophila morphogen bicoid by the torso receptor-mediated signal transduction cascade. Cell 74, 347–355. [DOI] [PubMed] [Google Scholar]
- Rubinstein BY, Mattingly HH, Berezhkovskii AM, Shvartsman SY, 2016. Long-term dynamics of multisite phosphorylation. Mol. Biol. Cell 27, 2331–2340. 10.1091/mbc.E16-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E, 1986. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev. Biol 113, 443–448. 10.1016/0012-1606(86)90179-X. [DOI] [PubMed] [Google Scholar]
- Seger R, Ahn NG, Posada J, Munar ES, Jensen AM, Cooper JA, Cobb MH, Krebs EG, 1992. Purification and characterization of mitogen-activated protein kinase activator(s) from epidermal growth factor-stimulated A431 cells. J. Biol. Chem 267, 14373–14381. [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS, 2000. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet 9, 489–501. [DOI] [PubMed] [Google Scholar]
- Sprenger F, Nüsslein-Volhard C, 1992. Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell 71, 987–1001. [DOI] [PubMed] [Google Scholar]
- Strecker TR, Halsell SR, Fisher WW, Lipshitz HD, 1989. Reciprocal effects of hyper- and hypoactivity mutations in the Drosophila pattern gene torso. Science 243, 1062–1066. 10.1126/science.2922596. [DOI] [PubMed] [Google Scholar]
- Tan Q, Brunetti L, Rousseaux MWC, Lu H-C, Wan Y-W, Revelli J-P, Liu Z, Goodell MA, Zoghbi HY, 2018. Loss of Capicua alters early T cell development and predisposes mice to T cell lymphoblastic leukemia/lymphoma. Proc. Natl. Acad. Sci. USA 115, E1511–E1519. 10.1073/PNAS.1716452115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Yoshimoto T, Nakamura T, 2017. A double-edged sword: the world according to Capicua in cancer. Cancer Sci. 108, 2319–2325. 10.1111/cas.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda R, Assimacopoulos S, Wilcoxon J, Taylor A, Feldman P, Suzuki-Hirano A, Shimogori T, Grove EA, 2010. FGF8 acts as a classic diffusible morphogen to pattern the neocortex. Development 137, 3439–3448. 10.1242/dev.055392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Güttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, Kirschner MW, Gygi SP, 2015. The Nuclear Proteome of a Vertebrate. Curr. Biol 25, 2663–2671. 10.1016/j.cub.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Paul S, Trieu KG, Dent LG, Froldi F, Forés M, Webster K, Siegfried KR, Kondo S, Harvey K, Cheng L, Jiménez G, Shvartsman SY, Veraksa A, 2016. Minibrain and Wings apart control organ growth and tissue patterning through down-regulation of Capicua. Proc. Natl. Acad. Sci. USA 113, 10583–10588. 10.1073/pnas.1609417113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Lai F, Beckedorff F, Zhang A, Pastori C, Shiekhattar R, 2017. Integrator orchestrates RAS/ERK1/2 signaling transcriptional programs. Genes Dev. 31, 1809–1820. 10.1101/gad.301697.117. [DOI] [PMC free article] [PubMed] [Google Scholar]