Abstract
The thirteen nuclear cleavages that give rise to the Drosophila blastoderm are some of the fastest known cell cycles [1]. Surprisingly, the fertilized egg is provided with at most one-third of the dNTPs needed to complete the thirteen rounds of DNA replication [2]. The rest must be synthesized by the embryo, concurrently with cleavage divisions. What is the reason for the limited supply of DNA building blocks? We propose that frugal control of dNTP synthesis contributes to the well-characterized deceleration of the cleavage cycles and is needed for robust accumulation of zygotic gene products. In support of this model, we demonstrate that when the levels of dNTPs are abnormally high, nuclear cleavages fail to sufficiently decelerate, the levels of zygotic transcription are dramatically reduced, and the embryo catastrophically fails early in gastrulation. Our work reveals a direct connection between metabolism, cell cycle, and zygotic transcription.
eTOC Blurb
Djabrayan et al demonstrate a direct connection between dNTP metabolism, cell cycle, and zygotic transcription at the Mid Blastula Transition in the Drosophila embryo. Dynamic restriction of dNTP levels is involved in slowing down of nuclear divisions in the syncytial blastoderm, robust accumulation of zygotic gene products, and morphogenesis.
Results and Discussion
The developing organism starts to gain its independence from maternal controls from the very first steps of embryogenesis. Of these steps, the most extensively studied is the so-called maternal-to-zygotic transition (MZT), when maternal mRNAs are degraded and zygotic gene expression is initiated on the genome scale [3]. An even earlier step towards independence occurs during the cleavage divisions leading to the MZT. This step is associated with a shift from using maternally deposited to newly synthesized deoxyribonucleotides (dNTPs), the building blocks for DNA polymerization [4]. Measurements of dNTP concentrations in fruit fly, frog, and sea urchin embryos reveal that maternal stores of dNTPs are sufficient for making only a fraction of the genomes that must be present at MZT, which implies that these embryos must synthesize dNTPs concurrently with DNA polymerization [2, 5, 6]. The rate limiting reaction in this process is catalyzed by Ribonucleotide Reductase (RNR), a highly conserved enzyme that is subject to multiple levels of allosteric regulation, including inhibition by dATP, one of the four end products of the dNTP synthesis pathway [7]. The essential role of de novo dNTP synthesis in early embryogenesis is demonstrated by the fact that the pre-MZT cleavages are arrested by injection of hydroxyurea, a chemical which interferes with the generation of tyrosine free radicals during the reduction of ribonucleotides by RNR [2].
Dynamic control of dNTP synthesis is especially impressive in Drosophila, where thirteen rounds of DNA replication during the pre-MZT cleavages generate ~6000 nuclei in only 2 hours [1]. The Drosophila egg is maternally supplied with one third of the dNTPs needed to form these nuclei; the rest of dNTPs are synthesized de novo, controlled by maternally deposited RNR [2]. Initially, RNR is allosterically inhibited by high levels of dATP, but as these levels are lowered by the first rounds of DNA replication, the inhibition is lifted and the enzyme begins to work, supplying the embryo with newly synthesized dNTPs [2]. Consistent with time-resolved measurements of dNTP concentrations in early embryos, this model of dNTP regulation predicts that the levels of all four dNTPs become limiting during the last three rounds of DNA replication prior to morphogenesis [2]. The model also readily explains why cleavages are halted when embryos are injected with either hydroxyurea or dATP. In both cases, the embryo runs out of dNTPs and fails to synthesize the genomes needed for proceeding to the next stage of development.
Interestingly, abnormally high dNTP concentrations caused by expression of feedback insensitive RNR also result in high embryonic lethality (Figure 1A, B, [2]). This suggests that the limited supply of dNTPs during the cleavage cycles is essential for normal embryogenesis. What is the reason for limiting the rate of dNTP synthesis, especially when the cleavage cycles are so fast? To address this question, we used live imaging to examine the early stages of development in embryos with abnormally high levels of dNTPs. We established that these embryos go through the correct number of cleavages without significant nuclear fallout (Figure S1A). This indicates that high dNTP concentrations do not cause significant levels of DNA damage, at least at this stage of embryogenesis [8]. Indeed, examination of cell death reporters did not reveal any signs of aberrant cell death until much later stages of development (Figure S1B–F). Moreover, these embryos cellularized with unaltered kinetics (data not shown) and proceeded through the first stages of gastrulation, including the formation of the cephalic furrow and invagination of the presumptive mesoderm tissue (Video S1A). In addition, increased levels of dNTPs do not interfere with axial patterning since the expression domains of key regulators of the anteroposterior, dorsoventral, and terminal patterning systems remained unchanged (Figure 1C,D and Figure S1G–J).
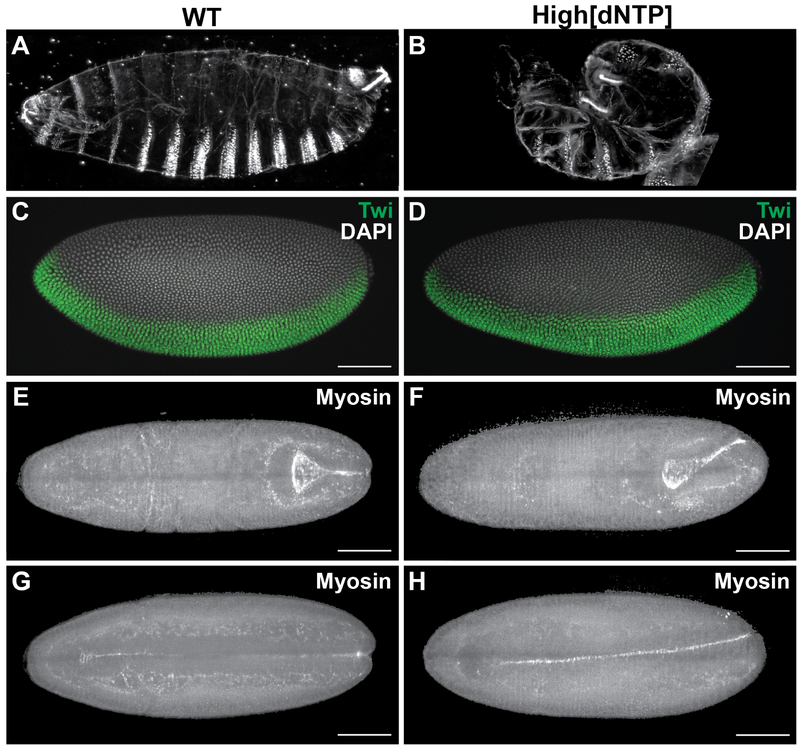
Figure 1: Constitutively high dNTP concentrations cause morphogenetic defects.
(A, B) Cuticle preparations of a wild type embryo (A) and an embryo with high dNTP concentrations (B). (C, D) Cellular blastoderm staged embryos stained with anti-Twist antibody (green). Both wild type embryos (G) and those with high dNTP concentrations (H) show the normal expression pattern. (E-H) Dorsal and ventral views of wild type embryo (E, G) and an embryo with high dNTP concentrations (F, H) expressing Sqh::mCherry to visualize myosin and midline formation, which is twisted in the embryo with high dNTP concentrations (F, H). Scale bars represent 30 m. See also Figure S1. See also Video S1
Thus, embryos with high levels of dNTPs are cellularized and patterned normally, and succeed in completing mesoderm invagination. Yet, all of these embryos fail at the stage of germband extension. During germband extension in wild type embryos, the ventral midline, which is formed by mesoderm invagination, maintains its position at the midsagittal plane (Figure 1E,G, Video S1B). In contrast, the ventral midline in embryos with high levels of dNTP veers off course (Figure 1F,H, Video S1C). This deviation from the normal trajectory disrupts future steps of morphogenesis. Most critically, the extended germband cannot retract, which explains why embryos developing with high levels of dNTPs acquire a characteristic twisted shape (Figure 1B). Note that similar morphogenetic defects are observed in a number of patterning mutants [9–11], but since the spatial expression domains of the key patterning genes appear unchanged, a different mechanism must be at play. We hypothesized that morphogenetic defects can arise as a consequence of the quantitative changes in the transcription of zygotic genes within their normal expression domains.
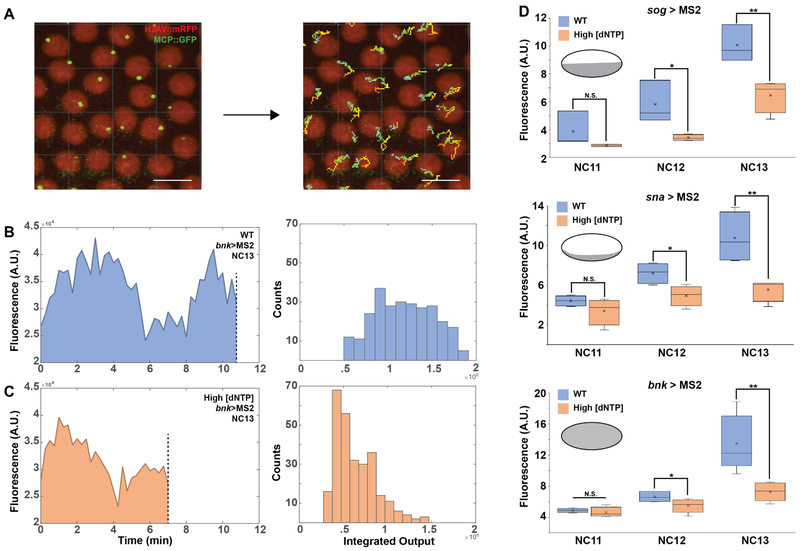
To test whether increased dNTP concentrations can lead to quantitative changes in zygotic transcript accumulation, we quantified MS2 reporter activity during the last three nuclear cleavage cycles for three of the early transcribed genes sog, sna and bnk [12, 13]. These genes are also essential for proper morphogenesis [9–11]. In these experiments, we took advantage of the MS2-MCP reporter system which inserts 24 repeats of the MS2 sequence, which upon transcription forms a hairpin secondary structure that is capped by the MCP::GFP protein [14]. In this way, transcriptional activity can be seen as the fluorescence intensity over time. We segmented and tracked the MS2-MCP::GFP signal in individual nuclei during the interphases of the cleavage cycles (Figure 2A, Video S2A, B). We defined the total transcriptional output for each nucleus during the corresponding interphase by calculating the area under the curve for the time course of fluorescence intensity (Figure 2B, C left, Figure S2A–C). By tracking multiple nuclei, we obtained the empirical distribution functions for the transcriptional output for each of the three genes and each of the analyzed cell cycles (Figure 2B, C right). The medians of these distributions from multiple embryos allowed us to compare the transcriptional outputs between wild type embryos and embryos developing at high concentrations of dNTPs. For all three genes we observed a significant reduction in transcriptional output during nuclear cycles (NC) 12 and 13 (Figure 2D).
Figure 2: Decreased transcriptional output in embryos with high dNTP concentrations.
(A) MS2 signal is segmented from live imaging datasets and converted into time traces. Scale bar represents 10 m. (B left, C left) Calculation of the integrated output for an individual nucleus during a given interphase. Signal spans the length of active transcription, beginning when the MS2 signal becomes visible and ends at mitosis, when transcription is terminated (dotted line). (B right, C right) Distribution functions of the outputs for multiple nuclei in a sample embryo. (D) Box plots representing median transcriptional output from wild type embryos and embryos with high dNTP concentrations for three early expressed genes: sog>MS2 (N = 4), sna>MS2 (N = 5) and bnk>MS2 (N = 5). X represents mean and bar represents median. Statistical test, t-Test. N.S. Not significant, * p < 0.05, ** p < 0.01. Insets show expression domain of respective regulatory regions. See also Figure S2. See also Video S2.
What can cause the observed reduction in transcriptional output? Earlier studies suggested that the accumulation of zygotic gene products in Drosophila requires progressive lengthening of the interphases during the nuclear cleavage divisions, to give sufficient time for transcript initiation and elongation [15–17]. Furthermore, studies in Xenopus which tracked dNTP levels at the MBT suggest that availability dNTPs tunes the cell cycle [6]. Additionally, studies in bacteria and yeast established that increased dNTP pools can accelerate progression through S-phase. We thus hypothesized that a similar mechanism might affect the duration of interphases during the cleavage divisions and, consequently, lead to the overall reduction of accumulated zygotic transcripts [18, 19]. Consistent with this idea, we found that when the overall transcriptional output was divided by the period of active transcription, the outputs of embryos with high dNTP concentrations became statistically indistinguishable from control embryos (Figure S2D–F). This result indicates that the difference in S-phase length, as opposed to general effects on transcriptional activity, accounts for the difference in total transcriptional output.
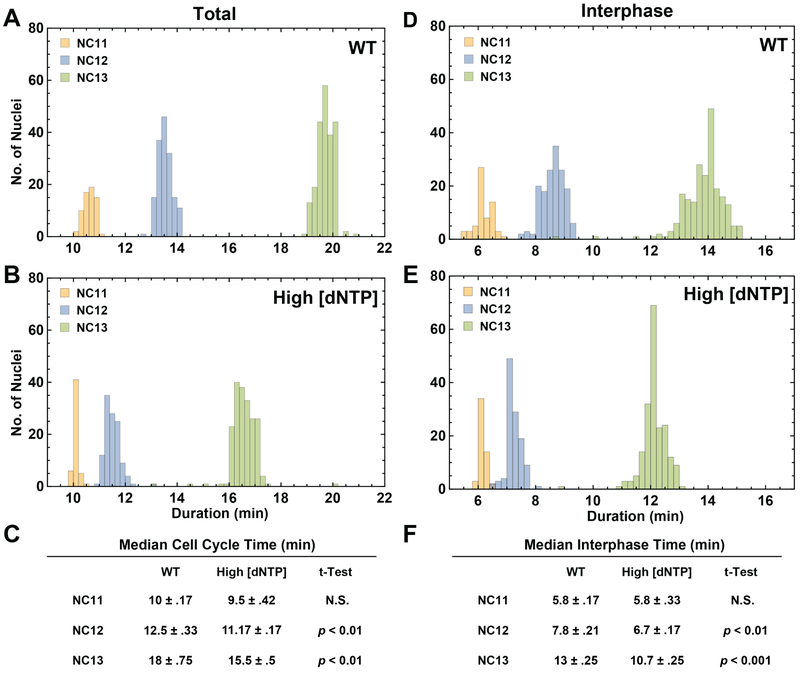
To evaluate our hypothesis more directly, we quantified the amount of time spent in a given phase of the nuclear cleavage cycles in wild type embryos and in embryos with high dNTP concentrations (see ‘Division Phase Labeling‘ section of Methods). Our analysis revealed a clear reduction in the total cell cycle times for nuclear cleavage cycles 12 and 13 (Figure 3A–C). Moreover, we established that almost all of the difference in total cell cycle time can be accounted for by a shorter interphase (Figure 3D–F). The durations of the other phases of the cell cycle remained unchanged at high levels of dNTPs (Figure S3A–C). These results are consistent with the observed effects on the overall transcriptional output and suggest that high levels of dNTPs can prevent the prolongation of S phase in the pre-gastrulation Drosophila embryo. Furthermore, we propose that this effect gives rise to the quantitative reduction of zygotic gene expression and may explain why the last nuclear cleavages proceed at limiting concentrations of dNTPs.
Figure 3: High dNTP concentrations interfere with cell cycle slowing.
(A, B) Histograms representing the total cell cycle times for nuclei tracked in a representative wild type embryo (A) and an embryo with high dNTP concentrations (B). Bins contain the number of nuclei for which the total cell cycle lasted a given amount of time in NC11, NC12 and NC13. (C) Median cell cycle times with interquartile range for wild type embryos (N = 5) and embryos with high dNTP concentrations (N = 5) whose nuclei were tracked from NC11 through NC13. (D, E) Histograms representing the interphase times for nuclei tracked in a representative wild type embryo (D) and an embryo with high dNTP concentrations (E). Bins contain the number of nuclei for which interphase lasted a given amount of time in NC11, NC12 and NC13. (F) Median interphase times with interquartile range for wild type embryos (N = 5) and embryos with high dNTP concentrations (N = 5) whose nuclei were tracked from NC11 through NC13. See also Figure S3. See also Video S3.
The S phase length is controlled by both origin usage and fork progression rate. Recent work has shown that the rapid nuclear cycles of the pre MBT embryo defer the formation of heterochromatin and late replicating regions [20], suggesting that high dNTP concentrations may affect origin usage. Using live imaging of embryos expressing a GFP-tagged form of the Proliferating Cell Nuclear Antigen (PCNA::GFP), which marks active replication forks, we did not observe a difference in origin usage, as indicated by the presence of late replicating regions, which form bright punctae at the end of S-phase (Video S2C, D, Figure S3D, E) [20, 21]. This suggests that the overall order of replication is maintained and that fork progression speed is faster. Additionally, the fact that the shortened interphases of cycles 12 and 13 are not as short as the earlier cycles demonstrates that dNTPs are clearly not the only regulators of the cleavage dynamics. Indeed, regulation of Cyclin B and histone levels, the physical hindrance of replication by DNA bound proteins and replication stress are additional inputs into the speed and progression of nuclear cycles during the MZT [15, 22–26].
While we have shown that shortened interphases interfere with transcription, the origins of the observed morphogenetic defects are yet to be fully elucidated. The earliest transcribed zygotic genes govern the process of cellularization [27]. Indeed, we observed significant reductions in the protein products of nullo and sry-a (Figure S2G–L), the key regulators of cellularization, which are highly transcribed during interphase 13 [28–30]. Conversely, we did not observe a difference in protein levels for Sna, whose activity is required only after the cellularization is completed (data not shown) [31]. Based on this, we speculate that defects in morphogenesis are caused by cumulative reduction in many zygotic gene products that control the mechanical properties of the newly formed epithelial tissue [32]. For example, we found that cell sizes are highly variable in cellular blastoderms of embryos developing at high levels of dNTPs (Figure S2M, N).
Finally, in addition to their roles in cellularization, zygotic gene expression is involved in the downregulation of maternally supplied cell cycle regulators such as Twe/CDC25, preventing mitosis during NC 14 [33–35]. Based on this, one can expect that quantitative changes in zygotic transcription at high levels of dNTPs may lead to extra divisions. Indeed, we observe that high levels of dNTPs result in higher levels of Twe/CDC25 during NC14 (Figure S3F). Furthermore, ~10% of embryos (5/42) exhibit a precocious entry into mitosis during NC14 (Figure S3G, H Video S3A–C). These observations further support the model whereby quantitative control of dNTP concentrations is essential for cell cycle slowing, robust accumulation of zygotic transcripts, and tissue morphogenesis.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Stanislav Y. Shvartsman (stas@princeton.edu)
Experimental Model and Subject Details
Adult flies were matured at 25°C on fresh yeast pas te for at least three and at most 14 days to collect embryos. All embryos used in this study were at developmental stages prior to gastrulation, which precluded the determination of the sex of embryos. All embryos were grown on apple juice plates at 25°C. All fly stocks were maintained by standard methods at 25°C, and were grown on a standard cornmeal, molasses, and yeast media. Fly media recipe: water (1726 mL), agar (11 g), potassium sodium tartrate (12 g), calcium chloride dihydrate (1 g), sucrose (43.35 g), dextrose (86.65 g), yeast (44 g), cornmeal (105 g), propionic acid (10 mL). Prepare as follows: measure water into kettle, mix in agar and bring to a boil to melt agar. Slowly add potassium sodium tartrate, calcium chloride dihydrate, sucrose, and dextrose, stirring as you add. Bring to a boil. Mix yeast and cornmeal with a little water, and add to kettle and stir. Boil 2 minutes. Cool to 80°C and add prop ionic acid. Stir very well.
Method Details
Maternal expression of RNRLD68N:
Virgin females of the MTD-Gal4 line were crossed to w;sp/cyo; UAS-RNRLD68N/TM6B males. The subsequent generation of adults were transferred into an embryo collection cage from which staged embryos could be collected for assay.
Cuticle Preparation:
Embryos were dechorionated after being aged for more than 24 h. Dechorionated embryos were shaken in methanol and heptane (1:1) and incubated overnight in medium containing lactic acid and Hoyer’s medium (1:1) at 65 °C. Embryos were imaged on a Nikon Eclipse Ni in dark field.
Immunostaining and FISH:
Embryos from Oregon R flies and flies with maternal expression of RNRLD68N were collected and allowed to develop until the cellular blastoderm stage.
For Immunofluorescence:
Antibodies used: Mouse anti-Twist (1:100), Mouse anti-Eve (1:100), Rabbit anti-GammaH2AV (1:500). Embryos were dechorionated in 50% bleach and fixed in 4% formaldehyde in PBS for 20 min. Embryos were then devitelinized in 1:1 Methanol and Heptane and stored in methanol. Following stepwise rehydration with PBST (.002% triton in PBS), embryos were blocked with 5% Normal Goat Serum and 10% Bovine Serum Albumin in PBST and then incubated at 4C°C overnight with primary a ntibodies. The following day after being washed in PBST, embryos were incubated with secondary antibodies for 1.5 hours at room temperature. Embryos were then mounted in Aqua-polymount which was then allowed to cure before imaging.
For FISH:
Embryos were dechorionated in 50% bleach and fixed in 4% formaldehyde in PBS for 20 min. Embryos were then incubated in 90% xylene for 1 hr and then treated with 80% acetone for 10 min at −20°C. Next, embryos were hybridized overnight at 60°C with antisense probes labeled with digoxigenin (DIG). Embryos with labeled probes were visualized using standard immunofluorescence technique. The following primary antibodies were used in this study: sheep anti-DIG (Roche; 1:200).
Light-sheet microscopy:
Imaging was done in the Keller lab with a custom built SiMView light-sheet microscope. Embryos from mothers expressing Sqh::mCherry as well as those maternally expressing UAS-RNRD68N were dechorionated in 50% bleach, washed and them prepared as in [40]. Embryos were volumetrically imaged with the 568nm laser at 30 second intervals, using the AutoPilot framework for sample-adaptive imaging [41].
Scanning Electron Microscopy:
Embryos were collected and allowed to develop until the cellular blastoderm stage and prepared according to [42]. Embryos were then dechorionated in 50% bleach and fixed in 25% Glutaraldehyde in PBS and Heptane in a scintillation vile for 25 minutes at room temperature. Embryos were then washed and manually devitelinized in PBS. Embryos were then dehydrated by stepping into ethanol (25%, 50%, 75%, 95%, and 100%) followed by incubation in 1:1 Ethanol Hexamethyldisilazane (HMS) for 10 minutes and finally 10 minutes in 100% HMS. Embryos were then allowed to dry before being coated with gold-palladium with a Denton Vacuum DeskII Sputterer at 45mAmps for 65s and imaged with a Hitachi TM-1000 desktop Scanning Electron Microscope.
Live imaging of nuclear divisions:
Embryos were collected from either wild type or flies maternally expressing RNRLD68N that also expressed H2AV::mRFP in order to visualize nuclei. Embryos were dechorionated with 50% bleach, washed and mounted on Biofoil membrane in halocarbon 27 oil. Embryos were then imaged on the Nikon Ti-E microscope with the Yokogawa spinning disc (CSU-21) module using the 561 laser to visualize nuclei at 10 second intervals.
Nucleus segmentation and feature extraction:
To extract all individual nuclei at each time point we first used the ‘pixel classification’ workflow of the publicly available image processing software Ilastik [43]. After segmentation of nuclei, we exported a variety of geometric and texture features generated for each nucleus as a part of the ‘object classification’ workflow. These features included the position, diameter, length of the skeleton, number of branches, average branch length, size in pixels, variance of intensity, convex hull area, mean defect displacement, object area, and convexity. For every embryo an array of these features was exported as a .csv to then be inputted into Matlab for tracking and phase identification.
Nucleus tracking:
To allow reconstruction of the temporal dynamics of each nucleus in the time-lapse, we designed a ‘nearest-neighbor’ based tracking algorithm using the position extracted via ilastik. Our algorithm was able to identify the exact time point of division of each nucleus, as well as the identity of the two daughter nuclei, thereby recovering the nucleus lineage tree of the embryo. Division identification was done as follows: after pairing each nucleus from time point t to its nearest neighbor from time t+1, daughter nuclei, and hence an event of division, were identified as unpaired nuclei from time point t+1. Then, each such daughter nucleus was paired to its nearest unpaired neighbor. Lastly, based on the identified division times we were able to label the nuclear cycle (NC) of each nucleus.
Division phase labeling via Dynamic Time Warping:
Dynamic time warping (DTW) is a well-known dissimilarity measure between a pair of sequences which, by stretching and compressing them locally, makes one resemble the other as much as possible [44]. Therefore, with proper quantitative temporal phenotypic description of each nucleus, DTW allows accurate matching of division phases between pairs of nuclei. To this end, we generated a quantitative phenotypic description for each nucleus in the form of a multidimensional feature vector (Fij)n, wherein rows and columns correspond to different features extracted via ilastik and successive time points, respectively. To achieve phase labeling we first calculated the DTW dissimilarity score between all pairs of tracked nuclei of the same NC. Then, we identified the nucleus with the minimal median dissimilarity score to all other nuclei from that NC, to be referred as the “template”, and manually labeled each of its time point with a division phase (i.e. Anaphase/Interphase/Prohase/Metaphase). Lastly, we propagated the phase labels from the template to every other nucleus from that NC based on the matching calculated by the DTW.
MS2 Data Analysis:
All MS2 data analysis after image processing was performed in MATLAB. Files containing dot intensity, frame number, and track ID were exported from Imaris as a .csv file and imported into MATLAB. All intensities and frame numbers were first sorted by track ID (each unique track ID corresponds to a single MS2 dot for all frames that it was detected), and the first frame for each track ID was normalized to zero, to account for the metasynchrony of mitotic divisions in nuclear cycles 11–13. Integrated intensity was calculated as the sum of all intensities in a single track, maximum intensity was defined as the maximum intensity that was reached for a single track, time to maximum intensity was defined as the number of frames between the start of detection to the detected intensity maximum (the first maximum, if this number was reached multiple times during the course of a track), and the track length was defined as the total number of frames that a dot was detected. All tracks for a single nuclear cycle were pooled and the median was taken for every reported value. These median values for multiple biological replicates were plotted in Microsoft Excel and further analyzed for statistical significance.
MS2 Constructs and transgenics:
Starting with the pBphi-evePromoter-24X MS2-yellow vector [14], a DNA fragment containing 206 bp upstream and 48 bp downstream of the bnk transcription start site was placed before the 24X MS2 repeat sequence via a convenient BamHI site. Within the bnk proximal promoter region are four Zelda binding sites, which likely drive the ubiquitous expression of bnk [12, 13]. Transgenic lines carrying the bnk-MS2 construct were generated by phiC31 integration in the 53B2 landing site (VK00018), Bloomington stock number 9736 [45, 46] by Best Gene, Inc.
Sequence of the bnk promoter region (TATATAA box and transcription start site in italics):
CTCCTTTGAGAAATCTTCCTTTTTTACTTTCATAGCTTAGGTAGTGATCTCAGGTAGTTTCCCGGAATTAAGTTAGGCTGGGTATCGCCTATCGGGATCGGCTACCTGAACTTTTGGCACCAGCTGCCGGGGGTGAAAACTGGACCAGGTAGTCTTTAGAgacgtcAGTGCACCtatataaGGTCTG CGTTCCTGGTCTCAACAGCaCATTCGTACTTCAACCGTCAACATGAGCATCAGCACTTTCAA CTTCCAggatcc
Sequence of the MS2 loops:
tacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcagatcctacggtacttattgccaagaaagcacgagcatcagccgtgcctccaggtcgaatcttcaaacgacgacgatcacgcgtcgctccagtattccagggttcatcggatct
Sequence of the yellow reporter gene (intron iitalics):
atgttccaggacaaagggtggattcttgtgaccctgatcaccttggtgacgccgtcttgggctgcttacaaacttcaggagcgatatagttggagccagctggactttgctttcccgaatacccgactaaaggaccaagctctggctagtggagattatattccgcaaaatgctctacctgttggagtcgaacactttggcaatcggttattcgtcactgttccccgctggcgtgatggtaagtggaagttaaatatgaagcccttggggagatcgtaaatgggacattcttacttagggcatcagagatatctgattgagtggttgacagttttatatggcttgtttgacatgatgtaaaaacacaaaattcatttagtttaggtattcaaataagagcttgttatttattttagaatttggagaacatttttttgtctttctacccttcttagaaaataatattgttttgtacaatttaattttaaactagtacagacgaaaaatgtatttttttatttgtatgccttttcaccattttggcagaggaataaatatgacaatatattttgagagcaccctcatgtaaaggttttagcgtggcgacctctcataaatccggttggtacctgcgcgttattttaacattttaaacaattaaccgttgtaaaatcgaagccaatagcatggcattggctttatactgtattaaattgtattatattaccatccgaattgtaaagacttcttcagggccgccacatagaaatggaaatccaatcacaaacaataacttatggcattagctattaaccgacgattagctgtcagttcaacaaatgtaagagtggcgaaatgtttaaatgcgaaggcattgttctgtgactcacgttttattattaatcacacaaatgattttgcttcaaaattatttggcttacacaataatcaaaatttttatgaaatagttgaaacacaaaactaggaaattttaaaaagcaatgaaactaaaaaacccaattgctaagattatatgatgcgcatacaaatacttcagtacgtctaggaatgctttcgatgattgattagtttttatgcatggcttacaattggtatttacacagaaaaacacggcgatgttaataaattttgtacatatgttcttaagcagtccgaaacacccaaacttctgactaaacttaaaaaaacacgctcttcaaggatatcttaatgtcacttataatgtaatggtattgtattattaagtatatcaaaatattctggcccagccttgaggtctctttttaaaaagatatcgactgactacctccagtcaatgaaataatagcccagaaggccgaatcggcaaaaaataaaccccaagttacggcaaacaaaacatagtgaaagttgtggcaaagtggaacatttaaaggcatgcttcaatggccatcgaagcaaatcaattagtcaaagcaaatcggtagtggcaacaacaggctacagaatacctataagtgacagttatggggtatgattaattataaatattatcattgaccaccaatgctgggctcaattggaaaaactattctatgaagatttgagtaaataaattttgatttaaaaaagcccatggttatcgcgacaactagctacgggacaagattactgtttaaaatcaagtgtgaaatatcaaaatcaaaatcggattccgatcgggaagttgtatccgattctgaaactaaaacacagaattgccaacattttccgatatcgactcagctcacgtatttcatacagattcattaggccaccagccattgaataatataccccagtcaattgagctactcgatagttgatcaacttagcttttgtcaacgagtgaacgcataaactactacatcaacgatatttgcggcccattccaagctaaaagttcatcttaattacaaataagattagaaaaaatatctgaatgaaaaaaatgttgagacatatttctttggaaaaggagaacctcaagacagtcgaaaaaattgtttacaatgaaaatgttgaaaatcatgaagcagataaatctgtcagttgcgaggttttaggactgaaagagcacatgtcaaaatataaatttgttcaaatactttatatttgactgaattagattgttattttaaaagttatgaattaaataaagattgaaaggtgcattatgctcaaatgtatatttatcgcaacccccggttactttgtaaagcaaaaacgcctggtttgatttttaagaagatgggtcggtaaatcgataaaagctatattttctggtcgttgcagtctcactcgcctgctataaaaacattaaaagttcccagaaacaataaatgtctttaaattcaattaacgaagaaataaagaaggaaaagaactggagcggaaatcggtcgaaatactgccaatggccacatatacatttaacagcgatatatggtatacatattgataatgatgtcagacgcaattgcttcagacggctaatgacatcgcaaattgcacgcaacttgcaatagtgccaattatgactgaagtacatatagccggggatcttttaacataaacttccagtagatgtacaagcagaaaaaagagccattagcacggcagttaccattgcttatgattccttgtgtccaaaataatgacaaataggtatataaataattaaatgccaaacataagcgattctaatttacctttacatctgtatgcatttacatattatccagaaaacagacagcgataacttgcaacattgcttagtataataatccaaagaaggaatttaggcagaaattccagttaattaaatattcaaaacaaactttatttagtgcctcaataatagtttggccctgctaattctcctattttattttttagggattccggccactctgacctatataaacatggaccgcagtttgacgggttcaccggagctaattccgtatccagattggcgctcaaatacagctggagattgcgccaacagtattaccactgcctaccgcattaaagtggatgagtgtggtcggctgtgggttttggacactggaaccgtgggcatcggcaataccaccactaatccgtgcccctatgcggtaaatgtctttgacttgaccacggatacgcgaattcggagatacgagctacctggcgtggacacaaatccaaatactttcatagctaacattgccgtggatataggcaaaaattgcgatgatgcatatgcctattttgccgatgaattgggatacggcttgattgcttactcctgggaactgaacaagtcctggagattctcggcacattcgtattttttccccgatccattgaggggcgatttcaatgtcgctggtattaacttccaatggggcgaggagggtatatttggtatgtccctttcgcccattcgatcggatggttatcgtaccctgtactttagtccgttagcaagtcatcgacaatttgccgtatccacgaggattttgagggatgaaaccaggacggaagatagctatcatgactttgttgccttagatgaacggggtccaaactcccataccacttcacgtgtgatgagcgatgatggaattgagctgttcaatttaatagatcaaaatgcagtgggttgctggcactcatcaatgccgtactcaccgcaatttcatggcattgtggatcgcgatgacgttggcttagtttttccggccgatgtgaaaattgatgagaacaaaaacgtttgggttctatccgataggatgcccgttttcttgctgtctgacttggattattcagatactaatttccgaatttacacggctcccttggccactttaattgagaatactgtgtgtgatttgaggaataacgcctatgggccgccaaataccgtttcaataccaaaacaagccgttttgccaatgggtccaccgttatatacgaaacaatatcgtcctgtcttgccacagaaacctcagaccagctgggcttcctcgccgcctcctccaagtcgcacttatttgcccgccaattcaggcaatgtagtctccagtattagtgtctctacaaattctgtgggtcctgcaggagtggaggtgccaaaggcctatattttcaaccagcacaacggcataaattacgagacaagtggtccccatctatttcccacccatcaacccgcccaaccgggtggccaggatggtgggttaaaaacttatgtgaatgcccgccaatctgggtggtggcatcatcagcatcaaggttaaggcg
Live imaging of MS2 reporter lines:
Virgin flies from w;nos>MCPGFP, H2AV::mRFP that were maternally expressing RNRLD68N were crossed with males with various MS2 reporter elements. Embryos were dechorionated with 50% bleach, washed and mounted on membrane in halocarbon oil. Embryos were then imaged on the Nikon Ti-E microscope with the Yokogawa spinning disc (CSU-21) module using 488 laser to visualize MS2 and 561 laser to visualize nuclei at 15 second intervals.
MS2 signal segmentation and extraction:
MS2 signal was segmented as spots in Imaris, using the following parameters: Expected particle size: 1 μm, Calculate point spread function in Z: 2 m. Identified spots were filtered based on StdDev of Intensity in the MS2 channel. Tracking was achieved with default settings. Tracks were then filtered by minimum length to eliminated errantly classified tracks as well as MS2 signals that were truncated during the time series. Track statistics such as position and sum intensity were exported as a .csv for further analysis with Matlab.
Detailed Genotypes:
Figure 1:
1A: Oregon R
1B: otu>gal-4/+; nos>gal-4/cyo; nos>gal-4/UAS-RNRLD68N
1C: Oregon R
1D: otu>gal-4/+; nos>gal-4/cyo; nos>gal-4/UAS-RNRLD68N
1E, G: w; Sqh::mCherry/cyo; UAS-RNRLD68N/Tm3
1F, H: otu>gal-4/+; nos>gal-4/Sqh::mCherry; nos>gal-4/UAS-RNRLD68N 1I: Oregon R
1J: otu>gal-4/+; nos>gal-4/cyo; nos>gal-4/UAS-RNRLD68N
Figure 2:
WT: w; H2AV::mRFP
High[dNTP]: otu>gal-4/+; nos>gal-4/H2AV::mRFP; nos>gal-4/UAS-RNRLD68N
Figure 3:
3E
WT: ♀ w; nos>MCPGFP, H2AV::mRFP X ♂ sog-MS2-yellow
High[dNTP]: ♀ otu>gal-4/+; nos>gal-4/nos>MCP::GFP, H2AV::mRFP; nos>gal-4/UASRNRLD68N X ♂ sog-MS2-yellow
3F
WT: ♀ w; nos>MCPGFP, H2AV::mRFP X ♂ sna-MS2-yellow
High[dNTP]: ♀ otu>gal-4/+; nos>gal-4/nos>MCP::GFP, H2AV::mRFP; nos>gal-4/UASRNRLD68N X ♂ sna-MS2-yellow
3G
WT: ♀ w; nos>MCPGFP, H2AV::mRFP X ♂ bnk-MS2-yellow
High[dNTP]: ♀ otu>gal-4/+; nos>gal-4/nos>MCP::GFP, H2AV::mRFP; nos>gal-4/UASRNRLD68N X ♂ bnk-MS2-yellow
Quantification and Statistical Analysis
Analysis of Transcriptional Output
Time traces were analyzed in Matlab. Average integrated outputs were compared in Microsoft Excel. T-test was used to compare outputs between WT and High [dNTP] embryos. Significance and N values (embryos) can be found in the legend for Figure 2. Comparison of time normalized transcriptional output can be found in the legend for Supplemental Figure 2.
Analysis of Protein Levels
Normalized intensities were measured in Fiji and entered into Excel worksheets for later processing. Bootstrapping was accomplished using the boostrap function in Matlab. Bootstrapped means and ratios were computed in Matlab. N values (embryos) and significance values can be found in the main text and the legend for Supplemental Figure 2.
Analysis of Cell Cycle Times
Cell cycle phases and times were classified and quantified using a custom script in Matlab. Histograms of nuclear cycle length were generated in Matlab. Values were imported into an Excel worksheet and compared using the t-Test. Significance values and N values (embryos) can be found in the main text and the legend for Figure 3. Significance (confidence intervals) for comparison of Prophase, Metaphase and Anaphase times can be found in the legend for Supplemental Figure 3.
Analysis of Twe Protein Levels
Average fluorescence intensities were measured in Fiji and values entered into an Excel worksheet for further processing in Matlab. 95% confidence intervals were calculated based on Standard Error and exponential fits were plotted using the Polyfit function in Matlab. N values (embryos) can be found in the legend for Supplemental Figure 3.
Supplementary Material
Video S1: Morphogenetic movements in WT and embryos with high dNTP levels. Related to Figure 1. (A) Time-lapse depicting an embryo with high dNTP concentrations expressing H2::mRFP undergoing gastrulation movements. (B, C) Time-lapse depicting embryos expressing Sqh::mCHerry to visualize the midline. Normal midline formation in wild type embryo (B) and formation of a twisted midline in an embryo with high dNTP concentrations (C).
Video S2: MS2 expression and replication dynamics. Related to Figure 2. (A, B) Time-lapse of MS2 construct expression in wild type (A) and an embryo with high dNTP concentrations (B) during interphase 13. H2::mRFP shows nuclei in red and MCP::GFP shows MS2 signal in green. (C, D) Time- lapse depicting PCNA::GFP activity during interphase 13 of a wild type (C) and an embryo with high levels of dNTPs (D).
Video S3: Embryos with high dNTP concentrations undergo extra division. Related to Figure 3. Time-lapse depicting a wild type (A) and two embryos with high levels of dNTPs Movie B, C) which undergo an extra round of mitosis.
Highlights.
Metabolic control of cell cycle dynamics
Decreasing dNTP levels slow down nuclear cleavages
Short nuclear cycles interfere with zygotic transcription
Acknowledgments
We thank Yonghyun Song, Amanda Amodeo, and Jasmin Imran-Alsous for critical reading of the manuscript. We thank Gabrielle Shvartsman and Rochelle Forni for technical assistance during the early stages of this work. We also thank Eric Wieschaus, Trudi Schüpbach, Robert Marmion, Shelby Blythe, and Andrei Chabes for numerous helpful discussions. We also thank Michael Levine for fly stocks. We also thank Gary Laevsky of the Molecular Biology Confocal Microscopy Core for expert imaging support. This work was supported by R01GM086537 from NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- 1.Foe VE, and Alberts BM (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci 61, 31–70. [DOI] [PubMed] [Google Scholar]
- 2.Song Y, Marmion RA, Park JO, Biswas D, Rabinowitz JD, and Shvartsman SY (2017). Dynamic Control of dNTP Synthesis in Early Embryos. Dev Cell 42, 301–308 e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tadros W, and Lipshitz HD (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- 4.Ferree PL, and Di Talia S (2017). For Embryos, Mother Can Only Take You So Far. Dev Cell 42, 203–205. [DOI] [PubMed] [Google Scholar]
- 5.Mathews CK (1975). Giant pools of DNA precursors in sea urchin eggs. Exp Cell Res 92, 47–56. [DOI] [PubMed] [Google Scholar]
- 6.Vastag L, Jorgensen P, Peshkin L, Wei R, Rabinowitz JD, and Kirschner MW (2011). Remodeling of the metabolome during early frog development. PLoS One 6, e16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordlund P, and Reichard P (2006). Ribonucleotide reductases. Annu Rev Biochem 75, 681–706. [DOI] [PubMed] [Google Scholar]
- 8.Iampietro C, Bergalet J, Wang X, Cody NA, Chin A, Lefebvre FA, Douziech M, Krause HM, and Lecuyer E (2014). Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev Cell 29, 468–481. [DOI] [PubMed] [Google Scholar]
- 9.Jurgens G, Wieschaus E, Nusslein-Volhard C, and Kluding H (1984). Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster : II. Zygotic loci on the third chromosome. Wilehm Roux Arch Dev Biol 193, 283–295. [DOI] [PubMed] [Google Scholar]
- 10.Nusslein-Volhard C, Wieschaus E, and Kluding H (1984). Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster : I. Zygotic loci on the second chromosome. Wilehm Roux Arch Dev Biol 193, 267–282. [DOI] [PubMed] [Google Scholar]
- 11.Wieschaus E, Nusslein-Volhard C, and Jurgens G (1984). Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster : III. Zygotic loci on the X-chromosome and fourth chromosome. Wilehm Roux Arch Dev Biol 193, 296–307. [DOI] [PubMed] [Google Scholar]
- 12.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, and Rushlow C (2011). Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, and Levine M (2014). Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci U S A 111, 10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar BA, Kiehle CP, and Schubiger G (1986). Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44, 365–372. [DOI] [PubMed] [Google Scholar]
- 16.Edgar BA, and Schubiger G (1986). Parameters controlling transcriptional activation during early Drosophila development. Cell 44, 871–877. [DOI] [PubMed] [Google Scholar]
- 17.Shermoen AW, and O’Farrell PH (1991). Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, and Pasero P (2012). dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu M, Dai X, Guo W, Ge Z, Yang M, Wang H, and Wang YP (2017). Manipulating the Bacterial Cell Cycle and Cell Size by Titrating the Expression of Ribonucleotide Reductase. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seller CA, Cho C-Y, and O’Farrell PH (2018). Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila. 450155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shermoen AW, McCleland ML, and O’Farrell PH (2010). Developmental control of late replication and S phase length. Curr Biol 20, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sibon OC, Laurencon A, Hawley R, and Theurkauf WE (1999). The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol 9, 302–312. [DOI] [PubMed] [Google Scholar]
- 23.Ji JY, Squirrell JM, and Schubiger G (2004). Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila. Development 131, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amodeo AA, Jukam D, Straight AF, and Skotheim JM (2015). Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci U S A 112, E1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blythe SA, and Wieschaus EF (2015). Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seller CA, and O’Farrell PH (2018). Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol 16, e2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieschaus E, and Sweeton D (1988). Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development 104, 483–493. [DOI] [PubMed] [Google Scholar]
- 28.Schweisguth F, Lepesant JA, and Vincent A (1990). The serendipity alpha gene encodes a membrane-associated protein required for the cellularization of the Drosophila embryo. Genes Dev 4, 922–931. [DOI] [PubMed] [Google Scholar]
- 29.Postner MA, and Wieschaus EF (1994). The nullo protein is a component of the actin-myosin network that mediates cellularization in Drosophila melanogaster embryos. J Cell Sci 107 (Pt 7), 1863–1873. [DOI] [PubMed] [Google Scholar]
- 30.Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, and Eisen MB (2011). Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol 9, e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leptin M (1991). twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev 5, 1568–1576. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, and Blankenship JT (2018). Differentially-dimensioned furrow formation by zygotic gene expression and the MBT. PLoS Genet 14, e1007174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar BA, and Datar SA (1996). Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev 10, 1966–1977. [DOI] [PubMed] [Google Scholar]
- 34.Di Talia S, She R, Blythe SA, Lu X, Zhang QF, and Wieschaus EF (2013). Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Curr Biol 23, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrell JA, and O’Farrell PH (2013). Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr Biol 23, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin AC, Kaschube M, and Wieschaus EF (2009). Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukaya T, Lim B, and Levine M (2016). Enhancer Control of Transcriptional Bursting. Cell 166, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang JK, Sherizen DE, Bhagat R, Manheim EA, and McKim KS (2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci 116, 3069–3077. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y, Iagovitina A, Ishihara K, Fitzgerald KM, Deplancke B, Papatsenko D, and Shvartsman SY (2013). Context-dependent transcriptional interpretation of mitogen activated protein kinase signaling in the Drosophila embryo. Chaos 23, 025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomer R, Khairy K, Amat F, and Keller PJ (2012). Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods 9, 755–763. [DOI] [PubMed] [Google Scholar]
- 41.Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, Myers EW, and Keller PJ (2016). Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol 34, 1267–1278. [DOI] [PubMed] [Google Scholar]
- 42.Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, and Wieschaus EF (2010). Integration of contractile forces during tissue invagination. J Cell Biol 188, 735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer C, Straehle C, Köthe U, and Hamprecht FA (2011). Ilastik: Interactive learning and segmentation toolkit. In 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. pp. 230–233. [Google Scholar]
- 44.El-Labban A, Zisserman A, Toyoda Y, Bird A, and Hyman A (2011). Dynamic time warping for automated cell cycle labelling. Microscopic Image Analysis with Applications in Biology, 580–583. [Google Scholar]
- 45.Groth AC, Fish M, Nusse R, and Calos MP (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Morphogenetic movements in WT and embryos with high dNTP levels. Related to Figure 1. (A) Time-lapse depicting an embryo with high dNTP concentrations expressing H2::mRFP undergoing gastrulation movements. (B, C) Time-lapse depicting embryos expressing Sqh::mCHerry to visualize the midline. Normal midline formation in wild type embryo (B) and formation of a twisted midline in an embryo with high dNTP concentrations (C).
Video S2: MS2 expression and replication dynamics. Related to Figure 2. (A, B) Time-lapse of MS2 construct expression in wild type (A) and an embryo with high dNTP concentrations (B) during interphase 13. H2::mRFP shows nuclei in red and MCP::GFP shows MS2 signal in green. (C, D) Time- lapse depicting PCNA::GFP activity during interphase 13 of a wild type (C) and an embryo with high levels of dNTPs (D).
Video S3: Embryos with high dNTP concentrations undergo extra division. Related to Figure 3. Time-lapse depicting a wild type (A) and two embryos with high levels of dNTPs Movie B, C) which undergo an extra round of mitosis.