PRX9 and PRX40 are essential class III peroxidases that act redundantly to cross-link extensins in tapetal cell walls, which prevents tapetum swelling and enables pollen development.
Abstract
Pollen and microspore development are essential steps in the life cycle of all land plants that generate male gametes. Within flowering plants, pollen development occurs inside of the anther. Here, we report the identification of two class III peroxidase-encoding genes, PEROXIDASE9 (PRX9) and PRX40, that are genetically redundant and essential for proper anther and pollen development in Arabidopsis (Arabidopsis thaliana). Arabidopsis double mutants devoid of functional PRX9 and PRX40 are male sterile. The mutant anthers display swollen, hypertrophic tapetal cells and pollen grains, suggesting disrupted cell wall integrity. These phenotypes lead to nearly 100%-penetrant pollen degeneration upon anther maturation. Using immunochemical and biochemical approaches, we show that PRX9 and PRX40 likely cross-link extensins to contribute to tapetal cell wall integrity during anther development. This work suggests that PRX9 and PRX40 encode Arabidopsis extensin peroxidases and highlights the importance of extensin cross-linking during pollen development.
INTRODUCTION
Pollen performs a critical step in the flowering plant life cycle by transporting the male gamete to the female ovule for fertilization. To do this, pollen must survive the journey from its origin in the anther to its destination atop the carpel. Wind-borne or insect-dispersed pollen may need to travel great distances to reach a suitable carpel. During its journey, pollen must endure a wide range of abiotic stresses, including UV radiation, extreme temperatures, and desiccation. To survive these challenges, pollen grains are protected by a unique and specialized cell wall.
The synthesis of the pollen wall was one of the most important innovations that enabled plant life on land. The pollen wall is highly structured and indispensable for pollen vitality. It is composed of two parts: an outer exine and an inner intine (Heslop-Harrison, 1968). The exine is composed of a uniquely robust polymer called sporopollenin. Many genes involved in the biosynthesis of sporopollenin have been identified, mostly through genetic studies, but the detailed chemical structure of the polymer is poorly understood (Quilichini et al., 2015). The intine resembles a conventional plant cell wall and consists of cellulose, hemicellulose, and pectin as well as cell wall–associated proteins (Knox et al., 1970). Pollen development occurs in the anther, concurrent with anther development.
Arabidopsis (Arabidopsis thaliana) anthers are specialized organs that are designed to produce and release pollen grains. Arabidopsis flowers have six anthers that are positioned atop anther filaments and encircle the female carpel. Each anther has four functionally equivalent lobes. Within each lobe, pollen develops in a chamber known as the locule. Locule walls are lined by a specialized tissue composed of secretory cells known as the tapetum. The tapetum is the innermost layer of the anther and provides nutrients to developing pollen grains. The tapetum is also thought to supply the sporopollenin precursors that later polymerize to form the pollen exine. At late stages of pollen development, the tapetum undergoes programmed cell death (PCD), depositing a mixture of protein and wax on the surface of the exine, known as tryphine (Heslop-Harrison, 1968).
Although the sequence of events in tapetum and pollen development has been well described, the exact biochemical processes involved remain poorly understood. In the model plant Arabidopsis, hundreds of evolutionarily conserved genes are specifically expressed in developing anthers, but only a few have been functionally characterized to date (Li et al., 2017). In this work, we sought to better understand the role of a subset of these genes encoding class III peroxidases.
Class III peroxidases are a large family of heme-iron–dependent peroxidases that arose specifically within land plants and have expanded widely since their emergence; the Arabidopsis genome contains 73 members (Tognolli et al., 2002; Welinder et al., 2002; Duroux and Welinder, 2003; Weng and Chapple, 2010). Many biochemical activities important for various aspects of land plant physiology have been attributed to class III peroxidases. For example, class III peroxidases oxidatively polymerize monolignols in the apoplast of the lignifying cells. Previous studies have found that PEROXIDASE2 (PRX2), PRX4, PRX17, PRX25, PRX52, PRX71, and PRX72 are involved in stem lignification in Arabidopsis (Herrero et al., 2013; Fernández-Pérez et al., 2015a, 2015b; Shigeto et al., 2015). In addition, class III peroxidases are also thought to polymerize other elements of the plant cell wall, including suberin and extensins (Bernards et al., 1999; Jackson et al., 2001). Conversely, some class III peroxidases may function to destabilize plant cell walls by generating hydroxyl radicals that cleave load-bearing cell wall polysaccharides (Liszkay et al., 2004). This is exemplified by PRX36, which serves as a seed mucilage extrusion factor by loosening the walls of epidermal cells in the Arabidopsis seed coat (Kunieda et al., 2013).
In this work, we identified two class III peroxidase-encoding genes, PRX9 and PRX40, that are required for proper development of tapetum and microspores in Arabidopsis. The prx9 prx40 double mutants exhibit distinctive tapetum swelling and enlarged developing pollen grains, ultimately leading to microspore degeneration and male sterility. We provide further evidence that PRX9 and PRX40 ensure proper establishment of the tapetum and microspore cell walls by cross-linking extensins.
RESULTS
PRX9 and PRX40 Are Specifically Expressed in the Tapetum of Developing Anthers
To identify uncharacterized metabolic enzymes involved in pollen and anther development, we performed a coexpression analysis using the known exine biosynthetic genes as search queries (Obayashi et al., 2007). This analysis uncovered only two class III peroxidase genes, PRX9 (AT1G44970) and PRX40 (AT4G16270), which are specifically expressed in early stages of flower development (Supplemental Figure 1; Winter et al., 2007; Waese and Provart, 2017). Neither PRX9 nor PRX40 has been the focus of any prior publication, but both genes have been implicated in root elongation and lateral root primordia formation (Tsukagoshi et al., 2010; Fernández-Marcos et al., 2017). Nevertheless, PRX9 and PRX40 have not been shown to have roles in flower development.
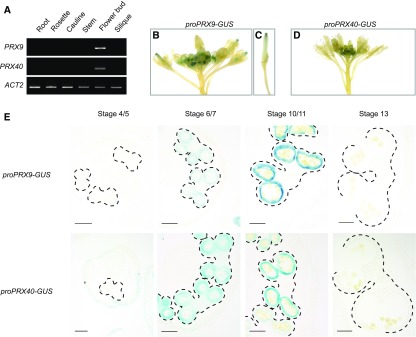
To experimentally examine the tissue specificity of PRX9 and PRX40 expression, we first profiled the transcript abundance of PRX9 and PRX40 across six different Arabidopsis tissue types by RT-PCR. This experiment verified that PRX9 and PRX40 are both specifically expressed in the flower buds, but not in the other five tissues examined (Figure 1A). To obtain higher spatial and temporal resolution for PRX9 and PRX40 expression, we generated promoter-β-glucuronidase (GUS) reporter lines for PRX9 and PRX40 and examined the tissue-specific GUS activities in six independent T1 transformants for each reporter construct. In both cases, all six replicates produced nearly identical results. Whereas PRX9 promoter-reporter activity was detected in both early-developing anthers and mature carpels, PRX40 promoter-reporter activity was exclusively observed in early-developing anthers (Figures 1B to 1D). Closer examination of the GUS-stained anther sections further established that PRX9 and PRX40 are specifically expressed in the tapetum (Figure 1E). Expression of PRX9 and PRX40 appeared to initiate at anther stage 6 and peak around anther stage 10. No staining was observed in later stages of anther development in which the tapetum had undergone PCD.
Figure 1.
PRX9 and PRX40 Are Expressed in the Tapetum of Developing Anthers.
(A) to (E) RT-PCR across a variety of Col-0 tissues shows PRX9 and PRX40 are specifically expressed in the flower buds (A). The ACT2 transcript was used as a positive control and could be amplified in all samples. Whole-mount images show reporter ProPRX9-GUS activity is detected in the early anthers (B) and mature carpel (C). ProPRX40-GUS reporter activity is detected in the early anthers (D). Images of anther sections of GUS reporter across development shows that PRX9 and PRX40 promoter activity is localized to the tapetum, is initiated by stage 6, and peaks around stage 10 (E). Promoter reporter activity is absent by anther dehiscence. Anthers are outlined with dashed lines. Bars = 50 μm.
PRX9 and PRX40 Are Genetically Redundant and Required for Male Fertility
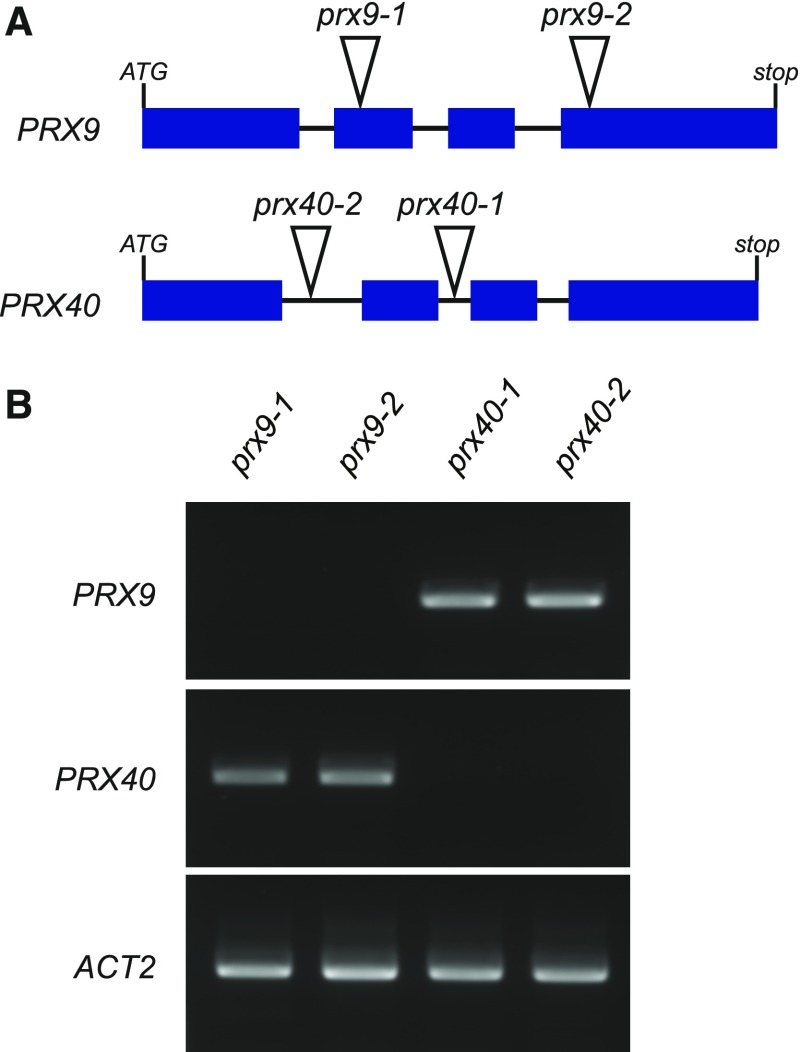
To probe the in vivo function of PRX9 and PRX40, we obtained two independent T-DNA insertion lines for each of the two genes. SALK_204557 (prx9-1) and SAIL_875_A09 (prx9-2) contain insertions in the second and fourth exons of PRX9, respectively, and SALK_031680 (prx40-1) and SALK_061827 (prx40-2) contain insertions in the second and first introns of PRX40, respectively (Figure 2A). To assess whether these insertion events resulted in loss-of-function mutations, we examined the expression of PRX9 and PRX40 in the T-DNA lines by RT-PCR (Figure 2B) and established that all the four T-DNA insertion events produced loss-of-function alleles.
Figure 2.
Molecular Characterization of PRX9 and PRX40 Insertion Lines.
(A) prx9-1 and prx9-2 contain T-DNA insertions in the second and fourth exons of PRX9, respectively. prx40-1 and prx40-2 contain T-DNA insertions in the second and first introns of PRX40, respectively. Blue rectangles represent exons, lines represent introns, and triangles indicate T-DNA insertion sites.
(B) RT-PCR showing full-length PRX9 cDNA could not be amplified from prx9-1 or prx9-2 mutants, while full-length PRX40 cDNA could not be amplified from prx40-1 or prx40-2 mutants. The ACT2 transcript was used as a positive control and could be amplified in all four samples.
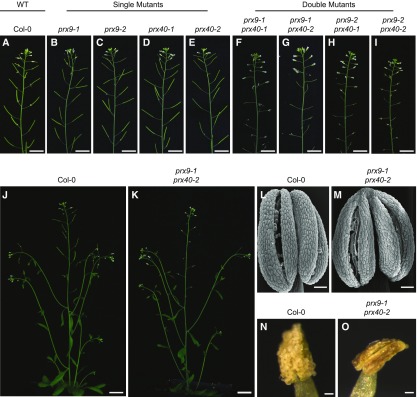
Single mutants of either prx9 or prx40 display no visible differences from the Columbia (Col-0) wild-type plants. In particular, the prx9 and prx40 mutants are fully fertile and develop fully elongated, seed-bearing siliques (Figures 3A to 3E).
Figure 3.
prx9 prx40 Double Mutants Are Male Sterile and Do Not Produce Viable Pollen.
(A) to (O) While the wild-type (A) and (J), single prx9 (B) and (C), and single prx40 (D) and (E) plants show no defects in fertility, all prx9 prx40 double mutants (F) to (I) and (K) are male sterile. Bars = 2 cm. Stage 13 wild-type (L) and prx9-1 prx40-2 (M) anthers dehisce normally. Bars = 50 μm. Stage 14 wild-type (N) anthers are coated in pollen grains. prx9-1 prx40-2 (O) anthers are devoid of pollen grains. Stages according to Sanders et al. (1999). Bars = 50 μm. WT, wild type.
Since none of the single mutants exhibited any visible phenotype, we hypothesized that PRX9 and PRX40 may be genetically redundant. To test this, we generated all four possible prx9 prx40 double mutants using the four single mutant alleles. In contrast to the single mutants, which displayed no obvious defects in fertility, all prx9 prx40 double mutant plants were completely sterile (Figures 3F to 3K). Siliques from double mutants were short and contained no seeds. The prx9 prx40 plants are likely male sterile as prx9 prx40 plants were readily fertilized by the wild-type pollen, but all attempts to fertilize the wild-type plants with prx9 prx40 pollen failed. Since two independent alleles are represented for each peroxidase gene, and all four double mutants displayed identical phenotypes, we conclude that PRX9 and PRX40 are genetically redundant and required for male fertility in Arabidopsis.
We then examined the flowers from the double mutant at various developmental stages by light microscopy. At stage 14 of anther development (Sanders et al., 1999), the wild-type anthers were coated with mature pollen grains, whereas prx9-1 prx40-2 anthers were devoid of pollen (Figures 3N and 3O). We also observed that anthers from the prx9-1 prx40-2 plants displayed defects in anther filament elongation (Supplemental Figure 2). However, this did not appear to be the primary cause of sterility, as prx9 prx40 anthers were incapable of depositing pollen on the stigma, even when manually crossed. Similar defects in anther filament extension have been previously observed in mutants of other important tapetal regulators, such as DYSFUNCTIONAL TAPETUM1, DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1, and ABORTED MICROSPORES (Sorensen et al., 2003; Zhang et al., 2006; Zhu et al., 2008). This is likely a secondary effect of tapetal dysfunction that is caused by an impairment of auxin production, which occurs in the tapetum during anther development (Cecchetti et al., 2008). Since indehiscence is one of the mechanisms that can lead to male sterility even when pollen grains are properly developed (Sanders et al., 1999; Hao et al., 2014; Yang et al., 2017), we examined stage 13 anthers (Sanders et al., 1999; Hao et al., 2014; Yang et al., 2017) by scanning electron microscopy. Anthers from both the wild-type and the prx9-1 prx40-2 plants clearly showed breakage along the stomiums, indicating that the male-sterility phenotype of prx9-1 prx40-2 is unlikely to be caused by indehiscence (Figures 3L and 3M).
PRX9 and PRX40 Play Important Roles in Maintaining Tapetum and Microspore Cell Wall Integrity
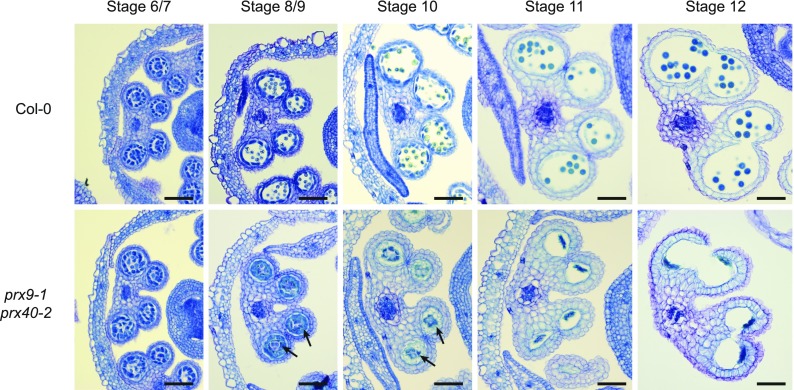
To identify the stage at which prx9-1 prx40-2 anther and pollen development first derails, we performed a histological comparison between the prx9-1 prx40-2 and the wild-type anthers over various developmental stages (Figure 4). At stages 5 and 6, prx9-1 prx40-2 and wild-type anthers were indistinguishable. Noticeable differences first appeared at stages 8 and 9, with the double mutant exhibiting ectopically swollen tapetal cells that invade into the locular space. The swollen tapetum persisted through stage 10 in prx9-1 prx40-2 plants. By stages 11 and 12, the tapetum had degenerated in both plants, but the pollen grains were shriveled and crushed in the prx9-1 prx40-2 anthers. The appearance of the tapetum swelling phenotype between stages 7 and 10 closely parallels PRX9 and PRX40 expression (Figure 1). These observations suggest that the male sterility of prx9-1 prx40-2 plants is likely caused by tapetum swelling (i.e., tapetum hypertrophy) and subsequent pollen degeneration.
Figure 4.
prx9 prx40 Anthers Display Tapetum Hypertrophy and Pollen Degeneration.
The wild-type (top row) and prx9-1 prx40-2 (bottom row) anther development from stages 5 to 12 (Sanders et al., 1999). Tapetum swelling in prx9-1 prx40-2 first appears at stage 8 and persists through stage 10, indicated by arrows. Tapetum PCD and anther dehiscence occur normally in both the wild type and prx9-1 prx40-2 during stages 11 and 12. Bars = 25 μm.
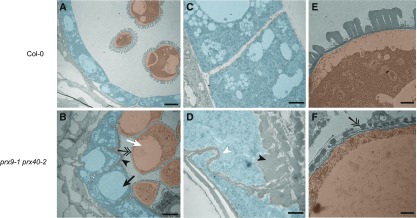
To further characterize the tapetum hypertrophy phenotype, we examined stage 9 anthers by transmission electron microscopy (Figure 5). Consistently, the prx9-1 prx40-2 sample displayed expanded tapetum cells with enlarged tapetal vacuoles, compared with the wild type. Microspores in prx9-1 prx40-2 anthers were also abnormally enlarged and vacuolated. The boundaries between tapetal cells in prx9-1 prx40-2 plants were jigsaw shaped, with one cell swelling into the neighboring cell, whereas the boundaries between tapetal cells in the wild-type plants were typically well defined and straight. This phenotype may be a manifestation of compromised tapetal cell wall integrity. Moreover, we observed aberrant distributions of electron-dense material in the swollen tapetum of prx9-1 prx40-2 anthers, reminiscent of sporopollenin typically observed at the outer wall of normally developed pollen. By stage 9, the wild-type microspores exhibit a fully developed exine. By contrast, only nexine and probaculae had formed on the microspore surface in the double mutant, with the majority of the sporopollenin-like material accumulated at the locular face of the tapetum. Finally, the middle layer was not crushed in prx9-1 prx40-2 plants. Taken together, this suggests that PRX9 and PRX40 play an important role in maintaining tapetal and microspore cell size and shape by modulating the cell wall.
Figure 5.
Stage 9 prx9-1 prx40-2 Anthers Contain Swollen, Hypervacuolated Tapetal Cells and Pollen Grains.
(A) to (F) The stage 9 wild-type (A) and prx9-1 prx40-2 (B) locules were compared by transmission electron microscopy. Tapetal cells are false colored in blue and developing pollen grains are false colored in orange. The prx9-1 prx40-2 tapetum is swollen and hypervacuolated (black arrow). Pollen grains are also swollen and hypervacuolated (white arrow). Electron-dense sporopollenin-like material accumulates on the locular face of the tapetum (black arrowhead). Bars = 4 μm. The wild-type tapetal cells (C) have rectangular boundaries. prx9-1 prx40-2 tapetal cells (D) have irregular, jigsaw-shaped boundaries (white arrowhead). Bars = 600 nm. The wild-type pollen grains (E) have a fully developed exine, whereas prx9-1 prx40-2 pollen grains (F) only have visible probaculae and nexine (double arrow). Bars = 600 nm.
PRX9 and PRX40 Are Active Peroxidases In Vitro
To probe the biochemical function of PRX9 and PRX40, we first tested whether PRX9 and PRX40 are catalytically active hydrogen peroxide (H2O2)–dependent peroxidases by heterologously expressing PRX9 and PRX40 in Escherichia coli. Although both proteins were highly expressed in E. coli, they were insoluble. We solubilized recombinant PRX9 and PRX40 by denaturing them in 8 M urea. After purification, we subjected the unfolded PRX9 and PRX40 to a peroxidase refolding screen using pyrogallol as a generic peroxidase substrate (Smith et al., 1990). Refolding conditions were tested for activity by the addition of pyrogallol and H2O2. Activity was observed by a colorimetric change associated with an increase in absorbance at 420 nm as pyrogallol was converted into purpurogallin. We optimized the refolding conditions for both PRX9 and PRX40 and observed H2O2-dependent peroxidase activity on pyrogallol for both enzymes (Supplemental Figure 3). This result demonstrates that PRX9 and PRX40 behave as typical class III peroxidases and are capable of completing the catalytic cycle in vitro.
PRX9 and PRX40 Are Likely Extensin Cross-Linking Peroxidases
Since the cell wall exerts a strong influence on the size and shape of plant cells, and since class III peroxidases are typically localized in the plant apoplast, we hypothesized that PRX9 and PRX40 may oxidize a component of the tapetum and microspore cell walls and thus reinforce them. Since the tapetum is not thought to be lignified or suberized, extensins stood out as a potential substrate for PRX9 and PRX40. Extensins are structural Hyp-rich glycoproteins in the cell wall that are known to regulate cell size and shape. They form large fibrillar networks and are thought to scaffold the assembly of the plant cell wall (Cannon et al., 2008). Previous studies showed that extensins may be intra- and intermolecularly cross-linked by class III peroxidases via Tyr residues (Fry, 1982; Brownleader et al., 1995, 2000 ; Brady et al., 1996). It is unclear why extensins must be cross-linked, but cross-linking is necessary for their function (Hall and Cannon, 2002; Ringli, 2010).
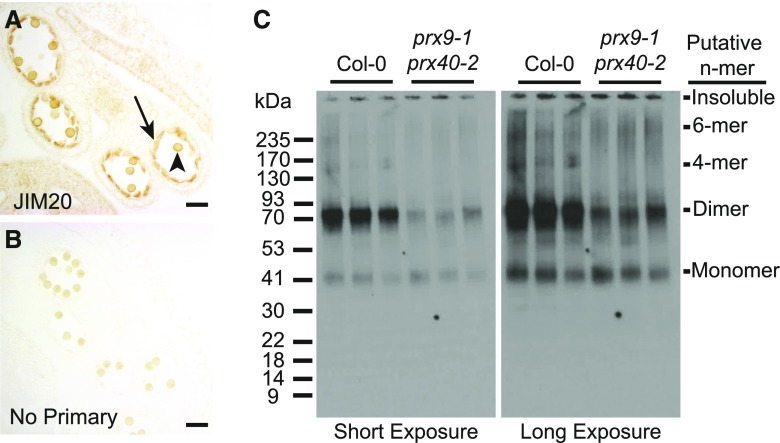
To assess the role of extensins in tapetal cell development, we first examined the accumulation of extensins in the wild-type anthers by immunostaining. We took advantage of John Innes Monoclonal Antibody 20 (JIM20), a well-characterized extensin-specific monoclonal antibody (Pattathil et al., 2010). Consistent with our hypothesis, JIM20 extensin epitopes were highly enriched in the wild-type tapetum (Figures 6A and 6B). Pollen walls also appeared to be stained, but it is difficult to determine the extent of staining because pollen grains are naturally pigmented.
Figure 6.
High Molecular Weight JIM20 Extensin Epitopes Accumulate in the Wild-Type, but Not prx9-1 prx40-2, Tapetal Cells.
(A) to (C) Extensins in the wild-type stage 10 anthers were examined by immunostaining with the JIM20 antibody (A) and compared with a control without a primary antibody (B). Staining was most obvious in the tapetum (black arrow) and may also be present in the pollen grain wall (black arrowhead). The apparent molecular weight of extensins was compared between three biological replicates of the wild-type and prx9-1 prx40-2 stages 10 to 12 (Sanders et al., 1999) anthers by immunoblot (C). Left panel is a short exposure and right panel is a long exposure of the same blot. Similar amounts of a putative ∼40-kD monomer are present in both samples. Substantially more putative ∼80-kD dimer is present in the wild-type anthers. Higher molecular weight species of ∼160 and 240 kD, corresponding to putative 4-mer and 6-mer, respectively, are present in the wild-type anthers but absent from prx9-1 prx40-2 anthers.
Since cross-linking increases the apparent molecular weight of extensins, we expected high molecular weight extensins to accumulate in the tapetum of the wild-type anthers. If PRX9 and PRX40 were the primary extensin peroxidases in tapetum, we would expect prx9-1 prx40-2 anthers to accumulate less high molecular weight extensins. To test this hypothesis, we performed an immunoblot, comparing protein extracts of the stage 10 (Sanders et al., 1999) wild-type and prx9-1 prx40-2 anthers (Figure 6C). The wild-type anthers accumulate a putative monomeric extensin at ∼40 kD, with higher molecular weight species also present. The most intense bands appeared to be at ∼80 kD, corresponding to the molecular weight of an extensin dimer. Additional bands at ∼160 and 240 kD were visible, likely corresponding to the molecular weight of a 4-mer and 6-mer, respectively. By comparison, prx9-1 prx40-2 anthers accumulated substantially less 80-kD extensins and the 160- and 240-kD bands were absent, even though prx9-1 prx40-2 anthers accumulated approximately the same amount of the putative monomeric species.
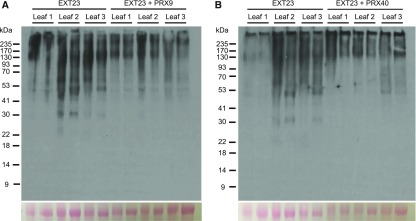
If PRX9 and PRX40 are truly extensin peroxidases, they would be sufficient to cross-link a model extensin in a reconstituted system. Because extensins produced in plant cells undergo extensive glycosylation prior to polymerization (Showalter and Basu, 2016), soluble extensins could not be produced in microbial hosts, making in vitro biochemical assays with physiologically relevant monomeric extension substrates intractable. As an alternative approach to test whether PRX9 and PRX40 are capable of cross-linking extensins, we transiently expressed EXTENSIN23 (EXT23 [AT5G19810]) in Nicotiana benthamiana leaves with or without peroxidase and examined the size distribution of JIM20 extensin epitopes by immunoblot (Figure 7). We chose EXT23 because it has been shown to be expressed in the anthers (Hruz et al., 2008), and its molecular weight after glycosylation would be roughly equal to the putative extensin monomer that we observed in anther extracts (Figure 6). Overall, we observed that higher molecular weight JIM20 epitopes accumulated in N. benthamiana leaves coexpressing either PRX9 (Figure 7A) or PRX40 (Figure 7B) than in those expressing EXT23 alone. Altogether, these data further support the biochemical function of PRX9 and PRX40 as extensin cross-linking peroxidases in planta.
Figure 7.
High Molecular Weight Extensins Accumulate in N. benthamiana Leaves When Coexpressed with either PRX9 or PRX40.
EXT23 was transiently expressed in N. benthamiana leaves, either with or without a peroxidase, and the apparent molecular weights of extensins were analyzed by immunoblotting with JIM20. In each blot, lanes 1 to 6 analyze samples from three independent leaves expressing EXT23. Two independent sample were analyzed from each leaf. Lanes 7 to 12 analyze samples from three independently transformed leaves coexpressing EXT23 with either PRX9 (A) or PRX40 (B). The Ponceau-stained Rubisco band is included below each blot to indicate total protein loaded.
The ext18 Mutant Partially Phenocopies prx9 prx40 Anthers
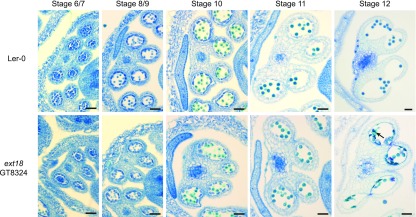
If extensin cross-linking by PRX9 and PRX40 is important for maintaining tapetum and microspore cell size, knockout mutants of the extensins that are substrates for these peroxidases should phenocopy the prx9 prx40 plants. EXT18 knockouts have previously been reported to cause defects in male fertility (Choudhary et al., 2015). The ext18 mutant was reported to exhibit decreased pollen viability, suggesting that EXT18 is likely expressed in the developing anther and may be cross-linked by PRX9 and PRX40. To assess the role of EXT18 in tapetum development, we performed a histological comparison between ext18 (genetrap 8324) and the Landsberg erecta-0 wild-type anther development (Figure 8). As previously reported, we observed degenerated pollen grains at later stages of anther development, but no differences in the shape or size of the tapetum at earlier developmental stages. As with EXT23, we observed a shift toward higher molecular weights when EXT18 was coexpressed with either PRX9 or PRX40 in N. benthamiana (Supplemental Figure 4). Since we did not observe tapetum hypertrophy in ext18 plants, additional extensins likely serve as redundant or more prominent substrates for PRX9 and PRX40 in the tapetum.
Figure 8.
ext18 Anthers Exhibit Pollen Degeneration, but Not Tapetum Hypertrophy.
The wild-type Landsberg erecta-0 (top row) and ext18 (bottom row) anther development from stages 5 to 12 (Sanders et al., 1999). The wild-type and ext18 are indistinguishable until stage 12 when numerous degenerated pollen grains are observed in ext18 anthers (black arrow). Bars = 25 μm.
PRX9 and PRX40 Are Evolutionarily Conserved Class III Peroxidases in Land Plants
Since PRX9 and PRX40 play an essential role in Arabidopsis anther development, we sought to predict whether orthologs are likely to be important in other plant species. To examine the evolution of PRX9 and PRX40, we performed comprehensive phylogenetic analyses of class III peroxidases across diverse land plant lineages. First, we performed a large-scale neighbor-joining phylogenetic analysis using 887 class III peroxidases collected from several reference model plants representing major land plant lineages (Supplemental Figure 5; Supplemental Data Set 1). This initial analysis identified six major phylogenetic clades of peroxidases, termed clades i through vi, that overall agree well with previously published phylogenies (Tognolli et al., 2002). By mapping known functions associated with class III peroxidases to the tree, we observed that the phylogeny does not separate peroxidases by function. For example, lignin peroxidases do not cluster into one unique clade but rather are distributed across clades i, iii, and vi. This reflects the well-recognized substrate promiscuity of class III peroxidases (Shigeto and Tsutsumi, 2016). Under low selective pressure to maintain substrate specificity, lignin peroxidase activities might have evolved multiple times in parallel during land plant evolution through gene duplication followed by neofunctionalization or subfunctionalization.
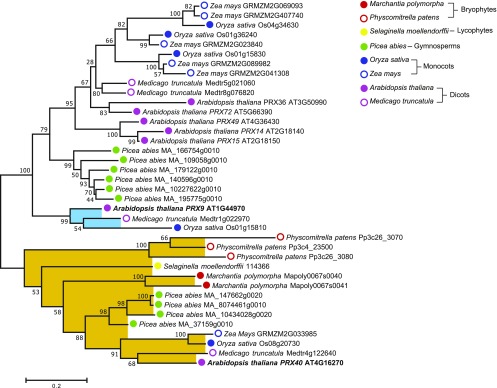
To gain insight into the deep evolutionary history of PRX9 and PRX40, we then performed a more focused maximum-likelihood phylogenetic analysis using only the sequences from clade i, which contains both PRX9 and PRX40 (Figure 9; Supplemental Data Set 2). This analysis revealed that PRX40 is highly conserved among all major lineages of land plants, with putative orthologs found in species that span bryophytes to angiosperms. By contrast, PRX9 falls into a smaller clade of orthologs that contains only angiosperm sequences and likely arose by gene duplication. These results suggest that PRX40 orthologs likely emerged in the common ancestor of all land plants during early land colonization 450 million years ago, whereas PRX9 emerged through parallel evolution much later within angiosperms.
Figure 9.
Maximum-Likelihood Phylogenetic Analysis of PRX9, PRX40, and Related Class III Peroxidases from Various Plant Lineages.
The analysis was generated with 1000 bootstrap replications using a subset of sequences from clade i (Supplemental Figure 4). Bootstrap values are indicated at nodes within the tree. The scale measures evolutionary distance in substitutions per amino acid. PRX9 is placed within a clade containing only angiosperm sequences, highlighted in blue. PRX40 is placed in a clade containing sequences spanning land plants from angiosperms to bryophytes, highlighted in orange. PRX9 and PRX40 branch labels are bolded.
DISCUSSION
PRX9 and PRX40 Function to Maintain Tapetum and Microspore Cell Wall Integrity during Anther Development
One of the most defining features of the prx9 prx40 phenotype is tapetum hypertrophy and subsequent male sterility. Tapetum hypertrophy is a defect in anther development that has been observed in many different contexts across multiple plant species. To our knowledge, tapetum hypertrophy was first reported in rice (Oryza sativa) as a stress response to cold (Satake and Hayase, 1970; Mamun et al., 2006) and was later observed in cytoplasmic male-sterile Capsicum annuum and in heat- or abscisic acid–treated Triticum aestivum (Horner and Rogers, 1974; Saini et al., 1984). These early studies established a connection between environmental stress and tapetum hypertrophy–associated male sterility; however, the mechanistic basis for this phenomenon has remained elusive.
In the model plant Arabidopsis, tapetum hypertrophy has not been seen as a stress phenotype but instead was reported as a developmental phenotype associated with a wide collection of mutants, including roxy1, roxy2, aborted microspores, male sterility1, defective in tapetal development and function1, dysfunctional tapetum1, myb33, myb65, bhlh010, bhlh089, bhlh091, kaonashi4/uneven pattern of exine1, gus negative1, gus negative4, and fat tapetum (Sanders et al., 1999; Wilson et al., 2001; Sorensen et al., 2002, 2003; Millar and Gubler, 2005; Zhang et al., 2006; Xing and Zachgo, 2008; Zhu et al., 2008, 2015; Phan et al., 2011; Suzuki et al., 2017). Besides KAONASHI4/UNEVEN PATTERN OF EXINE1, which is a glycosyl transferase, these genes encode regulatory genes and transcription factors or are unmapped mutant alleles. Hence, these mutants provided little insight into a mechanistic understanding of tapetum hypertrophy or how it is prevented in the course of normal anther development in Arabidopsis. PRX9 and PRX40 are likely downstream of these transcriptional regulators and directly influence tapetum size by cross-linking extensins in the cell wall.
PRX9 and PRX40 Are Likely Extensin Peroxidases
Extensins are a large family of highly repetitive Hyp-rich glycoproteins that are ubiquitous structural cell wall proteins in land plants (Lamport et al., 2011). Extensins contain alternating hydrophilic and hydrophobic stretches that allow them to self-assemble into dendritic networks (Cannon et al., 2008). The characteristic hydrophilic extensin motif consists of heavily glycosylated Ser-(Hyp)3-5 interspersed with hydrophobic Tyr-Val-Tyr (Lamport et al., 2011). Class III peroxidases act on these Tyr residues to form intra- and intermolecular cross-links of extensins.
The tapetal and microspore swelling phenotype seen in prx9 prx40 anthers is reminiscent of other extensin-associated phenotypes. The best-studied extensin in Arabidopsis is EXT3/ROOT-SHOOT-HYPOCOTYL-DEFECTIVE (RSH; Hall and Cannon, 2002). Similar to prx9 prx40 mutants, rsh mutants display defects in cell size and shape due to compromised cell walls. Cells within the developing embryos of rsh plants are disorganized and fail to form the characteristic heart shape (Hall and Cannon, 2002). Cells throughout seedling roots are also abnormally shaped and dysfunctional. Similarly, mutations in genes involved in extensin glycosylation display phenotypes associated with uninhibited cell expansion, including abnormally elongated roots and petioles, larger rosette leaves, and defects in root hair growth (Gille et al., 2009; Velasquez et al., 2011; Saito et al., 2014; Møller et al., 2017).
Our experimental data also indicate that PRX9 and PRX40 play a role in regulating tapetum and microspore size and shape primarily by their extensin cross-linking activities. We observed the accumulation of extensins in the developing tapetum by immunostaining. Furthermore, we observed that prx9 prx40 anthers failed to accumulate high molecular weight extensins, a direct consequence of extensin cross-linking in vivo. We also observed that coexpression of either PRX9 or PRX40 with EXT23 in N. benthamiana leaves resulted in the accumulation of higher molecular weight extensins. Finally, the pollen degeneration seen in ext18 plants partially phenocopied the prx9-1 prx40-1 phenotype.
Additionally, previous work has suggested that extensin disassembly is important for tapetal PCD. A papain-like Cys protease, CYSTEINE ENDOPEPTIDASE1 (CEP1), was previously found to be specifically expressed in the tapetum (Zhang et al., 2014). Papain-like Cys proteases have the unique ability to degrade extensins during plant PCD (Helm et al., 2008). Indeed, the Arabidopsis cep1 mutant displays delayed tapetal PCD, which may be partially explained by its inability to dismantle the extensin scaffold in the tapetum cell wall during late stages of anther development. Altogether, this evidence points to an important role for extensins in the tapetum and microspore cell walls.
PRX9 and PRX40 Substrates: Extensins and H2O2
Although EXT18 appears to be a substrate for PRX9 and PRX40, additional extensins are likely involved in the tapetum and microspore cell walls. EXT23 is expressed in the anthers and appears to be a substrate for transiently expressed PRX9 and PRX40 in N. benthamiana leaves. Additionally, three extensins or extensin-like genes were found to be upregulated in cep1 flower buds (Zhang et al., 2014), implying their role in the tapetum: EXT3/RSH (AT1G21310), AT5G25550, and EXT21 (AT2G43150). Of these three, only EXT3 and EXT21 contain the necessary Tyr residues for cross-linking and are therefore possible candidate substrates for PRX9 and PRX40. In addition to EXT18 (AT1G26250), EXT19 (AT1G26240) encodes another candidate extensin substrate. EXT19 is a tandem duplicate of EXT18 and the two share 84% sequence similarity at the amino acid level. EXT18 and EXT19 are arranged head to head in the genome, suggesting that they may be coregulated by a bidirectional promoter. The roles for these extensins in tapetal and pollen walls are likely redundant or partially overlapping. Future work is necessary to dissect the functions of individual extensins and the coordination of these extensions during anther development.
As a cosubstrate for class III peroxidases, H2O2 is required for the catalytic function of PRX9 and PRX40. H2O2 may be derived from multiple sources in planta. During plant defense and lignification, H2O2 is primarily derived from the reduction of molecular oxygen by the respiratory burst oxidase homologs (RBOHs) to superoxide (Marino et al., 2012; Lee et al., 2013) that is further converted to H2O2 by superoxide dismutase (SOD). The Arabidopsis genome encodes 10 RBOHs: RBOHA through RBOHJ. RBOHE was shown to be specifically expressed in the tapetum during anther development (Xie et al., 2014). Interestingly, the rbohe mutant also displays a mildly hypertrophic tapetum, degenerated pollen grains, and pollen grains with distorted morphologies. Synergistic effects in rbohe rbohc/rhd2 suggest that RBOHC may also play a role in generating tapetal H2O2. These phenotypes are consistent with RBOHE and RBOHC acting upstream of PRX9 and PRX40 by generating the superoxide that is subsequently converted to H2O2 by a putative SOD. At present, no tapetal SOD has been identified or characterized.
In summary, this work identifies PRX9 and PRX40 as critical genes for Arabidopsis anther development, likely through their extensin cross-linking activity. PRX40 is highly conserved among all land plants, suggesting that extensin cross-linking was an ancient and critical function for the colonization of land by early land plants. PRX9 likely arose later within angiosperms and descended from a duplication event that can be traced back to a PRX40 progenitor. At present, most of the 73 class III peroxidases in Arabidopsis have no assigned functions. Future studies may identify additional class III peroxidases as being involved in extensin cross-linking during other developmental processes in plants. Finally, this work sheds light on the mechanism of tapetum hypertrophy, a stress phenotype associated with reductions in crop yield of rice and wheat (Triticum aestivum; Satake and Hayase, 1970; Horner and Rogers, 1974; Saini et al., 1984; Mamun et al., 2006). As we continue to expand our understanding of the dynamic signaling and metabolic processes during anther development, we hope to improve our ability to develop stress-resistant crops.
METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were placed on water-saturated 3:1 soil:vermiculite supplemented with Osmocote (Greenhouse Megastore). Plants were grown under a 16-h-light/8-h-dark cycle with fluorescent light of 80 to 120 μmol photons m−2 s−1 at 22°C and 60% RH. T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center or the Cold Spring Harbor Laboratory and grown under the same conditions.
RT-PCR
Total RNA was extracted from ∼100 mg of fresh plant tissue with the RNeasy mini kit (Qiagen) and subjected to an on-column DNase digest to remove contaminating genomic DNA. cDNA was prepared using the SuperScript III Reverse Transcriptase kit (Invitrogen) and used as template for RT-PCR.
GUS Localization
The 507-bp PRX9 promoter region and 959-bp PRX40 promoter region were amplified out of Col-0 genomic DNA and ligated into BamHI-linearized pBI101.2 (Gibson Assembly). Assemblies were confirmed by DNA restriction digest and Sanger sequencing and were transformed into Agrobacterium tumefaciens GV3101 strain. Agrobacterium-mediated Arabidopsis transformation was performed using the floral dip method (Clough and Bent, 1998). T1 kanamycin-resistant transformants were selected on Murashige and Skoog/agar media containing 200 µg/mL Timentin (ticarcillin-clavulanate, Gold Bio) and 50 µg/mL kanamycin. After selection, kanamycin-resistant seedlings were transferred to soil and grown under greenhouse conditions.
For GUS staining, flower buds from adult plants were collected into ice-cold 90% (v/v) acetone and incubated for 20 min at room temperature. Samples were washed in staining buffer (50 mM sodium phosphate, pH 7.2, 0.2% (v/v) Triton X-100, and either 2 or 10 mM potassium ferrocyanide/potassium ferricyanide) and then incubated in staining solution (staining buffer containing 2 mM 5-Bromo-4-chloro-3-indolyl-β-D-glucuronide) on ice. Sample were infiltrated with staining solution under vacuum for 20 min, or until samples sank, and then incubated overnight at 37°C to allow the GUS staining reaction to proceed. The following day, samples were processed through a graded series into 50% (v/v) ethanol and placed in formalin-aceto-alcohol (50% (v/v) ethanol, 10% (v/v) glacial acetic acid, and 5% (w/v) formaldehyde) for fixation. Finally, samples were placed in 70% (v/v) ethanol for examination.
Genotyping
Approximately 1 mg of plant tissue was placed in 50 µL of Tris-EDTA buffer (10 mM Tris, pH 8, and 1 mM EDTA). Tissue was ground with a pestle, and 1 µL of the crude extract was used as a template for genotyping PCR. Genotyping primers were designed using the SIGnal Salk T-DNA primer design tool (http://signal.salk.edu/tdnaprimers.2.html; Supplemental Table).
Paraffin Histology and Histochemistry
Formalin-aceto-alcohol–fixed flower buds were transferred through a graded series into 100% ethanol and then through a graded series into 100% tert-butanol. Samples were then processed to paraplast and embedded in paraffin wax blocks. Then, 8- to 10-µm sections were collected with a microtome, transferred to slides, and dried overnight at 37°C. For GUS-stained samples, slides were deparaffinized with two changes of Histo-Clear (National Diagnostics) and cover slipped with Permount (Thermo Fisher Scientific). For toluidine blue staining, samples were deparaffinized with Histo-Clear, rehydrated, stained with 0.5% toluidine blue, dehydrated, and cover slipped with Permount.
Scanning Electron Microscopy
Pollen grains were collected from open flowers and transferred to carbon adhesive tabs (Electron Microscopy Sciences) that were placed on aluminum mounts (Electron Microscopy Sciences). Samples were sputter coated with Au/Pd with a Hummer sputter coater and imaged with JEOL 5600 LV scanning electron microscope. For scanning electron microscopy of anthers, samples were fixed in 2% glutaraldehyde, 3% paraformaldehyde, 0.1 M sodium cacodylate, pH 7.4, 5% Suc, and 0.01% Triton X-100 overnight. Fixed samples were dehydrated through a graded series into 100% ethanol, critical point dried in a critical point dryer (Tousimis), and sputter coated.
Transmission Electron Microscopy
Stage 12 flower buds were fixed in 2.5% glutaraldehyde and 3% paraformaldehyde with 5% Suc in 0.1 M sodium cacodylate buffer, pH 7.4, and 0.01% Triton X-100 overnight. Samples were postfixed in 1% OsO4 in veronal-acetate buffer. Samples were stained en block overnight with 0.5% uranyl acetate in veronal-acetate buffer, pH 6.0, dehydrated, and embedded in Embed-812 resin. Sections were cut on a Leica EM UC7 ultramicrotome with a Diatome diamond knife at a thickness setting of 50 nm and stained with 2% uranyl acetate and lead citrate. The sections were examined using a Field Electron and Ion Company Tecnai spirit at 80 kV and photographed with an AMT charge-coupled device camera.
Immunostaining
Ten-micrometer sections were collected from paraffin-embedded flower buds, deparaffinized with Histo-Clear, and transferred into water through a graded series of ethanol. Sections were equilibrated in PBS and then blocked with 3% dry nonfat milk. Next, 10 μL of undiluted JIM20 hybridoma supernatant (CarboSource) was added to the sections and incubated overnight at 4°C. Primary antibody was washed away with five exchanges of PBS and 10 μL of 1:10,000 mouse anti-rat IgM secondary antibody-horseradish peroxidase (catalog no. 31,476; Thermo Fisher Scientific) was added to sections and incubated at room temperature for 1 h. Secondary antibody was washed away with five exchanges of PBS. Next, 20 μL of 0.05% diaminobenzidine/0.015% H2O2 was added and allowed to incubate until sections showed robust staining. Reactions were stopped with a rapid dilution into MilliQ water and four subsequent exchanges with fresh MilliQ water.
Immunoblotting
Stages 10 to 12 anthers were dissected out of individual flower buds, frozen on dry ice, and ground in a microcentrifuge tube with a pestle. Next, 50 μL of extraction buffer (25 mM Tris, pH 8, and 4% SDS) was added, and the samples were mixed thoroughly with the pestle. Extracts were refrozen and stored at −20°C. Ten-microliter samples were separated on 3-(N-morpholino)-propanesulfonic acid-Tris SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% dry non-fat milk in Tris-buffered saline-Tween and incubated with 1:50 JIM20 overnight at 4°C. Membranes were washed five times with Tris-buffered saline-Tween and incubated with 1:10,000 mouse anti-rat IgM secondary antibody-horseradish peroxidase (Thermo Fisher Scientific). Immunostaining was visualized with the Enhance Chemiluminescence kit (Thermo Fisher Scientific).
Heterologous Protein Expression in Nicotiana benthamiana
Agrobacterium tumefaciens LBA4404 was transformed with pEAQ-HT vector constructs by electroporation (2.5 kV) and plated on yeast and mold (YM) agar (0.4 g of yeast extract, 10 g of mannitol, 0.1 g of sodium chloride, 0.2 g of magnesium sulfate [heptahydrate], 0.5 g of potassium phosphate [dibasic, trihydrate], 15 g of agar, and 1 L of MilliQ water, pH 7) with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin, and incubated for 2 d at 30°C. A 5-mL starter culture of YM medium with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin was inoculated with a clone of Agrobacterium tumefaciens LBA4404 containing the plasmid and incubated for 24 to 36 h at 30°C/225 rpm. The starter culture was used to inoculate a 35-mL culture of YM medium with 100 µg/mL rifampicin, 50 µg/mL kanamycin, and 100 µg/mL streptomycin, which was incubated for 24 h at 30°C/225 rpm. The 35-mL cultures were pelleted by centrifugation for 15 min at 5000 rpm at 4°C, the supernatant was discarded, and the cells were resuspended in PBS to give a final OD of 0.8. The Agrobacterium suspensions were syringe infiltrated into N. benthamiana, and the plants were placed in shade overnight. Infiltrated Nicotiana plants were grown in greenhouse conditions for 5 d before tissue was harvested and analyzed by immunoblot.
Recombinant Protein Expression and Refolding Optimization
PRX9 and PRX40 coding sequences were cloned without their predicted signal peptide sequences into the pHIS8-4 bacterial expression vector with an N-terminal His-tag and transformed into Escherichia coli BL-21(DE3). Next, 500 mL of transformed bacteria were grown in Terrific broth at 37°C/225 rpm until reaching an OD600 of 0.8. Cultures were induced with 500 μM isopropyl β-d-thiogalactoside and incubated overnight at 16°C/225 rpm. Cultures were pelleted by centrifugation for 15 min at 6000 rpm/4°C, and pellets were resuspended in 12 mL of lysis buffer (50 mM Tris, pH 8, 500 mM NaCl, and 30 mM imidazole). Resuspensions were divided into 1-mL aliquots, frozen, and stored at −80°C.
Resuspensions were thawed and lysed by sonication for 20 s. Lysates were centrifuged for 30 min at 16,000 rpm/4°C, and supernatants were discarded. Insoluble PRX9 and PRX40 were washed three times by resuspending the pellets in lysis buffer with 2 M urea, spinning for 30 min at 16,000 rpm/4°C, and discarding the supernatant. PRX9 and PRX40 were then solubilized in lysis buffer with 8 M urea and bound to nickel-nitrilotriacetic acid resin to separate them from bacterial cell material. Protein-bound nickel-nitrilotriacetic resin was diluted into elution buffer (50 mM Tris, pH 8, 500 mM NaCl, and 300 mM imidazole) with 8 M urea, and the resin was separated from the elution by centrifugation for 10 min at 10,000 rpm/4°C.
Refolding conditions were tested by diluting 10 μL of HisTrap elution into 190 μL of various refolding conditions in a 96-well plate format. All wells contained 10 mM MES, pH 6, 5 mM CaCl2, 5 μM hemin, and 0.7 mM GSSG. Concentrations of urea and GSH were varied. Wells were sealed with an adhesive plate cover, and the plate was incubated for 60 h in darkness at room temperature. Next, 50 μL of refolding reactions was added to 50 μL of pyrogallol reaction buffer (final concentration: 10 mM MES, pH 6, 50 mM pyrogallol, and 1 mM H2O2). The relative activity of refolding conditions was assessed by monitoring the formation of purpurogallin, which is yellow and absorbs light at 420 nm.
Phylogenetic Analysis
Protein sequences from Arabidopsis, Medicago truncatula, Oryza sativa, Zea mays, Selaginella moellendorffii, Physcomitrella patens, and Marchantia polymorpha genomes were collected from Phytozome (https://phytozome.jgi.doe.gov) by BLAST search. Picea abies sequences were collected by BLAST search from Congenie (http://congenie.org/). Incomplete sequences were manually excluded by eye, and sequences were aligned in MEGA6 (Tamura et al., 2013) with the MUSCLE algorithm (Edgar, 2004). For the large-scale phylogeny (Supplemental Figure 4), all 877 sequences were included. The tree was generated with the neighbor-joining method (Saitou and Nei, 1987). Evolutionary distances were calculated using the Jones–Taylor–Thornton matrix-based method (Jones et al., 1992) and are in units of number of amino acid substitutions per site.
For the maximum-likelihood tree (Figure 9), only the sequences from clade i of the neighbor-joining tree were included. Sequences were realigned in MEGA6 with MUSCLE. Positions in the alignment that were represented by only a single sequence were manually removed and the truncated sequences were realigned. A distance matrix was generated in MEGA6 and sequences with high divergence from PRX9 and PRX40 were manually removed. Several preliminary phylogenies were generated and sequences that could not be confidently placed on branches that related to PRX9 or PRX40 were manually removed. The final phylogeny was generated with the remaining sequences using the maximum-likelihood method based on the Whelan and Goldman model (Whelan and Goldman, 2001). A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 0.7211]). Branch lengths were measured in the number of substitutions per site.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers AT1G44970 (PRX9), AT4G16270 (PRX40), AT3G18780 (ACTIN2), AT5G19810 (EXT23), and AT1G26250 (EXT18).
Supplemental Data
Supplemental Figure 1. BAR eFP predicts PRX9 and PRX40 are expressed in early flower buds.
Supplemental Figure 2. Anther filaments from stage 14 prx9-1 prx40-2 flower are not fully elongated.
Supplemental Figure 3. Reconstituted enzyme activity can be obtained by solubilizing, purifying, and refolding protein from E. coli expressing PRX9 and PRX40.
Supplemental Figure 4. PRX9 and PRX40 cross-link EXT18 when transiently expressed in Nicotiana benthamiana.
Supplemental Figure 5. Neighbor-joining phylogenetic analysis of class III peroxidases across land plants.
Supplemental Table. List of primers used in this study.
Supplemental Data Set 1. Alignment of class III peroxidases protein sequences used for large-scale neighbor-joining phylogenetic analysis (Supplemental Figure 5).
Supplemental Data Set 2. Alignment of sequences used for maximum-likelihood analysis (Figure 9).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Nicki Watson at the Whitehead Institute Keck Imaging Facility for assistance with electron microscopy. This work was supported by the Pew Scholars Program in the Biomedical Sciences (27345), the Searle Scholars Program Kinship Foundation (15-SSP-162), and the National Science Foundation (CHE-1709616). J.R.J. was supported by the National Science Foundation Graduate Research Fellowship (1122374).
Author Contributions
J.R.J. and J.K.W. designed and analyzed all experiments. J.R.J. and W.C.D. performed the experiments. J.R.J. and J.K.W. wrote the article.
References
- Bernards M.A., Fleming W.D., Llewellyn D.B., Priefer R., Yang X., Sabatino A., Plourde G.L. (1999). Biochemical characterization of the suberization-associated anionic peroxidase of potato. Plant Physiol. 121: 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J.D., Sadler I.H., Fry S.C. (1996). Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: A new potential cross-link. Biochem. J. 315: 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownleader M.D., Ahmed N., Trevan M., Chaplin M.F., Dey P.M. (1995). Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 109: 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownleader M.D., Hopkins J., Mobasheri A., Dey P.M., Jackson P., Trevan M. (2000). Role of extensin peroxidase in tomato (Lycopersicon esculentum Mill.) seedling growth. Planta 210: 668–676. [DOI] [PubMed] [Google Scholar]
- Cannon M.C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B.L., Chen L., Lamport D.T., Chen Y., Kieliszewski M.J. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Falasca G., Costantino P., Cardarelli M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary P., Saha P., Ray T., Tang Y., Yang D., Cannon M.C. (2015). EXTENSIN18 is required for full male fertility as well as normal vegetative growth in Arabidopsis. Front. Plant Sci. 6: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Duroux L., Welinder K.G. (2003). The peroxidase gene family in plants: A phylogenetic overview. J. Mol. Evol. 57: 397–407. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M., Desvoyes B., Manzano C., Liberman L.M., Benfey P.N., Del Pozo J.C., Gutierrez C. (2017). Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytol. 213: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pérez F., Pomar F., Pedreño M.A., Novo-Uzal E. (2015a). The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol. Plant. 154: 395–406. [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez F., Vivar T., Pomar F., Pedreño M.A., Novo-Uzal E. (2015b). Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J. Plant Physiol. 175: 86–94. [DOI] [PubMed] [Google Scholar]
- Fry S.C. (1982). Isodityrosine, a new cross-linking amino acid from plant cell-wall glycoprotein. Biochem. J. 204: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Hänsel U., Ziemann M., Pauly M. (2009). Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. USA 106: 14699–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Q., Cannon M.C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., et al. (2014). Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 5: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Schmid M., Hierl G., Terneus K., Tan L., Lottspeich F., Kieliszewski M.J., Gietl C. (2008). KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am. J. Bot. 95: 1049–1062. [DOI] [PubMed] [Google Scholar]
- Herrero J., Fernández-Pérez F., Yebra T., Novo-Uzal E., Pomar F., Pedreño M.Á., Cuello J., Guéra A., Esteban-Carrasco A., Zapata J.M. (2013). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237: 1599–1612. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1968). Pollen wall development. The succession of events in the growth of intricately patterned pollen walls is described and discussed. Science 161: 230–237. [DOI] [PubMed] [Google Scholar]
- Horner H.T. Jr., Rogers M.A. (1974). A comparative light and electron microscopic study of microsporogenesis in male-fertile and cytoplasmic male-sterile pepper (Capsicum annuum). Can. J. Bot. 52: 435–441. [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P.A., Galinha C.I., Pereira C.S., Fortunato A., Soares N.C., Amâncio S.B., Pinto Ricardo C.P. (2001). Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol. 127: 1065–1076. [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Knox R.B., Heslop-Harrison J., Reed C. (1970). Localization of antigens associated with the pollen grain wall by immunofluorescence. Nature 225: 1066–1068. [DOI] [PubMed] [Google Scholar]
- Kunieda T., Shimada T., Kondo M., Nishimura M., Nishitani K., Hara-Nishimura I. (2013). Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25: 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D.T.A., Kieliszewski M.J., Chen Y., Cannon M.C. (2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Li D.-D., Xue J.-S., Zhu J., Yang Z.-N. (2017). Gene regulatory network for tapetum development in Arabidopsis thaliana. Front. Plant Sci. 8: 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszkay A., van der Zalm E., Schopfer P. (2004). Production of reactive oxygen intermediates (O(2)(.-), H(2)O(2), and (.)OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 136: 3114–3123, discussion 3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamun E.A., Alfred S., Cantrill L.C., Overall R.L., Sutton B.G. (2006). Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 30: 583–591. [DOI] [PubMed] [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. (2012). A burst of plant NADPH oxidases. Trends Plant Sci. 17: 9–15. [DOI] [PubMed] [Google Scholar]
- Millar A.A., Gubler F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller S.R., et al. (2017). Identification and evolution of a plant cell wall specific glycoprotein glycosyl transferase, ExAD. Sci. Rep. 7: 45341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35: D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., et al. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan H.A., Iacuone S., Li S.F., Parish R.W. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Grienenberger E., Douglas C.J. (2015). The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113: 170–182. [DOI] [PubMed] [Google Scholar]
- Ringli C. (2010). The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J. 63: 662–669. [DOI] [PubMed] [Google Scholar]
- Saini H.S., Sedgley M., Aspinall D. (1984). Development anatomy in wheat of male sterility induced by heat stress, water deficit or abscisic acid. Funct. Plant Biol. 11: 243–253. [Google Scholar]
- Saito F., Suyama A., Oka T., Yoko-O T., Matsuoka K., Jigami Y., Shimma Y.-I. (2014). Identification of novel peptidyl serine α-galactosyltransferase gene family in plants. J. Biol. Chem. 289: 20405–20420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Satake T., Hayase H. (1970). Male sterility caused by cooling treatment at the young microspore stage in rice plants: V. Estimations of pollen developmental stage and the most sensitive stage to coolness. Jpn. J. Crop. Sci. 39: 468–473. [Google Scholar]
- Shigeto J., Tsutsumi Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209: 1395–1402. [DOI] [PubMed] [Google Scholar]
- Shigeto J., Itoh Y., Hirao S., Ohira K., Fujita K., Tsutsumi Y. (2015). Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 57: 349–356. [DOI] [PubMed] [Google Scholar]
- Showalter A.M., Basu D. (2016). Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Front. Plant Sci. 7: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.T., Santama N., Dacey S., Edwards M., Bray R.C., Thorneley R.N., Burke J.F. (1990). Expression of a synthetic gene for horseradish peroxidase C in Escherichia coli and folding and activation of the recombinant enzyme with Ca2+ and heme. J. Biol. Chem. 265: 13335–13343. [PubMed] [Google Scholar]
- Sorensen A., Guerineau F., Canales-Holzeis C., Dickinson H.G., Scott R.J. (2002). A novel extinction screen in Arabidopsis thaliana identifies mutant plants defective in early microsporangial development. Plant J. 29: 581–594. [DOI] [PubMed] [Google Scholar]
- Sorensen A.-M., Kröber S., Unte U.S., Huijser P., Dekker K., Saedler H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Narciso J.O., Zeng W., van de Meene A., Yasutomi M., Takemura S., Lampugnani E.R., Doblin M.S., Bacic A., Ishiguro S. (2017). KNS4/UPEX1: A type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173: 183–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli M., Penel C., Greppin H., Simon P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616. [DOI] [PubMed] [Google Scholar]
- Velasquez S.M., et al. (2011). O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403. [DOI] [PubMed] [Google Scholar]
- Waese J., Provart N.J. (2017). The bio-analytic resource for plant biology. Methods Mol. Biol. 1533: 119–148. [DOI] [PubMed] [Google Scholar]
- Welinder K.G., Justesen A.F., Kjaersgård I.V., Jensen R.B., Rasmussen S.K., Jespersen H.M., Duroux L. (2002). Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269: 6063–6081. [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Chapple C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187: 273–285. [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Morroll S.M., Dawson J., Swarup R., Tighe P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H.-T., Wan Z.-Y., Li S., Zhang Y. (2014). Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Zachgo S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53: 790–801. [DOI] [PubMed] [Google Scholar]
- Yang C., Song J., Ferguson A.C., Klisch D., Simpson K., Mo R., Taylor B., Mitsuda N., Wilson Z.A. (2017). Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol. 175: 333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Liu D., Lv X., Wang Y., Xun Z., Liu Z., Li F., Lu H. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095. [DOI] [PubMed] [Google Scholar]
- Zhu E., You C., Wang S., Cui J., Niu B., Wang Y., Qi J., Ma H., Chang F. (2015). The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 83: 976–990. [DOI] [PubMed] [Google Scholar]
- Zhu J., Chen H., Li H., Gao J.-F., Jiang H., Wang C., Guan Y.-F., Yang Z.-N. (2008). Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55: 266–277. [DOI] [PubMed] [Google Scholar]